Abstract
Background:
Adenosine has an immunosuppressive and angiogenic modulation of the tumor microenvironment. This study aimed to explore the efficacy of single nucleotide polymorphisms (SNPs) in adenosine-related molecules for patients with metastatic colorectal cancer (mCRC) treated with bevacizumab-based chemotherapy.
Patients and Methods:
We analyzed genomic DNA extracted from 451 samples of three independent cohorts: a discovery cohort of 107 patients with FOLFIRI plus bevacizumab in FIRE-3 (NCT00433927); a validation cohort of 215 patients with FOLFIRI plus bevacizumab in TRIBE (NCT00719797); a control cohort of 129 patients with FOLFIRI plus cetuximab in FIRE-3. The relationship between the selected SNPs and clinical outcomes was analyzed.
Results:
In the discovery cohort, patients with any C allele in CD39 rs11188513 had significantly shorter median progression-free survival (11.3 vs 13.1 months, HR: 1.70, 95% CI: 1.04–2.77, P = 0.022) in univariate analysis and overall survival (OS) (27.4 vs 49.9 months, HR: 2.10, 95% CI: 1.07–4.10, P = 0.031) in uni- and multivariable analyses than those with the T/T variant. The significant association between CD39 rs11188513 and OS was confirmed in the validation cohort (25.8 vs 31.6 months, HR: 1.53, 95% CI: 1.09–2.15, P = 0.013). CD73 rs2229523 and A2BR rs2015353 in the discovery cohort, and CD39 rs2226163 in the validation cohort had significant correlations with OS in uni- and multivariable analyses. None of SNPs were significant in the cetuximab control cohort.
Conclusion:
Selected SNPs in the adenosine pathway may impact clinical outcome of mCRC patients treated with FOLFIRI plus bevacizumab.
Keywords: metastatic colorectal cancer, adenosine, SNP, bevacizumab, cetuximab: CD39, CD73, A2AR, A2BR
Micro abstract:
Adenosine has an immunosuppressive and angiogenic modulation of the tumor microenvironment. With the 451 metastatic colorectal cancer patients from phase III clinical trials (FIRE-3 and TRIBE), this work revealed that CD39 rs11188513, a SNP in the adenosine pathway, impacted clinical outcome of metastatic colorectal cancer patients treated with FOLFIRI plus bevacizumab.
Introduction
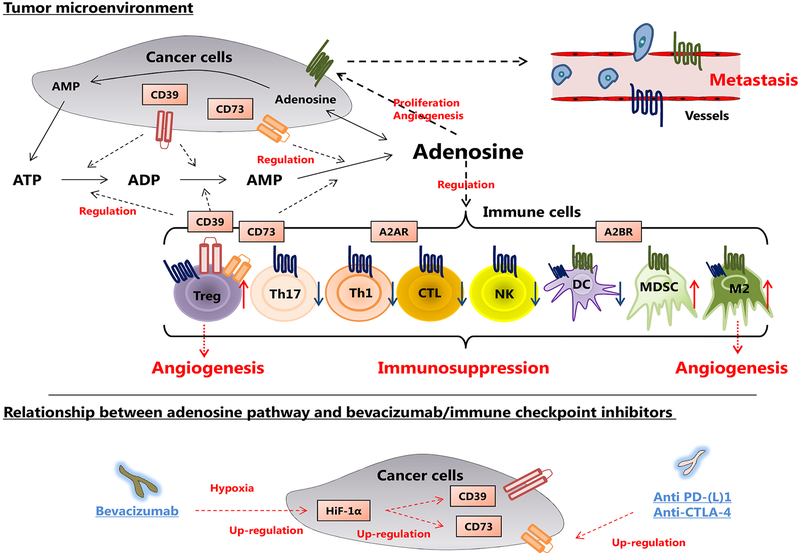
Adenosine has a potent immunosuppressive ability and is autonomously produced from ATP through ADP via CD39 and CD73, two ectonucleotidases, expressed mainly in the surface membrane of cancer cells, B cells or regulatory T cells (Tregs).1–3 Although extracellular ATP enhances immune cell chemotaxis and activities, extracellular adenosine rapidly acts on immune cells and leads to immunosuppression in tumor microenvironment.4 There are four receptors for extracellular adenosine (A1R, A2AR, A2BR, and A3R).5, 6 Among them, A2AR is predominantly present on the surface of the lymphocytes, and A2BR is mainly present in myeloid cells.7, 8 Furthermore, extracellular adenosine also acts on cancer cells through A2BR, and stimulates tumor growth and metastasis.9 (Figure 1) Consistent with these functions, previous reports have shown that the molecules of adenosine pathway can be prognostic factors and therapeutic targets in various cancers.10–12 Interestingly, blockade of adenosine pathway also increases the therapeutic efficacy of anti-PD-1 or anti-CTLA-4 in preclinical models, suggesting the expression levels of adenosine-related molecules could be biomarkers for efficacy of checkpoint inhibitors.13, 14
Figure 1.
Mechanism of adenosine pathway in tumor microenvironment. Adenosine is autonomously produced from ATP through ADP via CD39 and CD73 expressed mainly on the surface of cancer cells, B cells or Tregs. The work of extracellular adenosine is mainly divided into two directions. For immune cells, adenosine has a potent immunosuppressive ability through A2AR and A2BR. For cancer cells, adenosine stimulates tumor growth and metastasis through A2BR. Abbreviations: CTL, cytotoxic lymphocyte; DC, dendritic cell; NK, natural killer; M2, M2 macrophage; MDSC, myeloid derived suppressor cell; Th1, T helper 1; Th17, T helper 17; Treg, regulatory T cell.
Extracellular adenosine production is strongly activated in the hypoxic state. Importantly, hypoxia-induced HIF-1α increases extracellular adenosine production through upregulations of CD39 and CD73,15 leading to suppression of the cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells.16 (Figure 1) Given the critical effects of adenosine on immune conditioning and tumor angiogenesis, it is of clinical significance to examine the association between adenosine-related molecules and the effect of anti-VEGF therapy. Although some reports have shown that the expressions of adenosine-related molecules have strong association with survival of cancer patients,17, 18 the predictive or prognostic role of genetic changes within adenosine-related molecules remains unknown.
We hypothesized that the single nucleotide polymorphisms (SNPs) in adenosine-related molecules were associated with immune dysregulation and the efficacy of bevacizumab. Therefore, we investigated relationships between the SNPs and clinical outcomes in patients with metastatic colorectal cancer (mCRC) treated with bevacizumab-based chemotherapy, and with cetuximab-based chemotherapy as control. Our findings suggest that the selected SNPs in adenosine-related molecules may be biomarkers for bevacizumab-based chemotherapy as well as promising therapeutic targets in mCRC.
Materials and Methods
Baseline patients and study design
The study subjects were 451 patients with mCRC treated with chemotherapy. The patients underwent FOLFIRI plus bevacizumab or cetuximab as the first-line chemotherapy in two prospective, randomized, open-label, phase III clinical trials: FIRE-319 and TRIBE20. We selected the patients treated with FOLFIRI plus bevacizumab in FIRE-3 as the discovery cohort (n = 107), the patients treated with FOLFIRI plus bevacizumab in TRIBE as the validation cohort (n = 215), and the patients treated with FOLFIRI plus cetuximab in FIRE-3 as negative control cohort (n = 129), respectively. Patients without sufficient samples for analysis were excluded.
Selection of single nucleotide polymorphisms (SNPs) and genotyping
Polymorphisms underlying adenosine-related molecules; CD39 (ENTPD1: rs11188513, rs2226163), CD73 (NT5E: rs2229523), A2AR (ADORA2A: rs5751876), A2BR (ADORA2B: rs2015353), and HIF-1α (HIF1A: rs2057482, rs11549465), were selected using the following predefined criteria: (i) biological significance according to published literature review; (ii) a cut-off of minor allele frequency is at least 10% in Caucasians (in the Ensemble Genome Browser: https://www.ensembl.org/index.html ); and (iii) tag SNPs chosen by the HapMap genotype data with r2 threshold = 0.8: https://snpinfo.niehs.nih.gov/snpinfo/snptag.html. (Supplementary Table 1) Genomic DNA was extracted from peripheral whole blood derived from patients, using the QIAmp Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol (www.qiagen.com). The OncoArray was used for genotyping in this study (Illumina, San Diego, CA, USA).
Statistical analysis
The primary purpose of this study was to evaluate the associations of SNPs in adenosine-related molecules with tumor response, progression-free survival (PFS), and overall survival (OS). Patients were defined as responders when achieving complete or partial response and non-responders when stable or progressive disease occurred as defined by RECIST 1.1 criteria. The comparison of baseline patient characteristics between cohorts and the correlation between SNPs and tumor response were analyzed using Chi-square test. The Kaplan-Meier plots and log-rank tests were performed to evaluate the association between candidate SNPs and clinical outcome, PFS and OS. The Cox proportional hazards regression model and Wald tests were used to reevaluate the independent effect between candidate SNPs and PFS and OS. All statistical analyses were performed by SAS 9.4 (SAS Institute, Cary, NC, USA). All tests were two sided at a significant level of 0.05.
Results
Patients and tumor characteristics
Baseline characteristics of patients in the discovery (FIRE-3 FOLFIRI bevacizumab), validation (TRIBE FOLFIRI bevacizumab), and control (FIRE-3 FOLFIRI cetuximab) cohorts are shown in Supplementary Table 2. The median follow-up time was 26.7 months in the discovery cohort, 49.0 months in the validation cohort, and 29.2 months in the control cohort. The median PFS and OS were 11.6 months and 31.5 months in the discovery cohort, 9.7 months and 26.3 months in the validation cohort, and 12.8 months and 49.8 months in the control cohort, respectively. Compared to the TRIBE cohort, patients in the FIRE-3 cohort had higher median age (P = 0.006), higher ECOG performance status (P < 0.001), higher rates of primary tumor resection (P < 0.001), and much lower KRAS (P < 0.001) and RAS mutation rates (P < 0.001). The control cohort had more males than the other two cohorts (P = 0.013).
Predictive and prognostic values of the adenosine-related SNPs in the discovery cohort
Table 1 summarizes associations between the selected adenosine-related SNPs and clinical outcomes. Among the seven candidate SNPs, CD39 rs11188513, CD73 rs2229523, and A2BR rs2015353 were significantly associated with clinical outcome in the discovery cohort.
Table 1.
Associations between adenosine-related SNPs and clinical outcomes
| Tumor Response | Progression-free Survival | Overall Survival | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs | Genotype | n | Yes | No |
P
value |
Median months (95%CI) |
HR (95%CI) |
P
value |
Adjusted HR (95%CI) |
P
value |
Median months (95%CI) |
HR (95%CI) |
P
value |
Adjusted HR (95%CI) |
P
value |
| CD39rs11188513 | |||||||||||||||
| Discovery cohort | T/T | 42 | 27(64.3%) | 15(35.7%) | 0.92 | 13.1(9.9,16.9) | 1 (Reference) | 0.022 | 1 (Reference) | 0.080 | 49.9(28.1,NE) | 1 (Reference) | 0.012 | 1 (Reference) | 0.031 |
| Validation cohort | T/T | 77 | 46(63.0%) | 27(37.0%) | 0.30 | 9.6(8.8,11.7) | 1 (Reference) | 0.10 | 1 (Reference) | 0.052 | 31.6(21.1,42.7) | 1 (Reference) | 0.014 | 1 (Reference) | 0.013 |
| Control cohort | T/T | 47 | 34(77.3%) | 10(22.7%) | 0.99 | 11.8(9.0,14.1) | 1 (Reference) | 0.66 | 1 (Reference) | 0.35 | 37.5(27.1,67.4) | 1 (Reference) | 0.26 | 1 (Reference) | 0.12 |
| CD39 rs2226163 | |||||||||||||||
| Discovery cohort | Any A | 82 | 52(65.8%) | 27(34.2%) | 0.50 | 11.3(9.9,13.1) | 1 (Reference) | 0.53 | 1 (Reference) | 0.83 | 28.6(24.7,44.3) | 1 (Reference) | 0.21 | 1 (Reference) | 0.22 |
| Validation cohort | Any A | 165 | 92(57.1%) | 69(42.9%) | 0.44 | 10.3(9.2,11.0) | 1 (Reference) | 0.24 | 1 (Reference) | 0.20 | 25.8(22.3,28.6) | 1 (Reference) | 0.023 | 1 (Reference) | 0.024 |
| Control cohort | Any A | 103 | 73(76.0%) | 23(24.0%) | 0.68 | 12.8(10.0,14.1) | 1 (Reference) | 0.78 | 1 (Reference) | 0.72 | 52.0(42.1,67.4) | 1 (Reference) | 0.52 | 1 (Reference) | 0.34 |
| Combined CD39 rs11188513 and CD39 rs2226163 | |||||||||||||||
| Discovery cohort
(FOLFIRI + Bev) |
Group I | 24 | 14(58.3%) | 10(41.7%) | 0.65 | 11.6(8.6,20.5) | Reference | 0.049 | Reference | 0.14 | 49.9(21.5,NE) | Reference | 0.040 | Reference | 0.095 |
| Validation cohort
(FOLFIRI + Bev) |
Group I | 47 | 28(63.6%) | 16(36.4%) | 0.69 | 9.5(8.6,11.3) | Reference | 0.28 | Reference | 0.18 | 37.6(19.8,48.6) | Reference | 0.047 | Reference | 0.048 |
| Control cohort (FOLFIRI + Cet) |
Group I | 26 | 20(80.0%) | 5(20.0%) | 0.88 | 12.2(7.9,15.8) | Reference | 0.68 | Reference | 0.62 | 37.5(24.5,56.2) | Reference | 0.52 | Reference | 0.29 |
| CD73 rs2229523 | |||||||||||||||
| Discovery cohort | G/G | 46 | 26(59.1%) | 18(40.9%) | 0.30 | 10.3(9.2,13.5) | 1 (Reference) | 0.81 | 1 (Reference) | 0.46 | 23.7(17.6,44.3) | 1 (Reference) | 0.017 | 1 (Reference) | 0.026 |
| Validation cohort | G/G | 118 | 63(55.3%) | 51(44.7%) | 0.46 | 9.6(8.8,11.0) | 1 (Reference) | 0.68 | 1 (Reference) | 0.66 | 25.1(19.8,29.1) | 1 (Reference) | 0.68 | 1 (Reference) | 0.78 |
| Control cohort | G/G | 64 | 44(75.9%) | 14(24.1%) | 0.80 | 12.8(10.1,14.5) | 1 (Reference) | 0.49 | 1 (Reference) | 0.38 | 52.0(36.4,NE) | 1 (Reference) | 0.88 | 1 (Reference) | 0.47 |
| A2BR rs2015353 | |||||||||||||||
| Discovery cohort | Any C | 84 | 50(61.7%) | 31(38.3%) | 0.22 | 11.3(9.8,12.2) | 1 (Reference) | 0.055 | 1 (Reference) | 0.073 | 28.1(21.5,40.0) | 1 (Reference) | 0.008 | 1 (Reference) | 0.004 |
| Validation cohort | Any C | 168 | 94(58.0%) | 68(42.0%) | 0.56 | 9.5(8.8,10.8) | 1 (Reference) | 0.32 | 1 (Reference) | 0.66 | 26.5(21.1,32.0) | 1 (Reference) | 0.54 | 1 (Reference) | 0.61 |
| Control cohort | Any C | 99 | 71(76.3%) | 22(23.7%) | 0.46 | 11.8(9.3,13.6) | 1 (Reference) | 0.80 | 1 (Reference) | 0.94 | 42.8(37.5,60.7) | 1 (Reference) | 0.71 | 1 (Reference) | 0.94 |
P value was based on the Chi-square test for tumor response, log-rank test for progression-free survival and overall survival in the univariate analysis, and Wald test in the multivariable Cox proportional hazards regression model adjusting for sex, ECOG performance status, liver limited disease, and BRAF status in the discovery and control cohorts; adjusting for sex, age, ECOG performance status, primary tumor site, primary tumor resected, liver limited disease, adjuvant chemotherapy, BRAF status, and RAS status in the validation cohort.
Abbreviations: Bev, Bevacizumab; Cet, Cetuximab; SNP, single nucleotide polymorphism.
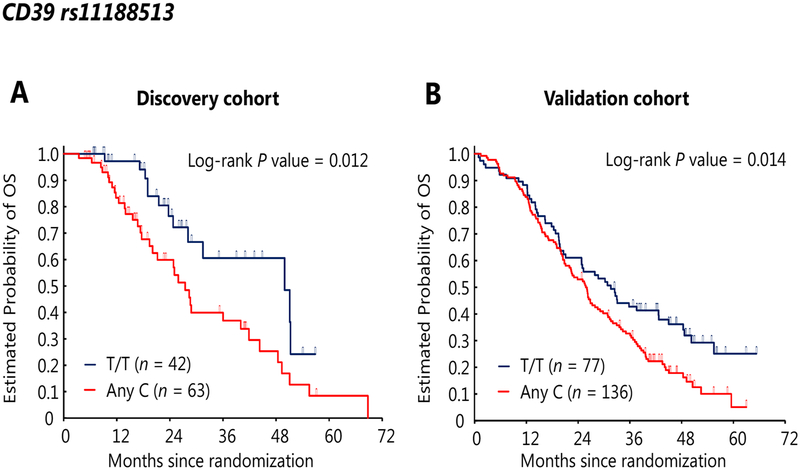
In univariate analysis, mCRC patients with any C allele in CD39 rs11188513 had significantly shorter median PFS [11.3 vs 13.1 months, hazard ratio (HR): 1.70, 95% confidence intervals (CI): 1.04–2.77, P = 0.022] and OS (27.4 vs 49.9 months, HR: 2.19, 95% CI: 1.15–4.14, P = 0.012) than those with the T/T variant. (Figure 2A) Other SNPs also had significant differences in OS: patients carrying any A allele in CD73 rs2229523 had significantly longer median OS (41.9 vs 23.7 months, HR: 0.50, 95% CI: 0.28–0.91, P = 0.017) than those with the G/G variant; patients carrying T/T variant in A2BR rs2015353 had significantly longer median OS (49.9 vs 28.1 months, HR: 0.34, 95% CI: 0.14–0.84, P = 0.008) than those with any C allele. (Supplementary Figure 1) In multivariable analysis, CD39 rs11188513 did not show a significant difference but a trend with PFS (HR: 1.60, 95% CI: 0.95–2.71, P = 0.080). Besides, all three SNPs remained significant for OS: CD39 rs11188513 (HR: 2.10, 95% CI: 1.07–4.10, P = 0.031), CD73 rs2229523 (HR: 0.49, 95% CI: 0.26–0.92, P = 0.026), A2BR rs2015353 (HR: 0.24, 95% CI: 0.09–0.64, P = 0.004).
Figure 2.
Overall survivals of patients with CD39 rs11188513 variants, T/T or any C (T/C or C/C), in the discovery cohort (A) and validation cohort (B).
Confirmation of the predictive and prognostic impacts of adenosine-related SNPs in the validation cohort
Among the three SNPs, CD39 rs11188513 was also significantly associated with survival in the validation cohort. In addition, CD39 rs2226163 was significantly associated with survivals. (Table 1)
Patients with any C allele in CD39 rs11188513 had significantly shorter median OS (25.8 vs 31.6 months, HR: 1.50, 95% CI: 1.08–2.09, P = 0.014) in univariate analysis, (Figure 2B) and with consistent results in multivariable analysis (HR: 1.53, 95% CI: 1.09–2.15, P = 0.013). CD39 rs11188513 did not show a significant difference but a trend with PFS in univariate (9.9 vs 9.6 months, HR: 1.30, 95% CI: 0.94–1.80, P = 0.100) and multivariable analysis (HR: 1.41, 95% CI: 1.00–1.98, P = 0.052). Furthermore, patients carrying the G/G variant in CD39 rs2226163 had significantly longer median OS in univariate (37.6 vs 25.8 months, HR: 0.63, 95% CI: 0.42–0.95, P = 0.023) and multivariable analysis (HR: 0.62, 95% CI: 0.41–0.94, P = 0.024). (Supplementary Figure 1) This SNP was also checked in the discovery cohort, and it had the same trend for OS in uni- and multivariable analysis. Patients carrying the G/G variant in CD39 rs2226163 had longer median OS in univariate (49.9 vs 28.6 months, HR: 0.62, 95% CI: 0.29–1.33, P = 0.21) and multivariable analysis (HR: 0.61, 95% CI: 0.28–1.35, P = 0.22).
Evaluation of the predictive and prognostic impacts of adenosine-related SNPs in the control cohort
In the control cohort, there was no evidence for associations of the identified SNPs: CD39 rs11188513, CD39 rs2226163, CD73 rs2229523, and A2BR rs2015353, with PFS or OS in uni- and multivariable analysis. Interestingly, opposite trends to the results in the discovery and validation cohorts were observed in the CD39 rs11188513 and CD39 rs2226163 SNPs, where patients with any C allele in CD39 rs11188513 had longer median OS in univariate (52.0 vs 37.5 months, HR: 0.70, 95% CI: 0.37–1.32, P = 0.26) and multivariable analysis (HR: 0.59, 95% CI: 0.31–1.14, P = 0.12), and patients with G/G allele in CD39 rs2226163 had shorter median OS in univariate (37.5 vs 52.0 months, HR: 1.27, 95% CI: 0.60–2.68, P = 0.52) and multivariable analysis (HR: 1.50, 95% CI: 0.65–3,46, P = 0.34). (Table 1)
Combination of CD39 rs11188513 and CD39 rs2226163
To further understanding the effect of two SNPs from CD39 gene, we combined CD39 rs11188513 and CD39 rs2226163 as the following schema: group I: patients with T/T in CD39 rs11188513 and G/G in CD39 rs2226163; group II: patients with T/T in CD39 rs11188513 and any A in CD39 rs2226163; group III: patients with any C in CD39 rs11188513 and any A in CD39 rs2226163. In the discovery cohort, group III patients showed significantly shorter PFS and OS with P = 0.049 and P = 0.040, respectively, in the univariate analysis; however, the effect was no longer significant in the multivariable analysis. In the validation cohort, patients in group III still showed significant association with shorter OS in both uni- and multivariable analyses (P = 0.047 and P = 0.048, respectively). This effect was not found in the control cohort. (Table 1)
Discussion
We tested the hypothesis that the adenosine-related SNPs are associated with the efficacy of bevacizumab-based chemotherapy in patients with mCRC. Our data showed that the SNPs in CD39, CD73, and A2BR were significantly associated with clinical outcome in mCRC patients treated with FOLFIRI plus bevacizumab in the first-line treatment. CD39 rs11188513 is a strong prognosticator, which was validated in three independent cohorts.
Although adenosine-related molecules have been examined in various cancers, its relationship with efficacy of anti-VEGF therapy has not been studied. Extracellular adenosine, produced via CD39 and CD73, not only stimulates cancer cells through A2BR but also regulates tumor-infiltrating immune cells through A2AR and A2BR.4 Consistent with this, loss of CD39, CD73, A2AR, or A2BR in mice models reportedly leads to activation of anti-tumor immunity and tumor resistance.8, 21−23 Furthermore, under hypoxic states, CD39 and CD73 upregulation through HIF-1α contributes to adenosine production,15, 16 and may therefore serve as a resistance mechanism to anti-angiogenic therapy. Interestingly, the Treg and M2 type macrophage reportedly stimulate angiogenesis through adenosine-A2AR/A2BR signals in mice models,24, 25 implying that the adenosine pathway is an important alternative pro-angiogenic pathway. These evidences are consistent with our data that adenosine pathway is not only prognostic but also predictive for bevacizumab-based chemotherapy.
To our knowledge, the role of CD39 polymorphisms in regulating angiogenesis and clinical outcome in cancer patients has not been previously reported. Both CD39 rs11188513 and CD39 rs2226163 are in the 3’-UTR region of the gene, and are considered binding sites of microRNA. Liu et al. reported that miR-155 expression level was proportional to peripheral CD39-expressed Tregs in sepsis patients,26 and Zhao et al. showed a close inverse relationship between miR-142–3p and CD39 expression level on Tregs in healthy mouse and human models.27 These indicate that SNPs in the binding site of miR-155 and miR-142–3p may be regulating the function of CD39. In addition, the two SNPs may work as tag SNPs and influence functional effects through related polymorphisms at other loci of CD39. In our current study, CD39 rs11188513 was associated not only with OS but also PFS. Although the influence of SNPs in CD39 on phenotypic change is still unknown, it is considered that the gain of function helps develop resistance to anti-VEGF-based chemotherapy.
Interestingly, our study showed that the SNPs in CD39 had opposite trends in patients treated with cetuximab-based chemotherapy. These findings are in consensus with data from Zhi et al., who showed that the expression levels of CD73 and EGFR had positive correlation in breast cancer clinical samples, and EGFR expression was decreased by suppressing not only CD73 or A2AR but also adenosine.28 In colorectal cancer, Wu et al. showed that adenosine increased the expression levels of EGFR.29 Furthermore, Cushman et al. demonstrated that high CD73 expression was associated with longer PFS in mCRC patients treated with cetuximab-based chemotherapy, using clinical samples from CALGB 80203 (n = 238),17 which is consistent with our data. The gain of function status from the CD39 SNPs may have better survival with cetuximab-based chemotherapy than with bevacizumab-based chemotherapy in mCRC patients.
The clinical significance of CD73 and A2BR polymorphisms in cancer patients also remains unknown. CD73 rs2229523 is a common missense SNP, which shows A1682G as a base pair change and Thr376Ala as an amino acid change. Similarly, A2BR rs2015353 is a common missense SNP, which shows T437C as a base pair change and Ser9Pro as an amino acid change. Figler et al. demonstrated that A2BR mRNA levels in macrophages and A2BR rs2015353 showed significant correlation with IL-6 production in diabetic patients, suggesting that the SNP can change the function of A2BR.30 Furthermore, this SNP may affect functional activity as a tag SNP. According to our experimental data, both CD73 rs2229523 and A2BR rs2015353 are strongly associated with prognosis in the discovery cohort. Further studies are necessary to confirm the clinical significance of these SNPs.
Based on accumulating evidences, this adenosine pathway can be crucial for cancer progression and metastasis. Clinical trials with inhibitors of adenosine pathway are ongoing. Although the antibodies for CD39 or A2BR are still in preclinical stage, an anti-CD73 mAb, MEDI944731 (NCT02503774), and antagonists for the A2AR, CPI-444 (NCT02655822) or PBF-50932 (NCT02403193), are currently in phase I clinical trials for solid cancers. Notably, activation of A2AR reportedly promotes the expression of CTLA-4 and PD-1 on T cells, and enhances immunosuppressive functions.13, 33 Conversely, anti-PD-1 therapy or adoptive T-cell therapy for cancer patients in turn stimulates the expression of CD73, suggesting a possible resistance mechanism.34 Recent reports also showed that anti-adenosine pathway therapy can cooperate with other existing immune checkpoint inhibitors such as anti-CTLA-4 or anti-PD1 in preclinical studies.13, 35, 36 Following the preclinical results, the above phase I clinical trials (NCT02503774, NCT02655822, and NCT0240319) are evaluating the clinical impact of combination of anti-CD73/anti-A2AR drugs and PD-1/ PD-L1 inhibitors. It would be warranted to examine if the selected SNPs may be potential biomarkers not only for the novel adenosine-related drugs but also for the existing immunotherapies.
Limitations need to be discussed. A selection bias cannot be excluded due to the retrospective study design. Although CD39 rs2226163, CD73 rs2229523 and A2BR rs2015353 were prognostic markers in one cohort, the results were not validated in another. This result may be due to the differences in baseline characteristics. (Supplementary Table 2) Nonetheless, our results convincingly support the past findings, which are from two prospective phase III clinical trials, FIRE-3 and TRIBE. Further, the SNPs were more strongly related to OS than PFS, and not to tumor response. Hence, it is unclear whether these SNPs are prognostic or predictive, and whether these results are attributed mainly to the effect of bevacizumab or subsequent immune modulation. Further functional studies and large-scale prospective studies are warranted to fully elucidate our results.
Conclusion
This is the first study to show the associations of genetic variants in CD39, CD73, and A2BR with clinical outcomes of mCRC patients treated with FOLFIRI plus bevacizumab. We have found that the patients with the C allele of CD39 rs11188513 had much worse OS than others in the bevacizumab cohort, and may have the survival benefits not from bevacizumab-based chemotherapy but from cetuximab-based chemotherapy as a first-line treatment. If our findings are validated prospectively, this SNP could be predictive for the mCRC patients who would benefit from bevacizumab-based chemotherapy.
Supplementary Material
Supplementary Figure 1. Overall survivals of patients with CD39 rs2226163 variants, G/G or any A (G/A or A/A), with CD73 rs2229523 variants, any A (G/A or A/A) or G/G, and with A2BR rs2015353 variants, T/T or any C (T/C or C/C), in the discovery cohort (A, C, E) and validation cohort (B, D, F), respectively.
Clinical practice point:
Adenosine has a potent immunosuppressive and angiogenic ability in the tumor microenvironment.
Although some reports have shown that the expressions of adenosine-related molecules have strong association with survival of cancer patients, the predictive or prognostic role of genetic changes within adenosine-related molecules remains unknown.
With the 451 metastatic colorectal cancer (mCRC) patients from phase III clinical trials (FIRE-3 and TRIBE), our study tested the hypothesis that the single nucleotide polymorphisms (SNPs) in adenosine-related molecules were associated with immune dysregulation and the efficacy of bevacizumab.
We investigated relationships between the SNPs and clinical outcomes in mCRC patients treated with bevacizumab-based chemotherapy, and with cetuximab-based chemotherapy as control.
The patients with any C allele in CD39 rs11188513 had significantly shorter median overall survival (OS), which was confirmed in the discovery and the validation cohort.
The patients with any A allele in CD73 rs2229523 had significantly longer median OS; the patients carrying T/T variant in A2BR rs2015353 had significantly longer median OS; and the patients carrying G/G variant in CD39 rs2226163 had significantly longer median OS, in one cohort, respectively.
The SNPs in CD39 had opposite results in the patients treated with bevacizumab-based chemotherapy and cetuximab-based chemotherapy.
Our results suggest that the selected SNPs in adenosine-related molecules may be biomarkers for bevacizumab-based chemotherapy as well as promising therapeutic targets in mCRC.
Funding:
R Tokunaga was supported by the Uehara Memorial Foundation. Martin D. Berger received a grant from the Swiss Cancer League (BIL KLS-333402 2014) and the Werner and Hedy Berger-Janser Foundation for cancer research. H.-J. Lenz was supported by the National Cancer Institute (grant number P30CA014089), the Gloria Borges WunderGlo Foundation-The Wunder Project, Dhont Family Foundation, San Pedro Peninsula Cancer Guild, Daniel Butler Research Fund, and Call to Cure Fund.
Abbreviations:
- CI
confidence interval
- CTL
cytotoxic T lymphocytes
- HR
hazard ratio
- mCRC
metastatic colorectal cancer
- NK cells
natural killer cells
- OS
overall survival
- PFS
progression-free survival
- SNP
single nucleotide polymorphism
- Treg
regulatory T cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Presentation:
We have not presented this article anywhere.
Conflict of interest:
The authors declare that they have no conflict of interest.
Ethical approval and ethical standards:
The clinical trial registry identifiers are NCT00433927 (FIRE-3) and NCT00719797 (TRIBE). Use of the clinical data and clinical samples for molecular analysis were approved by the institutional review boards of each participating institute and conducted in accordance with the Declaration of Helsinki and Good Clinical Practice Guidelines.
References
- 1.Hausler SF, Montalban del Barrio I, Strohschein J, et al. Ectonucleotidases CD39 and CD73 on OvCA cells are potent adenosine-generating enzymes responsible for adenosine receptor 2A-dependent suppression of T cell function and NK cell cytotoxicity. Cancer Immunol Immunother 2011; 60:1405–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deaglio S, Dwyer KM, Gao W, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med 2007; 204:1257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Resta R, Yamashita Y, Thompson LF. Ecto-enzyme and signaling functions of lymphocyte CD73. Immunol Rev 1998; 161:95–109. [DOI] [PubMed] [Google Scholar]
- 4.Yegutkin GG. Nucleotide- and nucleoside-converting ectoenzymes: Important modulators of purinergic signalling cascade. Biochim Biophys Acta 2008; 1783:673–94. [DOI] [PubMed] [Google Scholar]
- 5.Junger WG. Immune cell regulation by autocrine purinergic signalling. Nat Rev Immunol 2011; 11:201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christofi FL, Zhang H, Yu JG, et al. Differential gene expression of adenosine A1, A2a, A2b, and A3 receptors in the human enteric nervous system. J Comp Neurol 2001; 439:46–64. [DOI] [PubMed] [Google Scholar]
- 7.Beavis PA, Divisekera U, Paget C, et al. Blockade of A2A receptors potently suppresses the metastasis of CD73+ tumors. Proc Natl Acad Sci U S A 2013; 110:14711–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohta A, Gorelik E, Prasad SJ, et al. A2A adenosine receptor protects tumors from antitumor T cells. Proc Natl Acad Sci U S A 2006; 103:13132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mittal D, Sinha D, Barkauskas D, et al. Adenosine 2B Receptor Expression on Cancer Cells Promotes Metastasis. Cancer Res 2016; 76:4372–82. [DOI] [PubMed] [Google Scholar]
- 10.Young A, Ngiow SF, Barkauskas DS, et al. Co-inhibition of CD73 and A2AR Adenosine Signaling Improves Anti-tumor Immune Responses. Cancer Cell 2016; 30:391–403. [DOI] [PubMed] [Google Scholar]
- 11.Leclerc BG, Charlebois R, Chouinard G, et al. CD73 Expression Is an Independent Prognostic Factor in Prostate Cancer. Clin Cancer Res 2016; 22:158–66. [DOI] [PubMed] [Google Scholar]
- 12.Young A, Ngiow SF, Madore J, et al. Targeting adenosine in BRAF-mutant melanoma reduces tumor growth and metastasis. Cancer Res 2017. [DOI] [PubMed] [Google Scholar]
- 13.Allard B, Pommey S, Smyth MJ, et al. Targeting CD73 enhances the antitumor activity of anti-PD-1 and anti-CTLA-4 mAbs. Clin Cancer Res 2013; 19:5626–35. [DOI] [PubMed] [Google Scholar]
- 14.Beavis PA, Milenkovski N, Henderson MA, et al. Adenosine Receptor 2A Blockade Increases the Efficacy of Anti-PD-1 through Enhanced Antitumor T-cell Responses. Cancer Immunol Res 2015; 3:506–17. [DOI] [PubMed] [Google Scholar]
- 15.Synnestvedt K, Furuta GT, Comerford KM, et al. Ecto-5’-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia. J Clin Invest 2002; 110:993–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatfield SM, Kjaergaard J, Lukashev D, et al. Systemic oxygenation weakens the hypoxia and hypoxia inducible factor 1alpha-dependent and extracellular adenosine-mediated tumor protection. J Mol Med (Berl) 2014; 92:1283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cushman SM, Jiang C, Hatch AJ, et al. Gene expression markers of efficacy and resistance to cetuximab treatment in metastatic colorectal cancer: results from CALGB 80203 (Alliance). Clin Cancer Res 2015; 21:1078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai XY, Wang XF, Li J, et al. High expression of CD39 in gastric cancer reduces patient outcome following radical resection. Oncol Lett 2016; 12:4080–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinemann V, von Weikersthal LF, Decker T, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol 2014; 15:1065–75. [DOI] [PubMed] [Google Scholar]
- 20.Loupakis F, Cremolini C, Masi G, et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med 2014; 371:1609–18. [DOI] [PubMed] [Google Scholar]
- 21.Cekic C, Sag D, Li Y, et al. Adenosine A2B receptor blockade slows growth of bladder and breast tumors. J Immunol 2012; 188:198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckle T, Fullbier L, Wehrmann M, et al. Identification of ectonucleotidases CD39 and CD73 in innate protection during acute lung injury. J Immunol 2007; 178:8127–37. [DOI] [PubMed] [Google Scholar]
- 23.Stagg J, Divisekera U, Duret H, et al. CD73-deficient mice have increased antitumor immunity and are resistant to experimental metastasis. Cancer Res 2011; 71:2892–900. [DOI] [PubMed] [Google Scholar]
- 24.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity 2010; 32:593–604. [DOI] [PubMed] [Google Scholar]
- 25.Ren L, Yu Y, Wang L, et al. Hypoxia-induced CCL28 promotes recruitment of regulatory T cells and tumor growth in liver cancer. Oncotarget 2016; 7:75763–75773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Shi K, Chen M, et al. Elevated miR-155 expression induces immunosuppression via CD39(+) regulatory T-cells in sepsis patient. Int J Infect Dis 2015; 40:135–41. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J, Cao Y, Lei Z, et al. Selective depletion of CD4+CD25+Foxp3+ regulatory T cells by low-dose cyclophosphamide is explained by reduced intracellular ATP levels. Cancer Res 2010; 70:4850–8. [DOI] [PubMed] [Google Scholar]
- 28.Zhi X, Wang Y, Yu J, et al. Potential prognostic biomarker CD73 regulates epidermal growth factor receptor expression in human breast cancer. IUBMB Life 2012; 64:911–20. [DOI] [PubMed] [Google Scholar]
- 29.Wu R, Chen Y, Li F, et al. Effects of CD73 on human colorectal cancer cell growth in vivo and in vitro. Oncol Rep 2016; 35:1750–6. [DOI] [PubMed] [Google Scholar]
- 30.Figler RA, Wang G, Srinivasan S, et al. Links between insulin resistance, adenosine A2B receptors, and inflammatory markers in mice and humans. Diabetes 2011; 60:669–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geoghegan JC, Diedrich G, Lu X, et al. Inhibition of CD73 AMP hydrolysis by a therapeutic antibody with a dual, non-competitive mechanism of action. MAbs 2016; 8:454–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mediavilla-Varela M, Castro J, Chiappori A, et al. A Novel Antagonist of the Immune Checkpoint Protein Adenosine A2a Receptor Restores Tumor-Infiltrating Lymphocyte Activity in the Context of the Tumor Microenvironment. Neoplasia 2017; 19:530–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohta A, Kini R, Subramanian M, et al. The development and immunosuppressive functions of CD4(+) CD25(+) FoxP3(+) regulatory T cells are under influence of the adenosine-A2A adenosine receptor pathway. Front Immunol 2012; 3:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinhardt J, Landsberg J, Schmid-Burgk JL, et al. MAPK Signaling and Inflammation Link Melanoma Phenotype Switching to Induction of CD73 during Immunotherapy.Cancer Res 2017. [DOI] [PubMed] [Google Scholar]
- 35.Mittal D, Young A, Stannard K, et al. Antimetastatic effects of blocking PD-1 and the adenosine A2A receptor. Cancer Res 2014; 74:3652–8. [DOI] [PubMed] [Google Scholar]
- 36.Iannone R, Miele L, Maiolino P, et al. Adenosine limits the therapeutic effectiveness of anti-CTLA4 mAb in a mouse melanoma model. Am J Cancer Res 2014; 4:172–81. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Overall survivals of patients with CD39 rs2226163 variants, G/G or any A (G/A or A/A), with CD73 rs2229523 variants, any A (G/A or A/A) or G/G, and with A2BR rs2015353 variants, T/T or any C (T/C or C/C), in the discovery cohort (A, C, E) and validation cohort (B, D, F), respectively.