Abstract
Since the advent of induced pluripotent stem cell (iPSC) technology a decade ago, enormous progress has been made in stem cell biology and regenerative medicine. Human iPSCs have been widely used for disease modeling, drug discovery, and cell therapy development. Novel pathological mechanisms have been elucidated, new drugs originating from iPSC screens are in the pipeline, and the first clinical trial using human iPSC-derived products has been initiated. In particular, the combination of human iPSC technology with recent developments in gene editing and three-dimensional organoids makes iPSC-based platforms even more powerful in each area of their application, including precision medicine. In this overview, we will discuss the progress in applications of iPSC technology that are particularly relevant to drug discovery and regenerative medicine, in light of the remaining challenges and the emerging opportunities in the field.
Introduction
In 2006, a major technological breakthrough in science and medicine was made with the report that cells with gene expression/epigenetic profile and developmental potential that are similar to embryonic stem cells (ESCs) can be generated from somatic cells (such as fibroblasts) in mice by using a cocktail of four transcriptional factors1. These cells were termed induced pluripotent stem cells (iPSCs) and the four factors — Oct4, Sox2, Klf4 and c-Myc — were named “Yamanaka factors”. Just one year later, the generation of iPSCs from human fibroblasts was reported from two laboratories simultaneously2,3.
Human iPSC technology, which has evolved rapidly since 2007 (Box 1), has ushered in an exciting new era for the fields of stem cell biology and regenerative medicine, as well as disease modeling and drug discovery. Soon after the development of the technology, human iPSCs were rapidly applied to generate human ‘disease-in-a-dish’ models and used for drug screening for both efficacy and potential toxicities. Such approaches are now becoming increasingly popular, given the surge of interest in phenotypic screening and the advantages of human iPSCs in disease modeling, compared with traditional cellular screens. These advantages include their human origin, easy accessibility, expandability, ability to give rise to almost any cell types desired, avoidance of ethical concerns associated with human ESCs, and the potential to develop personalized medicine using patient-specific iPSCs. Furthermore, recent advances with gene-editing technologies — in particular the CRISPR/Cas9 technology — are enabling the rapid generation of genetically defined human iPSC-based disease models. iPSCs are also a key component of an emerging generation of more physiologically representative cellular platforms incorporating three dimensional (3D) architectures and multiple cell types.
Box 1 |. Evolution of human iPSC technology.
Since its beginning in 2006, iPSC technology has evolved rapidly. Because iPSCs were initially generated by introducing reprogramming factors using integrating viral vectors, such as retrovirus or lentivirus, there is a concern about clinical application of these iPSCs due to potential insertional mutagenesis that might be caused by integration of transgenes into the genome of host cells204. To make iPSCs clinically applicable, a variety of non-integrating methods have been developed to circumvent the risk of insertional mutagenesis and genetic alterations associated with retroviral and lentiviral transduction-mediated introduction of reprogramming factors205. These non-integrating methods include reprogramming using episomal DNAs206,207, adenovirus208, Sendai virus209, PiggyBac transposons210, minicircles211, recombinant proteins212, synthetic modified mRNAs213, microRNAs214,215, and small molecules216, although the small molecule approach is not applicable to human iPSC derivation yet. Among these approaches, episomal DNAs, synthetic mRNAs and sendai virus are commonly applied to derive integration-free iPSCs due to their relative simplicity and high efficiency185. The use of non-viral methods or non-integrating viruses could avoid genomic insertions, thus reducing the risk for translational application of iPSCs. Human iPSCs derived using these non-integrating approaches provide a cellular resource that is more relevant for clinical applications.
iPSC technology has also attracted considerable interest in its potential applicability for regenerative medicine. The first clinical study using human iPSC-derived cells was initiated in 2014, which used human iPSC-derived retinal pigment epithelial (RPE) cells to treat macular degeneration4, and was reported to have improved the patient’s vision5. Although the clinical study was subsequently put on hold due to the identification of two genetic variants in iPSCs of the patient, the trial is expected to resume6.
Clearly, human iPSC technology holds great promise for human disease modeling, drug discovery, and stem cell-based therapy, and this potential is only beginning to be realized. In this article, we overview the progress in each of the main applications of iPSCs in the decade since the discovery of the technology, featuring key illustrative examples, discussing remaining limitations and approaches to address them, and highlighting emerging opportunities.
iPSC-based disease modeling
Identifying pathological mechanisms underlying human diseases has a key role in discovering novel therapeutic strategies. Animal models have provided valuable tools for modeling human diseases, allowing the identification of pathological mechanisms at distinct developmental stages and in specific cell types in an in vivo setting. Moreover, in mice it is possible to develop in vitro iPSC-based disease models and the corresponding in vivo models in parallel. Comparing the phenotypes observed with corresponding in vitro and in vivo mouse models could provide a better understanding of the strength and limitations of in vitro human iPSC-based models.
However, significant species differences could prevent the recapitulation of full human disease phenotypes in animals such as mice, which are the most commonly used animal models. For example, although many transgenic mouse models have been created for Alzheimer’s disease, none has captured the entire spectrum of the human disease pathology, including considerable neuronal loss7,8. This is likely due to fundamental species differences between mouse and human neural cells. Thus, there is an urgent need to establish human disease modeling platforms to complement studies in animal models for biomedical research.
Disease modeling using primary patient-derived cells is helpful for studying the etiology of human diseases and developing therapeutic strategies for these diseases. However, the unavailability of expandable sources of primary cells from patients, especially hard-to-access cells such as brain cells and heart cells, is a critical limitation. Human iPSCs are therefore an attractive alternative because of the ease with which human diseases (particularly those with defined genetic causes) could in principle be modeled using iPSCs derived from easily accessible cell types, such as skin fibroblasts and blood cells from diverse patients. Because of their intrinsic properties of self-renewal and potential to differentiate into nearly any cell types in the body, patient-specific iPSCs could provide large quantities of disease-relevant cells and a variety of different cell types that were previously inaccessible, such as neurons and cardiomyocytes. Furthermore, because iPSCs can be derived from the relevant patients themselves, they could enable personalized disease modeling that would be a central part of precision medicine.
Both human ESCs and iPSCs have been used for modeling human genetic diseases. The earlier models were developed using ESCs9, but following the advent of human iPSC technology, human iPSCs have become the preferred option because of their availability and lack of potential ethical concerns associated with human ESCs. Human iPSCs are very similar to human ESCs. Both types of cells express human pluripotent factors and ESC surface markers, and exhibit developmental potential to differentiate into three germ layers2,3. Residual epigenetic memory of somatic cells could occur in iPSCs10–12, which may affect the differentiation potential of these cells13. Although the persistence of epigenetic memory of parental cells has been reported in iPSCs10–12, similar phenotypes have been reported in disease modeling using human ESCs and iPSCs in most cases13, validating the effectiveness of disease modeling using patient-derived iPSCs.
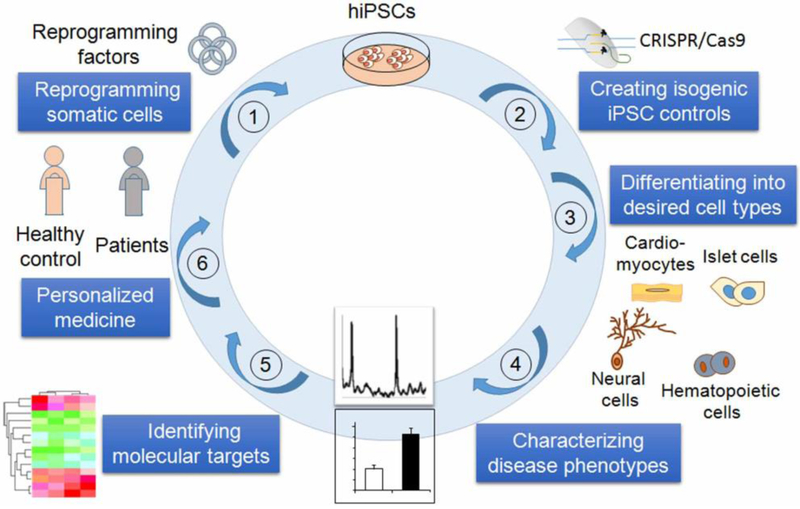
Disease modeling using human iPSCs starts by deriving iPSCs containing the disease-causing mutation(s) (Fig. 1). These cells are then differentiated into disease-relevant cell types. The resultant cells are used to reveal disease etiology and identify pathological mechanisms. In early studies of iPSC-based disease modeling, iPSCs derived from non-disease-affected individuals were used as controls for patient-derived iPSCs. However, like other cells, iPSCs have exhibited line-to-line variations, which complicates data interpretation because one has to distinguish the line-to-line variation from the true disease-relevant phenotypes.
Figure 1.
A schematic for human iPSC-based disease modeling. Human iPSCs are derived from individual patients and differentiated into specific cell types. To develop new therapies, the resultant cells are used to observe disease-specific phenotypes and identify novel pathological mechanisms,. Human iPSC-based disease modeling with patient-specific cells now provides an exciting new approach for the development of personalized diagnosis and medicine.
Rapidly developing genome editing technologies now enable the introduction of genetic changes into iPSCs in a site-specific manner, including correction of disease-causing gene mutations in patient-derived iPSCs and introduction of specific mutations into non-disease affected wild type (WT) iPSCs. These approaches allow the generation of genetically matched, isogenic iPSC lines with the introduced mutation as the sole variable, ensuring the reliable identification of the true pathology while avoiding the confusion with any disparities in genetic background or epiphenomena resulting from possible line-to-line variations. The isogenic iPSC controls will be especially important when modeling sporadic or polygenic diseases, in which phenotypic differences are expected to be small14.
The development of programmable site-specific nucleases, including the zinc-finger nuclease (ZFN)15,16, transcription activator-like effector nucleases (TALENs)17–19, and the CRISPR/Cas9 system20–22,23 (Table 1), has improved gene editing efficiency in human ESCs and iPSCs substantially by inducing DNA double-strand breaks at the site of gene modification. The CRISPR/Cas9 technology in particular has attracted much attention and gained wide usage in gene editing of human ESCs and iPSCs due to its simplicity in design and ease of use. This gene editing technology allows researchers to introduce disease-causing mutations to WT iPSCs and eliminate such mutations in patient iPSCs to create isogenic controls for iPSC-based disease modeling (Fig. 1).
Table 1.
Technology for gene editing of human ESCs and iPSCs
| System | Enzyme | Mode of action | References |
|---|---|---|---|
| ZFN | Zinc-finger nucleases | Custom zinc-finger protein DNA binding modules fused to the cleavage domain of the bacterial endonuclease FokI to induce site-specific DNA double-strand breaks (DSBs), followed by DNA repair through NHEJ to create small insertion and deletion mutations (Indels) or HDR to introduce precise nucleotide modifications. | Urnov et al. 2005217; Lombardo et al. 2007218; Hockemeyer et al. 200915; Zou et al. 200916 |
| TALEN | Transcription activator-like effector nucleases | Custom TALE protein DNA binding modules fused to the bacterial endonuclease FokI to induce site-specific DSBs, followed by DNA repair through NHEJ or HDR to introduce Indels or specific DNA mutations. | Hockemeyer et al. 201118 |
| CRISPR/Cas9 | WT Cas9, Cas9 nickase | RNA-guided site-specific DNA cleavage triggers NHEJ to create Indels or HDR to introduce precise DNA modifications. | Cho et al. 2013219; Cong et al. 201320; Jinek et al. 2013220 |
| CRISPR/Cas9 | Cas9 nickase | This approach combines a Cas9 nickase with paired sgRNA to create targeted DSBs. The paired nicking reduces off-target activity substantially (50 to 1,500-fold). | Ran et al. 201334 |
| CRISPR/Cas9 | eSpCas9 | Structure-guided protein engineering was used to create spCas9 variants with reduced off-target effect but robust on-target activity. | Slaymaker et al. 201635 |
| CRISPR/Cas9 | Cas9-VRER variant | This platform, called ‘CORRECT’, allows introduction of DNA modification in a precise mono-allelic or bi-allelic manner | Paquet et al. 201637 |
| CRISPR/Cas9/cytidine deaminase | Fusions of CRISPR/Cas9 and a cytidine deaminase | The base editing approach allows direct conversion of cytidine to uridine without the need for DSB and donor DNA template. | Komor et al. 201638 |
ZFN: Zinc-Finger nucleases; TALEN: Transcription activator-like effector nucleases; TALE: Transcription activator-like effector; NHEJ: Non-homologous end joining; HDR: homology-directed repair; sgRNA: small guide RNA; spCas9: Streptococcus pyogenes Cas9; eSpCas9: enhanced specificity spCas9; CORRECT: consecutive re-guide or re-Cas steps to erase CRISPR/Cas9-blocked targets; NgArgo: Natronobacterium gregoryi Argonaute; PAM: protospacer-adjacent motif (PAM).
However, a major challenge in applications using the CRISPR/Cas9 technology is the possibility of off-target effects. Nevertheless, although relatively high levels of off-target gene modifications by CRISPR/Cas9 have been described in cancer cell lines24, recent studies from multiple laboratories using whole genome sequencing (WGS) indicate that off-target gene modifications are rare in normal human cells, including human iPSCs and ESCs25–29. WGS using genomic DNAs isolated from the original iPSCs and corresponding gene-edited iPSCs, coupled with comprehensive bioinformatic analysis25,27–29, is useful for detecting off-target effects such as single nucleotide variants (SNVs) and insertions or deletions (indels), especially for cells that will be used for clinical applications. At present, WGS is expensive, but it is expected that the price will go down with continuous development of the technology. Alternative approaches for detecting off-target effects include exosome sequencing30 and targeted deep sequencing29. For targeted deep sequencing, one can search for potential off-target sites that are different from the on-target sites in the human genome using Cas-OFFinder (http://www.rgenome.net)31, an algorithm for identifying off-target sites, including off-target SNVs or indels.
Gene editing tools are also being continuously improved and refined, which may help address the issue of off-target effects. Originally CRISPR/Cas9 edits a genomic locus by inducing DNA double-strand breaks using a single guide RNA-directed wild type Cas9 nuclease. The nickase version of Cas9 (D10A mutant) directed by paired guide RNAs or the engineered Cas9 nuclease variants with enhanced specificity (eSpCas9) is now being used increasingly for genome editing32–34, because both have been shown to reduce off-target effects substantially while retaining rigorous on-target cleavage34,35. Furthermore, catalytically dead Cas9 (dCas9) fused with transcriptional activator or suppressor has been used to modulate transcription of endogenous genes (so-called CRISPRi or CRISPRa) or image genomic loci by fusing with a fluorescent protein32–34,36. Modifications of the CRISPR/Cas9 system also enable explicit introduction of DNA sequence changes in a precise mono-allelic or bi-allelic manner with high efficiency37. A recent development in base editing takes advantage of the fusion of CRISPR/Cas9 and a cytidine deaminase enzyme to allow direct conversion of cytidine to uridine without the need of double strand DNA break38. This new approach enhances gene editing efficiency and will further facilitate gene editing in human ESCs and iPSCs.
iPSC-based disease modeling is widely used for studying disorders caused by a single gene mutation (monogenic disorders) that have an early onset39,40, as the approach is ideally suited to such disorders — because iPSCs can be easily derived from patients with these disorders and differentiated into disease-relevant cells, such as neurons. Furthermore, given the relative immaturity of cells differentiated from iPSCs41, there is greater confidence that the phenotypes of cells differentiated from iPSCs provide a good model for diseases with an early onset versus late onset, for which cellular aging may be important in disease pathology41. For example, neurons differentiated from patient iPSCs were used to model spinal muscular atrophy (SMA), an early-onset disease caused by mutations in the gene encoding the survival motor neuron 1 (SMN1)39. Mutations in the SMN1 gene led to degeneration of motor neurons and subsequent muscular atrophy. Type 1 SMA patients usually show symptoms at 6 months from birth, with a rapid disease progression that kills them by the age of two42. In an initial iPSC-based disease modeling study39, iPSCs were derived from fibroblasts of a type 1 SMA patient and differentiated into a disease-relevant cell type, motor neurons. Reduced survival of motor neurons differentiated from patient iPSCs was observed, compared to that of motor neurons derived from an unaffected control. Moreover, the SMA patient-derived iPSCs were able to respond to valproic acid and tobramycin, two compounds known to induce SMN protein levels, by increasing SMN protein levels and SMN protein-containing ‘gems’39. This study provides a proof-of-principle that patient-derived iPSCs could be used to model early-onset genetic diseases and serve as potential drug-screening platforms.
Modeling diseases that have a late onset is more challenging, because cells differentiated from human iPSCs in general exhibit fetal-like properties41. However, induced cellular aging has been used to aid in successful modeling of late-onset diseases43–46. One way to induce aging in cells differentiated from human iPSCs is to treat cells with cellular stressors, including compounds that target mitochondrial function or protein degradation, such as pyraclostrobin and MG-13243,44,46,47. Another way to induce cellular aging is to ectopically express gene products that induce premature aging, such as progerin45. However, whether cellular stressors or progerin expression can elicit cellular aging through a mechanism that is similar to normal aging remains to be determined41. Moreover, recent studies indicate that cellular maturation and aging may be distinct events41,48. It remains unclear whether the cellular aging inducers can promote both cellular maturation and aging, as opposed to triggering cellular aging in immature cells48. Alternatively, the direct reprogramming approach that involves direct conversion of human fibroblasts into other lineage-specific cells, such as neurons, does not erase cellular aging markers49. Indeed, neurons derived from aged fibroblasts through direct reprogramming have been shown to maintain cellular age50, therefore offering an alternative cellular model to study age-related disorders. It is worth noting that there has also been success in promoting cellular maturation, such as by using improved formulation of cultured medium51 and neuron-astrocyte co-culture system52,53.
iPSCs also offer a new way to study sporadic diseases (the causes of which have not been identified in patients’ family histories or genetic mutations), which is important as the majority of patients with many diseases have sporadic forms of the disease. For example, in Alzheimer’s disease, 95% of patients fall under the sporadic category. Interestingly, analysis of iPSC-derived nerve cells from patients with sporadic Alzheimer’s disease identified several sporadic cases that exhibited the same phenotypes as familial Alzheimer’s disease with a specific gene mutation54 , which indicated that it may be possible to re-classify the sporadic condition using iPSCs. However, modeling sporadic diseases using iPSCs is generally more difficult than monogenic disorders because the phenotypic changes in such diseases are often thought to be induced by multiple small-effect genetic risk variants, in combination with environmental factors. Although iPSCs derived from patients with such diseases would contain disease-relevant risk variants, using iPSCs to model such diseases is complicated by line-to-line variation in genetic and epigenetic background. Such variation is more problematic for modeling sporadic diseases, because the phenotypes of the sporadic disease iPSC-derived cells are expected to be more subtle than for those derived from monogenic diease iPSCs.
Thus, a key question for human iPSC-based modeling of sporadic diseases is how to generate paired isogenic cell lines that only differ at relevant risk variants14. Recently, the CRISPR/Cas9 gene editing approach has been used to generate isogenic iPSC lines that differ at a PD-associated risk variant55. The ability to generate genetically controlled isogenic iPSC lines in which specific disease-associated genetic risk variants are the sole variable creates a well-controlled system, which in combination with an allele-specific assay has enabled robust dissection of a genetic risk variant for Parkinson’s disease55. This experimental paradigm could be applied to studying genetic risk factors associated with other diseases.
To date, many diseases have been studied using a single disease-relevant cell type derived from iPSCs. For example, iPSC-derived neurons have been used to model Alzheimer’s disease54,56–68 (Table 2) and Parkinson’s disease (Table 3)43–45,55,69–84. However, more than one cell type may be required to effectively model some diseases. Indeed, comparable efforts have been devoted to model schizophrenia using patient iPSC-derived neurons85–87 and neural progenitor cells88–92. To better recapitulate disease phenotypes, co-culture of more than one cell types may also be needed to study the interaction of different cell types. For example, astrocyte/neuron co-cultures have been used to model the pathology of amyotrophic lateral sclerosis (ALS)93–96. The co-culture system allowed the investigation of non-cell-autonomous aspects of disease pathology, which would otherwise be impossible with single cell types, such as neurons. Moreover, these studies enabled the identification of astrocytes as a critical cellular component contributing to motor neuron degeneration in ALS and provided a drug screening platform for ALS using patient iPSC-derived astrocytes93–96.
Table 2.
Patient iPSC-based modeling of Alzheimer’s disease (AD)
| AD patients for iPSC derivation | Gene mutation | Cell types analyzed | Phenotypes in the dish | References |
|---|---|---|---|---|
| fAD | PS1 (A246E) PS2 (N1411) |
Neurons | Increased amyloid β (Aβ) 42 secretion. | Yagi et al. 201156 |
| fAD and sAD | APPDp | Neurons | Elevated Aβ40 secretion; elevated tau phosphorylation; increased active GSK3β; increased number of endosomes. | Israel et al. 201257 |
| fAD and sAD | APP (E693Δ) APP (V717L) |
Cortical neurons | Increased Aβ42 secretion; elevated Aβ oligomers; reduced survival; vulnerability to oxidative stress. | Kondo et al. 201354 |
| fAD | PS1 (A79V) APP (K724N) |
Neurons | Elevated Aβ42/Aβ40 ratio. | Mertens et al. 201359 |
| fAD | PS1 (ΔE9) | Neurons | Elevated Aβ42/Aβ40 ratio; reduced γ-secretase activity. | Woodruff et al. 201358 |
| sAD | APOE (E3/E4) | Forebrain cholinergic neurons | Elevated Aβ42/Aβ40 ratio; increased vulnerability to glutamate-mediated cell death; increased intracellular calcium levels upon glutamate stimulation. | Duan et al. 201461 |
| fAD | PS1 mutation | Neurons | Increased Aβ42 secretion; impaired autophagic function | Lee et al. 201460 |
| fAD | PS1 (A246E) PS1 (H163R) PS1 (M146L) |
Neurons | Elevated Aβ42/Aβ40 ratio | Liu et al. 201464 |
| fAD | PS1 (A246E) | Neurons | Elevated Aβ42/Aβ40 ratio; increased expression of FOXG1, mGLUR1 and SYT1. | Mahairaki et al. 201463 |
| fAD | APP (V717I) | Forebrain neurons | Elevated Aβ42/Aβ40 ratio; altered APP subcellular localization; increased levels of total tau and p-tau. | Muratore et al. 201462 |
| sAD | - | Neurons | AD-related protein interaction network composed of APP and GSK3β | Hossini et al. 201566 |
| fAD | PS1 (Y115C) PS1 (M146I) PS1 (intron 4) APP (V717I) APPDp |
Cortical excitatory neurons | Elevated Aβ42 secretion; Increased intracellular levels of tau and p-tau. | Moore et al. 201567 |
| sAD | SOR1 variants | Neurons | Altered induction of SORL1 expression; altered Aβ peptide production. | Young et al. 201565 |
| fAD | APP (V717I) | Neurons and glia | High levels of Aβ and sAPPα secreted from both neuronal and glial cells, especially GABAergic neurons. | Liao et al. 201668 |
fAD: familial AD; sAD: sporadic AD; PS1: presenilin 1; PS2: presenilin 2; APP: amyloid-β precursor protein; APPDp: duplication of the amyloid-β precursor protein gene; p-tau: phosphorylated tau.
Table 3.
Patient iPSC-based modeling of Parkinson’s disease (PD)
| Patients for iPSC derivation | Gene mutation | Cell types analyzed | Phenotypes in the dish | References |
|---|---|---|---|---|
| fPD | SCNA triplication | DA neurons | Doubling the expression of α-synuclein protein. | Devine et al. 201170 |
| fPD | LRRK2 (G2019S) | DA neurons | Elevated oxidative stress response; increased sensitivity to stress-induced cell death. | Nguyen et al. 201143 |
| fPD | PINK1 (c.1366C>T; p.Q456X) or PINK 1 (c.509T>G; p.V170G) |
DA neurons | Impaired recruitment of Parkin to mitochondria; increased copy number of mitochondria; up-regulation of PGC1α. | Seibler et al. 201169 |
| fPD | PINK1 (Q456X); LRRK2 (G2019S) |
DA and non-DA neurons, & immature cells | Increased vulnerability to stress; dysfunction of mitochondrial. | Cooper et al. 201244 |
| fPD | PARKIN (exon 2–4 deletion); PARKIN (exon 6, 7 deletion) |
Neurons | Increased oxidative stress; activated Nrf2 pathway; abnormal mitochondrial morphology and turnover; elevated accumulation of α-synuclein. | Imaizumi et al. 201271 |
| fPD | PARKIN (exon 3, 5 deletion) PARKIN (exon 3 deletion) |
DA Neurons | Increased oxidative stress; reduced DA uptake; enhanced spontaneous release of DA. | Jiang et al. 201272 |
| fPD | LRRK2 (G2019S) | Neural stem cells | Increase sensitivity to stress; progressive impairment in nuclear envelope organization; defective self-renewal and neuronal differentiation. | Liu et al. 201246 |
| fPD & sPD | LRRK2 (G2019S) | DA neurons | Increase apoptosis; reduced neurite numbers and complexity; increased autophagic vacuoles. | Sanchez-Danes et al. 201273 |
| fPD | SCNA (A53T) SCNA triplication |
Cortical neurons | Increased nitrosative stress; elevated ER stress and ERAD substrates. | Chung et al. 201374 |
| fPD | PINK1 (Q456X) PINK1 (R275W) PARKIN (V324A) |
DA neurons | Increased neuronal death; degenerated dendrites; impaired AKT signaling. | Miller et al. 201345 |
| fPD | LRRK2 (G2019S) | DA neurons | Reduced neurite outgrowth; dysregulated autophagy system; increased cell death in response to neurotoxins; elevated α-synuclein protein level; dysregulation of genes related to dopaminergic neurodegeneration. | Reinhardt et al. 201365 |
| fPD | SCNA (A53T) | DA neurons | Elevated α-synuclein aggregation and Lewy body-like deposition; induced nitrosative/oxidative stress; increased vulnerability to mitochondrial toxin-induced cell death. | Ryan et al. 201376 |
| sPD | GBA1 (RecNcil/+) GBA1 (L444P/+) GBA1 (N370S/+) |
DA neurons | Elevated levels of α-synuclein and glucosylceramide; defective autophagic/lysosomal machinery; increased basal and induced calcium levels; enhanced vulnerability to ER stress. | Schondorf et al. 201478 |
| sPD | GBA (N370S/+) | DA neurons | Elevated levels of α-synuclein; reduced level of dopamine; induced expression of MAO-B; disrupted network activity. | Woodard et al. 201479 |
| fPD | SCNA (A53T) | Neurons | Decreased α-synuclein tetramers; increased neurotoxicity. | Dettmer et al. 201580 |
| fPD and sPD | LRRK2 (G2019S) | DA neurons | Hypermethylation in gene regulatory regions; reduced expression of transcription factors related to PD. | Fernandez-Santiago et al. 201581 |
| fPD | PARKIN (exon 3, 5 deletion) PARKIN (exon 3 deletion) |
TH+ or TH− neurons | Reduced neurite complexity; diminished microtubule stability; | Ren et al. 201582 |
| fPD | PARKIN (R42P; exon 3 deletion) PARKIN (exon 3, 4 deletion; 255A deletion) PARKIN (R275W) PARKIN (R42P) |
DA neurons | Reduced capacity to differentiate into DA neurons; altered mitochondrial volume fraction. | Shaltouki et al. 201583 |
| sPD | GBA1 mutation | DA neurons | Reduced dopamine storage and uptake; elevated levels of α-synuclein. | Aflaki et al. 201684 |
| sPD | SNP | Neurons | A PD-associated risk variant that regulates SCNA expression. | Soldner et al. 201655 |
DA neurons: dopaminergic neurons; SCNA: the gene encoding α-synuclein; PINK1: PTEN-induced putative kinase 1; LRRK2: leucine-rich repeat kinase 2; ERAD: endoplasmic reticulum-associated degradation; TH: tyrosine hydroxylase; GBA1: acid β-glucocerebrosidase; MAO-B: monoamine oxidase B.
The interactions between different cell types can be better modeled using 3D organoids. Organoids have been generated for multiple organs, including the brain, retina, intestine, kidney, liver, lung, and stomach, using both tissue stem cells and pluripotent stem cells from mice and humans97. Human iPSC-derived organoids have been developed for a variety of applications due to their resemblance to endogenous cell organization and organ structure, and are particularly useful because they allow the possibility to study cell-cell interactions in a cellular context that mimics human physiology and development. The 3D organoids have been used in modeling human organ development and diseases, testing therapeutic compounds, and cell transplantation98–114 (Table 4). Multiple cell types that are physiologically relevant can be generated in organoids following a spatial-temporal order. Moreover, cells generated in organoids can be functionally more mature than cells derived using directed differentiation protocols, due to the interaction of different cell types, such as neurons and astrocytes, in the 3D structure. Therefore, 3D organoids allow dissection of disease pathology in a developmentally relevant spatial-temporal context and have the potential to offer a drug response at the level of an organ, rather than at the level of individual cells.
Table 4.
human iPSC-derived organoids for modeling development and disease
| Starting cells | Organoids | Applications | References |
|---|---|---|---|
| Human ESCs and iPSCs | Cerebral organoids | Modeling human cortical development and microcephaly | Lancaster et al. 2013117 |
| Human ESCs and iPSCs | Cerebral organoids | Modeling human fetal neocortex development | Camp et al. 2015107 |
| Human iPSCs | Brain organoids | Modeling autism spectrum disorders | Mariani et al. 2015108 |
| Human ESCs and iPSCs | Brain organoids | Modeling Zika virus infection | Cugola et al. 2016109 |
| Human iPSCs | Brain-region-specific organoids | Modeling Zika virus infection. Future applications include modeling human brain development and diseases and drug testing. | Qian et al. 2016110 |
| Human iPSCs | Brain organoids | Modeling Zika virus infection | Garcez et al. 2016111 |
| Human iPSCs | Brain organoids | Modeling Seckel syndrome | Gabriel et al. 2016112 |
| Human ESCs and iPSCs | Cortical organoids | Modeling species difference | Otani et al. 2016113 |
| Human iPSCs | Cortical spheroids | Modeling cortical development and diseases | Pasca et al. 2015114 |
| Human iPSCs | Retinal organoids | Modeling glaucoma | Tucker et al. 2014106 |
| Human ESCs and iPSCs | Intestinal organoids | Studying intestinal development. Future applications include intestinal disease modeling. | Spence et al. 2011103 |
| Human ESCs and iPSCs | Small intestinal organoids | Transplantation to demonstrate responsiveness to systemic signals. Future applications include studying intestinal development and diseases. | Watson et al. 2014105 |
| Human iPSC | Liver bud | Organ-bud transplantation for regenerative medicine | Takebet al. 201399 |
| Human iPSCs | Cystic organoids | Modeling Alagille syndrome, polycystic liver disease and cystic fibrosis-associated cholangiopathy and drug validation | Sampaziotis et al. 2015100 |
| Human iPSCs | Cystic organoids | Studying biliary development and cystic fibrosis | Ogawa et al. 2015101 |
| Human iPSC | Lung organoids | Studying lung development. Future applications include lung disease modeling. | Dye et al. 2015102 |
| Human iPSCs | Kidney organoids | Future applications include kidney disease modeling, nephrotoxicity screening and cell therapy. | Takasato et al. 2015104 |
| Human ESCs and iPSCs | Gastric organoids | Modeling gastric development and H. pylori infection | McCracken et al. 201698 |
While 3D organoids provide highly promising tools for iPSC-based disease modeling, the organoid technology has limitations. One challenge is to create an organoid platform with increased efficiency and reproducibility as compared to traditional two dimensional cultures115. The recent application of miniaturized spinning bioreactors with 3D design has allowed the generation of forebrain organoids with high reproducibility110. The development of more standardized organoid culture medium, together with a more defined extracellular matrix, would further facilitate the generation of a highly reproducible organoid system that is more applicable for accurate disease modeling, drug discovery, and therapeutic development116. Another challenge is the lack of vascularization in the current organoid system97. Accordingly, organoids exhibit limited growth and maturation due to the lack of continuous nutrient supply. Spinning bioreactors and shaking culture platforms have been shown to provide better nutrient supply and improve the growth of organoids110,117. Co-culture with endothelial cells has allowed generation of vascular-like network in organoids99. Moreover, transplantation of in vitro generated human organoids into relevant sites of animal hosts facilitates vascularization and maturation of organoids. This transplantation approach may be applied when organoids with increased size and improved maturation are needed for the study.
iPSC-based drug discovery
Screening for efficacy.
Many drug screens are based on targets that are considered to be relevant to the disease mechanisms. However, the low success rates of compounds originating from target-based screening have led to greater interest in phenotypic screening118. This revival in phenotypic screening has been aided by the discovery of iPSCs for numerous reasons, including the scalability of iPSC production, which facilitates assay development, and their pluripotency, which allows differentiation into multiple disease-relevant cell types (especially those that are otherwise hard to access, such as neurons)119. Patient-derived iPSC models make it possible to recapitulate disease phenotypes and pathologies in a culture dish. Cells differentiated from patient-derived iPSCs could present molecular and cellular phenotypes. Whether the phenotype that is selected as readout for drug screen is truly relevant to the disease can be confirmed by gene editing approach if the gene responsible for disease phenotypes is known, and can be further validated in patient samples and/or animal models120. In addition to phenotypic screening, iPSCs can also be used for target-based screening. Using human iPSC models, many drug screens have been conducted and potential drug candidates have been identified using either phenotypic or target-based screening.
To obtain target cells with high purity on a large scale, purification and enrichment technologies using specific cell surface markers121,122, cell-specific promoters123 and microRNAs124 have been established. In the first report of large-scale drug screening using an iPSC-based disease model, neural crest precursors for autonomic neurons were sorted and purified from iPSCs derived from patients with familial dysautonomia, a monogenic early-onset disease that is characterized by degeneration of neurons in the sensory and autonomic nervous systems121. It is caused by mutations in the gene coding for the IkB kinase complex-associated protein (IKBKAP) that result in a splicing defect and production of a dysfunctional truncated protein. The screening was conducted using 6,912 compounds, and a compound known as SKF-86466 was found to improve disease-specific aberrant splicing. Interestingly, SKF-86466 was not effective in non-target cells, including iPSCs, fibroblasts, and lymphocytes. These results illustrated the advantage of iPSC-based drug screening to explore cell type-specific pathogenesis.
Burkhardt et al. performed disease modeling and drug screening using sporadic ALS patient-derived iPSCs125. The authors identified de novo aggregation of TAR DNA-binding protein 43 (TDP-43) in motor neurons of sporadic ALS patients, using TDP-43 aggregation as readout for a high-content drug screen to identify compounds that reduce TDP-43 aggregation125. The same research team also made effective use of patient-derived iPSC model of Alzheimer’s disease126. The authors identified a disease-relevant protein, extracellular tau (eTau), in the conditioned medium of cortical neurons derived from the iPSCs of an Alzheimer’s disease patient, generating a therapeutic antibody against eTau126. This disease-relevant protein would not have been discovered without using the human iPSC model. eTau causes neuronal hyperactivity and increases amyloid beta (Aβ) production. Using human iPSC models as a tool to identify disease-relevant targets could be a critical component for future drug development. Naryshkin et al.127 found that an SMA patient-derived iPSC model could be used to validate human- and disease-specific drug responsiveness after initial screening using a HEK293 cell line127. These compounds were then validated in patient-specific fibroblasts, and in motor neurons differentiated from patient-derived iPSCs that serve as a patient-specific and disease-relevant cellular model127. Finally, the hit compound was evaluated in a mouse model for in vivo activity127. This drug discovery approach includes a patient-derived iPSC model as one of the validation steps by taking advantage of the patient-specific and disease-relevant properties of motor neurons derived from patient iPSCs.
Overall, iPSC-based drug screening has been used to evaluate more than 1,000 compounds for several diseases (Table 5)121,125,128,129, and several clinical candidates have been identified (Table 6)126,127,130. However, these studies require considerable time (several weeks or more) to differentiate iPSCs into disease-relevant cell types. Although this may not seem long for phenotypic screening, a shorter differentiation period is preferable to avoid variation in cell quality. Therefore, faster and more stable differentiation methods that result in higher maturity and purity are being sought. An alternative approach is to perform drug screen using cells derived from direct conversion131,132. Direct conversion forces the target somatic cells (e.g. fibroblasts) to express cell-specific transcription factors and reprogram one somatic cell state to another somatic cell state without passing through the iPSC state49,132. Direct conversion has been used to reprogram myocardial cells, liver cells, neural cells, or other type of somatic cells from a different type of somatic cells, such as fibroblasts. As an advantage of direct conversion, authentic human neurons that reflect important aspects of cellular aging can be generated50. However, the non-renewable source of cells provided by this approach may not be applicable for large-scale drug screening. The forced expression of transcription factors also offers the potential to differentiate patient iPSCs much more rapidly. In a recent study, forced expression of MYOD1 (myogenic differentiation gene), a master regulator of skeletal muscle differentiation, was used to produce new cellular models of intractable muscle disease pathologies such as Miyoshi myopathy133 and Duchenne-type muscular dystrophy134.
Table 5.
Large scale drug screening using human iPSCs
| Disease | Cell type derived from iPSCs | Readout | # compounds screened | Hit rate (%) | Reference |
|---|---|---|---|---|---|
| Familial dysautonomia | Patient neural crest cells | Expression of IKBKAP | 6,912 | 0.6 | Lee et al. 2012121 |
| Alzheimer’s disease | Human cortical neurons | Cell death | Approximately 350 | ~5.4 | Xu et al. 2013128 |
| Motor neuron disease | Human neural precursor cells for the hit validation | Neurite length | 11,819 | 0.3 | Hoing et al 2012129 |
| ALS | Patient motor neurons | TDP-43 aggregates | 1,757 | 2.2 | Burkhardt et al. 2013125 |
| Fragile X syndrome | Patient NPCs | Expression of FMRP | 50,000 | 4.2 | Kaufmann et al. 2015221 |
| Fragile X syndrome | Patient NSCs | Expression of FMR1 | Approximately 5,000 | 0.12 | Kumari et al. 2015222 |
| Fragile X syndrome | Human NPCs | Expression of FMR1 | 1,134 | 0.17 | Li et al. 2016223 |
Table 6.
Drug from iPSC research in clinical trials
| Candidate drug | Disease | Mechanism | Formulation | Company | Reference |
|---|---|---|---|---|---|
| BMS-986168 (IPN-007) | Progressive supranuclear palsy (PSP) | Neutralizing eTau | Antibody | Bristol-Myers-Squibb | Bright et al. 2015126 |
| Ezogabine | ALS | Kv7.2/3 potassium channel agonist | Small molecule compound | GlaxoSmithKline | McNeish et al. 2015137 |
| RG7800 | SMA | Increasing SMN protein levels | Small molecule compound | Roche | Naryshkin et al. 2014127 |
An important point in pathology research using iPSCs is the nature of the control group. For genetic disorders, a control group can be created by conducting gene correction of the mutant allele in patient iPSCs. Comparisons between various groups of iPSCs (healthy, patient, and gene-corrected patient iPSCs) can be conducted to validate the results of drug screening119. iPSCs also make invaluable models in the case of sporadic diseases. In these cases, because no causal mutation is known, the nature of a control group is difficult to establish, but disease-relevant single-nucleotide polymorphisms (SNPs) can be considered instead65. As described in the “Perspective” section below, for future drug screening, sporadic disease iPSCs should allow for investigation of whether the disease is caused by genetic factors such as SNPs, somatic mutation/mosaicism, or epigenetic factors. These developments could further open the door to personalized drug screening using iPSCs135.
Another application is drug repositioning using disease-specific iPSCs. In drug repositioning, existing drugs already approved for specific diseases are tested to find new applications for other diseases. For example, a human iPSC model derived from achondroplasia patients with fibroblast growth factor receptor 3 (FGFR3) mutations showed that patient iPSCs did not differentiate well into cartilage tissue136. Using this model, a screen for molecules that rescue chondrogenically differentiated iPSCs from the defective cartilage phenotype identified several statins, which are approved drugs for cardiovascular disease. The same study found that statins could promote the growth of shortened limbs in a mouse model of FGFR3-linked disease. These results indicate that statins may be repositioned as candidate drugs for achondroplasia136. As another example of drug repositioning, the anti-epileptic drug ezogabine was found to be effective in an iPSC model of the motor neuron disease amyotrophic lateral sclerosis (ALS) and is now undergoing clinical trial137. In this study, the authors showed the effect of ezogabine on an iPSC model derived from not only ALS patients with mutations in the superoxide dismutase 1 (SOD1) gene, but also ALS patients with mutations in other genes linked to ALS, such as C9orf72 and FUS. It has also been demonstrated that ALS patient-derived iPSC motor neurons initially exhibit a hyper-excitable state, followed by a decrease in excitability138, suggesting that early intervention with ezogabine treatment may be required for the treatment of ALS patients. The observation of similar drug response in different patient groups allowed generalizing the drug responsiveness across ALS patient types. Drug discovery using patient iPSCs derived from multiple genetic forms is of great value, because it allows testing the drug responsiveness in a broad patient population. In contrast, it is hard to analyze the effect of a drug on multiple mouse models simultaneously.
Screening for toxicity.
The development of new drugs is enormously costly, mostly because of failures, particularly those in late-stage clinical trials, which are in part due to unanticipated side effects139,140. Many unpredicted adverse effects of new candidate drugs can occur, with cardiac and liver toxicity being of special concern. Consequently, there is considerable interest in approaches that could more effectively predict the likelihood of candidate drugs to cause serious side effects, thereby enabling the selection of candidates that are less likely to fail due to toxicity in late-stage trials.
Lethal arrhythmias with a QT prolongation account for 21% of total cardiac toxicities141. QT prolongation is an adverse effect related to human Ether-a-go-go Related Gene (hERG) channels. Cardiac safety testing has been mainly dependent on the hERG assay, because blocking the hERG current is considered to be associated with the deadly ventricular arrhythmia named torsades de pointes (or TdP). It has been discovered that 40–60% of drugs that inhibit hERG channel current do not cause QT prolongation142,143. These false positive results from the hERG assay have hindered the development of promising drugs. Preclinical strategies have been proposed to detect drug-induced electrophysiological cardiotoxicity using in vitro human ion channel assays, human-based in silico reconstructions, and human stem cell-derived cardiomyocytes144. Recent efforts have shown that multi-electrode arrays (MEA) assays using human iPSC-derived cardiomyocytes may offer a reliable, cost-effective surrogate for preclinical in vitro testing145 that could be used to assess pro-arrhythmic risk146.
For hepatotoxicity, hepatocyte cell lines or human primary hepatocytes are widely used. However, there are limitations to these models too, including cell resources, loss of function due to freezing-thawing, and lot-to-lot variation. Recently, human ESC/iPSC-derived hepatic cells were generated that express functional molecules such as CYP3A4 and uptake Indocyanine Green147 responding to known hepatotoxic drugs148. Functional 3D liver organ buds have also been reported, which may result in better drug screening99.
Finally, regarding the nervous system, a platform that assesses adverse drug effects using pluripotent stem cells is now being developed. To conduct such an assessment, the analysis of alterations in the gene expression of cells in the nervous system, such as neuronal cells, mesenchymal stem cells, and vascular endothelial cells derived from human ESCs in a culture dish has been proposed149.
Clinical applications using human iPSC products
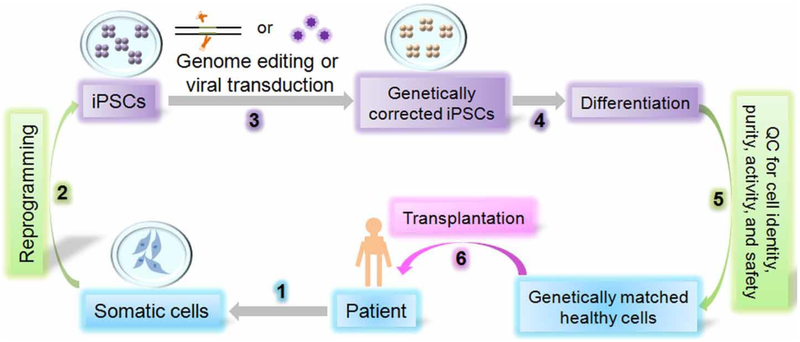
The potential of regenerative medicine based on the use of stem cells to promote endogenous regenerative processes or replace damaged tissues after cellular transplantation has attracted considerable interest. Since the discovery of human ESCs in 1998150 and of human iPSCs in 20072,3, the stem cell research community has continued to identify more suitable sources for exploring cell therapy and endogenous repair in humans. A general approach to develop iPSC-based cell therapy products is summarized in Fig. 2. Of the 13 clinical trials with stem cell therapy products being conducted currently, 8 are for ESC- and 1 for iPSC-derived retinal pigment epithelial (RPE) cells to treat macular degeneration, which causes the progressive deterioration of light-sensing photoreceptors in the eye (https://clinicaltrials.gov)151. In 2014, the first clinical study using human iPSC products was initiated by transplanting RPE sheets derived from the patient’s own iPSCs. The therapy has resulted in positive results, stopping macular degeneration and improving the vision of the patient. Although the trial was subsequently put on hold due to mutations observed in a second patient’s iPSCs4, it is expected to resume6. In addition, a recent study has demonstrated the feasibility of transplanting human ESC-derived cardiac progenitor cells embedded in a fibrin scaffold to patients with severe heart failure152.
Figure 2.
A schematic for human iPSCs-based cell therapy. Human iPSC-based cell therapy development usually includes the following steps: 1) Collect somatic cells from patients and culture somatic cells from affected patients; 2) Reprogram patient somatic cells into iPSCs; 3) Use genome editing technology or viral transduction method to repair patient iPSCs and turn them into genetically corrected iPSCs; 4) Differentiate the corrected iPSCs into desired cell types to serve as genetically matched healthy donor cells; 5) Perform quality control test for cell identity, purity, activity, and safety; and 6) Transplant the genetically matched healthy cells into patients for cell therapy.
However, there are several obstacles associated with iPSC-based therapy that will need to be addressed before routine clinical applications can begin153. One concern is the risk of tumorigenicity from ESCs and iPSCs154. Because pluripotent cells are maintained in culture for prolonged periods of time, they can accumulate karyotypic abnormalities, copy number variants, and loss of heterozygosity155. Hence prior to clinical use, iPSC-derived products need to be carefully screened for the lack of potentially risky genetic alterations155 and rigorously tested to ensure their purity, quality, and sterility. Increased knowledge on the basic biology of pluripotency induction and maintenance will also help us to reduce the risk of mutation development and genetic instability associated with human iPSC derivation and maintenance.
Although the products differentiated from iPSCs have not been shown to generate teratomas, it is critical to ensure the final product does not contain undifferentiated cells that have the potential to generate teratomas. Accordingly, improved protocols for differentiating human iPSCs into desired cell types with precise identity and cellular functions are needed. To this end, small molecule inhibitors that have been shown to induce selective and complete cell death of undifferentiated human pluripotent stem cells without affecting their differentiated derivatives have been identified156,157. Treatment of the iPSC-derived cellular product with these inhibitors may reduce the potential tumorigenicity. Another potential solution is to sort the iPSC-derived cells before transplantation through positive selection for desired cell types and negative selection against human ESC markers using fluorescence-activated cell sorting (FACS). Lastly, the risk of tumorigenecity can be tested in animal models prior to transplant. However, this approach may not be applicable to patients with rapid disease progression due to the long period of time associated with animal tests.
Compliance with good manufacturing practice (GMP) is mandatory before human transplantation of cell therapies. Once cells are safely delivered, ideally patients should be monitored for the development of potential tumors and activation of the immune system158. One approach for tumor monitoring may be to assess the enhanced angiogenesis that often accompanies teratoma formation, which can be detected using 64Cu-labeled cyclic arginine-glycine-aspartic acid tetramer (64Cu-DOTA-RGD4) radiotracer with positron emission tomography (PET) imaging159. Another approach may be to use a combination of serum biomarkers (e.g., carcinoembryonic antigen, α-fetoprotein, or human chorionic gonadotropin) and magnetic resonance imaging (MRI) screening as described recently160. However, it is worth noting that these approaches would be mostly useful at the preclinical stage, especially if they are already part of the imaging procedure required for evaluating an endpoint. Their feasibility and necessity for future human trials remain to be determined.
The lack of an effective method of inducing immune tolerance is a major roadblock for human ESC-based therapies. ESCs were once considered immune-privileged due to the low expression of major histocompatibility complex (MHC) class I, MHC II, and costimulatory molecules161. Although undifferentiated ESCs might be immune-privileged, their differentiated derivatives can trigger cellular and humoral immune responses162. By contrast, autologous iPSCs may avoid the high cost and serious side effects associated with lifelong immunosuppression required for allogeneic cell transplantation163. Despite some controversy over the immunogenicity of undifferentiated iPSCs164, recent studies demonstrate that differentiation of iPSCs could result in loss of immunogenicity165–167.
The application of cells derived from individual patients’ own iPSCs or iPSCs from matched donors may become a cornerstone of precision medicine, and has the important advantage that there should be no need for long-term immune suppression to preserve the transplante cells. Indeed, the first iPSC clinical trial used RPEs from autologous iPSCs derived from the patient. Using autologous iPSC products for personalized cell therapy seems ideal for orphan diseases, as massive cell banking is not required. However, for more common diseases, especially acute common diseases such as cerebrovascular accident (CVA) or myocardial infarction (MI), autologous iPSC therapy may not be practical for large number of patients given the high cost and lengthy period of time needed for careful validation of each cell line. For these reasons, the second phase of the iPSC-RPE trial in Japan will be employing allogeneic products168.
The allogeneic iPSC approach could also bring down the cost for iPSC-based cell therapy. Excluding high startup cost, each iPSC line costs ~$10,000–20,000 to produce169. Meeting cGMP requirements increases this cost substantially170. Costs are even higher, by approximately $800,000169, to generate an iPSC-derived tissue product suitable for clinical use (e.g., differentiation of iPSC-neuronal cells for CVA, iPSC-cardiomyocytes for MI, or iPSC-RPE cells for macular degeneration). Banking iPSCs for allogeneic transplant has the potential to reduce cost because one production may be used for multiple patients. To facilitate allogeneic transplant, the effectiveness of conventional immunosuppressive protocols and newer regimen of co-stimulatory blockers for inducing immunotolerance will need to be improved in preclinical and clinical settings171,172. Moreover, understanding how pluripotent stem cells interact with the immune system and why they may be more tolerance-inducing than other transplanted cells may lead to the identification of new immunosuppressive mechanisms and strategies163. Furthermore, transplantation to immune-privileged sites may serve as a possible strategy to overcome immune rejection. Incorporating recent advances in genome editing strategies to create universally accepted donor cells could be another alternative approach173.
The combination of the human iPSC platform with the recently developed gene editing and 3D organoid technologies could make human iPSCs an even more powerful cellular resource for stem cell-based cell therapy development. As a proof-of-principle, mouse iPSCs corrected through gene editing have been used to generate hematopoietic progenitors for successful treatment of sickle cell anemia in a mouse model174. Furthermore, the integration of genetically corrected human iPSCs with 3D organoids could allow tissues to be generated as sources for organ replacement therapies97. Indeed, human iPSC-derived liver organoids have been shown to successfully generate functional human liver-like tissues in transplanted mice in a proof-of-principle study99. However, there are still challenges to overcome for such approaches to become applicable in human cell therapy. For example, the potential off-target effects associated with gene editing need to be addressed, as do the limitations of organoids, as described in the section of “iPSC-based disease modeling”.
Perspectives
The discovery of iPSCs has provided a revolutionary new research platform for the study of diseases. In the ten years since the first iPSC report, great progress has been made in investigating disease mechanisms and potential treatments by combining human iPSCs with other new technologies, but several important issues remain to be addressed.
iPSC clones show variations in differentiation efficiency, including clones derived from the same person119. These variations are important to consider when selecting control groups for disease modeling studies. Applying CRISPR/Cas9 technology may help address this issue, as discussed above. Multiple reports have now shown that gene correction of an iPSC mutation improves the disease phenotype of differentiated cells175–178. In addition to correcting gene mutations in disease iPSCs, researchers have also successfully introduced gene mutations into healthy iPSCs87,88. Although several challenges for the combination of CRISPR/Cas9 technology with iPSC technology remain, including the off-target effects of CRISPR/Cas9 editing, the high cost of assaying for them, and the limited application of gene editing to genetic diseases with unknown disease-causing mutations or risk variants14, the potential of this combiniation to dissect disease mechanisms and to develop novel cell therapies is high. Moreover, CRISPR/Cas9 or CRISPRi-based genome-wide genetic screening179,180 in human iPSCs could open a new avenue for understanding basic biological mechanisms underlying human iPSC pluripotency, maintenance and differentiation.
Unlike mouse ESCs and iPSCs that represent the ‘naïve’ state and are homogeneous, human ESCs and iPSCs represent the ‘primed’ state and are heterogeneous in both cell population and differentiation potential181,182. Moreover, the reprogramming process can result in not only fully reprogrammed iPSCs but also partially reprogrammed cells, which may have different differentiation potential183,184. Therefore, human iPSCs need to be carefully selected and thoroughly characterized for their pluripotency before clinical applications185. Further basic research on reprogramming mechanisms may help researchers to develop methods that allow generation of a standardized human iPSC state, which would lead to reduced technical variability and enable the identification of true biological phenotypes. Besides clonal variation in iPSC differentiation efficiency, another obstacle for disease modeling is line-to-line variation in the maturation of differentiated cells. The acquisition of mature cells requires improved culture conditions and the use of fate conversion with gene regulation119. A recent technology, the microRNA switch124, is expected to increase the maturation quality of iPSC-differentiated cells and reduce clonal variation.
In conventional disease-modeling studies, cells are seeded in a two-dimensional plane. However, in vitro models with a 3D structure are closer to the physiological condition and thus may be better suited for the study of the disease pathology. Using disease-specific iPSCs, technologies for inducing the differentiation of several 3D structures, including those similar to the cortex, optic cup, Rathke’s pouch, cerebellum, and hippocampus, have been reported186–191. By taking advantage of the dynamic patterning and structural self-formation of complex organ buds, the construction of 3D structures and their corresponding networks has become a reality. Such a strategy is already being used in disease modeling for brain abnormalities117 and mental illness108. Similar to the self-organization of ectodermal tissue structures, endodermal tissue formation in 3D stem cell culture99 has been developed and applied to gastrointestinal disease modeling192. Physiological interactions among complex tissues from different lineages but with the same genetic background, such as the blood-brain barrier193 or the immune system194,195 within the organoid, could add new insights into normal physiology and diseases196,197.
Although several limitations exist in current 3D technology as detailed in the section of “human iPSC-based disease modeling”198,199, combining disease-specific iPSCs with 3D technology allows the examination of spatiotemporal cellular interactions that could reveal the physiological disease status, thus providing an unprecedented drug-screening platform and offering a new option for tissue-replacement therapy. However, transplantation of human stem cell-derived organoids into animals to derive human tissues or organs may bring new issues in biomedical ethics that warrant further attention200, such as the potential of transplanted human stem cells to mix with host cells and development in the nervous system and germlines of host animals.
iPSCs also provide a new way to study sporadic diseases. Prior to the development of iPSC technology, it was impossible to analyze sporadic diseases in cellular models, but now several studies have successfully modeled sporadic neurological diseases54,57,65,73,85,108,125,135,201,202. It has been hypothesized that the pathological mechanisms of sporadic diseases might be the same as familial ones. However, sporadic and familial diseases have significant differences, such as the age of onset and severity, as well as the pathology.
iPSC models suggest that even if the effect of each individual genetic risk is small, the combined effect may initiate and accelerate the development of the pathology of sporadic diseases. In addition, even if SNP genotyping only indicates a small risk factor, it could be an important one, and modeling the pathological phenotype with iPSC technology could lead to a re-classification of sporadic diseases. Such re-classifications could have important implications for drug development. iPSC modeling has the potential to identify drug-responsive patient subgroups, including those with sporadic diseases, which should improve the quality of clinical trials119. A large cohort analysis with medical records and genome information combined with patient iPSCs is also expected and so iPSC-derived cells could provide a far more precise analysis of the individual genes and proteins involved in the disease.
Accumulating information from disease iPSC research, in combination with patients’ personalized clinical experience, will aid “disease repositioning”, in which diseases are defined not by clinical but by cellular phenotypes. If analysis of the cellular phenotypes of in vitro iPSC models of clinically different diseases indicates that the phenotype is the same or similar, then a treatment that is effective in one condition may be effective in the others. For example, an iPSC model of bipolar disorder identified hyperexcitable neuronal cells201. Similar hyperexcitability was found in iPSC-derived motor neurons from a patient with amyotrophic lateral sclerosis203. Therefore, the same therapeutic agent may be effective for these clinically disparate but cellularly similar diseases. Accumulating data of cellular phenotypes of iPSC models from a cross-sectional variety of diseases may contribute to new stratifications and understanding of different diseases, which could also lead to new cross-sectional treatment approaches.
The development of iPSC technology has generated a powerful new way to both define and treat diseases. iPSCs represent a paradigm shift because they now allow us to directly observe and treat relevant patient cells. In particular, they have revealed new relationships in gene expression, which have broadened and deepened our understanding of disease development in patients with sporadic disease. Progress with other technologies, such as CRISPR/Cas9, 3D organoids, and microRNA switches, will further advance the already rapid pace of iPSC-based disease modeling and therapeutic development.
Acknowledgements
We thank Jianfei Chao, Li Li, Qiuhao Qu and E Tian for their help in preparing the figures, Peter Karagiannis and Chris Gandhi for critical reading and editing of the manuscript, Rosa Carrasco, Lupe Zaragoza, Ann Margaret Chrisney, Mansze Kong, Yoko Miyake, Noriko Endo, Rumi Ueno, and Rie Okuyama for their administrative support. This work was funded in part by the Herbert Horvitz Fellowship (Y.S.); Sidell Kagan Foundation (Y.S.); California Institute for Regenerative Medicine RB4–06277 (Y.S.), TRAN1–08525 (Y.S.), RT3–07798 (J.C.W.), and DR2A-05394 (J.C.W.); National Institutes of Health R01 HL130020 (J.C.W.) and R01 HL128170 (J.C.W.); the iPS Cell Research Fund (S.Y.); Center for iPSC production, the Program for Intractable Diseases Research utilizing Disease-specific iPS cells, Research Center Network for Realization of Regenerative Medicine from the Japan Agency for Medical Research and Development (AMED) (S.Y.); the grant for Core Center for iPS cell Research of Research Center Network for Realization of Regenerative Medicine from AMED (S.Y., H.I.); the Program for Intractable Diseases Research utilizing disease-specific iPS cells from AMED (H.I.); Research Project for Practical Applications of Regenerative Medicine from AMED (H.I); the Mochida Memorial Foundation for Medical and Pharmaceutical Research (H.I.); the Daiichi Sankyo Foundation of Life Science (H.I.).
Footnotes
Conflict of Interest
J.C.W. is a co-founder of Stem Cell Theranostics. S.Y. is a scientific advisor of iPS Academia Japan without salary. The other authors declare no conflict of interest.
References
- 1.Takahashi K and Yamanaka S Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, and Yamanaka S Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, and Thomson JA Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Kimbrel EA and Lanza R Current status of pluripotent stem cells: moving the first therapies to the clinic. Nat Rev Drug Discov 14, 681–692 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Scudellari M How iPS cells changed the world. Nature 534, 310–312 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Trounson A and DeWitt ND Pluripotent stem cells progressing to the clinic. Nat Rev Mol Cell Biol 17, 194–200 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Onos KD, Sukoff Rizzo SJ, Howell GR, and Sasner M Toward more predictive genetic mouse models of Alzheimer’s disease. Brain Res Bull 122, 1–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puzzo D, Gulisano W, Palmeri A, and Arancio O Rodent models for Alzheimer’s disease drug discovery. Expert Opin Drug Discov 10, 703–711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-David U, Kopper O, and Benvenisty N Expanding the boundaries of embryonic stem cells. Cell Stem Cell 10, 666–677 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, Huo H, Loh YH, Aryee MJ, Lensch MW, Li H, Collins JJ, Feinberg AP, and Daley GQ Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol 29, 1117–1119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, Yu J, Hebrok M, Hochedlinger K, Costello JF, Song JS, and Ramalho-Santos M Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol 13, 541–549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Nur O, Russ HA, Efrat S, and Benvenisty N Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell 9, 17–23 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Avior Y, Sagi I, and Benvenisty N Pluripotent stem cells in disease modelling and drug discovery. Nat Rev Mol Cell Biol 17, 170–182 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Hockemeyer D and Jaenisch R Induced Pluripotent Stem Cells Meet Genome Editing. Cell Stem Cell 18, 573–586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, Meng X, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, and Jaenisch R Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27, 851–857 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, Porteus MH, Joung JK, and Cheng L Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell 5, 97–110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, and Voytas DF Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, and Jaenisch R Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29, 731–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, and Zhang F A transcription activator-like effector toolbox for genome engineering. Nat Protoc 7, 171–192 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, and Zhang F Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, and Gersbach CA RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods 10, 973–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, and Zhang F Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, and Sander JD High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31, 822–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, and Ye Z Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 15, 12–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Cowan CA, Talkowski ME, and Musunuru K Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15, 27–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C, Ding L, Sun CW, Wu LC, Zhou D, Pawlik KM, Khodadadi-Jamayran A, Westin E, Goldman FD, and Townes TM Novel HDAd/EBV Reprogramming Vector and Highly Efficient Ad/CRISPR-Cas Sickle Cell Disease Gene Correction. Sci Rep 6, 30422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang CW, Lai YS, Westin E, Khodadadi-Jamayran A, Pawlik KM, Lamb LS Jr., Goldman FD, and Townes TM Modeling Human Severe Combined Immunodeficiency and Correction by CRISPR/Cas9-Enhanced Gene Targeting. Cell Rep 12, 1668–1677 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Park CY, Kim DH, Son JS, Sung JJ, Lee J, Bae S, Kim JH, Kim DW, and Kim JS Functional Correction of Large Factor VIII Gene Chromosomal Inversions in Hemophilia A Patient-Derived iPSCs Using CRISPR-Cas9. Cell Stem Cell 17, 213–220 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Firth AL, Menon T, Parker GS, Qualls SJ, Lewis BM, Ke E, Dargitz CT, Wright R, Khanna A, Gage FH, and Verma IM Functional Gene Correction for Cystic Fibrosis in Lung Epithelial Cells Generated from Patient iPSCs. Cell Rep 12, 1385–1390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bae S, Park J, and Kim JS Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, and Church GM RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominguez AA, Lim WA, and Qi LS Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol 17, 5–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, and Zhang F Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, and Zhang F Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, and Huang B Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, and Tessier-Lavigne M Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Komor AC, Kim YB, Packer MS, Zuris JA, and Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ebert AD, Yu J, Rose FF Jr., Mattis VB, Lorson CL, Thomson JA, and Svendsen CN Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 457, 277–280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, Menon J, Shimizu F, Viale A, Tabar V, Sadelain M, and Studer L Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 461, 402–406 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Studer L, Vera E, and Cornacchia D Programming and Reprogramming Cellular Age in the Era of Induced Pluripotency. Cell Stem Cell 16, 591–600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munsat TL and Davies KE International SMA consortium meeting. (26–28 June 1992, Bonn, Germany). Neuromuscul Disord 2, 423–428 (1992). [DOI] [PubMed] [Google Scholar]
- 43.Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, Gujar P, Kee K, Schule B, Dolmetsch RE, Langston W, Palmer TD, and Pera RR LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell 8, 267–280 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, Graziotto J, Sundberg M, McLean JR, Carrillo-Reid L, Xie Z, Osborn T, Hargus G, Deleidi M, Lawson T, Bogetofte H, Perez-Torres E, Clark L, Moskowitz C, Mazzulli J, Chen L, Volpicelli-Daley L, Romero N, Jiang H, Uitti RJ, Huang Z, Opala G, Scarffe LA, Dawson VL, Klein C, Feng J, Ross OA, Trojanowski JQ, Lee VM, Marder K, Surmeier DJ, Wszolek ZK, Przedborski S, Krainc D, Dawson TM, and Isacson O Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci Transl Med 4, 141ra190 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, Mandal PK, Vera E, Shim JW, Kriks S, Taldone T, Fusaki N, Tomishima MJ, Krainc D, Milner TA, Rossi DJ, and Studer L Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 13, 691–705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu GH, Qu J, Suzuki K, Nivet E, Li M, Montserrat N, Yi F, Xu X, Ruiz S, Zhang W, Wagner U, Kim A, Ren B, Li Y, Goebl A, Kim J, Soligalla RD, Dubova I, Thompson J, Yates J 3rd, Esteban CR, Sancho-Martinez I, and Izpisua Belmonte JC Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature 491, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearson BL, Simon JM, McCoy ES, Salazar G, Fragola G, and Zylka MJ Identification of chemicals that mimic transcriptional changes associated with autism, brain aging and neurodegeneration. Nat Commun 7, 11173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ho R, Sances S, Gowing G, Amoroso MW, O’Rourke JG, Sahabian A, Wichterle H, Baloh RH, Sareen D, and Svendsen CN ALS disrupts spinal motor neuron maturation and aging pathways within gene co-expression networks. Nat Neurosci 19, 1256–1267 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mertens J, Marchetto MC, Bardy C, and Gage FH Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience. Nat Rev Neurosci 17, 424–437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mertens J, Paquola AC, Ku M, Hatch E, Bohnke L, Ladjevardi S, McGrath S, Campbell B, Lee H, Herdy JR, Goncalves JT, Toda T, Kim Y, Winkler J, Yao J, Hetzer MW, and Gage FH Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell 17, 705–718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bardy C, van den Hurk M, Eames T, Marchand C, Hernandez RV, Kellogg M, Gorris M, Galet B, Palomares V, Brown J, Bang AG, Mertens J, Bohnke L, Boyer L, Simon S, and Gage FH Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci U S A 112, E2725–2734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song H, Stevens CF, and Gage FH Astroglia induce neurogenesis from adult neural stem cells. Nature 417, 39–44 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Tian E, Sun G, Sun G, Chao J, Ye P, Warden C, Riggs AD, and Shi Y Small-Molecule-Based Lineage Reprogramming Creates Functional Astrocytes. Cell Rep 16, 781–792 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y, Imamura K, Egawa N, Yahata N, Okita K, Takahashi K, Asaka I, Aoi T, Watanabe A, Watanabe K, Kadoya C, Nakano R, Watanabe D, Maruyama K, Hori O, Hibino S, Choshi T, Nakahata T, Hioki H, Kaneko T, Naitoh M, Yoshikawa K, Yamawaki S, Suzuki S, Hata R, Ueno S, Seki T, Kobayashi K, Toda T, Murakami K, Irie K, Klein WL, Mori H, Asada T, Takahashi R, Iwata N, Yamanaka S, and Inoue H Modeling Alzheimer’s Disease with iPSCs Reveals Stress Phenotypes Associated with Intracellular Abeta and Differential Drug Responsiveness. Cell Stem Cell 12, 487–496 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, Goldmann J, Myers RH, Young RA, and Jaenisch R Parkinson-associated risk variant in distal enhancer of alpha-synuclein modulates target gene expression. Nature 533, 95–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, and Suzuki N Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Hum Mol Genet 20, 4530–4539 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS, Carson CT, Laurent LC, Marsala M, Gage FH, Remes AM, Koo EH, and Goldstein LS Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature 482, 216–220 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woodruff G, Young JE, Martinez FJ, Buen F, Gore A, Kinaga J, Li Z, Yuan SH, Zhang K, and Goldstein LS The presenilin-1 DeltaE9 mutation results in reduced gamma-secretase activity, but not total loss of PS1 function, in isogenic human stem cells. Cell Rep 5, 974–985 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mertens J, Stuber K, Wunderlich P, Ladewig J, Kesavan JC, Vandenberghe R, Vandenbulcke M, van Damme P, Walter J, Brustle O, and Koch P APP processing in human pluripotent stem cell-derived neurons is resistant to NSAID-based gamma-secretase modulation. Stem Cell Reports 1, 491–498 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JK, Jin HK, Park MH, Kim BR, Lee PH, Nakauchi H, Carter JE, He X, Schuchman EH, and Bae JS Acid sphingomyelinase modulates the autophagic process by controlling lysosomal biogenesis in Alzheimer’s disease. J Exp Med 211, 1551–1570 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duan L, Bhattacharyya BJ, Belmadani A, Pan L, Miller RJ, and Kessler JA Stem cell derived basal forebrain cholinergic neurons from Alzheimer’s disease patients are more susceptible to cell death. Mol Neurodegener 9, 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muratore CR, Rice HC, Srikanth P, Callahan DG, Shin T, Benjamin LN, Walsh DM, Selkoe DJ, and Young-Pearse TL The familial Alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum Mol Genet 23, 3523–3536 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahairaki V, Ryu J, Peters A, Chang Q, Li T, Park TS, Burridge PW, Talbot CC Jr., Asnaghi L, Martin LJ, Zambidis ET, and Koliatsos VE Induced pluripotent stem cells from familial Alzheimer’s disease patients differentiate into mature neurons with amyloidogenic properties. Stem Cells Dev 23, 2996–3010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Q, Waltz S, Woodruff G, Ouyang J, Israel MA, Herrera C, Sarsoza F, Tanzi RE, Koo EH, Ringman JM, Goldstein LS, Wagner SL, and Yuan SH Effect of potent gamma-secretase modulator in human neurons derived from multiple presenilin 1-induced pluripotent stem cell mutant carriers. JAMA Neurol 71, 1481–1489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Young JE, Boulanger-Weill J, Williams DA, Woodruff G, Buen F, Revilla AC, Herrera C, Israel MA, Yuan SH, Edland SD, and Goldstein LS Elucidating molecular phenotypes caused by the SORL1 Alzheimer’s disease genetic risk factor using human induced pluripotent stem cells. Cell Stem Cell 16, 373–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hossini AM, Megges M, Prigione A, Lichtner B, Toliat MR, Wruck W, Schroter F, Nuernberg P, Kroll H, Makrantonaki E, Zouboulis CC, and Adjaye J Induced pluripotent stem cell-derived neuronal cells from a sporadic Alzheimer’s disease donor as a model for investigating AD-associated gene regulatory networks. BMC Genomics 16, 84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moore S, Evans LD, Andersson T, Portelius E, Smith J, Dias TB, Saurat N, McGlade A, Kirwan P, Blennow K, Hardy J, Zetterberg H, and Livesey FJ APP metabolism regulates tau proteostasis in human cerebral cortex neurons. Cell Rep 11, 689–696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liao MC, Muratore CR, Gierahn TM, Sullivan SE, Srikanth P, De Jager PL, Love JC, and Young-Pearse TL Single-Cell Detection of Secreted Abeta and sAPPalpha from Human IPSC-Derived Neurons and Astrocytes. J Neurosci 36, 1730–1746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seibler P, Graziotto J, Jeong H, Simunovic F, Klein C, and Krainc D Mitochondrial Parkin recruitment is impaired in neurons derived from mutant PINK1 induced pluripotent stem cells. J Neurosci 31, 5970–5976 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Devine MJ, Ryten M, Vodicka P, Thomson AJ, Burdon T, Houlden H, Cavaleri F, Nagano M, Drummond NJ, Taanman JW, Schapira AH, Gwinn K, Hardy J, Lewis PA, and Kunath T Parkinson’s disease induced pluripotent stem cells with triplication of the alpha-synuclein locus. Nat Commun 2, 440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Imaizumi Y, Okada Y, Akamatsu W, Koike M, Kuzumaki N, Hayakawa H, Nihira T, Kobayashi T, Ohyama M, Sato S, Takanashi M, Funayama M, Hirayama A, Soga T, Hishiki T, Suematsu M, Yagi T, Ito D, Kosakai A, Hayashi K, Shouji M, Nakanishi A, Suzuki N, Mizuno Y, Mizushima N, Amagai M, Uchiyama Y, Mochizuki H, Hattori N, and Okano H Mitochondrial dysfunction associated with increased oxidative stress and alpha-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol Brain 5, 35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang H, Ren Y, Yuen EY, Zhong P, Ghaedi M, Hu Z, Azabdaftari G, Nakaso K, Yan Z, and Feng J Parkin controls dopamine utilization in human midbrain dopaminergic neurons derived from induced pluripotent stem cells. Nat Commun 3, 668 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanchez-Danes A, Richaud-Patin Y, Carballo-Carbajal I, Jimenez-Delgado S, Caig C, Mora S, Di Guglielmo C, Ezquerra M, Patel B, Giralt A, Canals JM, Memo M, Alberch J, Lopez-Barneo J, Vila M, Cuervo AM, Tolosa E, Consiglio A, and Raya A Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson’s disease. EMBO Mol Med 4, 380–395 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chung CY, Khurana V, Auluck PK, Tardiff DF, Mazzulli JR, Soldner F, Baru V, Lou Y, Freyzon Y, Cho S, Mungenast AE, Muffat J, Mitalipova M, Pluth MD, Jui NT, Schule B, Lippard SJ, Tsai LH, Krainc D, Buchwald SL, Jaenisch R, and Lindquist S Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science 342, 983–987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reinhardt P, Schmid B, Burbulla LF, Schondorf DC, Wagner L, Glatza M, Hoing S, Hargus G, Heck SA, Dhingra A, Wu G, Muller S, Brockmann K, Kluba T, Maisel M, Kruger R, Berg D, Tsytsyura Y, Thiel CS, Psathaki OE, Klingauf J, Kuhlmann T, Klewin M, Muller H, Gasser T, Scholer HR, and Sterneckert J Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell 12, 354–367 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Ryan SD, Dolatabadi N, Chan SF, Zhang X, Akhtar MW, Parker J, Soldner F, Sunico CR, Nagar S, Talantova M, Lee B, Lopez K, Nutter A, Shan B, Molokanova E, Zhang Y, Han X, Nakamura T, Masliah E, Yates JR 3rd, Nakanishi N, Andreyev AY, Okamoto S, Jaenisch R, Ambasudhan R, and Lipton SA Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1alpha transcription. Cell 155, 1351–1364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hartfield EM, Yamasaki-Mann M, Ribeiro Fernandes HJ, Vowles J, James WS, Cowley SA, and Wade-Martins R Physiological characterisation of human iPS-derived dopaminergic neurons. PLoS One 9, e87388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schondorf DC, Aureli M, McAllister FE, Hindley CJ, Mayer F, Schmid B, Sardi SP, Valsecchi M, Hoffmann S, Schwarz LK, Hedrich U, Berg D, Shihabuddin LS, Hu J, Pruszak J, Gygi SP, Sonnino S, Gasser T, and Deleidi M iPSC-derived neurons from GBA1-associated Parkinson’s disease patients show autophagic defects and impaired calcium homeostasis. Nat Commun 5, 4028 (2014). [DOI] [PubMed] [Google Scholar]
- 79.Woodard CM, Campos BA, Kuo SH, Nirenberg MJ, Nestor MW, Zimmer M, Mosharov EV, Sulzer D, Zhou H, Paull D, Clark L, Schadt EE, Sardi SP, Rubin L, Eggan K, Brock M, Lipnick S, Rao M, Chang S, Li A, and Noggle SA iPSC-derived dopamine neurons reveal differences between monozygotic twins discordant for Parkinson’s disease. Cell Rep 9, 1173–1182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dettmer U, Newman AJ, Soldner F, Luth ES, Kim NC, von Saucken VE, Sanderson JB, Jaenisch R, Bartels T, and Selkoe D Parkinson-causing alpha-synuclein missense mutations shift native tetramers to monomers as a mechanism for disease initiation. Nat Commun 6, 7314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fernandez-Santiago R, Carballo-Carbajal I, Castellano G, Torrent R, Richaud Y, Sanchez-Danes A, Vilarrasa-Blasi R, Sanchez-Pla A, Mosquera JL, Soriano J, Lopez-Barneo J, Canals JM, Alberch J, Raya A, Vila M, Consiglio A, Martin-Subero JI, Ezquerra M, and Tolosa E Aberrant epigenome in iPSC-derived dopaminergic neurons from Parkinson’s disease patients. EMBO Mol Med 7, 1529–1546 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ren Y, Jiang H, Hu Z, Fan K, Wang J, Janoschka S, Wang X, Ge S, and Feng J Parkin mutations reduce the complexity of neuronal processes in iPSC-derived human neurons. Stem Cells 33, 68–78 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shaltouki A, Sivapatham R, Pei Y, Gerencser AA, Momcilovic O, Rao MS, and Zeng X Mitochondrial alterations by PARKIN in dopaminergic neurons using PARK2 patient-specific and PARK2 knockout isogenic iPSC lines. Stem Cell Reports 4, 847–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aflaki E, Borger DK, Moaven N, Stubblefield BK, Rogers SA, Patnaik S, Schoenen FJ, Westbroek W, Zheng W, Sullivan P, Fujiwara H, Sidhu R, Khaliq ZM, Lopez GJ, Goldstein DS, Ory DS, Marugan J, and Sidransky E A New Glucocerebrosidase Chaperone Reduces alpha-Synuclein and Glycolipid Levels in iPSC-Derived Dopaminergic Neurons from Patients with Gaucher Disease and Parkinsonism. J Neurosci 36, 7441–7452 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, McCarthy S, Sebat J, and Gage FH Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, and Lachman HM RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One 6, e23356 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, Kim NS, Yoon KJ, Shin J, Zhang C, Makri G, Nauen D, Yu H, Guzman E, Chiang CH, Yoritomo N, Kaibuchi K, Zou J, Christian KM, Cheng L, Ross CA, Margolis RL, Chen G, Kosik KS, Song H, and Ming GL Synaptic dysregulation in a human iPS cell model of mental disorders. Nature (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Murai K, Sun G, Ye P, Tian E, Yang S, Cui Q, Sun G, Trinh D, Sun O, Hong T, Wen Z, Kalkum M, Riggs AD, Song H, Ming GL, and Shi Y The TLX-miR-219 cascade regulates neural stem cell proliferation in neurodevelopment and schizophrenia iPSC model. Nat Commun 7, 10965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, Beaumont KG, Kim HJ, Topol A, Ladran I, Abdelrahim M, Matikainen-Ankney B, Chao SH, Mrksich M, Rakic P, Fang G, Zhang B, Yates JR 3rd, and Gage FH Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry 20, 361–368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Topol A, Zhu S, Hartley BJ, English J, Hauberg ME, Tran N, Rittenhouse CA, Simone A, Ruderfer DM, Johnson J, Readhead B, Hadas Y, Gochman PA, Wang YC, Shah H, Cagney G, Rapoport J, Gage FH, Dudley JT, Sklar P, Mattheisen M, Cotter D, Fang G, and Brennand KJ Dysregulation of miRNA-9 in a Subset of Schizophrenia Patient-Derived Neural Progenitor Cells. Cell Rep 15, 1024–1036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z, Makri G, Nauen D, Shin JH, Park Y, Chung R, Pekle E, Zhang C, Towe M, Hussaini SM, Lee Y, Rujescu D, St Clair D, Kleinman JE, Hyde TM, Krauss G, Christian KM, Rapoport JL, Weinberger DR, Song H, and Ming GL Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell 15, 79–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Han J, Kim HJ, Schafer ST, Paquola A, Clemenson GD, Toda T, Oh J, Pankonin AR, Lee BS, Johnston ST, Sarkar A, Denli AM, and Gage FH Functional Implications of miR-19 in the Migration of Newborn Neurons in the Adult Brain. Neuron 91, 79–89 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, and Przedborski S Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci 10, 615–622 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, and Eggan K Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci 10, 608–614 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marchetto MC, Muotri AR, Mu Y, Smith AM, Cezar GG, and Gage FH Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell 3, 649–657 (2008). [DOI] [PubMed] [Google Scholar]
- 96.Haidet-Phillips AM, Hester ME, Miranda CJ, Meyer K, Braun L, Frakes A, Song S, Likhite S, Murtha MJ, Foust KD, Rao M, Eagle A, Kammesheidt A, Christensen A, Mendell JR, Burghes AH, and Kaspar BK Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat Biotechnol 29, 824–828 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lancaster MA and Knoblich JA Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125 (2014). [DOI] [PubMed] [Google Scholar]
- 98.McCracken KW, Cata EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, and Wells JM Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, Aoyama S, Adachi Y, and Taniguchi H Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484 (2013). [DOI] [PubMed] [Google Scholar]
- 100.Sampaziotis F, Cardoso de Brito M, Madrigal P, Bertero A, Saeb-Parsy K, Soares FA, Schrumpf E, Melum E, Karlsen TH, Bradley JA, Gelson WT, Davies S, Baker A, Kaser A, Alexander GJ, Hannan NR, and Vallier L Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol 33, 845–852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ogawa M, Ogawa S, Bear CE, Ahmadi S, Chin S, Li B, Grompe M, Keller G, Kamath BM, and Ghanekar A Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol 33, 853–861 (2015). [DOI] [PubMed] [Google Scholar]
- 102.Dye BR, Hill DR, Ferguson MA, Tsai YH, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD, White ES, Deutsch GH, and Spence JR In vitro generation of human pluripotent stem cell derived lung organoids. Elife 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, Shroyer NF, and Wells JM Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, Chuva de Sousa Lopes SM, and Little MH Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015). [DOI] [PubMed] [Google Scholar]
- 105.Watson CL, Mahe MM, Munera J, Howell JC, Sundaram N, Poling HM, Schweitzer JI, Vallance JE, Mayhew CN, Sun Y, Grabowski G, Finkbeiner SR, Spence JR, Shroyer NF, Wells JM, and Helmrath MA An in vivo model of human small intestine using pluripotent stem cells. Nat Med 20, 1310–1314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tucker BA, Solivan-Timpe F, Roos BR, Anfinson KR, Robin AL, Wiley LA, Mullins RF, and Fingert JH Duplication of TBK1 Stimulates Autophagy in iPSC-derived Retinal Cells from a Patient with Normal Tension Glaucoma. J Stem Cell Res Ther 3, 161 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Brauninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, Knoblich JA, Lachmann R, Paabo S, Huttner WB, and Treutlein B Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A 112, 15672–15677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, Gerstein M, Grigorenko EL, Chawarska K, Pelphrey KA, Howe JR, and Vaccarino FM FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 162, 375–390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimaraes KP, Benazzato C, Almeida N, Pignatari GC, Romero S, Polonio CM, Cunha I, Freitas CL, Brandao WN, Rossato C, Andrade DG, Faria Dde P, Garcez AT, Buchpigel CA, Braconi CT, Mendes E, Sall AA, Zanotto PM, Peron JP, Muotri AR, and Beltrao-Braga PC The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534, 267–271 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon KJ, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Chickering M, Nauen D, Ho CY, Wen Z, Christian KM, Shi PY, Maher BJ, Wu H, Jin P, Tang H, Song H, and Ming GL Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 165, 1238–1254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, and Rehen SK Zika virus impairs growth in human neurospheres and brain organoids. Science 352, 816–818 (2016). [DOI] [PubMed] [Google Scholar]
- 112.Gabriel E, Wason A, Ramani A, Gooi LM, Keller P, Pozniakovsky A, Poser I, Noack F, Telugu NS, Calegari F, Saric T, Hescheler J, Hyman AA, Gottardo M, Callaini G, Alkuraya FS, and Gopalakrishnan J CPAP promotes timely cilium disassembly to maintain neural progenitor pool. Embo J 35, 803–819 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Otani T, Marchetto MC, Gage FH, Simons BD, and Livesey FJ 2D and 3D Stem Cell Models of Primate Cortical Development Identify Species-Specific Differences in Progenitor Behavior Contributing to Brain Size. Cell Stem Cell 18, 467–480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O’Rourke NA, Nguyen KD, Smith SJ, Huguenard JR, Geschwind DH, Barres BA, and Pasca SP Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods 12, 671–678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tzatzalos E, Abilez OJ, Shukla P, and Wu JC Engineered heart tissues and induced pluripotent stem cells: Macro- and microstructures for disease modeling, drug screening, and translational studies. Adv Drug Deliv Rev 96, 234–244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fatehullah A, Tan SH, and Barker N Organoids as an in vitro model of human development and disease. Nat Cell Biol 18, 246–254 (2016). [DOI] [PubMed] [Google Scholar]
- 117.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, and Knoblich JA Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vincent F, Loria P, Pregel M, Stanton R, Kitching L, Nocka K, Doyonnas R, Steppan C, Gilbert A, Schroeter T, and Peakman MC Developing predictive assays: the phenotypic screening “rule of 3”. Sci Transl Med 7, 293ps215 (2015). [DOI] [PubMed] [Google Scholar]
- 119.Inoue H, Nagata N, Kurokawa H, and Yamanaka S iPS cells: a game changer for future medicine. Embo j 33, 409–417 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Matsa E, Burridge PW, Yu KH, Ahrens JH, Termglinchan V, Wu H, Liu C, Shukla P, Sayed N, Churko JM, Shao N, Woo NA, Chao AS, Gold JD, Karakikes I, Snyder MP, and Wu JC Transcriptome Profiling of Patient-Specific Human iPSC-Cardiomyocytes Predicts Individual Drug Safety and Efficacy Responses In Vitro. Cell Stem Cell 19, 311–325 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee G, Ramirez CN, Kim H, Zeltner N, Liu B, Radu C, Bhinder B, Kim YJ, Choi IY, Mukherjee-Clavin B, Djaballah H, and Studer L Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nat Biotechnol 30, 1244–1248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Doi D, Samata B, Katsukawa M, Kikuchi T, Morizane A, Ono Y, Sekiguchi K, Nakagawa M, Parmar M, and Takahashi J Isolation of human induced pluripotent stem cell-derived dopaminergic progenitors by cell sorting for successful transplantation. Stem Cell Reports 2, 337–350 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, Yamamoto T, Adachi F, Kondo T, Okita K, Asaka I, Aoi T, Watanabe A, Yamada Y, Morizane A, Takahashi J, Ayaki T, Ito H, Yoshikawa K, Yamawaki S, Suzuki S, Watanabe D, Hioki H, Kaneko T, Makioka K, Okamoto K, Takuma H, Tamaoka A, Hasegawa K, Nonaka T, Hasegawa M, Kawata A, Yoshida M, Nakahata T, Takahashi R, Marchetto MC, Gage FH, Yamanaka S, and Inoue H Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med 4, 145ra104 (2012). [DOI] [PubMed] [Google Scholar]
- 124.Miki K, Endo K, Takahashi S, Funakoshi S, Takei I, Katayama S, Toyoda T, Kotaka M, Takaki T, Umeda M, Okubo C, Nishikawa M, Oishi A, Narita M, Miyashita I, Asano K, Hayashi K, Osafune K, Yamanaka S, Saito H, and Yoshida Y Efficient Detection and Purification of Cell Populations Using Synthetic MicroRNA Switches. Cell Stem Cell 16, 699–711 (2015). [DOI] [PubMed] [Google Scholar]
- 125.Burkhardt MF, Martinez FJ, Wright S, Ramos C, Volfson D, Mason M, Garnes J, Dang V, Lievers J, Shoukat-Mumtaz U, Martinez R, Gai H, Blake R, Vaisberg E, Grskovic M, Johnson C, Irion S, Bright J, Cooper B, Nguyen L, Griswold-Prenner I, and Javaherian A A cellular model for sporadic ALS using patient-derived induced pluripotent stem cells. Mol Cell Neurosci 56, 355–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bright J, Hussain S, Dang V, Wright S, Cooper B, Byun T, Ramos C, Singh A, Parry G, Stagliano N, and Griswold-Prenner I Human secreted tau increases amyloid-beta production. Neurobiol Aging 36, 693–709 (2015). [DOI] [PubMed] [Google Scholar]
- 127.Naryshkin NA, Weetall M, Dakka A, Narasimhan J, Zhao X, Feng Z, Ling KK, Karp GM, Qi H, Woll MG, Chen G, Zhang N, Gabbeta V, Vazirani P, Bhattacharyya A, Furia B, Risher N, Sheedy J, Kong R, Ma J, Turpoff A, Lee CS, Zhang X, Moon YC, Trifillis P, Welch EM, Colacino JM, Babiak J, Almstead NG, Peltz SW, Eng LA, Chen KS, Mull JL, Lynes MS, Rubin LL, Fontoura P, Santarelli L, Haehnke D, McCarthy KD, Schmucki R, Ebeling M, Sivaramakrishnan M, Ko CP, Paushkin SV, Ratni H, Gerlach I, Ghosh A, and Metzger F Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science 345, 688–693 (2014). [DOI] [PubMed] [Google Scholar]
- 128.Xu X, Lei Y, Luo J, Wang J, Zhang S, Yang XJ, Sun M, Nuwaysir E, Fan G, Zhao J, Lei L, and Zhong Z Prevention of beta-amyloid induced toxicity in human iPS cell-derived neurons by inhibition of Cyclin-dependent kinases and associated cell cycle events. Stem Cell Res 10, 213–227 (2013). [DOI] [PubMed] [Google Scholar]
- 129.Hoing S, Rudhard Y, Reinhardt P, Glatza M, Stehling M, Wu G, Peiker C, Bocker A, Parga JA, Bunk E, Schwamborn JC, Slack M, Sterneckert J, and Scholer HR Discovery of inhibitors of microglial neurotoxicity acting through multiple mechanisms using a stem-cell-based phenotypic assay. Cell Stem Cell 11, 620–632 (2012). [DOI] [PubMed] [Google Scholar]
- 130.Mullard A Stem-cell discovery platforms yield first clinical candidates. Nat Rev Drug Discov 14, 589–591 (2015). [DOI] [PubMed] [Google Scholar]
- 131.Wang H, Li X, Gao S, Sun X, and Fang H Transdifferentiation via transcription factors or microRNAs: Current status and perspective. Differentiation 90, 69–76 (2015). [DOI] [PubMed] [Google Scholar]
- 132.Pereira CF, Lemischka IR, and Moore K Reprogramming cell fates: insights from combinatorial approaches. Ann N Y Acad Sci 1266, 7–17 (2012). [DOI] [PubMed] [Google Scholar]
- 133.Tanaka A, Woltjen K, Miyake K, Hotta A, Ikeya M, Yamamoto T, Nishino T, Shoji E, Sehara-Fujisawa A, Manabe Y, Fujii N, Hanaoka K, Era T, Yamashita S, Isobe K, Kimura E, and Sakurai H Efficient and reproducible myogenic differentiation from human iPS cells: prospects for modeling Miyoshi Myopathy in vitro. PLoS One 8, e61540 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shoji E, Sakurai H, Nishino T, Nakahata T, Heike T, Awaya T, Fujii N, Manabe Y, Matsuo M, and Sehara-Fujisawa A Early pathogenesis of Duchenne muscular dystrophy modelled in patient-derived human induced pluripotent stem cells. Sci Rep 5, 12831 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Inoue H and Yamanaka S The use of induced pluripotent stem cells in drug development. Clin Pharmacol Ther 89, 655–661 (2011). [DOI] [PubMed] [Google Scholar]
- 136.Yamashita A, Morioka M, Kishi H, Kimura T, Yahara Y, Okada M, Fujita K, Sawai H, Ikegawa S, and Tsumaki N Statin treatment rescues FGFR3 skeletal dysplasia phenotypes. Nature 513, 507–511 (2014). [DOI] [PubMed] [Google Scholar]
- 137.McNeish J, Gardner JP, Wainger BJ, Woolf CJ, and Eggan K From Dish to Bedside: Lessons Learned While Translating Findings from a Stem Cell Model of Disease to a Clinical Trial. Cell Stem Cell 17, 8–10 (2015). [DOI] [PubMed] [Google Scholar]
- 138.Devlin AC, Burr K, Borooah S, Foster JD, Cleary EM, Geti I, Vallier L, Shaw CE, Chandran S, and Miles GB Human iPSC-derived motoneurons harbouring TARDBP or C9ORF72 ALS mutations are dysfunctional despite maintaining viability. Nat Commun 6, 5999 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Avorn J The $2.6 billion pill--methodologic and policy considerations. N Engl J Med 372, 1877–1879 (2015). [DOI] [PubMed] [Google Scholar]
- 140.DiMasi JA, Grabowski HG, and Hansen RW Innovation in the pharmaceutical industry: New estimates of R&D costs. J Health Econ 47, 20–33 (2016). [DOI] [PubMed] [Google Scholar]
- 141.Wilke RA, Lin DW, Roden DM, Watkins PB, Flockhart D, Zineh I, Giacomini KM, and Krauss RM Identifying genetic risk factors for serious adverse drug reactions: current progress and challenges. Nat Rev Drug Discov 6, 904–916 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lu HR, Vlaminckx E, Hermans AN, Rohrbacher J, Van Ammel K, Towart R, Pugsley M, and Gallacher DJ Predicting drug-induced changes in QT interval and arrhythmias: QT-shortening drugs point to gaps in the ICHS7B Guidelines. Br J Pharmacol 154, 1427–1438 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Doherty KR, Wappel RL, Talbert DR, Trusk PB, Moran DM, Kramer JW, Brown AM, Shell SA, and Bacus S Multi-parameter in vitro toxicity testing of crizotinib, sunitinib, erlotinib, and nilotinib in human cardiomyocytes. Toxicol Appl Pharmacol 272, 245–255 (2013). [DOI] [PubMed] [Google Scholar]
- 144.Gintant G, Sager PT, and Stockbridge N Evolution of strategies to improve preclinical cardiac safety testing. Nat Rev Drug Discov 15, 457–471 (2016). [DOI] [PubMed] [Google Scholar]
- 145.Harris K, Aylott M, Cui Y, Louttit JB, McMahon NC, and Sridhar A Comparison of electrophysiological data from human-induced pluripotent stem cell-derived cardiomyocytes to functional preclinical safety assays. Toxicol Sci 134, 412–426 (2013). [DOI] [PubMed] [Google Scholar]
- 146.Qu Y and Vargas HM Proarrhythmia Risk Assessment in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes Using the Maestro MEA Platform. Toxicol Sci 147, 286–295 (2015). [DOI] [PubMed] [Google Scholar]
- 147.Yamada T, Yoshikawa M, Kanda S, Kato Y, Nakajima Y, Ishizaka S, and Tsunoda Y In vitro differentiation of embryonic stem cells into hepatocyte-like cells identified by cellular uptake of indocyanine green. Stem Cells 20, 146–154 (2002). [DOI] [PubMed] [Google Scholar]
- 148.Takayama K, Inamura M, Kawabata K, Katayama K, Higuchi M, Tashiro K, Nonaka A, Sakurai F, Hayakawa T, Furue MK, and Mizuguchi H Efficient generation of functional hepatocytes from human embryonic stem cells and induced pluripotent stem cells by HNF4alpha transduction. Mol Ther 20, 127–137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schwartz MP, Hou Z, Propson NE, Zhang J, Engstrom CJ, Santos Costa V, Jiang P, Nguyen BK, Bolin JM, Daly W, Wang Y, Stewart R, Page CD, Murphy WL, and Thomson JA Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc Natl Acad Sci U S A 112, 12516–12521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, and Jones JM Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998). [DOI] [PubMed] [Google Scholar]
- 151.Sayed N, Liu C, and Wu JC Translation of Human-Induced Pluripotent Stem Cells: From Clinical Trial in a Dish to Precision Medicine. J Am Coll Cardiol 67, 2161–2176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Menasche P, Vanneaux V, Hagege A, Bel A, Cholley B, Cacciapuoti I, Parouchev A, Benhamouda N, Tachdjian G, Tosca L, Trouvin JH, Fabreguettes JR, Bellamy V, Guillemain R, Suberbielle Boissel C, Tartour E, Desnos M, and Larghero J Human embryonic stem cell-derived cardiac progenitors for severe heart failure treatment: first clinical case report. Eur Heart J 36, 2011–2017 (2015). [DOI] [PubMed] [Google Scholar]
- 153.Neofytou E, O’Brien CG, Couture LA, and Wu JC Hurdles to clinical translation of human induced pluripotent stem cells. J Clin Invest 125, 2551–2557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lee AS, Tang C, Rao MS, Weissman IL, and Wu JC Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med 19, 998–1004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lund RJ, Narva E, and Lahesmaa R Genetic and epigenetic stability of human pluripotent stem cells. Nat Rev Genet 13, 732–744 (2012). [DOI] [PubMed] [Google Scholar]
- 156.Lee MO, Moon SH, Jeong HC, Yi JY, Lee TH, Shim SH, Rhee YH, Lee SH, Oh SJ, Lee MY, Han MJ, Cho YS, Chung HM, Kim KS, and Cha HJ Inhibition of pluripotent stem cell-derived teratoma formation by small molecules. Proc Natl Acad Sci U S A 110, E3281–3290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ben-David U, Gan QF, Golan-Lev T, Arora P, Yanuka O, Oren YS, Leikin-Frenkel A, Graf M, Garippa R, Boehringer M, Gromo G, and Benvenisty N Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell Stem Cell 12, 167–179 (2013). [DOI] [PubMed] [Google Scholar]
- 158.Nguyen PK, Neofytou E, Rhee JW, and Wu JC Potential strategies for addressing the major clinical hurdles facing stem cell regenerative therapy. JAMA Cardiol In press, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cao F, Li Z, Lee A, Liu Z, Chen K, Wang H, Cai W, Chen X, and Wu JC Noninvasive de novo imaging of human embryonic stem cell-derived teratoma formation. Cancer Res 69, 2709–2713 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Riegler J, Ebert A, Qin X, Shen Q, Wang M, Ameen M, Kodo K, Ong SG, Lee WH, Lee G, Neofytou E, Gold JD, Connolly AJ, and Wu JC Comparison of Magnetic Resonance Imaging and Serum Biomarkers for Detection of Human Pluripotent Stem Cell-Derived Teratomas. Stem Cell Reports 6, 176–187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Koch CA, Geraldes P, and Platt JL Immunosuppression by embryonic stem cells. Stem Cells 26, 89–98 (2008). [DOI] [PubMed] [Google Scholar]
- 162.Drukker M, Katz G, Urbach A, Schuldiner M, Markel G, Itskovitz-Eldor J, Reubinoff B, Mandelboim O, and Benvenisty N Characterization of the expression of MHC proteins in human embryonic stem cells. Proc Natl Acad Sci U S A 99, 9864–9869 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pearl JI, Kean LS, Davis MM, and Wu JC Pluripotent stem cells: immune to the immune system? Sci Transl Med 4, 164ps125 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhao T, Zhang ZN, Rong Z, and Xu Y Immunogenicity of induced pluripotent stem cells. Nature 474, 212–215 (2011). [DOI] [PubMed] [Google Scholar]
- 165.de Almeida PE, Meyer EH, Kooreman NG, Diecke S, Dey D, Sanchez-Freire V, Hu S, Ebert A, Odegaard J, Mordwinkin NM, Brouwer TP, Lo D, Montoro DT, Longaker MT, Negrin RS, and Wu JC Transplanted terminally differentiated induced pluripotent stem cells are accepted by immune mechanisms similar to self-tolerance. Nat Commun 5, 3903 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhao T, Zhang ZN, Westenskow PD, Todorova D, Hu Z, Lin T, Rong Z, Kim J, He J, Wang M, Clegg DO, Yang YG, Zhang K, Friedlander M, and Xu Y Humanized Mice Reveal Differential Immunogenicity of Cells Derived from Autologous Induced Pluripotent Stem Cells. Cell Stem Cell 17, 353–359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Huang K, Liu P, Li X, Chen S, Wang L, Qin L, Su Z, Huang W, Liu J, Jia B, Liu J, Cai J, Pei D, and Pan G Neural progenitor cells from human induced pluripotent stem cells generated less autogenous immune response. Sci China Life Sci 57, 162–170 (2014). [DOI] [PubMed] [Google Scholar]
- 168.Team, Cell Stem Cell Editorial 10 Questions: Clinical Outlook for iPSCs. Cell Stem Cell 18, 170–173 (2016). [DOI] [PubMed] [Google Scholar]
- 169.Bravery CA Do human leukocyte antigen-typed cellular therapeutics based on induced pluripotent stem cells make commercial sense? Stem Cells Dev 24, 1–10 (2015). [DOI] [PubMed] [Google Scholar]
- 170.Jacquet L, Stephenson E, Collins R, Patel H, Trussler J, Al-Bedaery R, Renwick P, Ogilvie C, Vaughan R, and Ilic D Strategy for the creation of clinical grade hESC line banks that HLA-match a target population. EMBO Mol Med 5, 10–17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pearl JI, Lee AS, Leveson-Gower DB, Sun N, Ghosh Z, Lan F, Ransohoff J, Negrin RS, Davis MM, and Wu JC Short-term immunosuppression promotes engraftment of embryonic and induced pluripotent stem cells. Cell Stem Cell 8, 309–317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Swijnenburg RJ, Schrepfer S, Govaert JA, Cao F, Ransohoff K, Sheikh AY, Haddad M, Connolly AJ, Davis MM, Robbins RC, and Wu JC Immunosuppressive therapy mitigates immunological rejection of human embryonic stem cell xenografts. Proc Natl Acad Sci U S A 105, 12991–12996 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Meissner T, Strominger J, and WCowan C The universal donor stem cells: removing the immune barrier to transplantation using CRISPR/Cas9. The Journal of Immunology 194, Supplement 140.128 (2015). [Google Scholar]
- 174.Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, and Jaenisch R Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 318, 1920–1923 (2007). [DOI] [PubMed] [Google Scholar]
- 175.Hotta A and Yamanaka S From Genomics to Gene Therapy: Induced Pluripotent Stem Cells Meet Genome Editing. Annu Rev Genet 49, 47–70 (2015). [DOI] [PubMed] [Google Scholar]
- 176.Deleidi M and Yu C Genome editing in pluripotent stem cells: research and therapeutic applications. Biochem Biophys Res Commun 473, 665–674 (2016). [DOI] [PubMed] [Google Scholar]
- 177.Seah YF, El Farran CA, Warrier T, Xu J, and Loh YH Induced Pluripotency and Gene Editing in Disease Modelling: Perspectives and Challenges. Int J Mol Sci 16, 28614–28634 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Orqueda AJ, Gimenez CA, and Pereyra-Bonnet F iPSCs: A Minireview from Bench to Bed, including Organoids and the CRISPR System. Stem Cells Int 2016, 5934782 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, and Yusa K Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32, 267–273 (2014). [DOI] [PubMed] [Google Scholar]
- 180.Mandegar MA, Huebsch N, Frolov EB, Shin E, Truong A, Olvera MP, Chan AH, Miyaoka Y, Holmes K, Spencer CI, Judge LM, Gordon DE, Eskildsen TV, Villalta JE, Horlbeck MA, Gilbert LA, Krogan NJ, Sheikh SP, Weissman JS, Qi LS, So PL, and Conklin BR CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell 18, 541–553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Narsinh KH, Sun N, Sanchez-Freire V, Lee AS, Almeida P, Hu S, Jan T, Wilson KD, Leong D, Rosenberg J, Yao M, Robbins RC, and Wu JC Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J Clin Invest 121, 1217–1221 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hough SR, Thornton M, Mason E, Mar JC, Wells CA, and Pera MF Single-cell gene expression profiles define self-renewing, pluripotent, and lineage primed states of human pluripotent stem cells. Stem Cell Reports 2, 881–895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Miura K, Okada Y, Aoi T, Okada A, Takahashi K, Okita K, Nakagawa M, Koyanagi M, Tanabe K, Ohnuki M, Ogawa D, Ikeda E, Okano H, and Yamanaka S Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol 27, 743–745 (2009). [DOI] [PubMed] [Google Scholar]
- 184.Koyanagi-Aoi M, Ohnuki M, Takahashi K, Okita K, Noma H, Sawamura Y, Teramoto I, Narita M, Sato Y, Ichisaka T, Amano N, Watanabe A, Morizane A, Yamada Y, Sato T, Takahashi J, and Yamanaka S Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci U S A 110, 20569–20574 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Takahashi K and Yamanaka S A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol 17, 183–193 (2016). [DOI] [PubMed] [Google Scholar]
- 186.Sasai Y Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell 12, 520–530 (2013). [DOI] [PubMed] [Google Scholar]
- 187.Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, and Sasai Y Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci U S A 110, 20284–20289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, and Sasai Y Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785 (2012). [DOI] [PubMed] [Google Scholar]
- 189.Ozone C, Suga H, Eiraku M, Kadoshima T, Yonemura S, Takata N, Oiso Y, Tsuji T, and Sasai Y Functional anterior pituitary generated in self-organizing culture of human embryonic stem cells. Nat Commun 7, 10351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, and Sasai Y Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep 10, 537–550 (2015). [DOI] [PubMed] [Google Scholar]
- 191.Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, Takahashi J, Eiraku M, and Sasai Y Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat Commun 6, 8896 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dedhia PH, Bertaux-Skeirik N, Zavros Y, and Spence JR Organoid Models of Human Gastrointestinal Development and Disease. Gastroenterology 150, 1098–1112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Aday S, Cecchelli R, Hallier-Vanuxeem D, Dehouck MP, and Ferreira L Stem Cell-Based Human Blood-Brain Barrier Models for Drug Discovery and Delivery. Trends Biotechnol 34, 382–393 (2016). [DOI] [PubMed] [Google Scholar]
- 194.Goodridge HS Induced pluripotent stem cell-derived myeloid phagocytes: disease modeling and therapeutic applications. Drug Discov Today 19, 774–780 (2014). [DOI] [PubMed] [Google Scholar]
- 195.Smith MJ, Webber BR, Mohtashami M, Stefanski HE, Zuniga-Pflucker JC, and Blazar BR In Vitro T-Cell Generation From Adult, Embryonic, and Induced Pluripotent Stem Cells: Many Roads to One Destination. Stem Cells 33, 3174–3180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Huch M and Koo BK Modeling mouse and human development using organoid cultures. Development 142, 3113–3125 (2015). [DOI] [PubMed] [Google Scholar]
- 197.Sasai Y Cytosystems dynamics in self-organization of tissue architecture. Nature 493, 318–326 (2013). [DOI] [PubMed] [Google Scholar]
- 198.Li Y, Xu C, and Ma T In vitro organogenesis from pluripotent stem cells. Organogenesis 10, 159–163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yin X, Mead BE, Safaee H, Langer R, Karp JM, and Levy O Engineering Stem Cell Organoids. Cell Stem Cell 18, 25–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Inoue Y, Shineha R, and Yashiro Y Current Public Support for Human-Animal Chimera Research in Japan Is Limited, Despite High Levels of Scientific Approval. Cell Stem Cell 19, 152–153 (2016). [DOI] [PubMed] [Google Scholar]
- 201.Mertens J, Wang QW, Kim Y, Yu DX, Pham S, Yang B, Zheng Y, Diffenderfer KE, Zhang J, Soltani S, Eames T, Schafer ST, Boyer L, Marchetto MC, Nurnberger JI, Calabrese JR, Odegaard KJ, McCarthy MJ, Zandi PP, Alda M, Nievergelt CM, Mi S, Brennand KJ, Kelsoe JR, Gage FH, and Yao J Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 527, 95–99 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lord J, Lu AJ, and Cruchaga C Identification of rare variants in Alzheimer’s disease. Front Genet 5, 369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wainger BJ, Kiskinis E, Mellin C, Wiskow O, Han SS, Sandoe J, Perez NP, Williams LA, Lee S, Boulting G, Berry JD, Brown RH Jr., Cudkowicz ME, Bean BP, Eggan K, and Woolf CJ Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep 7, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yamanaka S Induced pluripotent stem cells: past, present, and future. Cell Stem Cell 10, 678–684 (2012). [DOI] [PubMed] [Google Scholar]
- 205.Hawley RG Does retroviral insertional mutagenesis play a role in the generation of induced pluripotent stem cells? Mol Ther 16, 1354–1355 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, and Thomson JA Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K, Shibata T, Kunisada T, Takahashi M, Takahashi J, Saji H, and Yamanaka S A more efficient method to generate integration-free human iPS cells. Nat Methods 8, 409–412 (2011). [DOI] [PubMed] [Google Scholar]
- 208.Stadtfeld M, Nagaya M, Utikal J, Weir G, and Hochedlinger K Induced pluripotent stem cells generated without viral integration. Science 322, 945–949 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Fusaki N, Ban H, Nishiyama A, Saeki K, and Hasegawa M Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci 85, 348–362 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, and Nagy A piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458, 766–770 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, Longaker MT, and Wu JC A nonviral minicircle vector for deriving human iPS cells. Nat Methods 7, 197–199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, and Kim KS Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 4, 472–476 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, and Rossi DJ Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, and Ying SY Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. Rna 14, 2115–2124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, and Mori M Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8, 633–638 (2011). [DOI] [PubMed] [Google Scholar]
- 216.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, Ge J, Xu J, Zhang Q, Zhao Y, and Deng H Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 341, 651–654 (2013). [DOI] [PubMed] [Google Scholar]
- 217.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, and Holmes MC Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651 (2005). [DOI] [PubMed] [Google Scholar]
- 218.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, Holmes MC, and Naldini L Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol 25, 1298–1306 (2007). [DOI] [PubMed] [Google Scholar]
- 219.Cho SW, Kim S, Kim JM, and Kim JS Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31, 230–232 (2013). [DOI] [PubMed] [Google Scholar]
- 220.Jinek M, East A, Cheng A, Lin S, Ma E, and Doudna J RNA-programmed genome editing in human cells. Elife 2, e00471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kaufmann M, Schuffenhauer A, Fruh I, Klein J, Thiemeyer A, Rigo P, Gomez-Mancilla B, Heidinger-Millot V, Bouwmeester T, Schopfer U, Mueller M, Fodor BD, and Cobos-Correa A High-Throughput Screening Using iPSC-Derived Neuronal Progenitors to Identify Compounds Counteracting Epigenetic Gene Silencing in Fragile X Syndrome. J Biomol Screen 20, 1101–1111 (2015). [DOI] [PubMed] [Google Scholar]
- 222.Kumari D, Swaroop M, Southall N, Huang W, Zheng W, and Usdin K High-Throughput Screening to Identify Compounds That Increase Fragile X Mental Retardation Protein Expression in Neural Stem Cells Differentiated From Fragile X Syndrome Patient-Derived Induced Pluripotent Stem Cells. Stem Cells Transl Med 4, 800–808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Li M, Zhao H, Ananiev GE, Musser MT, Ness KH, Maglaque DL, Saha K, Bhattacharyya A, and Zhao X Establishment of Reporter Lines for Detecting Fragile X Mental Retardation (FMR1) Gene Reactivation in Human Neural Cells. Stem Cells (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]