Abstract
Dictyostelium discoideum (D. discoideum) is a simple eukaryote with a unique life cycle in which it differentiates from unicellular amoebae into a fruiting body upon starvation. Reactive oxygen species (ROS) have been associated with bacterial predation, as well as regulatory events during D. discoideum development and differentiation. Coenzyme A (CoA) is a key metabolic integrator in all living cells. A novel function of CoA in redox regulation, mediated by covalent attachment of CoA to cellular proteins in response to oxidative or metabolic stress, has been recently discovered and termed protein CoAlation. In this study, we report that the level of CoA and protein CoAlation in D. discoideum are developmentally regulated, and correlate with the temporal expression pattern of genes implicated in CoA biosynthesis during morphogenesis. Furthermore, treatment of growing D. discoideum cells with oxidising agents results in a dose-dependent increase of protein CoAlation. However, much higher concentrations were required when compared to mammalian cells and bacteria. Increased resistance of D. discoideum to oxidative stress induced by H2O2 has previously been attributed to high levels of catalase activity. In support of this notion, we found that H2O2-induced protein CoAlation is significantly increased in CatA-deficient D. discoideum cells. Collectively, this study provides insights into the role of CoA and protein CoAlation in the maintenance of redox homeostasis in amoeba and during D. discoideum morphogenesis.
Keywords: Coenzyme A, Protein CoAlation, Oxidative stress, Reactive oxygen species (ROS), Dictyostelium discoideum, Morphogenesis
Highlights
-
•
D. discoideum cells are professional phagocytes and produce ROS for efficient bacterial killing.
-
•
D. discoideum cells are highly resistant to oxidative stress.
-
•
CoA biosynthetic genes are transcriptionally regulated during morphogenesis.
-
•
The level of CoA and protein CoAlation are developmentally regulated.
-
•
Oxidising agents induce protein CoAlation in D. discoideum cells.
1. Introduction
Coenzyme A (CoA) is a fundamental and ubiquitous cellular cofactor, and functions as a carbonyl-activating group and an acyl group carrier in diverse biological processes. CoA and its thioester derivatives (acetyl-CoA, malonyl-CoA, succinyl-CoA, HMG-CoA etc.) are involved in a wide range of biochemical reactions, including fatty acid metabolism, protein acylation, biosynthesis of amino acids and cholesterol, and the regulation of gene expression [1,2]. The CoA biosynthetic pathway is conserved across eukaryotes and prokaryotes and involves enzymatic conjugation of pantothenate (vitamin B5), cysteine and adenosine triphosphate (ATP) in five consecutive steps. It is modulated in different ways, including the expression of genes encoding for biosynthetic enzymes, regulation of their enzymatic activities, interconversion among CoA and its thioester derivatives and CoA degradation. Pantothenate kinase (Pank) is the primary and rate-limiting enzyme in CoA biosynthesis and its activity is regulated via feedback inhibition by CoA and its thioester derivatives [1,3]. The enzyme activity can also be modulated by the cell's energy status, as high levels of ATP can displace CoA or its thioesters and support Pank activity [4]. The activity of CoA synthase, which mediates the last two steps of CoA biosynthesis, is also regulated by phospholipids and signalling pathways induced by extracellular stimuli and stresses [[5], [6], [7], [8]]. Dysregulation of CoA biosynthesis and homeostasis has been linked to human pathologies such as cancer, diabetes, neurodegeneration and cardiac hypertrophy [1,3].
Recent studies from our laboratory have uncovered a novel unconventional function of CoA in redox regulation in mammalian and prokaryotic cells, which we termed protein CoAlation [9,10]. It is a reversible and widespread post-translational modification of proteins, involving covalent attachment of CoA via its thiol group to reactive surface exposed thiol groups of cysteine residues in response to oxidative and metabolic stress [11]. To study protein CoAlation we have developed: a) unique anti-CoA monoclonal antibodies; b) a robust procedure for the identification of CoAlated proteins via liquid chromatography tandem mass spectrometry (LC-MS/MS) and c) in vitro CoAlation and deCoAlation assays. To date, over one thousand CoAlated proteins have been identified in mammalian cells and tissues, and bacteria under various stress conditions [11].
D. discoideum is a soil-dwelling amoeba that has a unique life cycle with motile unicellular and multicellular phases. It is a powerful eukaryotic model organism for biomedical research, and is amenable to investigations of cellular processes, ranging from basic cell biology to growth and differentiation [12]. Single-celled D. discoideum amoebae consume bacteria by phagocytosis and proliferate by binary fission. Upon starvation, single vegetative cells cease growth and initiate a programme of multicellular development. This begins with aggregation of cells due to migration towards the secreted chemoattractant cyclic AMP. Aggregation leads to the creation of a tipped mound that further extends to form a finger. The finger forms a slug, which migrates thermotactically and phototactically. Finally, a fruiting body is formed which consists of terminally differentiated dead stalk cells and viable spore cells. The stalk cells are thought to help spore dispersal, when upon the availability of nutrients, they germinate into amoeba thus renewing the life cycle.
Being professional phagocytes, vegetative D. discoideum cells (or innate immune cells found in the slug) produce large quantities of toxic ROS for efficient bacterial killing [13,14]. It is thought that extensive exposure to ROS has led D. discoideum to evolve high levels of resistance to oxidative stress. Indeed, they express several different catalases and superoxide dismutases during vegetative growth, as well as at specific stages of the developmental cycle. Reactive oxygen species are also thought to play roles during the multicellular stages of development in D. discoideum. Manipulation of ROS levels can accelerate or inhibit aggregation [15]. Finally, terminally differentiated spore cells exhibit high levels of resistance to oxidative stress, which is dependent on the expression of a catalase specifically expressed in mature spore cells [16].
D. discoideum cells naturally encounter oxidative stress during unicellular growth, multicellular development and terminal spore cell differentiation and dormancy. Understanding the mechanisms underlying their high resistance to oxidative stress remains a key question. It thus represents an ideal system to explore the extent to which protein CoAlation, and its role in oxidative stress responses is conserved in eukaryotes. Here we demonstrate that genes involved in CoA biosynthesis are differentially expressed during growth and morphogenesis, and this pattern correlates with the level of CoA and protein CoAlation. We also found that exposure of D. discoideum to oxidative stress resulted in an increase of protein CoAlation. However, much higher concentrations of oxidising agents were required to induce protein CoAlation in D. discoideum, when compared to mammalian cells and bacteria [9,10]. Increased resistance to oxidative stress has been attributed to a very high level of catalase activity. Indeed, we found that H2O2-induced protein CoAlation is significantly increased in CatA-deficient D. discoideum cells. Taken together, these findings indicate that changes in the level of CoA and protein CoAlation are associated with redox regulation when D. discoideum cells are exposed to oxidative and metabolic stress.
2. Materials and methods
2.1. CoA antibodies
Anti-CoA monoclonal antibodies were previously reported [17]. The following antibodies and dilutions were used in Western blotting: anti-CoA, 1F10 (1:6000 dilution); anti-GSH (rabbit polyclonal, 1:2000 dilution, Abcam); and secondary goat anti-mouse immunoglobulin (IgG) conjugated to AlexaFluor680 (1:10000 dilution, Life Technologies) supplemented with 0.02% sodium dodecyl sulphate (SDS).
2.2. Cell culture
D. discoideum cells were cultured in suspension in axenic HL5 medium (ForMedium) supplemented with vitamin B12 (0.6 μg/ml) and folate (0.2 μg/ml) at 22 °C. Cells of the Ax3 strain were used in experiments involving treatment with oxidising agents. Morphogenesis studies were performed using Ax4 cells. HEK293 cells and HEK293 cells stably overexpressing Pank1β (HEK293/Pank1β) were maintained in DMEM (Lonza) supplemented with 50 U/ml penicillin, 0.25 μg/ml streptomycin (Lonza) and 10% foetal bovine serum (FBS, Hyclone) in 5% CO2 and 37 °C.
2.3. Oxidative stress induction
Cells in mid-log phase were collected by centrifugation for 4 min at 2200 rpm and washed three times to remove folate, which can act as antioxidant and neutralise oxidising agents [18]. Cells were re-suspended at a cell density of 2.5 × 106 cells/ml in vitamin-free HL5 media and shaken at 180 rpm at 22 °C for 60 min. This was followed by treatment for 30 min with diamide, H2O2 or t-butylhydroperoxide (TBH). Cells were collected via centrifugation at 3500 rpm and 4 °C and frozen on dry ice.
2.4. Cell development
Log phase D. discoideum cells were collected by centrifugation and washed with KK2 potassium phosphate buffer (20 mM K2HPO4, KH2PO4, pH6.8) before re-suspending at a density of 1 × 108 cells/ml in KK2. 3 × 107 cells were evenly spread over the surface of a freshly prepared 10 cm agar plate (20 ml of 1.5% agar in KK2) to induce starvation and initiate differentiation. Plates were incubated in a humid box at 22 °C in the dark before harvesting by centrifugation. Cells were instantly frozen in dry ice.
2.5. Cell lysis and Western blot analysis
Cell pellets were re-suspended in ice-cold lysis buffer (containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM tetra-sodium pyrophosphate, 50 mM sodium fluoride, 5 mM ethylenediaminetetraacetic acid (EDTA), and 1% Triton X-100) supplemented with freshly prepared 100 mM N-ethylmaleimide (NEM) and 2 x Protease Inhibitor Cocktail (PIC, Roche). To lyse spores, 0.1 mm diameter silica-zirconia beads (Biospec Products) and 2 mm Tungsten Carbide beads (Qiagen) were added and placed in a Tissue Lyser II (Qiagen) set at 30 Hz for 2 min. Total cell lysates were centrifuged for 20 min at 14000 rpm and 4 °C to remove the insoluble fraction. Protein concentration of the supernatant was measured by Bicinchoninic acid (BCA) Protein Assay Kit (Pierce ThermoFisher Scientific). Samples of cell lysates (∼40 μg of protein) were separated by SDS-PAGE on 10% TruPAGE Precast Gels and immunoblotted with anti-CoA mAb as previously described [9].
2.6. Analysis of CoA levels
2.6.1. Extraction of CoA and the recycling assay
The method to extract CoA from Bacillus megaterium was used in this study [19]. CoA extracts were re-suspended in 1 mM DTT and centrifuged for 5 min at 14 000 rpm and 20 °C. CoA level was measured by a modification of the recycling assay developed by Allred and Guy [20,21]. Sets of CoA standards (0.03 μM, 0.1 μM, 0.3 μM, 1 μM, 3 μM, 9 μM) were freshly prepared by serial dilutions of a standard stock solution in 10 mM potassium phosphate buffer (pH 7) and 1 mM DTT.
2.6.2. Normalisation of extracts through measurement of protein concentration
The level of CoA in cell cultures is commonly standardised per total protein [22]. To determine the total amount of protein in the sample from which CoA was extracted, the sample in extraction buffer was mixed with 6 M guanidine chloride and vortexed until the solution became transparent. The fruiting body sample was lysed in a Tissue Lyser II (Qiagen) set at 30 Hz for 2 min, using 0.1 mm diameter silica-zirconia beads (Biospec Products) and 2 mm Tungsten Carbide bead (Qiagen). The samples were centrifuged for 5 min at 14 000 rpm at RT and the BCA assay was used to measure protein concentration.
2.7. Statistical analysis
The concentration of CoA in the samples was extrapolated from the calibration curve and calculated as pmol/mg of total protein (mean ± standard error of the mean (SEM)). Paired t-test was used to compare level of CoA between developmental stages. N represents the number of independent experiments. Significant values were considered at p-value <0.05.
3. Results
3.1. Transcriptional regulation of CoA biosynthetic genes
Protein CoAlation has been suggested to play a role in oxidative stress responses. D. discoideum cells are likely to be exposed to oxidative stress at different stages of their life cycle. We therefore tested whether protein CoAlation may play a role in this system. Firstly, we examined the expression profile of genes required to produce CoA in D. discoideum at different stages of its life cycle (Fig. 1A). We found that all genes were developmentally regulated, with high expression levels in vegetative cells (Fig. 1B). Levels then fall upon starvation, but rise again during multicellular development. This pattern is most evident for Pank, which encodes the rate-limiting enzyme in the CoA biosynthetic pathway. Pank expression was highest in growing cells, then dropped significantly upon starvation, but gradually increased again during the developmental stages (Fig. 1B), reaching a peak during culmination that was comparable to that seen in vegetative cells. These findings support the idea that changes in cellular metabolism and redox regulation could modulate the production of CoA during morphogenesis.
Fig. 1.
The expression of genes involved in CoA biosynthesis, the level of CoA and protein CoAlation in D. discoideum are developmentally regulated. A. Schematic depicting the evolutionary conserved CoA biosynthetic pathway. B. Transcriptional profiling of genes involved in the CoA biosynthetic pathway during growth and development. C. Analysis of total CoA level at developmental stages by an enzymatic recycling assay. Data represent mean ± SEM. *p < 0.05 compared to mound, slug culminant and fruiting body stages, ≠ p < 0.05 compared to slug and culminant stages. D. Western blot analysis of protein CoAlation during vegetative growth and morphogenesis. Protein CoAlation was examined by immunoblotting with anti-CoA antibody.
3.2. Levels of CoA in D. discoideum cells
The expression pattern of genes required for CoA biosynthesis suggested that CoA levels may in turn be developmentally regulated. To test this hypothesis, we determined whether the total level of CoA in D. discoideum cells correlated with Pank mRNA levels. The extraction of CoA from D. discoideum was found to be efficient and experiments with internal standards showed that the recycling assay can measure CoA levels reliably in the picomolar range (Suppl. Figure 1A). The concentration of CoA in vegetative D. discoideum (45 pmol/mg total protein) cells was found to be comparable to that in HEK293 cells (18 pmol/mg total protein) and, as anticipated, less than in HEK293/Pank1β cells (381 pmol/mg total protein) (Suppl. Figure 1B). We next extracted and measured CoA from D. discoideum cells at different developmental stages. CoA levels were found to follow levels of Pank mRNA expression, decreasing after starvation but then rising again during multicellular development (Fig. 1C). CoA levels in the fruiting body stage (24 h of starvation) were statistically significantly higher than the CoA levels of slug, and culminant stages (17 h, and 21 h of starvation, respectively), which correlated with Pank mRNA expression profile.
3.3. Analysis of protein CoAlation during morphogenesis
Recent studies from our laboratory have revealed that the extent of protein CoAlation induced by oxidising agents and metabolic stress is determined by the level of CoA in cells or tissues [9]. Because there are significant fluctuations in CoA levels across different stages of morphogenesis, we immunoblotted these corresponding protein samples with anti-CoA antibody under non-reducing conditions. A distinctive pattern of CoA-modified proteins was observed at different developmental stages (Fig. 1D). Significant protein CoAlation was detected in vegetative cells, especially for a number of proteins in the lower molecular weight region. The signal intensity of these proteins around 50 kDa dropped after the initiation of starvation (aggregation stage), but then gradually increased to reach the maximum at the fruiting body stage. However, for other proteins, CoAlation increased during multicellular development, with a unique pattern evident upon fruiting body formation and spore cell differentiation (Fig. 1D).
3.4. Oxidising agents induce protein CoAlation in D. discoideum cells
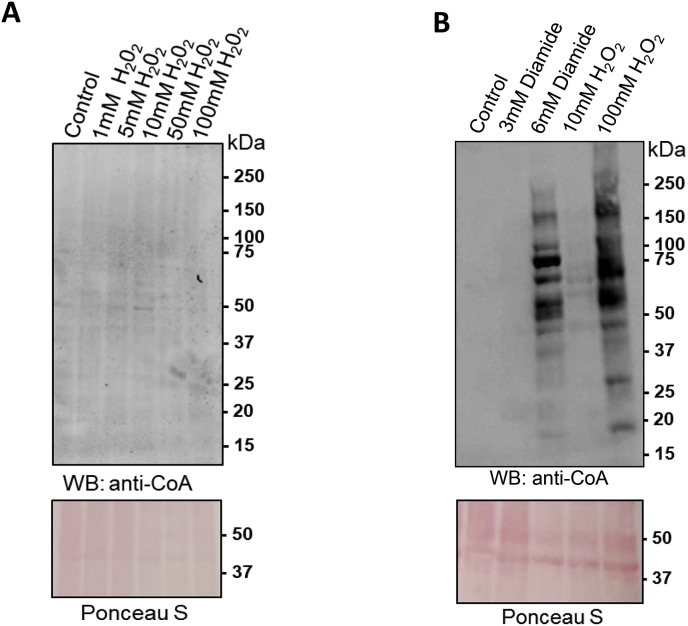
The expression of the Pank gene, the level of CoA and protein CoAlation all change during D. discoideum morphogenesis. Previous studies have revealed that protein CoAlation is induced in mammalian and bacterial cells exposed to oxidising agents [9,10]. We therefore investigated whether covalent protein modification by CoA is also associated with oxidative stress responses in D. discoideum. Taking into account that D. discoideum cells exhibit high resistance to oxidative stress [23], we examined protein CoAlation in a dose-dependent study using relatively high concentrations of oxidising agents. Growing cells were treated with H2O2, diamide or TBH. Cell lysates were prepared and protein extracts separated by SDS-PAGE. Western blot analysis with anti-CoA antibody revealed an increase in protein CoAlation with all treatments compared to control cells (Fig. 2 and Suppl. Figure 2). Levels of protein CoAlation were extensive when cells were treated with diamide, but much weaker with H2O2 and TBH even at very high doses (compared to those used in mammalian cells to induce oxidative stress). Immunoreactive bands were detected in control cells and cells exposed to 1 mM and 3 mM diamide, and protein CoAlation increased markedly when cells were treated with 6 mM or 9 mM diamide (Fig. 2A). In contrast, H2O2- and TBH-induced oxidative stress caused very weak protein CoAlation with up to 1M H2O2 and 0.3 M TBH in the culture medium (Fig. 2B and C). The above results suggest that D. discoideum cells employ different antioxidant defence mechanisms to cope with oxidative stress induced by diamide and H2O2.
Fig. 2.
Dose-dependent induction of protein CoAlation in D. discoideum cells in response to diamide (A, B), H2O2 (C, D) and TBH (E, F). Exponentially growing cells were treated or not treated with diamide, H2O2 and TBH at the indicated concentrations for 30 min. Cells were lysed and protein CoAlation was examined by immunoblotting with anti-CoA antibody.
3.5. Protein CoAlation is strongly induced in CatA-deficient cells treated with H2O2
To deal with redox active species, D. discoideum cells produce antioxidant enzymes, such as superoxide dismutase, catalase, and glutathione peroxidase. D. discoideum expresses two catalase genes (catA and catB), which are differentially regulated both temporally and spatially during morphogenesis [16]. The catA gene is expressed throughout growth and development, while the catB gene is only expressed at the fruiting body stage and spore formation. D. discoideum mutant cells with significantly reduced levels of catA mRNA and enzyme have been shown to exhibit markedly increased sensitivity to H2O2 [24]. We therefore compared the level of protein CoAlation in wild type and CatA deficient D. discoideum cells. As shown in Fig. 3A and B, protein CoAlation was markedly increased in CatA deficient cells treated with low doses of H2O2. These results indicate that CatA efficiently neutralises exogenous and endogenous H2O2, and protects D. discoideum cells from oxidative damage. Thus, in contrast to diamide, the involvement of low molecular weight (LMW) thiols, such as CoA, in antioxidant defence induced by H2O2 may normally be minimal.
Fig. 3.
Analysis of protein CoAlation in wild type and CatA deficient cells in response to TBH. Exponentially growing Ax3 (A) and CatA deficient (B) cells were treated or not treated with H2O2 at the indicated concentrations for 30 min. Cells were lysed and protein CoAlation was examined by immunoblotting with anti-CoA antibody.
4. Discussion
D. discoideum cells alternate between growing as single cells and developing as a multicellular organism after chemotactic aggregation induced by starvation. The expression of several thousand genes governs the programme of morphogenesis [25,26]. In this study we report the transcriptional regulation of genes involved in CoA biosynthesis during the life cycle of D. discoideum. We found that all genes in the CoA biosynthetic pathway are developmentally regulated, showing high expression levels in vegetative cells, then falling dramatically upon starvation, before gradually increasing again during multicellular development. This pattern is most apparent for Pank. Studies in mammalian cells have revealed Pank is a rate-limiting enzyme in the CoA biosynthetic pathway, as overexpression of Pank1β alone is sufficient to significantly increase CoA production in Cos7 and HEK293 cells [9,27]. Similarly, measurements of total level of CoA during normal growth and development in D. discoideum revealed a close correlation with the level of transcripts for CoA biosynthetic genes, especially Pank.
Why is the level of CoA in D. discoideum developmentally regulated? During vegetative growth, D. discoideum cells require large quantities of CoA to produce a diverse range of thioester derivatives (acetyl CoA, malonyl CoA etc.) which play a central role in catabolic and anabolic processes. The findings of this study suggest that CoA, in addition to its well-established functions in cellular metabolism, is also involved in redox regulation in D. discoideum. One reason for this may be that D. discoideum cells act as professional phagocytes that use ROS to kill bacteria and obtain nutrients for growth. Indeed, high levels of ROS production have been observed in vegetative cells upon LPS stimulation [13]. Furthermore, the extent of protein CoAlation changes during growth and morphogenesis, and this pattern correlates with phases of the life cycle of D. discoideum where redox regulation is likely to be necessary for innate immune cell function or the regulation of morphogenesis [14]. Indeed, ROS production is required for innate immune cell function, whilst low concentrations of ROS have been implicated in regulatory signalling pathways and numerous physiological processes during morphogenesis of a variety of systems [15]. Exposure to oxidative stress is thus likely to be a normal feature of the D. discoideum life cycle, and the ability of these cells to withstand oxidative stress by employing different antioxidant defence strategies, including LMW thiols, is likely crucial for their success.
We propose that covalent protein modification by CoA during the life cycle of D. discoideum has the potential to become an integral part of redox sensing and regulation by: a) modulating the activity, stability and subcellular localisation of modified proteins; b) generating a unique binding motif for intra- and inter-molecular interactions; and c) promoting the formation of regulatory signalling complexes. Finally, protein CoAlation may also contribute to maintaining metabolic dormancy of terminally differentiated spore cells which are highly resistant to diverse environmental stress conditions, including oxidative damage.
Acknowledgments
This study was supported by BBSRC (BB/L010410/1), National Academy of Sciences of Ukraine (0110U000692) and Wellcome Trust Investigator Award (095643/A/11/Z).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2019.02.031.
Transparency document related to this article can be found online at https://doi.org/10.1016/j.bbrc.2019.02.031.
Contributor Information
Christopher R.L. Thompson, Email: christopher.thompson@ucl.ac.uk.
Ivan Gout, Email: i.gout@ucl.ac.uk.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Transparency document
References
- 1.Leonardi R., Zhang Y.M., Rock C.O., Jackowski S. Coenzyme A: back in action. Prog. Lipid Res. 2005;44:125–153. doi: 10.1016/j.plipres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Martinez D.L., Tsuchiya Y., Gout I. Coenzyme A biosynthetic machinery in mammalian cells. Biochem. Soc. Trans. 2014;42:1112–1117. doi: 10.1042/BST20140124. [DOI] [PubMed] [Google Scholar]
- 3.Theodoulou F.L., Sibon O.C.M., Jackowski S., Gout I. Coenzyme A and its derivatives: renaissance of a textbook classic. Biochem. Soc. Trans. 2014;42:1025–1032. doi: 10.1042/BST20140176. [DOI] [PubMed] [Google Scholar]
- 4.Leonardi R., Jackowski S. Biosynthesis of pantothenic acid and coenzyme A. EcoSal Plus. 2007;2:1–28. doi: 10.1128/ecosalplus.3.6.3.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breus O., Panasyuk G., Gout I.T., Filonenko V., Nemazanyy I. CoA Synthase is phosphorylated on tyrosines in mammalian cells, interacts with and is dephosphorylated by Shp2PTP. Mol. Cell. Biochem. 2010;335:195–202. doi: 10.1007/s11010-009-0255-6. [DOI] [PubMed] [Google Scholar]
- 6.Gudkova D., Panasyuk G., Nemazanyy I., Zhyvoloup A., Monteil P., Filonenko V., Gout I. EDC4 interacts with and regulates the dephospho-CoA kinase activity of CoA synthase. FEBS Lett. 2012;586:3590–3595. doi: 10.1016/j.febslet.2012.08.033. [DOI] [PubMed] [Google Scholar]
- 7.Nemazanyy I., Panasyuk G., Zhyvoloup A., Panayotou G., Gout I.T., Filonenko V. Specific interaction between S6K1 and CoA synthase: a potential link between the mTOR/S6K pathway, CoA biosynthesis and energy metabolism. FEBS Lett. 2004;578:357–362. doi: 10.1016/j.febslet.2004.10.091. [DOI] [PubMed] [Google Scholar]
- 8.Zhyvoloup A., Nemazanyy I., Panasyuk G., Valovka T., Fenton T., Rebholz H., Wang M.L., Foxon R., Lyzogubov V., Usenko V., Kyyamova R., Gorbenko O., Matsuka G., Filonenko V., Gout I.T. Subcellular localization and regulation of coenzyme a synthase. J. Biol. Chem. 2003;278:50316–50321. doi: 10.1074/jbc.M307763200. [DOI] [PubMed] [Google Scholar]
- 9.Tsuchiya Y., Peak-Chew S., Newell C., Miller-Aidoo S., Mangal S., Zhyvoloup A., Baković J., Malanchuk O., Pereira G., Kotiadis V., Szabadkai G., Duchen M., Campbell M., Rodriguez Cuenca S., Vidal-Puig A., James A., Murphy M.P., Filonenko V., Skehel M., Gout I. Protein CoAlation: a redox-regulated protein modification by coenzyme A in mammalian cells. Biochem. J. 2017;2:2489–2508. doi: 10.1042/BCJ20170129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsuchiya Y., Zhyvoloup A., Bakovic J., Thomas N., Yi Kun Yu B., Das S., Orengo C., Newell C., Ward J., Saladino G., Comitani F., Gervasio F.L., Malanchuk O.M., Khoruzhenko A.I., Filonenko V., Peak-Chew S.Y., Skehel M., Gout I. Protein CoAlation and antioxidant function of Coenzyme A in prokaryotic cells. Biochem. J. 2018;475:1909–1937. doi: 10.1042/BCJ20180043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gout I. Coenzyme A, protein CoAlation and redox regulation in mammalian cells. Biochem. Soc. Trans. 2018;46:721–728. doi: 10.1042/BST20170506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Annesley S.J., Fisher P.R. Dictyostelium discoideum—a model for many reasons. Mol. Cell. Biochem. 2009;329:73–91. doi: 10.1007/s11010-009-0111-8. [DOI] [PubMed] [Google Scholar]
- 13.AU - Zhang X., AU - Soldati T. Detecting, visualizing and quantitating the generation of reactive oxygen species in an amoeba model system. JoVE. 2013 doi: 10.3791/50717. e50717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X., Zhuchenko O., Kuspa A., Soldati T. Social amoebae trap and kill bacteria by casting DNA nets. Nat. Commun. 2016;7:10938. doi: 10.1038/ncomms10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloomfield G., Pears C. Superoxide signalling required for multicellular development of Dictyostelium. J. Cell Sci. 2003;116:3387–3397. doi: 10.1242/jcs.00649. [DOI] [PubMed] [Google Scholar]
- 16.Garcia M.X.U., Alexander H., Mahadeo D., Cotter D.A., Alexander S. The Dictyostelium discoideum prespore-specific catalase B functions to control late development and to protect spore viability. Biochim. Biophys. Acta. 2003;1641:55–64. doi: 10.1016/s0167-4889(03)00064-8. [DOI] [PubMed] [Google Scholar]
- 17.Malanchuk O.M., Panasyuk G.G., Serbin N.M., Gout I.T., Filonenko V.V. Generation and characterization of monoclonal antibodies specific to Coenzyme A. Biopolym. Cell. 2015;31:187–192. [Google Scholar]
- 18.Sroka J., Madeja Z., Michalik M., Przestalski S., Korohoda W. Folic acid, ascorbic acid and sodium selenite restore the motility of Dictyostelium discoideum inhibited by triethyllead. Toxicology. 2002;180:275–292. doi: 10.1016/s0300-483x(02)00419-5. [DOI] [PubMed] [Google Scholar]
- 19.Setlow B., Setlow P. Levels of acetyl coenzyme A, reduced and oxidized coenzyme A, and coenzyme A in disulfide linkage to protein in dormant and germinated spores and growing and sporulating cells of Bacillus megaterium. J. Bacteriol. 1977;132:444–452. doi: 10.1128/jb.132.2.444-452.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allred J.B., Guy D.G. Determination of coenzyme A and acetyl CoA in tissue extracts. Anal. Biochem. 1969;29:293–299. doi: 10.1016/0003-2697(69)90312-1. [DOI] [PubMed] [Google Scholar]
- 21.Tsuchiya Y., Pham U., Gout I. Methods for measuring CoA and CoA derivatives in biological samples: table 1. Biochem. Soc. Trans. 2014;42:1107–1111. doi: 10.1042/BST20140123. [DOI] [PubMed] [Google Scholar]
- 22.Shurubor I.Y., D'Aurelio M., Clark-Matott J., Isakova P.E., Deryabina I.Y., Beal F.M., Cooper J.A., Krasnikov F.B. Determination of coenzyme a and acetyl-coenzyme a in biological samples using HPLC with UV detection. Molecules. 2017;22 doi: 10.3390/molecules22091388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoch B., Begum R. Biochemical basis of the high resistance to oxidative stress in Dictyostelium discoideum. J. Biosci. 2003;28:581–588. doi: 10.1007/BF02703333. [DOI] [PubMed] [Google Scholar]
- 24.Garcia M.X., Foote C., van Es S., Devreotes P.N., Alexander S., Alexander H. Differential developmental expression and cell type specificity of Dictyostelium catalases and their response to oxidative stress and UV-light. Biochim. Biophys. Acta. 2000;1492:295–310. doi: 10.1016/s0167-4781(00)00063-4. [DOI] [PubMed] [Google Scholar]
- 25.Loomis W.F., Shaulsky G. Developmental changes in transcriptional profiles. Dev. Growth Differ. 2011;53:567–575. doi: 10.1111/j.1440-169X.2010.01241.x. [DOI] [PubMed] [Google Scholar]
- 26.Rosengarten R.D., Santhanam B., Fuller D., Katoh-Kurasawa M., Loomis W.F., Zupan B., Shaulsky G. Leaps and lulls in the developmental transcriptome of Dictyostelium discoideum. BMC Genomics. 2015;16:294. doi: 10.1186/s12864-015-1491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rock C.O., Calder R.B., Karim M.A., Jackowski S. Pantothenate kinase regulation of the intracellular concentration of coenzyme A. J. Biol. Chem. 2000;275:1377–1383. doi: 10.1074/jbc.275.2.1377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.