Abstract
The Polycomb group (PcG) protein family is a well-known group of epigenetic modifiers. We used zebrafish to investigate the role of Rnf2, the enzymatic subunit of PRC1. We found a positive correlation between loss of Rnf2 and upregulation of genes, especially of those whose promoter is normally bound by Rnf2. The heart of rnf2 mutants shows a tubular shaped morphology and to further understand the underlying mechanism, we studied gene expression of single wildtype and rnf2 mutant hearts. We detected the most pronounced differences at 3 dpf, including upregulation of heart transcription factors, such as tbx2a, tbx2b, and tbx3a. These tbx genes were decorated by broad PcG domains in wildtype whole embryo lysates. Chamber specific genes such as vmhc, myh6, and nppa showed downregulation in rnf2 mutant hearts. The marker of the working myocard, nppa, is negatively regulated by Tbx2 and Tbx3. Based on our findings and literature we postulate that loss of Rnf2-mediated repression results in upregulation and ectopic expression of tbx2/3, whose expression is normally restricted to the cardiac conductive system. This could lead to repression of chamber specific gene expression, a misbalance in cardiac cell types, and thereby to cardiac defects observed in rnf2 mutants.
Introduction
Proper establishment of cellular identity and subsequent cell type maintenance is crucial during embryonic development and tissue homeostasis. Defects in this complex process can result in disease and/or lethality. Therefore, it is important to study these processes in the context of an in vivo system. Modifications of the DNA as well as the associated histones, affect the accessibility of the DNA for the transcriptional machinery. Epigenetic modifiers of the Polycomb group (PcG) protein family are well-known transcriptional silencers, which place specific histone marks1. PcG proteins can assemble in two Polycomb Protein Complexes (PRCs): PRC1 and PRC2. PcG proteins were first identified in Drosophila, in which mutations of PcG genes resulted in homeotic transformation, by deregulating homeotic (hox) genes2. In zebrafish the PRC1 core subunits are Rnf2, a Pcgf-family member, a Cbx protein, and a Phc-protein3. The core-components of PRC2 are Eed, Suz12, and Ezh1/-24. The canonical view is that PRC2 is first recruited to the chromatin and the enzymatic subunit Ezh2 trimethylates lysine 27 of histone H3 (H3K27me3). PRC1 is recruited to H3K27me3 via its subunit Cbx5. The PRC1 subunit Rnf2 mono-ubiquitinylates H2AK119 (H2AK119ub), via its RING-domain and this mark stabilizes H3K27me36. Both H3K27me3 and H2AK119ub are epigenetic marks associated with transcriptional repression7–10. H3K27me3 represses gene expression by changing the chromatin structure and by antagonizing the H3K27ac mark, which is a mark known to be present at active enhancers10,11. Because H3K27me3 and H3K27ac reside at the same amino acid of the same histone tail, they are mutually exclusive. Furthermore, recent studies showed that the recruitment of PRC1 to the chromatin can also be H3K27me3-independent12,13. PRC1 variants that contain subunits which have a DNA-binding domain are described to be involved in this process14. H3K27me3-independent recruitment of PRC1 can repress gene expression by different mechanism: by condensation of the chromatin structure, by preventing RNA polymerase II elongation, and by recruiting PRC213,15–17. However, much still remains unknown about the interplay between PRC1 and H3K27me3.
Studies using systems in which PRC1 is disrupted indicate a crucial role in cellular differentiation across species. Mice have two homologous of the RING-domain containing proteins that both can assemble in PRC1: Rnf2 (Ring1b) and Ring1 (Ring1a)18. Loss of Rnf2 in mice results in developmental arrest during gastrulation19. Murine Ring1 homozygous mutants are viable20, and similar to PcG mutants in Drosophila, Ring1 heterozygous mice display homeotic transformations and skeletal defects21. In mice, the loss of Ring1/Rnf2 postnatally results in dental defects, but no lethality, when the mice are studied up to 17 days22. Additionally, studies in mouse embryonic stem cells showed that Rnf2 and Ring1 are essential for maintaining cells in a pre-mature state, by repressing genes involved in differentiation pathways23,24. In zebrafish, only one Ring1 orthologue is identified, which shows most homology with Rnf23. Therefore, ablation of Rnf2 in zebrafish results in loss of functional PRC1 and the H2AK119ub mark8. Zinc-finger nuclease induced rnf2 null-mutant zebrafish embryos and rnf2 morphant embryos gastrulate normally, which makes it possible to study development in the absence of Rnf28,25. Rnf2 morphants have an overall normal morphology and, although their primitive erythropoiesis was largely unaffected, the number of hematopoietic stem and thrombocytes was shown to be smaller at 36 hpf25. An rnf2 mutant allele has been generated, and the rnf2 mutation results in pre-mature stop codon. These rnf2 null-mutant zebrafish embryos show lethality around 4–5 dpf and display defects in terminal differentiation of the pectoral fins, likely due to interference with Fgf-signaling8. In addition, it was found that Cranial Neural Crest (CNC) cells do not properly differentiate into chondrocytes in rnf2 mutants, resulting in cartilage malformation in the head26. These defects in pectoral fin and chondrocyte development upon loss of Rnf2 both arise during terminal tissue differentiation.
To study the role of PRC1 and PRC2 during embryogenesis is challenging due to lethality of mutants in many species before gastrulation19,27. Therefore, in this study, rnf2 mutant zebrafish embryos are used to investigate the effects of loss of Rnf2 on development by studying the transcriptome and correlate this to the Rnf2 binding pattern in wildtype embryos at 3 dpf. We find an important regulatory role for Rnf2 at the chromatin level. The loss of Rnf2 results in upregulation of the genes normally occupied by Rnf2; these include genes associated with transcriptional regulation. In order to gain insight in a tissue specific role of Rnf2 we studied the heart in more detail. Transcriptome analysis of single hearts of wildtype and rnf2 mutant embryos at 1, 2, and 3 dpf indicates that at 1 and 2 dpf the transcriptional difference between wildtype and rnf2 mutant hearts are minor and at 3 dpf these differences are more prominent. At 3 dpf upregulation of transcription factors like tbx2a, tbx2b, and tbx3a was detected and, in addition, a downregulation of cardiac chamber genes, such as nppa was observed. We suggest that the upregulation of the tbx transcription factors is a direct consequence of the loss of Rnf2-mediated repression and we hypothesize that these transcription factors are responsible for the downregulation of chamber genes, resulting in malformation of the rnf2 mutant hearts. This finding sheds new light on the molecular mechanisms underlying heart development and the role of Rnf2 during vertebrate embryogenesis.
Results
Phenotypical differences between rnf2 mutant and wildtype zebrafish embryos
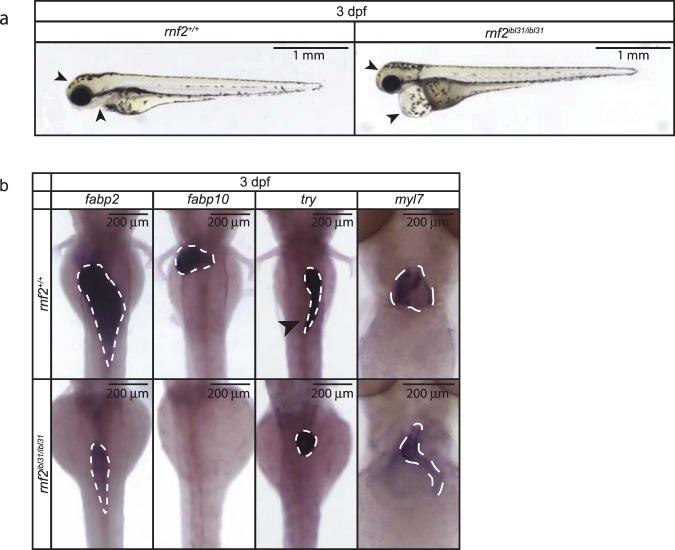
To gain additional insight in the role of PRC1 in development we used previously identified rnf2ibl31/ibl31 (referred to as rnf2) mutant zebrafish, which harbor a mutation in the enzymatic subunit of PRC1. A 14 base pair deletion in the rnf2 gene, results in a premature stop codon, and the mutant embryos were shown to lack rnf2 gene expression at 3 dpf8. We observed the same pleiotropic phenotype in rnf2 mutants, as reported before8. This includes motility defects, defects in craniofacial development, the lack of pectoral fins, and a pronounced heart edema (Fig. 1a).
Figure 1.
Zygotic rnf2 mutant zebrafish embryos show a pleiotropic phenotype. (a) Lateral view of wildtype embryos (left panel) and rnf2 mutant embryos (right panel) at 3 dpf. The rnf2 mutants show a pleiotropic phenotype, including motility problems, craniofacial defects (arrowheads), lack of pectoral fins, and a pronounced heart edema (arrowheads). Not all phenotypes are visible in the pictures. Scale bar = 1 mm. (b) Expression of tissue-specific markers was assessed by WISH at 3 dpf in wildtype and rnf2 mutant embryos. fabp2: intestinal marker, fabp10: liver marker, try: exocrine pancreas marker (arrowhead indicates pancreatic lobe), and myl7: cardiomyocyte marker. Scale bar is 200 µm.
Immunohistochemistry for Rnf2 in wildtype siblings shows that expression of Rnf2 protein at 2 dpf is mainly detected anteriorly and in the notochord (Supplementary Fig. S1, left panel). The rnf2 mutants lack Rnf2 protein at 2 dpf (Supplementary Fig. S1, right panel).
To obtain more insight in the pleiotropic phenotype we assessed the expression of four organ markers by whole mount in situ hybridization (WISH) in wildtype and rnf2 mutant embryos at 3 dpf. Results from WISH for fatty acid-binding protein type 2 (fabp2), an intestinal marker, suggests a smaller intestine in the rnf2 mutants compared to the wildtypes (Fig. 1b). Expression of the liver specific marker fatty acid-binding protein type 10 (fabp10) is present in wildtypes, whilst it cannot be detected in rnf2 mutants. This suggests that liver terminal differentiation is abrogated in rnf2 mutants. The terminal differentiation marker of the exocrine pancreas, trypsin (try), is present in both wildtype and rnf2 mutant embryos. However, the shape of the exocrine pancreas is different in rnf2 mutants: the pancreatic lobe is not detected. Lastly, the expression of the cardiomyocyte marker myosin light chain 7 (myl7) was assessed. Wildtype embryos show pronounced myl7 expression in the atrium and the ventricle of the heart. Expression of myl7 is detected in rnf2 mutant embryos at 3 dpf; however, the expression pattern of myl7 in rnf2 mutants indicates malformation of the heart. The rnf2 mutant heart shows a stringy morphology and appears smaller based on the myl7 expression pattern (Fig. 1b).
Rnf2 binds the same targets as H3K27me3 and H3K27me3 deposition is present in rnf2 mutants
We next studied the role Rnf2 on the molecular level by identifying its binding on the chromatin, which was not yet assessed in zebrafish. We performed Rnf2 and H3K27me3 chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) at 3 dpf in wildtype and rnf2 mutant embryos. The canonical pathway describes that the H2AK119ub mark, placed by PRC1 (Rnf2), stabilizes H3K27me328. Additionally, PRC1 variants, containing different subunits than canonical PRC1, have been proposed to be able to recruit PRC212,13,29. Interestingly, numerous studies also reported no effect on H3K27me3 upon loss of PRC123,30–34. Therefore, we studied Rnf2 and H3K27me3 binding patterns and thereby the potential functional redundancy in PRC1 and PRC2 in zebrafish.
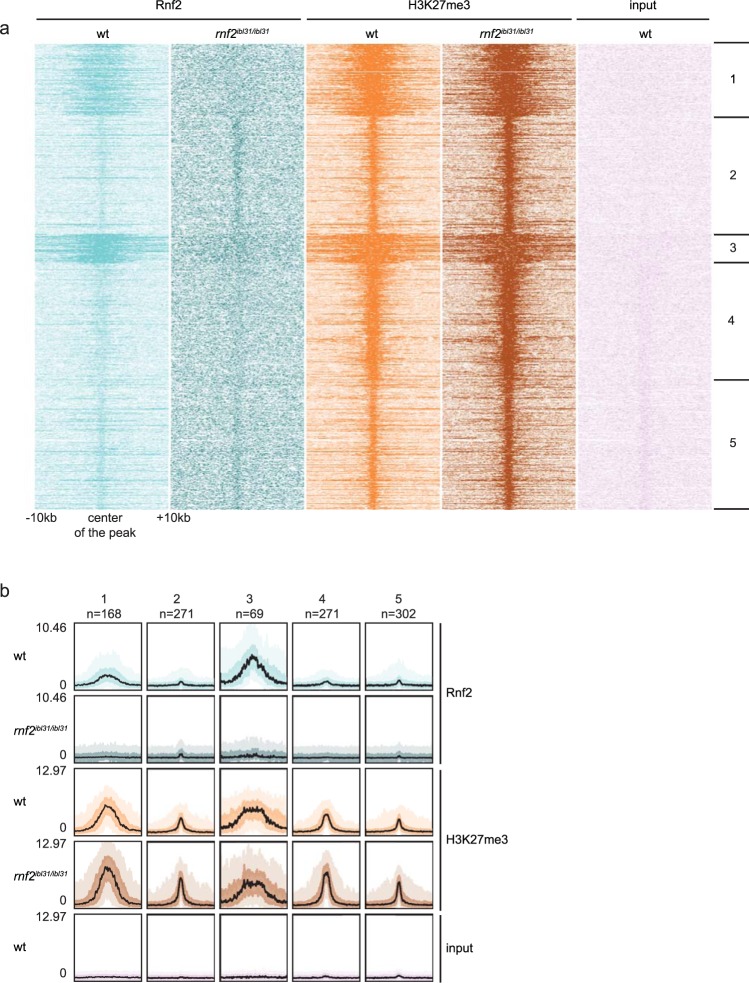
To allow quantitative normalization and to demonstrate the efficiency of the method, we added Drosophila melanogaster spike-in chromatin during the ChIP-seq procedure35. After ChIP-seq, k-means clustering revealed five different classes of binding of Rnf2 and H3K27me3 at promoter regions (Fig. 2a). The first cluster represents Rnf2 and H3K72me3 positive promoters. The second, fourth, and fifth cluster contain promoter regions that are positive for H3K27me3 and show close to background levels for Rnf2. The third cluster contains broad PcG domains, in which both Rnf2 and H3K27me3 are present. The intensity of the peaks for Rnf2 and H3K27me3 was analyzed and visualized with bandplots (Fig. 2b). Rnf2 is present at the chromatin in wildtype embryos and is at around background levels in rnf2 mutants. The levels of H3K27me3 are similar in rnf2 mutants compared to wildtypes. H3K27me3 presence has been retained upon loss of Rnf2, which could suggest that its deposition does not rely on Rnf2 (Fig. 2b).
Figure 2.
Rnf2 has a similar DNA-binding pattern as H3K27me3 and the presence of H3K27me3 mark is retained upon loss of Rnf2. (a) Heatmaps showing k-means clustering of Rnf2 and H3K27me3 ChIP-seq peaks in promoter regions in wildtype embryos and rnf2 mutant embryos at 3 dpf. In the figure 20 kb regions, with a viewpoint around the center of all peaks, are shown. (b) Bandplot showing the intensities of the ChIP-seq peaks for Rnf2 and H3K27me3 in wildtypes and rnf2 mutants at 3 dpf, and the input of the five clusters defined by k-means clustering. The number of peak regions included in each cluster is depicted above the plot. The black line indicates the median, the intense color 50% of the peaks, and the light color 90% of the peaks.
Loss of Rnf2 is associated with upregulation of genes decorated by broad PcG domains
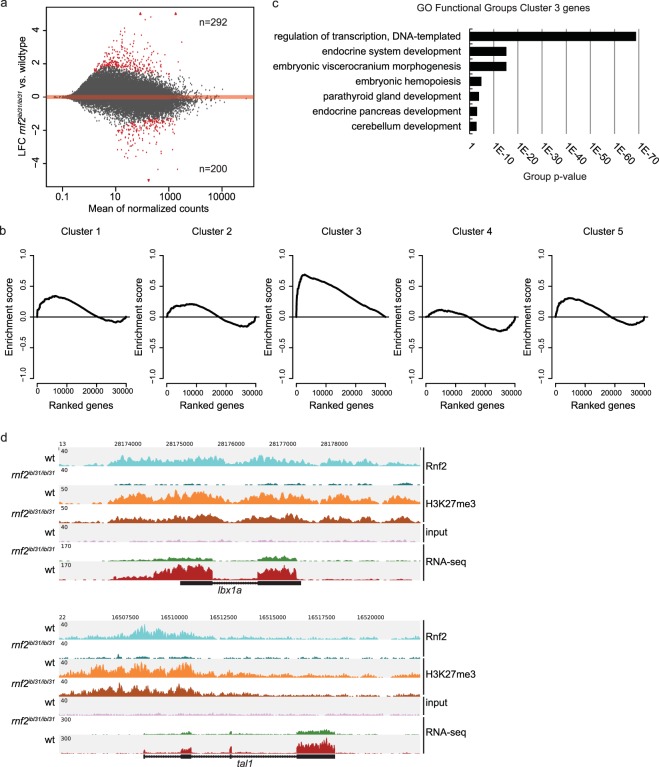
Presence of Rnf2 and H3K27me3 on the chromatin has a repressive effect on the underlying genes7–10. Therefore, we compared the transcriptome of wildtype embryos and rnf2 mutant embryos at 3 dpf. Our data shows both up- and downregulated genes upon loss of Rnf2 (Fig. 3a). In total, 492 genes were found to be differentially expressed (LFC ≥ 1; padj ≤ 0.1). Of these, 292 were identified to be upregulated and 200 genes to be downregulated in rnf2 mutant embryos (Supplementary Table S1). Hierarchical clustering confirms good homology of the replicates (Euclidian distance; Supplementary Fig. S2a). The organ markers tested by WISH, were also studied in the whole embryo RNA-sequencing dataset. A downregulation of marker expression in rnf2 mutants was observed for fabp2, fabp10, and myl7. Expression of try was not found to be affected by the loss of Rnf2 (Supplementary Fig. S2b).
Figure 3.
Loss of Rnf2 is directly associated with upregulation of gene expression. (a) MA-plot of differentially expressed genes between wildtypes and rnf2 mutants at 3 dpf. Significantly differentially expressed genes |Log2FC ≥ 1|; padj < 0.1 are highlighted in red. In total 292 genes are upregulated and 200 genes are downregulated. (b) Gene Set Enrichment Analysis (GSEA) for the genes whose promoters belong to the five clusters defined in Fig. 2a. Cluster 1, 2, 4, and 5 show a non-significant distribution of the genes based on their expression changes upon the rnf2 mutation. Genes belonging to the promoters from cluster 3 are enriched for upregulation upon mutation of rnf2. p-value = < 0.001; NES = 2.04. (c) Gene ontology of biological processes analysis of the 112 genes belonging to cluster 3. (d) ChIP-seq and RNA-seq coverage at the lbx1a gene and the tal1 gene in 3 dpf zebrafish embryos. Light blue: Rnf2 ChIP-seq tracks in wildtypes. Teal: Rnf2 ChIP-seq tracks in rnf2 mutants. Orange: H3K27me3 ChIP-seq track in wildtypes. Brown: H3K27me3 ChIP-seq track in rnf2 mutants. Lilac: wildtype input. Green: RNA-seq track in wildtypes. Red: RNA-seq track in rnf2 mutants.
The promoters of the five different clusters identified after ChIP-seq (Fig. 2a) have been linked to the genes they regulate. The expression of these genes was analyzed by Gene Set Enrichment Analysis (GSEA). We tested the differences in expression between rnf2 mutants and wildtypes in the five clusters (genes within the five clusters are enlisted in Supplementary Table S2). Genes belonging to the promoters of cluster 1, 2, 4, and 5 do not show significant enrichment for differential regulation upon the rnf2 mutation (p-value = 0.176, 0.679, 0.605, and 0.249, respectively). Cluster 3 contains 69 promoter regions, regulating 112 genes (‘cluster 3 genes’). This cluster is significantly enriched with genes that are upregulated in the rnf2 mutant embryos at 3 dpf (p-value < 0.001) (Fig. 3b). Gene ontology analysis shows that these ‘cluster 3 genes’ are associated with regulation of transcription and embryonic organ development (Fig. 3c). Examples of tracks of two ‘cluster 3 genes’ are shown in Fig. 3d. These genes are decorated by Rnf2 and H3K27me3 and the RNA-seq tracks indicate a significant upregulation of gene expression in rnf2 mutants at 3 dpf. In total, 24 genes were identified in cluster 3 that were significantly upregulated in rnf2 mutant embryos which are decorated by Rnf2 in the promoter region of wildtype embryos at 3 dpf (LFC ≥ 1; padj ≤ 0.1) (Supplementary Table S3).
The cardiac phenotype in rnf2 mutant zebrafish embryos shows looping defects
To gain insight into a tissue specific role of Rnf2 during development, we performed more extensive studies on the heart, which was one of the organs severely affected upon the rnf2 mutation. The heart is a well-studied organ in zebrafish, due to its similarity in cardiac development to other species and its regenerative capacities. Since we observe a severe cardiac phenotype upon loss of Rnf2, we studied a potential role of Rnf2-mediated regulation of heart development.
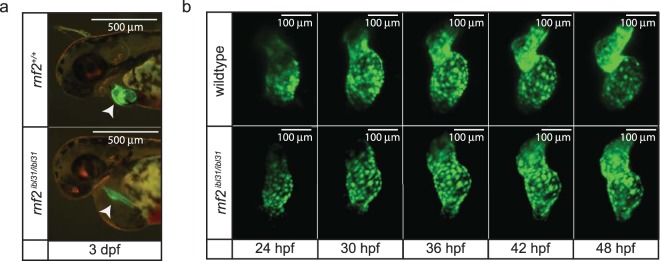
We analyzed the development of the heart of wildtype and rnf2 mutant embryos at 3 dpf in a Tg(myl7::GFP) background. This transgene specifically marks cardiomyocytes36. During normal cardiac development, cardiac looping ensures the proper positioning of the atrium and ventricle and it marks the transition from a linear heart tube to a two-chambered heart separated by the atrioventricular canal (AVC), which occurs between 28 and 50 hours post fertilization37. In contrast to the wildtype situation, the hearts of rnf2 mutant embryos display defective looping morphogenesis, resulting in a stringy heart phenotype with no clear defined cardiac chambers or AVC at 3 dpf (Fig. 4a). To investigate the underlying developmental dynamics, we used high speed selective plane illumination microscopy (SPIM) to time-lapse image wildtype and rnf2 mutant embryos from 1 to 2 dpf (Fig. 4b). Indeed, whereas wildtype embryos showed cardiac looping around 36 hpf the mutants fail to loop properly (Fig. 4b).
Figure 4.
Zebrafish rnf2 mutant embryos show cardiac looping defects. (a) Fluorescent images of wildtype and rnf2 mutant sibling embryos in a Tg(myl7::GFP) background at 3 dpf. Scale bar is 500 µm. (b) Stills of live-imaging by light sheet microscopy of wildtype and rnf2 mutant hearts in a Tg(myl7::GFP) background starting at 1 dpf. Scale bar is 100 µm.
Single HeartsRNA-seq highlights differences between wildtype and rnf2 mutant hearts over time
We next aimed at getting a better understanding of the molecular mechanisms underlying the heart defects detected in rnf2 mutants. To start with, we show that disturbing the epigenetic repressor Rnf2 shows a global effect on gene expression, especially of genes decorated by Rnf2 in wildtype embryos (Fig. 3a). In order to address this finding and in connection with the observed cardiac phenotype, we took genes positive for Rnf2 in their promoter, as identified by ChIP-seq (n = 206), and all genes detected by RNA-seq (n = 32,266) and searched for the presence of heart transcription factors (n = 18) within these two groups38, finding a significant enrichment of this category (chi-squared test; p-value < 0.001).
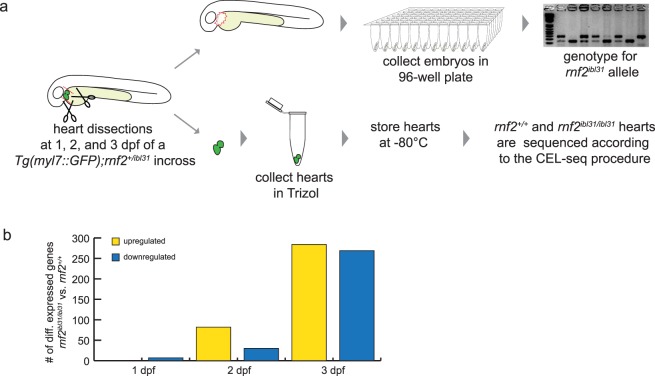
To gain more detailed insight into the role of Rnf2 in cardiac development, single embryonic hearts, both from rnf2 mutant and wildtype, serve as a useful model. The transcriptome of single hearts dissected from wildtype and rnf2 mutants was assessed, using a low-input RNA-seq method based on CEL-seq39–42. This method is very suitable for the number of cells present in a single embryonic heart, which is between 150 and 350 cells at 1 to 3 dpf41,43,44. We performed manual dissection of single embryonic hearts at 1, 2, and 3 dpf and prepared the individual rnf2 mutant and wildtypes dissected hearts for RNA-sequencing (Single HeartsRNA-seq, Fig. 5a). In total 63 single hearts were sequenced (Supplementary Fig. S3a). Samples were filtered based on the number of mRNAs they express, and genes were filtered based on the number of samples that express them (see Materials and Methods and Supplementary Fig. S3b). In total 5 samples were excluded and at least 8 replicates remained per genotype per developmental time point (Supplementary Fig. S3c). After this, gene counts were normalized to avoid, among others, differences derived from unequal amounts of cardiac tissue developed by the rnf2 mutant and wildtype embryos. Hierarchical clustering based on Euclidian Distances at the 3 different developmental time points indicates that differences between wildtype and rnf2 mutant hearts are minor at 1 and 2 dpf and more pronounced at 3 dpf (Supplementary Fig. S3d–f). This is also reflected in the number of genes that are differentially expressed at these different time points (Fig. 5b). At 1 dpf no genes were found to be upregulated and 7 genes were significantly downregulated (|Log2FC > 0|; padj < 0.01). This number increased to 82 upregulated and 30 downregulated genes at 2 dpf. At 3 dpf 284 genes were detected to be significantly upregulated and 269 genes to be significantly downregulated in rnf2 mutant heart. Overall, we observed that the number of differentially expressed genes between wildtype and rnf2 mutant hearts increased over time (Fig. 5b, Supplementary Table S4).
Figure 5.
Single HeartsRNA-seq is used to assess transcriptional differences between wildtype and rnf2 mutant hearts over time. (a) Workflow of Single HeartsRNA-seq. Zebrafish hearts were manually dissected at 1, 2, and 3 dpf as described previously85. The remaining tissue was used for genotyping and the rnf2 mutant and wildtype hearts were sequenced. (b) Differentially expressed genes |Log2FC > 0|; padj < 0.01 between rnf2 mutant and wildtype hearts are visualized in the bar graphs. Yellow: upregulated (up) in rnf2 mutants compared to wildtypes. Blue: downregulated (dn) in rnf2 mutants compared to wildtypes.
Cardiac chamber identity is disrupted in rnf2 mutants at 3 dpf
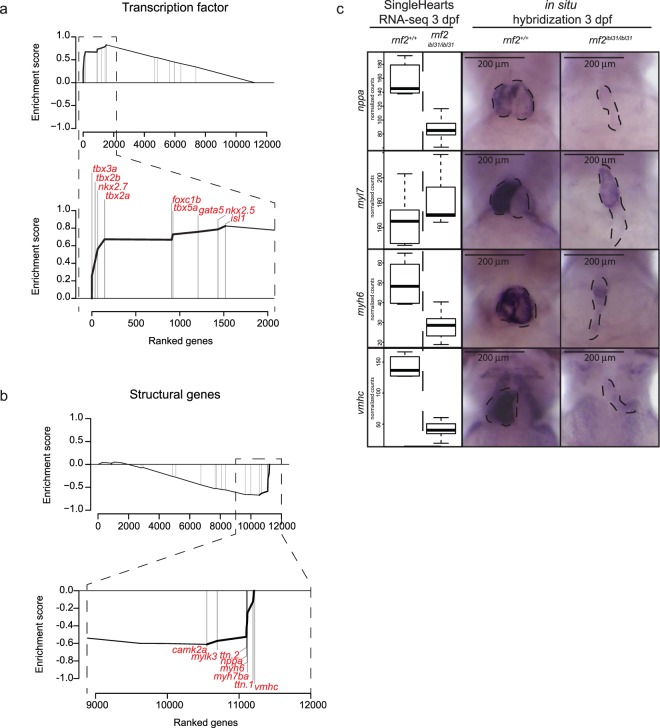
Recently, Hill et al. generated a hand-curated list of cardiac markers38. This list contains genes that are expressed in the developing heart and includes 26 annotations about the function and the location of these genes. These 26 annotations were studied for the gene expression changes of the genes falling into these categories. We used GSEA using the Single HeartsRNA-sequencing results of 3 dpf hearts (Fig. 6a). This analysis revealed that rnf2 mutant hearts are enriched for genes expressed in the atrioventricular canal (FDR q-value = 0.089; NES = 1.46) and for heart transcription factors (FDR q-value = 0.001; NES = 1.96; Fig. 6a). These two annotations show overlap in the genes they contain. Three transcription factors that repress myocardial genes (tbx2a, tbx2b, and tbx3a) are present in the top 4 of genes that are upregulated in rnf2 mutant hearts. Single HeartsRNA-seq results at 1, 2, and 3 dpf indicate that the expression of tbx3a is significantly upregulated at 2 dpf and that expression of tbx2a, tbx2b, and tbx3a is significantly upregulated at 3 dpf in rnf2 mutants (|Log2FC > 0|; padj < 0.01, Supplementary Fig. S4).
Figure 6.
Cardiac chamber identity is disrupted in rnf2 mutants at 3 dpf. (a) GSEA using the list of cardiac genes reported by Hill et al.38 indicates that the group of transcription factors is significantly enriched in being upregulated in rnf2 mutant hearts at 3 dpf as detected by Single HeartsRNA-seq (FDR q-value = 0.001; NES = 1.96). The leading transcription factors (n = 9) are listed in the zoom of the GSEA plot. (b) GSEA indicates the annotation ‘Structural Group’ genes (n = 23) to be enriched for downregulation upon the rnf2 mutation in hearts at 3 dpf. The leading genes (n = 8) are listed in the zoom of the GSEA plot. FDR q-value = 0.054; NES = −1.67. (c) Normalized counts as found by Single HeartsRNA-seq for nppa, myl7, myh6, and vmhc at 3 dpf in wildtype and rnf2 mutant hearts with their accompanying whole mount in situ hybridizations. Scale bar is 200 µm.
The GSEA using the 3 dpf Single HeartsRNA-sequencing results additionally indicated an enrichment for downregulation of structural genes (FDR q-value = 0.054; NES = −1.67; Fig. 6b) and myocardial genes (FDR q-value = 0.043; NES −1.62). The group of genes downregulated upon the rnf2 mutation is enriched in structural genes such as: vmhc, ttn.1, myh7ba, myh6, nppa, ttn.2, mylk3, and camk2a (Fig. 6b). Amongst the genes that are leading there is the marker for the working myocard nppa, the atrium marker myh6, and the ventricle marker vmhc. We analyzed the expression of nppa, myl7, myh6, and vmhc by Single HeartsRNA-seq and additionally tested the spatio-temporal expression of these genes by whole mount in situ hybridization (WISH) (Fig. 6c, Supplementary Fig. S5). At 1 and 2 dpf the expression differences between wildtype and rnf2 mutant hearts for these four genes are relatively small. We found nppa and vmhc to be significantly downregulated in rnf2 mutant hearts at 1 dpf and 2 dpf, respectively. Single HeartsRNA-seq results at 3 dpf indicate that the expression of nppa, myh6, and vmhc is significantly decreased in rnf2 mutants (|Log2FC > 0|; padj < 0.01, Fig. 6c).
Discussion
Rnf2 mutants show a pleiotropic phenotype
In this study zygotic rnf2 mutant embryos (rnf2ibl31/ibl31) are used as a model for loss of PRC1 and H2AK119ub8,26. These mutant embryos display defects in the maintenance of cellular identity and organ integrity8,26. Rnf2 is the only catalytic subunit of PRC1 in zebrafish and therefore disrupting Rnf2 is informative of PRC1’s putative role in zebrafish development.
The rnf2 mutation results in a pleiotropic phenotype in zebrafish embryos at 3 dpf8. A multitude of genes and processes were described to be affected upon the loss of Rnf28,26, however no genome-wide molecular expression datasets are available for these mutants. Since Rnf2 is an epigenetic modifier, ChIP-sequencing can give insight in its mode of action and RNA-sequencing can elucidate its downstream effects. With bright field microscopy and WISH analyses we studied the rnf2 mutant phenotype on a global level. Next to the described phenotypes of rnf2 mutants8,26, we observed a heart edema accompanied by a tubular heart. WISH analyses suggest developmental organ defects of the intestine, pancreas, and liver at 3 dpf. If the observed differences are due to organ defects, they could result from defects in differentiation, cell proliferation, or tissue maintenance, as well as a combination of these three, as PcG proteins are described to play a role in these processes45. In this study we focused on heart development.
H3K27me3 is retained upon rnf2 mutation
H2AK119ub, the mark deposited by PRC1, and more specifically, by its enzymatic subunit Rnf2, is described to stabilize H3K27me39. In addition, also a role for PRC1 in the recruitment of PRC2 is proposed12,13. Therefore, loss of H3K27me3 was considered to occur in the absence of Rnf2. However, many studies also reported that the loss of PRC1 does not affect H3K27me3 deposition23,30–34. We used our PRC1 null model (rnf2 mutants) to study this process in vivo and show that H3K27me3 deposition at 3 dpf is retained upon loss of Rnf2/PRC1, which is thus in line with the majority of these previous reports23,30–34. Based on our data one could even argue that the rnf2 mutation results in a slight increase in H3K27me3 levels. We study whole embryo lysates and therefore this slight increase can be due to more H3K27me3 deposition within the cell, however it can also be due to changes in the abundance of cells that repress these genes via an H3K27me3-mediated manner. This could also be the reason why we do not detect a loss of H3K27me3 in the absence of Rnf2, as changes in individual cells will be overshadowed by the signal detected in the bulk of all cells analyzed.
The rnf2 mutation results in derepression of genes decorated by Rnf2
A subset of genes was found to lose Rnf2-mediated repression and this subset was detected to be significantly upregulated in rnf2 mutants; however, they retain H3K27me3-mediated repression at the whole embryo lysate level. A potential explanation for gene upregulation, whilst they are decorated by H3K27me3, could be derived from the type of sample that has been used. The samples are lysates from whole embryos at 3 dpf, in which many cell types are present and the sequencing results give an average of the signal coming from across all those different cell types. Therefore, overall gene upregulation could be caused by cells in which these genes are not repressed by H3K27me3.
The role of epigenetics in heart development
The role of epigenetics in cardiac development has gained more attention over the years, as reviewed by Vallaster et al., and Shirai et al.46,47. Zebrafish hearts are of high interest due to their regenerative capacity and epigenetics is also implied to be important in this process, as reviewed Quaife-Ryan et al.48. Therefore, it is of essence to better understand the role of epigenetics in heart tissue specification, maintenance, and regeneration. To unravel important regulators of cardiac development, studies have been performed that aimed to make a roadmap of the transcriptome and epigenome during myocardial differentiation49,50. Other approaches focus on chromatin remodelers and the identification of enhancers to gain insight in the role of epigenetics in heart development51,52. Many histone modifiers are described to play a role during cardiac development, such as histone deacetylases, HDACs53,54, H3K4me3 methyltransferase55, and also the PRC2-component Ezh256–58.
The PRC1-variant containing Mel18 was described to be essential for specification of mesodermal cell fate, by preventing alternative lineage commitment29. In line with that, the PRC1-component Bmi1 was shown to act as a barrier during cardiac reprogramming in mouse cells59. Interestingly, a role in cardiac development in vivo for both Mel18 and Bmi1 has not been established60,61. That Mel18-PRC1 and Bmi1-PRC1 do not play a role in cardiac development does not exclude the possibility that any of the other PRC1 variants do. A role for Rnf2 in cardiac development has not been described in vivo, so far. In our current study we show that depletion of Rnf2 affects heart morphology and gene expression during zebrafish development.
A study on H3K4me3 methyltransferases in zebrafish heart development has shown that a decrease in H3K4me3 results in a linear shaped heart, similar to the rnf2 mutants. However, in that study the heart markers myl7, vmhc, and myh6 were reported not to be affected at 2 dpf upon decrease of H3K4me355. This is in contrast to the rnf2 mutants, as we observe at 2 dpf significant downregulation of vmhc expression. A previous study from our lab using maternal zygotic ezh2 mutants also shows a linear shaped heart upon the mutation of the catalytic subunit of PRC2. Altogether, these results indicate that a similar phenotype can have different causes and different epigenetic signatures, as both a loss of H3K4me3, H3K27me3, and H2AK119ub result in a linear shaped heart phenotype55,58.
Repression of tbx-genes by Rnf2 is important for cardiac development
At early stages (1 and 2 dpf) we found the expression of cardiac genes in rnf2 mutant hearts to be more or less similar to wildtype hearts, therefore we hypothesize that maternal rnf2 RNA and Rnf2 protein are sufficient for correct cell specification in the rnf2 mutants. On top of that, as most PcG proteins, Rnf2 is expected to be mostly involved in tissues maintenance rather than tissue specification8,19,23,26,58.
Since we observe an upregulation of tbx-genes in a system in which we mutate a transcriptional repressor, we analyzed our Rnf2 ChIP-seq results on whole embryo lysates and found that the tbx3a gene is bound by Rnf2 and decorated by H3K27me3 in the wildtype situation. This observation strongly hints towards Polycomb-mediated regulation of Tbx3. Literature describes that this is not zebrafish-specific, since Pcl2 (PRC2 subunit) knock-out murine ESC show an upregulation of Tbx362. Research has revealed that when Tbx2/3 forms a complex with Gata and Nkx, it locally represses chamber myocardial gene expression and thereby enhances the formation of the conduction system63. Tbx2/3 are described to directly repress, amongst others, the myocardial gene nppa to allow for the formation of the conductive system in the heart64,65. Zebrafish rnf2 mutants show a malformed heart and our 3 dpf Single HeartsRNA-seq dataset shows that nppa is significantly downregulated, which we validated by WISH experiments. Tbx2/3 overexpression in mouse embryos results in a heart looping defect and the lack of cardiac chambers66,67. In these murine embryos the chamber-myocardial gene program is not correctly set up. Interestingly, the rnf2 mutant zebrafish embryos also show a heart in which the atrium and ventricle are not well developed.
Furthermore, Tbx5 competes with Tbx2/3 to form a complex with Nkx and Gata, and this complex has an activating effect on gene expression66. Early stages of rnf2 mutants show correct tbx5 levels, most likely due to the maternal load of rnf28. However, at 3 dpf we detect tbx5a to be upregulated in the heart upon the rnf2 mutation and whole embryo lysate ChIP-seq indicates that the tbx5a gene is decorated by Rnf2 and H3K27me3. Overexpression of tbx5 in vitro has been reported to represses proliferation and cell growth68, which is in line with our observations. In contrast, rnf2 mutant zebrafish were reported to have greatly reduced expression of tbx5 at the pectoral fin mesenchyme, which is correlated to the absence of pectoral fins in these mutants8 and interestingly, the loss of Tbx5 results in a stringy heart in zebrafish embryos69.
These studies and our observations indicate that tight regulation of tbx2a, tbx2b, tbx3a, and tbx5a is required for proper heart development. Since three Tbx2/3 variants are overexpressed in the rnf2 mutant heart to a larger extent than tbx5a, we suggest that overexpression of tbx2/3 is the main driver of the observed myocardial phenotype. We therefore postulate that the overall downregulation of myocardial genes is the result of inadequate activation or maintenance of the chamber-myocardial gene expression program, which results in defects in maintenance of cell identity. Studies by others indicate that Rnf2 is an important player in the maintenance of tissue integrity in a wide variety of systems, and the zebrafish data on single hearts from rnf2 mutants adds to this list8,19,23,26. We hypothesize that the molecular pathway that allows for the formation of the conductive system is partially regulated by Rnf2 and that this ensures the correct balance in chamber and conductive cell identity within the heart. Disruption in this balance results in defects in cardiac development and functioning.
Methods
Zebrafish genetics and strains
Zebrafish (Danio rerio), were housed at 27.5 °C in a 14/10 h light/dark cycle. The evening before spawning, one male and one female were placed into a tank with a divider and the following morning, at the moment the light switched on, the fish were placed together for breeding. Embryos were collected and staged according to Kimmel et al.70. The rnf2ibl31/ibl31 zebrafish were out-crossed with wildtype (TLF) or with Tg(myl7::GFP)8,36. All methods were carried out in accordance with relevant guidelines and regulations of national animal welfare laws.
Genotyping
DNA was purified from embryos or from caudal fin tissue, taken from anesthetized adult zebrafish. Genotype analysis was performed by PCR using the primer set forward: 5′-TCTAAGCGCTCTCTTCGTCCAGA-3′ and reverse: 5′-ACAAGAGGATTTGTAACAAAGCCG-3′, followed by digestion of the PCR product with restriction enzyme Taq1 to identify the rnf2ibl31/ibl31 allele8. The agarose gel was imaged using the Gel Doc XR+ Imaging System (Bio-Rad), in combination with Image Lab Software (Bio-Rad). After acquisition, the image color was inverted and the levels were adjusted, using Photoshop, to visualize all bands for correct genotyping.
Whole mount in situ hybridization
Dechorionated embryos were fixed overnight at 4 °C in 4% PFA (Aurion, 151710) in PBST (PBS with 0.1% Tween-20), after which they were gradually transferred to and stored in 100% methanol. To prevent probe trapping, the heart edema of 3 dpf rnf2 mutant embryos was pierced with watchmaker forceps (INOX5). Embryos were treated with proteinase K. Whole mount in situ hybridization was performed as described previously71. Trypsin and fabp10 probes were generated by PCR from cDNA from 1 dpf wildtype embryos using the following primers: forward trypsin CAGG CCCTTTAGTGAGGGTTAATT TGTCTGCTGCTCACTGGTAC; reverse trypsin CAGG TAATACGACTCACTATAGGG GTCCTTGCCTCCCTCCATAA. Forward fabp10 CAGG CCCTTTAGTGAGGGTTAATT GTTGAGCTTCTCCAGAAAGCATG, reverse fabp10 CAGG TAATACGACTCACTATAGGG GATCATGGTGGTTCCTCCGA. T7 polymerase was used to generate the anti-sense probes. After WISH the embryos were mounted in 4% methylcellulose and imaged by light microscopy on a Leica MZFLIII, equipped with a DFC450 camera. The embryos were genotyped after imaging.
Immunostainings
Dechorionated embryos were fixed overnight in 4% PFA in PBST at 4 °C. After fixation, embryos were gradually transferred to and stored in 100% methanol. Before immunostaining embryos were transferred stepwise to PBST. Rabbit anti-Ring1b antibody from Cell Signaling Technology was used (RING1B Cell Signaling D22F2 1:4000). Antibody incubation was followed by a secondary antibody and subsequent DAB staining (EnVision+ System-HRP (DAB) k4010). The embryos were mounted in 4% methylcellulose and imaged by light microscopy on a Leica MZFLIII, equipped with a DFC450 camera. The embryos were genotyped after imaging.
ChIP-sequencing
Embryos from a rnf2 heterozygous incross were sorted for the rnf2 pleiotropic phenotype at 3 dpf. These phenotypical mutants are used for the rnf2 mutant sample. The rnf2 mutation has a 100% penetrance and no false positives have been detected by genotyping after phenotypic screening. As controls we used a wildtype strain from the same genetic background. Pools of 80 to 100 embryos of 3 dpf were fixed, deyolked, and homogenized using pestles and sonicated to release and isolate the chromatin. Chromatin was stored at −80 °C. For spike-in experiments 30 µg zebrafish chromatin was mixed with Drosophila spike-in chromatin and incubated overnight with H2Aγ and RING1B antibodies (H2Aγ Active Motif 104597; RING1B cell signaling D22F2) or with H2Aγ and H3K27me3 antibodies (H2Aγ Active Motif 104597; Millipore 07–449). After overnight antibody incubation, the chromatin was extensively washed. The resulting ChIP-DNA was used as input for KAPA-HYPERprep library preparation. Libraries were paired-end sequenced (43 bp read-length) on an Illumina NextSeq500 platform. For wildtype and rnf2 mutant samples, two and three replicates were used, respectively.
ChIP-sequencing analyses
In order to avoid biases due to differences in the efficiency of the sequencing runs, spike-in ChIP-seq reads were mapped to the Drosophila melanogaster genome v6 using bwa mem version 0.7.1572 with default settings. Multimapping reads were excluded using samtools version 1.3.173 and duplicated reads were removed with Picard (http://broadinstitute.github.io/picard/). Once we obtained the number of reads mapped per sample, we normalized them based on the sample of each experiment with lowest number of mapped reads, by removing random reads accordingly. After this, remaining ChIP-seq reads were mapped to the GRCz10/danRer10 using bwa mem version 0.7.1572 with default settings. Multimapping reads were excluded using samtools version 1.3.173 and duplicated reads were removed with Picard (http://broadinstitute.github.io/picard/). Peaks were called using MACS 2.1.1.2016030974 relative to the input track using the options -f BAMPE -g 1.3e9 -q 1e-2–broad –broad-cutoff 1e-1. Peaks 1 kb or closer from each other were merged, and H3K27me3 peaks narrower than 100 nt were discarded. Intersecting peaks were considered for replicates using GenomicRanges75. Peaks overlapping peaks called in the input track were excluded. Clustering of peaks was done using the union of the remaining peaks found in wildtype Rnf2 and H3K27me3 ChIPs and rnf2 mutant H3K27me3 ChIP, considering only those overlapping promoter regions (400 nt upstream–100 nt downstream of the transcription start site). Clustering and visualization of the peaks (i.e. heatmaps, bandplots and profiles) was done using fluff version 2.1.376. GO term analysis for biological process on genes of cluster 3 was performed with Cytoscape 3.3.0 using the ClueGO 2.2.4 plugin with default settings. To reduce redundancy in biological process GO terms, GO term fusion and GO term grouping was applied and the groups were plotted with group p-value corrected with Bonferroni step down.
Gene set enrichment analyses
For whole embryo lysates, gene set enrichment analyses were performed using the GSEA software version 3.0 from the Broad Institute77 using default parameters, comparing gene counts from rnf2 mutants and wildtypes, normalized with DESeq2 1.28.078. As gene sets, genes belonging to the promoter regions of the five different clusters were considered. For single hearts, gene counts from 3 dpf wildtype and rnf2 mutant samples were compared after data normalization with Monocle version 2.4.079. As gene sets, a hand-curated list of heart markers38 classified by “Annotation” was used.
RNA-sequencing of whole embryo lysates
Embryos from a rnf2 heterozygous incross were sorted for the rnf2 phenotype at 3 dpf. These phenotypical mutants are used for the rnf2 mutant sample. As controls a wildtype strain from the same genetic background was used. Pools of 11 to 23 embryos of 3 dpf were homogenized in TRIzol. The ZYMO RNA microprep kit was used to isolate RNA and treat the samples with DNAseI. Subsequently, 750 ng RNA was used as starting material. rRNA was depleted using the Illumina RiboZero kit, followed by fragmentation, cDNA synthesis, and KAPA-HYPERprep library preparation. Libraries were paired-end sequenced (43 bp read-length) on an Illumina NextSeq500 platform. For wildtype and rnf2 mutant samples, eight and seven replicates were used, respectively.
RNA-sequencing analyses
RNA-seq reads were mapped to the D. rerio genome (GRCz10/danRer10) with the Ensembl gene annotation v87 using STAR80 version 2.5.2b with default parameters and –quantMode on “GeneCounts” to obtain quantification of expression levels. Analysis of differentially expressed genes was done with DESeq2 1.28.078 after removing the batch effect on all samples with RUVSeq 1.10.081.
Fluorescent imaging
Embryos from a Tg(myl7::GFP);rnf2 heterozygous incross were anaesthetized in MS-222 and embedded in 1.5% low-melting-point agarose (Sigma). The embryos were imaged by light microscopy on a Leica MZFLIII, equipped with a DFC450 camera. The embryos were genotyped after imaging.
SPIM-imaging
Embryos from a Tg(myl7::GFP)_rnf2 heterozygous incross were injected with α-Bungarotoxin at the one-cell stage82. As controls we used embryos from a Tg(myl7::GFP) incross, which were also injected with α-Bungarotoxin at the one-cell stage82. At 1 dpf the embryos were embedded for SPIM imaging in 1.5% low-melting-point agarose (Sigma) in FEP tubes (Bola, S1815-04). We used the custom build multidirectional selective plane illumination microscopy (mSPIM) as described before83. Photos were taken with a 20-minute interval and the images were synchronized. The stages in which both the ventricle and atrium are dilated were used for data visualization by Imaris software (bitmap). The embryos were genotyped after imaging.
Single HeartsRNA-sequencing
Hearts were manually dissected from 1, 2, or 3 dpf Tg(myl7::GFP) positive embryos from an rnf2 heterozygous incross using watchmaker forceps (INOX5) and placed into Eppendorf LoBind tubes with TRIzol (Ambion), rapidly frozen in liquid nitrogen, and stored at −80 °C prior to further processing. The remainder of the embryos was individually collected in methanol and used for genotyping. RNA was extracted from the wildtype and rnf2 mutant hearts using TRIzol reagent (Ambion) according to the manufacturer’s manual. After RNA extraction, pellets were resuspended with barcoded primers. Primers consisted of a 24 bp polyT stretch, a 4 bp random barcode, a unique 8 bp sample-specific barcode, the 50 Illumina adaptor (as used in the TruSeq small RNA kit), and a T7 promoter for in vitro transcription84. The RNA samples were subsequently reverse transcribed, pooled, and in vitro transcribed for linear amplification with the MessageAmpII kit (Ambion) according to the CEL-seq protocol42. Illumina sequencing libraries were prepared with the TruSeq small RNA sample prep kit (Illumina) and sequenced single-end at 75 bp read length on an Illumina NextSeq. 500 instrument.
Single HeartsRNA-sequencing analyses
Raw reads were processed to obtain expression levels following the pipeline designed for the CEL-Seq method42 using the scripts available at https://github.com/yanailab/CEL-Seq-pipeline. Data normalization and differential gene expression analyses were done with Monocle version 2.4.079. Samples were filtered according to +/−2 SD cut-off in the amounts of mRNA that is detected in the sample after log transformation. Genes expressed in at least 25% of the samples at a given developmental stage were used for differential gene expression analysis and GSEA.
Supplementary information
Acknowledgements
We thank M. Hellinga and S.C. Steenbeek for technical support; Dr. Anna Pavlina Haramis from the Institute of Biology in Leiden for providing the rnf2ibl31/ibl31 zebrafish, Dr. Sylvia Boj from the Hubrecht Institute for providing the fabp2 probe, and the Bakkers laboratory from the Hubrecht Institute for providing the myl7, nppa, myh6, and vmhc probes. We thank Prof. Dr. Ir. H.G. Stunnenberg from the Radboud University for suggestions and critically reading the manuscript, Prof. Dr. G.J.C. Veenstra from the Radboud University for suggestions, and the animal caretakers for zebrafish husbandry. We thank the members of the Kamminga laboratory from the Radboud University, the Bakkers laboratory at the Hubrecht Institute, and the Huisken laboratory at the Max Planck Institute of Molecular Cell Biology and Genetics for suggestions and discussions. The work was funded by the Innovative Research scheme of the Netherlands Organisation for Scientific research (www.nwo.nl, NWO-Vidi 864.12.009, L.M.K.).
Author Contributions
N.D.C. designed and performed experiments, analyzed the ChIP- and RNA-seq data, wrote and edited the manuscript. D.M.E. analyzed the ChIP- and RNA-seq data, edited the manuscript. M.M. performed experiments and edited the manuscript. M.A. performed experiments and edited the manuscript. D.E.M.d.B. assisted with experiments and edited the manuscript. K.M.A. performed experiments and edited the manuscript. J.H. assisted with experiments and edited the manuscript. J.B. designed experiments and edited the manuscript. L.M.K. designed and performed experiments, acquired funding, wrote and edited the manuscript, and supervised the work. All authors read and approved the final manuscript.
Data Availability
The ChIP-seq data and RNA-seq data from whole embryo lysates and the single hearts discussed in this manuscript have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE114038.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40867-1.
References
- 1.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–43. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 2.Simon JA, Kingston RE. Occupying Chromatin: Polycomb Mechanisms for Getting to Genomic Targets, Stopping Transcriptional Traffic, and Staying Put. Mol. Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Faou P, Völkel P, Angrand P-O. The zebrafish genes encoding the Polycomb repressive complex (PRC) 1. Gene. 2011;475:10–21. doi: 10.1016/j.gene.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–70. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 5.Kaustov L, et al. Recognition and specificity determinants of the human cbx chromodomains. J. Biol. Chem. 2011;286:521–9. doi: 10.1074/jbc.M110.191411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luis NM, Morey L, Di Croce L, Benitah SA. Polycomb in stem cells: PRC1 branches out. Cell Stem Cell. 2012;11:16–21. doi: 10.1016/j.stem.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 8.van der Velden YU, Wang L, van Lohuizen M, Haramis A-PG. The Polycomb group protein Ring1b is essential for pectoral fin development. Development. 2012;139:2210–20. doi: 10.1242/dev.077156. [DOI] [PubMed] [Google Scholar]
- 9.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat. Rev. Cancer. 2009;9:773–84. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 10.Lindeman LC, et al. Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos. Int. J. Dev. Biol. 2010;54:803–13. doi: 10.1387/ijdb.103081ll. [DOI] [PubMed] [Google Scholar]
- 11.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalb R, et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014;21:569–71. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 13.Blackledge NP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farcas AM, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper, S. et al. Targeting Polycomb to Pericentric Heterochromatin in Embryonic Stem Cells Reveals a Role for H2AK119u1 in PRC2 Recruitment. CellReports 1–15, 10.1016/j.celrep.2014.04.012 (2014). [DOI] [PMC free article] [PubMed]
- 16.Stock JK, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 17.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–7. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 18.Vidal M. Role of polycomb proteins Ring1A and Ring1Bin the epigenetic regulation of gene expression. Int. J. Dev. Biol. 2009;53:355–370. doi: 10.1387/ijdb.082690mv. [DOI] [PubMed] [Google Scholar]
- 19.Voncken JW, et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. USA. 2003;100:2468–73. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Napoles M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 2004;7:663–76. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 21.del Mar Lorente M, et al. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development. 2000;127:5093–100. doi: 10.1242/dev.127.23.5093. [DOI] [PubMed] [Google Scholar]
- 22.Lapthanasupkul P, et al. Ring1a/b polycomb proteins regulate the mesenchymal stem cell niche in continuously growing incisors. Dev. Biol. 2012;367:140–153. doi: 10.1016/j.ydbio.2012.04.029. [DOI] [PubMed] [Google Scholar]
- 23.van der Stoop P, et al. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS One. 2008;3:e2235. doi: 10.1371/journal.pone.0002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Endoh M, et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–24. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 25.Yu M, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol. Cell. 2012;45:330–43. doi: 10.1016/j.molcel.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Velden YU, Wang L, Querol Cano L, Haramis A-PG. The polycomb group protein ring1b/rnf2 is specifically required for craniofacial development. PLoS One. 2013;8:e73997. doi: 10.1371/journal.pone.0073997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Carroll D, et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 2001;21:4330–6. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao R, et al. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science (80-.). 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 29.Morey L, et al. Polycomb Regulates Mesoderm Cell Fate-Specification in Embryonic Stem Cells through Activation and Repression Mechanisms. Cell Stem Cell. 2015;17:300–15. doi: 10.1016/j.stem.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Illingworth RS, et al. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev. 2015;29:1897–902. doi: 10.1101/gad.268151.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bratzel F, López-Torrejón G, Koch M, Del Pozo JC, Calonje M. Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 2010;20:1853–9. doi: 10.1016/j.cub.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 32.Eskeland R, et al. Ring1B Compacts Chromatin Structure and Represses Gene Expression Independent of Histone Ubiquitination. Mol. Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leeb M, et al. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–76. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokobayashi S, et al. PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature. 2013;495:236–40. doi: 10.1038/nature11918. [DOI] [PubMed] [Google Scholar]
- 35.Egan B, et al. An alternative approach to ChIP-Seq normalization enables detection of genome-wide changes in histone H3 lysine 27 trimethylation upon EZH2 inhibition. PLoS One. 2016;11:e0166438. doi: 10.1371/journal.pone.0166438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C-J, Tu C-T, Hsiao C-D, Hsieh F-J, Tsai H-J. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- 37.Bakkers J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc. Res. 2011;91:279–88. doi: 10.1093/cvr/cvr098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill, J. T., Demarest, B., Smith, M., Gorsi, B. & Yost, H. J. Heart morphogenesis gene regulatory networks revealed by temporal expression analysis. Development (2017). [DOI] [PMC free article] [PubMed]
- 39.Kruse, F., Junker, J. P., van Oudenaarden, A. & Bakkers, J. Tomo-seq. Biophysical Methods in Cell Biology135 (Elsevier Ltd, 2016). [DOI] [PubMed]
- 40.Junker JP, et al. Genome-wide RNA Tomography in the zebrafish embryo. Cell. 2014;159:662–75. doi: 10.1016/j.cell.2014.09.038. [DOI] [PubMed] [Google Scholar]
- 41.Wu C-C, et al. Spatially Resolved Genome-wide Transcriptional Profiling Identifies BMP Signaling as Essential Regulator of Zebrafish Cardiomyocyte Regeneration. Dev. Cell. 2015;36:36–49. doi: 10.1016/j.devcel.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 42.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–73. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 43.de Pater E, et al. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136:1633–41. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strate I, Tessadori F, Bakkers J. Glypican4 promotes cardiac specification and differentiation by attenuating canonical Wnt and Bmp signaling. Development. 2015;142:1767–76. doi: 10.1242/dev.113894. [DOI] [PubMed] [Google Scholar]
- 45.Comet I, Helin K. Revolution in the Polycomb hierarchy. Nat. Publ. Gr. 2014;21:573–575. doi: 10.1038/nsmb.2848. [DOI] [PubMed] [Google Scholar]
- 46.Vallaster M, Vallaster CD, Wu SM. Epigenetic mechanisms in cardiac development and disease. Acta Biochim. Biophys. Sin. (Shanghai). 2012;44:92–102. doi: 10.1093/abbs/gmr090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shirai, M., Takihara, Y. & Morisaki, T. In Etiology and Morphogenesis of Congenital Heart Disease: From Gene Function and Cellular Interaction to Morphology 305–312, 10.1007/978-4-431-54628-3_43 (Springer, 2016). [PubMed]
- 48.Quaife-Ryan GA, Sim CB, Porrello ER, Hudson JE. Resetting the epigenome for heart regeneration. Semin. Cell Dev. Biol. 2016;58:2–13. doi: 10.1016/j.semcdb.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 49.Paige SL, et al. A Temporal Chromatin Signature in Human Embryonic Stem Cells Identifies Regulators of Cardiac Development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wamstad JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–20. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alexander, J. M. et al. Brg1 modulates enhancer activation in mesoderm lineage commitment. Development 1418–1430, 10.1242/dev.109496 (2015). [DOI] [PMC free article] [PubMed]
- 52.He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. USA. 2011;108:5632–7. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S, Lee JW, Lee S-K. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev. Cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J-D, et al. Proper Activity of Histone H3 Lysine 4 (H3K4) Methyltransferase Is Required for Morphogenesis during Zebrafish Cardiogenesis. Mol. Cells. 2015;38:580–6. doi: 10.14348/molcells.2015.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He A, et al. Polycomb Repressive Complex 2 Regulates Normal Development of the Mouse Heart. Circ. Res. 2012;110:406–415. doi: 10.1161/CIRCRESAHA.111.252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delgado-Olguín P, et al. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat. Genet. 2012;44:343–347. doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.San, B. et al. Normal formation of a vertebrate body plan and loss of tissue maintenance in the absence ofezh2. Sci. Rep. 6 (2016). [DOI] [PMC free article] [PubMed]
- 59.Zhou Y, et al. Bmi1 Is a Key Epigenetic Barrier to Direct Cardiac Reprogramming. Cell Stem Cell. 2016;18:382–95. doi: 10.1016/j.stem.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akasaka T, et al. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development. 2001;128:1587–97. doi: 10.1242/dev.128.9.1587. [DOI] [PubMed] [Google Scholar]
- 61.van der Lugt NM, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8:757–69. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 62.Li X, et al. Mammalian polycomb-like Pcl2/Mtf2 is a novel regulatory component of PRC2 that can differentially modulate polycomb activity both at the Hox gene cluster and at Cdkn2a genes. Mol. Cell. Biol. 2011;31:351–64. doi: 10.1128/MCB.00259-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dupays L, Kotecha S, Angst B, Mohun TJ. Tbx2 misexpression impairs deployment of second heart field derived progenitor cells to the arterial pole of the embryonic heart. Dev. Biol. 2009;333:121–131. doi: 10.1016/j.ydbio.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 64.Washkowitz AJ, Gavrilov S, Begum S, Papaioannou VE. Diverse functional networks of Tbx3 in development and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012;4:273–83. doi: 10.1002/wsbm.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Greulich F, Rudat C, Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc. Res. 2011;91:212–222. doi: 10.1093/cvr/cvr112. [DOI] [PubMed] [Google Scholar]
- 66.Christoffels VM, et al. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev. Dyn. 2004;229:763–770. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- 67.Hoogaars WMH, et al. TBX3 and its splice variant TBX3+ exon 2a are functionally similar. Pigment Cell Melanoma Res. 2008;21:379–387. doi: 10.1111/j.1755-148X.2008.00461.x. [DOI] [PubMed] [Google Scholar]
- 68.Hatcher CJ, Basson CT. Getting the T-box dose right. Nat. Med. 2001;7:1185–1186. doi: 10.1038/nm1101-1185. [DOI] [PubMed] [Google Scholar]
- 69.Garrity DM, Childs S, Fishman MC. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development. 2002;129:4635–4645. doi: 10.1242/dev.129.19.4635. [DOI] [PubMed] [Google Scholar]
- 70.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 71.Houwing S, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 72.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 00, 1–3 (2013).
- 73.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lawrence M, et al. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Georgiou G, van Heeringen SJ. fluff: exploratory analysis and visualization of high-throughput sequencing data. PeerJ. 2016;4:e2209. doi: 10.7717/peerj.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Swinburne IA, Mosaliganti KR, Green AA, Megason SG. Improved long-term imaging of embryos with genetically encoded α-bungarotoxin. PLoS One. 2015;10:1–15. doi: 10.1371/journal.pone.0134005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mickoleit M, et al. High-resolution reconstruction of the beating zebrafish heart. Nat. Methods. 2014;11:919–922. doi: 10.1038/nmeth.3037. [DOI] [PubMed] [Google Scholar]
- 84.Grün D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat. Methods. 2014;11:637–40. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 85.Noël ES, et al. A Nodal-independent and tissue-intrinsic mechanism controls heart-looping chirality. Nat. Commun. 2013;4:2754. doi: 10.1038/ncomms3754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-seq data and RNA-seq data from whole embryo lysates and the single hearts discussed in this manuscript have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE114038.