Abstract
Protopanaxadiol (PPD), a ginseng metabolite generated by the gut bacteria, was shown to induce colorectal cancer cell death and enhance the anticancer effect of chemotherapeutic agent 5-FU. However, the mechanism by which PPD promotes cancer cell death is not clear. In this manuscript, we showed that PPD activated p53 and ER stress and induced expression of BH3-only proteins Puma and Noxa to promote cell death. Induction of Puma by PPD was p53-dependent while induction of Noxa was p53-independent. On the other hand, PPD also induced pro-survival mechanisms including autophagy and expression of Bcl2 family apoptosis regulator Mcl-1. Inhibition of autophagy or knockdown of Mcl-1 significantly enhanced PPD-induced cell death. Interestingly, PPD inhibited expression of genes involved in fatty acid and cholesterol biosynthesis and induced synergistic cancer cell death with fatty acid synthase inhibitor cerulenin. As PPD-induced ER stress was not significantly affected by inhibition of new protein synthesis, we suggest PPD may induce ER stress directly through causing lipid disequilibrium.
Keywords: Ginseng, p53, ER stress, fatty acid synthase FASN, Protopanaxadiol PPD, BH3-only protein Puma and Noxa
Introduction
Natural products are a potentially valuable source for the development of new anti-cancer drugs (da Rocha et al., 2001; Mann, 2002). American ginseng is a very popular herb in the United States and has been reported to have a wide variety of biological activities including immunomodulatory, anti-inflammatory and anti-tumor effects. After oral ingestion, the parent compounds of ginseng are metabolized by the intestinal microbiome to create compound K (CK), a major ginseng metabolite with greater anticancer effects than the parent compounds (Wang et al., 2012). Interestingly, CK can be further converted by gut bacteria to protopanaxadiol (PPD), which was also found to be produced by gut bacteria from ginseng extracts or parent compound Rb1 (Hasegawa et al., 1996; Bae et al., 2000; Wang et al., 2015).
We previous reported that PPD induced colorectal cancer cell death and enhanced the anticancer effects of 5-FU in vivo without showing obvious toxicity (Wang et al., 2015; Zhang et al., 2015). However, the mechanism by which PPD promotes cancer cell death was not clear. In mammalian cells, the Bcl-2 family of proteins are key regulators of cell death by modulating mitochondrial integrity (Cory and Adams, 2002). The BH3-only subfamily of proteins, such as Noxa, Bim, Puma, promote mitochondria outer membrane permeability and induce cell death (Puthalakath and Strasser, 2002). The BH3-only subfamily proteins are antagonized by the Bcl-2 family of proteins, including Bcl-2, Bcl-XL, and MCl-1, which promote survival (Cory and Adams, 2002; Thomenius and Distelhorst, 2003). The expression of BH3-only proteins are induced by different stress signals, including p53, endoplasmic reticulum (ER) stress, reactive oxygen species (ROS), etc to induce cell death (Oda et al., 2000; Nakano and Vousden, 2001; Puthalakath et al., 2007; Zhang et al., 2011; Jin et al., 2014).
The ER is an intracellular organelle with important roles in lipid biosynthesis, intracellular membrane homeostasis, and proper folding and modification of secreted and transmembrane proteins. The accumulation of misfolded proteins in the ER triggers the activation of the ER unfolded protein response (UPRER), which include three distinct branches: IRE1, ATF6, and PERK (Walter and Ron, 2011). Activation of UPRER induces XBP1 splicing, increases expression of ER chaperones and synthesis of lipid, and inhibits global translation to reduce the influx of new proteins into the ER (Walter and Ron, 2011). Therefore, UPRER activation can promote survival by re-establishing ER homeostasis. However, excessive or prolonged ER stress will induce cell death (Tabas and Ron, 2011).
Interestingly, recent studies showed that lipid disequilibrium can induce UPRER without disturbing ER proteostasis and that mutant ER stress sensors lacking their luminal unfolded protein stress-sensing domain still retained responsiveness to lipid disequilibrium (Hou et al., 2014; Volmer and Ron, 2015). These results suggest that UPRER can be activated independently by disturbances in either the protein or the lipid in the ER.
In this report, we showed that PPD activated p53, ER stress, and autophagy and increased expression of both pro-apoptotic BH3 proteins Puma and Noxa as well as the pro-survival protein Mcl-1. Furthermore, PPD inhibited expression of fatty acid and cholesterol biosynthetic genes and induced synergistic cell death with fatty acid synthase inhibitor. These results provide novel insights into the role of PPD in lipid metabolism, ER stress, and cell death induction.
Materials and methods
PPD source and quality.
PPD was synthesized and purified as described (Wang et al., 2015). The HPLC-determined purity was 95.3%.
Cell culture, chemicals and reagents.
Human colorectal cancer cells HCT116 were obtained from the American Type Culture Collection. The p53−/− HCT116 cells were described previously (Bunz et al., 1999; Li et al., 2011). HCT116 cells were maintained in DMEM medium supplemented with 5% fetal bovine serum (FBS, Hyclone Laboratories), 50 IU of penicillin/streptomycin (Gemini Bio-Products) and 2 mmol/l of L-glutamine (Invitrogen) in a humidified atmosphere with 5% CO2 at 37 C. The Chloroquine (CQ), Cerulenin, and Mevastatin were obtained from Sigma.
Western blot analysis.
After HCT116 cells were treated as indicated, the cells were harvested and washed twice by PBS and lysed in RIPA buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 1 mM phenylmethlsulfonyl fluoride, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium vanadate, 1 μg/ml leupeptin). Antibodies used are listed below: β-actin antibody (Cell Signaling, 4967S), LC3A/B antibody (Cell Signaling, 4108S), Grp78 antibody (Cell Signaling, 3177S), p53 antibody (Santa Cruz SC-126), Puma antibody (Proteintech, 55120–1-AP), and Mcl-1 antibody (Santa Cruz, SC-819 and SC-12756). Immunoblot was detected by Li-Cor Odyssey image reader or with ECL western blot detection kit.
FACS analysis.
For the FACS analysis, 2×105 cells/well were seeded into 6-well plates. Samples were prepared based on the instructions provided by the Annexin V Apoptosis Kit (BD Biosciences). Briefly, after cells were treated as indicated, floating cells in the medium and adherent cells were collected following 48 h of PPD treatment. The cells were stained with annexin V-FITC and propidium iodide (PI) according to the manufacturer’s instructions. Untreated cells were used as the control for double staining. Cells were analyzed immediately using a FACScan flow cytometer.
Microarray data analysis.
The data from 6 microarrays of control or PPD treated samples was obtained and normalized as described previously (Zhang et al., 2015). The normalized data was analyzed using ArrayTools Version 4.3 to identify genes significantly affected by 20–25 μM of PPD treatment. Replicates were averaged by geometric mean. P values were computed using t tests on the logarithms of gene expression intensities. Genes involved in fatty acid and cholesterol biosynthesis were shown in Table 1.
Table 1.
Microarray data showing PPD reduced expression of genes involved in fatty acid and cholesterol biosynthesis.
| Gene name | P-value | PPD/Control ratio | Function |
|---|---|---|---|
| Fatty acid synthase (FASN) | 0.00045 | 0.57 | Fatty acid biosynthesis |
| stearoyl-CoA desaturase (delta-9-desaturase) (SCD) | 0.001 | 0.68 | Fatty acid biosynthesis |
| Acetyl-CoA acetyltransferase 2 (ACAT2) | 0.00024 | 0.31 | Cholesterol biosynthesis |
| 3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS) | 3.05e-05 | 0.53 | Cholesterol biosynthesis |
| 24-dehydrocholesterol reductase (DHCR24) | 5.16e-05 | 0.57 | Cholesterol biosynthesis |
| 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) | 8.36e-05 | 0.58 | Cholesterol biosynthesis |
| 7-dehydrocholesterol reductase (DHCR7) | 0.000942 | 0.61 | Cholesterol biosynthesis |
RNA isolation and RT-PCR.
Total RNA was extracted with the RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen). cDNA was synthesized using M-MLV reverse transcriptase from Promega. Primer pairs used for RT-PCR are: human XBP1, 5′-GGAGTTAAGACAGCGCTTGG-3′ (sense) and 5′-ACTGGGTCCAAGTTGTCCAG-3′ (antisense); human Puma, 5′-CAGACTGTGAATCCTGTGCT-3′ (sense) and 5′-ACAGTATCTTACAGGCTGGG-3′ (anti-sense); human Noxa, 5′-GCTCCAGCAGAGCTGGAAGT-3′ (sense) and 5′-GCAGTCAGGTTCCTGAGCAG-3′ (antisense); human BCL-XL, 5′-CCTTTGCCTAAGGCGGATTT-3′ (sense) and 5′-GCTCACTCACTGAGTCTCGT-3′ (antisense); human MCL1, 5′-AGAAAGCTGCATCGAACCATT-3′ (sense) and 5′-CAGCTCCTACTCCAGCAAC-3′ (antisense); human GAPDH, 5′-CTCTGACTTCAACAGCGACAC-3′ (sense) and 5′-CATACCAGGAAATGAGCTTGACAA-3′ (antisense).
Lentiviral preparation and transduction.
The pLKO.1 lentiviral sh-RNA expression system was used to generate shRNA constructs. The sequences of shRNA used in this study included the following: sh-GFP (5′-ACGTCTATATCATGGCCGACA-3′), sh-Puma (5′-CGGACGACCTCAACGCACA-3′), sh-Noxa (5′-GTAATTATTGACACATTTCTT-3′), sh-Mcl-1–1 (5′-CGGGACTGGCTAGTTAAAC-3′), sh-Mcl-1–2 (5’-GCTAAACACTTGAAGACCATA-3’). Viral packaging was done according to the previously described protocol (Li et al., 2010). Briefly, expression plasmids pCMV-dR8.91 and pCMV-VSV-G were cotransfected into 293T cells using the calcium phosphate method at 20:10:10 μg (for a 10-cm dish). The transfection medium containing calcium phosphate and plasmid mixture was replaced with fresh complete medium after incubation for 5 hr. Media containing virus was collected 48 hr after transfection and then concentrated using 20% sucrose buffer at 20,000 g for 4 hr. The virus pellet was re-dissolved in the proper amount of complete growth medium and stocked at −80°C. Cells were infected with the viruses at the titer of 100% infection in the presence of Polybrene (10 μg/ml) for 48 hr, and were treated as desired.
Results
PPD kills colorectal cancer cells through induction of BH3 family of pro-apoptosis regulators Puma and Noxa
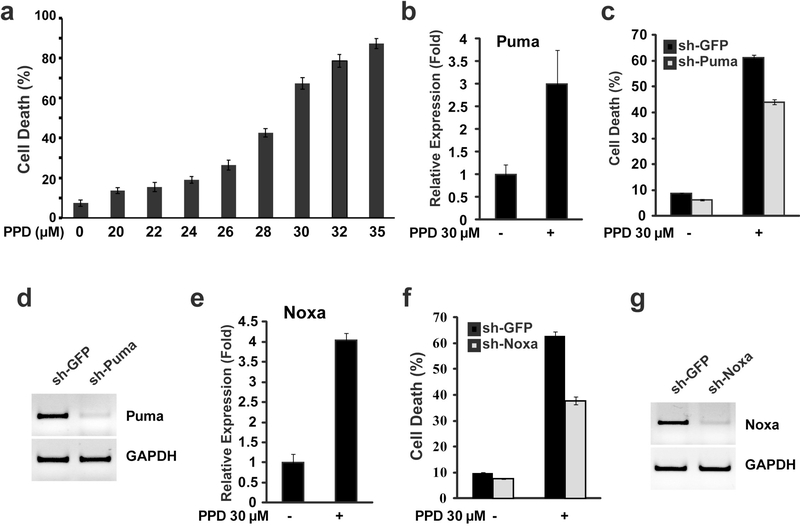
Our previous study showed that moderate level of PPD (20 μM) induced low level of cell death by itself but can significantly enhance 5-FU-induced cell death (Wang et al., 2015). To characterize how PPD modulates cell death, we first determined cell death induction by different levels of PPD. We found that PPD induced cell death in a dose-dependent manner with 50% cell death induced by 28.6 μM of PPD (Fig. 1a). Since 30 μM of PPD induced cell death in a large majority of cells (Fig. 1a), we used this concentration of PPD to determine the cell death regulators involved. Bcl-2 family of pro- and anti-apoptosis regulators are critical regulators of cell death in mammalian cells. To determine whether BH3-family of cell death regulators may regulate PPD induced cell death, we determined whether PPD induced expression of BH3 family of proteins. Two BH3 family members, Puma and Noxa, were found to have significantly induced expression (Fig. 1b, 1e). To determine whether Puma and Noxa contribute to PPD-induced cell death, shRNA constructs was used to knockdown Puma or Noxa (Fig. 1d and 1g). Knockdown of either Puma or Noxa can partially reduce PPD-induced cell death (Fig. 1c and 1f), indicating that both Puma and Noxa contribute to PPD-induced cell death.
Figure 1.
BH3-only proteins Puma and Noxa mediated PPD-induced cell death. (a) HCT116 cells were treated with different concentration of PPD for 48h, and then the cell death was determined. (b) HCT116 cells were treated with PPD (30μM) for 24h, and then the expression levels of Puma were determined by qRT-PCR. (c) HCT116 cells were infected with sh-Puma construct or sh-GFP control, and were treated with PPD (30μM) for 48 h, and then the cell death was determined. (d) RT-PCR showing knockdown of Puma expression by sh-Puma construct. (e) HCT116 cells were treated with PPD (30μM) for 24h, and then the expression levels of Noxa were determined by qRT-PCR. (f) HCT116 cells were infected with sh-Noxa construct or sh-GFP control, and were treated with PPD (30μM) for 48 h, and then the cell death was determined. (g) RT-PCR showing knockdown of Noxa expression by sh-Noxa construct.
P53 regulates the induction of Puma but not Noxa and contributes to PPD-induced cell death.
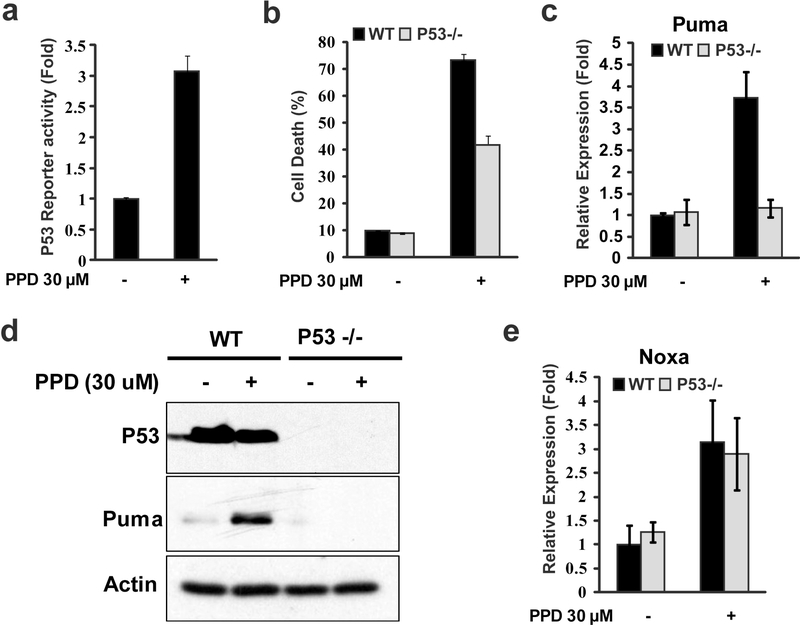
Both Puma and Noxa are BH3 family of pro-apoptosis regulators induced by p53 (Oda et al., 2000; Nakano and Vousden, 2001) and ginsenosides have been previous shown to activate p53 (Li et al., 2011; Zhang et al., 2013). Indeed, increased p53 reporter activity was induced by PPD (Fig. 2a), indicating PPD induced p53 activation. To determine whether p53 activation contributes to PPD-induced cell death, we compared PPD-induced cell death in p53 WT and p53 mutant HCT116 cells (Bunz et al., 1999; Li et al., 2011). While PPD induced significant levels of cell death in both p53 WT and p53 mutant HCT116 cells, PPD-induced cell death in p53 mutant HCT116 cells were significantly lower (Fig. 2b). These results suggest PPD-induced cell death were partially mediated by p53 activation.
Figure 2.
P53 regulated PPD-induced Puma expression and contributed to PPD-induced cell death in HCT116 cells. (a) HCT116 cells were transfected with P53 reporter gene for 24 h and then treated with PPD for 24h, and then the luciferase activity was measured. (b) HCT116 (P53+/+) and HCT116 (P53−/−) cells were treated with PPD (30μM) for 48 h, and then the cell death was determined. (c and d) HCT116 (P53+/+) and HCT116 (P53−/−) cells were treated with PPD (30 μM) for 24 h, and then the Puma mRNA (c) and protein level (d) were determined by RT-PCR and western blots, respectively. (e) HCT116 (P53+/+) and HCT116 (P53−/−) cells were treated with PPD (30 μM) for 24 h, and then Noxa expression level were determined by RT-PCR.
We further investigated whether PPD-induced Puma and Noxa expression was mediated by p53. Interestingly, Puma was induced by PPD in p53 WT cells but not in p53 mutant cells (Fig. 2c). The induction of Puma by PPD in WT by not p53 mutant HCT116 cells was also confirmed by western blots (Fig. 2d) In contrast, Noxa was similarly induced by PPD in both p53 WT and p53 mutant cells (Fig. 2e). These results showed that PPD-induced Puma expression was p53-dependent while PPD-induced Noxa was p53-independent.
PPD induces ER stress and autophagy. Inhibition of autophagy enhanced PPD-induced cell death
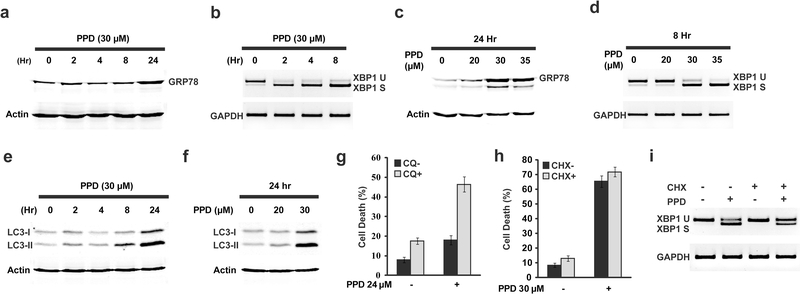
To characterize additional mechanisms that contribute to PPD-induced cell death, we determined whether PPD can induce ER stress and autophagy since our previous studies showed that ER stress can induce cell death in part through upregulating Noxa (Jin et al., 2012; Jin et al., 2014). XBP1 splicing was rapidly induced by PPD (Fig. 3b). In addition, GRP78, a UPR target gene, was also significantly induced at around 24 hours after PPD treatment (Fig. 3a). The level of UPR induction by PPD is concentration dependent. High levels of UPR was induced at 30 μM of PPD, a concentration of PPD that induced high levels of cell death while low levels of UPR was induced at 20 μM of PPD, a concentration of PPD that induced very low levels of cell death (Fig. 3c-d). Therefore PPD-induced cell death is correlated with its induction of ER stress.
Figure 3.
PPD induced endoplasmic reticulum stress and autophagy in HCT116 cells. (a) Western blot showing the induction of GRP78 after HCT116 cells were treated with PPD (30μM) at different time points. (b) RT-PCR analysis showing the induction of XBP1 splicing after HCT116 cells were treated with PPD (30μM) at different time points. (c) Western blot analysis showing the induction of GRP78 protein after HCT116 cells were treated with different concentrations of PPD for 24 hours. (d) RT-PCR analysis showing the induction of XBP1 splicing after HCT116 cells were treated with different concentrations of PPD for 8 hours. (e) LC3-I/II protein levels were determined by western blot analysis after HCT116 cells were treated with 30 μM of PPD for the indicated periods. (f) HCT116 cells were treated with different concentrations of PPD for 24 hours, and LC3-I/II protein levels were determined by western blot analysis. (g) Cell death analysis of HCT116 cells treated with 24 μM PPD for 48 hours in the presence or absence 20 μM of Chloroquine (CQ). (h) HCT116 cells were treated with 30 μM PPD for 48 hours in the presence or absence of cycloheximide (10 μg/ml), and then cell death was determined. (i) HCT116 cells were treated with 30 μM PPD for 8 hours in the presence or absence of cycloheximide (10 μg/ml), the XBP1 splicing was determined by RT-PCR.
In addition to ER stress, PPD also increased cleaved form of LC3 (Fig. 3e-f), indicating PPD also induced autophagy. To determine whether PPD-induced autophagy affects its induction of cell death, we tested the effect of inhibiting autophagy with chloroquine (CQ). Inhibition of autophagy with CQ significantly increased PPD-induced cell death (Fig. 3g). These results showed that PPD-induced autophagy protects cells from death.
ER stress are generally induced by the accumulation of unfolded protein in the ER and can be relieved by inhibiting protein synthesis. Our previous study showed that addition of cycloheximide (CHX) with falcarindiol or the related compound Oplopantriol A, which induce ER stress by interfering with the ubiquitin/proteasome pathway, dramatically inhibited ER stress and inhibited cell death induced by these compounds (Jin et al., 2012; Jin et al., 2014). We similarly tested whether inhibiting protein synthesis can similarly affect PPD-induced ER stress and cell death. Interestingly, CHX treatment only slightly decreased PPD-induced ER stress (Fig. 3i) and failed to inhibit PPD-induced cell death (Fig. 3h). These results suggest that PPD-induced ER stress and cell death is not very sensitive to the change in ER protein load.
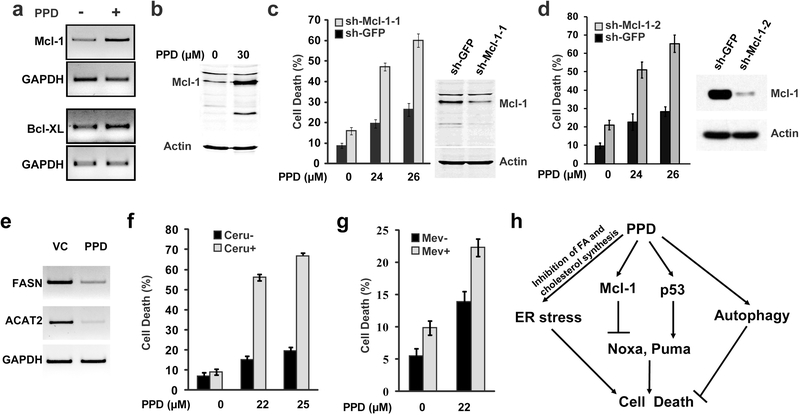
As BH3 family of pro-apoptotic regulators Puma and Noxa were induced by PPD and contribute to PPD-induced cell death, inhibiting the synthesis of these pro-apoptotic regulators would be expected to inhibit cell death. Therefore, we tested the idea whether PPD may also induce Bcl2 family of anti-apoptotic regulators. We found that PPD induced Mcl-1 and Bcl-XL expression (Fig. 4a) and increased Mcl-1 protein levels (Fig. 4b). Knockdown of Mcl-1 with two independent sh-Mcl-1 constructs significantly enhanced PPD-induced cell death (Fig. 4c-d). Taken together, our results suggest that PPD activated both pro-death as well as pro-survival signaling pathways and induced expression of both pro-apoptotic and anti-apoptotic regulators. In addition, PPD- induced ER stress appears to be distinct from that induced by FAD and OPT, which are due to the accumulation of unfolded proteins as a consequences of inhibiting the ubiquitin/proteasome pathway (Jin et al., 2012; Jin et al., 2014).
Figure 4.
Knockdown of anti-apoptotic protein Mcl-1 or inhibiting fatty acid synthesis shows synergistic effect with PPD-induced cancer cell death. (a) HCT116 cells were treated with 30 μM PPD for 24 hours, and then the expression levels of anti-apoptotic proteins Mcl-1 and Bcl-XL were determined by RT-PCR. (b) HCT116 cells were treated with 30 μM PPD for 20 hours, and then the level of Mcl-1 protein was determined by western blot analysis. (c) HCT116 cells infected with sh-Mcl1–1 or sh-GFP control virus. Cell death was determined after 48 hours of PPD treatment and western blots showing knockdown of Mcl-1 by sh-Mcl-1–1 construct. (d) HCT116 cells infected with sh-Mcl-1–2 or sh-GFP control virus. Cell death was determined after 48 hours of PPD treatment and western blots showing knockdown of Mcl-1 by sh-Mcl-1–2 construct. (e) HCT116 cells were treated with 30 μM PPD for 24 hours, and then the expression levels of FASN and ACAT2 were determined by RT-PCR. (f) Cell death was determined after HCT116 cells were treated with different concentrations of PPD for 48 hours in the presence or absence of the 5 μM Cerulenin. (g) Cell death was determined after HCT116 cells were treated with PPD (22 μM) for 48 hours in the presence or absence of the 10 μM Mevastatin. (h) A model summarizing the cellular mechanisms that modulate PPD-induced cell death.
PPD significantly decreases FASN expression and synergizes with FAS inhibitor to kill cancer cells.
Since UPR signaling in the ER can potentially be induced directly by lipid perturbation in addition to the accumulation of unfolded protein (Hou et al., 2014; Volmer and Ron, 2015), we tested whether PPD may affect lipid metabolism by determining the expression of genes involved in fatty acid and cholesterol synthesis. Analysis of microarray data revealed that PPD significantly affected expression of genes involved in biosynthesis of steroids, PPAR signaling, metabolism of pyruvate and glycerophospholipid. In particular, PPD significantly inhibited expression of genes involved in fatty acid synthesis such as FASN and SCD as well as genes involved in cholesterol synthesis such as ACAT2, HMGCS, HMGCR, DHCR24 and DHCR7 (Table 1). Indeed, significant reduced expression of FASN as well as ACAT2 was confirmed by RT-PCR (Fig. 4e). These results supports the idea that PPD inhibiting de novo fatty acid and cholesterol synthesis, which may affect normal lipid metabolism.
To determine whether the observed effect on lipid metabolism affects PPD-induced cell death, we tested the effect of inhibitors of fatty acid or cholesterol synthesis inhibitor on PPD-induced cell death. Interestingly, while 22–25 μM of PPD alone only induced low levels of cell death, addition of 5 μM of FAS inhibitor cerulenin dramatically increased PPD-induced cell death even though this concentration of cerulenin alone did not induce significant cell death (Fig. 4f). On the other hand, while inhibition of cholesterol synthesis with 10 μM Mevastatin also increased PPD-induced cell death, the observed increase was mostly additive instead of synergistic (Fig. 4g).
Discussion
Our results show that ginseng metabolite PPD activates three stress signaling pathways: p53, ER stress, and autophagy (Fig. 4h). Activation of p53, which induces expression of Puma, contributes to PPD-induced cell death (Fig. 2). In contrast, induction of autophagy promotes survival and protects cells from PPD-induced cell death (Fig. 3e-g). One the other hand, ER stress could potentially either protect or promote PPD-induced cell death depending on the level of ER stress induced. It is known that moderate levels of ER will reduce general protein synthesis and induce transcriptional programs to promote homeostasis while prolonged excessive ER stress will promote cell death (Walter and Ron, 2011). This is consistent with our observation that moderate levels of PPD induced low levels of ER stress and low levels of cell death while high levels of PPD induced high levels of ER stress and high levels of cell death.
While it is well established that accumulation of misfolded proteins in the ER will induce ER stress, recent studies suggest ER stress can also be induced directly by lipid disequilibrium (Hou et al., 2014; Volmer and Ron, 2015). We suggest that PPD-induced ER stress is mediated by its effect on disrupting the normal lipid equilibrium instead of accumulation of misfolded protein. First, we showed that PPD significantly inhibited expression of genes involved in both fatty acid synthesis and cholesterol synthesis, which may cause lipid disequilibrium and induce ER stress. Indeed, inhibition of fatty acid synthase was also shown to induce ER stress and cell death (Little et al., 2007). Second, we found that PPD-induced ER stress did not significantly respond to changes in new protein synthesis or ER chaperon levels. While inhibiting protein synthesis with CHX dramatically inhibited FAD and OPT-induced ER stress (Jin et al., 2012; Jin et al., 2014), CHX did not significantly inhibit PPD-induced ER stress or cell death. In addition, overexpression of Grp78, which significantly decreased FAD and OPT-induced ER stress and cell death (Jin et al., 2012; Jin et al., 2014), did not significantly affect cell death induced by PPD (data not shown).
This study revealed that PPD activated both pro-survival mechanisms such as autophagy and Mcl-1 as well as the pro-death mechanisms such as p53, Puma, and Noxa (Fig. 4h). Whether a cell will survive or undergo cell death will likely be determined by the relative strength of the pro-survival and the pro-death mechanisms. This is consistent with the observed low level of cell death induced by moderate levels of PPD. Furthermore, it is predicted that PPD-induced cell death will be enhanced by increasing the pro-death mechanisms or decreasing the pro-survival mechanisms. Consistent with this, knockdown of Mcl-1, which decreased PPD-induced pro-survival mechanism, significantly enhanced cell death induced by moderate levels of PPD (Fig. 4c). On the other hand, high levels of PPD, which induced high levels of ER stress, induced extensive cell death by increasing pro-death mechanisms. Similarly, treatment of moderate levels of PPD with FAS inhibitor will likely lead to more severe disruption of lipid equilibrium and induce extensive cell death by increasing the pro-death mechanisms.
Acknowledgments:
This work is supported in part by a grant from National Institute of Health NIH R01 GM120046.
Footnotes
Conflict of interests: The authors declare that they have no conflicts of interest with the contents of this article.
References
- Bae EA, Park SY, Kim DH. 2000. Constitutive beta-glucosidases hydrolyzing ginsenoside Rb1 and Rb2 from human intestinal bacteria. Biol Pharm Bull 23: 1481–1485. [DOI] [PubMed] [Google Scholar]
- Bunz F, Hwang PM, Torrance C, et al. 1999. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest 104: 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory S, Adams JM. 2002. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer 2: 647–656. [DOI] [PubMed] [Google Scholar]
- da Rocha AB, Lopes RM, Schwartsmann G. 2001. Natural products in anticancer therapy. Curr Opin Pharmacol 1: 364–369. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Sung JH, Matsumiya S, Uchiyama M. 1996. Main ginseng saponin metabolites formed by intestinal bacteria. Planta Med 62: 453–457. [DOI] [PubMed] [Google Scholar]
- Hou NS, Gutschmidt A, Choi DY, et al. 2014. Activation of the endoplasmic reticulum unfolded protein response by lipid disequilibrium without disturbed proteostasis in vivo. Proc Natl Acad Sci U S A 111: E2271–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin HR, Liao Y, Li X, et al. 2014. Anticancer compound Oplopantriol A kills cancer cells through inducing ER stress and BH3 proteins Bim and Noxa. Cell Death Dis 5: e1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin HR, Zhao J, Zhang Z, et al. 2012. The antitumor natural compound falcarindiol promotes cancer cell death by inducing endoplasmic reticulum stress. Cell Death Dis 3: e376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gordon GM, Du CH, Xu J, Du W. 2010. Specific killing of Rb mutant cancer cells by inactivating TSC2. Cancer Cell 17: 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Zhao J, Wang CZ, et al. 2011. Ginsenoside Rh2 induces apoptosis and paraptosis-like cell death in colorectal cancer cells through activation of p53. Cancer Lett 301: 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JL, Wheeler FB, Fels DR, Koumenis C, Kridel SJ. 2007. Inhibition of fatty acid synthase induces endoplasmic reticulum stress in tumor cells. Cancer Res 67: 1262–1269. [DOI] [PubMed] [Google Scholar]
- Mann J 2002. Natural products in cancer chemotherapy: past, present and future. Nat Rev Cancer 2: 143–148. [DOI] [PubMed] [Google Scholar]
- Nakano K, Vousden KH. 2001. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 7: 683–694. [DOI] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, et al. 2000. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288: 1053–1058. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, O’Reilly LA, Gunn P, et al. 2007. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 129: 1337–1349. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, Strasser A. 2002. Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ 9: 505–512. [DOI] [PubMed] [Google Scholar]
- Tabas I, Ron D. 2011. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 13: 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomenius MJ, Distelhorst CW. 2003. Bcl-2 on the endoplasmic reticulum: protecting the mitochondria from a distance. J Cell Sci 116: 4493–4499. [DOI] [PubMed] [Google Scholar]
- Volmer R, Ron D. 2015. Lipid-dependent regulation of the unfolded protein response. Curr Opin Cell Biol 33: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334: 1081–1086. [DOI] [PubMed] [Google Scholar]
- Wang CZ, Du GJ, Zhang Z, et al. 2012. Ginsenoside compound K, not Rb1, possesses potential chemopreventive activities in human colorectal cancer. Int J Oncol 40: 1970–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CZ, Zhang Z, Wan JY, et al. 2015. Protopanaxadiol, an active ginseng metabolite, significantly enhances the effects of fluorouracil on colon cancer. Nutrients 7: 799–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lopez H, George NM, Liu X, Pang X, Luo X. 2011. Selective involvement of BH3-only proteins and differential targets of Noxa in diverse apoptotic pathways. Cell Death Differ 18: 864–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Du GJ, Wang CZ, et al. 2013. Compound K, a Ginsenoside Metabolite, Inhibits Colon Cancer Growth via Multiple Pathways Including p53-p21 Interactions. Int J Mol Sci 14: 2980–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li Z, Wu X, et al. 2015. TRAIL pathway is associated with inhibition of colon cancer by protopanaxadiol. J Pharmacol Sci 127: 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]