Abstract
LIM domain binding protein 1 (LDB1) is a protein cofactor that participates in several multiprotein complexes with transcription factors that regulate mouse forebrain development. Since Ldb1 null mutants display early embryonic lethality, we used a conditional knockout strategy to examine the role of LDB1 in early forebrain development using multiple Cre lines. Loss of Ldb1 from E8.75 using Foxg1Cre caused a disruption of midline boundary structures in the dorsal telencephalon. While this Cre line gave the expected pattern of recombination of the floxed Ldb1 locus, unexpectedly, standard Cre lines that act from embryonic day (E)10.5 (Emx1Cre) and E11.5 (NesCre) did not show efficient or complete recombination in the dorsal telencephalon by E12.5. Intriguingly, this effect was specific to the Ldb1 floxed allele, since three other lines including floxed Ai9 and mTmG reporters, and a floxed Lhx2 line, each displayed the expected spatial patterns of recombination. Furthermore, the incomplete recombination of the floxed Ldb1 locus using NesCre was limited to the dorsal telencephalon, while the ventral telencephalon and the diencephalon displayed the expected loss of Ldb1. This permitted us to examine the requirement for LDB1 in the development of the thalamus in a context wherein the cortex continued to express Ldb1. We report that the somatosensory VB nucleus is profoundly shrunken upon loss of LDB1. Our findings highlight the unusual nature of the Ldb1 locus in terms of recombination efficiency, and also report a novel role for LDB1 during the development of the thalamus.
Keywords: Cre recombinase activity, forebrain, inefficient floxing, Ldb1, somatosensory thalamus
Significance Statement
The role of transcriptional co-factor LIM domain binding protein 1 (LDB1) in mouse forebrain development was examined using a floxed Ldb1 line and standard Cre driver lines Foxg1Cre, Emx1Cre, and NesCre. Foxg1Cre revealed that LDB1 is a key regulator of early telencephalic midline development. Curiously, the floxed Ldb1 locus appeared to be selectively resistant to Cre-mediated recombination in the dorsal telencephalon using Emx1Cre and NesCre. Recombination improved with time in the case of Emx1Cre. NesCre recombined the floxed Ldb1 locus efficiently in the ventral telencephalon and in the diencephalon, where a critical requirement for this factor in the development of the somatosensory VB nucleus of the thalamus was revealed. Our findings highlight the importance of assessing the extent of recombination when interpreting conditional loss-of-function phenotypes.
Introduction
LIM domain binding protein 1 (LDB1; also called NLI/CLIM2) is a protein cofactor with an impressive list of binding partners, and is found in multiprotein complexes in several systems including the nervous system and the hematopoietic system, associated with transcription factors of the GATA, bHLH, LIM-HD, and OTX families (Hobert and Westphal, 2000; Matthews and Visvader, 2003; Love et al., 2014). LDB1 has domains that interact with its partners, and also a dimerization domain that permits it to serve as a “bridge,” bringing together multi-protein complexes associated with each LDB1 monomer (Jurata and Gill, 1997; Milán and Cohen, 1999; Van Meyel et al., 1999, 2000; Thaler et al., 2002). LDB1, therefore, functions as a nuclear adaptor forming tetrameric, hexameric, or other higher-order protein complexes with these transcription factors, thereby enabling their function (Matthews and Visvader, 2003; Love et al., 2014).
Murine Ldb1 is widely expressed in the entire embryo starting from the earliest stages of development (Bach et al., 1997; Visvader et al., 1997; Mukhopadhyay et al., 2003). Ldb1 null mutants die between embryonic day (E)9.5 and E10.5 and display truncated anterior head structures and loss of heart and foregut formation, indicating a crucial early role in developmental processes (Mukhopadhyay et al., 2003). Ldb2, a related family member found in several vertebrate species (Agulnick et al., 1996; Bach et al., 1997; Matthews and Visvader, 2003), has a more restricted expression in the developing brain, limited to the cortical hem and antihem at E12.5, and is expressed in layer 5 neurons starting from late embryonic stages (Bach et al., 1997; Bulchand et al., 2003), where it regulates aspects of corticospinal motor neuron differentiation together with Ldb1 (Leone et al., 2017).
Of the transcription factors that interact with LDB1, LIM-HD proteins LHX2 and LHX5 play crucial roles in regulating early dorsal telencephalic development. LHX2 has been identified as a cortical selector. Lhx2 null mutants lack both hippocampus and neocortex, while the non-cortical fates of hem and antihem are expanded (Bulchand et al., 2001; Mangale et al., 2008). LHX5 is critical for medial telencephalic patterning, which is grossly disrupted in Lhx5 null mutants (Zhao et al., 1999). Another transcription factor, OTX2, is required for the normal development and maintenance of the choroid plexus (Johansson et al., 2013) and is also known to act in a complex with LDB1 (Costello et al., 2015). These studies strongly motivate an examination of whether the functions of these genes in regulating the early development of the cortical primordium require LDB1.
We used a Foxg1Cre driver and a floxed Ldb1 line to examine the early stages of dorsal telencephalic patterning, and discovered specific deficits in telencephalic midline formation. Using a NestinCre (NesCre) line, we also discovered severe and specific defects in the thalamus in the absence of Ldb1. Finally, our data identified an apparent resistance to recombination of the floxed Ldb1 allele that is selective to the dorsal telencephalon when either NesCre or Emx1Cre is used. Therefore, this study not only identifies novel functions of LDB1 in early forebrain development, but also brings out a hitherto unreported shortcoming of conditional gene deletion strategies using standard Cre lines.
Materials and Methods
Mice
All animal protocols were approved by the Institutional Animal Ethics Committee according to regulations devised by the Committee for the Purpose of Control and Supervision of Experiments on Animals. The Foxg1Cre line was obtained from Susan McConnell (Hebert and McConnell, 2000). Mice carrying the floxed Ldb1 line were obtained from Paul Love, NIH, and Susan McConnell, Stanford, with the kind permission of Yangu Zhao, NIH. Noon of the day of vaginal plug was designated as E0.5. Mouse embryos of either sex were harvested at E12.5, E15.5, E17.5, and postnatal day (P)0. Controls used for each experiment were age-matched littermates. ISH for each marker was performed in more than or equal to three biological replicates. Some of the embryos examined were also heterozygous for the Ldb2 null allele, but these were indistinguishable from embryos that carried both wild-type alleles, likely due to the very limited expression of Ldb2 in the brain (Bulchand et al., 2003; Leone et al., 2017).
The Emx1Cre line (strain name: B6.Cg-Emx1tm1(cre)Krj/J; stock number: 005628; Gorski et al., 2002) is the same as the Emx1CreKJ in Shetty et al. (2013). The NestinCre line is from JAX labs (strain name: B6N.Cg-Tg(Nes-cre)1Kln/CjDswJ; stock number: 019103). The GFP reporter mTmG (strain name: Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo; stock number: 007676) and Ai9 (strain name: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J; stock number: 007909) reporter lines were also obtained from JAX labs. The floxed Lhx2 line used in this study was obtained from Edwin Monuki, University of California (Mangale et al., 2008).
Embryos used for sections in Figures 1, 3–6 did not carry any reporter allele.
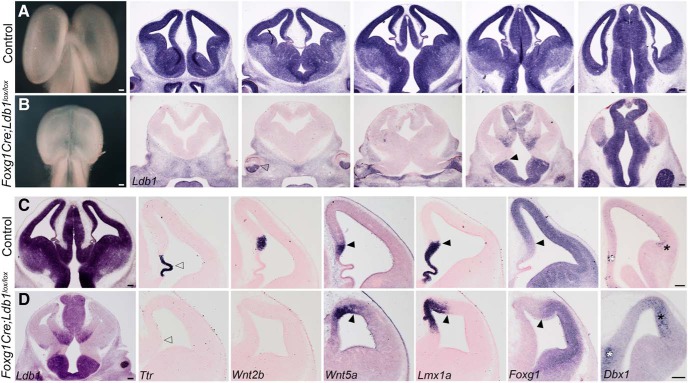
Figure 1.
Early deletion of Ldb1 using Foxg1Cre results in reduced telencephalic size and disrupted telencephalic midline patterning. A, B, Whole brains and coronal sections at E12.5 from control (A) and Foxg1Cre;Ldb1lox/lox mutants (B) reveal a greatly reduced telencephalon on loss of Ldb1. Ldb1 is expressed in the entire control telencephalon and thalamus at E12.5 (A) but is undetectable in the mutant telencephalon (B). The expression boundaries of Ldb1 in the retina (open arrowhead, B) and the diencephalon (black arrowhead, B) are consistent with the reported activity of Foxg1Cre (Hebert and McConnell, 2000). C, D, E12.5 sections from control (C) and Foxg1Cre;Ldb1lox/lox mutant (D) brains. The expression of choroid plexus marker Ttr is lost in the mutant (open arrowheads, C, D). The mutant does not express hem marker Wnt2b but displays an expanded expression of Wnt5a and Lmx1a. Lmx1a labels both hem and choroid plexus in control sections. Foxg1 is expressed in a manner complementary to Lmx1a in control and mutant sections. Black arrowheads mark the cortex-hem boundary (C, D). Dbx1 is expressed in the septum (white asterisks) and antihem (black asterisks), both of which are expanded on loss of LDB1 (C, D). Scale bars: 100 μm.
Figure 3.
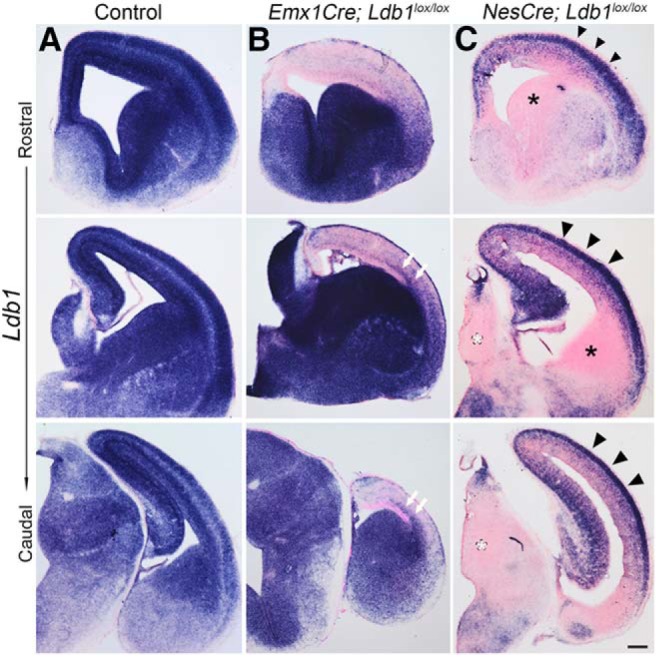
Recombination efficiency of the Ldb1 locus improves with time in the case of Emx1Cre but not NesCre. A–C, Expression of Ldb1 mRNA in E15.5 control (A), Emx1Cre;Ldb1lox/lox (B), and NesCre;Ldb1lox/lox (C) brains at three rostro-caudal levels. Emx1Cre;Ldb1lox/lox brains display low Ldb1 expression in the dorsal telencephalon, with no medio-lateral expression gradient. White arrows in B mark a spur of Ldb1 expression consistent with the migrating interneuron stream that originates in the Ldb1-expressing ventral telencephalon. In contrast, NesCre;Ldb1lox/lox brains display robust expression in the cortical plate and in the hippocampus (black arrowheads), but the ganglionic eminences (black asterisks) and thalamus (white asterisks) appear to have lost Ldb1 expression. Scale bars: 100 μm.
Figure 6.
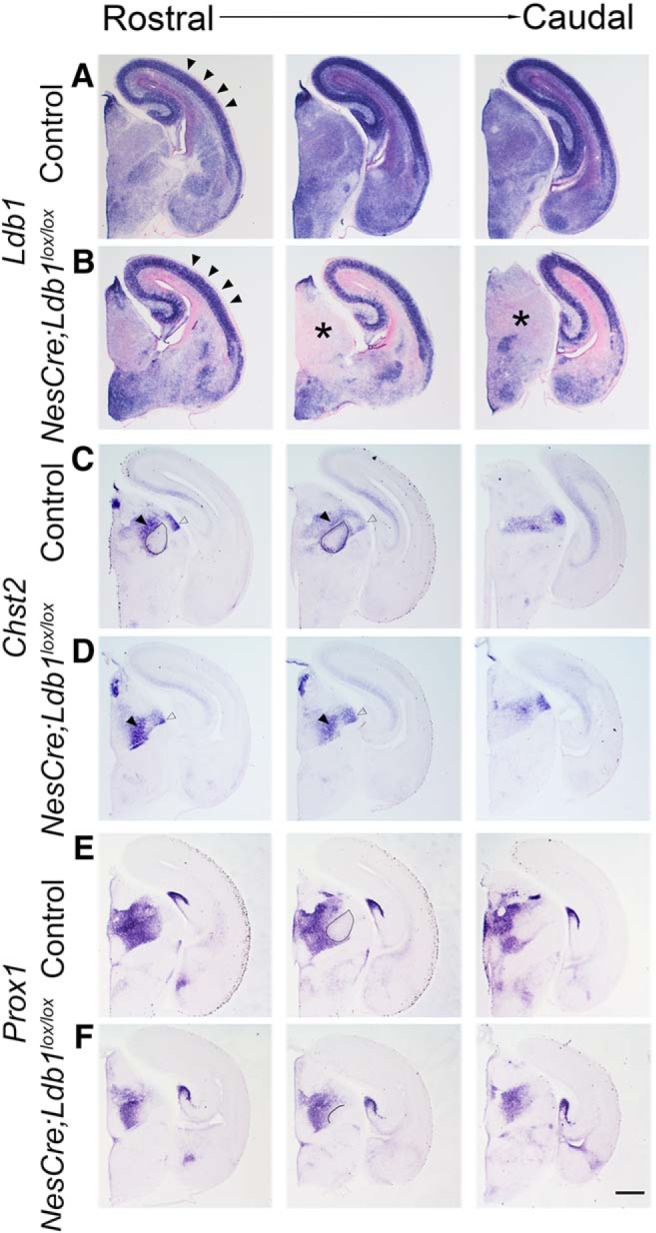
Multiple markers reveal shrinkage of the somatosensory VB on loss of LDB1. A–F, Sections from E17.5 control (A, C, E) and NesCre;Ldb1lox/lox (B, D, F) embryos at three rostro-caudal levels. A, B, Expression of Ldb1 mRNA in E17.5 control (A) and NesCre;Ldb1lox/lox (B) brains. In the mutant, Ldb1 expression is undetectable in a broad region of the dorsal thalamus (asterisks, B), while expression in the cortical plate (black arrowheads, A, B) and hippocampus persists. C–F, Chst2 expression is seen at the perimeter of the VB nucleus (ovals, C), and Prox1 expression excludes the VB nucleus leaving a distinct negative zone (oval, E). These features are not revealed in mutant sections (D, F), in which only a small Prox1 negative domain is seen (solid line, F). Black and open arrowheads identify the PO and the dLGN, respectively, which appear to be present in both control and mutant brains. Scale bars: 500 μm.
Genotyping of the various alleles was performed by PCR using the following primers:
Ldb1-F: 5′-CTTATGTGACCACAGCCATGCATGCATGTG-3′
Ldb1-R: 5′-CAGCAAACGGAGGAAACGGAAGATGTCAG-3′
Foxg1Cre-F: 5’AGTATTGTTTTGCCAAGTTCTAAT-3′
Foxg1Cre-R: 5’TCCTATAAGTTGAATGGTATTTTG-3′
Emx1Cre-F: 5′-ATTTGCCTGCATTACCGGTC-3′
Emx1Cre-R: 5′-ATCAACGTTTTCTTTTCGG-3′
NestinCre-F: 5′-ATTTGCCTGCATTACCGGTC-3′
NestinCre-R: 5′-ATCAACGTTTTCTTTTCGG-3′
Lhx2-F: 5′-ACCGGTGGAGGAAGACTTTT-3′
Lhx2-R: 5′-CAGCGGTTAAGTATTGGGACA-3′
Sample preparation and in situ hybridization
For in situ hybridization, the mouse embryos were harvested in PBS, fixed in 4% paraformaldehyde (PFA), equilibrated in 30% sucrose made in 4% PFA, and sectioned at 16 μm on a freezing microtome.
In situ hybridization was performed as follows: the sections were fixed in 4% PFA, washed in PBS, and treated with proteinase K (1 µg/ml). Hybridization was performed overnight at 70°C in hybridization buffer (4× SSC, 50% formamide, and 10% SDS) containing different antisense RNA probes. Post-hybridization washes were performed at 70°C in solution X (2× SSC, 50% formamide, and 1% SDS). These were followed by washes in 2× SSC, 0.2× SSC, and then Tris-buffered saline–1% Tween 20 (TBST). The sections were incubated in anti-digoxigenin Fab fragments (Roche) at 1:5000 in TBST overnight at 4°C. The color reaction was performed using NBT/BCIP (Roche) in NTMT [100 mM NaCl, 100 mM Tris (pH 9.5), 50 mM MgCl2, and 1% Tween 20] according to the manufacturer’s instructions.
Probe preparation
All probes were prepared by in vitro transcription using a kit from Roche as per manufacturer’s instructions. Templates for Ldb1 exon1-9, Lhx2 exon2-3, SERT, Chst2, and Prox1 were generated by PCR using specific primers from E15 (for Ldb1 exon 1-9 and Lhx2 exon2-3) and P7 (for SERT, Chst2, and Prox1) mouse cDNA (T7 polymerase promoter sequence was added to the reverse primer sequence). Templates for the other probes were generated from respective plasmid DNA by restriction enzyme digestion. Plasmids used were kind gifts from Forbes D. Porter (Lhx2), Elizabeth Grove (Lhx9, Neurog2), Alessandra Pierani (Dbx1), Cliff Ragsdale (Wnt2b), Kathleen Millen (Lmx1a), and Eseng Lai (Foxg1).
Primers for PCR-generated probes:
Ldb1 exon1-9-F: TACCCACCTACATACCTGGA
Ldb1 exon1-9-R: TGAGAGTGGAATTGGACAGC
Lhx2 exon2-3-F: CGCGGATCCACCATGCCGTCCATCAGC
Lhx2 exon2-3-R: TAATACGACTCACTATAGGG
SERT-F: CAAAACGTCTGGCAAGGTGG
SERT-R: CATACGCCCCTCCTGATGTC
Chst2-F: CATCTTTGGGGCAGCCACTA
Chst2-R: CGAAAGGCTTGGAGGAGGAG
Prox1-F: GCAGGCCTACTATGAGCCAG
Prox1-R: TTTGACCACCGTGTCCACAA
Results
Ldb1 is ubiquitously expressed throughout the embryonic forebrain (Bulchand et al., 2003). We used three standard Cre driver lines to examine the stage-wise roles of LDB1 in early forebrain development, Foxg1Cre, Emx1Cre, and NesCre. Foxg1Cre acts from E8.75 in the entire telencephalon and in a limited portion of the diencephalon (Hébert and McConnell, 2000). Emx1Cre action is specific to the dorsal telencephalon, and complete recombination is expected from E10.5 (Gorski et al., 2002). NesCre activity initiates a day later, from E11.5, and is active in radial glia in the entire central nervous system (Tronche et al., 1999). We generated a set of male mice each carrying a particular Cre driver and at least one Ldb1 floxed allele, and crossed them with Ldb1lox/lox or Ldb1lox/lox;Ai9 females.
Intact brains from E12.5 Foxg1Cre;Ldb1lox/lox embryos appeared distinctly smaller, with a poorly developed midline, compared to controls (Fig. 1A,B). We examined Ldb1 mRNA expression in these brains in a series of rostro-caudal sections, and observed the expected, well-characterized pattern of recombination for Foxg1Cre (Hébert and McConnell, 2000), such that the entire telencephalon did not display detectable Ldb1 expression, but most of the diencephalon and the ventral half of the retina was spared (Fig. 1A,B). In coronal sections, the midline deficits in Foxg1Cre;Ldb1lox/loxembryos were obvious (Fig. 1C,D). The choroid plexus appeared to be missing, both in terms of morphology and Ttr expression (open arrowheads; Fig. 1C,D). The cortical hem lacked Wnt2b expression, but the expression of two other hem-specific genes, Wnt5a and Lmx1a, appeared expanded, and Foxg1, a known suppressor of hem fate (Muzio and Mallamaci, 2005; Godbole et al., 2017), was excluded from this region (black arrowheads; Fig. 1C,D). The septum (white asterisk) and the antihem (black asterisk), both identified by Dbx1 expression, also appeared expanded in the Foxg1Cre;Ldb1lox/lox mutant.
Since early loss of LDB1 results in profound patterning defects, we used the later-acting Emx1Cre and NesCre lines to examine LDB1 function in the early development of the cortex and hippocampus. The Ai9 reporter was used to reveal the domain of Cre activity in Emx1Cre;Ldb1lox/lox;Ai9 and NesCre;Ldb1lox/lox;Ai9 embryos examined at E12.5. The Emx1Cre driver is known to recombine floxed alleles in the dorsal telencephalon starting from E10, and recombination is complete by E10.5 (Gorski et al., 2002; Shetty et al., 2013). Unexpectedly, a medio-lateral gradient of Ldb1 expression was seen in the dorsal telencephalon of Emx1Cre;Ldb1lox/lox;Ai9 embryos, suggesting that the floxed Ldb1 alleles may not be completely recombined in the lateral telencephalon Fig. 2B, open arrowhead). We compared these results with embryos carrying the NesCre driver, which is known to be effective from E11.5 (Chen et al., 2015). In NesCre;Ldb1lox/lox;Ai9, we discovered that the floxed Ldb1 appeared to be poorly recombined in the dorsal telencephalon by E12.5; however, the ventral telencephalon (vtel) and diencephalon (asterisks) displayed the expected loss of Ldb1 expression. In contrast, the Ai9 reporter that was also present in each of these embryos displayed robust region-appropriate expression, restricted to the dorsal telencephalon in the case of Emx1Cre, and in the entire telencephalon and diencephalon in the case of NesCre.
Figure 2.
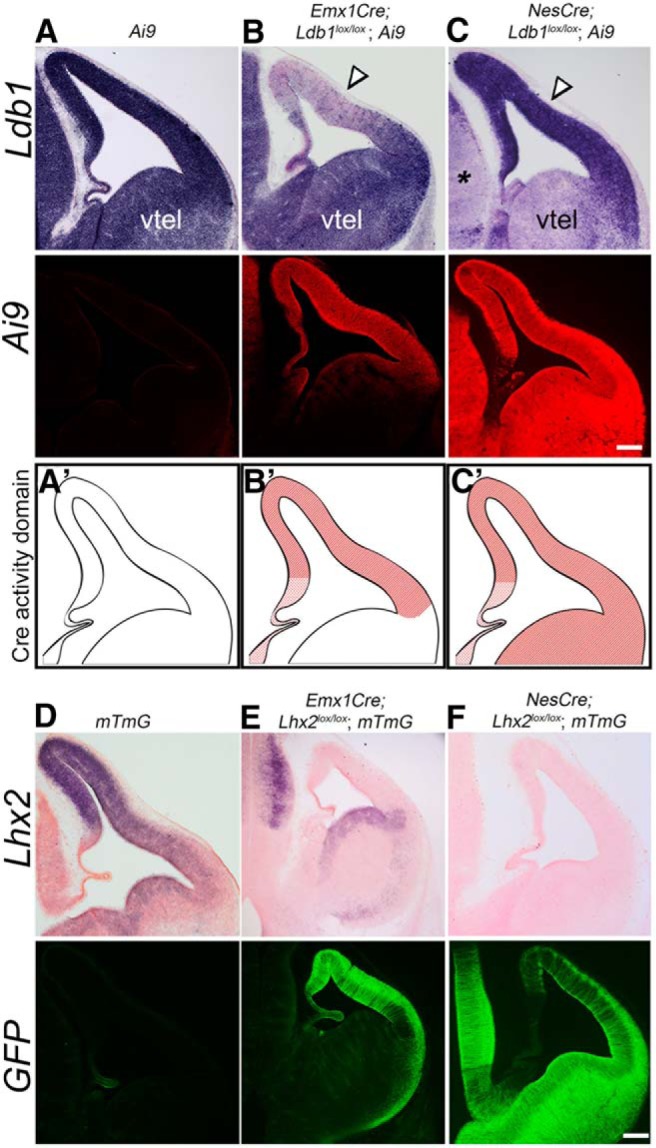
Two standard Cre drivers display differential recombination of the Ldb1 locus in the dorsal telencephalon. A–C, Expression of Ldb1 mRNA and the Ai9 reporter in E12.5 control (A), Emx1Cre;Ldb1lox/lox;Ai9 (B), and NesCre;Ldb1lox/lox;Ai9 (C) brains. A’–C’, Cartoons indicating the domain of Cre activity in the respective conditions. In each condition, Ai9 fluorescence faithfully reports Cre activity in the expected domains. However, Ldb1 expression is seen in a medio-lateral gradient in the dorsal telencephalon of Emx1Cre;Ldb1lox/lox;Ai9 embryos (B) and persists in the entire dorsal telencephalon in NesCre;Ldb1lox/lox;Ai9 brains (C). In contrast, the ventral telencephalon (vtel) and the diencephalon (black asterisk) display the expected loss of Ldb1 expression. D–F, Expression of Lhx2 mRNA and mTmG (GFP) reporter in E12.5 control (D), Emx1Cre;Lhx2lox/lox;mTmG (E), and NesCre;Lhx2lox/lox;mTmG (F) brains. Lhx2 is recombined and its expression is undetectable in the dorsal telencephalon of Emx1Cre;Lhx2lox/lox;mTmG brains, and mTmG reporter displays a complementary pattern, consistent with the activity domain for Emx1Cre (E). NesCre;Lhx2lox/lox;mTmG brains display no detectable Lhx2 expression and widespread expression of the mTmG reporter, consistent with the activity domain of NesCre (F). The control brains display autofluorescence in the green channel in the region of the choroid plexus which is an artifact. Scale bars: 100 μm.
Both the Emx1Cre and NesCre drivers are standard lines widely used in the literature. We tested whether they display the expected pattern of activity in another conditional line available to us that was homozygous for floxed Lhx2 alleles (Mangale et al., 2008) together with a GFP reporter line (“mTmG,” Muzumdar et al., 2007). Both these loci displayed the expected spatial recombination pattern: Lhx2 expression was undetectable in the dorsal telencephalon of Emx1Cre;Lhx2lox/lox;mTmG embryos at E12.5, and reporter GFP expression was robust in the same region; the ventral telencephalon and diencephalon displayed no apparent recombination of either the Lhx2 or the mTmG locus. Likewise, NesCre;Lhx2lox/lox;mTmG embryos displayed complete Lhx2 recombination and no detectable expression in the telencephalon or diencephalon, and GFP reporter expression in both structures. Together with the Ai9 data (Fig. 2A–C), this indicated that the Emx1Cre and NesCre drivers were working normally in our hands. For subsequent experiments, we used Ldb1lox/lox mice that did not carry any reporter.
We tested whether the efficiency of recombination of the floxed Ldb1 gene in the dorsal telencephalon improved with time. This appeared to be at least partially true for Emx1Cre;Ldb1lox/lox embryos, in which the recombination in the dorsal telencephalon appeared to improve by E15.5 at all rostro-caudal levels of sectioning. Only the lateral-most portion continued to display Ldb1 expression (Fig. 3B, white arrows), which could be due to migrating interneurons from the Ldb1-expressing ventral telencephalon. In contrast, in NesCre;Ldb1lox/loxembryos, the neocortex and hippocampus continued to display robust Ldb1 expression (Fig. 3C, black arrowheads), although the ganglionic eminences and thalamus appeared completely recombined (Fig. 3C, black and white asterisks, respectively).
In summary, in contrast to the Foxg1Cre driver which acts from E8.75, the Ldb1 floxed allele appeared to be inefficiently recombined in the dorsal telencephalon by Cre drivers acting from E10.5 (Emx1Cre) and E11.5 (NesCre), and this did not ameliorate with time in the dorsal telencephalon in the case of the later acting driver NesCre. However, the efficient recombination of the Ldb1 floxed locus in the developing thalamus of NesCre;Ldb1lox/loxembryos offered the opportunity of examining the role of LDB1 in the development of this structure without the complication of simultaneous loss of Ldb1 in the neocortex. We therefore focused our attention on the diencephalon and thalamus.
First, we ascertained that the region of the diencephalon from which the thalamus arises did indeed lose Ldb1 expression by E12.5. Indeed, in NesCre;Ldb1lox/loxembryos, Ldb1 expression was undetectable in this region at all rostro-caudal levels (Fig. 4B, black asterisks), although expression in the dorsal telencephalon persisted. We examined the expression of Neurog2, which is enriched in progenitors (Fig. 4C, white dashed line), and the expression tapers off in lateral domains where postmitotic neurons reside. In the mutant embryos, Neurog2 expression appeared to intensify in this lateral domain (Fig. 4C,D, white asterisks). We also examined Lhx9 expression, which delineates three domains: expression is weak to undetectable in the medial progenitor domain, high in the adjacent domain containing postmitotic neurons, but undetectable in an extreme lateral domain (Fig. 4E, black lines). In the mutant diencephalon, Lhx9 expression delineates only two domains, the medial progenitor domain lacking expression and a continuous lateral domain that displays expression (Fig. 4E,F, open arrowheads). The diencephalon appeared smaller in the mutant compared with control brains. Together, these results suggested a fundamental defect in diencephalic development on loss of LDB1.
Figure 4.
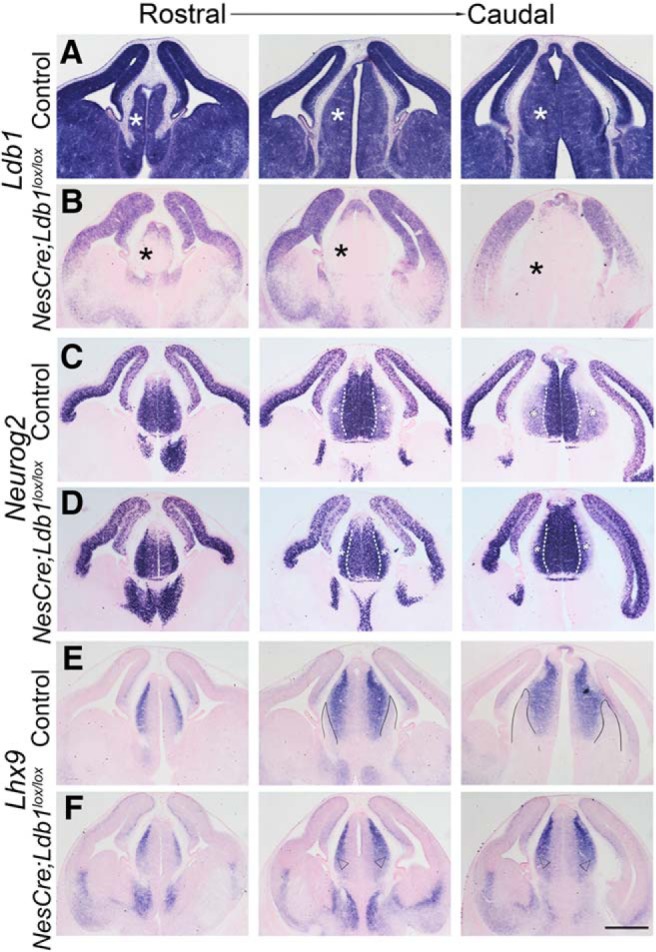
LDB1 is required for patterning the dorsal thalamus. A, B, Expression of Ldb1 mRNA in E12.5 control (A) and NesCre;Ldb1lox/lox (B) brains at three rostro-caudal levels. In the mutant, Ldb1 expression is undetectable in a broad region of the dorsal thalamus of the mutant brains (black asterisks, B). C–F, Serial sections from the same brains in A, B probed for the expression of Lhx9 and Neurog2 at three rostro-caudal levels. In the mutant, the medial domain of intense Neurog2 expression (white dashed lines, C, D) expands laterally at mid and caudal levels (white asterisks, C, D). At the same levels of sectioning, Lhx9 expression, which is normally not seen in a lateral domain (solid black lines, E), expands in the mutant to fill this domain (open arrowheads, F). Scale bars: 500 μm.
To explore the consequences of this early disruption of diencephalic development, we examined the thalamus of E17.5 NesCre;Ldb1lox/loxembryos with a battery of markers that are expressed in different sensory thalamic nuclei (Yuge et al., 2011). We first examined SERT expression, which identifies primary sensory nuclei (Lebrand et al., 1998). Whereas the control sections displayed intense SERT expression in the somatosensory ventrobasal (VB) nucleus (Fig. 5A, ovals), this nucleus appeared to be dramatically shrunken and was hard to identify in all but the most caudal levels of NesCre;Ldb1lox/loxbrains (Fig. 5B, open arrowhead).
Figure 5.
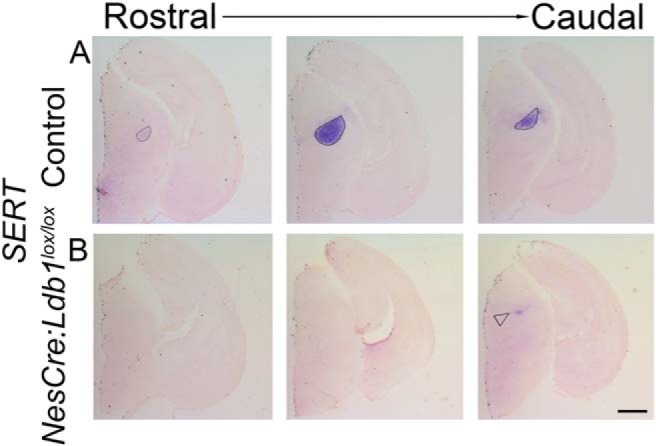
The somatosensory VB nucleus is profoundly shrunken on loss of LDB1. A, B, Expression of SERT mRNA in the VB nucleus in control (ovals, A) and NesCre;Ldb1lox/lox (B) brains at three rostro-caudal levels. In the mutant, SERT expression is limited to a very narrow region in caudal sections, a severely diminished VB (open arrowhead, B). Scale bars: 500 μm.
We examined additional markers of the embryonic thalamus, Chst2 and Prox1, to better understand the nature of the VB shrinkage. Since thalamic nuclei are small structures, we used one hemisphere of each brain to examine Ldb1 expression, and the contralateral hemisphere to examine thalamic markers in serial sections. Ldb1 expression was lost in the thalamus (asterisks) and persisted in the cortex (Fig. 6A,B, arrowheads). Chst2 and Prox1 expression delineates the border of the VB in control sections (Fig. 6C,E, ovals) but failed to exclude a territory consistent with the VB. However, Chst2 expression was seen in the adjacent posteromedial thalamic nucleus (PO; solid arrowheads) and dorsal lateral geniculate nucleus (dLGN; open arrowheads) of both control and mutant brains, suggesting that the deficit may be specific to the VB (Fig. 6C–F). Unfortunately, the NesCre;Ldb1lox/lox mutants die at birth, so it was not possible to analyze the mutant thalamus at postnatal stages. However, we were able to harvest one mutant brain by monitoring the dam during delivery, and we examined adjacent sections of the thalamus for the expression of SERT, Chst2, and Prox1 (Fig. 7). As at E17.5, it is not possible to identify the VB by SERT expression, and neither Chst2 nor Prox1 expression delineates this structure on loss of LDB1.
Figure 7.
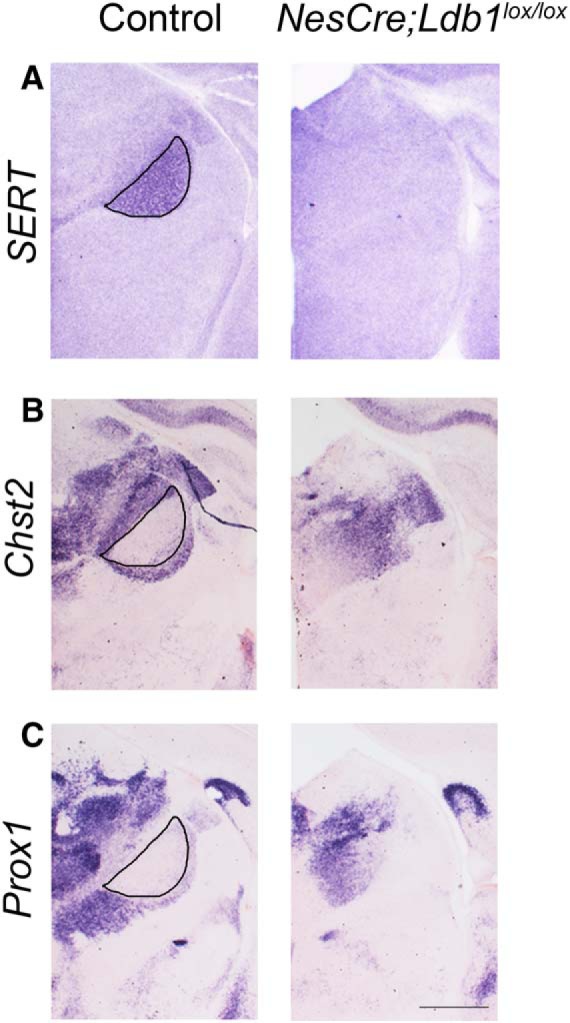
The VB is undetectable at P0 in LDB1 mutant brains. A–C, Sections from P0 control and NesCre;Ldb1lox/lox brains. A, SERT expression reveals the VB nucleus in control brains (black outline), which is undetectable in the mutant. B, C, Chst2 and Prox1 expression is seen at the perimeter of the VB nucleus in control (black outlines), but not in mutant brains. Scale bars: 500 μm.
In summary, these data indicate that the VB critically requires LDB1 for its development and appears to be missing when Ldb1 is disrupted in the thalamus using NesCre.
Discussion
Using a conditional gene disruption strategy, we demonstrate for the first time the requirement of Ldb1 in the development of the primary somatosensory VB nucleus of the thalamus. Since the neocortex displays strong Ldb1 expression due to the apparent inability of NesCre to effectively delete the floxed Ldb1 allele gene in this tissue, we had the fortuitous advantage of a system in which the thalamus, but not the cortex, lacks Ldb1. This allowed us to specifically examine any thalamic patterning defects that may have manifested independent of any cortex-derived influences; in any case, corticothalamic innervation does not enter the thalamus by E17.5 (Jacobs et al., 2007), making it more likely that the apparent loss of the VB is due to an autonomous requirement for LDB1. Furthermore, VB neurons are born at E11.5 (Angevine, 1970), so it is unlikely that the near-complete absence of the VB at E17.5 is due to a developmental delay in the production of these neurons. Chst2 and Prox1 expression appears to identify the adjacent PO, and Chst2 also marks the dLGN in the mutant, indicating that the defect neither includes other somatosensory nuclei nor other primary sensory nuclei. This suggests that the VB may have a specific requirement for LDB1 for its development. Since such a defect has not been reported on loss of any known binding partners of LDB1 such as LHX2, LHX9, or NEUROG2 (Seibt et al., 2003; Lakhina et al., 2007; Marcos-Mondéjar et al., 2012), it remains an open question as to which factor(s) LDB1 complexes with to regulate VB development.
In contrast to the VB phenotype, the defects seen on early loss of LDB1 using Foxg1Cre appear to be consistent with a combination of the Lhx2 and Lhx5 mutant phenotypes. This is expected, since LDB1 is a known co-factor of LIM domain-containing transcription factors (Matthews and Visvader, 2003). The expansion of the hem and antihem seen on loss of LDB1 is similar to that reported in Lhx2 null mutants (Bulchand et al., 2001; Mangale et al., 2008; Roy et al., 2013). Whereas Lhx2 null embryos also have an expanded choroid plexus (Monuki et al., 2001), the complete absence of the choroid plexus on loss of LDB1 is similar to the phenotype reported for loss of Lhx5 (Zhao et al., 1999). Loss of the choroid plexus is also seen in animals in which Otx2 is disrupted either constitutively or conditionally (Matsuo et al., 1995; Johansson et al., 2013). OTX2 is known to act in a complex with LDB1 (Costello et al., 2015); therefore, the loss of the choroid plexus we report may be explained by the disruption of a complex containing LDB1 and either or both of LHX5 and OTX2.
Finally, it is an unexplained conundrum as to why the Ldb1 locus, which is expressed in the entire forebrain throughout development (Bulchand et al., 2003), and therefore accessible in terms of the binding of transcriptional machinery, appears to be selectively resistant to recombination in the dorsal telencephalon from E10.5/E11.5, but not earlier. Very few studies have used the Ldb1lox/lox line to examine LDB1 function in the developing telencephalon. Zhao et al. (2014) used the Nkx2.1Cre driver and reported effective recombination in ventral telencephalic structures such as the medial ganglionic eminence (MGE) and the preoptic area (POA). This is consistent with our results in which we demonstrate that NesCre recombines the floxed Ldb1 locus effectively in the ventral telencephalon. Leone et al. (2017) used the same Emx1Cre driver (Gorski et al., 2002) as in our study to inactivate Ldb1 in the dorsal telencephalon. They report complete loss of Ldb1 expression from the mutant cortex at postnatal day 4. We notice a progressive improvement of recombination such that the dorsal telencephalon is largely negative for Ldb1 expression by E15.5, which is consistent with the findings of Leone et al. (2017). One study that reported incomplete Ldb1 inactivation (Tzchori et al., 2009) found it to be due to lack of expression of the Cre driver in the cells that apparently escaped recombination (Narkis et al., 2012). In contrast, the Cre drivers in our study are expressed and effectively recombine other floxed lines (Lhx2lox/lox, mTmG, Ai9) in the dorsal telencephalon, but seem to be specifically inefficient at recombining the floxed Ldb1 allele in this structure. One concern is that the presence of the Ai9 reporter could result in a competition for Cre recombinase, and the floxed Ldb1 locus may not be favored for recombination over the reporter. However, except for Figure 2, none of the embryos used for the other figures carried the Ai9 reporter, so this does not explain the results. A mutation of the loxP sites could potentially make recombination of the Ldb1 locus less likely. However, since Ldb1 is efficiently recombined in the dorsal telencephalon by Foxg1Cre (Fig. 1), such a scenario is unlikely. Furthermore, any such mutation would have to arise only in the dorsal telencephalon, since the ventral telencephalon is efficiently recombined by NesCre. Another explanation could be that recombination does in fact take place, but existing Ldb1 mRNA transcripts may persist due to the presence of RNA stabilizing proteins. The temporal regression of recombination efficiency, Foxg1Cre>Emx1Cre>NesCre, correlating with the timing of action of the Cre lines, could be explained by the progressive accumulation of such an RNA stabilizing protein specific to the dorsal telencephalon. Further studies are needed to examine these issues.
In summary, the incomplete recombination that we observe in the dorsal telencephalon seems to be a region-specific and temporally dynamic phenomenon that is unique to the Ldb1 locus and has not been previously reported in any conditional mutant line. Our findings underscore the importance of examining the stage and extent of recombination when interpreting conditional loss of function phenotypes.
Acknowledgments
Acknowledgements: We thank Edwin S. Monuki for the kind gift of the Lhx2lox/lox line; Yangu Zhao, Paul Love, and Susan McConnell for the Ldb1lox/lox line; and Susan McConnell for the Foxg1Cre line. We also thank Forbes D. Porter (Lhx2), Elizabeth Grove (Lhx9, Neurog2), Alessandra Pierani (Dbx1), Cliff Ragsdale (Wnt2b), Kathleen Millen (Lmx1a), and Eseng Lai (Foxg1) for gifts of plasmid DNA used for generating RNA probes; Shital Suryavanshi and the animal house staff of the Tata Institute for Fundamental Research (TIFR) for excellent support; Tanja Vogel for helpful discussions; and the support and mentorship of Medha Rajadhyaksha (V.K.).
Synthesis
Reviewing Editor: Esther Stoeckli, University of Zurich
Decisions are customarily a result of the Reviewing Editor and the peer reviewers coming together and discussing their recommendations until a consensus is reached. When revisions are invited, a fact-based synthesis statement explaining their decision and outlining what is needed to prepare a revision will be listed below. The following reviewer(s) agreed to reveal their identity: Alexandre Dayer, Guillermina Lopez-Bendito.
Thank you for the revisions made on your manuscript. We have discussed the changes and came to the conclusion that we are satisfied with the revised version in most aspects. Obviously, a mechanistic explanation of the failure of Ldb1 recombination in the dorsal telencephalon would be desirable but we are aware that this would go beyond the scope of the manuscript. Still, we feel that reporting this observation is of interest to the community, as it calls for respective controls in many future studies. We would like to come back to one issue that we raised in response to your initial submission that was not addressed in your revision. The images reporting SERT expression are very misleading, as they suggest a broad expression of SERT in the mutant versus the control mice. You explain the difference in staining by 'overdevelopment' of the staining. However, we do not see a need for this. Please provide adequate images for this group, maybe along with sense controls.
References
- Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H (1996) Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature 384:270–272. 10.1038/384270a0 [DOI] [PubMed] [Google Scholar]
- Angevine JB (1970) Time of neuron origin in the diencephalon of the mouse. An autoradiographic study. J Comp Neurol 139:129–187. 10.1002/cne.901390202 [DOI] [PubMed] [Google Scholar]
- Bach I, Carrière C, Ostendorff HP, Andersen B, Rosenfeld MG (1997) A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev 11:1370–1380. 10.1101/gad.11.11.1370 [DOI] [PubMed] [Google Scholar]
- Bulchand S, Grove EA, Porter FD, Tole S (2001) LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech Dev 100:165–175. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Subramanian L, Tole S (2003) Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev Dyn 226:460–469. 10.1002/dvdy.10235 [DOI] [PubMed] [Google Scholar]
- Chen Q, Arai D, Kawakami K, Sawada T, Jing X, Miyajima M, Hirai S, Sakaguchi K, Furushima K (2015) EphA4 regulates the balance between self-renewal and differentiation of radial glial cells and intermediate neuronal precursors in cooperation with FGF signaling. PLoS One 10:e0126942. 10.1371/journal.pone.0126942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello I, Nowotschin S, Sun X, Mould AW, Hadjantonakis AK, Bikoff EK, Robertson EJ (2015) Lhx1 functions together with Otx2, Foxa2, and Ldb1 to govern anterior mesendoderm, node, and midline development. Genes Dev 29:2108–2122. 10.1101/gad.268979.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbole G, Shetty AS, Roy A, Chen B, Miyoshi G, Tole S (2017) Hierarchical genetic interactions between FOXG1 and LHX2 regulate the formation of the cortical hem in the developing telencephalon. Development 145:dev154583. 10.1242/dev.154583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JLR, Jones KR (2002) Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci 22:6309–6314. 10.1523/JNEUROSCI.22-15-06309.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert JM, McConnell SK (2000) Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol 222:296–306. 10.1006/dbio.2000.9732 [DOI] [PubMed] [Google Scholar]
- Hobert O, Westphal H (2000) Functions of LIM-homeobox genes. Trends Genet 16:75–83. 10.1016/s0168-9525(99)01883-1 [DOI] [PubMed] [Google Scholar]
- Jacobs EC, Campagnoni C, Kampf K, Reyes SD, Kalra V, Handley V, Xie YY, Hong-Hu Y, Spreur V, Fisher RS, Campagnoni AT (2007) Visualization of corticofugal projections during early cortical development in a tau-GFP-transgenic mouse. Eur J Neurosci 25:17–30. 10.1111/j.1460-9568.2006.05258.x [DOI] [PubMed] [Google Scholar]
- Johansson PA, Irmler M, Acampora D, Beckers J, Simeone A, Gotz M (2013) The transcription factor Otx2 regulates choroid plexus development and function. Development 140:1055–1066. 10.1242/dev.090860 [DOI] [PubMed] [Google Scholar]
- Jurata LW, Gill GN (1997) Functional analysis of the nuclear LIM domain interactor NLI. Mol Cell Biol 17:5688–5698. 10.1128/MCB.17.10.5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhina V, Falnikar A, Bhatnagar L, Tole S (2007) Early thalamocortical tract guidance and topographic sorting of thalamic projections requires LIM-homeodomain gene Lhx2. Dev Biol 306:703–713. 10.1016/j.ydbio.2007.04.007 [DOI] [PubMed] [Google Scholar]
- Lebrand C, Cases O, Wehrlé R, Blakely RD, Edwards RH, Gaspar P (1998) Transient developmental expression of monoamine transporters in the rodent forebrain. J Comp Neurol 401:506–524. [DOI] [PubMed] [Google Scholar]
- Leone DP, Panagiotakos G, Heavner WE, Joshi P, Zhao Y, Westphal H, McConnell SK (2017) Compensatory actions of Ldb adaptor proteins during corticospinal motor neuron differentiation. Cereb Cortex 27:1686–1699. 10.1093/cercor/bhw003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love PE, Warzecha C, Li LQ (2014) Ldb1 complexes: the new master regulators of erythroid gene transcription. Trends Genet 30:1–9. 10.1016/j.tig.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PRV, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV, Li Y, Flanagan LA, Tole S, Monuki ES (2008) Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science 319:304–309. 10.1126/science.1151695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos-Mondéjar P, Peregrín S, Li JY, Carlsson L, Tole S, López-Bendito G (2012) The lhx2 transcription factor controls thalamocortical axonal guidance by specific regulation of robo1 and robo2 receptors. J Neurosci 32:4372–4385. 10.1523/JNEUROSCI.5851-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S (1995) Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev 9:2646–2658. 10.1101/gad.9.21.2646 [DOI] [PubMed] [Google Scholar]
- Matthews JM, Visvader JE (2003) LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep 4:1132–1137. 10.1038/sj.embor.7400030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milán M, Cohen SM (1999) Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and Apterous confers activity and capacity for regulation by dLMO. Mol Cell 4:267–273. 10.1016/S1097-2765(00)80374-3 [DOI] [PubMed] [Google Scholar]
- Monuki ES, Porter FD, Walsh CA (2001) Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-lhx2 pathway. Neuron 32:591–604. 10.1016/S0896-6273(01)00504-9 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M, Teufel A, Yamashita T, Agulnick AD, Chen L, Downs KM, Schindler A, Grinberg A, Huang S, Dorward D, Westphal H (2003) Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development 130:495–505. 10.1242/dev.00225 [DOI] [PubMed] [Google Scholar]
- Muzio L, Mallamaci A (2005) Foxg1 confines Cajal-Retzius neuronogenesis and hippocampal morphogenesis to the dorsomedial pallium. J Neurosci 25:4435–4441. 10.1523/JNEUROSCI.4804-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L (2007) A global double-fluorescent Cre reporter mouse. Genesis 45:593–605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Narkis G, Tzchori I, Cohen T, Holtz A, Wier E, Westphal H (2012) Isl1 and Ldb Co-regulators of transcription are essential early determinants of mouse limb development. Dev Dyn 241:787–791. 10.1002/dvdy.23761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, de Melo J, Chaturvedi D, Thein T, Cabrera-Socorro A, Houart C, Meyer G, Blackshaw S, Tole S (2013) LHX2 is necessary for the maintenance of optic identity and for the progression of optic morphogenesis. J Neurosci 33:6877–6884. 10.1523/JNEUROSCI.4216-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibt J, Schuurmans C, Gradwhol G, Dehay C, Vanderhaeghen P, Guillemot F, Polleux F (2003) Neurogenin2 specifies the connectivity of thalamic neurons by controlling axon responsiveness to intermediate target cues. Neuron 39:439–452. 10.1016/S0896-6273(03)00435-5 [DOI] [PubMed] [Google Scholar]
- Shetty AS, Godbole G, Maheshwari U, Padmanabhan H, Chaudhary R, Muralidharan B, Hou PS, Monuki ES, Kuo HC, Rema V, Tole S (2013) Lhx2 regulates a cortex-specific mechanism for barrel formation. Proc Natl Acad Sci USA 110:E4913–E4921. 10.1073/pnas.1311158110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL (2002) LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell 110:237–249. 10.1016/S0092-8674(02)00823-1 [DOI] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schütz G (1999) Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet 23:99–103. 10.1038/12703 [DOI] [PubMed] [Google Scholar]
- Tzchori I, Day TF, Carolan PJ, Zhao Y, Wassif CA, Li L, Lewandoski M, Gorivodsky M, Love P, Porter FD, Westphal H, Yang Y (2009) LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development 136:1375–1385. 10.1242/dev.026476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meyel DJ, O’Keefe DD, Jurata LW, Thor S, Gill GN, Thomas JB (1999) Chip and Apterous physically interact to form a functional complex during Drosophila development. Mol Cell 4:259–265. 10.1016/S1097-2765(00)80373-1 [DOI] [PubMed] [Google Scholar]
- Van Meyel DJ, O’Keefe DD, Thor S, Jurata LW, Gill GN, Thomas JB (2000) Chip is an essential cofactor for apterous in the regulation of axon guidance in Drosophila . Development 127:1823–1831. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Mao X, Fujiwara Y, Hahm K, Orkin SH (1997) The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc Natl Acad Sci USA 94:13707–13712. 10.1073/pnas.94.25.13707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuge K, Kataoka A, Yoshida AC, Itoh D, Aggarwal M, Mori S, Blackshaw S, Shimogori T (2011) Region-specific gene expression in early postnatal mouse thalamus. J Comp Neurol 519:544–561. 10.1002/cne.22532 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Sheng HZ, Amini R, Grinberg A, Lee E, Huang S, Taira M, Westphal H (1999) Control of hippocampal morphogenesis and neuronal differentiation by the LIM homeobox gene Lhx5. Science 284:1155–1158. 10.1126/science.284.5417.1155 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Flandin P, Vogt D, Blood A, Hermesz A, Westphal H, Rubenstein J (2014) Ldb1 is essential for development of Nkx2.1 lineage derived GABAergic and cholinergic neurons in the telencephalon. Dev Biol 385:94–106. 10.1016/j.ydbio.2013.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]