Abstract
Increasing antibiotic resistance has led to the development of new strategies to combat bacterial infection. Anti-virulence strategies that impair virulence of bacterial pathogens are one of the novel approaches with less selective pressure for developing resistance than traditional strategies that impede viability. In this study, a small molecule CM14 [N-(4-oxo-4H-thieno[3,4-c]chromen-3-yl)-3-phenylprop-2-ynamide] that inhibits the activity of HlyU, a transcriptional regulator essential for the virulence of the fulminating human pathogen Vibrio vulnificus, has been identified. Without affecting bacterial growth or triggering the host cell death, CM14 reduces HlyU-dependent expression of virulence genes in V. vulnificus. In addition to the decreased hemolysis of human erythrocytes, CM14 impedes host cell rounding and lysis caused by V. vulnificus. Notably, CM14 significantly enhances survival of mice infected with V. vulnificus by alleviating hepatic and renal dysfunction and systemic inflammation. Biochemical, mass spectrometric, and mutational analyses revealed that CM14 inhibits HlyU from binding to target DNA by covalently modifying Cys30. Remarkably, CM14 decreases the expression of various virulence genes of other Vibrio species and thus attenuates their virulence phenotypes. Together, this molecule could be an anti-virulence agent against HlyU-harboring Vibrio species with a low selective pressure for the emergence of resistance.
Introduction
Traditional strategies to combat bacterial infection are mostly dependent on the use of antibiotics that inhibit bacterial viability. However, inhibition of viability leads to the inevitable emergence of strains resistant to antibiotics. The emergence and spread of antibiotic-resistant bacteria have become a threat to public health by reducing the effectiveness of present antibiotics, and thus these are a major cause for the rising healthcare costs1–3. This situation leads to an imminent need for the development of new strategies to impede the virulence, rather than viability, of bacterial pathogens4,5. Anti-virulence strategies disarm the pathogens, thereby rendering them harmless and more susceptible to immune clearance6–8. Compared to strategies that target viability, anti-virulence strategies may impose less selective pressure for the emergence of resistant strains2, and even further diminish the risk of commensal bacteria elimination9,10. Considerable works have been conducted to develop anti-virulence strategies, such as the inhibition of expression, secretion, or activity of virulence factors2,8.
Vibrio species generally inhabit in diverse marine environments. As an emerging cause of bacterial infection, some pathogenic Vibrio species infect humans and lead to a variety of clinical symptoms11,12. For example, Vibrio vulnificus can cause life-threatening septicemia and necrotizing fasciitis with high mortality rates in susceptible individuals13. Vibrio parahaemolyticus is a leading cause of seafood-borne gastroenteritis worldwide, resulting in diarrhea, nausea, fever, and chills14. Vibrio cholerae, a causative agent of watery diarrhea, is responsible for large outbreaks of cholera in various countries15, and Vibrio alginolyticus causes otitis and superficial wound infections in humans16. Although many antibiotics such as quinolones and tetracyclines have been applied for the treatment of Vibrio infection11,17, the recent reports of antibiotic resistant Vibrios threaten the efficacies of these antibiotics as treatment options18,19. In an effort to develop anti-virulence strategies against pathogenic Vibrio species, small molecules targeting virulence of Vibrio species have been identified20–25. However, very little is known about the molecular mechanisms of the compounds.
HlyU is a conserved transcriptional regulator required for the activation of various virulence genes in Vibrio species14,26–28. For example, V. vulnificus HlyU induces the expression of vvhA, rtxA, and plpA encoding hemolysin, multifunctional-autoprocessing repeats-in-toxin (MARTX) toxin, and phospholipase A2, respectively, by directly binding to the promoter region26,29,30. Similarly, V. parahaemolyticus HlyU directly induces the expression of exsA, which is essential for the type III secretion system 1 (T3SS1)14. The hemolysin VvhA lyses erythrocytes, damages endothelial cells, and induces inflammatory cell infiltration31,32. The MARTX toxin causes host cell rounding by dysregulating actin cytoskeleton and antagonizes phagocytic activity of host immune cells33–36. The secretory phospholipase A2 PlpA contributes to the lysis and necrotic death of host cells30. T3SS1 directly delivers multiple cytopathic and cytotoxic effector proteins into the host cells37. Host tissue destruction and inflammation caused by these virulence factors promote the survival, dissemination, and pathogenesis of V. vulnificus and V. parahaemolyticus in mice30,38,39. Accordingly, a deletion mutation of hlyU significantly attenuated virulence of the bacteria against human epithelial HeLa cells or mice14,29. Therefore, inhibition of the HlyU activity could be a plausible anti-virulence strategy against these Vibrio species.
In the present study, we performed high-throughput screening of 8,385 compounds and identified a small-molecule inhibitor of HlyU, CM14, that significantly inhibited the HlyU activity in V. vulnificus. CM14 reduced the expression of HlyU-regulated virulence genes, attenuating the virulence-related phenotypes of V. vulnificus in vitro, ex vivo, and in a mouse model. Biochemical analysis indicated that CM14 prevents HlyU binding to its target promoter DNA. Further mass spectrometric and mutational analyses revealed that a part of CM14 covalently modifies Cys30, a well-conserved residue of HlyU proteins in Vibrio species, both in vitro and in vivo. Remarkably, CM14 decreased the expression of virulence genes and showed anti-virulence effects against other pathogenic Vibrio species, without affecting the bacterial growth.
Results
Identification of CM14 as an inhibitor of the HlyU activity
To identify a specific inhibitor of HlyU, we constructed an Escherichia coli reporter strain containing pKK1306 (carrying an arabinose-inducible hlyU of V. vulnificus) and pZW1608 (carrying a promoterless lux operon fused to a promoter PVVMO6_00539)40. Because the VVMO6_00539 gene is directly repressed by HlyU (Fig. 1a; Supplementary Fig. S1a,b), the resulting E. coli strain remains non-luminescent in an arabinose-containing media unless a potential hit molecule inhibits either the expression or function of HlyU (Fig. 1a). By using this HlyU-repressed lux reporter system instead of the HlyU-activated system, we could eliminate the false identification of luciferase-inhibiting and/or luminescence-absorbing molecules as hits. Due to the lack of a previously discovered ligand or a putative ligand-binding site in HlyU, a random chemical library containing 8,385 small molecules was screened using the E. coli reporter strain. From the screening, three hit molecules (1025E12, 1030B04, and 1040E12) were identified as putative HlyU inhibitors (Fig. 1b). These hit molecules were reexamined using the V. vulnificus reporter strains containing the same reporter plasmid pZW1608 (Fig. 1c) or pZW1609 (Fig. 1d), respectively. In contrast to pZW1608, pZW1609 carries the promoterless lux operon fused to a promoter of the rtxA gene, PrtxA, which is directly induced by HlyU26. With each of the hit molecules, the wild-type V. vulnificus containing pZW1608 was more luminescent than the negative control (dimethyl sulfoxide, DMSO) (Fig. 1c), while V. vulnificus containing pZW1609 was less luminescent than the negative control (Fig. 1d). The use of these two distinct V. vulnificus reporter strains verified that the hit inhibitor molecules function directly on HlyU, not on other components such as a luciferase enzyme.
Figure 1.
High-throughput screening for HlyU inhibitors. (a) Schematic demonstration of high-throughput screening of small molecules. An E. coli reporter strain contains pKK1306 expressing HlyU under arabinose-inducible promoter PBAD and pZW1608 carrying the luxCDABE genes under HlyU-repressed promoter PVVMO6_00539. (b–d) Each bar represents RLU of the E. coli reporter strain (b) and V. vulnificus reporter strains containing pZW1608 (c) or pZW1609 (d) in the presence of hit molecules as indicated. Error bars represent the standard deviation (SD) from biological triplicates. Statistical significance was determined by multiple comparisons after one-way analysis of variance (ANOVA) (***p < 0.0005). 1025E12, 1030B04, and 1040E12, hit molecules; Positive, RLUs from E. coli without arabinose (b) or V. vulnificus hlyU mutant (c,d); Negative, RLUs from E. coli with arabinose (b) or V. vulnificus wild type (c,d); RLU, relative luminescence unit.
Among the hit molecules, 1025E12, N-(4-oxo-4H-thieno[3,4-c]chromen-3-yl)-3-phenylprop-2-ynamide (C20H11NO3S, molecular weight of 345.37) was most effective in the HlyU inhibition, and thus selected as a HlyU inhibitor and renamed ‘CM14’ (Fig. 2a). The structure of CM14 was confirmed by 1H NMR, 13C NMR, and mass spectrometric analyses (see Supplementary Information Methods). The HlyU activities were assessed using the wild-type V. vulnificus containing pZW1609 in the presence of various concentrations of CM14, and the half maximal effective concentration (EC50) of the molecule was determined as 30.97 μM (Fig. 2b). It is noteworthy that CM14 in the range of 20 to 200 μM did not alter the HlyU levels in V. vulnificus cells (Fig. 2c), suggesting that CM14 inhibits the activity rather than the cellular levels of HlyU. In addition, CM14 did not affect the growth of V. vulnificus (up to 2 mM) and was not toxic to the human epithelial INT-407 cells (up to 500 μM) (Fig. 2d,e). Therefore, these results suggested that CM14 is a small-molecule inhibitor of HlyU activity having a potential to be developed as an anti-virulence agent against V. vulnificus.
Figure 2.
CM14 inhibits the HlyU activity without affecting V. vulnificus growth. (a) The chemical structure of CM14, N-(4-oxo-4H-thieno[3, 4-c]chromen-3-yl)-3-phenylprop-2-ynamide. (b) The EC50 of CM14 inhibiting HlyU activity was calculated as described in the Methods section from three independent experiments. (c) Clear lysate samples of the wild type and hlyU mutant cells grown along with various concentrations of CM14 or DMSO (control) were resolved by SDS-PAGE, after which HlyU and DnaK were detected by Western blot analysis. Molecular size markers (Bio-Rad) are shown in kDa. Representative images of Western blot for HlyU and DnaK are shown, and the full-length blots are presented in Supplementary Figure S7. (d) Growth of the V. vulnificus strains along with various concentrations of CM14 or 2% DMSO (control) was monitored at 1 h intervals using a microplate reader. (e) Cytotoxicity was determined using LDH activities released from INT-407 cells incubated at 37 °C for 3 h with various concentrations of CM14, the wild-type V. vulnificus (at an MOI of 10), or 2% DMSO (control). The cytotoxicity was expressed using the LDH activity from the cells completely lysed by 5% Triton X-100 as 100%. Error bars represent the SD from the representative of three independent experiments. Statistical significance was determined by one-way ANOVA (e) (***p < 0.0005; ns, not significant; ND, not detected). WT, wild type; hlyU, hlyU mutant; MOI, multiplicity of infection.
CM14 reduces the HlyU-dependent virulence gene expression in vitro
Next, we examined if CM14 affects the expression of vvhA, rtxA, and plpA in V. vulnificus. Consistent with the previous result that CM14 inhibits HlyU activity, the transcript levels of vvhA, rtxA, and plpA of the wild-type V. vulnificus strain were significantly reduced in the presence of the molecule at 20 μM (Fig. 3a; WT + DMSO vs. WT + CM14). The reduced expression levels of the genes were close to those of the hlyU mutant strain ZW141 (Fig. 3a; WT + CM14 vs. hlyU + DMSO). We further investigated whether the reduced expression of the virulence genes is reflected in the virulence-related phenotypes. It was reported that V. vulnificus VvhA has a hemolytic activity against erythrocytes31. Thus, we compared the hemolytic activities in the culture supernatants of the V. vulnificus strains grown in the presence or absence of CM14. When incubated with human erythrocytes, the culture supernatant of the wild-type V. vulnificus grown in the presence of DMSO control showed robust hemolytic activity (Fig. 3b). In contrast, the culture supernatant of the wild-type V. vulnificus grown in the presence of CM14 exhibited significantly reduced (at 20 μM) or nearly no hemolytic activities (at 50 μM) similar to that of the hlyU mutant (Fig. 3b). Collectively, these results indicated that the effect of CM14 on the decreased expression of virulence genes is also represented as a reduced virulence-related phenotype of V. vulnificus in vitro.
Figure 3.
Effects of CM14 on the virulence-related phenotypes of V. vulnificus. (a,b) The V. vulnificus strains grown along with CM14 as indicated or DMSO (control) were harvested and fractionated for further analyses. (a) The transcript levels of vvhA, rtxA, and plpA in the total RNA of the cells were quantified by qRT-PCR and expressed using each transcript level of the wild type in the presence of DMSO as 1. (b) Hemolytic activities of the culture supernatants were determined against human erythrocytes and expressed using complete hemolysis by 5% Triton X-100 as 100%. (c) Morphological changes of HeLa cells infected with the V. vulnificus strains along with CM14 (50 μM) or DMSO (control) were photographed. Scale bars, 100 μm. (d) Cytotoxicity was determined using LDH activities released from INT-407 cells infected with the V. vulnificus strains along with CM14 as indicated and expressed using the LDH activity from the cells completely lysed by 5% Triton X-100 as 100%. Error bars represent the SD from three independent experiments (a,b) and from the representative of three independent experiments (d). Statistical significance was determined by the Student’s t-test (a) and by one-way ANOVA (b,d) (***p < 0.0005; **p < 0.005; *p < 0.05; ns, not significant). WT, wild type; hlyU, hlyU mutant.
CM14 attenuates the virulence of V. vulnificus ex vivo
The effects of CM14 on the V. vulnificus-mediated cytopathic changes of the host cells were assessed ex vivo. Since CM14 significantly decreased the rtxA transcript level in V. vulnificus (Fig. 3a), we first examined whether the molecule prevents the actin cytoskeleton dysregulation primarily caused by the MARTX toxin35,36. To this end, we monitored a rapid rounding phenotype of the HeLa cells infected with the V. vulnificus strains in the presence or absence of CM14. HeLa cells became round at 1 h post infection of the wild type41 (Fig. 3c; WT + DMSO). However, the rounding of HeLa cells was significantly attenuated in the presence of CM14 at 50 μM (Fig. 3c; WT + CM14), and thus the morphology of the cells was comparable to that of the cells with phosphate buffered saline (PBS, vehicle control) or the hlyU mutant (Fig. 3c; PBS + DMSO or hlyU + DMSO).
Furthermore, the effects of CM14 on the cytotoxicity of V. vulnificus were evaluated. For this purpose, lactate dehydrogenase (LDH) release from the INT-407 cells infected with the bacteria was determined. As shown in Fig. 3d, CM14 reduced LDH release from the cells infected with the wild-type V. vulnificus in a dose-dependent manner. Notably, 100 μM of CM14 almost abolished the LDH-releasing activity of V. vulnificus (Fig. 3d). Taken together, these results revealed that CM14 successfully attenuates the cytopathicity and cytotoxicity of V. vulnificus ex vivo.
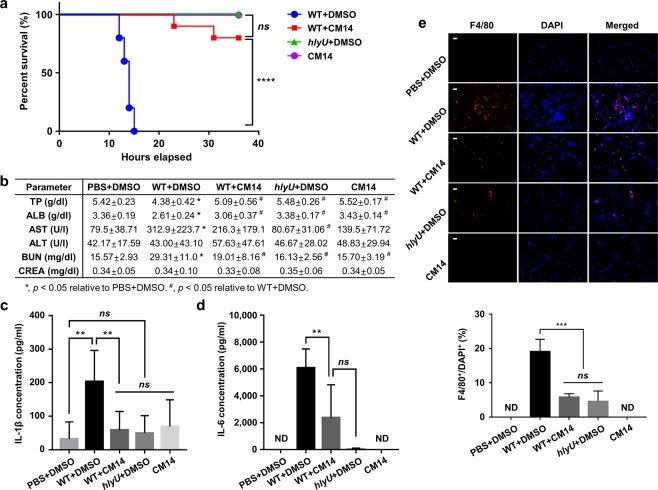
CM14 attenuates the pathogenesis of V. vulnificus in mice
To investigate the in vivo efficacy of CM14, mortality of mice infected with V. vulnificus was evaluated with or without co-administration of the molecule (Fig. 4a). All of the mice infected subcutaneously with the wild type strain were succumbed within 15 h post infection (Fig. 4a; WT + DMSO). In contrast, 80% of the mice survived until the end of experiment (36 h post infection) when CM14 was co-administered at 1.125 mM concentration (1.4 mg/kg body weight) (Fig. 4a; WT + CM14). These results revealed that co-administration of CM14 significantly prolonged the survival of mice infected with V. vulnificus (p < 0.0001, log rank test). Markedly, the survival rate of the mice infected with the wild type in the presence of CM14 was not statistically different from that of mice infected with the hlyU mutant (Fig. 4a; hlyU + DMSO). The combined results indicated that CM14 effectively inhibits the pathogenesis of V. vulnificus during murine infection.
Figure 4.
Effects of CM14 on the survival, pathophysiological changes, and inflammatory responses of mice infected with V. vulnificus. (a) Survival of the mice anesthetized with isoflurane and subcutaneously injected with wild type (n = 10), wild type with CM14 (n = 10), hlyU mutant (n = 5) at doses of 7.5 × 105 colony forming unit (CFU), or CM14 alone (n = 5, control). (b–e) The mice, injected as described in (a), were sacrificed at 7 h post infection to obtain blood and skin tissue samples. (b) The levels of TP, ALB, AST, ALT, BUN, and CREA in the blood plasma of each group [WT + DMSO (n = 10), WT + CM14 (n = 8), hlyU + DMSO (n = 6), DMSO (n = 6, control), and CM14 (n = 6, control)] were determined by blood biochemical analysis. The data represent the mean ± SD. Statistical significance was determined by multiple comparisons after one-way ANOVA (*p < 0.05 relative to PBS + DMSO; #p < 0.05 relative to WT + DMSO). (c,d) The cytokine levels of IL-1β (c) and IL-6 (d) in the blood plasma of each group (n = 7) were quantified by enzyme-linked immunosorbent assay (ELISA). (e) Infiltration of macrophages at the injection sites was determined using skin tissue samples that were immune-stained with F4/80 antibody (for macrophages, red) and DAPI (for nucleus, blue) for counter staining. The percentage of F4/80+ cells in DAPI+ cells was analyzed by using MetaMorph software. Scale bars, 10 μm (n = 4). Error bars represent the SD. Statistical significance was determined by log rank test (a) and by multiple comparisons after one-way ANOVA (c–e) (****p < 0.0001; ***p < 0.0005; **p < 0.005; *p < 0.05; ns, not significant; ND, not detected). WT, wild type; hlyU, hlyU mutant.
To examine not only survival but also pathophysiological changes, especially in the degrees of hepatic and renal dysfunction, we analyzed the biochemical parameters in the blood of the mice infected with V. vulnificus in the presence or absence of CM14. When mice were infected with the wild type (WT + DMSO), the blood plasma levels of total protein (TP) and albumin (ALB) were decreased, while the levels of aspartate aminotransferase (AST) and blood urea nitrogen (BUN) were increased, compared to the uninfected control mice injected with the vehicle (Fig. 4b; PBS + DMSO). However, the levels of biochemical parameters in mice infected with wild type in the presence of CM14 (WT + CM14) were comparable to those in the control groups such as mice injected with the vehicle or hlyU mutant (Fig. 4b; PBS + DMSO or hlyU + DMSO). The levels of alanine aminotransferase (ALT) and creatine (CREA) did not show any significant differences among the groups in the conditions tested (Fig. 4b).
Since severe inflammation is accompanied with V. vulnificus infection38,42, we next assessed immune responses in the V. vulnificus-infected mice either co-administered with or without CM14. The pro-inflammatory cytokines interleukin (IL)-1β and IL-6 levels in mouse blood plasma were significantly elevated upon infection of the wild type (Fig. 4c,d; PBS + DMSO vs. WT + DMSO). However, co-administration of CM14 alleviated the secretion of these pro-inflammatory cytokines (Fig. 4c,d; WT + CM14). Consistent with this, the recruitment of F4/80+ macrophages to the infection site was also reduced by the administration of CM14 (Fig. 4e). Remarkably, the percentage of F4/80+ cells over 4’,6-diamidino-2-phenylindole (DAPI)+ cells at the site infected with the wild type in the presence of CM14 was not significantly different from that with the hlyU mutant (Fig. 4e). Meanwhile, CM14 did not appear to be toxic to mice, as the levels of blood parameters and macrophage infiltration of the mice injected with CM14 were comparable to those of the mice injected with the vehicle (Fig. 4b to e; CM14 vs. PBS + DMSO). Furthermore, none of the mice injected with CM14 died (Fig. 4a). Taken together, these results indicated that CM14 attenuates the virulence of V. vulnificus in vivo and is not toxic toward mice.
CM14 inhibits the binding of HlyU to its target promoter DNA
As a transcriptional regulator, HlyU functions by binding directly to its target DNA14,26,30,43. Thus, we examined whether CM14 inhibits the activity of HlyU by altering the DNA binding of HlyU. Electrophoretic mobility shift assays (EMSAs) revealed that HlyU bound to the target PrtxA DNA and resulted in a retarded band of the DNA-HlyU complex in a HlyU concentration-dependent manner (Fig. 5a, DMSO). When 20 μM of CM14 was added, however, the HlyU binding to the DNA decreased, as less amount of retarded bands were detected compared to the DMSO-added control (Fig. 5a; CM14). In contrast, a random molecule that showed no HlyU-inhibiting activity in the screening did not affect HlyU binding to the DNA (Fig. 5a; Control). To determine the effect of CM14 on the dissociation constant (Kd) for HlyU, additional EMSA experiments were performed (Fig. 5b,c). Based on the concentration of HlyU required to bind 50% of the DNA probe, the Kd for HlyU without CM14 was estimated as 25.16 nM, while that with 2.5 µM of CM14 was estimated as 54.83 nM (Fig. 5d), indicating that the molecule significantly affects the equilibrium between free and DNA-bound HlyU proteins in the binding reaction. Indeed, the addition of increasing amounts of CM14 resulted in a concentration-dependent inhibition of HlyU binding to the DNA, and 50 μM of CM14 completely abolished the formation of the DNA-HlyU complex (Fig. 5e). Together, the results suggested that inhibition of HlyU binding to its target DNA is a possible mechanism of CM14.
Figure 5.
EMSAs of the HlyU and the rtxA regulatory DNA complexes. (a) HlyU protein (50 or 100 nM) was added to the radioactively-labeled PrtxA DNA (5 nM) along with either DMSO (10%), CM14 (20 μM), or a random chemical (20 μM, control), and the complexes were separated by electrophoresis on a 6% nondenaturing polyacrylamide gel. (b,c) The labeled probe DNA was mixed with increasing amounts of HlyU as indicated, and then the complexes were separated as described above. (d) The relative affinities of HlyU in the presence or absence of CM14 were compared using the data from (b,c). The concentration of bound DNA was calculated and plotted against the concentration of HlyU added. Each arrow points to the position of half-maximal binding corresponding to the Kd. (e) HlyU protein (100 nM) was added to the labeled probe DNA along with increasing concentrations of CM14 as indicated, and then the complexes were separated as described above. The cropped gel images are shown, and the full-length gels are presented in Supplementary Figure S8a to d. B, bound DNA; F, free DNA.
Chemical modification of HlyU by CM14
The possible mechanism of CM14 to inhibit the DNA-binding activity of HlyU was further investigated at a molecular level. To this end, tandem mass spectrometric analysis was performed for the CM14-treated HlyU sample. Figure 6a clearly showed that the Cys30 residue (C#) in the HlyU peptide, RLQILC#MLHNQELSVGELCAK, was covalently modified by the moiety with molecular mass of 130.042 Da, indicating that a certain part of CM14, probably consisting of C9H7O, is attached to the Cys30 of HlyU. Importantly, this modification seems to occur in vivo as well, because the freshly purified HlyU protein from the CM14 (50 μM)-treated E. coli cells also revealed the same result (Supplementary Fig. S2a). To verify this modification on the Cys30, a mutant HlyU protein with Cys to Ser substitution at Cys30 (HlyUC30S) was prepared and reacted with CM14. When the resulting mixture was analyzed by tandem mass spectrometry, a spectrum corresponding to the HlyU peptide containing a substituted serine, but not containing the covalently modified moiety, was detected (Supplementary Fig. S2b), indicating that the thiol group of Cys30 is important for the covalent modification. Consistent with this, the mutant HlyUC30S became resistant to CM14, as supported by the observations that the DNA-binding activity of HlyUC30S was less affected by the molecule in vitro (Fig. 6b) and that the expression of rtxA was not attenuated by the molecule in vivo (Fig. 6c).
Figure 6.
Chemical modification of the Cys30 residue of HlyU by CM14. (a) MS/MS spectrum of Cys30-modified peptide (RLQILC#MLHNQELSVGELCAK) from the CM14-treated HlyU protein. C# indicates the mass shift of C9H6O (# = +130.042 Da) by the cysteine modification. Both N- and C-terminal fragment ion series are represented as b and y series, respectively (e.g. b5, b6, b7… and y1, y2, y3…), and the annotated fragment ions are marked in the inserted peptide sequence. The observed precursor ion (monoisotopic m/z 862.440) in the inserted high resolution MS spectrum matched exactly with a theoretical m/z (862.439). (b) EMSA of the wild-type (WT), C30S mutant, or C96S mutant HlyU proteins and the radioactively-labeled PrtxA DNA complexes. The HlyU proteins (100 nM) were added to the probe DNA (5 nM) along with either 10% DMSO (control) or CM14 (10 μM) as indicated, and then the complexes were separated as described in Fig. 5. The cropped gel image is shown, and the full-length gel is presented in Supplementary Figure S8e. B, bound DNA; F, free DNA. (c) The hlyU mutants containing plasmids expressing wild-type or mutant HlyUs as indicated were grown along with CM14 (20 μM) or DMSO (control). The rtxA transcript levels in the total RNA of the cells were quantified by qRT-PCR and expressed using the transcript level of each group in the presence of DMSO as 1. Error bars represent the SD from biological triplicates. Statistical significance was determined by the Student’s t-test (*p < 0.05; ns, not significant). (d) Structural comparison of the CM14-treated HlyU (green, PDB code: 5ZNX) and the apo-HlyU (magenta, PDB code: 3JTH). The distances between the DNA-binding helices (𝛼4) are indicated. (e) Electron density map around Cys30 of the CM14-treated HlyU structure. The 2Fo–Fc (blue mesh) and the Fo–Fc (green mesh) maps are contoured at 1.5 σ and 4.4 σ, respectively. (f,g) Close-up views around Cys30 and Cys96 of the apo-HlyU (f) and the CM14-treated HlyU (g). The distances between sulfur atoms of the two cysteine residues are indicated.
According to the previously determined crystal structure of HlyU, there is another Cys residue, Cys96, near the Cys30 (Supplementary Fig. S2c, PDB code: 3JTH). To examine the role of Cys96 on the CM14-mediated modification of Cys30, this residue was also substituted with Ser. The resulting HlyUC96S was also resistant to CM14 in vitro and in vivo, as was HlyUC30S (Fig. 6b,c; HlyUC96S). Notably, however, Cys96 residue was detected unmodified in the above tandem mass spectrometric analysis of CM14-treated HlyU sample. Taken together, the results indicated that CM14 reacts with the thiol group of Cys30 of HlyU via a putative chemical reaction involving Cys96, and consequently inhibits the DNA-binding activity of HlyU.
To gain insights into the structural influence of CM14 on HlyU, we determined the crystal structure of CM14-treated HlyU protein at 2.1 Å resolution and compared it with the previously determined apo-HlyU structure44 (PDB code: 3JTH) (Fig. 6d,e). The overall structure of the CM14-treated HlyU is similar to that of apo-HlyU (Fig. 6d). However, there is an extra electron density map around Cys30 of the CM14-treated HlyU suggesting a potential chemical modification of Cys30 (Fig. 6e). Although the moiety attached to Cys30 is partially visible presumably due to the high flexibility, this observation is consistent with the above result that CM14 modifies the Cys30 of HlyU (Fig. 6a). Notably, further comparison revealed that CM14 induces a conformational change of HlyU, thereby substantially decreasing the distance between Cys30 and Cys96 from 8.4 Å to 4.1 Å (Fig. 6f,g). In addition, we found that the distance between two DNA-binding 𝛼-helices (𝛼4) in HlyU dimer by 2.9 Å (Fig. 6d), which may account for the impaired DNA-binding activity of HlyU (Fig. 5).
CM14 exhibits anti-virulence effects against other Vibrio species
HlyU proteins are highly conserved in Vibrio species and show high degree of sequence similarity. Especially, the residues Cys30 and Cys96 are well conserved in HlyU homologues of common pathogenic Vibrios, including V. parahaemolyticus, V. alginolyticus, and V. cholerae43 (Supplementary Fig. S3). Thus, we hypothesized that CM14 would be effective against other Vibrio species harboring HlyU homologue. Unfortunately, the homologues of rtxA and vvhA are absent in V. parahaemolyticus and V. alginolyticus, while the plpA homologue is present. However, the plpA homologues have not been reported to be regulated by HlyU. Accordingly, we examined the expression of exsA in V. parahaemolyticus which is directly induced by HlyU14. As expected, CM14 significantly reduced the exsA expression in V. parahaemolyticus (Fig. 7a). Since ExsA positively regulates multiple T3SS1-associated genes14, we further examined the expression of T3SS1 genes37,45 in the presence or absence of CM14. Again, the expression of tested T3SS1 genes, such as vp1668, vopQ, vopS, and vopR was significantly attenuated by CM14 treatment (Fig. 7a). Moreover, this molecule reduced the cytotoxicity of V. parahaemolyticus against the INT-407 cells in a dose-dependent manner (Fig. 7b).
Figure 7.
CM14 is effective in attenuating virulence of other Vibrio species. (a) V. parahaemolyticus was grown in T3SS1 inducing condition for 3 h along with CM14 (20 μM) or DMSO (control). The transcript levels of exsA, vp1668, vopQ, vopS, and vopR in the total RNA of the cells were quantified by qRT-PCR and expressed using each transcript level in the presence of DMSO as 1. (b) Cytotoxicity was determined using LDH activities released from INT-407 cells infected with V. parahaemolyticus at an MOI of 10 along with CM14 as indicated for 2 h and expressed using the LDH activity from the cells completely lysed by 5% Triton X-100 as 100%. (c) V. alginolyticus was grown to A600 of 0.5 along with CM14 (20 μM) or 2% DMSO (control). The transcript levels of each gene in the total RNA of the cells were quantified by qRT-PCR and expressed using each transcript level in the presence of DMSO as 1. (d) Cytotoxicity of V. alginolyticus was determined as described in (b), except that V. alginolyticus infection was performed for 4 h. (e,f) V. cholerae grown to A600 of 0.5 along with CM14 (20 μM) or 2% DMSO (control) were harvested and fractionated for further analyses. (e) The transcript levels of hlyA, tlh, and rtxA in the total RNA of the cells were quantified by qRT-PCR and expressed using each transcript level in the presence of DMSO as 1. (f) Hemolytic activities of the culture supernatants of V. cholerae. Ten microliters of the concentrated supernatants were spotted onto 7% horse blood agar plate. Three different culture supernatants were spotted and monitored after incubation at 37 °C for 24 h. Error bars represent the SD from more than three biological replicates (a,c,e) and from the representative of three independent experiments (b,d). Statistical significance was determined by the Student’s t-test (a,c,e) and by one-way ANOVA (b,d) (***p < 0.0005; **p < 0.005; *p < 0.05; ns, not significant).
Next, the effects of CM14 on V. alginolyticus and V. cholerae were examined. Since V. alginolyticus possesses T3SS which is particularly similar to that of V. parahaemolyticus16, we assumed that HlyU may also regulate T3SS in V. alginolyticus. In V. cholerae, HlyU activates the expression of hlyA by directly binding to the promoter region43. As shown in Fig. 7c to f, CM14 markedly inhibited the expression of exsA and T3SS genes (val1668, vopQ, vopS, and vopR) in V. alginolyticus and two divergently transcribed hemolysin genes (hlyA and tlh) in V. cholerae, thereby attenuating cytotoxicity or hemolytic activity of the Vibrios. Notably, CM14 did not hamper the growth of V. parahaemolyticus, V. alginolyticus, and V. cholerae (Supplementary Fig. S4), as in the case of V. vulnificus (Fig. 2d).
Discussion
Numerous bacterial genes encoding virulence factors required for overall success in the pathogenesis have been identified46,47. Many of these genes are coordinately regulated by a common global regulatory protein(s) to obtain their effective cooperation during infection48,49. Therefore, inhibiting the activity of global regulatory proteins is a promising strategy that can prevent the production of virulence factors simultaneously and thereby impede bacterial pathogenesis efficiently1,2,6. HlyU homologue in Vibrio species is a key regulatory protein that induces the expression of various virulence genes, suggesting that it could be an attractive target to develop the anti-virulence strategies against the pathogenic Vibrios. In the present study, we have identified and characterized a small molecule, CM14, that specifically inhibits HlyU activity, thus attenuating the pathogenesis of V. vulnificus without suppressing its growth. As expected, it also attenuated virulence phenotypes of other pathogenic Vibrios.
Among the genes regulated by HlyU in V. vulnificus, the expressions of VVMO6_00539 and VVMO6_03281 which are directly repressed by the protein (Supplementary Fig. S1a to c) were significantly induced in the presence of CM14 (Supplementary Fig. S1d,e). These results indicated that CM14 inhibits HlyU activity regardless of its regulatory mode, and also suggested that the molecule functions at a stage of HlyU binding to the target promoter DNA rather than other stages such as interaction of the protein with RNA polymerase. Indeed, the EMSA results revealed that CM14 directly inhibits DNA-HlyU interaction (Fig. 5). This inhibitory mode of action is advantageous in controlling pathogenesis of the bacteria because it blocks the production of virulence factors at the earliest step2,5.
To the best of our knowledge, CM14 is the first compound that covalently modifies HlyU and inhibits the virulence of V. vulnificus in a mammalian infection model. Although two compounds, fursultiamine hydrochloride and 2′,4′-dihydroxychalcone, have been identified as HlyU inhibitors, their mode of action was barely demonstrated24,25. Moreover, both of them failed to show in vivo efficacy in an animal model, and the latter even impeded bacterial growth at the low concentration of 15 μM. From the structural point of view, compared to the two compounds, CM14 is endowed with a novel keto-alkyne moiety that is required for the covalent modification of Cys30 in HlyU (see below).
Acute failures of liver and kidney in V. vulnificus-infected patients are the key pathophysiological features associated with fatal death50,51. Our study revealed that the inhibition of HlyU activity by CM14 suppressed the hepatic and renal dysfunction (Fig. 4b) and subsequently increased the survival rate of mice infected with V. vulnificus (Fig. 4a). In addition, our data showed that CM14 reduces both the production of pro-inflammatory cytokines in the blood plasma (Fig. 4c,d) and the massive recruitment of macrophages to the infection site (Fig. 4e). Because the MARTX toxin and VvhA induce pro-inflammatory cytokine production in mice38 and these cytokines trigger the recruitment of immune cells such as macrophages52,53, the in vivo results indicate that CM14 alleviates the clinical manifestations related to the V. vulnificus-induced septicemia by down-regulating the virulence factors. Since these virulence factors are also crucial for the invading pathogen to combat against residing immune cells and thus to proliferate/disseminate in the host30,33,38, V. vulnificus cells attenuated by the molecule might be readily cleared out of the mice.
Given the clear mass spectrometric evidence and biochemical data (Fig. 6a to c), we concluded that the Cys30 residue of HlyU was covalently modified with a certain part of CM14 consisting of C9H7O, and the Cys96 residue participated in this modification reaction. Based on these observations, we propose a possible chemical reaction mechanism for the covalent modification of HlyU by CM14 (Fig. 8; see the blue dashed box on the right). In the proposed reaction, the sulfur atom of Cys96 may first attack a carbon atom of the carbon-carbon triple bond of CM14. The second attack by the sulfur atom of Cys30 would release the amine group with the bulky rings, remaining a part with the phenyl group of CM14. Subsequently, a nucleophile (e.g. His92; Supplementary Fig. S2c) around the reaction site would cleave the sulfur-carbon bond between Cys96 and the remaining part of CM14, and the carbon is protonated, leaving the C9H7O moiety on Cys30.
Figure 8.
Proposed molecular mechanism underlying the CM14-mediated inhibition of HlyU binding to target DNA. A possible reaction mechanism for the Cys30 modification of HlyU by CM14 is shown in a blue dashed box on the right. First, the sulfur atom of Cys96 of HlyU reacts with a carbon atom (asterisk) of CM14, a Michael reaction acceptor site. Then, a sulfur atom of Cys30 of HlyU attacks a carbonyl carbon of CM14 and releases an amine group with bulky rings. Subsequently, a nucleophile (Nu, e.g. His92) around the reaction site cleaves the sulfur-carbon bond between Cys96 and the remaining part of CM14, protonating the carbon to create the carbon-carbon double bond. The remaining part of CM14 (represented by a green rounded box) is covalently linked to the sulfur atom of Cys30. Active HlyU can bind to target promoter DNA, leading to the production of virulence factors and making V. vulnificus fully virulent. In contrast, inactivation of HlyU by CM14 inhibits the DNA binding of HlyU, resulting in reduced expression of HlyU-regulated virulence genes. These events eventually attenuate the virulence of V. vulnificus.
Notably, CM14 seems specific for HlyU among various thiol-dependent transcriptional regulators, because only the HlyU regulon was differentially regulated by CM14 in the whole transcriptome sequencing analysis (Supplementary Fig. S5a). Indeed, samples of WT + CM14, hlyU + DMSO, hlyU + CM14 were clustered into a certain group that is distinct from the WT + DMSO samples in a principal component analysis (Supplementary Fig. S5b). We thus hypothesize that the bulky rings of CM14 may be involved in the specific interaction with HlyU at the early steps of binding, but the details of interactions including binding constant remain to be studied in the future. The effects of CM14 on thiol groups of other proteins such as those in the host should also be clarified by future studies.
Nonetheless, how does this modification affect the DNA-binding activity of HlyU protein? Intriguingly, a previous simulation study on the V. cholerae HlyU protein revealed that a distance between Cys38 and Cys104, which correspond to the Cys30 and Cys96 of V. vulnificus HlyU, respectively, has a correlation with the target DNA binding. Specifically, the distance between Cys38 and Cys104 is 8.67 Å when the protein is expected to bind to a target DNA43. From the comparison of the crystal structure of CM14-treated HlyU with that of apo-HlyU (Fig. 6d), we found that the distance between Cys30 and Cys96 residues was significantly shortened from 8.4 Å to 4.1 Å upon CM14 treatment (Fig. 6f,g). Furthermore, the distance between two DNA-binding 𝛼-helices (𝛼4) in HlyU dimer was also decreased by 2.9 Å (Fig. 6d). Altogether, the results indicate that CM14-mediated Cys30 modification substantially changes the HlyU conformation, and thus inhibits HlyU binding to target DNA (Figs 5 and 8).
Increasing number of studies have reported small molecules that can inhibit the activity or expression of virulence factors without affecting bacterial growth. For instance, Virstatin precludes dimerization of V. cholerae ToxT and prevents the expression of cholera toxin and toxin coregulated pilus54,55. Similarly, LED209 inhibits QseC activity, reducing the QseC-dependent virulence gene expression and virulence of multiple Gram-negative pathogens56,57. ITC-12 covalently modifies a cysteine residue of LasR, inhibits quorum sensing-mediated gene expression, and attenuates virulence of Pseudomonas aeruginosa58. Ebselen binds to an active cysteine residue in the cysteine protease domain and thereby inhibits the autoproteolytic cleavage of TcdA and TcdB, the Clostridium difficile major toxins59. Interestingly, CM14, in addition to ITC-12 and Ebselen, also covalently modifies Cys30 of V. vulnificus HlyU (Fig. 6a), supporting the present idea that cysteine residues, along with their scarcity and enhanced reactivity, can be good targets for the development of selective inhibitors of proteins60.
CM14 successfully inhibited the expression of various virulence genes in Vibrio species, including vvhA, rtxA, and plpA of V. vulnificus (Fig. 3a), T3SS1 genes of V. parahaemolyticus (Fig. 7a), T3SS genes of V. alginolyticus (Fig. 7c), and hlyA and tlh of V. cholerae (Fig. 7e). Consistent with the previous report that the promoter region of rtxA in V. cholerae is not directly bound by the HlyU protein61, the expression of rtxA in V. cholerae was not affected by CM14 (Fig. 7e). This is noteworthy because it further supports that CM14 specifically affects the HlyU protein. Nevertheless, these results suggest that CM14 has a broad-spectrum anti-virulence effect against pathogenic Vibrio species harboring HlyU homologue to regulate the expression of diverse virulence genes.
In conclusion, we identified a small molecule CM14 which inhibits HlyU activity by covalently modifying Cys30 and thus attenuates the virulence of Vibrio species. CM14 exhibited its anti-virulence effect even at the post-infection treatment, although it was ex vivo case (Supplementary Fig. S6). Further studies are needed to explore the potential of CM14 as a therapeutic agent against V. vulnificus infection, including the evaluation of CM14 analogues with improved bioavailability. Since CM14 does not hamper the bacterial growth, it would present no or low selective pressure for the development of resistance.
Methods
Strains, plasmids, culture conditions, and high-throughput screening
The strains and plasmids used in this study are listed in Supplementary Table S1. E. coli and V. vulnificus strains were grown in Luria-Bertani (LB) medium and LB supplemented with 2% (w/v) NaCl (LBS) at 37 °C and 30 °C, respectively. V. parahaemolyticus, V. alginolyticus, and V. cholerae were grown in LBS, tryptic soy broth supplemented with 1% (w/v) NaCl, and LB, respectively, at 37 °C. For T3SS1 inducing condition, V. parahaemolyticus was grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 1% fetal bovine serum (FBS)45. Bacterial growth was monitored spectrophotometrically at 600 nm (A600). HeLa cells originated from the American Type Culture Collection were maintained at 37 °C with 5% CO2 in DMEM containing 10% FBS, 50 μg/ml penicillin and 50 μg/ml streptomycin. For infection experiments, the cells were washed with pre-warmed PBS and kept in fresh DMEM. The small molecule libraries (each dissolved in 100% DMSO at 1 mM) were kindly provided by the Korea Chemical Bank (http://www.chembank.org). Detailed descriptions on construction of the E. coli reporter strain, information related to the high-throughput screening and results are provided in Supplementary Information Methods and Supplementary Table S2.
Determination of the EC50 of CM14
To determine EC50 (the concentration of CM14 inhibiting the HlyU activity by 50%), the wild-type V. vulnificus reporter strain containing pZW1609 (Supplementary Table S1), a HlyU-activated reporter plasmid, was exposed to various concentrations (10−10 to 10−3 M) of CM14. Luminescence and growth (A600) of the reporter strain were measured after 1.5 h incubation using a microplate reader (Infinite™ M200 microplate reader, Tecan, Männedorf, Switzerland), and RLUs were calculated by dividing luminescence with A60062. The HlyU activities were expressed using the RLU observed in the absence of CM14 (in the presence of 2% DMSO) as 100%. The EC50 was calculated by plotting the relative HlyU activities versus the CM14 concentrations using GraphPad Prism 7.0 (GraphPad Software, San Diego, CA).
Western blot and transcript analyses
The V. vulnificus strains along with CM14 or 2% DMSO were grown to A600 of 0.5 and used to analyze either the HlyU protein or the vvhA, rtxA, and plpA transcript levels. HlyU and DnaK in the cell lysates were detected using rabbit anti-V. vulnificus HlyU antibody and mouse anti-E. coli DnaK antibody (Enzo lifescience, Farmingdale, NY) by Western blot analysis. Expression of specific genes was determined by qRT-PCR with a pair of specific primers (Supplementary Table S3). Relative expression levels of each gene were calculated by using the 16S rRNA expression level as the internal reference for normalization.
Virulence assays
To determine hemolytic activity in vitro, the V. vulnificus strains grown to A600 of 1.0 along with CM14 or 2% DMSO (control) were harvested and fractionated into cells and supernatants by centrifugation. The culture supernatants were purified through PuradiscTM 25 mm syringe filter (pore size 0.2 μm; GE healthcare, Menlo Park, CA) and concentrated using Amicon Ultra-15 (cut-off 10 kDa; Millipore, Temecula, CA). An aliquot of the supernatants was mixed with an equal volume of human erythrocytes (10% in PBS; Innovative Research, Novi, MI) and incubated at 37 °C for 3 h. The hemolytic activity was measured by spectrophotometry as described previously63.
Two different assays were performed to determine cytopathicity and cytotoxicity of the V. vulnificus strains ex vivo. To examine the cytopathic changes, HeLa cells grown in a µ-slide 4-well plates (Ibidi, Germany) were infected with the V. vulnificus strains at an MOI of 2 along with 50 μM of CM14 or 1% DMSO (control). After 1 h incubation at 37 °C, the cells were washed and fixed, and nuclei and actin of the cells were stained with Hoechst® 33342 (final 5 μg/ml; Thermo Fisher Scientific, Waltham, MA) and with rhodamine-phalloidin (one unit per microscope slide; Thermo Fisher Scientific), respectively. Cell morphological changes were photographed using a laser scanning confocal microscope (C2plus, Nikon, Japan) and analyzed using NIS-Elements software (Nikon). To examine cytotoxicity, the monolayers of INT-407 cells (HeLa cell-derived epithelial cells) grown in a 96-well tissue culture plate (Nunc, Roskilde, Denmark) were infected with V. vulnificus strains at an MOI of 10 along with various concentrations of CM14 or 1% DMSO (control). After 2.5 h incubation at 37 °C, the LDH activities in the supernatant were measured as described previously30.
Mouse infection assays
All manipulations for mouse infection assay were performed following the National Institutes of Health Guidelines for Humane Treatment and approved by the Animal Care and Use Committee of Seoul National University (SNU-170417-26-2). Mouse mortality, blood biochemical parameters, pro-inflammatory cytokine production, and macrophage infiltration were evaluated to determine the virulence of V. vulnificus in vivo. For the mouse mortality test, the V. vulnificus strains grown to A600 of 0.5 were harvested and suspended in PBS to 7.5 × 106 CFU/ml. Groups of Institute of Cancer Research (ICR) female mice (7-week-old, specific-pathogen-free; Orient Bio, Seongnam, Republic of Korea) were injected with 100 μl of the bacterial suspension along with CM14 (to achieve 1.4 mg/kg body weight) or 10% DMSO subcutaneously under the dorsal skin. Survival of the mice was monitored for 36 h as described previously30.
To examine the levels of blood biochemical parameters, pro-inflammatory cytokine production, and macrophage infiltration to the injection sites, the mice injected as described above were sacrificed at 7 h post infection to obtain blood and skin tissue samples, respectively. For blood biochemical analysis, the blood samples were collected using cardiac puncture in heparin-coated tube (IDEXX Laboratories, Westbrook, ME) and analyzed as described previously30. Breifly, the levels of TP, ALB, AST, ALT, BUN, and CREA in the blood plasma were measured by using a biochemistry autoanalyzer (Hitachi 7180 autoanalyzer, High-Technologies Corp., Tokyo, Japan). The remaining blood samples were fractionated by centrifugation for 10 min at 1,000 × g to obtain the blood plasma. Cytokine levels of IL-1β and IL-6 in the blood plasma were determined by ELISA using commercially available ELISA kits for IL-1β (R&D systems, Minneapolis, MN) and IL-6 (AbFrontier, Seoul, Republic of Korea). For immunohistochemical analysis, the mouse skin tissue samples around injection sites were embedded in optimum cutting temperature (O.C.T.) compound (Sakura Finetek, Torrance, CA) and stored at −80 °C. Frozen tissue samples were cryo-sectioned to a 20-μm thickness and then mounted on silane-coated slides (Muto Pure Chemicals, Tokyo, Japan). Tissue samples on slides were fixed with 80% acetone for 10 min, washed twice with PBS, and blocked in 5% normal goat serum (Sigma-Aldrich, St. Louis, MO) for 20 min. Slides were incubated with F4/80 antibody (1:100 dilution; Santa Cruz, Paso Robles, CA) for 2 h at room temperature. After washing three times with PBS, the slides were incubated with Alexa Fluor 488®-conjugated goat anti-rabbit secondary antibody (1:200 dilution; Thermo Fisher Scientific) for 1 h. Subsequently, all slides were incubated with DAPI solution (5 μg/ml; Thermo Fisher Scientific) in PBS for 5 min at room temperature. All immunofluorescence images were obtained by Eclipse Ts2® fluorescence microscopy (Nikon, Tokyo, Japan), and colocalization of F4/80 with DAPI was analyzed by MetaMorph software (Universal Imaging, West Chester, PA).
Protein purification, site-directed mutagenesis, and EMSA
The purification of recombinant HlyU was performed by affinity chromatography followed by size exclusion chromatography. Site-directed mutagenesis was performed using QuikChange Site-Directed Mutagenesis Kit as described previously64. For EMSA, the 264-bp [γ-32P]ATP-labeled DNA fragment of PrtxA was amplified and incubated with the purified HlyU. Electrophoretic analysis of the DNA-protein complexes was performed as described previously30. When necessary, various concentrations of CM14 or DMSO were added to reaction mixture before incubation. As a control, a chemical randomly chosen from libraries that had no HlyU-inhibiting activity was added to the reaction mixture instead of CM14.
Mass spectrometric analysis of the HlyU modification
The gel slices corresponding to HlyU protein treated with CM14 were destained and followed by in-gel reduction and alkylation of cysteine residues. The resulting samples were washed, digested by sequencing-grade trypsin, subjected to C18-SPE clean up, and reconstituted for LC-MS/MS analysis. The acquired datasets from LC-MS/MS experiment were initially searched to find the unknown cysteine modification and subjected to MS-GF + analysis65 to confirm the cysteine modification.
Crystallization, structure determination, and refinement
HlyU protein was incubated with CM14 for 0.5 h at 4 °C and crystallized in a precipitation solution containing 0.1 M HEPES (pH 8.0), 20% (w/v) polyethylene glycol (PEG) 4 K and 10% (v/v) 2-propanol by hanging-drop vapor diffusion method at 14 °C. The HlyU-CM14 crystals were flash-frozen using 20% (w/v) sorbitol as a cryoprotectant in a nitrogen stream at −173 °C. An X-ray diffraction dataset was collected at Pohang Accelerator Laboratory beamline 5 C. The structure was determined and refined at a 2.1 Å resolution with an R factor of 23.8% and an Rfree of 26.8%. Further details on the structure determination and refinement are given in Supplementary Table S4.
Statistical analysis
Statistical analyses were performed as indicated in figure legends using GraphPad Prism 7.0 (GraphPad Software). For mouse lethality, mouse infection experiments were repeated twice to ensure reproducibility.
Supplementary information
Acknowledgements
We thank the Korea Chemical Bank for generously providing the small-molecule libraries. We also thank Professor Seok, Yeong-Jae, Seoul National University, for help in the V. cholerae experiments. This work was supported by National Research Foundation of Korea, funded by the Ministry of Science, ICT, and Future Planning (2017R1E1A1A01074639) and the Institute of Planning and Evaluation for Technology (IPET) of the Ministry of Agriculture, Food, and Rural Affairs (710012-03-2-SB110) to S.H.C, and by the Ewha Womans University Research Grant of 2018 to B.S.K.
Author Contributions
Z.-W.L., B.S.K., N.-C.H., H.J.H., and S.H.C. conceived, designed, and coordinated the research. Z.-W.L., B.S.K., K.K.J., Y.-J.B., S.K., Y.H.J., H.J.L., J.-S.K., J.K., and P.K.S. performed the experiments and acquired the data. Z.-W.L., B.S.K., S.K., N.-C.H., Y.H.J., H.J.L., H.J.H., J.-S.K., J.K., L.S.J., M.H.K., and S.H.C. analyzed the data. Z.-W.L., B.S.K., N.-C.H., H.J.H., J.-S.K., and S.H.C. wrote the paper. All the authors participated in discussions of the results and reviewed the final draft.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zee-Won Lee and Byoung Sik Kim contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39554-y.
References
- 1.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 2.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 3.Smith RD, Coast J. Antimicrobial resistance: a global response. B World Health Organ. 2002;80:126–133. [PMC free article] [PubMed] [Google Scholar]
- 4.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol R. 2010;74:417–433. doi: 10.1128/Mmbr.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickey SW, Cheung GYC, Otto M. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nature Reviews Drug Discovery. 2017;16:457–471. doi: 10.1038/nrd.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol. 2014;12:300–308. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 8.Johnson BK, Abramovitch RB. Small molecules that sabotage bacterial virulence. Trends Pharmacol Sci. 2017;38:339–362. doi: 10.1016/j.tips.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McFarland LV. Antibiotic-associated diarrhea: epidemiology, trends and treatment. Future Microbiol. 2008;3:563–578. doi: 10.2217/17460913.3.5.563. [DOI] [PubMed] [Google Scholar]
- 10.Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 11.Baker-Austin C, et al. Vibrio spp. infections. Nat Rev Dis Primers. 2018;4:8. doi: 10.1038/s41572-018-0005-8. [DOI] [PubMed] [Google Scholar]
- 12.Newton A, Kendall M, Vugia DJ, Henao OL, Mahon BE. Increasing rates of Vibriosis in the United States, 1996-2010: review of surveillance data from 2 systems. Clin Infect Dis. 2012;54:S391–S395. doi: 10.1093/cid/cis243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker-Austin C, Oliver JD. Vibrio vulnificus: new insights into a deadly opportunistic pathogen. Environ Microbiol. 2018;20:423–430. doi: 10.1111/1462-2920.13955. [DOI] [PubMed] [Google Scholar]
- 14.Getz, L. J. & Thomas, N. A. The transcriptional regulator HlyU positively regulates exsA expression leading to type III secretion system-1 activation in Vibrio parahaemolyticus. J Bacteriol., 10.1128/JB.00653-17 (2018). [DOI] [PMC free article] [PubMed]
- 15.Perez-Reytor, D. et al. Role of non-coding regulatory RNA in the virulence of human pathogenic Vibrios. Front Microbiol7, 10.3389/fmicb.2016.02160 (2017). [DOI] [PMC free article] [PubMed]
- 16.Zhao Z, et al. Autophagy is induced by the type III secretion system of Vibrio alginolyticus in several mammalian cell lines. Arch Microbiol. 2011;193:53–61. doi: 10.1007/s00203-010-0646-9. [DOI] [PubMed] [Google Scholar]
- 17.Wong, K. C., Brown, A. M., Luscombe, G. M., Wong, S. J. & Mendis, K. Antibiotic use for Vibrio infections: important insights from surveillance data. Bmc Infect Dis15, 10.1186/s12879-015-0959-z (2015). [DOI] [PMC free article] [PubMed]
- 18.Elmahdi S, DaSilva LV, Parveen S. Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: A review. Food Microbiol. 2016;57:128–134. doi: 10.1016/j.fm.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Scarano C, et al. Antibiotic resistance of Vibrio species isolated from Sparus aurata reared in Italian mariculture. New Microbiol. 2014;37:329–337. [PubMed] [Google Scholar]
- 20.Brackman, G. et al. Cinnamaldehyde and cinnamaldehyde derivatives reduce virulence in Vibrio spp. by decreasing the DNA-binding activity of the quorum sensing response regulator LuxR. Bmc Microbiol8, 10.1186/1471-2180-8-149 (2008). [DOI] [PMC free article] [PubMed]
- 21.Lee BC, Lee A, Jung JH, Choi SH, Kim TS. In vitro and in vivo anti-Vibrio vulnificus activity of psammaplin A, a natural marine compound. Mol Med Rep. 2016;14:2691–2696. doi: 10.3892/mmr.2016.5522. [DOI] [PubMed] [Google Scholar]
- 22.Kim JR, et al. Resveratrol modulates RTX toxin-induced cytotoxicity through interference in adhesion and toxin production. Eur J Pharmacol. 2010;642:163–168. doi: 10.1016/j.ejphar.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 23.Packiavathy I, Sasikumar P, Pandian S, Veera Ravi A. Prevention of quorum-sensing-mediated biofilm development and virulence factors production in Vibrio spp. by curcumin. Appl Microbiol Biot. 2013;97:10177–10187. doi: 10.1007/s00253-013-4704-5. [DOI] [PubMed] [Google Scholar]
- 24.Imdad, S., Chaurasia, A. K. & Kim, K. K. Identification and validation of an antivirulence agent targeting HlyU-regulated virulence in Vibrio vulnificus. Front Cell Infect Mi8, 10.3389/fcimb.2018.00152 (2018). [DOI] [PMC free article] [PubMed]
- 25.Imdad, S., Batool, N., Pradhan, S., Chaurasia, A. K. & Kim, K. K. Identification of 2′,4′-dihydroxychalcone as an antivirulence agent targeting HlyU, a master virulence regulator in Vibrio vulnificus. Molecules23, 10.3390/molecules23061492 (2018). [DOI] [PMC free article] [PubMed]
- 26.Liu M, Alice AF, Naka H, Crosa JH. HlyU protein is a positive regulator of rtxA1, a gene responsible for cytotoxicity and virulence in the human pathogen Vibrio vulnificus. Infect Immun. 2007;75:3282–3289. doi: 10.1128/Iai.00045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams SG, Attridge SR, Manning PA. The transcriptional activator HlyU of Vibrio cholerae: nucleotide sequence and role in virulence gene expression. Mol Microbiol. 1993;9:751–760. doi: 10.1111/j.1365-2958.1993.tb01735.x. [DOI] [PubMed] [Google Scholar]
- 28.Li L, Mou XY, Nelson DR. HlyU is a positive regulator of hemolysin expression in Vibrio anguillarum. J Bacteriol. 2011;193:4779–4789. doi: 10.1128/Jb.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YR, et al. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect Immun. 2003;71:5461–5471. doi: 10.1128/Iai.71.10.5461-5471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang KK, et al. Identification and characterization of Vibrio vulnificus plpA encoding a phospholipase A2 essential for pathogenesis. J Biol Chem. 2017;292:17129–17143. doi: 10.1074/jbc.M117.791657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray LD, Kreger AS. Purification and characterization of an extracellular cytolysin produced by Vibrio Vulnificus. Infect Immun. 1985;48:62–72. doi: 10.1128/iai.48.1.62-72.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray LD, Kreger AS. Mouse skin damage caused by cytolysin from Vibrio Vulnificus and by V. vulnificus infection. J Infect Dis. 1987;155:236–241. doi: 10.1093/infdis/155.2.236. [DOI] [PubMed] [Google Scholar]
- 33.Lo HR, et al. RTX toxin enhances the survival of Vibrio vulnificus during infection by protecting the organism from phagocytosis. J Infect Dis. 2011;203:1866–1874. doi: 10.1093/infdis/jir070. [DOI] [PubMed] [Google Scholar]
- 34.Kim, B. S., Gavin, H. E. & Satchell, K. J. F. Distinct roles of the repeat-containing regions and effector domains of the Vibrio vulnificus multifunctional-autoprocessing repeats-in-toxin (MARTX) toxin. Mbio6, 10.1128/mBio.00324-15 (2015). [DOI] [PMC free article] [PubMed]
- 35.Dolores JS, Agarwal S, Egerer M, Satchell KJF. Vibrio cholerae MARTX toxin heterologous translocation of beta-lactamase and roles of individual effector domains on cytoskeleton dynamics. Mol Microbiol. 2015;95:590–604. doi: 10.1111/mmi.12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y, et al. N-epsilon-Fatty acylation of Rho GTPases by a MARTX toxin effector. Science. 2017;358:528–530. doi: 10.1126/science.aam8659. [DOI] [PubMed] [Google Scholar]
- 37.Broberg CA, Calder TJ, Orth K. Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 2011;13:992–1001. doi: 10.1016/j.micinf.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeong HG, Satchell KJF. Additive function of Vibrio vulnificus MARTX(Vv) and VvhA cytolysins promotes rapid growth and epithelial tissue necrosis during intestinal infection. Plos Pathog. 2012;8:e1002581. doi: 10.1371/journal.ppat.1002581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pineyro P, et al. Development of two animal models to study the function of Vibrio parahaemolyticus type III secretion systems. Infect Immun. 2010;78:4551–4559. doi: 10.1128/Iai.00461-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, B. S. et al. QStatin, a selective inhibitor of quorum sensing in Vibrio Species. Mbio9, 10.1128/mBio.02262-17 (2018). [DOI] [PMC free article] [PubMed]
- 41.Satchell, K. J. F. Multifunctional-autoprocessing repeats-in-toxin (MARTX) Toxins of Vibrios. Microbiol Spectr3, 10.1128/microbiolspec.VE-0002-2014 (2015). [DOI] [PMC free article] [PubMed]
- 42.Shin SH, Shin DH, Ryu PY, Chung SS, Rhee JH. Proinflammatory cytokine profile in Vibrio vulnificus septicemic patients’ sera. Fems Immunol Med Mic. 2002;33:133–138. doi: 10.1111/j.1574-695X.2002.tb00582.x. [DOI] [PubMed] [Google Scholar]
- 43.Mukherjee D, Pal A, Chakravarty D, Chakrabarti P. Identification of the target DNA sequence and characterization of DNA binding features of HlyU, and suggestion of a redox switch for hlyA expression in the human pathogen Vibrio cholerae from in silico studies. Nucleic Acids Res. 2015;43:1407–1417. doi: 10.1093/nar/gku1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishi K, et al. Crystal structure of the transcriptional activator HlyU from Vibrio vulnificus CMCP6. Febs Lett. 2010;584:1097–1102. doi: 10.1016/j.febslet.2010.02.052. [DOI] [PubMed] [Google Scholar]
- 45.Zhou XH, Shah DH, Konkel ME, Call DR. Type III secretion system 1 genes in Vibrio parahaemolyticus are positively regulated by ExsA and negatively regulated by ExsD. Mol Microbiol. 2008;69:747–764. doi: 10.1111/j.1365-2958.2008.06326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finlay BB, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol R. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu HJ, Wang AHJ, Jennings MP. Discovery of virulence factors of pathogenic bacteria. Curr Opin Chem Biol. 2008;12:93–101. doi: 10.1016/j.cbpa.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 48.Cotter PA, DiRita VJ. Bacterial virulence gene regulation: An evolutionary perspective. Annu Rev Microbiol. 2000;54:519–565. doi: 10.1146/annurev.micro.54.1.519. [DOI] [PubMed] [Google Scholar]
- 49.Miller JF, Mekalanos JJ, Falkow S. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science. 1989;243:916–922. doi: 10.1126/science.2537530. [DOI] [PubMed] [Google Scholar]
- 50.Chou TNK, et al. Predictors of mortality in skin and soft-tissue infections caused by Vibrio vulnificus. World J Surg. 2010;34:1669–1675. doi: 10.1007/s00268-010-0455-y. [DOI] [PubMed] [Google Scholar]
- 51.Huang, K. C., Tsai, Y. H., Huang, K. C. & Lee, M. S. Model for end-stage liver disease (MELD) score as a predictor and monitor of mortality in patients with Vibrio vulnificus necrotizing skin and soft tissue infections. Plos Neglect Trop D9, 10.1371/journal.pntd.0003720 (2015). [DOI] [PMC free article] [PubMed]
- 52.Kumar H, Kawai T, Akira S. Pathogen Recognition by the Innate Immune System. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 53.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 54.Hung DT, Shakhnovich EA, Pierson E, Mekalanos JJ. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science. 2005;310:670–674. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 55.Shakhnovich EA, Hung DT, Pierson E, Lee K, Mekalanos JJ. Virstatin inhibits dimerization of the transcriptional activator ToxT. P Natl Acad Sci USA. 2007;104:2372–2377. doi: 10.1073/pnas.0611643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rasko DA, et al. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Curtis, M. M. et al. QseC inhibitors as an antivirulence approach for Gram-negative pathogens. Mbio5, 10.1128/mBio.02165-14 (2014). [DOI] [PMC free article] [PubMed]
- 58.Amara N, et al. Covalent inhibition of bacterial quorum sensing. J Am Chem Soc. 2009;131:10610–10619. doi: 10.1021/ja903292v. [DOI] [PubMed] [Google Scholar]
- 59.Bender, K. O. et al. A small-molecule antivirulence agent for treating Clostridium difficile infection. Sci Transl Med7, 10.1126/scitranslmed.aac9103 (2015). [DOI] [PMC free article] [PubMed]
- 60.Lagoutte R, Patouret R, Winssinger N. Covalent inhibitors: an opportunity for rational target selectivity. Curr Opin Chem Biol. 2017;39:54–63. doi: 10.1016/j.cbpa.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 61.Wang, H. X., Ayala, J. C., Benitez, J. A. & Silva, A. J. RNA-Seq analysis identifies new genes regulated by the histone-like nucleoid structuring protein (H-NS) affecting Vibrio cholerae virulence, stress response and chemotaxis. Plos One10, 10.1371/journal.pone.0118295 (2015). [DOI] [PMC free article] [PubMed]
- 62.Jang KK, Gil SY, Lim JG, Choi SH. Regulatory characteristics of Vibrio vulnificus gbpA gene encoding a mucin-binding protein essential for pathogenesis. J Biol Chem. 2016;291:5774–5787. doi: 10.1074/jbc.M115.685321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lim JG, Choi SH. IscR is a global regulator essential for pathogenesis of Vibrio vulnificus and induced by host cells. Infect Immun. 2014;82:569–578. doi: 10.1128/IAI.01141-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bang YJ, et al. OxyR2 functions as a three-state redox switch to tightly regulate production of Prx2, a peroxiredoxin of Vibrio vulnificus. J Biol Chem. 2016;291:16038–16047. doi: 10.1074/jbc.M115.710343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim S, Pevzner PA. MS-GF plus makes progress towards a universal database search tool for proteomics. Nat Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.