Abstract
The rapidly growing field of microbiome research presents a need for better methods of monitoring gut microbes in vivo with high spatial and temporal resolution. We report a method of tracking microbes in vivo within the gastrointestinal tract by programming them to incorporate non-standard amino acids (NSAA) and labeling them via click chemistry. Using established machinery constituting an orthogonal translation system (OTS), we engineered Escherichia coli to incorporate p-azido-L-phenylalanine (pAzF) in place of the UAG (amber) stop codon. We also introduced a mutant gene encoding for a cell surface protein (CsgA) that was altered to contain an in-frame UAG codon. After pAzF incorporation and extracellular display, the engineered strains could be covalently labeled via copper-free click reaction with a Cy5 dye conjugated to the dibenzocyclooctyl (DBCO) group. We confirmed the functionality of the labeling strategy in vivo using a murine model. Labeling of the engineered strain could be observed using oral administration of the dye to mice several days after colonization of the gastrointestinal tract. This work sets the foundation for the development of in vivo tracking microbial strategies that may be compatible with non-invasive imaging modalities and are capable of longitudinal spatiotemporal monitoring of specific microbial populations.
Keywords: Curli Fibers, Non-standard Amino Acid, Click Chemistry, Microbiome Imaging
Introduction
Studies of interactions between resident microbes and their hosts are revealing a variety of previously unknown mechanisms by which microbes influence human physiology. The human gastrointestinal (GI) tract, which contains both the highest diversity and number of microbes in the human body, is a dynamic environment whose microbial composition can be affected by many factors, including: diet, age, and immune profile1, 2 Such changes have been linked to a wide range of disease states, including inflammatory bowel disease, obesity, resistance to pathogens, diabetes, and neurodegenerative disease1–3. These correlations have led to a need for reliable methods to monitor host-microbe interactions. A great deal has been learned about the compositional changes of the gut microbiome from DNA and RNA sequencing of fecal and tissue samples4–8 This approach can be non-invasive, and yields significant information about many bacterial species simultaneously, but does not give precise information about the spatiotemporal localization or density of specific microbes in the host. Other techniques, like fluorescence in situ hybridization (FISH) and immunofluorescence staining of microbes from harvested tissues, offer more spatial information9, 10. However, these methods are inherently invasive and require tissue biopsies in larger animals, or can be performed at the endpoints of small animal studies, which hinders longitudinal studies and real-time tracking. Therefore, less invasive tools to track the spatiotemporal dynamics of gut microbes in vivo would be useful for microbiome study.
The need for such tools is further underscored by recent efforts to use living microbes as orally deliverable therapeutics and diagnostics11–16. Unlike traditional drugs, whose pharmacokinetics and pharmacodynamics (PK/PD) can be tracked experimentally with medical imaging techniques, or predicted based on well-established models, microbial PK/PD is comparatively harder to characterize. Indeed, microbes exhibit a range of context-dependent active processes, including self-replication, chemotaxis, and biochemical communication with other cells in their environment, that make it difficult to determine their bio-distribution in a time-resolved manner. Thus, if living microbes are to be treated as sophisticated versions of drugs or drug delivery vehicles, we must develop better tools to track their trafficking and interactions with their surroundings using medically-relevant imaging techniques.
There are some techniques to track labeled microbes in vivo. One common method involves genetically engineering microbes to express luciferase or fluorescent proteins so that they can be imaged using in vivo imaging techniques17–19. However, the poor tissue permeability of optical wavelengths limits the use of such techniques to small animals. Additionally, since most of the GI tract is anaerobic, low oxygen availability can inhibit the proper function of certain fluorophores and chromophores and limit the signal that is produced20. Furthermore, the spatial resolution of optical imaging on small animals reveals little about the location of the microbes in the gut or in other organs. Another recent example used click chemistry to fluorescently label anaerobic bacteria displaying azido sugars on their surface21–23. This enabled pre-labeled bacteria to be tracked during their transit through the GI tract. However, the technique required metabolic incorporation of azido sugars in the bacteria prior to ingestion, so progeny cells were no longer labeled, precluding the longitudinal tracking of the microbes beyond 24 hours.
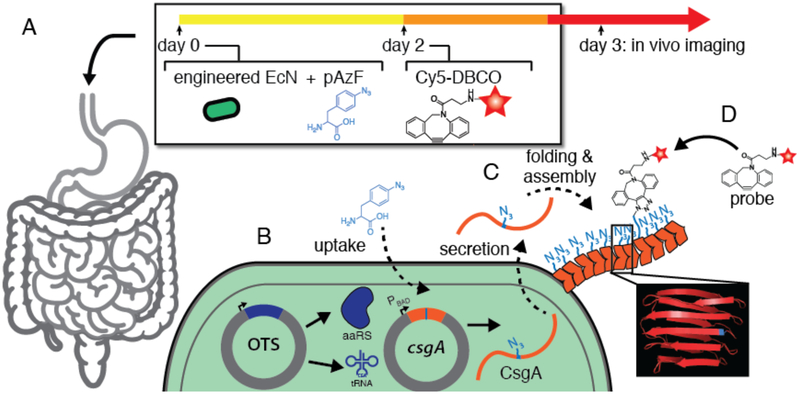
An ideal in vivo tracking system for microbes in humans and other large animals should satisfy two primary criteria. First, the spatial localization of the microbes should be ascertainable with some precision using medically-relevant and widely available imaging modalities. Second, it should be feasible to track the microbes longitudinally over the course of several days to months. This line of reasoning led us to a design wherein a biorthogonal click reaction is used to target an imaging probe to microbes that can stably display an appropriate click-able functional group. The imaging probe could then be delivered orally at any time point after microbial ingestion. The versatility of the click reaction would also make this approach adaptable to multiple imaging modalities, depending on the need. Here we report a proof of concept study that satisfies the criteria stated above. The click-able functional group was introduced by virtue of an orthogonal translation system (OTS) that enables the probiotic Escherichia coli Nissle 1917 (EcN) to incorporate the NSAA p-azido-phenylalanine (pAzF) at UAG (amber) stop codons24. The strain also harbors a gene expressing an extracellularly displayed protein, csgA, containing an in-frame UAG codon mutation. Together, these modifications enable the bacteria to incorporate and display the bioorthogonal chemical handle during residence in the GI tract that can be targeted by a click-able imaging probe of choice (Figure 1).
Figure 1. Tracking of engineered bacteria in vivo using non-standard amino acid incorporation,
(A) Bacteria, pAzF, and imaging probe (Cy5-DBCO) are delivered orally in sequence as shown. (B) The bacteria have been engineered to express an orthogonal translation system (OTS) and a gene containing an in-frame UAG codon (csgA, orange). (C) Bacteria residing in the gastrointestinal tract incorporate pAzF and display it on their surface through the secretion and assembly of extracellular curli fibers (orange chevrons) at any point during their residency. (D) The displayed azide functional group can be targeted by an imaging probe in order to label the engineered bacterial population.
Results
Azide-containing proteins are successfully secreted and assembled extracellularly from engineered E. coli Nissle 1917
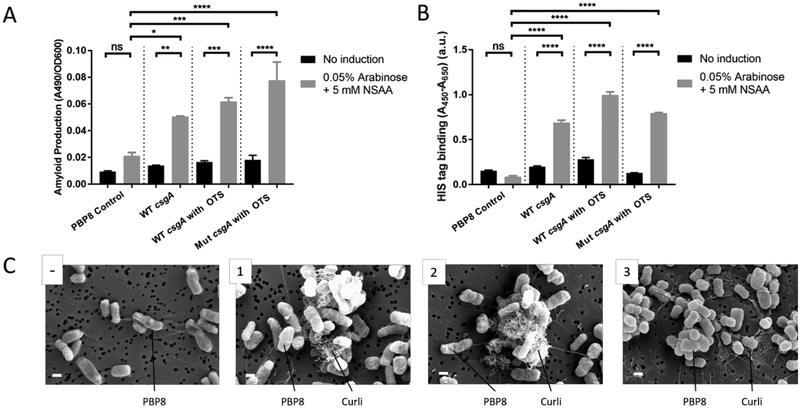
In order for the incorporated pAzF to be accessible to imaging probes, we reasoned that it had to be displayed extracellularly. We incorporated the requisite in-frame UAG codon into the csgA gene, which encodes for curli fibers. Curli are a biofilm matrix component native to E. coli that are produced through the secretion and self-assembly of the CsgA protein into cell-anchored amyloid fibers25, 26. We created two plasmid constructs - pBbB8k-wt and pBbB8k-mut that contained either the wild-type (wt) or mutant csgA (mut csgA), respectively, in the same cistron as the other curli genes necessary for secretion and assembly, all under the control of an inducible promoter (PBAD). The mut csgA was generated in such a way that the UCA codon that normally encodes for serine at position 89 in the CsgA amino acid sequence was mutated to an amber stop codon (UAG) (Figure S1), which can be recognized by the OTS to incorporate the pAzF in this location during translation. S89 was chosen for the location of the mutation because the native serine residue is predicted to be surface-accessible in the assembled amyloid state, based on multiple structural models of the parallel β-sheet structure of CsgA27–29. Hence, we reasoned that the location of the mutation would interfere neither with CsgA’s structure nor its assembly into amyloid fibers. A separate plasmid construct (pEVOL-pAzF) harbored the OTS, encoding for a tRNA and an aminoacyl tRNA synthetase that worked together to incorporate pAzF specifically and orthogonally at UAG codons. All in vitro experiments were performed with an EcN-derived strain called PBP8, harboring chromosomal deletions of all the curli genes (EcN::ΔcsgBACDEFG). The genotype of PBP8 was confirmed by sequencing (Figure S2) and its phenotype was confirmed by decreased staining with the amyloid-specific dye, Congo Red30, 31, compared to the parent EcN strain under conditions known to produce curli fibers (Figure S3)32, 33.
PBP8 co-transformed with pEVOL-pAzF and either pBbB8k-wt or pBbB8k-mut successfully produced amyloid fibers in the presence of inducer and pAzF, as confirmed by a quantitative Congo Red (CR) binding assay. In contrast, when any of the necessary components - OTS, csgA mut gene, or inducer - were omitted, less CR binding was observed (Figure 2A). The identity of the extracellular amyloid was confirmed with a whole-cell filtration ELISA that measured the C-terminal His-6 tag on both wt and mutant CsgA (Figure 2B). Electron microscopy of induced cultures further confirmed the ability of the co-transformed PBP8 to produce amyloid fibers with the same morphological features as wild-type curli fibers (Figure 2C). Notably, induced mutants in the absence of pAzF produced a truncated version of CsgA, which could also assemble into amyloid fibers (Figure S4). However, overproduction of this truncated CsgA could lead to slower growth of the engineered cells, which did not happen when the cells were induced in the presence of pAzF (Figure S5).
Figure 2. Engineered bacteria can produce mutant curli fiber.
(A) Amyloid production assay for various strains, based on Congo Red binding, with and without induction. (B) Whole-cell filtration ELISA assays with anti-HIS antibody detection confirms the presence of extracellular HIS-tagged curli fibers under appropriate conditions. Ordinary one way ANOVA with Tukey’s multiple comparison test n=3, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 (C) Scanning electron microscopy of PBP8 transformed with various plasmid combinations: (−) no plasmid (1) pBb8k-wt (containing wild-type csgA) (2) pBb8k-wt and pEVOL-pAzF (containing OTS) (3) pBb8k-mut (containing mutant csgA) and pEVOL-pAzF. All samples with curli genes successfully express curli fibers including the mutant curli fibers (scale bar = 1 μm)
Overproduction of recombinant proteins can be metabolically taxing, hindering the ability of engineered bacteria to compete in the stringent environment of the gut34, 35. We used growth curves of engineered EcN strains to evaluate fitness. We observed little variation in doubling times between the parent strain and those harboring either plasmid with and without induction (Figure S5), suggesting that neither OTS nor curli expression negatively affects growth.
Engineered curli fibers containing non-standard amino acids can perform click chemistry in vitro
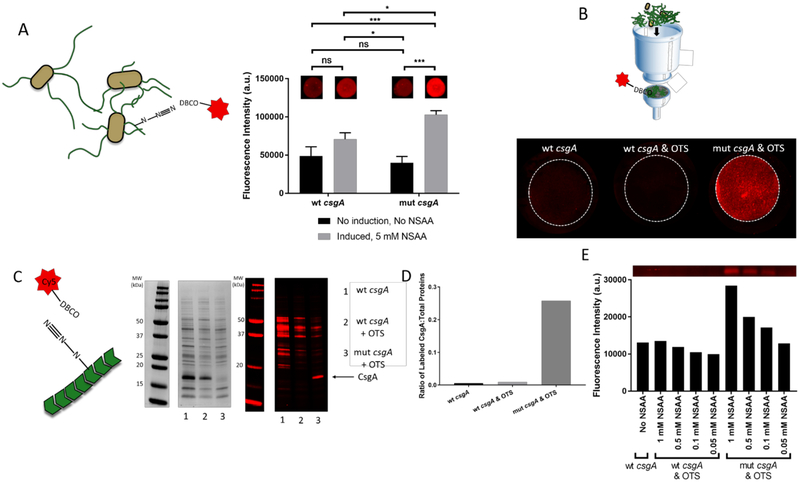
To determine the ability of the mutant curli fibers to perform click chemistry labeling, we exposed induced broth cultures to a Cy5-dibenzocyclooctyl (DBCO) conjugate, a probe designed to react with pAzF. In this case, the cycloalkyne group of DBCO is able to undergo copper-free click chemistry with the azide moiety of pAzF36. After exposure to the probe conjugate, the bacterial cells were pelleted and washed by resuspending several times in PBS, then aliquots were spotted onto a nitrocellulose membrane and imaged. As expected, PBP8 cultures induced to express mutant curli gave the highest fluorescence signal. However, induced cultures producing wild-type curli exhibited increased signal over the background, suggesting that non-specific adsorption of the Cy5 dye to curli fibers could be a factor in determining labeling efficiency (Figure 3A). The click reaction yield was not affected significantly by Cy5-DBCO concentrations above 20 μM and labeling times above 1 hour, underscoring the robustness of the azide-DBCO reaction for rapid labeling under a range of conditions (Figure S6).
Figure 3. In vitro click chemistry-dependent labeling of engineered curli fibers,
(A) Fluorescence signal from cell cultures spotted onto nitrocellulose membranes and labeled with Cy5-DBCO. Ordinary one way ANOVA with Tukey’s multiple comparison test n=3, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 (B) Fluorescence signal after labeling of curli amyloid mats after cell removal. (C) SDS-PAGE analysis of lysed cells after induction and semi-purification of curli fibers. Coomassie blue stained gel (left) and fluorescence signal from unstained gel (right). (D) Ratio of labeled CsgA to total labeled proteins from various experiment conditions (E) Fluorescence intensity of CsgA variants as a function of pAzF concentration.
To test the labeling efficiency of the mutant curli fibers while removing the effects of non-specific binding to the bacterial cells themselves, we performed labeling reactions on crudely purified curli fibers. We employed a previously established isolation protocol to obtain curli fiber mats in the absence of cells37. The filter membrane-supported curli mats were incubated with Cy5-DBCO, washed, and imaged (Figure 3B). The fluorescence results indicate that the dye bound specifically to the curli fiber mats only when the mutant fibers, OTS, and pAzF were present.
Click labeling is specific to mutant curli fibers
We selected the OTS designed for pAzF incorporation at UAG sites (pEVOL) for several reasons. First, pAzF and the pEVOL system are well-characterized and have been shown to have high incorporation efficiency into mutant proteins24, 35 Second, UAG is the rarest codon in the E. coli genome, with only 366 instances in EcN. Notably, efforts have been made to increase the specificity and efficiency of NSAA incorporation by replacing all native instances of UAG with a synonymous stop codon across the E. coli genome, permitting the deletion of RF-1, which terminates translation at UAG codons38. Since we did not use a recoded strain for this work, there remained the possibility of pAzF incorporation at other genes terminated by UAG in the presence of the OTS. Although it was unclear how much this would affect the success of the in vivo labeling scheme, we wanted to characterize the level of pAzF incorporation in proteins other than CsgA and demonstrate the preferential binding of the dye to the mutant CsgA. We performed SDS-PAGE analysis on semi-purified labeled lysed cells to determine the distribution of dye across the PBP8 proteome (Figure 3C). In the Coomassie-stained gel, the semi-purified proteins from cultures expressing wild-type curli with and without the OTS showed stronger CsgA bands near 17 kDa, compared to those expressing mutant CsgA (Figure 3C). This is perhaps the result of inefficient translation of full-length CsgA, since pAzF incorporation must compete with premature termination from the cell’s native release factor-1 (RF-1). The fluorescence-imaged gel showed that cells expressing mut csgA and the OTS exhibited higher intensity for the CsgA band compared to other background proteins in the lysate, whereas those expressing wild-type fibers showed higher non-specific labeling of other proteins. Expressed as a ratio of fluorescence intensities between (CsgA):(all other proteins), the labeling specificity for CsgA was 45x higher when the mut csgA gene was present compared to the wt csgA gene, in the presence of the OTS (Figure 3D). Even with the lower absolute amount of mutant CsgA, we observed much higher dye labeling efficiency for the mutant CsgA compared to wild-type CsgA. Furthermore, the fluorescence intensity of the CsgA band in the gel increases in a dose-dependent manner with pAzF concentration, with labeling being indistinguishable from background signal at a concentration of 0.05 mM (Figure 3E).
Labeling efficiency of engineered bacteria
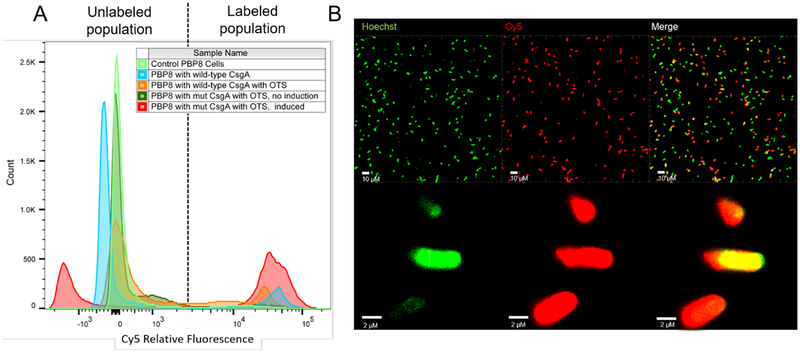
We assessed the labeling efficiency on a per cell basis by subjecting bacterial cultures that had been induced to produce curli fibers and exposed to Cy5-DBCO to flow cytometry analysis. This confirmed that there was some dye association with cells producing wild-type CsgA both without (17.9% labeling) and with (26.2% labeling) the OTS present. However, more than 63.8% of PBP8 cells producing mutant CsgA in the presence of the OTS and the NSAA were labeled (Figure 4A, Figure S7). The non-specific labeling appeared to be dependent on curli production, since it was present at a lower level in the case of untransformed or non-induced PBP8 cells, corroborating the results shown in Figure 3B. Confocal microscopy qualitatively supported the flow cytometry data, with more than half of the cells exhibiting a signal for the Cy5 dye (Figure 4B).
Figure 4. Labeling efficiency of engineered bacteria,
(A) Flow cytometry histogram of cell cultures under various conditions after exposure to Cy5-DBCO. (B) Confocal fluorescent micrographs of PBP8 harboring pBb8k-mut and pEVOL-pAzF after induction and labeling.
Engineered probiotic bacteria with mutant curli fibers are able to perform click chemistry in vivo in murine model
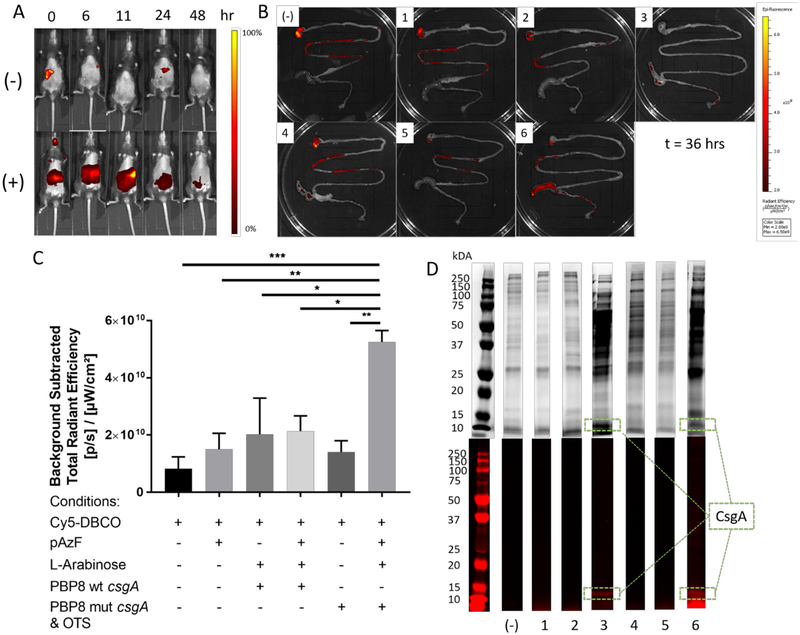
In order to demonstrate the feasibility of performing the labeling reaction in vivo, we administered the engineered EcN to mice via oral gavage and induced the expression of mutant curli using arabinose and pAzF in their drinking water. Cy-5-DBCO was administered orally 42 hours after administering the bacteria, and Cy5 fluorescence signals from the abdominal area were detected using an in vivo imaging system (IVIS) over the course of 48 hours (Figure 5A). Fluorescence intensity remained high in mice that had been administered the engineered PBP8 up to 24 hours after dye administration. Control experiments with no bacterial administration showed signal decay after 6 hours. However, subsequent experiments with non-induced bacteria made it difficult to distinguish click chemistry-based labeling from non-specific dye binding and tissue autofluorescence (Figure S8). We, therefore, sought to demonstrate the specificity of the labeling reaction using ex vivo methods and additional experimental conditions (i.e. with and without bacteria administration, dye administration, and pAzF administration). Based on the previous experiments, we selected 36 hours post-dye administration as an experimental endpoint that would ensure that even non-specifically bound dye would have been cleared from the GI tract. The in vivo experimental scheme is depicted in Figure S9. After harvesting of the GI tracts, Cy5 signal was measured using the IVIS. Significant tissue/food autofluorescence was observed from the stomach, so analysis was confined to the cecum and lower intestine. Qualitative and quantitative analysis showed that the experimental group containing all the necessary components (bacteria, mutant CsgA, pAzF, and inducer) showed a distinct increase in fluorescence intensity measured at 675 nm in the cecum, which is the primary location of E. coli in the mice39, compared to any of the other experimental conditions (Figure 5B,C). Low intensities were observed in the cecum of mice that had been administered bacteria producing wild-type curli fibers, which correlated to the non-specific dye adsorption observed in vitro (Figure 5B, panels 3 and 4). Conditions in which pAzF was administered without the necessary machinery for incorporation exhibited cecal fluorescence intensities that were not significantly higher than that observed for dye administration alone, suggesting that non-specific incorporation of pAzF by host cells or other microbes using native machinery did not contribute significantly to the signal observed for the desired click labeling. Finally, we performed SDS-PAGE analysis of homogenized cecum tissue in order to determine if the fluorescence signals that we observed from the lower GI tract were actually from the click-labeled CsgA protein (Figure 5D). Coomassie staining revealed protein bands at the expected size for mutant CsgA (14.7 kDa) in group 3 (induced PBP8, wild-type curli) and group 6 (induced PBP8, mutant curli). Fluorescence analysis of the gels before Coomassie staining revealed that the observed signal could be attributed to these same bands, with higher intensities for the mutant curli. No other labeled proteins were observed, confirming the reaction specificity for the incorporated pAzF probe and engineered bacteria.
Figure 5. Engineered bacteria are labeled via click chemistry in vivo.
(A) Fluorescence imaging of live mice fed with either PBS only (−), or PBP8 harboring pBbk8-mut and pEVOL-pAzF (+). The mice were given arabinose and pAzF to induce mutant curli expression 2 days prior to the start of the experiment. Images were taken at a different time points after Cy5-DBCO administration at time = 0 hours. (B) Representative fluorescence images of harvested mouse GI tracts, collected 36 hours after Cy5-DBCO administration under various conditions: (−) negative control with no bacteria or dye administration, (1) no bacteria, (2) no bacteria, but with pAzF in drinking water, (3) PBP8 expressing wt-csgA, (4) PBP8 expressing wt-csgA with pAzF in water, (5) non-induced PBP8 expressing mut-csgA with OTS (6) PBP8 expressing mut-csgA with OTS. (C) Quantification of Cy5 fluorescence signals from the lower GI tracts. Ordinary one way ANOVA with Tukey’s multiple comparison test n=3, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 (D) SDS-PAGE analysis of homogenized cecal contents stained with Coomassie blue (top) and unstained fluorescent imaging (bottom). Lane markers correspond to numbering in part B.
Discussion
Most existing methods for studying the spatiotemporal dynamics within the gut microbiome rely on analytical techniques that are either indirect (e.g. fecal bioinformatics), or highly invasive (e.g. histology). Less invasive methods to track microbes in live animals would benefit fundamental microbiome research as well as the development of living microbial therapeutics and diagnostics. Here we describe a new method to label engineered bacteria in vivo using bio-orthogonal click chemistry. This technique enables monitoring of microbial localization several days after ingestion and should be compatible with any “click-able” imaging probe.
This work shows a basic proof-of-concept that such a labeling strategy can be successfully performed. However, there are several ways in which the technique could be improved upon. The in vitro results suggest that inefficient translation of the mutant CsgA, even in the presence of the OTS, could limit the amount of CsgA that was produced. Although we did not find this to prevent the proof-of-concept demonstration, a recoded E. coli strain might significantly improve the NSAA incorporation efficiency by removing the possibility for premature termination via RF-1. The labeling efficiency on a per microbe basis in vitro was also only ~60%, which we speculate was related to the well-documented nature of arabinose-inducible promoter systems40. This suggests that further optimization of promoter systems could also contribute to higher signals in vivo. Detachment of curli fibers from bacteria in the gut could also complicate interpretation of the imaging results, so other proteins bound directly to the cell surface could be worth exploring as alternative labeling targets. Other cell-surface proteins as labeling targets could also ameliorate the non-specific dye labeling observed for assembled CgsA. Autofluorescence of the upper GI tract and long clearance times for the Cy5-DBCO dye also hindered effective in vivo imaging of the engineered bacteria. Other imaging probes with further red-shifted emissions or probes that incorporate a “turn-on” function upon undergoing the click reaction could improve signal-to-noise ratios. With regard to the potential toxicity of pAzF as in vivo, there is limited data. It is worth noting that copper-free chemistry using Cy5-DBCO has been performed in live mice and rat with no apparent toxicity41–44. We did not observe any apparent toxicity in our short term study, though a more thorough toxicity study may have to be performed in the future to assess the safety of pAzF in mouse models.
The approach we demonstrate here could enable the development of analytical techniques with new capabilities. For example, the in-frame UAG mutation could be used to report specifically on the production of other extracellularly displayed proteins. Furthermore, the proliferation of OTS systems compatible with different NSAAs and organisms could enable a multiplexed approach to tracking multiple microbes simultaneously. By changing the nature of the probe, one could envision longitudinal, non-invasive imaging of microbial populations in humans or large animals using clinical imaging modalities, like MRI or PET, which enhance tissue penetration in the way that fluorescence or luminescence signals cannot. Excitement for such prospects must be tempered with the requirement that the cellular population to be tracked using this technique must be genetically engineered to enable NSAA incorporation. However, with the meteoric rise of CRISPR/Cas9-based techniques to rapidly modify genomes and the continuing development of new OTS, this could become increasingly accessible in the near future.
Ongoing studies in our lab are focused on the development of a second generation system that, in addition to addressing the issues raised above, would rely on incorporation of the OTS and mutant genes in to the bacterial chromosome so that antibiotic selection would not be required during in vivo deployment. Overall, our approach enables a wider selection of imaging probes compared to conventional microbe tracking methods, such as fluorescent labelling or luminescent proteins expression. Non-protein dyes or inorganic probes can be utilized because they are provided externally and do not have to rely on oxygen-dependent maturation chemistry, like some fluorescent proteins. Alternatively, the incorporation and display of the azide bearing amino acid could be tied to other environmental sensing mechanisms relevant to the gut11, 45, leading to diagnostics that would report on disease states with spatial resolution within the GI tract. Furthermore, the approach may be useful for tracking the progression of infections in model systems.
Experimental Procedures
Cell strains and plasmids
The divergent curli operon regions consisting of csgBAC and csgEFG were PCR isolated from E. coli K12 substr. W3110 and cloned by overlap extension into the pBbB8k plasmid, to create a single operon, csgBACEFG, under the control of the araBAD promoter. The cloning of the curli operon was performed in an analogous manner to our previously published work37. A six-histidine tag (HisTag) was added to the C-terminus of CsgA to allow for immunodetection (pBbB8k-WT) (See plasmid map in figure S10). A control plasmid was constructed by cloning the malE gene encoding for the maltose binding protein (MBP) from the W3110 genome into the pBbB8k plasmid under the control of the araBp promoter (pBbB8k-MBP).
To allow for the incorporation of the NSAA, a mutation encoding for the amber stop codon (UAG) was inserted within the open-reading frame of CsgA (pBbB8k-Mut csgA). Using site-directed mutagenesis, we directed the amber mutation in place of a UCA codon, which normally encodes for serine at amino acid position 89 of CsgA (See DNA sequences in supporting information 1 and 2).
The orthogonal translation system (OTS) plasmid pEVOL-pAzF24 was obtained from the Church Lab with slight modification where the chloramphenicol resistance was swapped to spectinomycin resistance and the sequences were codon optimized.
Protein expression was performed in a curli operon deletion mutant, PBP8, an E. coli strain derived from Nissle 1917 (ECN). PBP8 strain was constructed using lambda red recombineering method described previously by Datsenko and Wanner46. In brief, the chloramphenicol acetyltransferase (CAT) cassette was constructed with 500 bp homology arms upstream and downstream of the curli operon using SOEing PCR. The CAT cassette was transformed into electrocompetent EcN with pre-induced lambda red genes from the pKD46 plasmid using 0.5% arabinose (Sigma) during the growth step of electrocompetent cell preparation. The transformant was expanded using SOC media at 30°C shaking incubator for 3 hours to promote the genomic replacement of the curli operon with CAT cassette. The transformant was then plated onto lysogeny broth (LB) agar plates (Sigma) containing 25 μg/mL chloramphenicol (RPI) and incubated overnight at 37°C. Colonies that are resistant to chloramphenicol were selected and verify the curli operon knockout by colony PCR and genomic DNA sequencing comparing to wild-type EcN.
Mice
Female 6- to 8-week-old C57BL/6 mice were purchased from Charles River Laboratories. Mice were housed in SPF conditions with sterile food and water ad libitum. The animal chow included LabDiets 5K67 and the non-fluorescent food LabDiets 5V5R. Mice were maintained in sterile vinyl isolators equipped with food, water and bedding in the Harvard Medical School animal facility. Before any experiment, mice had at least one week to acclimatize to the facility environment. All experiments were conducted in accordance with US National Institutes of Health guidelines and approved by the Harvard Medical Area Standing Committee on Animals.
In vitro expression of curli fibers
PBP8 cells transformed with a pBbB8k plasmid were streaked onto LB agar plates containing 50 μg/mL kanamycin (Teknova). PBP8 cells transformed both with a pBbB8k plasmid and with the OTS plasmid were streaked onto LB agar plates containing both 50 μg/mL kanamycin and 50 μg/mL spectinomycin (RPI). Colonies were picked from the plates and 5 mL cultures were inoculated (in LB containing the appropriate antibiotics). Cultures were grown overnight at 37°C. The overnight cultures were diluted 100-fold in fresh LB medium containing the appropriate antibiotics, and cultured at 37°C until they reached an optical density (OD) at 600nm of 0.6 to 0.8. Arabinose was added to the bacterial cultures at a final concentration of 0.05 % to induce protein expression. If NSAA incorporation was desired, the NSAA p-azido-L-phenylalanine (pAzF) (Toronto Research Chemicals) was also added to the cultures to a final concentration ranging between 1 mM and 5mM. Higher concentrations affected cell growth. Protein expression was allowed to occur at 37°C overnight.
Quantitative Congo Red binding assay
The quantitative CR binding assay was adapted from a previously published protocol32. In brief, one mL of testing bacteria culture was centrifuged at 8,000 rpm for 10 minutes. The pellet was resuspended gently in 0.025 mM CR in phosphate buffered saline (PBS), incubated at 25 °C for 10 minutes and centrifuged at 15,000 rpm for 10 minutes. 150 μL of the supernatant was transferred to a 96-well plate with clear bottom (Corning) and the absorbance at 490 nm was read using a BioTek H1 microplate reader. The amount of CR binding was calculated by subtracting the measured absorbance from the 0.025 mM CR in PBS control absorbance and normalized by the OD600 of the culture.
Whole-cell filtration ELISA
The whole-cell filtration ELISA assay was adapted from a previously published protocol47, 48. In brief, the testing bacteria cultures were chilled on ice for at least 20 minutes and diluted to OD600 of 0.3 using tris-buffered saline (TBS) (Thermo Fisher Scientific). 200 μL of the diluted samples were added to a Multiscreen-GV 96-well filter plate (0.22 μm pore size; EMD Millipore) and filtered. The sample wells were washed three times with a wash buffer (TBS, 0.1% Tween-20 (Sigma)) and blocked by incubating with 1% bovine serum albumin (BSA) (Sigma) and 0.01% H2O2 (Sigma) in wash buffer for 1.5 hours at 37 °C. The 3-time wash step was repeated. The wells were incubated with 50 μL of anti-HIS antibody-horseradish peroxidase (HRP) conjugated (1:200 dilution) (Thermo Fisher Scientific) for 2 hours at 25 °C and washed 3 times. 100 μL of Ultra-TMB (3,3’,5,5’-tetramethylbenzidine) ELISA substrate (Thermo Fisher Scientific) was added to each well and incubated for 20 minutes at 25 °C. Then, 50 μL of 2 M H2SO4 (Alfa Aesar) was added to the wells to stop the reaction. 100 μL of this reaction was transferred to a 96-well plate where the absorbance at 450 nm and 650 nm was measure. The relative amount of displayed HIS tag was obtained by subtracting absorbance at 450 nm with absorbance at 650 nm.
Electron microscopy
Cells expressing curli fibers (with and without pAzF) were imaged by scanning electron microscopy (SEM) to assess the formation of fibers. 100 μl of cell cultures were filtered onto Nucleopore filter membranes (0.22 um pore size, GE Healthcare Bio-Sciences) under vacuum. The samples were fixed with 2 % (m/v) glutaraldehyde and 2 % (m/v) paraformaldehyde in 0.1 M sodium cacodylate buffer for 2 hours at room temperature. The membranes were then gently washed with water, and the solvent was gradually exchanged to ethanol with an increasing ethanol 15-minute incubation step gradient (25 %, 50 %, 75 % and 100 % (v/v) ethanol). The samples were dried in a critical point dryer, placed onto SEM sample holders using silver adhesive (Electron Microscopy Sciences), and sputtered until they were coated in a 5 nm layer of Pt/Pd. Imaging was performed using a Zeiss Ultra 55 Field Emission SEM.
Growth curves and doubling times
The cells were grown in LB broth containing appropriate antibiotics overnight, diluted 1:100 in fresh LB Broth with antibiotics, and grown in a 1mL 96 well plate at 37°C and shaken at 900 rpm. Once the cells reached an OD600 of 0.6, they were again diluted 1:100 in fresh LB Broth in a 250mL 96 well plate. Half the cells were induced with 0.05% Arabinose and incubated with 5 mM pAzF. The other half of the cells were used as negative controls and were not induced with Arabinose and weren’t exposed to pAzF. Cells expressing the curli operon, or, as controls, PBP8 cells, were used. After this preparation, the 96 well plate was placed in a Biotek Kinetics Machine, which measured OD600 every 5 minutes and shook the plate on medium speed for 24 hours. The data were then plotted and the linear slope during exponential growth was used to calculate doubling times. Because curli-expression cells derivatives had more irregular exponential growths than control cells, we used the first exponential growth “hump” of curli variants to determine doubling times.
In vitro labeling of click microbes
Cells expressing curli fibers were pelleted at 4000xg for 10 min. The cells were resuspended in phosphate buffer saline (PBS) containing 1 % (m/v) bovine serum albumin (BSA). The centrifugation and resuspension process was repeated 3 times to clean the cells from any excess free NSAA present in the culture medium. The appropriate volume for the experiments was taken (200 μl of OD 5 per sample), pelleted and incubated at OD600 of10 in 100 μL Cy5-DBCO (5 μM) in PBS + 1% BSA for 2 hours at 37°C with agitation. The labeled cells were pelleted again and resuspended in PBS containing 3% (m/v) BSA. This washing step was repeated 3 times to remove unbound dye.
Labeling was assessed by spotting 10 μl of labeled cells on a nitrocellulose membrane and detecting the fluorescence of Cy5 using a FluorChem™ M system (Protein Simple).
Labeling of simulated curli fiber mats
Curli fiber mats from bacteria in different conditions were created by following published protocol37. In brief, we used E. coli PQN4 lab strain, which has been previously characterized to produce a high amount of curli materials37. Following curli expression protocol, we induced PQN4 wild type curli, wild type curli with OTS and mutant curli with OTS by adding 0.05% arabinose and 0.1 mM pAzF in the culture media. After the induction period, guanidinum chloride (GdmCl) (Sigma) was added to the cultures to 0.8 M final concentration and incubated for 1–2 hours at 4 °C prior to filtration. 30 to 50 mL of the GdmCl treated cultures were vacuum-filtered onto polycarbonate filter membranes (47 mm diameter, 10 μm pore size, EMD Millipore). The filter membranes were incubated with 5 mL of 8 M GdmCl for 5 minutes and vacuum filtered to eliminate the cell debris, and rinsed with 5 mL of deionized water 3 times. Then, the membranes were incubated with 5 mL of nuclease solution (1.5 U/ml of Benzonase, EMD Millipore, with 2 mM MgCl2 (Sigma)) for 10 minutes and vacuum filtered to remove remaining nucleic acids, and rinsed with 5 mL of deionized water 3 times. Then, the semipurified curli fiber mats on the filters were incubated with 2 mL 5 μM Cy5-DBCO for 1 hour and rinsed with 5 mL of deionized water 3 times to remove unbound dyes, before imaging with a FluorChem™ M system (Protein Simple).
Purification and identification of proteins labeled in vitro
500mL of pre-labeled cells at OD600 1 were pelleted at 4000xg for 20 min and resuspended in 30mL of lysis buffer (50 mM phosphate buffer, 300 mM NaCl, 7M guanidinium hydrochloride). A protease inhibitor cocktail was added, and the suspension was stored at − 20 °C overnight. The suspension was thawed on ice and sonicated to fully lyse the cells (40 % amplitude, 3×25s ON, 35s OFF, QSONICA Q700 Sonicator). After centrifugation at 10,000xg for 30 min, the supernatant was applied to a NiNTA beads and incubated 2 h with agitation at room temperature in the presence of 10 mM imidazole. The beads were then transferred into the plastic columns. His-tag purification was performed by washing the column 3 times with 40 mM imidazole, and eluting his-tagged CsgA proteins with 500 mM imidazole (both in 50 mM phosphate buffer with 100 mM NaCl).
The eluate was concentrated using 3kDa Amicon ultracentrifugal filters (EMD Millipore) and ran on a 4–20% SDS-PAGE gel. Prior to staining with Coomassie blue, the Cy5 fluorescence within the gel was detected.
Flow cytometry and confocal microscopy
Flow cytometry was used to determine the fraction of cells that were labeled. The cells were further diluted to an OD600 of 0.01 per mL in PBS and analyzed with a BD LSRFortessa Flow Cytometer System (BD Biosciences) using the APC channel for Cy5 fluorescence detection. For confocal microscopy the cells were also stained with 5 μM Hoechst 33342 (Thermo Fischer Scientific) in 1x PBS for 1 h and washed 3 times with 1 mL of PBS. Subsequently, the cells were fixed in 2% paraformaldehyde for 10 min at 25 °C and washed 3 times with 1 mL of PBS. 10 μl of cells were deposited on a glass slide for imaging with a Leica SP5 X MP Inverted Confocal Microscope (Leica Microsystems).
In vivo tracking of click microbes
Prior to the experiment, mice were given a non-fluorescent food (alfalfa-free) for at least 5 days to minimize background autofluorescence. At 12 hours prior to oral gavage of bacteria, mice were fasted and given special water containing antibiotics (1 g/L kanamycin, 1 g/L spectinomycin), inducer (10 g/L L-(+)-arabinose) and pAzF (5 mM) based on their assigned experimental conditions. The inducer concentration was chosen based on comparison to previously published examples of engineered bacteria in mice16, 49. Then, the starting culture of PBP8 transformed with pBbB8k-WT was expanded 1:100 dilution in LB containing 50 μg/mL kanamycin, while that of PBP8 transformed with pBbB8k-Mut and pEVOL-pAzF was expanded 1:100 dilution in LB containing 50 μg/mL kanamycin and spectinomycin to OD600 of 0.5. The log-phase cultures were centrifuged at 4000 g for 15 minutes at 4°C and resuspended in 20% sucrose (OmniPur) in PBS to OD600 of 10. Once the mice have been fasted for 12 hours, 100 μL of the prepared log-phase bacteria were administered to the mice via oral gavage.
The alfalfa-free food was returned to the mice as well as the special water to allow proliferation and protein induction of engineered bacteria inside the mouse gut. Meanwhile, the mouse’s abdominal hair was removed to allow the upcoming IVIS imaging. 30 hours after the bacteria gavage, mice were fasted for another 12 hours to remove the food from the GI tract.10 hours after that, the NSAA and arabinose containing water was switched into antibiotic-only water to prepare the mice for dye administration.
Two hours after the water has been switched and the food has been withheld, 100 μL of 100 μM of Cy5-DBCO in 20% sucrose in PBS are administered to the mice by oral gavage. The alfalfa-free food and antibiotic water were returned to the mice. Then, mice were imaged under anesthesia using IVIS Lumina II (PerkinElmer) at t = 0 (before the gavage), 6, 11, 24, and 48 hours. The IVIS instrument is equipped with 10 narrow-band excitation filters (30 nm bandwidth) and 4 broadband emission filters (60- and 75-nm bandwidth). At time point, the mice were imaged using excitation filter at 675 nm, and 500 nm for background subtraction, and emission filter at 695–770 nm, with field of view (FOV) = D (12.5 cm), fstop = 2 and medium binning. Living Image software version 4.3.1/4.4 was used for image analysis.
Whole GI tract ex vivo fluorescence imaging
The mouse experiment was performed in the same manner as the in vivo tracking experiment up to the dye administration. 36 hours following the dye administration, the mice were euthanized to harvest the whole GI tract for ex vivo fluorescence imaging. The GI tract was placed on the lid of 150×25 mm polystyrene tissue culture plate (Falcon) and imaged using IVIS Lumina II with the same setting as the in vivo imaging. After imaging, the GI tract was kept at −20°C for protein extraction. The images were then analyzed using ImageJ software for the fluorescence signal from the lower GI tract.
Whole cecum fluorescence and protein analysis
The GI tract specimens used in the ex vivo fluorescence imaging were further analyzed for curli and Cy5-DBCO interaction. The mouse GI tracts were removed from −20°C. The cecum from each sample was excised, transferred to a 15-ml falcon tube, and crushed to homogenize. 3 mL of PBS was added to each sample, supplemented with 4 μL of benzonase nuclease (Novagen) and 8 μL of 1 M magnesium sulfate (MgSO4) (Sigma). The samples were incubated at 37°C overnight.
Each sample was transferred to 2-ml microtube and centrifuged at 14,000 rpm for 10 minutes to extract the supernatant. The supernatant samples were transferred to another set of tubes, and concentrated using 10-kDa Amicon Ultra 0.5 mL centrifugal filters (Millipore) at 10,000 rpm centrifugation for 15 minutes twice. The pellet samples were resuspended in 200 μL of PBS. Both supernatant and pellet samples were diluted five times, mixed with 2X dyeless Laemli buffer in 1:1 ratio, incubated at 95°C for 5 minutes and run on 12% SDS-PAGE gel (Bio-Rad) at 200 Volts for 30 minutes.
After the gel electrophoresis, the gels were rinsed with water and imaged to detect the fluorescence of Cy5 using a FluorChem™ M system (Protein Simple). Then, the gels were stained with Coomassie Brilliant Blue to detect the presence of proteins and compared the size against standard protein ladder (Bio-Rad).
Supplementary Material
Acknowledgments
This work made use of the Harvard Center for Nanoscale Systems (CNS) and Harvard Medical School ICCB-Longwood Screening Facility, and the Wyss Institute for Biologically Inspired Engineering. We would like to thank Alexis Rovner, Amanda Graveline, Andyna Vernet, Frank Urena, Thomas Ferrante, Garry Cuneo and Franziska Bahl for their help. P.P. thankfully acknowledges the royal Thai government scholarship. N.-M.D.C. gratefully acknowledges a Postdoctoral research fellowship from the Fonds de Recherche Nature et Technologies du Québec (FRQNT). J.J.K thankfully acknowledges the Harvard College Program for Research in Science and Engineering (PRISE) and the Harvard College Research Program (HCRP) fellowships. This work was supported by National Institutes of Health (1R01DK110770–01A1) and the Wyss Institute for Biologically Inspired Engineering.
Footnotes
The authors have no competing financial interests to report.
References
- 1.Clemente JC, Ursell LK, Parfrey LW, and Knight R (2012) The Impact of the Gut Microbiota on Human Health: An Integrative View, Cell 148, 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch SV, and Pedersen O (2016) The Human Intestinal Microbiome in Health and Disease, New England Journal ofMedicine 375, 2369–2379. [DOI] [PubMed] [Google Scholar]
- 3.Tamboli CP, Neut C, Desreumaux P, and Colombel JF (2004) Dysbiosis in inflammatory bowel disease, Gut 53, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, Knight R, and Ley RE (2014) Conducting a Microbiome Study, Cell 158, 250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas V, Clark J, and Doré J (2015) Fecal microbiota analysis: an overview of sample collection methods and sequencing strategies, Future Microbiology 10, 1485–1504. [DOI] [PubMed] [Google Scholar]
- 6.Jeffery IB, Toole PW, Öhman L, Claesson MJ, Deane J, Quigley EMM, and Simrén M (2012) An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota, Gut 61, 997. [DOI] [PubMed] [Google Scholar]
- 7.Aziz Q, Doré J, Emmanuel A, Guarner F, and Quigley EMM (2013) Gut microbiota and gastrointestinal health: current concepts and future directions, Neurogastroenterology & Motility 25, 4–15. [DOI] [PubMed] [Google Scholar]
- 8.Ashton JJ, Colquhoun CM, Cleary DW, Coelho T, Haggarty R, Mulder I, Batra A, Afzal NA, Beattie RM, Scott KP, and Ennis S (2017) 16S sequencing and functional analysis of the fecal microbiome during treatment of newly diagnosed pediatric inflammatory bowel disease, Medicine 96, e7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Earle Kristen A., Billings G, Sigal M, Lichtman Joshua S., Hansson Gunnar C., Elias Joshua E., Amieva Manuel R., Huang Kerwyn C., and Sonnenburg Justin L. (2015) Quantitative Imaging of Gut Microbiota Spatial Organization, Cell Host & Microbe 78, 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tropini C, Earle KA, Huang KC, and Sonnenburg JL (2017) The Gut Microbiome: Connecting Spatial Organization to Function, Cell Host & Microbe 21, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riglar DT, Giessen TW, Baym M, Kerns SJ, Niederhuber MJ, Bronson RT, Kotula JW, Gerber GK, Way JC, and Silver PA (2017) Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation, Nature Biotechnology 35, 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yaung SJ, Deng L, Li N, Braff JL, Church GM, Bry L, Wang HH, and Gerber GK (2015) Improving microbial fitness in the mammalian gut by in vivo temporal functional metagenomics, Molecular Systems Biology 11, 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang IY, Koh E, Wong A, March JC, Bentley WE, Lee YS, and Chang MW (2017) Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models, Nature Communications 8, 15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandenbroucke K, de Haard H, Beirnaert E, Dreier T, Lauwereys M, Huyck L, Van Huysse J, Demetter P, Steidler L, Remaut E, Cuvelier C, and Rottiers P (2009) Orally administered L. lactis secreting an anti-TNF Nanobody demonstrate efficacy in chronic colitis, Mucosal Immunology 3, 49. [DOI] [PubMed] [Google Scholar]
- 15.Duan FF, Liu JH, and March JC (2015) Engineered Commensal Bacteria Reprogram Intestinal Cells Into Glucose-Responsive Insulin-Secreting Cells for the Treatment of Diabetes, Diabetes 64, 1794–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danino T, Prindle A, Kwong GA, Skalak M, Li H, Allen K, Hasty J, and Bhatia SN (2015) Programmable probiotics for detection of cancer in urine, Science Translational Medicine 7, 289ra284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell-Valois FX, and Sansonetti PJ (2014) Tracking bacterial pathogens with genetically-encoded reporters, FEBSLetters 588, 2428–2436. [DOI] [PubMed] [Google Scholar]
- 18.Karimi S, Ahl D, Vågesjö E, Holm L, Phillipson M, Jonsson H, and Roos S (2016) In Vivo and In Vitro Detection of Luminescent and Fluorescent Lactobacillus reuteri and Application of Red Fluorescent mCherry for Assessing Plasmid Persistence, PLOS ONE 11, e0151969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhee K-J, Cheng H, Harris A, Morin C, Kaper JB, and Hecht GA (2011) Determination of spatial and temporal colonization of enteropathogenic E. coli and enterohemorrhagic E. coli in mice using bioluminescent in vivo imaging, Gut Microbes 2, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belas R, Mileham A, Cohn D, Hilmen M, Simon M, and Silverman M (1982) Bacterial Bioluminescence: Isolation and Expression of the Luciferase Genes from Vibrio harveyi, Science 218, 791–793. [DOI] [PubMed] [Google Scholar]
- 21.Geva-Zatorsky N, Alvarez D, Hudak JE, Reading NC, Erturk-Hasdemir D, Dasgupta S, von Andrian UH, and Kasper DL (2015) In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria, Nat Med 21, 1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Zhu Y, and Chen X (2017) Selective Imaging of Gram-Negative and Gram-Positive Microbiotas in the Mouse Gut, Biochemistry 56, 3889–3893. [DOI] [PubMed] [Google Scholar]
- 23.Hudak JE, Alvarez D, Skelly A, von Andrian UH, and Kasper DL (2017) Illuminating vital surface molecules of symbionts in health and disease, Nature Microbiology 2, 17099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young TS, Ahmad I, Yin JA, and Schultz PG (2010) An Enhanced System for Unnatural Amino Acid Mutagenesis in E. coli, Journal ofMolecular Biology 595, 361–374. [DOI] [PubMed] [Google Scholar]
- 25.Barnhart MM, and Chapman MR (2006) Curli Biogenesis and Function, Annual review ofmicrobiology 60, 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanco LP, Evans ML, Smith DR, Badtke MP, and Chapman MR (2012) Diversity, biogenesis and function of microbial amyloids, Trends in Microbiology 20, 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collinson SK, Parker JMR, Hodges RS, and Kay WW (1999) Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae11Edited by W. Baumeister, Journal ofMolecular Biology 290, 741–756. [DOI] [PubMed] [Google Scholar]
- 28.Tian P, Boomsma W, Wang Y, Otzen DE, Jensen MH, and Lindorff-Larsen K (2015) Structure of a Functional Amyloid Protein Subunit Computed Using Sequence Variation, Journal of the American Chemical Society 157, 22–25. [DOI] [PubMed] [Google Scholar]
- 29.DeBenedictis EP, Ma D, and Keten S (2017) Structural predictions for curli amyloid fibril subunits CsgA and CsgB, RSC Advances 7, 48102–48112. [Google Scholar]
- 30.Marcus A, Sadimin E, Richardson M, Goodell L, and Fyfe B (2012) Fluorescence Microscopy Is Superior to Polarized Microscopy for Detecting Amyloid Deposits in Congo Red-Stained Trephine Bone Marrow Biopsy Specimens, American Journal of Clinical Pathology 158, 590–593. [DOI] [PubMed] [Google Scholar]
- 31.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, and Hultgren SJ (2002) Role of Escherichia coli Curli Operons in Directing Amyloid Fiber Formation, Science 295, 851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Botyanszki Z, Tay PKR, Nguyen PQ, Nussbaumer MG, and Joshi NS (2015) Engineered catalytic biofilms: Site-specific enzyme immobilization onto E. coli curli nanofibers, Biotechnology and Bioengineering 112, 2016–2024. [DOI] [PubMed] [Google Scholar]
- 33.Monteiro C, Saxena I, Wang X, Kader A, Bokranz W, Simm R, Nobles D, Chromek M, Brauner A, Brown RM, and Römling U (2009) Characterization of cellulose production in Escherichia coli Nissle 1917 and its biological consequences, Environmental Microbiology 11, 1105–1116. [DOI] [PubMed] [Google Scholar]
- 34.Rosano GL, and Ceccarelli EA (2014) Recombinant protein expression in Escherichia coli: advances and challenges, Frontiers in Microbiology 5, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amiram M, Haimovich AD, Fan C, Wang Y-S, Aerni H-R, Ntai I, Moonan DW, Ma NJ, Rovner AJ, Hong SH, Kelleher NL, Goodman AL, Jewett MC, Söll D, Rinehart J, and Isaacs FJ (2015) Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids, Nature biotechnology 55, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, and Bertozzi CR (2010) Copper-free click chemistry in living animals, Proceedings of the National Academy ofSciences 107, 1821–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorval Courchesne N-M, Duraj-Thatte A, Tay PKR, Nguyen PQ, and Joshi NS (2017) Scalable Production of Genetically Engineered Nanofibrous Macroscopic Materials via Filtration, ACS Biomaterials Science & Engineering 5, 733–741. [DOI] [PubMed] [Google Scholar]
- 38.Lajoie MJ, Rovner AJ, Goodman DB, Aerni H-R, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, Rohland N, Schultz PG, Jacobson JM, Rinehart J, Church GM, and Isaacs FJ (2013) Genomically Recoded Organisms Expand Biological Functions, Science 542, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu S, Chen D, Zhang J-N, Lv X, Wang K, Duan L-P, Nie Y, and Wu X-L (2013) Bacterial Community Mapping of the Mouse Gastrointestinal Tract, PLOS ONE 8, e74957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khlebnikov A, Risa Ø, Skaug T, Carrier TA, and Keasling JD (2000) Regulatable Arabinose-Inducible Gene Expression System with Consistent Control in All Cells of a Culture, Journal ofBacteriology 182, 7029–7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang S-W, Lee S, Na JH, Yoon HI, Lee D-E, Koo H, Cho YW, Kim SH, Jeong SY, Kwon IC, Choi K, and Kim K (2014) Cell Labeling and Tracking Method without Distorted Signals by Phagocytosis of Macrophages, Theranostics 4, 420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie R, Dong L, Du Y, Zhu Y, Hua R, Zhang C, and Chen X (2016) In vivo metabolic labeling of sialoglycans in the mouse brain by using a liposome-assisted bioorthogonal reporter strategy, Proceedings of the National Academy of Sciences of the United States ofAmerica 113, 5173–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rong J, Han J, Dong L, Tan Y, Yang H, Feng L, Wang Q-W, Meng R, Zhao J, Wang S-Q, and Chen X (2014) Glycan Imaging in Intact Rat Hearts and Glycoproteomic Analysis Reveal the Upregulation of Sialylation during Cardiac Hypertrophy, Journal of the American Chemical Society 136, 17468–17476. [DOI] [PubMed] [Google Scholar]
- 44.Xie R, Dong L, Huang R, Hong S, Lei R, and Chen X (2014) Targeted Imaging and Proteomic Analysis of Tumor-Associated Glycans in Living Animals, Angewandte Chemie International Edition 53, 14082–14086. [DOI] [PubMed] [Google Scholar]
- 45.Kotula JW, Kerns SJ, Shaket LA, Siraj L, Collins JJ, Way JC, and Silver PA (2014) Programmable bacteria detect and record an environmental signal in the mammalian gut, Proceedings of the National Academy of Sciences 111, 4838–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Datsenko KA, and Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products, Proceedings of the National Academy of Sciences of the United States ofAmerica 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen PQ, Botyanszki Z, Tay PKR, and Joshi NS (2014) Programmable biofilm-based materials from engineered curli nanofibres, Nature Communications 5, 4945. [DOI] [PubMed] [Google Scholar]
- 48.Duraj-Thatte AM, Praveschotinunt P, Nash TR, Ward FR, and Joshi NS (2018) Modulating bacterial and gut mucosal interactions with engineered biofilm matrix proteins, Scientific Reports 8, 3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grespi F, Ottina E, Yannoutsos N, Geley S, and Villunger A (2011) Generation and Evaluation of an IPTG-Regulated Version of Vav-Gene Promoter for Mouse Transgenesis, PLoS ONE 6, e18051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.