Abstract
Therapeutic monoclonal antibodies (mAbs) are largely based on the immunoglobulin G1 (IgG1) scaffold and many elicit a cytotoxic cell-mediated response by binding Fc γ receptors. Core fucosylation, a prevalent modification to the asparagine(N)-linked carbohydrate on the IgG1 crystallizable fragment (Fc), reduces Fc γ receptor IIIa (CD16a) binding affinity and mAb efficacy. We determined IgG1 Fc fucosylation reduced CD16a affinity by 1.7 ± 0.1 kcal/mol when compared to afucosylated IgG1 Fc, however, CD16a N-glycan truncation decreased this penalty by 1.2 ± 0.1 kcal/mol or 70%. Fc fucosylation restricted the manifold of conformations sampled by displacing the CD16a Asn162-glycan which impinges upon the linkage between the αmannose(1-6)βmannose residues and promoted contacts with the IgG Tyr296 residue. Fucosylation also impacted IgG1 Fc structure as indicated by changes in resonance frequencies and nuclear spin relaxation observed by solution NMR spectroscopy. The effects of fucosylation on IgG1 Fc may account for the remaining 0.5 ± 0.1 kcal/mol penalty of fucosylated IgG1 Fc binding CD16a when compared to afucosylated IgG1 Fc. Our results indicated the CD16a Asn162-glycan modulates antibody affinity indirectly through reducing the volume sampled, as opposed to a direct mechanism with intermolecular glycan-glycan contacts previously proposed to stabilize this system. Thus, antibody engineering to enhance intermolecular glycan-glycan contacts will likely provide limited improvement and future designs should maximize affinity by maintaining CD16a Asn162-glycan conformational heterogeneity.
Keywords: Immunoglobulin G1, glycobiology, asparagine-linked carbohydrate
Introduction
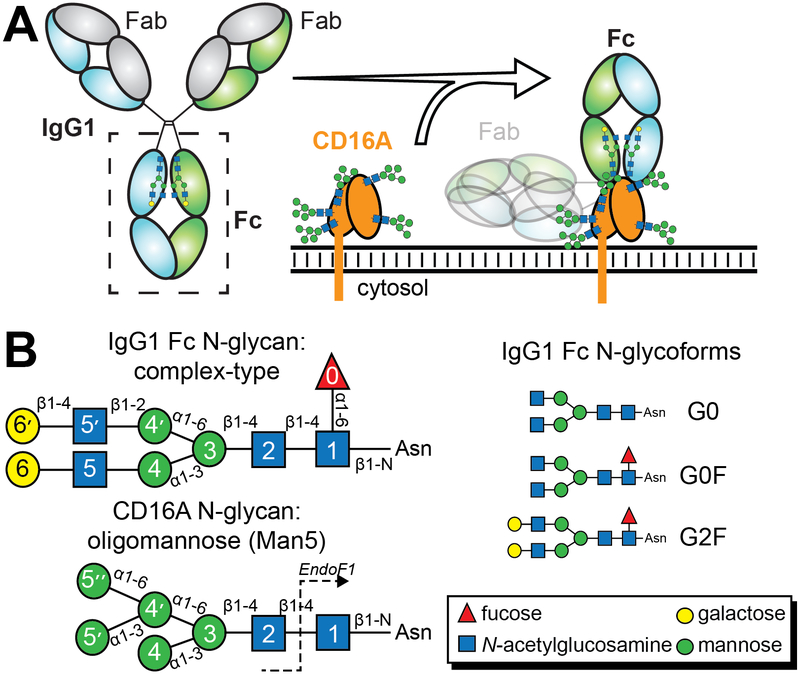
Antibodies bind to a pathogen or diseased tissue by recognizing specific surface features. The collection of antibodies on a target promotes the clustering of antibody receptors on the surface of leucocytes, triggering cell activation and a protective response 1–3. Therapeutic monoclonal antibodies (mAbs) of the immunoglobulin G (IgG) isotype are developed to target specific features of a cancerous tissue or dysregulated immune cells. IgG recognizes targets through the antigen binding fragments (Fabs) and binds multiple types of Fc γ receptors (FcγRs) expressed on leukocytes with the crystallizable fragment (Fc) to elicit a cell-mediated response to treat leukemia, lymphomas, tumors, autoimmune disease and other diseases (Figure 1). The efficacy of these therapies is likely directly related to the affinity between FcγRIIIa / CD16a and the antibody, triggering an antibody-dependent cell-mediated cytotoxic response (ADCC) 4, 5. Thus, treatment efficacy will benefit from mAbs with enhanced affinity for CD16a.
Figure 1.
N-glycans are required for IgG1 Fc to bind CD16a. (a) Both IgG and CD16a are N-glycosylated and multiple N-glycans contribute to affinity. (b) Cartoon schematics for the IgG1 Fc and CD16a N-glycans; individual carbohydrate residues are defined in the inset.
A major advance in mAb development came when Shields et al. discovered antibodies lacking a prevalent post-translational modification to the conserved and essential Fc Asn297-linked carbohydrate (N-glycan), core fucosylation, bound CD16a with 50-fold greater affinity than typical human IgG1s (~95% core fucosylated) and elicited ADCC at lower concentrations 6–8. This discovery stimulated the development of glycoengineered mAbs with higher affinity for CD16a that differ only in carbohydrate composition 9, 10. The body also modulates antibody fucosylation: Kapur et al. observed patients with severe neonatal alloimmune thrombocytopenia produced anti-platelet antibodies with very low fucosylation (<10%); furthermore, the degree of antigen-specific anti-platelet antibody fucosylation correlated inversely with immune response and disease severity 11. For these reasons it is important to determine how tuning the IgG1 Fc N-glycan composition imparts a relevant increase in CD16a binding affinity.
The composition of bulk serum IgG1 Fc N-glycans is widely studied as a biomarker for disease or aging (12–15, among others) and the discovery of antigen-specific IgG N-glycan composition, including fucosylation levels <10%, indicates the possibility of glycoengineering conducted by plasma cells or the clonal selection process to advantage responses to certain antibodies 11, 16–19. The predominant IgG1 Fc N-glycans in human serum are of a biantennary complex-type with variable levels of galactose and N-acetylneuraminic acid at the non-reducing termini (Figure 1; 20). The Fc N-glycan restricts conformational mobility of the Fc C’E loop which contains the Asn297 site of glycosylation and forms polypeptide-polypeptide contacts with FcγRs to form a complex with relatively high affinity, depending on the FcγR (low nM–μM; 21–23).
CD16a is likewise glycosylated, with five N-glycosylation sites on the extracellular antibody binding domain. Our lab determined that CD16a isolated from NK cells contains a substantial amount of under-processed oligomannose (23%) and hybrid-type glycans (22%) glycoforms with far less complex-type (55%) than CD16a expressed in human embryonic kidney 293F cells (73–82% complex-type; 24). Furthermore, CD16a displaying Man5 oligomannose N-glycans bound IgG1 Fc 12-fold tighter than CD16a with complex-type N-glycans 24. Of the five CD16a N-glycans, two, at Asn45 and Asn162, impact IgG1 binding affinity. Shibata-Koyama et al. reported the Asn45-glycan reduced affinity towards antibody, and Ferrara et al. reported that the Asn162-glycan was important for high affinity antibody binding and sensed the presence of antibody fucosylation 25, 26. The presence of a fucose residue on IgG1 Fc did not create a steric clash to disrupt the interface formed by polypeptide residues nor did fucose impart a change to the IgG1 Fc structure. Using X-ray crystallography, two groups determined similar structures of CD16a in complex with afucosylated IgG1 Fc that showed the CD16a Asn162-glycan contacted the IgG1 Fc Asn297-glycan 27, 28. These authors suggested IgG1 lacking the core fucose residue bound CD16a with higher affinity than fucosylated IgG1 because the CD16a Asn162-glycan formed stabilizing contacts with the IgG1 Fc Asn297 glycan that were disrupted by the fucose residue.
The identification of stabilizing carbohydrate-carbohydrate contacts suggested a novel mechanism for receptor-ligand interactions capable of sensing ligand N-glycan composition. Furthermore, the nature of these observed intermolecular carbohydrate-carbohydrate interactions were unlike other examples and consisted of mostly stabilizing H-bonds with some van der Waals contacts 27, 28. Carbohydrate polymers including cellulose or chitin are predominantly stabilized by hydrophobic interactions mediating carbohydrate-carbohydrate interactions and are well characterized 29–31. N-glycan stabilization, when observed, occurs largely through hydrophobic forces 32, including dispersion forces 33. One example of N-glycan stabilization is observed with IgG1 Fc. Mobility of the IgG1 Fc N-glycan is restricted by interactions with side chains from two aromatic residues, F241 and F243, and restricting N-glycan motion increases CD16a affinity 21. The appearance of carbohydrate-carbohydrate interactions in structures of the CD16a:IgG1 Fc complex were surprising based on the aforementioned precedents. We began by investigating the energetic contribution of intermolecular carbohydrate-carbohydrate interactions to the CD16a:IgG1 Fc complex.
Results
Two CD16a N-glycans impact affinity
Our binding analyses confirmed the role of the CD16a Asn162-glycan: the CD16a N162Q (Man5 glycoform) mutation eliminates N-glycosylation at position 162 and decreased affinity for IgG1 Fc with the G0F N-glycan by 3.5-fold (Table 1). Affinity improved after removing the Asn38, Asn74 and Asn169 N-glycans. Shibata-Koyama et al. reported that removing the Asn45-glycan from CD16a expressed with complex-type N-glycans enhanced affinity 26. However, we determined that removing the Asn45-glycan from CD16a expressed with Man5 N-glycans decreased affinity. Furthermore, CD16a with both the N45 and N162-glycans removed bound with even less affinity for IgG1 Fc than either singly-mutated CD16a (Table 1). It is possible that differences in CD16a glycan composition are responsible for the opposing reports on the contribution of the Asn45-glycan to affinity; it was recently established that CD16a N-glycan composition impacts affinity (Table 1 and 24, 34). Recent solution NMR and molecular dynamics studies showed that the Asn45-glycan(Man5) stabilizes CD16a through intramolecular protein-carbohydrate interactions, likely contributing to antibody binding affinity35. Complex-type N-glycans likely have a similar capacity to impact CD16a structure, though it is unclear how differences in N-glycan composition impact antibody binding affinity.
Table 1.
SPR analysis of the CD16a IgG1 Fc interactions. Glycoform analysis of fuc. IgG1 Fc, fuc. IgG1 Fc, and CD16a is shown in Supplemental Figures 1 and 2.
| CD16a (Man5 glycoform) | Fc G0F KD (nM) |
± err | ||||
|---|---|---|---|---|---|---|
| wt | 18 | 1 | ||||
| N38Q/N74Q/N169Q | 11 | 1 | ||||
| N45Q | 32 | 4 | ||||
| N162Q | 64 | 3 | ||||
| N45Q/N162Q | 290 | 10 | ||||
| N38Q/N45Q/N74Q/N169Q | 360 | 40 | ||||
| CD16a N-glycoform | fuc. IgG1 Fc KD (nM) |
± err | afuc. IgG1 Fc KD (nM) |
± err | ||
| Man5 | 57 | 9 | 2.5 | 0.2 | ||
| Man9 | 64 | 8 | 4 | 0.7 | ||
| complex-type | 245 | 30 | 38 | 9.0 | ||
| CD16a (Man5 glycoform) | Fc G0F KD (nM) |
± err | Fc G2F KD (nM) |
± err | Fc G0 KD (nM) |
± err |
| wt | 18 | 1 | 9.9 | 0.3 | 2.4 | 0.2 |
| wt-EF1a | 3.7 | 0.2 | 2.0 | 0.1 | 1.7 | 0.1 |
| N38Q/N74Q/N169Q | 11 | 1 | 5.2 | 0.5 | 1.4 | 0.3 |
| N38Q/N74Q/N169Q-EF1 | 2.9 | 0.4 | 1.6 | 0.1 | 1.3 | 0.1 |
CD16a following treatment with Endoglycosidase F1.
Truncating the CD16a N-glycans
Multiple groups reported that distal residues of the Asn162-glycan, notably the (2, 3 and 4)Man residues, are essential for high affinity binding of afucosylated IgG1 Fc through contacts with the Fc Asn297-glycan 25, 26. If the CD16a Asn162-glycan forms stabilizing contacts with Fc, truncating the CD16a N-glycan to remove these residues should reduce affinity. In fact, truncating the N-glycans of either wild-type CD16a (Man5 glycoform) or the N38Q/N74Q/N169Q CD16a variant (Man5 glycoform) to a single (1)GlcNAc residue increased affinity to the afucosylated IgG1 Fc glycoforms by 1.1 to 1.4 fold (Table 1). This result indicated that CD16a with truncated N-glycans bound with comparable affinity to afucosylated IgG1 Fc as CD16a with complete Man5 N-glycans because the distal portions of the Asn162 and Asn45-glycans had a negligible or small negative impact on binding affinity.
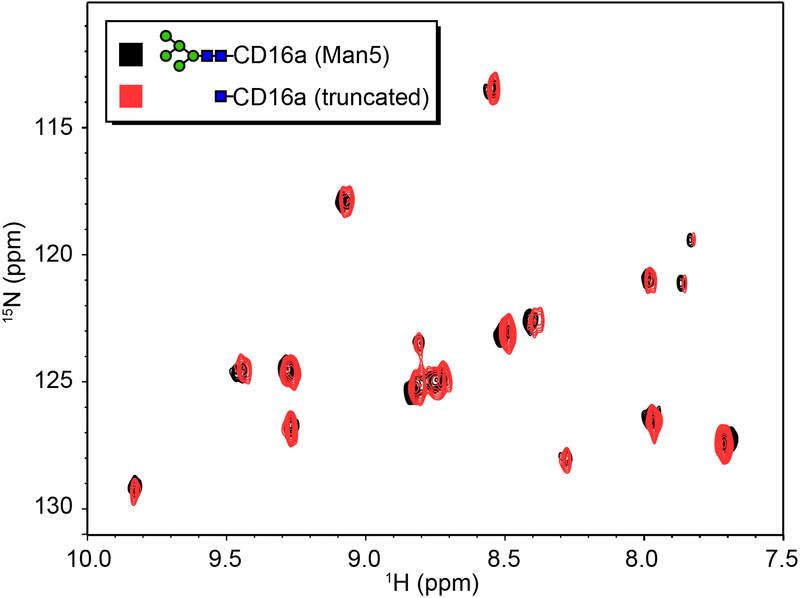
It is formally possible that N-glycan truncation allowed CD16a to sample conformations that bound IgG1 Fc with higher affinity, and overcome the impact of removing residues that contribute to affinity through stabilizing intermolecular N-glycan contacts. NMR spectra of 15N[Tyr,Phe]-CD16a with either Man5 or truncated (1)GlcNAc N-glycans reveals 1H and15N and are all less than 0.02 and 0.2 ppm, respectively, as shown in Figure 2. Furthermore, recent evidence from our group demonstrated that endoglycosidase F1 treatment of CD16a did not alter the chemical environment experienced by the (1)GlcNAc C1 and H1 nuclei in contrast to trypsin proteolysis which dramatically altered the environment 35. Therefore, N-glycan truncation does not impact CD16a structure proximal to the Phe, Tyr or (1)GlcNAc residues and it is unlikely that N-glycan truncation substantially alters other aspects of CD16a structure. Expression of CD16a variants containing a single N-glycan failed to provide enough material for binding affinity measurements.
Figure 2.
Predominant conformations sampled by CD16a are unaltered by N-glycan truncation. A 1H-15N heteronuclear single quntum coherence spectrum of 15N[Tyr,Phe]-CD16a. Seventeen peaks are visible and correspond to eight Tyr and eight Phe residues in the CD16a construct.
If the CD16a Asn162(Man5)-glycan forms stabilizing contacts with afucosylated Fc and core fucosylation of Fc disrupts those contacts, truncating the CD16a N-glycan to remove these residues should reduce affinity for afucosylated Fc to a value that is expected to be similar to fucosylated Fc. Furthermore, truncating the CD16a N-glycan should not impact affinity for fucosylated IgG1 Fc. However, CD16a N-glycan truncation slightly increased affinity towards afucosylated IgG1 Fc as noted and dramatically increased affinity towards fucosylated IgG1 Fc from three to five-fold (Table 1). As a result, truncating the CD16a N-glycans reduced the impact of IgG1 Fc core fucosylation from an eight-fold decrease in affinity upon fucosylation (G0 v. G0F for binding CD16a with Man5 N-glycans; Table 1) to a two-fold decrease (G0 v. G0F for binding CD16a with truncated N-glycans). This result further indicates residues on distal portions of the CD16a N162-glycan sense the fucosylation status of IgG1 Fc and reduce affinity towards fucosylated IgG1 Fc. Re-examination of the data in Table 1 for afucosylated IgG1 Fc are consistent with this interpretation, the CD16a with the larger Man9 oligomannose N-glycans binds slightly weaker than CD16a with the smaller Man5 N-glycans (errors for the fucosylated IgG1 Fc species are overlapping and do not allow a similar comparison).
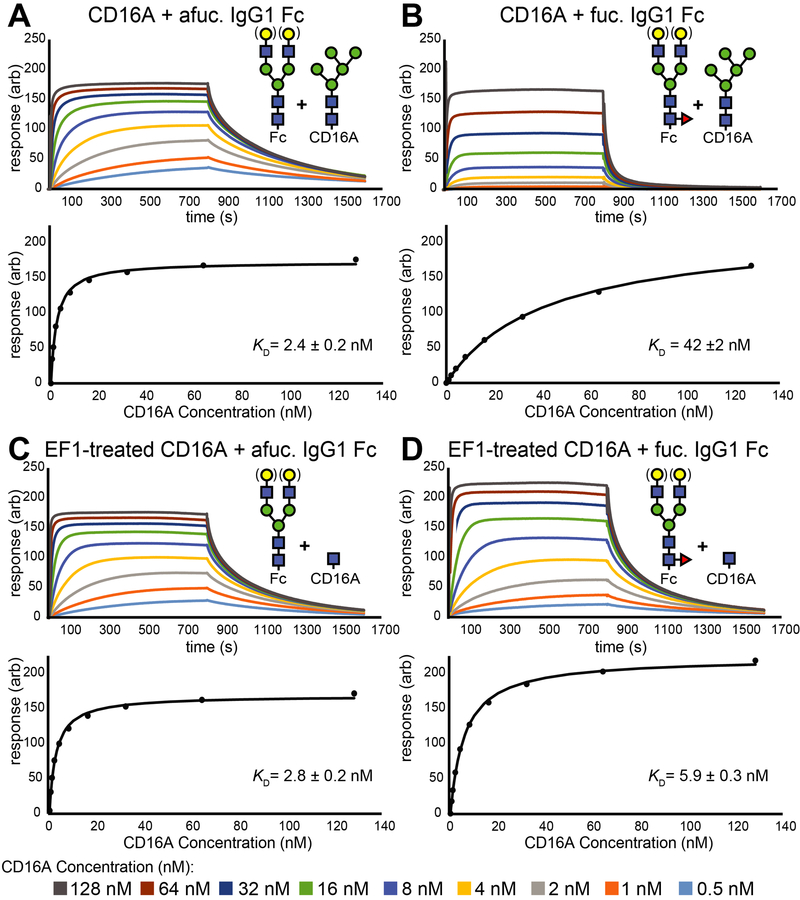
The preceding binding experiments utilized pure IgG1 Fc N-glycoforms, but in serum the glycoforms vary to a greater degree. To evaluate the impact of mixed N-glycan species, we prepared two pools of IgG1 Fc to compare the major species found in human sera: fucosylated IgG1 Fc with predominantly G0F/G1F/G2F and afucosylated IgG1 Fc with the G0F/G1F/G2F forms. Galactose residues were evenly distributed and IgG1 Fc contained N-glycans with one to four galactose residues (Supplemental Figure 2). Affinity measurements observed with SPR and ITC using these heterogeneous materials were comparable to those collected with pure N-glycoforms and revealed that CD16a N-glycan truncation had a minimal impact on CD16a affinity for afucosylated IgG1 Fc and increased affinity for fucosylated IgG1 Fc by 1.7 ± 0.1 kcal/mol (Figure 3 and Supplemental Figures 3&4). The ITC experiments demonstrated an increase of enthalpy upon truncating the N-glycan, which is inconsistent with stabilizing intermolecular contacts mediated by carbohydrates. The increase in enthalpy was largely mitigated by an increase in the magnitude of TΔS. Though the ITC experiments also provided estimates of entropy and enthalpy, it is difficult to determine from these experiments how conformational entropy of the CD16a N162-glycan impacts binding affinity for fucosylated or afucosylated antibodies because the CD16a used in these measurements displayed five N-glycans and it is not possible to separate the contribution from conformational or solvent entropy.
Figure 3.
Representative SPR sensograms for CD16a N-glycoforms binding IgG1 Fc. Response units from each sensogram were extracted once binding reached a steady state level to generate the binding curves shown on the bottom sections of each panel. Binding curves result from fitting the observed data shown as individual points in these plots.
Crystallography of the IgG1 Fc:CD16a complex
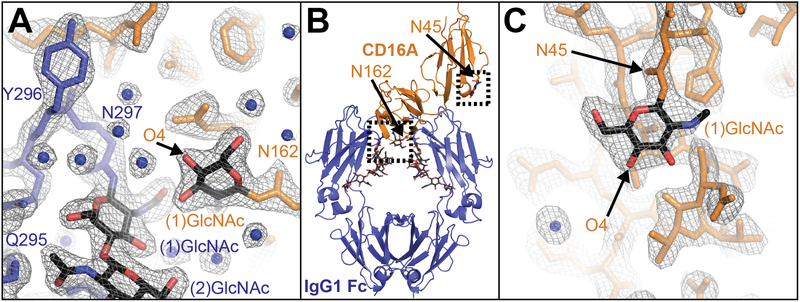
Previous X-ray diffraction studies identified a stabilizing interaction between the CD16a (N38Q/N74Q/N169Q) Asn162-glycan and the IgG1 Fc Asn297-glycan. We expect that if intermolecular glycan-glycan contacts are an important stabilizing feature of the IgG Fc:CD16a interaction that contacts will be preserved in all structures with glycosylated CD16a. Furthermore, we expect the highest affinity glycoform would be stabilized by glycan-glycan interactions. We crystallized CD16a (N38Q/N74Q/N169Q) with Man5 oligomannose N-glycans to mimic a glycoform that was found on CD16a expressed by human NK cells and bound afucosylated IgG1 Fc tighter than CD16a with either Man9 oligomannose N-glycans or complex-type N-glycans (Table 1, Supplemental Figure 1 and 24). This Man5 CD16a glycoform is different from complexes of IgG1 Fc with CD16a with Man9 oligomannose N-glycans 27 or complex-type N-glycans 28 previously reported. Crystals of the afucosylated IgG1 Fc (with G0, G1 and G2 N-glycans):CD16a(Man5) complex diffracted to 2.26 Å in the C 2 space group with high completeness (Supplemental Table 1). Molecular replacement revealed a high quality structural model with similar refinement statistics to the previous reports (Rwork 0.192, Rfree 0.237; Figure 4). This new model proved highly similar to previously determined models with RMSD values of 0.94 (PDB id 3sgj 27), 1.03 (3sgk 27), and 0.96 Å (3ay4 28) with small differences in the orientation of CD16a domain I which does not contact the Fc. In contrast to the previous reports, only the (1)GlcNAc residue of the CD16a Asn162 N-glycan appeared in density maps (Figure 4A). It was surprising that we did not observe density for the distal portions of the Asn162-glycan including the (2)GlcNAc and (3)Man residues if these residues form important stabilizing contacts.
Figure 4.
Structure of CD16a N38Q/N74Q/N169Q (Man5) in complex with afucosylated IgG1 Fc. (a) The binding interface shows an interaction between the Fc Asn297-glycan and clear density around the (1)GlcNAc residue from the CD16a Asn162-glycan scaled to σ=1.5. (b) An overall view of the complex. (c) The Asn45 glycan of CD16a shows clear density for only the (1)GlcNAc residue scaled to σ=1.0.
Simulation of the CD16a complex with IgG1 Fc
The preceding binding and structural analyses represent an important view of CD16a function but do not provide detailed information regarding mechanism at the atomic scale. X-ray crystallography elucidates atomic-level detail, but offers limited insight into fundamental, functionally-relevant motions. Computational simulations bridge this gap to provide a view into an ensemble of accessible conformations with atomic precision.
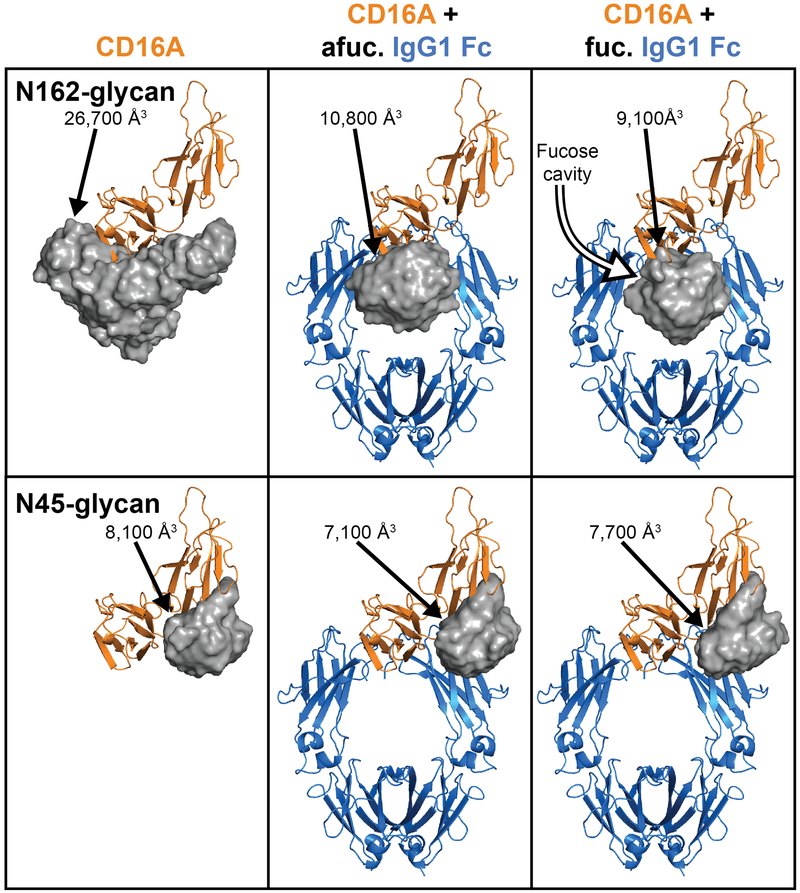
All-atom, solvated simulations of the CD16a:IgG1 Fc complex revealed motion of protein and carbohydrate components using IgG1 Fc Asn297-glycans with and without core fucosylation. CD16a N-glycan coordinates from X-ray crystallography for fucosylated and afucosylated complexes with significant portions of the carbohydrates observed provided starting positions for the simulations (pdbs 3sgj and 3sgk 27). Protein 2°, 3° and 4° structural elements showed limited conformational variability in the simulations, likely reflecting the stability of CD16a (N38Q/N74Q/N169Q) and IgG1 Fc, relatively high affinity and the simulation timescales (0.25 – 1 μs; Supplemental Figure 5). The CD16a Asn162-glycan, however, sampled a large 26,700 Å3 volume in a simulation of unliganded CD16a (Figure 5). The sampled volume diminished by 60% upon complexation with afucosylated IgG1 Fc. The addition of core fucose residues to the IgG1 Fc N-glycan reduced the volume sampled by an additional 16%, forming a cavity around the (0)fucose residue and the Asn162 N-glycan shifted towards the opposite Fc chain. This conformation sampling is unlike the CD16a Asn45-glycan that formed extensive interactions with the CD16a polypeptide surface and sampled a smaller area with much less variability between ligand states (average of 7,600 Å3; Figure 5). A duplicate set of 250 ns simulations revealed comparable results. RMSD values for each N-glycan residue showed a similar trend and support these observations (Supplemental Figure 6). This result indicated the (0)fucose residue altered the preferred conformations sampled by the Asn162 glycan, introducing a fucose-dependent energetic penalty upon binding IgG1 Fc.
Figure 5.
Molecular dynamics simulations of CD16a reveal the space sampled by CD16a N-glycans. The top row shows a grey surface representation for the area samples in simulations for the CD16a Asn162 N-glycan. The bottom row shows the same features for the CD16a Asn45 N-glycan. The volumes sampled by each glycan are indicated.
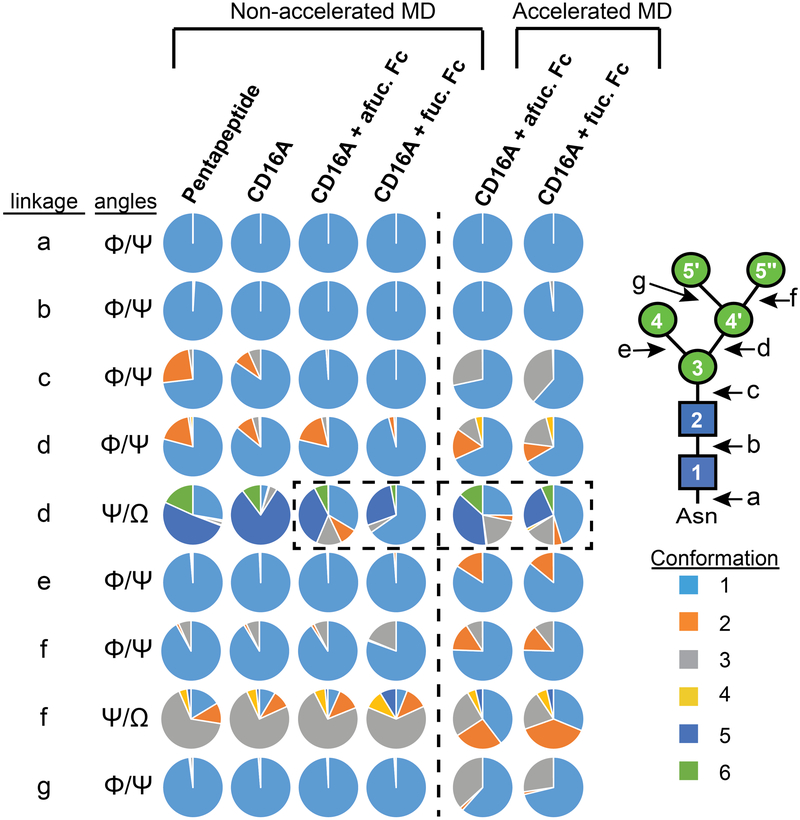
Conformations sampled by the Asn162-glycan demonstrated a role for glycan conformational sampling in antibody binding affinity. Each glycosidic linkage sampled a defined set of rotamers; rotamers for each linkage were defined using a 1 μs simulation of the Asn162-glycan on a five-residue CD16a glycopeptide angle (rotamers are defined in Supplemental Figure 7). This glycopeptide glycan sampled the greatest number of unique conformations (305). Sampled conformations decreased to 226 conformations resulting from steric constraints once attached to the CD16a polypeptide (the rotamer binning approach accounted for 99.95% of all observations). The N162-glycan on CD16a in complex with afucosylated IgG1 Fc sampled 103 unique states (99.76%), and the glycan on CD16a in complex with fucosylated IgG1 Fc sampled 75 unique states (100.00%). GlcNAc and mannose in the pyranose form behave largely as rigid units, with N-glycan flexibility limited to glycosidic linkages between residues characterized by Φ and Ψ dihedral angles. Linkages to primary alcohols (on the C6 of mannose) provide an additional degree of freedom with Φ, Ψ and Ω angles. Thus, differences in N-glycan conformation will be reflected in these dihedral angles.
Dihedral angles sampled by multiple residues in the Asn162-glycan shifted following binding to IgG1 Fc (Figure 6 & Supplemental Figure 7). One linkage, however, accounted for the majority of differences between binding afucosylated and fucosylated IgG1 Fc: Ψ and Ω between the (4’) αmannose and (3) βmannose residues (Figure 6 & Supplemental Figure 7-5). Though N162-glycans from all simulations sampled identical conformations of this linkage, the proportions differed. This became evident after analyzing the N162-glycan conformations sampled in the simulation of CD16a that sampled a predominant conformation (#5, 81%) that was sampled at a lower rate (36%) in the simulation of the complex with afucosylated IgG1 Fc and less still (28%) with fucosylated IgG1 Fc (Figure 6 and Supplemental Table 2). We believe this represents a penalty for burying the N162-glycan upon complexation that becomes exacerbated with antibody fucosylation, further restricting the available volume. A compensatory change is evident from the increased sampling of conformation #1 in the simulation with fucosylated IgG1 Fc (65%) compared to afucosylated (35%) that is lightly sampled in the simulation of CD16a alone (4%). The Fc Asn297-(0)fucose residue was proximal to the CD16a Asn162 (2)GlcNAc and (3)Man residues (pdb-3sgj; 27). The conformational restriction occurred from a steric clash between the fucose and (3)Man residues, displacing the entire Asn162 glycan to restrict conformations sampled by the (3)-(4’) linkage (Supplemental Figure 8). Fucosylation did not appear to impact the mobility of the IgG1 Fc Asn297-glycan when in complex with CD16a to a large extent (Supplemental Figure 9). A second set of 250 ns simulations revealed comparable results.
Figure 6.
A linkage in the CD16a Asn162-glycan explains the conformation difference between binding afucosylated Fc and fucosylated Fc. The area for each color in the discs represents the proportion of a distinct conformational state sampled by the Asn162-glycan in all-atom simulations. The dashed box highlights differences in conformations sampled by the linkage connecting the 3 and 4' mannose residues. Conformation states for each linkage are defined in Supplemental Figure 7.
Few contacts between the CD16a Asn162-glycan and IgG1 Fc Asn297-glycans formed during the simulations. An analysis of H-bond interactions from all snapshots of the simulations showed no single interchain Asn162-glycan/Asn297-glycan contact formed for more than 20% of the experiment (Supplemental Figure 10, orange bars). Significant intrachain Asn162-glycan contacts, including (1)GlcNAc O3-(2)GlcNAc O5 and (2)GlcNAc O3-(3)Man O5, stabilized the first three N-glycan residues and are consistent with contacts in β(1–4) linked oligosaccharides including cellulose (Supplemental Figure 10, yellow bars; 30). It is possible that the simulation parameters treated van der Waals interactions in an inappropriate manner, leading to a loss of Asn162-glycan stabilization. However, the IgG1 Fc-Asn297 N-glycan that is stabilized primarily through van der Waals interactions against the surface of the Fc Cγ2 domain 21, 36 formed the expected long-lasting interactions with IgG1 Fc residues F241 and F243 (Supplemental Figure 9).
Accelerated molecular dynamics simulations
One limitation of all-atom simulations for large complexes is an inability to sample extended timescales using conventional sampling and desktop computational resources, limiting the ability to observe motions that might occur on the time scale of hundreds of ns to tens of μs. We performed accelerated molecular dynamics simulations (aMD) using the same starting coordinates as the previous 250 ns all-atom simulations to allow for the sampling of a broader conformation range, if the simulation time for the previous examples were limiting. AMD reduces the average potential and dihedral energies and allows the evolution of a single copy of the molecule of interest without any prior knowledge of conformations, energy barriers or saddle points 37. Reducing barrier energies, however, may promote the formation of unreasonable or unlikely conformations. During the simulation of fucosylated IgG1 Fc in complex with CD16a, the IgG1 Fc Cγ3 domains adopted an unexpected, extended conformation. Though this may occur in vitro, the Cγ3 dimer is the most stable region of IgG1 Fc and unfolded conformations are likely sampled rarely (Supplemental Figure 11). Based on this result, we limited our analyses of the aMD simulations to the first 5000 sampled steps of each simulation that displayed well-defined protein secondary and tertiary structural elements.
Interestingly, the CD16a N162-glycan did not access any new states linking Man5 N-glycan monosaccharide residues indicating that the conventional MD approach captured a high degree of rotation around each individual linkage and that the rotamer binning approach was sufficient to capture appropriate local N-glycan conformational states from the aMD simulations. However, the N162-glycan did sample new combinations of linkage orientations. The N162-glycan from CD16a in complex with afucosylated and fucosylated IgG1 Fc accessed 517 and 570 distinct states, respectively. Furthermore, the rotamer binning approach, as applied to the aMD simulations accounted for the majority of observations (98.58% and 99.44%, respectively). The predominant conformations observed at most linkages were similar between the simulations that included fucosylated IgG1 Fc and afucosylated IgG1 Fc with the exception of the Ψ and Ω dihedral angles between the (4’) αmannose and (3) βmannose residues. For these dihedral angles, differences between the two simulations emerged. Though the precise percentage of each population was different when comparing conventional and aMD simulations of similarly glycosylated IgG1 Fc, the aMD simulations recapitulated differences between fucosylated and afucosylated IgG1 Fc. It is worthwhile to highlight another linkage in the glycan, between the (5’’) αmannose and (4’) αmannose residues, has an identical α1-6 linkage but does not contact IgG1 Fc. In this instance there is no discernable difference between simulations with fucosylated or afucosylated IgG1 Fc.
It is unclear why two independent groups observed intermolecular N-glycan contacts by X-ray crystallography and we observed neither corresponding electron density nor Asn162-glycan dependent stabilization upon binding by SPR, ITC or in the computational simulations. It is possible the conformation of the CD16a Asn162-glycan differed based on composition and crystal contacts restricted conformations in the previous cases. It is also possible stabilizing interactions are formed from low energy conformations and magnified by cryopreservation or the effects of these contacts are lessened in solution at physiological temperature and sampled only transiently during the simulations.
The role of fucose in IgG1 Fc structure
The presence of a fucose residue reduced ΔG for CD16a(Man5) binding IgG1 Fc by 1.7 ± 0.1 kcal/mol with the predominant, but not entire, contribution attributable to the CD16a Asn162-glycan (1.2 ± 0.1 kcal/mol; Supplemental Figures 3 & 4). Thus, an additional, unidentified feature other than the Asn162-glycan must contribute the remaining 0.5 ± 0.1 kcal/mol. Matsumiya and coworkers noted that fucosylating IgG1 Fc changed resonance frequencies for Tyr296 and Tyr300 in the Fc C’E loop, but not six other Tyr residues in other Fc regions, indicating minor Fc structural changes occur upon fucosylation 38. We also observed IgG1 Fc fucosylation-dependent effects on binding to CD32, which does not contain an N-glycan at a position analogous to CD16a Asn162 but does bind in a similar mode 23. These results indicated IgG1 Fc fucosylation impacts IgG1 Fc structure in a region and in a manner that is poised to impact FcγR affinity.
We probed the effect of core fucosylation on IgG1 Fc structure and motion in the absence of receptor by observing the resonance frequencies of (1)GlcNAc and (0)fucose nuclei with solution NMR spectroscopy. IgG1 Fc with a truncated N-glycan makes an ideal glycoform to study with solution NMR because this form is sensitive to fucosylation and will lead to simpler spectra of IgG1 Fc expressed in the presence of [13C]-glucose 22, 39, 40. IgG1 Fc with a single (1)GlcNAc residue at Asn297 showed a clear peak for the anomeric 1H-13C correlation and the other expected correlations in a 1H-13C HSQC spectrum of IgG1 Fc expressed in the presence of [13CU]-glucose (Supplemental Figures 12–13). A similar experiment using IgG1 Fc truncated to an N-glycan composition consisting of a (1)GlcNAc and (0)fucose residue revealed similar peaks that were displaced. It is possible that the chemical modification of the (1)GlcNAc residue with a fucose addition explained these changes in the spectra by changing through-bond interactions. However, treating the samples with trypsin to destroy intramolecular glycan/polypeptide contacts but preserve the (1)GlcNAc-(0)fucose glycosidic linkage showed very slight differences in resonance frequency position, in particular for the anomeric correlations, indicating the addition of a fucose residue altered the chemical environment of the (1)GlcNAc H1 and C1 nuclei by perturbing interactions with the Fc polypeptide. Correlations of the anomeric nuclei are most sensitive to intramolecular glycan/polypeptide interactions due to the proximity to the glycosidic linkage to Asn297, the predominant rotatable moiety in this carbohydrate residue. Furthermore, the linewidth of the IgG1 Fc (1)GlcNAc H1/C1 correlation with fucose attached was broader than the linewidth for the afucosylated IgG1 Fc (1)GlcNAc H1/C1 correlation, indicating the possibility of different rates of motion associated with the (1)GlcNAc residues.
Measurements of relaxation rates for signals in an NMR experiment may provide information regarding N-glycan motion faster than the rotational correlation time (expect ~20 ns 41; R1 and R2) or in the μs–ms timescale (R2 only; see 42 and references therein for a review). Measurements of R1 and R2 for the (1)GlcNAc C4 nucleus were consistent with different rates of motion between fucosylated and afucosylated samples. The R1 for afucosylated Fc was greater than fucosylated Fc (1.28 ± 0.01 s−1 vs. 1.08 ± 0.05 s−1) and smaller for R2 (51 ± 4 s−1 vs. 73 ± 6 s−1). These changes are consistent with faster motion of the (1)GlcNAc residue on the ns timescale without fucose. Slow μs–ms motions also appear to contribute to the unexpectedly large R2 values (based on the R1 values). These results are consistent with previous measurements of nuclei in galactose and sialic acid residues on the non-reducing termini of IgG1 Fc 41, 43.
The results from NMR experiments indicated fucosylation slows the motion of the (1)GlcNAc residue, further reducing affinity for CD16a. This result is consistent with the previous report revealing changes of Fc backbone amide peaks resulting from N-glycan fucosylation 38. From these spectra it is difficult to determine which Fc residues are impacted by fucose, however, Matsumiya et al. suggest the conformation of Tyr296 is important for CD16a affinity and is in a position to be affected by the (0)fucose residue; Tyr296 residue is immediately adjacent to the Asn297 site of N-glycan attachment 38.
Discussion
Here we present a model for the role of fucose in antibody/CD16 interactions: IgG1 Fc fucose disrupts the conformations sampled by the CD16a Asn162-glycan, exerting a negative impact of CD16a affinity for fucosylated IgG1 Fc relative to afucosylated IgG1. It is clear the CD16a Asn162-glycan senses IgG1 Fc fucosylation; truncating the CD16a N-glycans increases affinity for fucosylated IgG1 Fc and removes the sensitivity to core fucosylation of the IgG1 Fc Asn297-glycan. Fucose has additional, though less significant, impacts on IgG1 Fc structure and motion, slowing the motion of the Asn297 glycan and further reducing CD16a affinity. Our model indicates antibody engineering efforts to enhance the intermolecular carbohydrate-carbohydrate contacts will likely have limited impact, and engineering IgG to avoid impingment on the area sampled by the CD16a Asn162-glycan in complex or to truncate the Asn162-glycan is a promising pathway to increase affinity for CD16a.
The results presented in this manuscript are generally consistent with multiple aspects of the previous studies on the IgG1 Fc:CD16a complex that IgG1 Fc fucosylation disrupts the CD16a N162-glycan, leading to a reduction in binding affinity. Here we propose a different mechanism. The previous studies reported antibody fucosylation disrupted a single conformation of the CD16a N162-glycan conformation 27, 28. Based on our results we believe that antibody fucosylation restricts the manifold of conformations sampled by the CD16a N162-glycan with no state forming extensive intermolecular contacts with IgG1 Fc. Another recent report applied replica exchange MD simulations to study the complex of IgG1 Fc with CD16a. Sakae et al. showed that the Man3 and Man4 residues of the CD16a N162-glycan, in a complex-type biantennary glycoform, were consistently 1–3 Å closer to the IgG1 Fc (1)GlcNAc residue when the IgG1 Fc fucose residue was absent, though they did not report Van der Waals or hydrogen-bond contacts formed in the simulations nor whether these contacts were maintained in a way that would be expected for a stabilizing interaction 44. This approach is consistent with our simulations as the IgG1 Fc fucose and Y296 residues together displace the N162-glycan (Supplemental Figure 8). Furthermore, Sakae et al. also showed that the N162-glycan on the CD16a complex with fucosylated IgG1 Fc sampled greater volume and had a greater RMSF value than the N162-glycan on the CD16a:fucosylated IgG1 Fc complex. Thus, the N162 glycan in the simulations by Sakae et al. and introduced in this manuscript revealed that the N162 N-glycan is highly mobile in the CD16a:IgG1 Fc complex. Sakae et al. concluded that IgG1 Fc fucosylation increased N-glycan conformational fluctuation for a complex-type biantennary CD16a N162-glycan. Our results do not allow a definitive evaluation of this hypothesis; we observed more conformations sampled by the CD16a N162-glycan in aMD simulations with fucosylated compared to afucosylated IgG1 Fc (570 v. 517) but less with the conventional MD approach (75 v. 103). Here we determined that IgG1 fucosylation reduces the volume available to the CD16a N162-glycan in complex with IgG1 Fc, and perturbs the conformations sampled by a Man5 N-glycan.
The activity of the CD16a Asn162-glycan is the second example of an N-glycan indirectly modulating ligand binding affinity. Adding residues at the non-reducing termini of the IgG1 Fc Asn297-glycan increases CD16a affinity by stabilizing the motion of the carbohydrate and the Fc C’E loop residues at the binding interface 21. Both the CD16a Asn162-glycan and the IgG1 Fc Asn297-glycan influence affinity, but neither is directly bound by the other protein, as is the case for the recognition of carbohydrate ligands by a lectin. These types of intermolecular interactions are challenging to identify. However, N-glycans are pervasive modifications and important interactions involving N-glycans are potentially common features of glycoprotein interactions. Changing carbohydrate composition is potentially widely utilized and offers the capacity for a rapid and tunable response, in this case modifying the threshold of an immune response that does not require changes to protein coding genes.
Materials and Methods
All materials were purchased from Sigma-Aldrich unless otherwise specified.
Protein expression and purification
Human IgG1 Fc (residues 216–447) was expressed using the HEK293F cells grown in Freestyle293 medium (Life Technologies) as previously described 21. Afucosylated IgG1 Fc was expressed using culture medium supplemented with 250 μM 2-deoxy-2-fluoro-l-fucose (Santa Cruz Biotech)45. Plasmids encoding the CD16a variants Asn45Q, Asn162Q, Asn45Q/Asn162Q, N38Q/N74Q/N169Q were prepared according to the QuickChange protocol (Agilent Technologies) and confirmed by DNA sequencing (ISU DNA facility). CD16a (residues 19–193, V158 allotype) was expressed using the HEK293F or HEK293S(lec1−/−) cell lines with Freestyle293 medium (Life Technologies) as previously described 21, 39. CD16a with Man9 oligomannose N-glycans was expressed by supplementing the expression medium with 5 μM kifunensine (Cayman Chemical). Following purification, proteins were exchanged into 20 mM 3-morpholinopropane-1-sulfonic acid (MOPS), 100 mM sodium chloride, pH 7.2. CD16a was stored at −80 °C in 25% glycerol (v/v). 13C labeled protein was prepared by supplementing the expression medium with [13C6]-d-glucose as previously described 39. CD16a N-glycans were analyzed following derivatization with procainamide and HILIC-ESI-MS as previously described 24.
IgG1 Fc N-glycan remodeling in vitro
The IgG1 Fc glycovariants with homogenous N-glycans (G0, G0F and G2F) were also used for a previous study that reports high resolution analysis of the Fc N-glycans following in vitro remodelling (Figure 3 in ref 23). Agalactosylated IgG1 Fc (G0F) was prepared by adding 5 μL of 3 U/mL Streptococcus pneumoniae β−1,4 galactosidase to purified glycoprotein (2 mg at 12.5 mg/mL) and incubated overnight in the dark at room temperature. Afucosylated IgG1 Fc (G0) was prepared as described above for G0F except starting with material from expression supplemented with 250 μM 2-deoxy-2-fluoro-l-fucose. Digalactosylated IgG1 Fc (G2F) was prepared by incubating 5 mg of IgG1 Fc at 16 mg/mL, 20 mM MOPS, 100 mM sodium chloride, 20 mM manganese chloride, 10 mM UDP-galactose, 2 μL of 100 mU/mL β−1,4 galactosyltransferase (GalT), pH 7.2 at 37 °C for 24 h. Equal amounts of GalT and UDP-galactose were added after 24 h and the incubation was repeated. Following the glycan modification reactions, all samples were exchanged into a buffer containing 20 mM MOPS and 100 mM sodium chloride, pH 7.2. Glycan remodeling was verified using MALDI-TOF MS as described previously 46 as well as spectra showing conversion of the material utilized herein 23.
Preparation of endoglycosidase F1-treated CD16a
Endoglycosidase F1 was expressed and purified as described 22 then coupled to agarose beads using the Amino-Link Plus Immobilization kit (ThermoSci). Endoglycosidase F1-agarose beads were stored at 4°C in 20 mM MOPS, 100 mM sodium chloride, pH 7.2. Beads were added to a solution containing 1.4 mg/mL CD16a in 100 mM potassium phosphate, pH 6.0 and incubated with end-over-end mixing at 4 °C for 18 h. To isolate the remodeled CD16a, the solution was pipetted onto a MicroBio Spin column (BioRad) and gentle pressure applied to push the CD16a containing solution through leaving the beads behind, which were subsequently washed in buffer containing 20 mM MOPS, 100 mM sodium chloride, pH 7.2 and stored at 4°C.
Preparation of IgG1 Fc with truncated N-glycans
Human IgG1 Fc glycovariants derived from HEK293F (+fuc) and HEK293S (afuc) were remodeled by endoglycosidases S 47 and F1 48 respectively, which cleave after the first sugar residue of the Asn297 chain. Each glycovariant of IgG1 Fc was exchanged into 100 mM potassium phosphate, pH 6.0 using an 10 kDa cutoff Amicon Ultracentrifugation filter. Both reactions were carried out at an IgG1 Fc concentration of 115 μM with enzyme added at a 1/50 molar ratio at RT and incubated for 18 h.
Binding analysis using surface plasmon resonance
IgG1 Fc was immobilized on a CM5 chip surface (GE Life Sciences) by performing standard amine coupling procedures on a Biacore T100 instrument. The carboxymethyl dextran surface was activated by 1:1 mixture of 0.4 M 1-ethyl-3(3-dimethylaminopropyl)carbodiimide hydrochloride and 0.1 M N-hydroxy-succinimide for 7 min at a flow rate of 5 μl/min. IgG Fc was applied to the chip at 1 μg/mL in 10 mM sodium acetate, pH 5.0 buffer and at a flow rate of 5 μl/min. Residual functional sites were deactivated by washing with 1 M ethanolamine, pH 8.5 for 7 min. Final immobilization response units were between 400–700. Flow line 1 was used as a blank with no IgG1 Fc immobilized on all sensor chips. All SPR measurements were performed at 25°C. The binding analyses were performed with binding buffer containing 20 mM MOPS, 100 mM sodium chloride, 1 μM bovine serum albumin and 0.05% P20 surfactant (v/v, GE Life Sciences), pH 7.2. The CM5 chip surface was regenerated by a 100 mM glycine, pH 3.0 wash for 30 s to remove bound receptor. A minimum of one replication for each condition was collected on different days. Representative results are shown.
Isothermal Titration Calorimetry
CD16a was thawed and exchanged into a buffer containing 20 mM MOPS 100 mM sodium chloride, pH 7.2 using a 5 mL Sephadex G25 column. IgG Fc variants were exchanged into a buffer containing 20 mM MOPS 100 mM sodium chloride, pH 7.2 prior to an experiment using an Amicon 10 kDa cutoff Ultra centrifugal filter (Millipore). Protein concentrations were determined using the calculated molar extinction coefficients (75,000 M−1cm−1 for Fc dimer, and 64,205 M−1cm−1 for CD16a-GFP fusion) for each protein on a NanoDrop 2000c spectrophotometer (ThermoScientific.) All binding experiments were performed on a GE MicroCal 200 instrument with the following settings: reference power = 1, stirring speed = 500 rpm, and 30 injections (1 × 0.2 μL, 29 × 1 μL for all experiments). CD16a (16 – 26 μM) was loaded in the sample cell and IgG1 Fc was loaded into the syringe (117 – 250 μM). Each set of experiments also contained a heat of dilution experiment, for these experiments the relevant IgG1 Fc variant (in the syringe) was titrated into a cell containing 20 mM MOPS, 100 mM sodium chloride, pH 7.2. All data analysis was conducted with the Origin software using a single site model (GE LifeSciences).
Intact glycoprotein analysis by LC-ESI/MS
Protein (10 μL, 0.1 mg/mL in double distilled water) was applied to a C4 column and eluted from an Agilent 1260 liquid chromatography system with variable relative concentrations of Buffer A (0.1 % formic acid in water; v/v) and Buffer B (0.1 % formic acid in acetonitrile; v/v) with a constant flow rate of 0.1 mL/min: 1 mL 95% A plus 5% B, then 0.5 mL 100% B, and final 0.5 mL 95% A plus 5% B. ESI was conducted on a Q-Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Scientific) with positive polarity at 35.0 eV, and a scan range of 700–4000 m/z. Data was displayed and processed using ProteoWizard 3.0.9220 (http://proteowizard.sourceforge.net) and mMass 49.
Crystallization of the IgG Fc and CD16a(Man5) complex
For crystallization, the N-terminal His8-GFP tag of CD16a (N38Q/N74Q/N169Q) expressed in lec1−/− was removed by TEV protease digestion (1:50 TEV:GFP-CD16a ratio) in a buffer containing 50 mM Tris(hydroxymethyl)aminomethane, 100 mM sodium chloride, 0.5 mM Ethylenediaminetetraacetic acid (EDTA), pH 8.0, at RT overnight in the dark. CD16a was then purified by passing over a Ni-NTA column (QIAGEN). Flow through fractions were exchanged into a buffer containing 20 mM MOPS, 100 mM sodium chloride, pH 7.2 using a 10 kDa molecular weight cutoff Amicon Ultra centrifugal filters (Millipore). TEV-cleaved CD16a was mixed with IgG Fc at a molar ratio of 1:1.5 (CD16a:Fc) and applied to a Superdex200 size exclusion column (GE Healthcare) pre-equilibrated with 20 mM MOPS, 100 mM sodium chloride, pH 7.2. Factions containing the Fc:CD16a complex were identified by SDS-PAGE, pooled, and concentrated to 20 mg/ml. Initial crystallization screens were performed by hanging drop vapor diffusion method at 18°C. Small, dispersed, rod-like crystals were obtained with various buffers and salts in two weeks. The initial crystal hits obtained in 16% PEG 20K (w/v), 0.1 M Bis-Tris-Propane, pH 7.5. Pyramid shaped crystals were obtained in one week after microseeding in crystallization solution containing 1:1 mixture of protein to reservoir solution of 16% PEG 20K (w/v), 0.1M Bis-Tris-Propane pH 7.5 and 0.1M potassium thiocyanate.
X-ray diffraction and data processing
Pyramid shaped crystals were cryoprotected with a quick soak in 20% Ethylene glycol (v/v), 20% PEG 20K (w/v), 0.1M Bis-Tris-Propane, pH 7.5, and 0.1M potassium thiocyanate. Diffraction data were collected at Argonne National Labs on beamline 23-ID-B using a MAR300 detector. Phases were determined by molecular replacement using Phenix 50 and a Fc:FcγRIIIa model (PDB: 3SGK 27). Final refinement was performed with NCS and TLS restraints using Refmac5 in the CCP4 package 51.
Molecular dynamics simulations of CD16a
All simulations were performed with a desktop computer running the GPU version of Amber14 (pmemd.cuda 52) and the Amber force fields ff12SB and GLYCAM_06j-1 53 as previously described 35. CD16a (N38Q/N74Q/N169Q) was simulated for 1 μs, in complex with IgG1 Fc G0 or G0F for 250 ns (12,500 frames). A pentapeptide fragment containing a N-glycan at position 3 was simulated for 1 μs. CUDA acceleration of Amber14 (pmemd.cuda) permitted rapid simulation with an in house desktop system equipped with two Nvidia GTX960 cards in about 30 days per simulation. Simulations were performed in duplicate. Conformational analysis was performed using VMD and custom scripts to extract appropriate information from each frame.
Accelerated MD (aMD) simulations were performed using Amber16 (pmemd.cuda) and the Amber force fields ff14SB and GLYCAM-06EPb with TIP5 waters and equilibrated in the same manner as the non-accelerated structures. Boost parameters for the dihedral and potential energies were calculated as described 37. The average dihedral and potential energies were calculated from the respective all-atom simulations described above. For the IgG1 G0-CD16a complex the inputs used are as follows: average dihedral = 7,965 kcal/mol, average potential energy = −417410 kcal/mol, total atoms = 221028, and residues = 638. The resultant boosts for the IgG1 Fc G0–CD16a complex are then: E(dihedral) = 10198 kcal/mol, α (dihedral) = 446.6 kcal/mol, E (total) = −373205 kcal/mol, and α (total) = 44205 kcal/mol. For the IgG1 G0F-CD16a complex the inputs used are as follows: average dihedral = 8000 kcal/mol, average potential energy = −418383 kcal/mol, total atoms = 222123, and residues = 640. The resultant boosts for the IgG1 Fc G0F-CD16a complex are then: E(dihedral) = 10240 kcal/mol, α (dihedral) = 448 kcal/mol, E (total) = −373958 kcal/mol, and α (total) = 44424 kcal/mol.
NMR Spectroscopy
NMR spectra were obtained using a 700-MHz Bruker Avance II spectrometer or an 800-MHz Avance III spectrometer; both spectrometers were equipped with 5 mm cryogenically cooled probes and resultant spectra initially analyzed using Topspin 3. Experiments on IgG1 Fc were collected at 50 °C in a buffer containing 20 mM MOPS, 100 mM sodium chloride and 10% D20 (v/v), pH 7.4. 15N-[Tyr,Phe] CD16A was expressed in HEK293S cells as described previously 39. Briefly, [15N]-Tyr and [15N]-Phe were added to the culture media at 100 mg/L each during protein expression. NMR data for CD16a were collected at 25°C. CD16a samples were exchanged into a buffer containing 20 mM sodium phosphate, pH 7.4, 100 mM sodium chloride, 0.05 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) and 10% D2O (v/v) with a concentration between 100–200 μM in 200 μL. Spectra were processed using NMRpipe 54 and visualized using NMRViewJ (One Moon Scientific). All spectra were referenced to 4,4-dimethyl-4-silapentane-1-sulfonic acid (0.07 ppm).
Supplementary Material
Acknowledgment
We thank D. Fulton of the Iowa State University Biomolecular NMR Facility for assistance running experiments and J. Nott of the ISU Protein Facility for assistance with mass spectrometry. This material is based upon work supported by the National Institutes of Health under Award No. R01 GM115489 (NIGMS), the Roy J. Carver Department of Biochemistry, Biophysics & Molecular Biology at Iowa State University, and by the Department of Energy for synchrotron time at the Argonne National Laboratory Advanced Photon Source (GUP-48455). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Institutes of Health.
Footnotes
Supporting Information Available: Two supplemental tables and thirteen supplemental figures are available free of charge via the internet at http://pubs.acs.org.
Accession codes: X-ray data and models were deposited as PDB as entry 5VU0
References
- 1.Bakema JE, and van Egmond M (2014) Fc receptor-dependent mechanisms of monoclonal antibody therapy of cancer, Curr. Top. Microbiol. and Immunol. 382, 373–392. [DOI] [PubMed] [Google Scholar]
- 2.Shevtsov M, and Multhoff G (2016) Immunological and Translational Aspects of NK Cell-Based Antitumor Immunotherapies, Fron. Immunol. 7, 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeap WH, Wong KL, Shimasaki N, Teo EC, Quek JK, Yong HX, Diong CP, Bertoletti A, Linn YC, and Wong SC (2016) CD16 is indispensable for antibody-dependent cellular cytotoxicity by human monocytes, Sci. Rep. 6, 34310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, and Watier H (2002) Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor Fc gamma RIIIa gene, Blood 99, 754–758. [DOI] [PubMed] [Google Scholar]
- 5.Weng WK, and Levy R (2003) Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma, J. Clin. Oncol. 21, 3940–3947. [DOI] [PubMed] [Google Scholar]
- 6.Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, Weikert SH, and Presta LG (2002) Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity, J.Biol Chem. 277, 26733–26740. [DOI] [PubMed] [Google Scholar]
- 7.Nose M, and Wigzell H (1983) Biological significance of carbohydrate chains on monoclonal antibodies, Proc. Natl. Acad. Sci. U.S.A. 80, 6632–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao MH, and Morrison SL (1989) Studies of aglycosylated chimeric mouse-human IgG. Role of carbohydrate in the structure and effector functions mediated by the human IgG constant region, J. Immunol. 143, 2595–2601. [PubMed] [Google Scholar]
- 9.Hiatt A, Bohorova N, Bohorov O, Goodman C, Kim D, Pauly MH, Velasco J, Whaley KJ, Piedra PA, Gilbert BE, and Zeitlin L (2014) Glycan variants of a respiratory syncytial virus antibody with enhanced effector function and in vivo efficacy, Proc. Natl. Acad. Sci. U.S.A 111, 5992–5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tobinai K, Klein C, Oya N, and Fingerle-Rowson G (2016) A Review of Obinutuzumab (GA101), a Novel Type II Anti-CD20 Monoclonal Antibody, for the Treatment of Patients with B-Cell Malignancies, Adv. Ther. 34, 324–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapur R, Kustiawan I, Vestrheim A, Koeleman CA, Visser R, Einarsdottir HK, Porcelijn L, Jackson D, Kumpel B, Deelder AM, Blank D, Skogen B, Killie MK, Michaelsen TE, de Haas M, Rispens T, van der Schoot CE, Wuhrer M, and Vidarsson G (2014) A prominent lack of IgG1-Fc fucosylation of platelet alloantibodies in pregnancy, Blood 123, 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parekh RB, Dwek RA, Sutton BJ, Fernandes DL, Leung A, Stanworth D, Rademacher TW, Mizuochi T, Taniguchi T, Matsuta K, and et al. (1985) Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG, Nature 316, 452–457. [DOI] [PubMed] [Google Scholar]
- 13.Russell AC, Simurina M, Garcia MT, Novokmet M, Wang Y, Rudan I, Campbell H, Lauc G, Thomas MG, and Wang W (2017) The N-glycosylation of immunoglobulin G as a novel biomarker of Parkinson’s disease, Glycobiology 27, 501–510. [DOI] [PubMed] [Google Scholar]
- 14.Yu X, Wang Y, Kristic J, Dong J, Chu X, Ge S, Wang H, Fang H, Gao Q, Liu D, Zhao Z, Peng H, Pucic Bakovic M, Wu L, Song M, Rudan I, Campbell H, Lauc G, and Wang W (2016) Profiling IgG N-glycans as potential biomarker of chronological and biological ages: A community-based study in a Han Chinese population, Medicine 95, e4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vuckovic F, Kristic J, Gudelj I, Teruel M, Keser T, Pezer M, Pucic-Bakovic M, Stambuk J, Trbojevic-Akmacic I, Barrios C, Pavic T, Menni C, Wang Y, Zhou Y, Cui L, Song H, Zeng Q, Guo X, Pons-Estel BA, McKeigue P, Leslie Patrick A, Gornik O, Spector TD, Harjacek M, Alarcon-Riquelme M, Molokhia M, Wang W, and Lauc G (2015) Association of systemic lupus erythematosus with decreased immunosuppressive potential of the IgG glycome, Arthritis Rheumatol. 67, 2978–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ackerman ME, Crispin M, Yu X, Baruah K, Boesch AW, Harvey DJ, Dugast AS, Heizen EL, Ercan A, Choi I, Streeck H, Nigrovic PA, Bailey-Kellogg C, Scanlan C, and Alter G (2013) Natural variation in Fc glycosylation of HIV-specific antibodies impacts antiviral activity, J. Clin. Inv. 123, 2183–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonneveld ME, Natunen S, Sainio S, Koeleman CA, Holst S, Dekkers G, Koelewijn J, Partanen J, van der Schoot CE, Wuhrer M, and Vidarsson G (2016) Glycosylation pattern of anti-platelet IgG is stable during pregnancy and predicts clinical outcome in alloimmune thrombocytopenia, Br. J. Haematol. 174, 310–320. [DOI] [PubMed] [Google Scholar]
- 18.Selman MH, de Jong SE, Soonawala D, Kroon FP, Adegnika AA, Deelder AM, Hokke CH, Yazdanbakhsh M, and Wuhrer M (2012) Changes in antigen-specific IgG1 Fc N-glycosylation upon influenza and tetanus vaccination, Mol. Cell. Proteomics 11, M111 014563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahan AE, Jennewein MF, Suscovich T, Dionne K, Tedesco J, Chung AW, Streeck H, Pau M, Schuitemaker H, Francis D, Fast P, Laufer D, Walker BD, Baden L, Barouch DH, and Alter G (2016) Antigen-Specific Antibody Glycosylation Is Regulated via Vaccination, PLoS Pathog. 12, e1005456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnold JN, Wormald MR, Sim RB, Rudd PM, and Dwek RA (2007) The impact of glycosylation on the biological function and structure of human immunoglobulins, Annu. Rev. Immunol. 25, 21–50. [DOI] [PubMed] [Google Scholar]
- 21.Subedi GP, Hanson QM, and Barb AW (2014) Restricted motion of the conserved immunoglobulin G1 N-glycan is essential for efficient Fc gamma RIIIa binding, Structure 22, 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subedi GP, and Barb AW (2015) The Structural Role of Antibody N-Glycosylation in Receptor Interactions, Structure 23, 1573–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subedi GP, and Barb AW (2016) The immunoglobulin G1 N-glycan composition affects binding to each low affinity Fc gamma receptor, MAbs 8, 1512–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel KR, Roberts JT, Subedi GP, and Barb AW (2018) Restricted processing of CD16a/Fc gamma receptor IIIa N-glycans from primary human NK cells impacts structure and function, J. Biol. Chem. 293, 3477–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrara C, Stuart F, Sondermann P, Brunker P, and Umana P (2006) The carbohydrate at FcgammaRIIIa Asn-162. An element required for high affinity binding to non-fucosylated IgG glycoforms, J. Biol. Chem. 281, 5032–5036. [DOI] [PubMed] [Google Scholar]
- 26.Shibata-Koyama M, Iida S, Okazaki A, Mori K, Kitajima-Miyama K, Saitou S, Kakita S, Kanda Y, Shitara K, Kato K, and Satoh M (2009) The N-linked oligosaccharide at Fc gamma RIIIa Asn-45: an inhibitory element for high Fc gamma RIIIa binding affinity to IgG glycoforms lacking core fucosylation, Glycobiology 19, 126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrara C, Grau S, Jager C, Sondermann P, Brunker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, Umana P, and Benz J (2011) Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose, Proc. Natl. Acad. Sci. U.S.A 108, 12669–12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizushima T, Yagi H, Takemoto E, Shibata-Koyama M, Isoda Y, Iida S, Masuda K, Satoh M, and Kato K (2011) Structural basis for improved efficacy of therapeutic antibodies on defucosylation of their Fc glycans, Genes Cells 16, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deringer VL, Englert U, and Dronskowski R (2016) Nature, Strength, and Cooperativity of the Hydrogen-Bonding Network in alpha-Chitin, Biomacromolecules 17, 996–1003. [DOI] [PubMed] [Google Scholar]
- 30.Nishiyama Y, Langan P, and Chanzy H (2002) Crystal structure and hydrogen-bonding system in cellulose Ibeta from synchrotron X-ray and neutron fiber diffraction, J. Am. Chem. Soc. 124, 9074–9082. [DOI] [PubMed] [Google Scholar]
- 31.Popov D, Buleon A, Burghammer M, Chanzy H, Montesanti N, Putaux J-L, Potocki-Veronese G, and Riekel C (2009) Crystal Structure of A-amylose: A Revisit from Synchrotron Microdiffraction Analysis of Single Crystals Macromolecules 42, 1167–1174. [Google Scholar]
- 32.Barb AW, Borgert AJ, Liu M, Barany G, and Live D (2010) Intramolecular glycan-protein interactions in glycoproteins, Methods Enz. 478, 365–388. [DOI] [PubMed] [Google Scholar]
- 33.Chen W, Enck S, Price JL, Powers DL, Powers ET, Wong CH, Dyson HJ, and Kelly JW (2013) Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins, J. Am. Chem. Soc. 135, 9877–9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayes JM, Frostell A, Cosgrave EF, Struwe WB, Potter O, Davey GP, Karlsson R, Anneren C, and Rudd PM (2014) Fc gamma receptor glycosylation modulates the binding of IgG glycoforms: a requirement for stable antibody interactions, J. Proteome Res. 13, 5471–5485. [DOI] [PubMed] [Google Scholar]
- 35.Subedi GP, Falconer DJ, and Barb AW (2017) Carbohydrate-Polypeptide Contacts in the Antibody Receptor CD16A Identified through Solution NMR Spectroscopy, Biochemistry. 56, 3174–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank M, Walker RC, Lanzilotta WN, Prestegard JH, and Barb AW (2014) Immunoglobulin G1 Fc domain motions: implications for Fc engineering, J. Mol. Biol. 426, 1799–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierce LC, Salomon-Ferrer R, Augusto F. d. O. C., McCammon JA, and Walker RC (2012) Routine Access to Millisecond Time Scale Events with Accelerated Molecular Dynamics, J. Chem. Theory Comput. 8, 2997–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsumiya S, Yamaguchi Y, Saito J, Nagano M, Sasakawa H, Otaki S, Satoh M, Shitara K, and Kato K (2007) Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1, J. Mol. Biol. 368, 767–779. [DOI] [PubMed] [Google Scholar]
- 39.Subedi GP, Johnson RW, Moniz HA, Moremen KW, and Barb A (2015) High Yield Expression of Recombinant Human Proteins with the Transient Transfection of HEK293 Cells in Suspension, J. Visualized Exp, e53568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kao D, Danzer H, Collin M, Gross A, Eichler J, Stambuk J, Lauc G, Lux A, and Nimmerjahn F (2015) A Monosaccharide Residue Is Sufficient to Maintain Mouse and Human IgG Subclass Activity and Directs IgG Effector Functions to Cellular Fc Receptors, Cell Rep. 13, 2376–2385. [DOI] [PubMed] [Google Scholar]
- 41.Barb AW, and Prestegard JH (2011) NMR analysis demonstrates immunoglobulin G N-glycans are accessible and dynamic, Nat. Chem. Biol. 7, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barb AW (2017) Quantifying Carbohydrate Motions Through Solution Measurements: Applications to Immunoglobulin G Fc, In NMR in Glycoscience and Glycotechnology (Kato K, and Peters T, Eds.), pp 208–227, Royal Society of Chemistry, Cambridge, UK. [Google Scholar]
- 43.Barb AW, Meng L, Gao Z, Johnson RW, Moremen KW, and Prestegard JH (2012) NMR characterization of immunoglobulin G Fc glycan motion on enzymatic sialylation, Biochemistry 51, 4618–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakae Y, Satoh T, Yagi H, Yanaka S, Yamaguchi T, Isoda Y, Iida S, Okamoto Y, and Kato K (2017) Conformational effects of N-glycan core fucosylation of immunoglobulin G Fc region on its interaction with Fcgamma receptor IIIa, Sci. Rep. 7, 13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okeley NM, Alley SC, Anderson ME, Boursalian TE, Burke PJ, Emmerton KM, Jeffrey SC, Klussman K, Law CL, Sussman D, Toki BE, Westendorf L, Zeng W, Zhang X, Benjamin DR, and Senter PD (2013) Development of orally active inhibitors of protein and cellular fucosylation, Proc. Natl. Acad. Sci. U.S.A 110, 5404–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barb AW, Brady EK, and Prestegard JH (2009) Branch-specific sialylation of IgG-Fc glycans by ST6Gal-I, Biochemistry 48, 9705–9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collin M, and Olsen A (2001) EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG, EMBO J. 20, 3046–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trimble RB, and Tarentino AL (1991) Identification of distinct endoglycosidase (endo) activities in Flavobacterium meningosepticum: endo F1, endo F2, and endo F3. Endo F1 and endo H hydrolyze only high mannose and hybrid glycans, J. Biol. Chem. 266, 1646–1651. [PubMed] [Google Scholar]
- 49.Niedermeyer TH, and Strohalm M (2012) mMass as a software tool for the annotation of cyclic peptide tandem mass spectra, PLoS One 7, e44913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr. D 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, and Vagin AA (2011) REFMAC5 for the refinement of macromolecular crystal structures, Acta Crystallogr. D 67, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Case DA, Babin V, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham TE, Darden TA, Duke RE, Gohlke H, Goetz AW, Gusarov S, Homeyer N, HJanowski P, Kaus J, Kolossvary I, Kovalenko A, Lee TS, LeGrand S, Luchko T, Luo R, Madej B, Merz KM, Paesani F, Roe DR, Riotberg A, Sagui C, Salomon-Ferrer R, Seabrea G, Simmerling CL, Smith W, Swails J, Walker RC, Wang J, Wolf RM, Wu X, and Kollman PA (2014) AMBER14, University of California, San Francisco. [Google Scholar]
- 53.Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Foley BL, and Woods RJ (2008) GLYCAM06: a generalizable biomolecular force field. Carbohydrates, J. Comput. Chem. 29, 622–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes, J. Biomol. N.M.R. 6, 277–293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.