Abstract
It is well established that in bacteria, such as Escherichia coli, OxyR is a transcriptional regulator that mediates the response to H2O2 by activating the OxyR regulon, which consists of many genes that play vital roles in oxidative stress resistance. In Shewanella, OxyR regulates, however, in both reduced and oxidized states, the production of H2O2 scavengers, including major catalase KatB and NADH peroxidase AhpCF. Here we showed that the oxyR mutant carried a plating defect manifested as division arresting, a phenotype that can be completely suppressed by an OxyR variant constitutively existing in oxidized form (OxyRL197P). This effect of OxyRL197P could not be solely attributed to the increment in KatB production, since the suppression was also observed in the absence of KatB. Although expression of peroxidase CcpA was greatly activated by OxyRL197P, the contribution of the protein in alleviating plating defect was negligible. We eventually identified AhpCF as the critical factor, when produced at substantially elevated levels by OxyRL197P, to protect the cell from H2O2 attack. Our data indicate that AhpCF is a particularly important peroxidase in oxidative stress resistance in Shewanella, not only playing a compensatory role for catalase, but also by itself providing sufficient protection from killing of H2O2 generated abiotically.
Keywords: Shewanella, OxyR, oxidative stress, AhpCF, catalase
Introduction
Since the earth became an aerobic environment, one of the major challenges for living organisms has been the oxidative stress imposed by a variety of molecules that are produced by aerobic metabolism and by abiotic reaction. On the top of the list are reactive oxygen species (ROS), including superoxide (O2-), hydrogen peroxide (H2O2), and hydroxyl radical (OH), which cause damage to virtually all biomolecules such as DNA, RNA, lipid, and protein (Imlay, 2013). To adapt to or resist oxidative stress, both prokaryotes and eukaryotes have evolved many sophisticated antioxidant defense systems. In bacteria, cells employ enzymes like catalase, peroxidase, and superoxide dismutase to detoxify H2O2 and superoxide, respectively (Mishra and Imlay, 2012). Expression of the antioxidant defense systems is generally and concertedly controlled at the transcriptional level, a subject extensively studied in Escherichia coli (Imlay, 2015). OxyR, a transcriptional regulator of the LysR family, directly senses H2O2 via the oxidation of two conserved cysteine residues (Cys 199 and Cys 208 in E. coli) and the formation of an intramolecular disulfide bond (Zheng et al., 1998; Lee et al., 2004). In E. coli, oxidized OxyR activates the transcription of the antioxidant genes, including katG (hydrogen peroxidase I), ahpCF [Ahp, NADH peroxidase (originally named alkyl hydroperoxidase)], ccpA (cytochrome c peroxidase), dps (iron-sequestering protein), and oxyS (a small regulatory RNA) (Storz and Altuvia, 1994; Altuvia et al., 1997; Zheng et al., 2001; Khademian and Imlay, 2017). The Ahp system, consisting of AhpC and AhpF performing catalytic and AhpC-reactivating reactions, respectively, is the primary scavenger of microscale H2O2 (Seaver and Imlay, 2001), while KatG is a monofunctional catalase responsible for degrading a large amount of H2O2. AhpC contains two conserved cysteine residues to reduce H2O2 via the formation of a disulfide bond, which is subsequently recycled back by AhpF using NADH as reducing equivalent (Poole, 1996). CcpA, which is located in the periplasm, reduces H2O2 to water by using electrons from soluble cytochrome c (Atack and Kelly, 2007; Khademian and Imlay, 2017).
Although functioning as an activator, E. coli OxyR proteins in both oxidized and reduced forms possess DNA binding activity for a conserved binding motif comprising four regularly spaced ATAG elements (Toledano et al., 1994). OxyR is composed of a helix-turn-helix DNA binging domain (DBD) in the N-terminus and a regulatory domain in the C-terminus where lies the two conservative cysteine residues (Cys199 and Cys208). A C199S mutation locks OxyR in the reduced conformation and deprives of ability to activate oxyS (an untranslated regulatory RNA) transcription (Kullik et al., 1995; Zheng et al., 1998; Zhang et al., 2002).
Shewanella, a group of facultative Gram-negative anaerobes, inhabit in redox-stratified environments and are renowned for their remarkable respiratory abilities (Fredrickson et al., 2008). In recent years, members of the genus regarded as emerging pathogens for human and sea animals have been expanding (Janda and Abbott, 2014). Studies of Shewanella, mostly on genus representative S. oneidensis, suggest that the response to oxidative damage is quite different from that of E. coli. S. oneidensis is substantially more sensitive to H2O2 than E. coli, in concert with the high susceptibility to UV and ionizing radiation (Ghosal et al., 2005; Jiang et al., 2014; Li et al., 2014; Shi et al., 2015). In S. oneidensis, the primary catalase for H2O2 degradation is encoded by the katB gene; the KatB loss results in a plating defect on lysogeny broth (LB) plates (Jiang et al., 2014; Shi et al., 2015). This coincides with a similar scenario with an E. coli oxyR null mutant, whose plating defect is attributed to the lack of catalase induction (Christman et al., 1989). Unlike E. coli OxyR, the S. oneidensis counterpart functions as both a repressor and an activator for the katB gene (Jiang et al., 2014). As a result, compared to the wild-type, an oxyR null mutant produces KatB constitutively at levels between the conditions unchallenged and challenged by H2O2 (Shi et al., 2015; Wan et al., 2018). Despite this, the S. oneidensis oxyR mutant still carries a plating defect, which is even more severe than that of the katB mutant (Shi et al., 2015; Wan et al., 2017). Thus, there must be unknown factors responsible for the plating defect phenotype resulting from the OxyR loss in S. oneidensis.
In this report, we found that OxyRL197P, an OxyR variant that is locked into the oxidized state (Wan et al., 2018), and OxyR functioned, similarly, in alleviating plating defect of ΔoxyR but not of ΔkatB. In ΔoxyR, OxyRL197P, the same as OxyR, activated expression of katB as a main means to scavenge H2O2, correcting the defect. In contrast, in ΔkatB where AhpCF became an exclusive factor for H2O2 removal whereas contribution of other catalases and peroxidases was negligible, expression of ahpCF was elevated by OxyRL197P but not OxyR to levels sufficiently high to compensate for the KatB loss. These findings provide new insights into the complementary roles of H2O2-scavenging enzymes in S. oneidensis.
Materials and Methods
Bacterial Strains, Plasmids and Culture Conditions
All bacterial strains and plasmids used in this study are listed in Table 1. E. coli and S. oneidensis were grown in LB (containing 1% tryptone, 0.5% yeast extract, and 0.5% NaCl) under the aerobic condition at 37 and 30°C for genetic manipulation. When necessary, following chemicals were added to the growth medium: 2,6-diaminopimelic acid (DAP), 0.3 mM; ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; gentamycin, 15 μg/ml; and streptomycin, 100 μg/ml.
Table 1.
Bacterial strains and plasmid used in this study.
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | Host strain for cloning | Lab stock |
| WM3064 | ΔdapA, donor strain for conjugation | W. Metcalf, UIUC |
| S. oneidensis strains | ||
| MR-1 | Wild type | ATCC 700550 |
| HG1328 | ΔoxyR derived from MR-1 | Wan et al., 2017 |
| HG1070 | ΔkatB derived from MR-1 | Shi et al., 2015 |
| HG0956-8 | ΔahpCF derived from MR-1 | Shi et al., 2015 |
| HG2178 | ΔccpA derived from MR-1 | Shi et al., 2015 |
| HG2750 | ΔtolR derived from MR-1 | Gao et al., 2017 |
| HG0956-1070 | ΔkatBΔahpC derived from MR-1 | Shi et al., 2015 |
| HG1070-0725 | ΔkatBΔkatG-1 derived from MR-1 | This study |
| HG1070-4405 | ΔkatBΔkatG-2 derived from MR-1 | This study |
| HG1070-2178 | ΔkatBΔccpA derived from MR-1 | This study |
| Plasmids | ||
| pHGM01 | AprGmrCmr,att-based suicide vector | Jin et al., 2013 |
| pHG101 | Kmr, promoterless broad-host vector | Wu et al., 2011 |
| pHGEI01 | Kmr, integrative lacZ reporter vector | Fu et al., 2013 |
| pHGE-Ptac | Kmr,IPTG-inducible expression vector | Luo et al., 2013 |
| pHGE-Ptac-ccpA | Vector for inducible expression of CcpA | This study |
| pHGE-Ptac-ahpCF | Vector for inducible expression of ahpCF | This study |
| pHGEI01-PkatB | Reporter vector carrying PkatB-lacZ | This study |
| pHGEI01-PccpA | Reporter vector carrying PccpA-lacZ | This study |
| pHGEI01-Pdps | Reporter vector carrying Pdps-lacZ | This study |
| pHGEI01-PahpC | Reporter vector carrying PahpC-lacZ | This study |
| pHGEI01-PkatG-1 | Reporter vector carrying PkatG-1-lacZ | This study |
Mutagenesis, Complementation of Mutant Strains
In-frame deletion strains for S. oneidensis were constructed according to the att-based Fusion PCR method as described previously (Jin et al., 2013). In brief, two fragments flanking the target gene were amplified by PCR with primers containing attB and the gene specific sequence, which were linked by a linker sequence via second round of PCR. The fusion fragments were integrated into plasmid pHGM01 by using Gateway BP clonase II enzyme mix (Invitrogen). The resultant plasmid was introduced in E. coli WM3064 and transferred to S. oneidensis by conjugation. Integration of the mutagenesis constructs into the chromosome was selected by resistance to gentamycin and confirmed by PCR. Verified trans-conjugants were grown in LB broth without NaCl and plated on LB supplemented with 10% sucrose. Gentamycin-sensitive and sucrose-resistant colonies were screened by PCR for deletion of the target gene. To facilitate growth of mutants, catalase (from bovine liver, Sigma) was added onto plates at the final resolution step for genes critical for survival through ROS. Mutants were verified by sequencing the mutated regions.
Genetic complementation of mutants with an apparent phenotype was performed with plasmids pHG101 or pHGE-Ptac as described before (Wu et al., 2011; Luo et al., 2013). For complementation of genes next to their promoter, a fragment containing the gene of interest and its native promoter was generated by PCR and cloned into pHG101. For the remaining genes, the gene of interest was generated by PCR and introduced into pHGE-Ptac under the control of promoter Ptac. After sequencing verification, the resulting vectors were transferred into the relevant strains via conjugation.
Microscopy
Shewanella oneidensis were cultivated to the mid-logarithmic (OD600 of 0.4) and spotted onto a glass slide containing LB medium. Motic BA310 light microscope (Motic, Xiamen, China) was employed to observe the cell morphology. Micrographs were captured with a Moticam 2306 charged-coupled-device camera.
Spotting Assay
The spotting assay was used to evaluate the plating defect on LB plates. Cells of the log phase (OD600 of 0.4) were collected by centrifugation and adjusted to 109 cell/ml, which was set as the undiluted (dilution factor 0). Ten-fold serial dilutions were prepared with fresh LB medium. Five microliters of each dilution was spotted onto LB plates. The plates were incubated for 24 h or longer in dark before being photographed. All experiments were repeated at least three times.
Enzyme Activity Assay
Catalase and peroxidase activities were detected as described previously (Fridovich, 1984; Wayne and Diaz, 1986). For double staining to differentiate catalase and peroxidase, 2 ml of mid-log cultures were harvested by centrifugation and lysed by sonication. Forty microliters of total unboiled protein extract were loaded on a 10% non-denaturing polyacrylamide gel (PAGE) and run at 100 V at 4°C. After completion of the electrophoresis, the gel was washed in water and soaked in 100 ml of solution containing 0.01 ml of 30% H2O2, 50 mg of diaminobenzidine (Sigma) for 20 min. Then gel was washed by water and suspended in 5 mM H2O2, briefly washed in water and incubated in a solution containing 2% ferric chloride and 2% potassium ferricyanide. Catalase yielded clear bands on a green-stained background while peroxidase produced blue bands.
β-Galactosidase Activity Assay
β-Galactosidase activity assay was used to determine gene expression. The sequence in sufficient length (∼400 bp) upstream of gene of interest was amplified and inserted in front of the full-length E. coli lacZ gene in plasmid pHGEI01 (Fu et al., 2014). The resulting vector was verified by sequencing and then transformed into E. coli WM3064 and then conjugated with relevant S. oneidensis strains. Cells at the mid-log phase were collected and centrifugation, washed with PBS, and treated with lysis buffer (0.25 M Tris/HCl, 0.5% Triton X-100, pH 7.5) for 30 min. Soluble protein was collected by centrifugation and applied for enzyme assay by adding o-nitrophenyl-β-D-galactopyranoside (ONPG) (4 mg/ml). β-Galactosidase activity was determined by monitoring color development at 420 nM with a Synergy 2 Pro200 Multi-Detection Microplate Reader (Tecan), presented as Miller units.
Results
oxyR Mutant Cells Die Quickly on LB Plates Without Morphological Changes
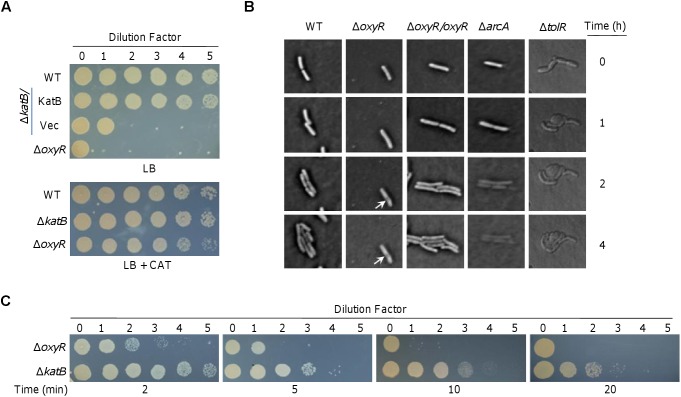
Deletion of the S. oneidensis oxyR gene (ΔoxyR) results in a serious plating defect (Jiang et al., 2014; Shi et al., 2015). Plating defect is a common phenotype that has been observed in many other bacteria, such as E. coli, whose OxyR proteins act as a positive regulator (Maciver and Hansen, 1996; Hahn et al., 2002). On LB plates, a drop (5 μl) of the culture containing 108 CFU/ml grew as the wild-type, whereas none of a 10-fold dilution series showed visible growth (Figure 1A). In order to figure out the nature of this phenotype, we visualized growth of cells of relevant S. oneidensis strains on LB plates under a phase-contrast microscope. After spotted on LB agar, the wild-type cells were able to divide (Figure 1B), leading to colony formation on LB plates (Figure 1A). In contrast, ΔoxyR cells stayed in the single-cell state for 4 h and even longer, implicating that the mutation deprives cells of ability to proliferate (Figure 1B).
FIGURE 1.
Characteristics of the plating defect of ΔoxyR and ΔkatB on LB plates. (A) Spotting assay. Cells grown to the mid-log phase (OD600 of ∼0.4) were regarded as undiluted (dilution factor 0, ∼108 CFU/ml) and were subjected to 10-fold series dilution. Five microliters of each dilution was dropped on LB plates. WT, the wild-type; Vec, empty vector. (B) Morphological observation under microscope. Cells of indicated strains at mid-log were spotted on slide with LB agar as described in the Materials and Methods. Pictures were captured at indicated times. The ΔarcA strain was incubated with 0.5 mM SDS. (C) Effects of catalase on rescuing viability of ΔoxyR and ΔkatB. Catalase (2,000 U) was applied to the drops at the indicated times. Experiments were performed at least three times, and representative results were shown.
The elongated ΔoxyR cells became damaged in the cell envelope at the middle after 2 h incubation (Figure 1B). To investigate whether the plating defect is due to, at least in part, impaired cell envelope, we performed a comparative study with arcA and tolR mutants. In S. oneidensis, ArcA and TolR are a global regulator implicated in diverse processes and a protein involved in peptidoglycan recycling and cell division, respectively; the consequences caused by their loss are the severe defect in the outer-membrane and peptidoglycan layer (Gao et al., 2008, 2017). As a result, ΔarcA cells are very sensitive to SDS and ΔtolR cells form cell-chain with blebs (Gao et al., 2017; Wan et al., 2017). Under the microscope, we observed that ΔarcA cells retained ability to divide in the presence of 0.5% SDS (Figure 1B). However, the newly generated cells died quickly with the cell border increasingly blurring, and eventually becoming invisible. This phenomenon indicates that the cell envelope is dissolved by SDS, consistent with the defect in the outer-membrane. Expectedly, ΔtolR cells in chain quickly formed blebs at the cell surface without stopping division. The difference in the manners of death among ΔoxyR, ΔarcA, and ΔtolR cells, implies that the oxyR mutation may not affect cell envelope integrity.
To determine whether the plating defect phenotype is due to cell death, we performed catalase-rescuing assays with the ΔoxyR culture diluted 100-fold, which could not grow on LB plates (Figure 1A). Before and after culture dropping, catalase solution was applied to culture droplets at different times. As reported before (Shi et al., 2015), when catalase was added to LB plates before dropping, the plating defect of ΔoxyR could be fully corrected (Figure 1C). In the case of the application after culture dropping, however, the effects of catalase on the culture droplets were time-dependent (Figure 1C). When catalase was added 2 min later, growth was largely recovered. The rescuing effects became less effective and undetectable when catalase was applied 5 and 10 min after the culture dropping, respectively. These data indicate that the S. oneidensis ΔoxyR cells are nearly immediately damaged by H2O2 on LB plates, further supporting that cell envelope integrity and cell division are not the primary cause for death because defects in cell envelope do not kill cells rapidly and are not affected by cell density (Zerbib, 2017).
A similar but less severe plating defect has also been observed with the loss of KatB (ΔkatB) (Shi et al., 2015) (Figure 1A). We therefore reasoned that catalase may rescue viability of the ΔkatB strain in a similar manner. Indeed, ΔkatB cells recovered ability to grow on LB plates if catalase was added in time (Figure 1C). Apparently, the rescuing effects of catalase on the ΔkatB strain appear more effective than on the ΔoxyR strain. This is expected because the plating defect of the ΔoxyR strain is more severe. Nevertheless, it is clear that both strains die of the same damage caused by H2O2, which is generated abiotically on LB plates (Shi et al., 2015). These data collectively indicate that the plating defects of ΔoxyR and ΔkatB strains are due to rapid cell death, rather than the inability to divide.
KatB Is Not an Exclusive Factor for the Rescuing Effect of OxyRL197P on the oxyR Mutant
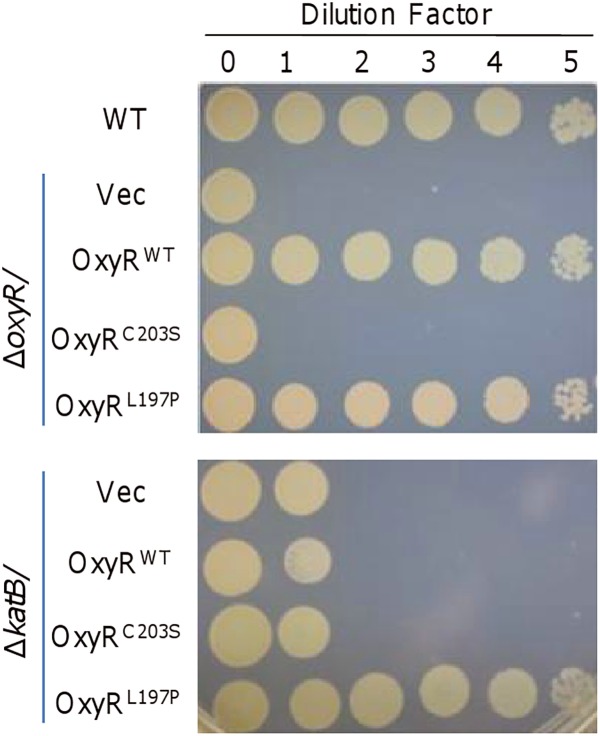
To further investigate into mechanisms for the plating defect of the oxyR mutant, we compared the complementary effects of three OxyR variants, OxyRWT (wild-type), OxyRC203S, and OxyRL197P (Binnenkade et al., 2014; Jiang et al., 2014). In S. oneidensis, OxyRWT proteins exist in both reduced and oxidized states, which could convert to each other through the formation of intramolecular disulfide bond between Cys-203 and Cys-212. While OxyRC203S is locked in reduced form (as a repressor only for katB and dps) because of the inability to form disulfide bond, OxyRL197P functions exclusively as an activator for all OxyR regulon members (Jiang et al., 2014; Wan et al., 2018). Plasmid pHG101 carrying the oxyR gene with respective mutations was used to produce OxyRC203S and OxyRL197P, whose expression was driven by the oxyR own promoter (Wu et al., 2011). Clearly, both OxyRWT and OxyRL197P could completely eliminate the plating defect of ΔoxyR but OxyRC203S failed (Figure 2).
FIGURE 2.
KatB is not an exclusive factor for the rescuing effect of OxyRL197P. The experiment was performed the same as described in Figure 1. Expression of OxyRC203S and OxyRL197P were driven by their own promoters in the background of ΔoxyR (upper) and ΔkatB (lower). Experiments were performed at least three times, and representative results were shown.
Given that the plating defect is observed in strains lacking OxyR or KatB and overproduction of KatB, which can be achieved with OxyRL197P, could greatly improve the viability of the ΔoxyR strain (Wan et al., 2018), it is conceivable that KatB would be the key factor for survival of S. oneidensis on LB plates. To test this, we compared effects of OxyRWT, OxyRC203S, and OxyRL197P on viability of the ΔkatB strain (Figure 2). Expectedly, OxyRC203S did not significantly alleviate the plating defect of ΔkatB. A similar result was obtained with OxyRWT, supporting that the KatB loss is critically responsible for the plating defect. However, in the presence of OxyRL197P, ΔkatB cells surprisingly displayed viability comparable to that of the wild-type (Figure 2). This phenomenon suggests that other unknown factors, which could be activated by OxyRL197P, are able to compensate for the KatB loss.
CcpA Overproduced by OxyRL197P Does Not Account for the Plating Defect
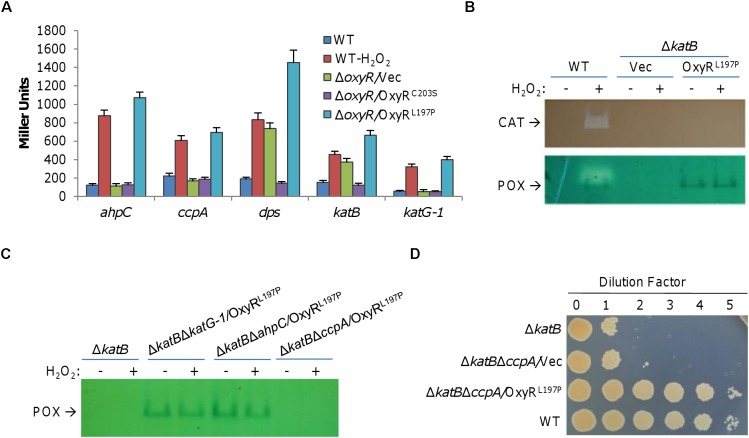
The predicted S. oneidensis OxyR regulon is rather small comparing to those reported in other bacteria, comprising five members (Wan et al., 2018). In addition to katB, ahpCF, and dps mentioned above, two remaining members are katG-1 (SO_0725) and ccpA, encoding catalase/peroxidase HPI and cytochrome c peroxidase, respectively. Single-gene mutants for all of these genes but katB are indistinguishable from the wild-type, with respect to growth in liquid LB and LB plates (Jiang et al., 2014; Shi et al., 2015). By using integrative lacZ-reporters (Jiang et al., 2014), we showed that ahpC, ccpA, and katG-1 required OxyRL197P for expression induction but were not affected by OxyRC203S (Figure 3A). In contrast, expression of both katB and dps was affected negatively and positively by OxyRC203S and OxyRL197P, respectively. Despite this difference, all predicted members of the OxyR regulon in the presence of OxyRL197P were expressed at drastically increased levels, validating that OxyRL197P functions as an activator only.
FIGURE 3.
CcpA overproduced by OxyRL197P does not account for the plating defect. (A) Expression of indicated genes in the presence of OxyRC203S and OxyRL197P. Cells grown to the mid-log phase were treated with 0.2 mM H2O2 for 10 min or not, and then harvested for the assays. For ΔoxyR strains, H2O2 treatment did not affect expression and results from treated samples were shown. β-Galactosidase assays were carried out with lacZ reporters and activities were reported as the mean ± SD (n ≥ 4). (B) Double staining analysis. Cells of the mid-log phase were collected before and after 0.2 mM H2O2 treatment for 10 min. Proteins of indicated cells lysates were separated by native PAGE and stained for catalase (seen as clear zone) and peroxidase (seen as dark band) activities as indicated. (C) Peroxidase staining verified expression of peroxidase. Cells were collected before and after 0.2 mM H2O2 treatment for 10 min. Proteins of indicated cells lysates were separated by native PAGE and stained for peroxidase. (D) CcpA is not the main cause for plating defect alleviating. Droplet assay was used to verify the impact of CcpA on plating defect. For B, C, and D, experiments were performed at least three times, and representative results were shown.
The causing agent of the plating defect is H2O2 that is generated abiotically on LB plates. As OxyRL197P may up-regulate expression of genes for diverse functions, we focused on proteins that may functionally replace KatB. To this end, we performed a double staining for both catalase and peroxidase in the wild-type cells producing OxyRL197P on native PAGE (Figure 3B). By this way, activity of catalase would be identified by a clear band, while peroxidase yields a blue band (Wayne and Diaz, 1986). Results revealed two bands: one for catalase, which is KatB as confirmed by the result with ΔkatB/OxyRL197P, and the other for peroxidase (Figure 3B). To identify this peroxidase, we knocked out all candidate genes for peroxidases from the ΔkatB background. As shown in Figure 3C, in the presence of OxyRL197P, the blue band remained in ΔkatBΔkatG-1 and ΔkatBΔahpC but disappeared in ΔkatBΔccpA, indicating that CcpA is the peroxidase. For validation, we placed the ccpA coding sequence under the control of isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible promoter Ptac within pHGE-Ptac and the expression vectors were introduced into the wild-type. Double staining revealed that the blue band became intensified with IPTG concentrations (Supplementary Figure S1). Thus, OxyRL197P greatly enhances the production of CcpA, which was the only peroxidase that could be detected by the double staining method.
CcpA is a periplasmic cytochrome c peroxidase, catalyzing the reduction of H2O2, which is regarded as an alternative terminal electron acceptor in bacteria (Schütz et al., 2011; Khademian and Imlay, 2017). Because many important cellular components outside the S. oneidensis cytoplasm are vulnerable to H2O2 (Shi et al., 2015), CcpA may be critical for ΔkatB survival. To test this, spotting assays of series dilution cultures prepared from relevant strains were performed (Figure 3D). Additional removal of CcpA (ΔkatBΔccpA) did not further reduce viability of KatB-deficient cells on LB plates. More importantly, the ΔkatBΔccpA strain with OxyRL197P, the same as ΔkatB, recovered ability to grow on LB plates (Figure 3D). These results rule out the possibility that CcpA is the peroxidase that is able to compensate for the KatB loss.
Ahp System Is Likely a Crucial Factor for Rescuing the Plating Defect
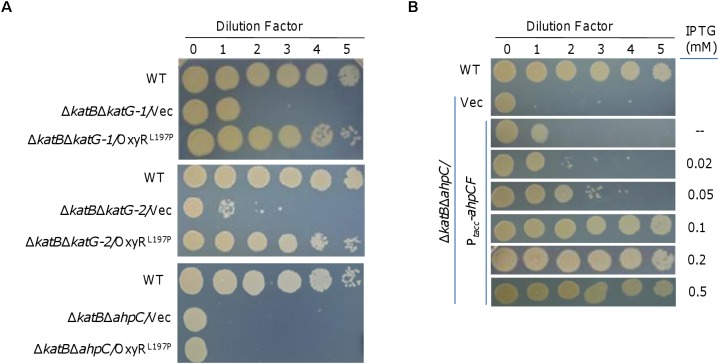
Given that KatB and CcpA are the only catalase and peroxidase identified by the staining method on native PAGE, it is clear that the method is not sufficiently sensitive to detect all H2O2-degrading proteins encoded in the S. oneidensis genome. In S. oneidensis, peroxidase Ahp plays a protective role against oxidative stresses imposed by both H2O2 and organic peroxides (Li et al., 2014), whereas physiological significance of dual-function (catalase/peroxidase) HPI, KatG-1, remains unknown (Jiang et al., 2014). Consistently, we found that the ΔkatBΔahpC strain was further impaired in viability on LB plates, whereas the ΔkatBΔkatG-1 strain displayed the plating defect the same as the ΔkatB strain (Figure 4A). It was worth mentioning that KatG-2 (SO_4405), another dual-function HPI according to the genome annotation whose expression is OxyR-independent (Jiang et al., 2014; Wan et al., 2018), played a negligible role in influencing the plating defect (Figure 4A).
FIGURE 4.
Ahp system is likely a crucial factor for rescuing the plating defect. (A) Spotting assay to screen for the crucial catalase and/or peroxidase that could correct the plating defect of ΔkatB. Mid-log cultures of indicated strains were subject to 10-fold serial dilution, and 5 μl of each dilution was spotted onto plates. (B) Effects of AhpCF expressed to varying levels on the plating defect of ΔkatBΔahpC. Expression of AhpCF was driven by IPTG-inducible Ptac. In both A and B, experiments were performed at least three times, and representative results were shown.
In order to identify the crucial catalase and/or peroxidase that could correct the plating defect of ΔkatB, we expressed OxyRL197P in the ΔkatBΔahpC, ΔkatBΔkatG-1, and ΔkatBΔkatG-2 strains and monitored viability on LB plates. The result revealed that OxyRL197P fully recovered viability of the ΔkatBΔkatG-1 and ΔkatBΔkatG-2 strains, but had no noticeable effect on the ΔkatBΔahpC strain (Figure 4A). These data suggest that Ahp is likely the peroxidase, when overproduced in the presence of OxyRL197P, that suppresses the plating defect of the ΔkatB strain.
To confirm that Ahp in excess could compensate for the loss of KatB, we placed the ahpCF operon under the control of IPTG-inducible promoter Ptac within pHGE-Ptac (Luo et al., 2013). It was immediately evident that the ahpCF operon expressed in trans had complementary effects on viability of the ΔkatBΔahpC strain (Figure 4B). In the absence of IPTG, viability was nearly restored to that of the ΔkatB strain because the promoter is slightly leaky (Luo et al., 2013; Shi et al., 2015; Meng et al., 2018). In the presence of IPTG less than 0.1 mM, viability of the ΔkatBΔahpC strain was found to be improved increasingly with IPTG levels (Figure 4B). A full restoration was achieved with IPTG at 1 mM and above. These data thus validate that Ahp in excess rescues the growth defect resulting from compromised H2O2 degrading capacity.
Discussion
The mechanism adopted by OxyR to cope with H2O2 stress is through activation of genes involved in adapting to and resisting oxidative stress, an understanding mostly built on the studies of the subject in E. coli (Imlay, 2013). In the absence of OxyR, expression of genes for major H2O2-degrading enzymes could not be activated, leading to plating defect, a well-documented common phenotype for oxyR mutants (Maciver and Hansen, 1996; Hahn et al., 2002). However, recent studies have revealed that OxyR proteins in some bacteria, including S. oneidensis, mediate expression of genes for major catalases and Ahp both positively and negatively (Loprasert et al., 2000; Tseng et al., 2003; Jiang et al., 2014; Wan et al., 2018). In this case, major H2O2-degrading enzymes are produced more in oxyR mutants than in the wild-type when cells are grown under normal conditions. Despite this, the OxyR loss still results in plating defect phenotype, at least in S. oneidensis (Jiang et al., 2014).
We have previously illustrated that the plating defect is due to H2O2 generated abiotically on LB plates (Shi et al., 2015). In this study, we uncovered that when properly diluted, cells lacking OxyR or major catalase KatB are unable to divide. Although membrane impairments are observed with extended incubation, they do not appear to be the cause for the plating defect. In S. oneidensis, KatB is the predominant force for H2O2 degradation, and its absence results in the plating defect. Given that effects of OxyR and KatB loss are highly similar, it is conclusive that the plating defect could be attributable to overall reduced H2O2-degrading capacity. In contrast, cells lacking Ahp, which is also a primary H2O2-degrading enzyme (Seaver and Imlay, 2001), are normal (Shi et al., 2015). Unlike catalase, Ahp decomposes multiple peroxides in addition to H2O2, including organic hydroperoxides (Niimura et al., 1995; Li et al., 2014). Despite this, it is clear that Ahp could not fully compensate for the loss of catalase in general, as shown here and in many other bacteria (Mongkolsuk et al., 2000; Seaver and Imlay, 2001; Cosgrove et al., 2007).
Given that the oxyR mutant produces KatB at levels higher than that in the wild-type grown under normal conditions, the catalase is not the exclusive factor for the plating defect (Shi et al., 2015). Indeed, we found here that Ahp is another critical factor for the plating defect. Clearly, enhanced production of Ahp, as a result of either OxyRL197P up-regulation or manipulated over-expression, is able to correct the defect resulting from the KatB loss. The difference in rescuing effects of OxyR and OxyRL197P on the plating defect of the katB mutant can be confidently explained by their redox states. OxyR proteins are always present in reduced and oxidized forms, which are in a dynamic equilibrium (Wan et al., 2018). When confronting H2O2 stress, OxyRoxi outcompetes OxyRred by higher affinity to target genes and activates expression. As OxyRWT could not be completely oxidized and OxyRL197P is locked in the oxidized state, the former is less effective in transcriptional activation as the latter.
Although the S. oneidensis genome encodes multiple H2O2-degrading enzymes, double staining for catalase and peroxidase has only identified activity of KatB and CcpA. Notably, activity for both enzymes is detected only in the wild-type cells challenged by H2O2 or with OxyRL197P. The failure to identify Ahp is likely a result of electron donors used in the analysis that could not serve as a cognate donor for Ahp (Trend et al., 2001). In parallel, activity of neither catalase nor peroxidase for HPI enzymes KatG-1 and KatG-2 is detected by the method, even in the presence of OxyRL197P. This may be readily explained by the lack of enzyme activity and extremely low expression, a scenario reported before (Jiang et al., 2014). Given that it is common that multiple enzymes for combating oxidative stress are encoded in bacteria and many of them remain functionally elusive (Mishra and Imlay, 2012), physiological impacts of KatG-1 and KatG-2 may still be worth investigation. For this, CcpA serves a good example. In S. oneidensis, CcpA is dispensable during aerobic growth since CcpA depletion leads to no phenotype (Jiang et al., 2014). However, the enzyme plays a protective role against oxidative stress under anaerobic conditions (Schütz et al., 2011). Similarly, CcpA is a potent degrader of H2O2 in anaerobic environment in E. coli (Khademian and Imlay, 2017). The mechanism underpinning this is that CcpA requires reductive activation for full activity, which depends on the absence of oxygen (Pulcu et al., 2012).
Author Contributions
FW conducted and performed the experiments. WS was involved with the microscope result. FW and JY contributed to data discussion and analysis. FW designed and supervised the study, and wrote the manuscript with HG.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This research was supported by National Natural Science Foundation of Zhejiang Province (LZ17C010001), the Fundamental Research Funds for the Central Universities, and by startup funds for FW.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00439/full#supplementary-material
References
- Altuvia S., Weinstein-Fischer D., Zhang A., Postow L., Storz G. (1997). A small, stable RNA induced by oxidative stress: roles as a pleiotropic regulator and antimutator. Cell 90 45–53. 10.1016/S0092-8674(00)80312-8 [DOI] [PubMed] [Google Scholar]
- Atack J. M., Kelly D. J. (2007). Structure, mechanism and physiological roles of bacterial cytochrome c peroxidases. Adv. Microb. Physiol. 52 73–106. 10.1016/S0065-2911(06)52002-8 [DOI] [PubMed] [Google Scholar]
- Binnenkade L., Teichmann L., Thormann K. M. (2014). Iron triggers λSo prophage induction and release of extracellular DNA in Shewanella oneidensis MR-1 biofilms. Appl. Environ. Microbiol. 80 5304–5316. 10.1128/AEM.01480-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman M. F., Storz G., Ames B. N. (1989). OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc. Natl. Acad. Sci. U.S.A. 86 3484–3488. 10.1073/pnas.86.10.3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove K., Coutts G., Jonsson I. M., Tarkowski A., Kokai-Kun J. F., Mond J. J., et al. (2007). Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 189 1025–1035. 10.1128/JB.01524-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson J. K., Romine M. F., Beliaev A. S., Auchtung J. M., Driscoll M. E., Gardner T. S., et al. (2008). Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6 592–603. 10.1038/nrmicro1947 [DOI] [PubMed] [Google Scholar]
- Fridovich I. (1984). Overview: biological sources of O2-. Methods Enzymol. 105 59–61. 10.1016/S0076-6879(84)05008-4 [DOI] [PubMed] [Google Scholar]
- Fu H., Chen H., Wang J., Zhou G., Zhang H., Zhang L., et al. (2013). Crp-dependent cytochrome bd oxidase confers nitrite resistance to Shewanella oneidensis. Environ. Microbiol. 15 2198–2212. 10.1111/1462-2920.12091 [DOI] [PubMed] [Google Scholar]
- Fu H., Jin M., Ju L., Mao Y., Gao H. (2014). Evidence for function overlapping of CymA and the cytochrome bc1 complex in the Shewanella oneidensis nitrate and nitrite respiration. Environ. Microbiol. 16 3181–3195. 10.1111/1462-2920.12457 [DOI] [PubMed] [Google Scholar]
- Gao H., Wang X., Yang Z. K., Palzkill T., Zhou J. (2008). Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genomics 9:42. 10.1186/1471-2164-9-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T., Meng Q., Gao H. (2017). Thioesterase YbgC affects motility by modulating c-di-GMP levels in Shewanella oneidensis. Sci. Rep. 7:3932. 10.1038/s41598-017-04285-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal S., Hemminger J. C., Bluhm H., Mun B. S., Hebenstreit E. L., Ketteler G., et al. (2005). Electron spectroscopy of aqueous solution interfaces reveals surface enhancement of halides. Science 307 563–566. 10.1126/science.1106525 [DOI] [PubMed] [Google Scholar]
- Hahn J. S., Oh S. Y., Roe J. H. (2002). Role of OxyR as a peroxide-sensing positive regulator in Streptomyces coelicolor A3(2). J. Bacteriol. 184 5214–5222. 10.1128/JB.184.19.5214-5222.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A. (2013). The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11 443–454. 10.1038/nrmicro3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A. (2015). Diagnosing oxidative stress in bacteria: not as easy as you might think. Curr. Opin. Microbiol. 24 124–131. 10.1016/j.mib.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda J. M., Abbott S. L. (2014). The genus Shewanella: from the briny depths below to human pathogen. Crit. Rev. Microbiol. 40 293–312. 10.3109/1040841X.2012.726209 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Dong Y., Luo Q., Li N., Wu G., Gao H. (2014). Protection from oxidative stress relies mainly on derepression of OxyR-dependent KatB and Dps in Shewanella oneidensis. J. Bacteriol. 196 445–458. 10.1128/JB.01077-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M., Jiang Y., Sun L., Yin J., Fu H., Wu G., et al. (2013). Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PLoS One 8:e75610. 10.1371/journal.pone.0075610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademian M., Imlay J. A. (2017). Escherichia coli cytochrome c peroxidase is a respiratory oxidase that enables the use of hydrogen peroxide as a terminal electron acceptor. Proc. Natl. Acad. Sci. U.S.A. 114 E6922–E6931. 10.1073/pnas.1701587114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullik I., Stevens J., Toledano M. B., Storz G. (1995). Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J. Bacteriol. 177 1285–1291. 10.1128/jb.177.5.1285-1291.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Lee S. M., Mukhopadhyay P., Kim S. J., Lee S. C., Ahn W. S., et al. (2004). Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat. Struct. Mol. Biol. 11 1179–1185. 10.1038/nsmb856 [DOI] [PubMed] [Google Scholar]
- Li N., Luo Q., Jiang Y., Wu G., Gao H. (2014). Managing oxidative stresses in Shewanella oneidensis: intertwined roles of the OxyR and OhrR regulons. Environ. Microbiol. 16 1821–1834. 10.1111/1462-2920.12418 [DOI] [PubMed] [Google Scholar]
- Loprasert S., Fuangthong M., Whangsuk W., Atichartpongkul S., Mongkolsuk S. (2000). Molecular and physiological analysis of an OxyR-regulated ahpC promoter in Xanthomonas campestris pv. phaseoli. Mol. Microbiol. 37 1504–1514. 10.1046/j.1365-2958.2000.02107.x [DOI] [PubMed] [Google Scholar]
- Luo Q., Dong Y., Chen H., Gao H. (2013). Mislocalization of Rieske protein PetA predominantly accounts for the aerobic growth defect of Tat mutants in Shewanella oneidensis. PLoS One 8:e62064. 10.1371/journal.pone.0062064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver I., Hansen E. J. (1996). Lack of expression of the global regulator OxyR in Haemophilus influenzae has a profound effect on growth phenotype. Infect Immun. 64 4618–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q., Yin J., Jin M., Gao H. (2018). Distinct nitrite and nitric oxide physiology in Escherichia coli and Shewanella oneidensis. Appl. Environ. Microbiol. 84:e0559–18. 10.1128/AEM.00559-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S., Imlay J. (2012). Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch. Biochem. Biophys. 525 145–160. 10.1016/j.abb.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Whangsuk W., Vattanaviboon P., Loprasert S., Fuangthong M. (2000). A Xanthomonas alkyl hydroperoxide reductase subunit C (ahpC) mutant showed an altered peroxide stress response and complex regulation of the compensatory response of peroxide detoxification enzymes. J. Bacteriol. 182 6845–6849. 10.1128/JB.182.23.6845-6849.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y., Poole L. B., Massey V. (1995). Amphibacillus xylanus NADH oxidase and Salmonella typhimurium alkyl-hydroperoxide reductase flavoprotein components show extremely high scavenging activity for both alkyl hydroperoxide and hydrogen peroxide in the presence of S. typhimurium alkyl-hydroperoxide reductase 22-kDa protein component. J. Biol. Chem. 270 25645–25650. 10.1074/jbc.270.43.25645 [DOI] [PubMed] [Google Scholar]
- Poole L. B. (1996). Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 2. Cystine disulfides involved in catalysis of peroxide reduction. Biochemistry 35 65–75. 10.1021/bi951888k [DOI] [PubMed] [Google Scholar]
- Pulcu G. S., Frato K. E., Gupta R., Hsu H. R., Levine G. A., Hendrich M. P., et al. (2012). The diheme cytochrome c peroxidase from Shewanella oneidensis requires reductive activation. Biochemistry 51 974–985. 10.1021/bi201135s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz B., Seidel J., Sturm G., Einsle O., Gescher J. (2011). Investigation of the electron transport chain to and the catalytic activity of the diheme cytochrome c peroxidase CcpA of Shewanella oneidensis. Appl. Environ. Microbiol. 77 6172–6180. 10.1128/AEM.00606-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver L. C., Imlay J. A. (2001). Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183 7173–7181. 10.1128/JB.183.24.7173-7181.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Wan F., Mao Y., Gao H. (2015). Unraveling the mechanism for the viability deficiency of Shewanella oneidensis oxyr null mutant. J. Bacteriol. 197 2179–2189. 10.1128/JB.00154-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G., Altuvia S. (1994). OxyR regulon. Methods Enzymol. 234 217–223. 10.1016/0076-6879(94)34088-9 [DOI] [PubMed] [Google Scholar]
- Toledano M. B., Kullik I., Trinh F., Baird P. T., Schneider T. D., Storz G. (1994). Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 978 897–909. 10.1016/S0092-8674(94)90702-1 [DOI] [PubMed] [Google Scholar]
- Trend M. A., Jorgensen M. A., Hazell S. L., Mendz G. L. (2001). Oxidases and reductases are involved in metronidazole sensitivity in Helicobacter pylori. Int. J. Biochem. Cell Biol. 33 143–153. 10.1016/S1357-2725(00)00085-6 [DOI] [PubMed] [Google Scholar]
- Tseng H. J., McEwan A. G., Apicella M. A., Jennings M. P. (2003). OxyR acts as a repressor of catalase expression in Neisseria gonorrhoeae. Infect Immun. 71 550–556. 10.1128/IAI.71.1.550-556.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F., Kong L., Gao H. (2018). Defining the binding determinants of Shewanella oneidensis OxyR: implications for the link between the contracted OxyR regulon and adaptation. J. Biol. Chem. 293 4085–4096. 10.1074/jbc.RA117.001530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F., Shi M., Gao H. (2017). Loss of OxyR reduces efficacy of oxygen respiration in Shewanella oneidensis. Sci. Rep. 7:42609. 10.1038/srep42609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne L. G., Diaz G. A. (1986). A double staining method for differentiating between two classes of mycobacterial catalase in polyacrylamide electrophoresis gels. Anal. Biochem. 157 89–92. 10.1016/0003-2697(86)90200-9 [DOI] [PubMed] [Google Scholar]
- Wu L., Wang J., Tang P., Chen H., Gao H. (2011). Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis. PLoS One 6:e21479. 10.1371/journal.pone.0021479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbib D. (2017). “Bacterial Cell Envelopes: Composition, Architecture, and Origin,” in Handbook of Electroporation, ed. Miklavcic D. (Cham: Springer International Publishing; ), 1–20. [Google Scholar]
- Zhang A., Wassarman K. M., Ortega J., Steven A. C., Storz G. (2002). The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9 11–22. 10.1016/S1097-2765(01)00437-3 [DOI] [PubMed] [Google Scholar]
- Zheng M., Aslund F., Storz G. (1998). Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279 1718–1721. 10.1126/science.279.5357.1718 [DOI] [PubMed] [Google Scholar]
- Zheng M., Wang X., Doan B., Lewis K. A., Schneider T. D., Storz G. (2001). Computation-directed identification of OxyR DNA binding sites in Escherichia coli. J. Bacteriol. 183 4571–4579. 10.1128/JB.183.15.4571-4579.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.