Abstract
The oral cavity contains different types of microbial species that colonize human host via extensive cell-to-cell interactions and biofilm formation. Candida albicans—a yeast-like fungus that inhabits mucosal surfaces—is also a significant colonizer of subgingival sites in patients with chronic periodontitis. It is notable however that one of the main infectious agents that causes periodontal disease is an anaerobic bacterium—Porphyromonas gingivalis. In our study, we evaluated the different strategies of both pathogens in the mutual colonization of an artificial surface and confirmed that a protective environment existed for P. gingivalis within developed fungal biofilm formed under oxic conditions where fungal cells grow mainly in their filamentous form i.e. hyphae. A direct physical contact between fungi and P. gingivalis was initiated via a modulation of gene expression for the major fungal cell surface adhesin Als3 and the aspartic proteases Sap6 and Sap9. Proteomic identification of the fungal surfaceome suggested also an involvement of the Mp65 adhesin and a “moonlighting” protein, enolase, as partners for the interaction with P. gingivalis. Using mutant strains of these bacteria that are defective in the production of the gingipains—the proteolytic enzymes that also harbor hemagglutinin domains—significant roles of these proteins in the formation of bacteria-protecting biofilm were clearly demonstrated.
Subject terms: Microbiology, Diseases
Introduction
Candida albicans, a commensal yeast-like fungus that commonly colonizes human mucosal surfaces1 often becomes an opportunistic pathogen in immunocompromised patients, thereby causing recurrent mucosal infections or life-threatening disseminated infections with high mortality rates2. During colonization or infections, C. albicans adheres to host surfaces and exploits an ability of fungal cells to switch their morphology between yeast and hyphal forms. The latter form promotes the formation of a structured microbial community, the biofilm, that increases a resistance of the fungal cells to antimicrobial agents or to environmental changes3. Notably also, the microbial biofilm formed by C. albicans can consist not only of this single fungal species but also contain numerous different species of bacteria, depending on the affected host niche4.
The oral cavity contains the highest diversity of microorganisms among all human microbial habitats5 and encompasses two types of functional surfaces, the mucosal surface and the teeth. Moreover, the various sites of the oral cavity can represent different conditions in terms of nutrient and oxygen availability The microorganisms that are early colonizers of the salivary pellicle on the tooth surface are streptococci, with Streptococcus mutans as the main bacterial pathogen involved in dental caries6 and some bridging microorganisms such as Fusobacterium nucleatum7. Both can be involved in the interactions with hyphal filaments of C. albicans, promoting co-colonization of these surfaces by yeast8.
Notably however, biofilm that extends below the gum line becomes subgingival plaque, where more pathogenic, Gram-negative anaerobes such as Porphyromonas gingivalis can arise9. P. gingivalis is the main infectious agent of periodontal disease, which involves an inflammatory process with many states and stages, ranging from easily curable gingivitis to irreversible severe periodontitis10. Moreover, it is believed that these bacteria play major roles as etiological factors or synergistic contributors in the multifaceted stimulation of the host immune system that can tip the scales towards development of chronic or autoimmune pathological states such as heart diseases, respiratory diseases, the osteoporosis or arthritis11.
Patients with chronic periodontitis show a significantly higher level of colonization with Candida spp. at subgingival sites than healthy individuals12. Moreover, C. albicans has been also frequently isolated from periodontal pockets13–16. It has also been reported that some C. albicans genotypes are unique to subgingival isolates, an observation that may reflect a selection process for C. albicans cells that are better adapted to this more anaerobic environment or to a coexistence with pathogens in plaque biofilm associated with periodontitis17.
Despite the extensive exploration of fungal-bacterial coexistence in prior studies, research concerning the interactions of C. albicans and P. gingivalis, particularly in the context of polymicrobial infections, is still at its infancy and in the context of periodontal disease has been nearly completely overlooked. Nevertheless, recent studies suggest that C. albicans, residing in different niches of the host organism, creates biofilms that provide a hypoxic microenvironment inside the biofilm community and support the growth of some anaerobic bacteria, even under apparently aerobic conditions that are normally toxic to them18. A gradient of oxygen concentration down to ca. 4% has been observed in C. albicans-formed biofilm, confirming the heterogeneous nature of this structure. In turn, P. gingivalis in a continuous culture system, depending on hemin availability, can tolerate low levels of oxygen (6–10%), and still reach steady-state growth. However, some of the physiologic processes of P. gingivalis have been found to be modulated19, thereby documenting a quick adaptation of bacteria to these new conditions. Moreover, C. albicans cells can survive and form biofilms under anaerobic and nutrient-limited conditions, and thus may pose a treatment challenge20. Another mechanism for protection of anaerobic bacteria by C. albicans cells was proposed for Clostridium difficile by Leeuwen and coworkers21 who suggested a reduction of oxygen level in the microbial environment as the consequence of antioxidant production or metabolic changes of C. albicans cells contacting these bacteria. However, in such microbial community the formation of fungal biofilm is suppressed and the bacterial adherence to C. albicans hyphae, rarely formed, is limited to their tips.
In the light of these data, we here examined a possible protection of the gingival anaerobe - P. gingivalis - through the formation of mixed biofilm with C. albicans inside which the bacteria can find better conditions for survival. We identified also the main bacterial and fungal surface proteins involved in mutual microbial interactions between these two potentially dangerous colonizers of the human oral cavity.
Results
Viability and adhesion properties of fungal (C. albicans) and bacterial (P. gingivalis) cells during the formation of different types of biofilms
To identify biofilm-favoring conditions, three different models of microbe contact were applied. In first two (sequential) a single-species biofilm (either fungal or bacterial) was developed prior to its colonization by the second microorganism. This model simulated different pathological conditions that occur during periodontal infections, where a dominant biofilm can be colonized by a second invading microbe. The third model (simultaneous), which was probably less likely to arise in vivo, involved the common contact of both pathogens that competed or cooperated in the colonization of the new surface. In all models, both anoxic or normoxic conditions were considered. In addition, as gingipains belong to the main virulence factors for P. gingivalis, we also compared the response of a wild strain of these bacteria with mutant strains lacking the genes that encode these proteolytic enzymes.
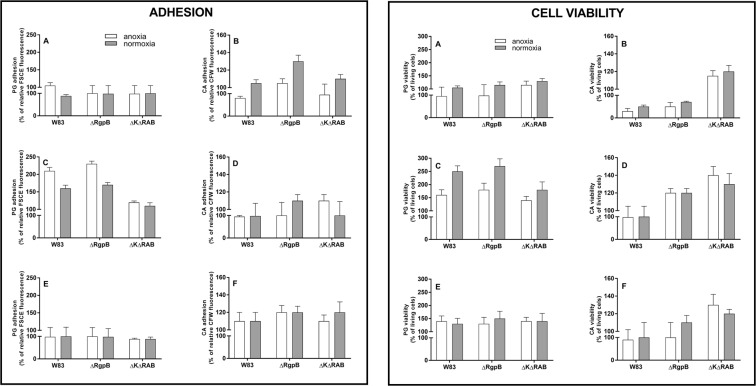
We first considered the bacteria as the initial surface colonizer that formed a stabile biofilm for 24 hours under anaerobic conditions and then enabled contact with C. albicans cells in the presence or absence of oxygen (Fig. 1, both panels, A, B). The stability of the bacterial biofilms formed by wild or mutant strains was unaffected by C. albicans colonization under both anoxic and normoxic conditions (left panel, A). However, bacterial biofilm viability increased by 20% in the presence of C. albicans cells in a normoxic environment (right panel, A). The viability of fungal cells was reduced to 35% by contact with the P. gingivalis wild type strain biofilm under anoxic conditions that favored bacterial growth and total gingipain activity (right panel, B). This observation suggests a dominant role of gingipains in mixed biofilm creation, where the proteolytic activity as well as agglutinin properties of these proteins could be essential. A reduction of gingipain activity in P. gingivalis cells by elimination of RgpB (∆RgpB) resulted in a 35% increase of fungal cell adhesion at normoxia (left panel, B). On the other hand, the elimination of all gingipains (∆K∆RAB) influenced mainly the fungal cell viability at both type of conditions but the adhesion, especially under anoxia was lower compared to the contact with wild type bacteria. It can be the first evidence for the importance of agglutinin domains of RgpA and Kgp in such interactions.
Figure 1.
Adhesion (left panel) and viability (right panel) of P. gingivalis (PG) and C. albicans (CA) cells that form a dual-species biofilm (A,B) Sequential model type I. A biofilm was formed by CFSE-stained bacteria for 24 h at 37 °C in Schaedler medium under anoxic conditions and this mono culture was covered with C. albicans cells and further propagated for 3 h at 37 °C in RPMI medium under both anoxic and normoxic conditions. After biofilm washing, the fungal cells were visualized by CFW staining. Adhesion was determined fluorometrically. Cell viability was determined after the biofilm had formed by cell scratching and propagation on the agar plate in media and under conditions appropriate for each species. (C,D) Sequential model type II. A biofilm was formed by C. albicans cells for 24 h at 37 °C in RPMI medium under normoxic conditions, and then settled with bacteria for 3 h at 37 °C in RPMI medium under anoxic and normoxic conditions. Further analyses were performed as described in (A,B). (E,F) Simultaneous model. A dual-species biofilm was formed by bacterial and fungal contact at the cell surface at the same time for 3 h at 37 °C in RPMI medium under anoxic and normoxic conditions. Further analyses were performed as described in A and B. All experiments (A–F) were performed with five repetitions in triplicate. The data shown are relative to a mono-species biofilm propagated under the same conditions. Data were presented as means ± SD (standard deviation) and analyzed using GraphPad Prism software (GraphPad, LaJolla, CA).
In the second model, where the fungal biofilm was developed under aerobic conditions for 24 h prior to contact with bacterial cells, with a domination of hyphal form of fungal cells, the bacteria exerted a marginal impact on them. However, wild type P. gingivalis under anoxic conditions lowered the fungal adhesion to the artificial surface, probably due to a degradation of fungal surface proteins (left panel, D). Nevertheless, bacteria were able to positively modulate the fungal viability, with the most pronounced effect observed during their contact under anoxic conditions with mutant strains defective in active gingipain production (right panel, D). On the other hand, bacteria presented more beneficial effects towards the fungal surfaces in terms of adhesion properties and viability (c.a. 200% increase), which were observed under anoxic and normoxic conditions for the wild type strain and also for the mutant strain partially lacking proteolytic activity (∆RgpB). However, no significant adhesion or viability enhancement was observed for the mutant lacking all gingipain activities and adhesive domains of RgpA and Kgp, again indicating the strong interactions between gingipains and fungal compounds located on the hyphal surface (both panels, C).
The findings for the third model of microbial co-adhesion, where both microorganisms contacted the artificial surface at the same time, corresponded to those obtained for the developed fungal biofilm under normoxia, with a protective effect of fungal biofilm on bacterial cell viability (right panel, E).
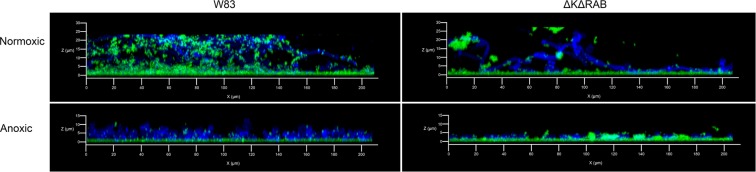
The aforementioned observations concerning microbial adhesion were confirmed by the analysis of the formed biofilms using confocal microscopy (Fig. 2). Under anoxic conditions, the bacterial cells were located mainly at the flat biofilm surfaces. However, during fast hypha development of C. albicans at normoxic conditions, the bacterial cells adhered directly to the growing hyphae. This effect was significantly reduced in the case of the mutant ∆K∆RAB.
Figure 2.
Microscopic evaluation of the sequential type II biofilm. The biofilm was prepared on poly-L-lysine coated glass slides as described for Fig. 1. CFSE-stained bacteria (green) and CFW-labeled fungal cells (blue) were incubated for 24 h at room temperature, then fixed with 3.8% paraformaldehyde.
Taken together, these results indicated that a developed fungal biofilm can serve as a protective environment for P. gingivalis and enable these bacteria to survive under unfavorable normoxia conditions. A question from this that remained to be resolved was the identity of the microbial surface components that could be responsible for this effect.
Expression changes in the genes encoding selected fungal virulence factors in mixed biofilms
The microorganisms that develop into an interspecies biofilm often change the type or levels of the expressed virulence factors employed, due to mutual interactions or regulatory mechanisms. Such changes were observed previously during biofilm formation between C. albicans and Streptococcus gordoni8 or Streptococcus oralis22–24.
To verify the relevance of these factors in biofilm formation between P. gingivalis and C. albicans, we analyzed changes in the expression level of the genes encoding representative adhesins belonging to Als family - Als3 and Als7, the cell surface adhesin - Hwp1, and a “moonlighting” protein ubiquitously expressed on the C. albicans cell surface - enolase (Eno1)25. We considered also the possible involvement of genes encoding proteases known to be preferentially secreted in yeast or hyphal forms of C. albicans (Sap3 and Sap6, respectively) and the surface-located and ubiquitously present Sap923. For these analyses, we applied the model of simultaneously formed biofilm after 3 hours of mutual contact under anoxic and normoxic conditions of the wild type P. gingivalis strain W83 and its mutant deprived of all gingipains (∆K∆RAB). The data (Fig. 3) showed a significant (10-fold) increase in ALS3 gene expression, mainly under normoxic conditions, in contact with the ∆K∆RAB mutant of P. gingivalis. A similar effect, although with a lower intensity was observed for HWP1 expression (5-fold increase). In contrast, the genes encoding Sap proteases were more active under anoxic conditions, with 3-fold increase in the expression of SAP9 encoding gene. Again, this phenomenon seemed to be slightly more intense during the contact of C. albicans cells with the mutant strain (∆K∆RAB). The expression of the ENO1 gene, encoding the essential glycolytic enzyme, enolase, was unchanged during 3 hours of fungal cell contact with any bacterial cells.
Figure 3.
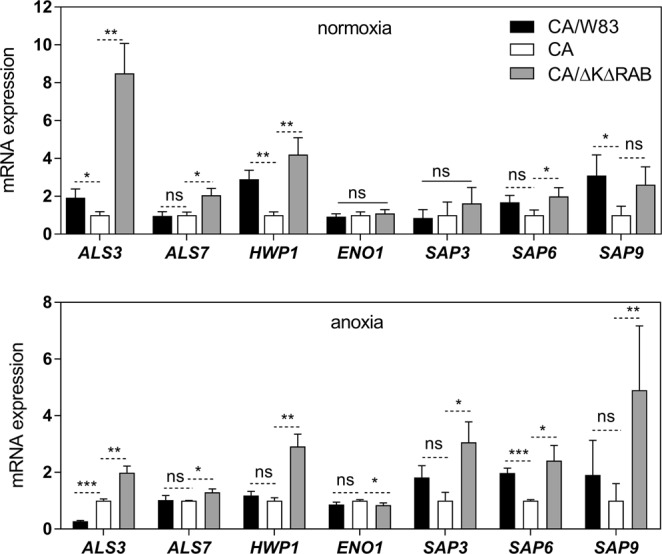
Changes in the expression levels of selected proteinous virulence factors in C. albicans during contact with P. gingivalis. After a 3 h contact between C. albicans and P. gingivalis cells in the simultaneous model of interaction under anoxic and normoxic conditions, total RNA was extracted from fungal cells using TRI reagent treatment. Real time quantitative PCR was then performed with a SYBR green detection assay. The selected genes encoding fungal surface protein Hwp1, adhesins of the agglutinin-like family, Als3 and Als7, aspartic proteases, Sap3, Sap6 and Sap9, and a cytosolic/‘moonlighting’ protein enolase (Eno1) were analyzed. The CT values (the average threshold cycle) were normalized to that of the ATC1 gene. To quantify gene expression, comparative CT was used and the relative expression was determined using the 2−ΔΔCT method71. Results are representative of three independent experiments and are expressed as means ± SD (standard deviation); n = 3. *P < 0.05; **P < 0.01; ***P < 0.001. The analysis was performed using GraphPad Prism software (GraphPad, LaJolla, CA, USA) with an one-way ANOVA test.
Differences between the fungal surface proteomes in mono- and dual-species (with P. gingivalis) biofilms
The significant expression changes identified for genes encoding different virulence factors of C. albicans that formed biofilm with P. gingivalis led us to investigate the surfaceome of C. albicans cells located within a mixed biofilm (Table 1). Under anoxic conditions, where P. gingivalis activity dominates, an overproduction of only three fungal proteins was identified compared to the surface proteins exposed in the mono-species biofilm. These were Eno1 (about 11-fold excess), Mp65 (9.4-fold excess) and Eng1 (2.5-fold excess). Although Eno1 is primarily a cytosolic enzyme involved in the glycolytic pathway, it has also been identified among candidal biofilm components26. Moreover, it has also been documented25 that extracellular enolase is not derived from simple fungal cell lysis but is actively secreted by both the yeast and hyphal forms of C. albicans cells, although the mechanism remains unknown. On the other hand, Mp65 is a cell wall mannoprotein of C. albicans with putative β-glucanase activity. It harbors multiple glycosylation sites and contains an RGD motif involved in surface adhesion. It is required for hyphal morphogenesis and belongs to the major targets of immune response in humans, similarly to enolase27. Eng1 is a cytosolic or periplasm-located protein often secreted into the media but its function outside the cell is unknown28.
Table 1.
Mass spectrometry analysis of C. albicans proteins identified at the cell surface after growth in mixed-species biofilm with P. gingivalis.
| Accession number | Protein name | Normoxia | Anoxia | ||
|---|---|---|---|---|---|
| W83 | ∆K∆RAB | W83 | ∆K∆RAB | ||
| O94049 | Acetyl-coenzyme A synthetase 1 (Acs1) | 6.0 | nd | nd | nc |
| P46273 | Phosphoglycerate kinase (Pgk1) | 4.6 | nd | nd | nd |
| P83776 | Hexokinase-2 (Hxk2) | 4.4 | nd | nd | nd |
| P30575 | Enolase 1 (Eno1) | 3.0 | 2.5 | 11.2 | 5.4 |
| P40953 | Chitinase 2 (Cht2) | 3.0 | 5.2 | nd | 4.0 |
| Q5AIR7 | Endo-1,3(4)-β-glucanase 1 (Eng1) | 2.6 | 1.9 | 2.5 | 1.4 |
| Q5AD07 | Cell surface Cu-only superoxide dismutase 5 (Sod5) | 2.5 | nd | nd | nd |
| P43076 | pH-responsive protein 1 (Phr1) | 2.2 | nd | nd | nd |
| P0CU38 | Agglutinin-like protein 2 (Fragment) (Als2) | 2.2 | 9.3 | nd | 14.8 |
| Q59XX2 | Cell surface mannoprotein MP65 (Mp65) | 1.6 | 5.5 | 9.4 | 6.9 |
| P43067 | Alcohol dehydrogenase 1 (Adh1) | 1.4 | nd | nd | 5.9 |
| Q5AFA2 | Extracellular glycosidase (Crh11) | 1.2 | 1.7 | nd | nd |
| Q59L12 | Agglutinin-like protein 3 (Als3) | 1.1 | 7.0 | nd | nd |
| Q59TP1 | Cell wall protein RTB1 (Rbt1) | 1.0 | 2.5 | nd | nd |
| Q5A8T4 | Agglutinin-like protein 1(Als1) | 0.4 | 3.4 | nd | nd |
| Q92211 | Glyceraldehyde-3-phosphate dehydrogenase (Tdh1) | 0.2 | nd | nd | 0.9 |
| Q5AMT2 | Glucan 1,3-β-glucosidase (Bgl2) | nd | 3.9 | nd | nd |
| Q9UWF6 | Lysophospholipase 1 (Plb1) | nd | 3.1 | nd | nd |
| P0CY35 | Elongation factor 1-α 1 (Tef1) | nd | nd | nd | 2.7 |
5 × 108 C. albicans cells were grown with 5 × 109 P. gingivalis W83 or ∆K∆RAB cells in RPMI 1640 medium for 24 hours at 37 °C under normoxic or anoxic conditions. Proteins were identified with LC-MS/MS after cell surface shaving with trypsin and additional digestion of released protein fragments for 24 hours. The resulting lists of peaks were used to search against the SwissProt protein database. Protein ranking with indicated fold changes according to the averaged NSAFs was created after comparison of C. albicans surface-exposed proteins released from cells grown in mixed-species biofilms to the sample containing fungal surface-exposed proteins of C. albicans grown in a single-species biofilm. Markedly enhanced protein presence at the surface of the fungal cells is highlighted in bold (nc, not comparable; nd, not detected).
The fungal surfaceome identified in the biofilm formed by C. albicans with a P. gingivalis mutant strain bereft of gingipains was also shown to contain a typical agglutinin-like adhesin, Als2 (14.8-fold increase), cytosol-derived alcohol dehydrogenase (Adh1; 5.9-fold increase), chitinase (Cht2; 4-fold increase) and elongation factor Tef1 (2.7-fold increase). All of these proteins were previously identified from candidal biofilm structure29.
Under the aerobic conditions preferred by C. albicans for growth, the increase in the amounts of proteins listed above was not as significant as at anoxia (2–6-fold), but additional cytosolic proteins were detected (Pgk1, Hxk2), probably resulting from fungal cell degradation by P. gingivalis W83 proteases. This possibility was supported by the expression of typical cell wall-damage response proteins, Phr1 and Sod530. Moreover, analysis of the biofilm formed between fungal and ∆K∆RAB cells under normoxic conditions additionally confirmed such a possibility, as in this case we did not observe any changes in the expression of proteins involved in fungal cell wall protection and none of the previously identified proteins typically functioning only in the cytosol were observed. What is also important to note is that a significant increase in the presence of agglutinin-like adhesins Als2, Als3 and Als1 (9.5, 7 and 3.5-fold increase, respectively) was observed compared to mono-species biofilm. These results suggest that these proteins could be involved in the interaction with P. gingivalis cells. Notably however, bacterial cells fully equipped with gingipains probably use different or additional fungal proteins during mutual contact within a biofilm. This suggestion comes from the prior observation that Als proteins were diminished in biofilm formed for a prolonged period and seemed to be more potently degraded by bacterial proteolytic enzymes, mainly gingipains (data not presented).
Confirmation of the interactions between selected candidal cell wall proteins and P. gingivalis cells
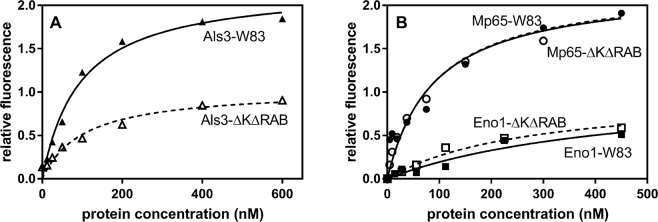
On the basis of our results suggesting the importance of the Als protein family, Mp65 cell wall protein and Eno1 in dual-species biofilm formation, we tested for possible interactions of these proteins with the wild type W83 strain of P. gingivalis and its mutant strain - ∆K∆RAB, using FACS analysis of bacterial cells after contact with FITC-labeled fungal proteins. The results presented in Fig. 4A showed that Als3 tightly interacts with the cell surface of both P. gingivalis strains. The elimination of gingipains however caused a significant reduction in the Als3-binding level. This result clearly indicated an involvement of gingipains in the interaction with fungal adhesin Als3, although other targets for Als3 would necessarily have had to exist.
Figure 4.
Binding of the fungal surface proteins Als3, Mp65 and Eno1 to P. gingivalis cells. Purified and fluorescein-stained fungal proteins were tested for binding to P. gingivalis wild type and mutant (∆K∆RAB) strains using FACS analysis. The results are representative of three independent experiments performed in triplicate. The fitting curve was generated using GraphPad software.
Two additionally identified fungal proteins, Mp65 and Eno1, also bound strongly to the P. gingivalis surface but no difference was observed between the binding properties of the wild type and mutant strains towards both fungal proteins (Fig. 4B). This could indicate that gingipains are not involved in these interactions or that there are other targets for these proteins which compensated for the interactions of the mutant cells.
Characteristics of RgpA complexes formed with the fungal cell wall proteins Als3, Mp65 and Eno1
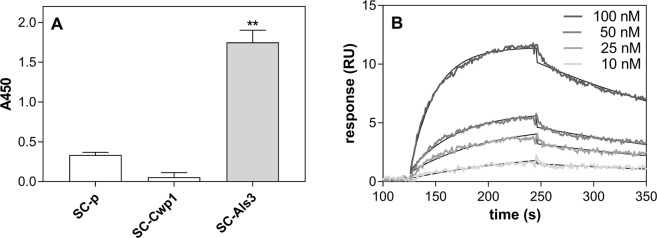
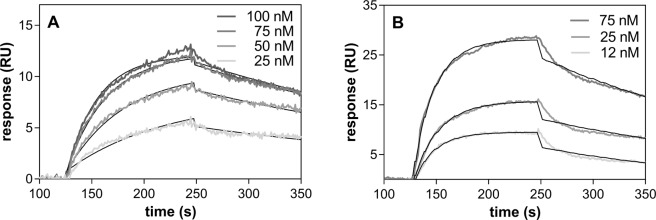
To further evaluate the gingipain role in the interaction with fungal biofilm proteins, a purified representative enzyme, RgpA, was used that contains both protease and agglutinin-like domains in its structure. The RgpA interaction with Als3 was first verified by testing its direct binding to S. cerevisiae cells with overexpressed Als3 exposed on the cell surface. To eliminate possible nonspecific binding of RgpA to fungal proteins, its binding to S. cerevisiae transformed with pBC542 plasmid alone or with cloned yeast CWP1 was analyzed for comparison31. The results of this analysis clearly supported our previous finding that gingipains are the target for Als3 (Fig. 5A). Moreover, the quantitative analysis of the Als3-RgpA interaction was performed, using SPR measurements, where Als3, isolated from the fungal surface was immobilized on the CM5-chip and tested for its interaction with RgpA present in solution poured over the chip surface at an increasing protein concentration range (Fig. 5B). The interaction was characterized by a dissociation constant (Kd) of 9.96 nM, which is strong enough to consider RgpA as an important determinant of candidal interaction with P. gingivalis cells within the biofilm structure.
Figure 5.
Interaction of RgpA with Als3. (A) The interaction of purified RgpA with S. cerevisiae containing surface overexpressed Als3 and Cwp1 or an empty pBC542 vector. The results are representative of three independent experiments and are expressed as means ± SD; n = 3. **P < 0.01 (by ANOVA with a Tukey test). (B) Interaction of RgpA with Als3 immobilized on the surface of a CM5 chip and analyzed by SPR measurements. The collected SPR data were analyzed using BIA evaluation software version 4.1 (GE Healthcare) using a simultaneous fitting of constants for the association rate (ka) and dissociation rate (kd) to the 1:1 Langmuir binding model with a baseline drift.
The role of Mp65 and Eno1, commonly identified as mixed biofilm components, in terms of possible interactions with gingipains was not clear as both wild type and a mutant strain defective in these protease productions interacted to the similar degree with these proteins. Although the targets in both P. gingivalis strains could possibly be different, we checked for possible interactions between RgpA and both fungal proteins using the SPR method. Both fungal proteins were bound to the chip surface thereby simulating their natural context within a biofilm. For the classical fungal adhesin, Mp65, the complex formed with RgpA was characterized by a Kd of 9.44 nM, i.e., the binding strength was similar to that for Als3 (Fig. 6A). On the other hand, we surprisingly found that Eno1 bound to RgpA with a 3-fold higher affinity than typical adhesins, with a Kd of 2.97 nM (Fig. 6B). We speculated therefore that a large amount of enolase detected within fungal biofilm could serve as additional support for the interactions between cells in a mixed microbial biofilm. However, the issue of which additional proteins exposed on the surface of the P. gingivalis mutant strain could be involved in the interaction with enolase and Mp65, remained to be clarified. The biology underlying this is likely to be quite complex due to the fact that gingipains are also involved in cell surface organization32.
Figure 6.
Interaction of the C. albicans proteins Mp65 and Eno1 with RgpA. RgpA binding to Mp65 and Eno1 was analyzed by SPR method using C. albicans proteins immobilized on a CM5 chip surface (ca. 300 RU) and contacting RgpA at a specified concentration range. Three independent experiments were performed in each case in triplicate. The data were analyzed using BIA evaluation software version 4.1 (GE Healthcare) using the 1:1 Langmuir binding model with a baseline drift.
Discussion
The oral cavity is the most diverse niche for microbial species including bacteria and fungi. Moreover, this environment undergoes continuous transformations, depending on the host age (i.e. from first teeth to dentures), diet, variable flow of saliva, pH, oxygen fluctuation, or prolonged use of drugs, especially antibiotics. To survive under such variable conditions, microorganisms develop biofilm communities and use diverse interaction mechanisms to protect or to dominate in these structures33.
C. albicans belongs to the most common oral fungal commensals. Although detected in oral microflora of healthy hosts in low amounts compared to bacteria, its colonization properties increase significantly in immunocompromised patients, especially if the environment favors the development of a hyphal morphology of the fungus and biofilm formation34. At sites of invasive fungal infection, the influx and activity of human host immune cells contribute to the development of hypoxic conditions, to which C. albicans cells can be well adapted as has been documented for fungal colonization of the human gastrointestinal tract35. Major C. albicans hypoxia responses affect the expression of genes producing transcripts, mainly hypha-specific, involved in cell wall and membrane structures, glycolysis and fermentation36. The broad adaptation abilities of C. albicans enable this fungus to colonize different niches in the oral cavity, creating mixed biofilms with aerobic, intermediate and anaerobic bacteria where mutualistic or commensal relationships are adopted to increase the resistance to the host immune defense or antimicrobial therapy37.
Recent evidence has suggested that C. albicans cells developing into a biofilm can cooperate with strictly anaerobic bacteria under aerobic conditions, thereby allowing these bacteria to survive and proliferate in an unfavorable environment and possibly induce a disease-associated phenotype18,38. Although the mechanisms of such anaerobic micro-niche formation by yeast are still not fully recognized, it was postulated that they could be related to the high level of O2 consumption by yeast39.
In order to evaluate and verify the different possible modes of microbial coexistence we applied three different experimental models to analyze the relationships between C. albicans and P. gingivalis cells forming a mixed biofilm. Despite prior evidence for the presence of Candida species in subgingival plaques or periodontal pockets40, it is difficult to attribute a definitive role for Candida spp. in the etiology of periodontitis in which P. gingivalis seems to be the keystone pathogen whose adhesion and viability could not be influenced significantly by contacting fungal cells. In contrast, C. albicans viability was found to be strongly dependent on the proteolytic potential of P. gingivalis, regardless of its adhesive properties. Moreover, the joint growth and competition for surface settlement under aerobic conditions favors C. albicans cell adhesion and hypha development that supports bacterial cell viability. Under anoxic conditions, preferred by bacteria for growth, fungal cell survival was still found to be dependent on gingipain activity. Contrary to this, when the surface is first settled by C. albicans cells that have formed a mature biofilm in which the hyphal form of the fungus is dominant and creates hypoxic conditions within the biofilm, P. gingivalis presented with increased adhesion, accompanied by a significant increase in cell viability, especially under normoxic conditions. These observations point to fungal biofilm as a source of potential receptors for P. gingivalis cell surface components and support the hypothesis that these biofilms form a protective environment for bacterial growth.
The preliminary gene expression analysis in our current study revealed that the expression of genes encoding the main fungal adhesins increased significantly under normoxia, whereas the aspartic protease-encoding genes, among which SAP5 and SAP6 have been identified as the major biofilm-related genes41, seemed to be mainly involved in fungal cell protection or defense against the bacterial neighborhood under anoxic conditions.
The following surface components of P. gingivalis were previously identified to be involved in cell-to-cell interactions: lipopolysaccharides (LPS), fimbriae, internalines and cysteine proteinases, i.e. gingipains42. Bacterial LPS can modulate Candida biofilm depending on the contacting bacterial species and contact duration43. Long fimbriae composed of FimA are mainly involved in co-aggregation with Actinomyces viscosus44, Treponema denticola45, Streptococcus gordonii46 and Streptococcus oralis47. However, no such interactions of FimA were detected in contact with C. albicans cells in a recent report48, where the authors presented also that P. gingivalis utilizes the InlJ internalin family protein to interact tightly with C. albicans hyphae within the biofilm structure. On the other hand, Kamaguchi et al. (2003) demonstrated that P. gingivalis vesicles, containing gingipains, strongly promote co-aggregation between S. aureus and filamentous form of C. albicans cells, suggesting a presence of a specific receptor for these proteins on the surface of both microorganisms and indicating a possible function of gingipains as bridging molecules in multispecies biofilms within the oral cavity49. These proteases, including arginine specific-gingipains, RgpA and RgpB, and a lysine specific gingipain Kgp are involved in hemagglutination50.
Within the structure of RgpA and Kgp, catalytic and hemagglutinin adhesive domains (Hgp44, Hgp15, Hgp17, Hgp27) have been identified, the latter of which are absent from RgpB51. The Hgp domains of gingipains are involved in the adherence to components of the intercellular matrix52 and in the binding of hemoglobin50 as well as in the binding and further activation of the human kinin-generating system53. Moreover, the importance of the Hgp44 domains of HRgpA and Kgp in the co-aggregation of P. gingivalis with T. denticola has also been documented54, as has the direct influence of Kgp on the composition and structure of a mixed biofilm with Tanerella forsythia55 and its indirect role in A. actinomycetemcomitans biofilm detachment56.
In our present experiments, we analyzed the differences in the interactions between wild type and gingipain-deprived P. gingivalis strains and concluded that the structure of gingipains, especially RgpA and Kgp, and the special contribution of their adhesive domains, could be particularly important for the cooperation with the C. albicans cell surface during biofilm formation. This hypothesis was additionally supported by our finding of a significant reduction of fungal cell viability under anoxia. Such close cell contact mediated by gingipains, which are proteins with both adhesive and proteolytic properties, could explain the observed changes in mixed biofilm characteristics.
During C. albicans hypha development within a biofilm, the structure becomes more prone to bacterial co-adhesion as the fungal cells expose several compounds on their surface that are susceptible to nonspecific, mostly hydrophobic or electrostatic interactions with bacteria. These surface molecules include, among others, surface located glucans and mannans, and also more deeply located chitin which will form an interaction if the bacteria approach the fungi closely enough57. Moreover, it has been reported that specific interactions with fungal mannoproteins can also occur depending on whether the nature of the relationship with bacteria is antagonistic or synergistic38. A good example of such a partner for synergistic interactions is Als3, a protein that is glycosylphosphatidylinositol-linked to the fungal cell surface and that belongs to the Als protein family uniquely expressed on hyphae. Its N-terminal region interacts with the SspB protein exposed on the surface of S. gordonii cells and has been thought to drive, at least in part, the formation of dual species biofilm58. Als3 has also been proposed to be a receptor for a S. aureus interaction that facilitates bacterial invasion into host tissues leading to systemic infections59.
In contact with P. gingivalis under anoxic and normoxic conditions, we here identified the involvement also of other proteins of the Als family, namely Als1–3. Surprisingly, the increased presence of these three proteins was detected mainly on the cell surface of fungi in contact with the P. gingivalis ∆K∆RAB mutant, with a lower surface expression observed when the P. gingivalis wild type strain was in contact. On the other hand, the Als3 binding capacity for wild type bacterial cells was found in our current analysis to be two-fold higher compared to the mutant strain, clearly indicating the involvement of gingipains in the interactions with Als3. This was further confirmed by the observation of complex formation between purified Als3 and RgpA, characterized by a low Kd (c.a. 10 nM), which was determined for the first time by our current analysis. Moreover, we could hypothesize that locally produced gingipains, being in close contact with fungal cells, can degrade the adhesins during prolonged mutual interactions. Such a conclusion was supported by our in silico analysis indicating many sites within Als3 structures that are prone to gingipain degradation. A degradation experiment we performed in vitro with purified proteins at optimal conditions also confirmed such an effect (data not presented). However, Als3 exposed on the fungal cell surface could be much less readily accessed and less sensitive to degradation due to glycosylation. Moreover, glycosylation of the Als proteins could also be responsible for its intriguing interactions with diverse types of bacterial proteins. Nevertheless, the role of the mannosyl portion of these proteins in the interaction with bacteria remains to be elucidated.
Another fungal cell wall protein overproduced during contact with P. gingivalis cells under both types of oxygen-availability conditions is mannoprotein Mp65, important for hyphal morphogenesis, membrane cell wall stability and organization, and, thus, biofilm formation. The RGD motif present in the Mp65 structure is directly involved in the adhesion properties of this protein27. In contact with P. gingivalis, we found that Mp65 was able to interact with RgpA at a similar affinity to Als3. Moreover, the analysis in silico and in vitro indicated a very definite stability of Mp65 against gingipain action (data not shown). However, it is also possible that Mp65 targets another or additional interaction partner(s) on the surface of the mutant bacterial strain.
Another fungal protein produced in increased amount upon contact with P. gingivalis cells, mainly under anoxic conditions but also, albeit marginally, under normoxia, was enolase. It is the most abundantly expressed cytosolic enzyme and is considered a multifunctional protein, with a predominant and classical role in the glycolytic pathway60 but also a factor that “moonlights” at the cell surface. As a surface protein, where it appears via a yet unknown translocation process, enolase was identified previously during C. albicans biofilm formation26 and was shown to be a partner for the interactions with human host proteins such as plasminogen and kininogen, mediate the interaction with extracellular matrix proteins like fibronectin, and promote a humoral immune response25,61. Recently, it was demonstrated that Eno1 at the fungal cell surface possesses transglutaminase activity, suggesting its involvement in the formation of covalent cross-links between cell wall proteins and chitin and/or glucan62, an effect that could stabilize a biofilm structure. Moreover, the role of bacterial enolase in interspecies interactions has been documented for Lactobacillus jensenii and Neisseria gonorrhea63. Our current study revealed the binding of Eno1 to both, mutant and wild P. gingivalis strains, suggesting the possibility that several bacterial partners exist for this protein. However, our more precise analysis of the binding between Eno1 and RgpA showed a 3-fold stronger affinity than that detected for the RgpA-Als3 complex. It could be suggested from this that the observed significant increase in enolase production, especially under anoxic conditions, is not only a metabolic response to an unfavorable environment but also has consequences for pathogen interactions. The increased enolase surface exposition did not correlate with previously detected expression of its encoding gene, probably owing to different timing of these events in our experiments or because the surface function of enolase can be regulated via more advanced processes that probably depend on many environmental stimuli. It is worth noting in this regard that surface-located fungal enolase is also vulnerable to citrullination by the peptidyl arginine deiminase produced by P. gingivalis with a possible consequence for protecting biofilm formation between C. albicans and P. gingivalis64.
Finally, an enzyme of interest that was identified from our present analysis to change its expression significantly on the fungal cells during contact with P. gingivalis was chitinase. This enzyme is associated with the yeast-to-hyphae transition and plays the role in the maintenance and architecture of C. albicans biofilm. Its altered presence in the mixed species biofilm suggests that the formation of these microbial communities is a diverse and dynamic process that attributes new properties to both pathogens and allows them to colonize new niches.
Our current analysis thus extends the understanding of P. gingivalis biofilm-driven chronic inflammatory disease that has been previously correlated with an increased risk of aspiration pneumonia, atherosclerosis, diabetes and rheumatoid arthritis65. Moreover, our findings in relation to the mutual interactions in biofilms that change over the contact time and respond to the host colonizing niche to protect the mutual coexistence of fungal and bacterial pathogens against antimicrobial therapy, creates new challenges in the future search for effective treatments of related diseases.
Materials and Methods
Bacterial growth conditions
P. gingivalis W83 (ATCC® BAA-308™) and gingipain mutants: ∆RgpB, lacking gingipain RgpB, and an isogenic gingipain-null mutant, ∆K∆RAB (lacking the presence of RgpA, RgpB and Kgp) were obtained as described previously65 and grown under anaerobic conditions (90% N2, 5% CO2, 5% H2) at 37 °C on blood agar plates or in liquid Schaedler broth supplemented with hemin (5 µg/ml;), L-cysteine (50 µg/ml), menadione (0.5 µg/ml), and additionally with tetracycline (1 µg/ml) in the case of ∆K∆RAB. Bacteria from the overnight cultures were centrifuged (4500 × g, 10 min) and the bacterial pellet was washed three times and resuspended in phosphate-buffered saline, pH 7.4 (PBS).
Fungal growth conditions
C. albicans strain 3147 (ATCC® 10231™) was purchased from American Type Culture Collection (Manassas, VA) and cultured aerobically at 30 °C on a YPD plate containing agar. The cells were then cultured for 4 h at 30 °C in YPD liquid medium on an orbital shaker (170 rpm) to achieve mid-logarithmic growth. The cells were then harvested by centrifugation (3000 × g, 5 min) and the cell pellet was washed twice with sterile PBS. Saccharomyces cerevisiae cells serving as a surrogate host for C. albicans Als3 (pBC542-als3lg) and S. cerevisiae Cwp1 (pBC542-cwp1), as well as the BY4742 strain transformed with pBC542 were obtained and cultured as described previously31.
Models of dual-species biofilm formation
Mixed species biofilm was prepared on the surface of flat-bottomed 96-well plastic plates (Corning Corporation, Corning, NY). In this present study, three different models of infection were applied to investigate C. albicans and P. gingivalis interactions. The term sequential model refers to surface infection, where one of the organisms plays a role as a first colonizer (C. albicans or P. gingivalis, respectively) and was grown overnight at optimal conditions after which the second coloniser was settled.
In sequential model type I, the P. gingivalis biofilm was first formed by settling the bacterial cells on the microplate wells (1 × 108 cells/well) in Schaedler medium followed by an overnight incubation at 37 °C under anaerobic conditions maintained using a GENbox jar with a Genbox anaer generator (bioMérieux S.A., Marcy l’Etoile, France). C. albicans cells (5 × 105) were then added and both organisms were cultivated for 3 and 24 hours at 37 °C under either normoxic or anoxic conditions. After this period, the supernatants were removed and each well was washed with PBS and further analysed as described below.
In sequential model type II, the suspension of C. albicans cells (5 × 105 cells/well) in RPMI 1640 medium was added to each well of the 96-well plate and the fungal cells were grown overnight under normoxic conditions at 37 °C. Subsequently, P. gingivalis cells (1 × 107 and 1 × 108 cells/well) were added and both microorganisms were cultivated for 3 and 24 hours at 37 °C under normoxic or anoxic conditions. After gentle washing of the cells, the adhesion and microorganism viability within the biofilm were examined.
In the simultaneous model, C. albicans (5 × 105 cells/well), and P. gingivalis (1 × 107 and 1 × 108 cells/well) cells were settled on the plate at the same time and competed for the surface under anoxic and normoxic conditions. After 3 and 24 h of coincubation, the unbound cells were removed by gentle washing of the plate with PBS and the formed biofilm was analysed as determined below.
In parallel with each of these mixed species biofilm models, monospecies biofilms were also developed under the same conditions to serve as an adequate reference.
Staining of bacterial and fungal cells
Before the preparation of biofilm, P. gingivalis cells were labelled with CellTraceTM CFSE staining probe (5(6)-carboxyfluorescein N-hydroxysuccinimidyl ester; Thermo Fisher Scientific, Waltham, MA) in the dark at 37 °C for 40 minutes. After washing out the excess unbound tracer, the labelled bacteria were used for biofilm formation. C. albicans cells were stained for 5 min with Calcofluor White solution (CFW, Sigma-Aldrich) at a final concentration of 1 µg/ml just prior to microscopic analysis or fluorescence measurements.
Determination of microbial cell adhesion and viability
The adhesion of bacterial and fungal cells within the formed dual-species biofilms was examined as follow. After gentle washing with PBS, the fluorescence intensity of CFSE-labelled P. gingivalis cells at 390/440 nm (excitation/emission) and CFW stained C. albicans cells at 480/520 nm was determined using a Synergy™ H1 Microplate Reader (BioTek Instruments, Winooski, VT). To test the viability of biofilm-forming microbial cells, a colony-forming unit (CFU) assay was used. C. albicans cells were grown aerobically at 30 °C for 24 hours on YPD agar plates, whereas P. gingivalis cells were cultured anaerobically at 37 °C for 7 days on blood agar plates. After incubation, the numbers of colonies were counted in four replicates from each sample.
Biofilm visualization with confocal scanning laser microscopy
The imaging of single- and dual-species biofilms was conducted as a Z-stack of images, using a Zeiss LSM 710 confocal laser scanning microscope set on Zeiss Axio Observer Z1 (Carl Zeiss, Oberkochen, Germany) with a 40× 1.4 NA oil immersion objective and with ZEN black version 8.10.484 software. For this purpose, 1 × 106 C. albicans cells in 400 μl of RPMI 1640 were seeded into the chamber of Eppendorf Cell Imaging Coverglasses of 170 μm thickness (Eppendorf, Hamburg, Germany), coated with 0.05% poly-L-lysine and cultured under aerobic conditions at 37 °C for 16 hours to form the biofilm. After biofilm washing, 1 × 108 CFSE-labeled P. gingivalis cells in 400 μl of RPMI 1640 were subsequently added to the chambers and further incubated at 37 °C for 24 hours under aerobic or anaerobic conditions, as described above. The biofilms were then stained with CFW and analyzed microscopically using an excitation wavelength of 405 nm to visualize CFW-stained fungal cells, and 488 nm to detect bacterial cells labeled with CFSE.
Expression of genes encoding selected yeast virulence factors
After 3 hours of mutual contact between both microbes under anoxia and normoxia, total RNA was extracted with a TRI®Reagent (Sigma Aldrich) in accordance with the protocol provided by the manufacturer. For cDNA synthesis, each 20-µl reaction mixture consisted of 2 µg of RNA, 0.5 µg of oligonucleotide (dT)18 primer and 200 U of MLV reverse transcriptase (Moloney murine leukemia virus reverse transcriptase; Promega, Madison, WI). Quantitative PCR was performed with the SYBR green-based detection assay and the StepOnePlus real-time PCR system (Applied Biosystems, Waltham, MA). Per 10 µl of reaction mixture, 2 µl of cDNA, 0.2 µl of forward and reverse primers (10 µM) and 5 µl of SYBR KAPA master mix (Kapa Biosystems, Wilmington, MA) were used. PCR amplification was carried out using the primer sets specified in Table 2, with ATC1 as a reference, and under the following conditions: 1 cycle of 95 °C for 10 min and 40 cycles of 95 °C for 20 s, 48–57 °C (depending on the analyzed gene) for 20 s, and 72 °C for 30 s. Samples were amplified in triplicate. Data analysis was performed using real-time PCR system Sequence Detection software (version 1.4; Applied Biosystems).
Table 2.
Primer sets.
| Gene | Primer sets | |
|---|---|---|
| ALS3 | 5′-TGCTGGTGGTTATTGGCAAC-3′ | (FR) |
| 5′-GTCGCGGTTAGGATCGAATG-3′ | (RV) | |
| ALS7 | 5′-TGTTGGCCCTGTTTACAACG-3′ | (FR) |
| 5′-ACCGGGACTGGCAATAGTAC-3′ | (RV) | |
| SAP3 | 5′-ATTCTCCAGGGTTTGTTGCTT-3′ | (FR) |
| 5′-CCAGCTTGACATGAAACTTGAG-3′ | (RV) | |
| SAP6 | 5′- CCCGTTTTGAAATTAAATATGCTGATGG-3′ | (FR) |
| 5′- GTCGTAAGGAGTTCTGGTAGCTTCG-3′ | (RV) | |
| SAP9 | 5′- ATGGTTCATTGGACATGACT-3′ | (FR) |
| 5′- TTAAATCGTGGGACATAACC-3′ | (RV) | |
| HWP1 | 5′- TCAACTGCTCAACTTATTGCT-3′ | (FR) |
| 5′- GCTTCCTCTGTTTCACCTTG-3′ | (RV) | |
| ENO1 | 5′- AAACCCAGAATCCGACCCATC-3′ | (FR) |
| 5′- GACCCAAGCATCCCAGTCATC-3′ | (RV) | |
| ATC1 | 5′-GATTTTGTCTGAACGTGGTAACAG-3′ | (FR) |
| 5′- GGAGTTGAAAGTGGTTTGGTCAATAC-3′ | (RV) |
Identification of C. albicans surface-exposed proteins, after growth in single- or dual-species biofilms with P. gingivalis
Aliquots of 5 × 108 C. albicans cells were grown with 5 × 109 P. gingivalis W83 or ∆K/∆RAB cells in 10 ml of RPMI 1640 medium for 24 hours at 37 °C with gentle agitation, under normoxia or anoxia. Fungal surface-exposed proteins were analyzed as described earlier64,66, using cell surface shaving with trypsin and a shotgun proteomic approach, with a liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS)-based protein identification. Protein ranking with an indicated fold change referring to the averaged normalized spectral abundance factors (NSAFs) was created after comparison of C. albicans surface-exposed proteins from cells grown in mixed-species biofilms with P. gingivalis with the sample containing fungal surface-exposed proteins of C. albicans grown in single-species biofilm.
Microbial protein isolation and purification
C. albicans enolase was purified from yeast cells cultured in YPD medium at 30 °C for 16 hours, the cell wall mannoprotein 65 (Mp65) from post-culture supernatants of C. albicans cells grown in an amino acid liquid synthetic medium, pH 6.867 at 37 °C for 72 hours and the agglutinin-like sequence protein 3 (Als3) from the whole mixture of candidal surface proteins extracted with β-1,6-glucanase (Takara Bio Inc., Otsu, Shiga, Japan) from the cell walls of C. albicans cultivated in hyphal forms in RPMI 1640 medium at 37 °C for 72 hours. A combination of ion-exchange chromatography (a Pharmacia Biotech Resource Q column, GE Healthcare, Uppsala, Sweden) and gel filtration (a Pharmacia Biotech Superdex 200 HR 10/30 column) was used for protein purification, as described previously61.
The purification of HRgpA was performed using the culture fluid from a HG66 strain of P. gingivalis according to the method described previously where gel filtration and arginine-Sepharose chromatography were applied68. Purified proteins were identified with LC-MS/MS analysis as described previously69.
Analysis of the interactions between purified microbial proteins and P. gingivalis and S. cerevisiae cells
To evaluate the possible presence of bacterial receptors for fungal purified proteins - Als3, Mp65 and Eno1 - wild type P. gingivalis cells and mutant counterparts defective in gingipain production were suspended in RPMI medium at a density of 106 cells/ml and incubated in the presence of FITC-labeled fungal proteins at a broad concentration range, for 1.5 h at 37 °C, under anoxic and normoxic conditions. After incubation, the P. gingivalis cells were washed three times with PBS and analyzed by flow cytometry (FACSCalibur, BD, NJJ). For each sample, 5 × 105 cells were collected and analyzed using a 488 nm laser and FITC signal detector. The intensity of fluorescence was calculated using FlowJo 8.7 software.
To verify interactions between RgpA and the fungal adhesin Als3, the binding of 100 nM biotin-labeled RgpA for 1.5 hours at 37 °C to the S. cerevisiae cells (3 × 107 yeast cell per tube) with heterologous expression of Als3 on the yeast cell surface was analyzed. This was compared to negative control S. cerevisiae transformed with pBC542 alone and yeast cells carrying the CWP1 gene of S. cerevisiae in a pBC542 vector31. After incubation, the unbound material was washed out and the yeast cells were mixed with solution of streptavidin-conjugated horseradish peroxidase in PBS and incubated in the dark for 1 hour at room temperature. After washing, the cells were transferred to new tubes and the bound biotinylated bacterial protein was measured after treatment with 3,3′5,5′ tetramethylbenzidine as a substrate for HRP70.
Determination of the binding parameters for complexes of bacterial (RgpA) and fungal (Als3, Mp65, Eno1) proteins using surface plasmon resonance (SPR) measurements
SPR measurements were performed at 25 °C using a Biacore 3000 (GE Healthcare, Milwaukee, WI) with a running buffer (RB) containing 10 mM HEPES, 150 mM NaCl, and 0.005% surfactant (v/v), pH 7.4. C. albicans proteins, Eno1, Als3 and Mp65 were immobilized onto a CM5 sensor chip (GE Healthcare) in accordance with the supplier’s instructions using a standard amine-coupling method. Immobilization was performed in 10 mM sodium acetate buffer to reach 300 resonance units (RU) for Eno1 and Mp65 (pH 4.0 and 4.,5, respectively), and 360 RU for Als3 (pH 4.0). To test the binding of P. gingivalis RgpA, solutions of this bacterial protein prepared in RB at concentrations of between 10–100 nM were injected over the chip surface containing the fungal protein of interest at 20 μl/min flow rate and with 2 minutes of contact time for the association and 2 minutes for the dissociation phase. The collected data were analyzed using BIA evaluation software version 4.1 (GE Healthcare). The dissociation and association rate constants (kd and ka) and the equilibrium dissociation constants (KD) were calculated from the global fit of a simple (1:1) Langmuir model with a baseline drift.
Acknowledgements
This work was supported by the National Science Centre of Poland (grant number 2015/17/B/NZ6/02078 to MRK).
Author Contributions
M.R.K. designed the project; D.B., K.M., G.Z., I.C., J.K. contributed with the biofilm analysis; D.S., B.P. purified the proteins; J.K.K., O.B. performed M.S. experiments; J.K.K., D.S., M.Z. performed the protein binding experiments, A.H.N., L.C.D. prepared S. cerevisiae mutant strains; M.Z., D.B., Z.B., Z.R. performed confocal microscopy analysis; M.R.K., D.B., J.K.K., M.Z., D.S., J.P., A.K. performed data analysis; M.R.K., D.B., J.K.K., M.Z., D.S. wrote the manuscript. All authors read and approved the final manuscript.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Agrawal A, Singh A, Verma R, Murari A. Oral candidiasis: An overview. J. Oral Maxillofac. Pathol. 2014;18:81. doi: 10.4103/0973-029X.141325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med. Mycol. 2007;45:321–346. doi: 10.1080/13693780701218689. [DOI] [PubMed] [Google Scholar]
- 3.Gulati M, Nobile CJ. Candida albicans biofilms: development, regulation and molecular mechanisms. Microbes Infect. 2016;18:310–321. doi: 10.1016/j.micinf.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nobile CJ, Johnson AD. Candida albicans biofilms and human disease. Annu. Rev. Microbiol. 2015;69:71–92. doi: 10.1146/annurev-micro-091014-104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huse SM, Ye Y, Zhou Y, Fodor A. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One. 2012;7:e34242. doi: 10.1371/journal.pone.0034242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metwalli KH, Khan SA, Krom BP, Jabra-Rizk MA. Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS Pathog. 2013;9:e1003616. doi: 10.1371/journal.ppat.1003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He X, et al. Adherence to Streptococci facilitates Fusobacterium nucleatum integration into an oral microbial community. Microb. Ecol. 2012;63:532–542. doi: 10.1007/s00248-011-9989-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bamford CV, et al. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect. Immun. 2009;77:3696–704. doi: 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuboniwa M, Lamont RJ. Subgingival biofilm formation. Periodontol. 2010;2000(52):38–52. doi: 10.1111/j.1600-0757.2009.00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 2015;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussain M, Stover CM, Dupont A. P. gingivalis in periodontal disease and atherosclerosis - scenes of action for antimicrobial peptides and complement. Front. Immunol. 2015;6:45. doi: 10.3389/fimmu.2015.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urzúa B, et al. Yeast diversity in the oral microbiota of subjects with periodontitis: Candida albicans and Candida dubliniensis colonize the periodontal pockets. Med. Mycol. 2008;46:783–793. doi: 10.1080/13693780802060899. [DOI] [PubMed] [Google Scholar]
- 13.McManus BA, et al. Enrichment of multilocus sequence typing clade 1 with oral Candida albicans isolates in patients with untreated periodontitis. J. Clin. Microbiol. 2012;50:3335–3344. doi: 10.1128/JCM.01532-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melton JJ, et al. Recovery of Candida dubliniensis and other Candida species from the oral cavity of subjects with periodontitis who had well-controlled and poorly controlled type 2. diabetes: A pilot study. Spec. Care Dent. 2010;30:230–234. doi: 10.1111/j.1754-4505.2010.00159.x. [DOI] [PubMed] [Google Scholar]
- 15.Miranda TT, et al. Diversity and frequency of yeasts from the dorsum of the tongue and necrotic root canals associated with primary apical periodontitis. Int. Endod. J. 2009;42:839–844. doi: 10.1111/j.1365-2591.2009.01601.x. [DOI] [PubMed] [Google Scholar]
- 16.Sardi JCO, Duque C, Höfling JF, Gonçalves RB. Genetic and phenotypic evaluation of Candida albicans strains isolated from subgingival biofilm of diabetic patients with chronic periodontitis. Med. Mycol. 2012;50:467–75. doi: 10.3109/13693786.2011.633233. [DOI] [PubMed] [Google Scholar]
- 17.Pizzo G, et al. Genotyping and antifungal susceptibility of human subgingival Candida albicans isolates. Arch. Oral Biol. 2002;47:189–196. doi: 10.1016/s0003-9969(01)00114-5. [DOI] [PubMed] [Google Scholar]
- 18.Fox EP, et al. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr. Biol. 2014;24:2411–6. doi: 10.1016/j.cub.2014.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diaz PI, Rogers AH. The effect of oxygen on the growth and physiology of Porphyromonas gingivalis. Oral Microbiol. Immunol. 2004;19:88–94. doi: 10.1046/j.0902-0055.2003.00121.x. [DOI] [PubMed] [Google Scholar]
- 20.Dumitru R, et al. In vivo and in vitro anaerobic mating in Candida albicans. Eukaryot. Cell. 2007;6:465–472. doi: 10.1128/EC.00316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Leeuwen PT, et al. Interspecies interactions between Clostridium difficile and Candida albicans. mSphere. 2016;1(6):e00187–16. doi: 10.1128/mSphere.00187-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavalcanti IMG, Cury DB, Jenkinson AA, Nobbs HF. A. H. Interactions between Streptococcus oralis, Actinomyces oris, and Candida albicans in the development of multispecies oral microbial biofilms on salivary pellicle. Mol. Oral Microbiol. 2017;32:60–73. doi: 10.1111/omi.12154. [DOI] [PubMed] [Google Scholar]
- 23.Dutton, L. C., Jenkinson, H. F., Lamont, R. J. & Nobbs, A. H. Role of Candida albicans secreted aspartyl protease Sap9 in interkingdom biofilm formation. Pathog. Dis. 74 (2016). [DOI] [PMC free article] [PubMed]
- 24.Klotz SA, et al. Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med. Mycol. 2007;45:363–370. doi: 10.1080/13693780701299333. [DOI] [PubMed] [Google Scholar]
- 25.Silva RC, et al. Extracellular enolase of Candida albicans is involved in colonization of mammalian intestinal epithelium. Front. Cell. Infect. Microbiol. 2014;4:66. doi: 10.3389/fcimb.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas DP, Bachmann SP, Lopez-Ribot JL. Proteomics for the analysis of the Candida albicans biofilm lifestyle. Proteomics. 2006;6:5795–804. doi: 10.1002/pmic.200600332. [DOI] [PubMed] [Google Scholar]
- 27.Sandini S, et al. The MP65 gene is required for cell wall integrity, adherence to epithelial cells and biofilm formation in Candida albicans. BMC Microbiol. 2011;11:106. doi: 10.1186/1471-2180-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorgo AG, et al. Mass spectrometric analysis of the secretome of Candida albicans. Yeast. 2010;27:661–672. doi: 10.1002/yea.1775. [DOI] [PubMed] [Google Scholar]
- 29.Rajendran R, et al. Extracellular DNA release confers heterogeneity in Candida albicans biofilm formation. BMC Microbiol. 2014;14:303. doi: 10.1186/s12866-014-0303-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martchenko M, Alarco A-M, Harcus D, Whiteway M. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell. 2004;15:456–67. doi: 10.1091/mbc.E03-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nobbs AH, Margaret Vickerman M, Jenkinson HF. Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot. Cell. 2010;9:1622–1634. doi: 10.1128/EC.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li N, Collyer CA. Gingipains from Porphyromonas gingivalis — complex domain structures confer diverse functions. Eur. J. Microbiol. Immunol. 2011;1:41–58. doi: 10.1556/EuJMI.1.2011.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenkinson HF. Beyond the oral microbiome. Environ. Microbiol. 2011;13:3077–3087. doi: 10.1111/j.1462-2920.2011.02573.x. [DOI] [PubMed] [Google Scholar]
- 34.Lohse MB, Gulati M, Johnson AD, Nobile CJ. Development and regulation of single- and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 2018;16:19–31. doi: 10.1038/nrmicro.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desai PR, van Wijlick L, Kurtz D, Juchimiuk M, Ernst JF. Hypoxia and temperature regulated morphogenesis in Candida albicans. PLoS Genet. 2015;11:e1005447. doi: 10.1371/journal.pgen.1005447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grahl N, Shepardson KM, Chung D, Cramer RA. Hypoxia and fungal pathogenesis: to air or not to air? Eukaryot. Cell. 2012;11:560–70. doi: 10.1128/EC.00031-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shirtliff ME, Peters BM, Jabra-Rizk MA. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett. 2009;299:1–8. doi: 10.1111/j.1574-6968.2009.01668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janus MM, et al. Candida albicans alters the bacterial microbiome of early in vitro oral biofilms. J. Oral Microbiol. 2017;9:1–10. doi: 10.1080/20002297.2016.1270613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wesolowski J, Hassan RYA, Hodde S, Bardroff C, Bilitewski U. Sensing of oxygen in microtiter plates: a novel tool for screening drugs against pathogenic yeasts. Anal. Bioanal. Chem. 2008;391:1731–7. doi: 10.1007/s00216-008-1947-6. [DOI] [PubMed] [Google Scholar]
- 40.Barros LM, et al. Genetic diversity and exoenzyme activities of Candida albicans and Candida dubliniensis isolated from the oral cavity of Brazilian periodontal patients. Arch. Oral Biol. 2008;53:1172–1178. doi: 10.1016/j.archoralbio.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 41.Winter, M. B. et al. Global identification of biofilm-specific proteolysis in Candida albicans. MBio7 (2016). [DOI] [PMC free article] [PubMed]
- 42.Gerits E, Verstraeten N, Michiels J. New approaches to combat Porphyromonas gingivalis. biofilms. J. Oral Microbiol. 2017;9:1300366. doi: 10.1080/20002297.2017.1300366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bandara HMHN, Lam OLT, Watt RM, Jin LJ, Samaranayake LP. Bacterial lipopolysaccharides variably modulate in vitro biofilm formation of Candida species. J. Med. Microbiol. 2010;59:1225–34. doi: 10.1099/jmm.0.021832-0. [DOI] [PubMed] [Google Scholar]
- 44.Goulbourne PA, Ellen RP. Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J. Bacteriol. 1991;173:5266–74. doi: 10.1128/jb.173.17.5266-5274.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashimoto M, Ogawa S, Asai Y, Takai Y, Ogawa T. Binding of Porphyromonas gingivalis fimbriae to Treponema denticola dentilisin. FEMS Microbiol. Lett. 2003;226:267–71. doi: 10.1016/S0378-1097(03)00615-3. [DOI] [PubMed] [Google Scholar]
- 46.Lamont RJ, Bevan CA, Gil S, Persson RE, Rosan B. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol. Immunol. 1993;8:272–6. doi: 10.1111/j.1399-302x.1993.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 47.Maeda K, et al. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect. Immun. 2004;72:1341–8. doi: 10.1128/IAI.72.3.1341-1348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sztukowska, M. N. et al. Community Development between Porphyromonas gingivalis and Candida albicans Mediated by InlJ and Als3. MBio9 (2018). [DOI] [PMC free article] [PubMed]
- 49.Kamaguchi, A. et al. Effect of Porphyromonas gingivalis vesicles on coaggregation of Staphylococcus aureus to oral Microorganisms. Curr. Microbiol. 47 (2003). [DOI] [PubMed]
- 50.Shi Y, et al. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 1999;274:17955–60. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 51.Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr. Protein Pept. Sci. 2003;4:397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- 52.O’Brien-Simpson NM, et al. An immune response directed to proteinase and adhesin functional epitopes protects against Porphyromonas gingivalis-induced periodontal bone loss. J. Immunol. 2005;175:3980–9. doi: 10.4049/jimmunol.175.6.3980. [DOI] [PubMed] [Google Scholar]
- 53.Rapala-Kozik M, et al. Adsorption of components of the plasma kinin-forming system on the surface of Porphyromonas gingivalis involves gingipains as the major docking platforms. Infect. Immun. 2011;79:797–805. doi: 10.1128/IAI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ito R, Ishihara K, Shoji M, Nakayama K, Okuda K. Hemagglutinin/Adhesin domains of Porphyromonas gingivalis play key roles in coaggregation with Treponema denticola. FEMS Immunol. Med. Microbiol. 2010;60:251–60. doi: 10.1111/j.1574-695X.2010.00737.x. [DOI] [PubMed] [Google Scholar]
- 55.Bao K, et al. Role of Porphyromonas gingivalis gingipains in multi-species biofilm formation. BMC Microbiol. 2014;14:258. doi: 10.1186/s12866-014-0258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haraguchi A, Miura M, Fujise O, Hamachi T, Nishimura F. Porphyromonas gingivalis gingipain is involved in the detachment and aggregation of Aggregatibacter actinomycetemcomitans biofilm. Mol. Oral Microbiol. 2014;29:131–43. doi: 10.1111/omi.12051. [DOI] [PubMed] [Google Scholar]
- 57.Ovchinnikova ES, Krom BP, Busscher HJ, van der Mei HC. Evaluation of adhesion forces of Staphylococcus aureus along the length of Candida albicans hyphae. BMC Microbiol. 2012;12:281. doi: 10.1186/1471-2180-12-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bamford CV, Nobbs AH, Barbour ME, Lamont RJ, Jenkinson HF. Functional regions of Candida albicans hyphal cell wall protein Als3 that determine interaction with the oral bacterium Streptococcus gordonii. Microbiology. 2015;161:18–29. doi: 10.1099/mic.0.083378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlecht LM, et al. Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology. 2015;161:168–181. doi: 10.1099/mic.0.083485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henderson B, Martin A. Bacterial Virulence in the Moonlight: Multitasking Bacterial Moonlighting Proteins Are Virulence Determinants in Infectious Disease. Infect. Immun. 2011;79:3476–3491. doi: 10.1128/IAI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seweryn K, et al. Kinetic and thermodynamic characterization of the interactions between the components of human plasma kinin-forming system and isolated and purified cell wall proteins of Candida albicans. Acta Biochim. Pol. 2015;62:825–35. doi: 10.18388/abp.2015_1142. [DOI] [PubMed] [Google Scholar]
- 62.Reyna-Beltrán E, et al. The Candida albicans ENO1 gene encodes a transglutaminase involved in growth, cell division, morphogenesis, and osmotic protection. J. Biol. Chem. 2018;293:4304–4323. doi: 10.1074/jbc.M117.810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spurbeck RR, Arvidson CG. Lactobacillus jensenii surface-associated proteins inhibit Neisseria gonorrhoeae adherence to epithelial cells. Infect. Immun. 2010;78:3103–11. doi: 10.1128/IAI.01200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karkowska-Kuleta, J. et al. The activity of bacterial peptidylarginine deiminase is important during formation of dual-species biofilm by periodontal pathogen Porphyromonas gingivalis and opportunistic fungus Candida albicans. Pathog. Dis. 76 (2018). [DOI] [PMC free article] [PubMed]
- 65.Benedyk M, et al. Gingipains: Critical Factors in the Development of Aspiration Pneumonia Caused by Porphyromonas gingivalis. J. Innate Immun. 2016;8:185–98. doi: 10.1159/000441724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karkowska-Kuleta J, Zajac D, Bochenska O, Kozik A. Surfaceome of pathogenic yeasts, Candida parapsilosis and Candida tropicalis, revealed with the use of cell surface shaving method and shotgun proteomic approach. Acta Biochim. Pol. 2015;62:807–19. doi: 10.18388/abp.2015_1140. [DOI] [PubMed] [Google Scholar]
- 67.Lee KL, Buckley HR, Campbell CC. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–53. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 68.Potempa, J. & Nguyen, K.-A. Purification and characterization of gingipains. In Current Protocols in Protein Science 21.20.1–21.20.27, 10.1002/0471140864.ps2120s49 (John Wiley & Sons, Inc., 2007). [DOI] [PubMed]
- 69.Karkowska-Kuleta J, et al. Binding of human plasminogen and high-molecular-mass kininogen by cell surface-exposed proteins of Candida parapsilosis. Acta Biochim. Pol. 2017;64:391–400. doi: 10.18388/abp.2017_1609. [DOI] [PubMed] [Google Scholar]
- 70.Karkowska-Kuleta J, Kozik A, Rapala-Kozik M. Binding and activation of the human plasma kinin-forming system on the cell walls of Candida albicans and Candida tropicalis. Biol. Chem. 2010;391:97–103. doi: 10.1515/BC.2009.145. [DOI] [PubMed] [Google Scholar]
- 71.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.