Abstract
Mitochondria are organized as tubular networks in the cell and undergo fission and fusion. Although several of the molecular players involved in mediating mitochondrial dynamics have been identified, the precise cellular cues that initiate mitochondrial fission or fusion remain largely unknown. In fission yeast (Schizosaccharomyces pombe), mitochondria are organized along microtubule bundles. Here, we employed deletions of kinesin-like proteins to perturb microtubule dynamics and used high-resolution and time-lapse fluorescence microscopy, revealing that mitochondrial lengths mimic microtubule lengths. Furthermore, we determined that compared with WT cells, mutant cells with long microtubules exhibit fewer mitochondria, and mutant cells with short microtubules have an increased number of mitochondria because of reduced mitochondrial fission in the former and elevated fission in the latter. Correspondingly, upon onset of closed mitosis in fission yeast, wherein interphase microtubules assemble to form the spindle within the nucleus, we observed increased mitochondrial fission. We found that the consequent rise in the mitochondrial copy number is necessary to reduce partitioning errors during independent segregation of mitochondria between daughter cells. We also discovered that the association of mitochondria with microtubules physically impedes the assembly of the fission protein Dnm1 around mitochondria, resulting in inhibition of mitochondrial fission. Taken together, we demonstrate a mechanism for the regulation of mitochondrial fission that is dictated by the interaction between mitochondria and the microtubule cytoskeleton.
Keywords: mitochondria, microtubule, in vivo imaging, Schizosaccharomyces pombe, cell biology, cytoskeleton, molecular motor, mitosis, Dnm1, mitochondrial dynamics
Introduction
Mitochondria are double-membraned organelles whose functions range from ATP production to calcium signaling. Inside cells, mitochondrial form is dynamic and transitions from tubular networks to fragmented entities depending on the activity of the mitochondrial fission and fusion machinery. The major mitochondrial fission protein is dynamin-related Drp1 GTPase (1) (Dnm13 in yeast (2, 3)). Multimeric Drp1 rings assemble around the mitochondrial membrane and utilize the energy from GTP hydrolysis to catalyze the constriction and fission of mitochondria (4, 5). Fusion of mitochondria requires two sets of proteins, namely Opa1 (6) for the inner membrane (Mgm1 in yeast (7)) and Mfn1/2 (8) for the outer membrane (Fzo1 in yeast (9)).
The requirement for dynamic mitochondria has been attributed to two primary reasons, namely quality control and energy production (10). Larger/longer mitochondria resulting from fusion are hypothesized to be capable of producing more energy, whereas shorter/smaller mitochondria formed following a fission event are likely to undergo mitophagy (11). In the latter case, fission could serve as an efficient mechanism to segregate and eliminate damaged mitochondria. Dysfunction of fission and fusion processes has been implicated in neurodegeneration (12, 13), cancer (14), and cardiomyopathies (15) among a host of metabolic disorders.
In mammalian cells, mitochondria are transported along microtubule tracks by the activity of motor proteins kinesin-1 and dynein (16). Kinesin-1 and dynein bind to the outer membrane of mitochondria via the Miro–Milton complex (17–19) and move mitochondria in the anterograde and retrograde directions, respectively. In neuronal cells, an increase in calcium levels results in the attachment of kinesin-1 motor to mitochondria via syntaphilin, which inhibits the ATPase activity of kinesin and hence leads to stationary mitochondria on neuronal microtubules (20). About 70% of mitochondria in neuronal cells have been visualized in this stationary state (21). In contrast to mammalian cells, mitochondria in fission yeast do not undergo motor-driven movement along microtubules (22, 23). However, the protein Mmb1 has been identified to associate mitochondria with dynamic microtubules (24). Upon microtubule depolymerization using methyl benzimidazol-2-yl-carbamate (MBC), mitochondria have been observed to undergo fragmentation (3, 24, 25). Additionally, mitochondrial dynamics and partitioning in fission yeast have been observed to be actin/myosin-independent processes (3), contrary to the mechanism of mitochondrial partitioning in budding yeast (26).
Cells utilize several strategies to reduce partitioning error of organelles during mitosis, such as ordered segregation mediated by spindle poles or increasing copy numbers of organelles prior to cell division (27). In the latter, homogeneous distribution of the multiple organelle copies serves to decrease partitioning error. Mitochondrial inheritance has been observed to be microtubule-dependent in mammalian cells (28). In fission yeast, mitochondrial partitioning during cell division has been proposed to be mediated by attachment of mitochondria with spindle poles (22, 29, 30), similar to the segregation of endosomes, lysosomes, and Golgi bodies in mammalian cells (31, 32). However, only a portion of the observed mitochondria associated with the spindle poles (22). Additionally, increased mitochondrial fragmentation upon the onset of mitosis has also been observed (3), perhaps indicating a binomial partitioning or independent segregation mechanism for mitochondrial distribution wherein each mitochondrion in the mother has a 50% likelihood of being partitioned into either of the daughter cells by virtue of symmetric division and without the involvement of active processes.
Although the molecular players that effect fission and fusion have been identified in several systems, the cellular signals that regulate these events are largely elusive. Here, we demonstrate that mitochondria piggyback on dynamic microtubules to selectively undergo fission when microtubules depolymerize. Reorganization of interphase microtubules into the nucleus when cells prepare for division also provide the cue for increased mitochondrial fission. We quantified the number of mitochondria in mother cells immediately after formation of mitotic spindle within the nucleus and the number of mitochondria in the resulting daughter cells and confirmed that the partitioning was indeed a good fit to a binomial distribution, indicating that an independent segregation mechanism serves to distribute mitochondria into the two daughter cells. We determined that the presence of long and stabilized microtubules was inhibitory to unopposed fission even when Dnm1 was overexpressed. Finally, we discovered that microtubule-bound mitochondria were unlikely to undergo fission due to the unavailability of space between microtubules and mitochondria for the formation of the Dnm1 ring.
Results
Perturbation of microtubule dynamics leads to changes in mitochondrial numbers
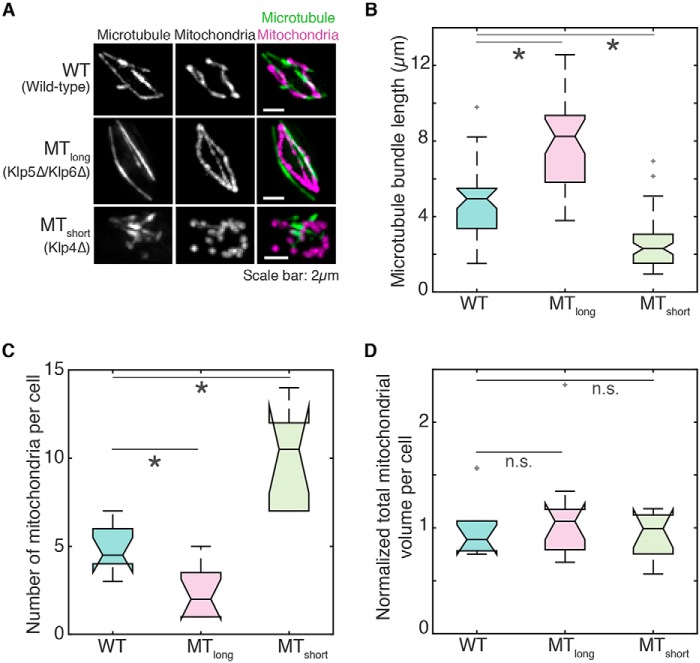
We observed that mitochondria underwent increased fission upon microtubule depolymerization but did not observe their subsequent aggregation as reported previously (Fig. S1, A–C). Instead, the fragmented mitochondria were mobile and frequently in close contact with each other (Fig. S1, B and C, and Movie S1). Because the depolymerization of microtubules had a direct effect on mitochondrial fission, we set out to study the consequence of modification of microtubule dynamics on mitochondrial dynamics. To this end, we visualized the mitochondria and microtubules of fission yeast strains carrying deletions of antagonistic kinesin-like proteins, Klp5/Klp6 and Klp4, in high-resolution deconvolved images (Fig. 1A and Movies S2–S4).
Figure 1.
Mitochondrial number is inversely proportional to microtubule length. A, maximum intensity projections of deconvolved Z-stack images of microtubules (left), mitochondria (center), and their composite (right) in WT (top; strain KI001; see Table S1), Klp5Δ/Klp6Δ (MTlong; strain G3B; see Table S1), and Klp4Δ (MTshort; strain G5B; see Table S1) cells. B, box plot of the length of antiparallel microtubule bundles in WT, MTlong, and MTshort cells (n = 40, 37, and 63 bundles, respectively). In all box plots that appear in this study, the central line indicates the median, and notches represent the 95% confidence interval of the median. C, box plot of the number of mitochondria per cell in WT, MTlong, and MTshort cells (n = 10, 12, and 13 cells, respectively). D, box plot of the total volume of mitochondria per cell in WT, MTlong, and MTshort cells normalized to mean total WT mitochondrial volume (n = 10, 12, and 13 cells, respectively). In B–D, light gray crosses represent outliers, an asterisk represents significance (p < 0.05), and n.s. indicates no significant difference (one-way ANOVA, Tukey's honestly significant difference procedure).
The heteromeric Klp5/Klp6 motor is required for maintenance of interphase microtubule length by promoting catastrophe at microtubule plus ends (33, 34). Cells lacking Klp5 and Klp6 exhibited long microtubule bundles (“MTlong”; Fig. 1B) as reported previously due to a decreased catastrophe rate (33). In contrast, Klp4 is required for polarized growth in fission yeast and has been suggested to promote microtubule growth (35, 36). As a result, in the absence of Klp4, microtubule bundles were only about half the length of WT bundles (“MTshort”; Fig. 1B).
As in WT cells, mitochondria in Klp5Δ/Klp6Δ were in close contact with the microtubule, whereas we observed reduced association between the short microtubules and mitochondria in Klp4Δ cells (Fig. 1A and Movies S3 and S4). Although WT cells had 4.9 ± 0.4 (mean ± S.E.) mitochondria per cell, we observed that Klp5Δ/Klp6Δ contained only 2.3 ± 0.4 (mean ± S.E.). In contrast, Klp4Δ cells had 10 ± 0.9 mitochondria per cell (mean ± S.E.; Fig. 1C). This indicated that the number of mitochondria per cell was inversely related to the length of microtubule bundle. However, the decrease in the number of mitochondria in Klp5Δ/Klp6Δ cells and increase in Klp4Δ cells were not at the expense of mitochondrial volume because the net mitochondrial volume in both cases was comparable with WT mitochondrial volume (Fig. 1D), with individual mitochondrial volumes changing to compensate for the difference in mitochondrial numbers among WT, Klp5Δ/Klp6Δ, and Klp4Δ cells (Fig. S1D).
Cells with short microtubules undergo increased fission
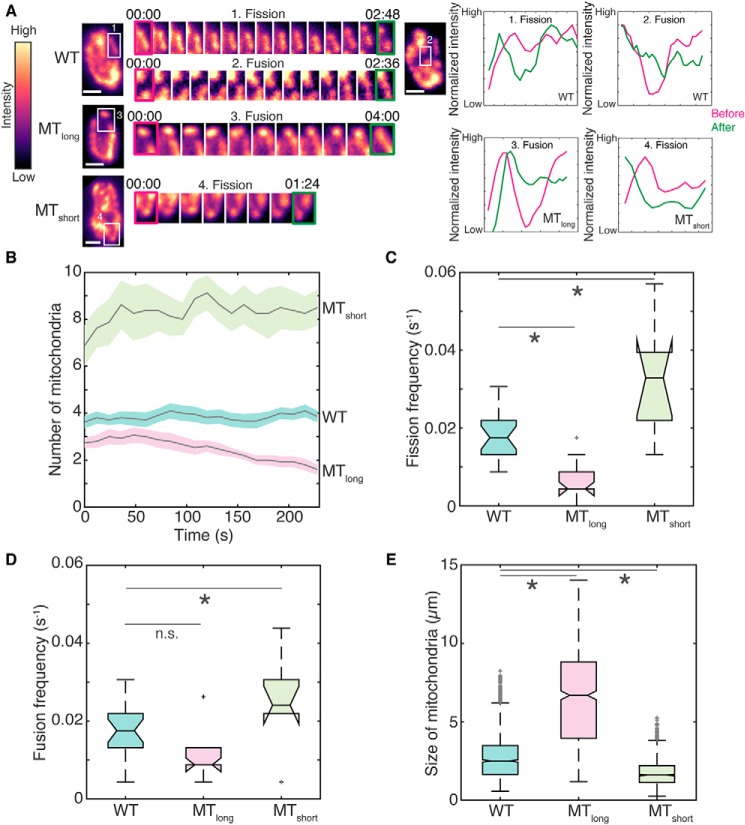
To understand the difference in mitochondrial numbers in WT, Klp5Δ/Klp6Δ, and Klp4Δ cells, we acquired and analyzed time-lapse videos at the single-mitochondrion level in all three cases (Fig. 2A and Movies S5–S7). Similar to our observations from high-resolution images, we measured 3.6 ± 0.2, 2.7 ± 0.2, and 6.9 ± 0.8 mitochondria (mean ± S.E.) in WT, Klp5Δ/Klp6Δ, and Klp4Δ cells, respectively (Figs. 1C and 2B). Analysis of evolution of these mitochondrial numbers revealed no significant changes over time (Fig. 2B). Additionally, WT cell exhibited ∼1 fission and ∼1 fusion event every minute on average, whereas Klp5Δ/Klp6Δ cells exhibited a fission frequency that was half that of WT, and Klp4Δ mitochondria had a fission frequency that was almost double that of WT (Fig. 2C). The fusion frequency of mitochondria in Klp4Δ cells was slightly higher than in WT and Klp5Δ/Klp6Δ cells (Fig. 2D), likely due to the increased number of mitochondria in Klp4Δ cells that could participate in fusion.
Figure 2.
Microtubule length determines fission frequency of mitochondria. A, montage of maximum intensity projected confocal Z-stack images of WT (strain KI001; see Table S1), Klp5Δ/Klp6Δ (MTlong; strain G3B; see Table S1), and Klp4Δ (MTshort; strain G5B; see Table S1) cells represented in the intensity map indicated to the left of the images. The insets (white box) and their montages on the right of the images are representative fission and fusion events in WT (1 and 2), fusion event in MTlong (3), and fission event in MTshort cell (4). Time is indicated as minutes:seconds above the montage of the insets. The normalized intensity along the mitochondrion in the inset before (magenta) and after (green) the fission or fusion event is indicated in plots to the right of the montages. Scale bars represent 2 μm. B, evolution of mitochondrial number over time indicated as mean (solid gray line) and S.E. (shaded region) for WT, MTlong, and MTshort cells (n = 21, 15, and 8 cells, respectively. C, box plot of the fission frequency of mitochondria per second in WT, MTlong, and MTshort cells (n = 21, 15, and 8 cells, respectively). D, box plot of the fusion frequency of mitochondria per second in WT, MTlong, and MTshort cells (n = 21, 15, and 8 cells, respectively). E, box plot of the size of mitochondria in WT, MTlong, and MTshort cells, calculated as the length of the major axis of an ellipse fitted to each mitochondrion (n = 1613, 739, and 1326 mitochondria, respectively). In C–E, light gray crosses represent outliers, an asterisk represents significance (p < 0.05), and n.s. indicates no significant difference (one-way ANOVA in C and D and Kruskal–Wallis test in E, Tukey's honestly significant difference procedure).
As observed in the high-resolution deconvolved images, we also measured significant differences in the size and morphology of the mitochondria in WT, Klp5Δ/Klp6Δ, and Klp4Δ cells (Fig. 2E and Fig. S2). Mitochondrial sizes reflected microtubule bundle lengths, with the largest mitochondria in Klp5Δ/Klp6Δ cells and the smallest in Klp4Δ cells. WT cells predictably had mitochondrial sizes between that of Klp4Δ and Klp5Δ/Klp6Δ cells (Fig. 2E and Fig. S2).
Cells devoid of microtubules undergo fission with increased frequency but show unaltered fusion frequencies
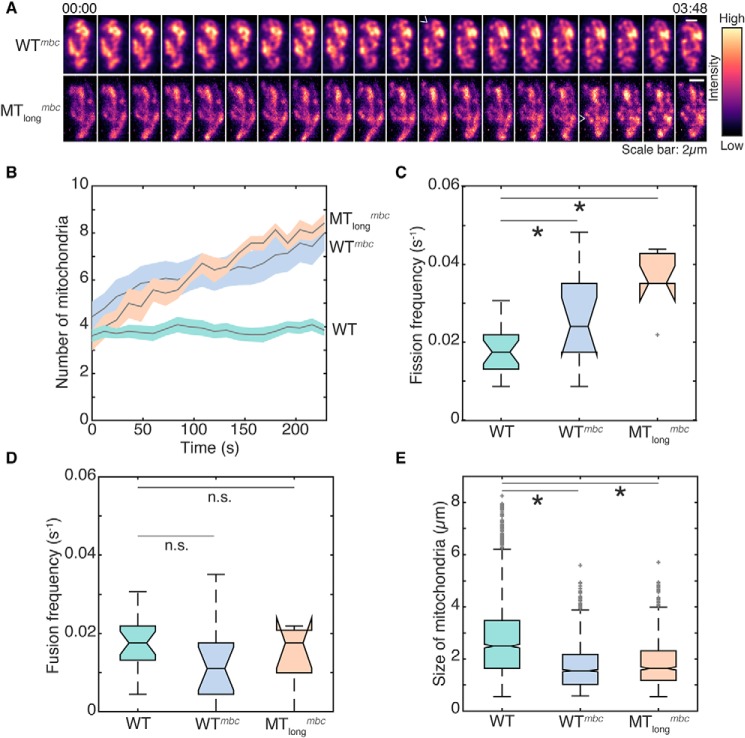
Although the Klp5Δ/Klp6Δ cells containing long microtubules had the same total volume of mitochondria as WT cells (Fig. 1D), their mitochondria appeared less fragmented. So, to specifically test the role of microtubules in dictating mitochondrial fission, we depolymerized microtubules in WT and Klp5Δ/Klp6Δ cells and measured mitochondrial dynamics in time-lapse movies (Fig. 3A and Movies S8 and S9). In both WT and Klp5Δ/Klp6Δ cells, upon microtubule depolymerization, we observed a progressive increase in the number of mitochondria (Fig. 3B). We next quantified the fission and fusion events in WT and Klp5Δ/Klp6Δ cells treated with MBC. We measured that the fission frequency of mitochondria in MBC-treated cells was doubled when compared with untreated control cells (Fig. 3C). At the same time, the mitochondrial fusion frequency remained unchanged (Fig. 3D), indicating that the immediate consequence of the loss of microtubules was increased fission, without concomitant changes in mitochondrial fusion. Additionally, upon MBC treatment, we measured mitochondrial sizes and morphologies that were reminiscent of Klp4Δ cells (Fig. 3E and Fig. S3, A–C).
Figure 3.
Microtubule depolymerization induces increased fission in WT and Klp5Δ/Klp6Δ cells. A, montage of maximum intensity projected confocal Z-stack images of MBC-treated wildtype (“WTmbc”; strain KI001; see Table S1) and Klp5Δ/Klp6Δ (“MTlongmbc”; strain G3B; see Table S1) cells represented in the intensity map indicated to the right of the images. White open arrowheads point to representative fission events. 00:00 indicates time (minutes:seconds) 2 min after addition of MBC. B, evolution of mitochondrial number with time indicated as mean (solid gray line) and S.E. (shaded region) for WT, WTmbc, and MTlongmbc cells (n = 21, 14, and 7 cells, respectively). C, box plot of the fission frequency of mitochondria per second in WT, WTmbc, and MTlongmbc cells (n = 21, 14, and 7 cells, respectively). D, box plot of the fusion frequency of mitochondria per second in WT, WTmbc, and MTlongmbc cells (n = 21, 14, and 7 cells, respectively). E, box plot of the size of mitochondria in WT, WTmbc, and MTlongmbc cells, calculated as the length of the major axis of an ellipse fitted to each mitochondrion (n = 1613, 1765, and 886 mitochondria, respectively). In C–E, light gray crosses represent outliers, an asterisk represents significance (p < 0.05), and n.s. indicates no significant difference (one-way ANOVA in C and D and Kruskal–Wallis test in E, Tukey's honestly significant difference procedure). Note that the WT data represented in this figure is identical to the WT data plotted in Fig. 2 and Fig. S2 and has been reused for comparison.
An increase in oxidative stress via reactive oxygen species levels has also been described to induce mitochondrial fission (37). However, we measured no difference in reactive oxygen species levels among WT, Klp5Δ/Klp6Δ, and Klp4Δ cells (Fig. S3D).
An independent segregation mechanism enables mitochondrial partitioning during mitosis
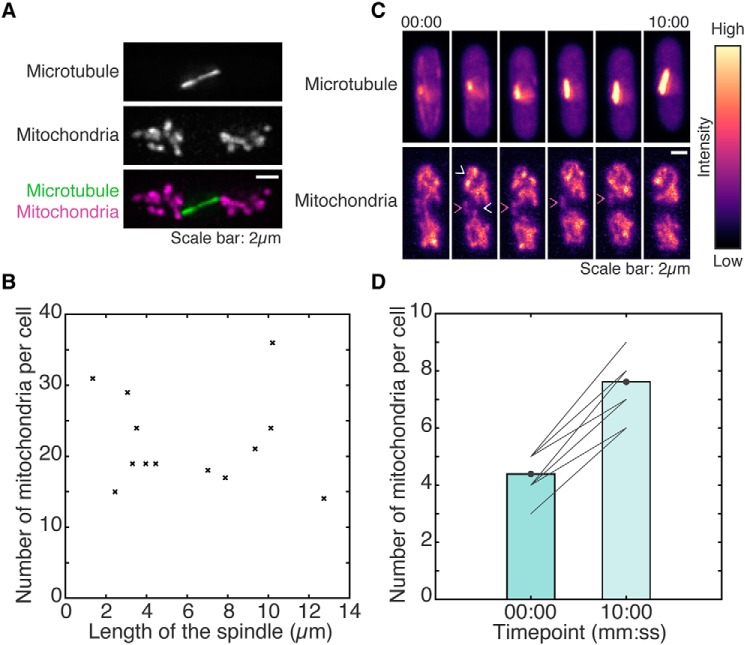
We next sought to understand the biological role for increased mitochondrial fission upon microtubule depolymerization. Fission yeast undergoes closed mitosis, wherein the nuclear envelope does not undergo breakdown during cell division (38). Upon onset of mitosis in fission yeast, the interphase microtubules that were previously in the cytoplasm are reorganized to form the spindle inside the closed nucleus. This natural situation mimics the depolymerization of microtubules via the chemical inhibitor MBC. Therefore, we set out to study the changes in the mitochondrial network upon cell entry into mitosis. We first obtained high-resolution deconvolved images of the microtubule and mitochondria in fission yeast cells undergoing cell division (Fig. 4A, Fig. S4A, and Movie S10). We observed that dividing WT cells had ∼4× the number of mitochondria in interphase cells (Fig. 4B). Moreover, similar to what was seen in cells lacking microtubules or Klp4 (Fig. 1A), mitochondria in dividing cells were shorter and more rounded (Fig. 4A and Fig. S4A). There was no relationship between length of the mitotic spindle and the number of mitochondria (Fig. 4B and Fig. S4A), indicating that the increased fission likely occurred fairly early upon entry into mitosis. Analysis of time-lapse videos of WT cells before and 10 min after entry into mitosis revealed a doubling of mitochondrial numbers in this time (Fig. 4, C and D, and Movie S11). The fragmented mitochondria appeared more mobile and were able to traverse distances of ∼1 μm in the cell (Fig. 4C). In this same period of time, nondividing interphase cells did not show any change in mitochondrial numbers (Fig. S4, B and C).
Figure 4.
Mitotic cells contain several short mitochondria. A, maximum intensity projections of deconvolved Z-stack images of the microtubules (top), mitochondria (center), and their composite (bottom) of a WT cell (strain KI001; see Table S1) undergoing division. B, scatter plot of the length of the mitotic spindle versus the number of mitochondria per cell in dividing cells (n = 13 cells). C, montage of maximum intensity projected confocal Z-stack images of the microtubules (top) and mitochondria (bottom) in a WT cell undergoing cell division represented in the intensity map indicated to the right of the images. White open arrowheads point to representative fission events. Magenta arrowheads point to a representative mobile, fragmented mitochondrion. Time is indicated above the images in minutes:seconds (mm:ss). D, bar plot of the mean number of mitochondria per cell before (00:00) and 10 min after (10:00) the onset of mitosis. Solid gray lines represent data from individual cells (n = 16 cells).
An increase in mitochondrial numbers prior to cell division could aid in increasing the likelihood of equal partitioning of mitochondria between daughter cells, given a binomial partitioning or independent segregation mechanism. To test whether mitochondria in our system underwent independent segregation (27), with each mitochondrion in the mother cell having a 50% chance of segregating to either of the future daughter cells during mitosis, we tested the fit of our data to a binomial distribution (39) using a χ2 test as described previously (40). Our data (Table S2) did not differ significantly from a binomial distribution with a χ2 statistic of 7.1846 with 3 degrees of freedom and p = 0.0662, indicating that mitochondrial partitioning in fission yeast is achieved by independent segregation during cell division. The increase in mitochondrial numbers upon onset of mitosis also served to reduce the partitioning error of mitochondria between the daughter cells as predicted by independent segregation (Fig. S4D).
Cells with long microtubules are protected from unopposed mitochondrial fission
The mitochondrial fission protein in yeast is a dynamin-related GTPase, Dnm1 (3). Dnm1 brings about the fission of mitochondria by self-assembling into rings or spirals around the mitochondrial outer membrane and utilizing its GTPase activity to effect the scission (4). In the absence of Dnm1, mitochondria are organized as extended, fused “nets” (3, 41), which do not undergo fission even in the absence of microtubules (Fig. S5A). Furthermore, in Klp4Δ cells, which typically contain several short mitochondria (Fig. 1A), absence of Dnm1 results in a single large, fused mitochondrion (Fig. S5B). Therefore, all mitochondrial fission in Schizosaccharomyces pombe is reliant on the activity of Dnm1. Additionally, during mitosis, cells lacking Dnm1 that contained a single large mitochondrion relied on the cytokinesis of the mother cell to also split the mitochondrion into the daughter cells (Fig. S5C).
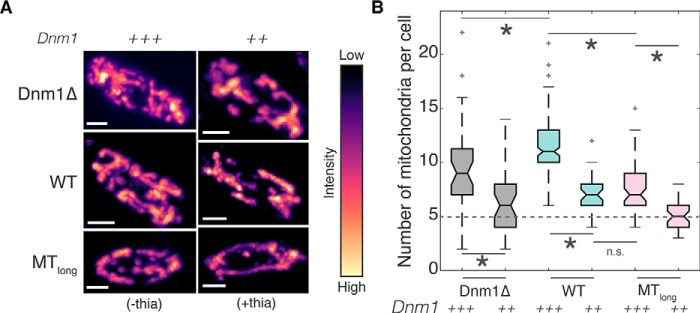
Taken together, our results suggest that association of mitochondria with microtubules inhibits mitochondrial fission. Previously, cryoelectron tomographic analysis in fission yeast indicated a preferred separation distance of ∼20 nm for mitochondria associated with microtubules (42). So too cryoelectron microscopy in budding yeast revealed that Dnm1 assembled into rings with an outer diameter of 129 nm and lumen diameter of 89 nm, resulting in ∼20-nm-high structures around mitochondria (43). Therefore, mitochondria associated with microtubules might have insufficient space to accommodate the Dnm1 ring around their diameter, thereby inhibiting fission due to simple physical constraints. To test this hypothesis, we first transformed Dnm1Δ, WT, and Klp5Δ/Klp6Δ cells with a plasmid expressing Dnm1 under the control of the nmt1 promoter (see Table S1) and visualized the mitochondria (Fig. 5A). These cells exhibit high overexpression of Dnm1 in the absence of thiamine and low overexpression in the presence of 0.05 μm thiamine in the culture medium. We counted the number of mitochondria present in these cells and estimated that Dnm1Δ cells contained 9.5 ± 0.7 and 6.4 ± 0.5 mitochondria (mean ± S.E.) with high overexpression and low overexpression of Dnm1, respectively (Fig. 5B). WT cells highly overexpressing Dnm1 had 11.6 ± 0.2 mitochondria (mean ± S.E.), which is twice that of WT cells expressing normal levels of Dnm1 (Fig. 5B). The increase in mitochondrial numbers in these cells is due to the increase in Dnm1 numbers, which likely accelerates the kinetics of the Dnm1 ring formation and hence mitochondrial fission. In WT cells that were grown in the presence of thiamine, the mitochondrial number was 7 ± 0.3 (mean ± S.E.), presumably because of low overexpression of Dnm1.
Figure 5.
Association with microtubules prevents unopposed fission of mitochondria. A, maximum intensity projections of deconvolved Z-stack images of mitochondria in cells transformed with functional untagged Dnm1 (plasmid pREP41-Dnm1; see Table S1) in Dnm1Δ cells (see Table S1), WT (strain FY7143; see Table S1), and Klp5Δ/Klp6Δ cells (MTlong; strain FY20823; see Table S1) represented in the intensity map indicated to the right of the images. High overexpression of Dnm1 is indicated with +++ (−thia), and low overexpression with ++ (+thia). Scale bars represent 2 μm. B, box plot of mitochondrial numbers in Dnm1Δ, WT, and MTlong cells with high overexpression (+++) or low expression of Dnm1 (++) (n = 37, 36, 149, 37, 63, and 41 cells, respectively). The mean mitochondrial number in WT cells expressing a normal amount of Dnm1 is depicted by the dashed line. Light gray crosses represent outliers, an asterisk represents significance (p < 0.05), and n.s. indicates no significant difference (Kruskal–Wallis test, Tukey's honestly significant difference procedure).
Interestingly, in Klp5Δ/Klp6Δ cells with high and low overexpression of Dnm1, we counted only 7.5 ± 0.3 and 5.2 ± 0.2 mitochondria (mean ± S.E.), respectively. In Klp5Δ/Klp6Δ, the presence of longer microtubules than in WT cells possibly prevented increased fission of mitochondria even when cells overexpressed Dnm1. In fact, in Klp5Δ/Klp6Δ cells where there was low overexpression of Dnm1, the mitochondrial number was comparable with that in WT cells expressing normal levels of Dnm1 (Fig. 5B).
We additionally used cells expressing mCherry-tagged microtubules, GFP-tagged Dnm1, and MitoTracker-stained mitochondria and visualized the localization of Dnm1 with respect to the microtubules and mitochondria in the same cells (Fig. S6A). Although 89.8% of Dnm1 spots colocalized with the mitochondria that were not bound to the microtubule (106 of 118 spots, n = 18 cells), only 10.2% (12 of 118 spots, n = 18 cells) localized on mitochondria that were attached to the microtubule. Although these results indicated an inhibitory role for microtubule–mitochondrion interactions in Dnm1 assembly, the nonfunctionality of fluorescently tagged Dnm1, which has been documented in other literature (3), prevented direct visualization of fission of mitochondria by Dnm1 in these cells.
Dissociation of mitochondria from microtubules leads to unopposed fission
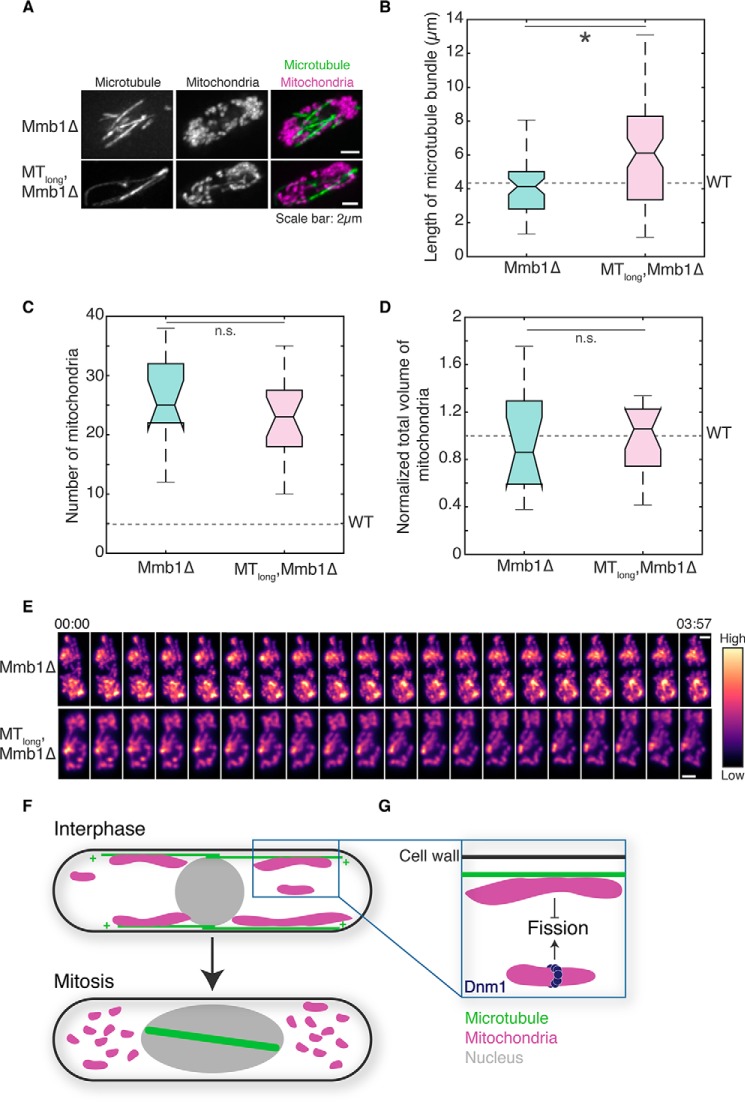
To further test the hypothesis that Dnm1 assembly was physically impeded on microtubule-bound mitochondria, we sought to move microtubules and mitochondria apart without perturbing the microtubule cytoskeleton. To this end, we used cells devoid of the protein that links mitochondria with microtubules in fission yeast, Mmb1 (Fig. 6A and Movies S12 and S13). First, we measured microtubule bundle lengths in Mmb1Δ cells and Klp5Δ/Klp6Δ-Mmb1Δ cells. We did not observe a significant difference in microtubule bundle lengths between WT and Mmb1Δ cells (Fig. 6B), but Klp5Δ/Klp6Δ-Mmb1Δ cells exhibited microtubules that were significantly longer than those in WT and Mmb1Δ cells, comparable with Klp5Δ/Klp6Δ cells.
Figure 6.
Deletion of Mmb1 results in unopposed fission in WT and MTlong cells. A, maximum intensity projections of deconvolved Z-stack images of microtubules (left), mitochondria (center), and their composite (right) in Mmb1Δ cells (top; strain PT2244; see Table S1) and Klp5Δ/Klp6Δ-Mmb1Δ cells (“MTlong, Mmb1Δ”; bottom, strain VA076; see Table S1). B, box plot of microtubule bundle length in Mmb1Δ (n = 54 microtubules from 13 cells) and Klp5Δ/Klp6Δ-Mmb1Δ cells (n = 81 microtubules from 25 cells). The mean microtubule bundle length in WT cells is depicted by the dashed line. C, box plot of mitochondrial numbers in Mmb1Δ and Klp5Δ/Klp6Δ-Mmb1Δ cells (n = 14 and 20, cells respectively). The mean mitochondrial number in WT cells is depicted by the dashed line. D, box plot of mitochondrial volume in Mmb1Δ and Klp5Δ/Klp6Δ-Mmb1Δ cells (n = 14 and 20 cells, respectively) normalized to mean total mitochondrial volume in WT cells. The mean mitochondrial volume in WT cells is depicted by the dashed line. E, montage of maximum intensity projected confocal Z-stack images of the mitochondria in a Mmb1Δ cell (top) and Klp5Δ/Klp6Δ-Mmb1Δ cell (MTlong, Mmb1Δ; bottom). The intensity map is indicated to the right of the images. Time is indicated above the images in minutes:seconds. Scale bars represent 2 μm. In B–D, an asterisk represents significance (p < 0.05), and n.s. represents no significant difference (one-way ANOVA, Tukey's honestly significant difference procedure). F, model of mitochondrial dynamics mediated by microtubule dynamics. Microtubules polymerize and depolymerize at their plus ends (+). Absence of microtubule bundles in the cytoplasm during cell division enables the fragmentation of mitochondria. G, when mitochondria are bound to microtubules, Dnm1 assembly is inhibited. Upon microtubule depolymerization, this inhibition is alleviated, and Dnm1 can effectively mediate scission of mitochondria.
Next, we counted 26.2 ± 1.8 (mean ± S.E.) mitochondria in these cells, which is significantly higher than that in WT cells (Fig. 6C). In the case of Klp5Δ/Klp6Δ-Mmb1Δ cells, although microtubules were significantly longer than in WT cells (Fig. 6B), the absence of the linker between microtubules and mitochondria resulted in extensive fission of mitochondria.
We counted 23.3 ± 1.4 (mean ± S.E.) mitochondria in Klp5Δ/Klp6Δ cells lacking Mmb1Δ (Fig. 6C), which was not significantly different from Mmb1Δ cells. Additionally, the total mitochondrial volume in both Mmb1Δ and Mmb1Δ-Klp5Δ/Klp6Δ cells was also unchanged compared with WT cells, indicating a change in morphology of mitochondria but not biogenesis (Fig. 6D). Time-lapse images of mitochondria in Mmb1Δ and Mmb1Δ-Klp5Δ/Klp6Δ cells revealed mobile mitochondrial fragments, consistent with the movement expected for mitochondria that are not bound to microtubules (Fig. 6E). Therefore, by separating mitochondria from microtubules, we alleviated the inhibition of Dnm1 assembly on the mitochondria, thereby promoting mitochondrial fission. Fig. S6B contains a summary of the mitochondrial parameters measured in this study.
Discussion
We discovered that association with microtubules determines mitochondrial dynamics and thereby mitochondrial morphology (Fig. 6F). Although previous studies discounted the role of motors in the determination of mitochondrial positioning in fission yeast (22, 23, 25, 44), we have identified kinesin-like proteins that regulate mitochondrial morphology through their control over microtubule length. The association of mitochondria with microtubules mediated by the protein Mmb1 was found to be necessary and sufficient to maintain equilibrium between fission and fusion of mitochondria, with loss of Mmb1 leading to unopposed fission.
In mammalian cells, mitochondria utilize microtubule-associated machinery primarily to traverse the cell. However, mitochondrial dynamics have been observed to be related to microtubule-assisted processes. For instance, in HeLa cells, Drp1 recruitment to the mitochondria was observed to be dependent on the motor protein cytoplasmic dynein (45). Mobile mitochondria on microtubules have also been seen to participate in kiss-and-run fusions that are transient (46). However, a direct role for the microtubule cytoskeleton in the maintenance of mitochondrial dynamics has not yet been explored.
We observed that fission yeast cells fragment mitochondria by emptying the cytoplasm of its microtubules upon onset of mitosis, thereby facilitating independent segregation (39) of the several small mitochondria between future daughter cells. The increase in mitochondrial numbers prior to cell division likely serves to create a well-mixed, homogeneous cytoplasm that is primed for binomial partitioning of mitochondria (Fig. 6F). Mitochondrial fragmentation also reduces the partitioning error between the daughter cells (40) because mitochondrial numbers of 5 or less, which is typical of WT interphase cells, exhibit errors between ∼45 and 100% (Fig. S4D). Although mammalian cells also exhibit fragmented mitochondria during mitosis (47, 48), it has not yet been conclusively demonstrated that partitioning to daughter cells is achieved by an independent segregation mechanism. As we observed here, in cells containing mutants of the mitochondrial fission protein, mammalian cells utilize the cytokinesis machinery to partition mitochondria between daughter cells (49). However, this mechanism of mitochondrial partitioning in Drp1-mutant cells is less uniform because mitochondria exist as a large network at the time of division (50).
We observed that the presence of long microtubules was inhibitory for fission of mitochondria even while Dnm1 was overexpressed. WT cells contained the same total volume of mitochondria as cells with long microtubules but exhibited increased mitochondrial fission in the presence of increased Dnm1 levels. In disease states such as cancer (51) and neurodegeneration (52), mitochondrial fragmentation has been linked with overexpression of Drp1 in mammalian cells. In this work, we have identified that cells that present increased microtubule stabilization are able to overcome the effect of Dnm1 overexpression.
By dissociating mitochondria from microtubules without perturbing the microtubule cytoskeleton, we demonstrated that the failure of Dnm1 ring assembly in the presence of microtubules was the nexus between microtubule dynamics and mitochondrial dynamics (Fig. 6G). This finding could have interesting implications for the interactions between microtubules and mitochondria in other cell types, including in neurons where, at any given time, a majority of the mitochondria are seemingly stably attached to the microtubule cytoskeleton (21). Future studies in these cells will illuminate the consequence of microtubule binding on mitochondrial dynamics.
Alternatively, increased fission upon microtubule depolymerization might be brought about by phosphorylation of the mitochondrial fission protein Dnm1. Mammalian Drp1 undergoes phosphorylation at Ser-616 mediated by Cdk1/cyclin B during cell division and increases mitochondrial fission (47). However, fission yeast Dnm1, however, lacks this phosphorylation site. Drp1 also contains another phosphorylation site at Ser-637, which is conserved in Dnm1. However, PKA-mediated phosphorylation at Ser-637 has been shown to inhibit fission by Drp1 (53), whereas Ca2+/calmodulin-dependent protein kinase α–dependent phosphorylation of Drp1 Ser-637 in rat hippocampal neurons was shown to increase fission (54). To date, yeast Dnm1 has not been demonstrated to undergo phosphorylation at this site.
Another possibility is the existence of an unidentified Dnm1-activating factor that is sequestered by microtubules. This could explain the increase in mitochondrial fragmentation upon microtubule depolymerization. However, in our experiments with cells lacking Mmb1, we induced mitochondrial fragmentation by dissociating mitochondria from microtubules without affecting the microtubule cytoskeleton. Therefore, it is unlikely that mitochondrial fission is promoted by a factor released upon microtubule depolymerization. In conclusion, we have discovered a novel mechanism of regulation of mitochondrial fission by the physical association of mitochondria with microtubules.
Experimental procedures
Strains and media
The fission yeast strains used in this study are listed in Table S1. All the strains were grown in yeast extract medium or Edinburg minimal medium (EMM) (55) with appropriate supplements at a temperature of 30 °C. Cells that were transformed with plasmid pREP41-Dnm1 or pREP41-Dnm1-Cterm-GFP (see Table S1) were cultured in EMM with appropriate supplements and 0.05 μm thiamine for partial induction of the nmt1 promoter. Strains VA076 and VA084 were constructed by crossing PT2244 with FY20823 and Dnm1Δ with G5B, respectively (see Table S1), following the random spore analysis protocol (55).
Plasmid transformation
Transformation of strains was carried out using the improved protocol for rapid transformation of fission yeast (56). In brief, cells were grown overnight to log phase in low-glucose EMM, pelleted, and washed with distilled water. The cells were then washed in a solution of lithium acetate/EDTA (100 mm lithium acetate, 1 mm EDTA, pH 4.9) and resuspended in the same solution. 1 μg of plasmid DNA was added to the suspension followed by addition of lithium acetate/PEG (40% (w/v) PEG, 100 mm lithium acetate, 1 mm EDTA, pH 4.9) and then incubated at 30 °C for 30 min in a shaking incubator. This was followed by a heat shock of 15 min at 42 °C. Thereafter, cells were pelleted down, resuspended in TE solution (10 mm Tris-HCl, 1 mm EDTA, pH 7.5), and plated onto selective EMM plates.
Preparation of yeast for imaging
For imaging mitochondria, fission yeast cells were grown overnight in a shaking incubator at 30 °C, washed once with distilled water, and stained with 200 nm MitoTracker Orange CMTMRos (Thermo Fisher Scientific, catalog number M7510) dissolved in EMM for 20 min. Following this, cells were washed thrice with EMM and then allowed to adhere on lectin-coated (Sigma-Aldrich, catalog number L2380) 35-mm confocal dishes (SPL Life Sciences, catalog number 100350) for 20 min. Unattached cells were then removed by washing with EMM. In experiments where mitochondria were not imaged, staining with MitoTracker was omitted.
Microtubule depolymerization
For depolymerization of microtubules, cells were treated with MBC (carbendazim; 97%; Sigma-Aldrich). A stock solution with a concentration of 25 mg/ml was prepared in DMSO and later diluted to a working concentration of 25 μg/ml in EMM.
Microscopy
Confocal microscopy was carried out using the InCell Analyzer-6000 (GE Healthcare) with 60×/0.7 numerical aperture objective fitted with an sCMOS 5.5MP camera having an x-y pixel separation of 108 nm. For GFP and MitoTracker Orange imaging, 488 and 561 nm laser lines and 525/20 and 605/52 nm bandpass emission filters, respectively, were used. Time lapses for visualization of mitochondrial dynamics were captured by obtaining five Z-stacks with a 0.5-μm step size every 12 s. Deconvolution was performed in images obtained using a Deltavision RT microscope (Applied Precision) with a 100×, oil-immersion 1.4 numerical aperture objective (Olympus, Japan). Excitation of fluorophores was achieved using InsightSSI (Applied Precision) and corresponding filter selection for excitation and emission of GFP and MitoTracker Orange. Z-stacks with 0.3-μm step sizes encompassing the entire cell were captured using a CoolSnapHQ camera (Photometrics) with 2 × 2 binning. The system was controlled using softWoRx 3.5.1 software (Applied Precision), and the deconvolved images were obtained using the built-in setting for each channel.
3D visualization of deconvolved images
3D views of the microtubules and mitochondria in Movies S2–S4, S8–S10, S12, and S13 were obtained from deconvolved images captured in the Deltavision microscope using Fiji's “3D project” function, with the brightest point projection method and 360° total rotation with 10° rotation angle increment.
Estimation of volume of mitochondria
Mitochondrial volume was estimated in Fiji by integrating the areas of mitochondria in thresholded 3D stacks of cells in fluorescence deconvolved images obtained using the Deltavision RT microscope. The total volume was then normalized to the mean total mitochondrial volume of WT cells. Individual mitochondrial volumes were estimated in the same fashion.
Analysis of mitochondrial dynamics
Individual mitochondria were identified in each frame of the time lapse obtained in confocal mode of the GE InCell Analyzer after projecting the maximum intensity of the 3D stack encompassing the cell followed by mean filtering and visualization in Fiji's “mpl-magma” lookup table. Following identification of mitochondria, the “Measure” function of Fiji was used to obtain the circularity, aspect ratio, and parameters of the fitted ellipse. The length of the major axis of the ellipse fitted to a mitochondrion was defined as the size of that mitochondrion. The size, circularity, and aspect ratio were estimated for mitochondria at each frame and each time point. Fission and fusion frequencies of mitochondria were estimated by counting the number of mitochondria identified during each frame of the time lapse. The difference in number of mitochondria from one frame to the next was counted, with an increase being counted as a fission event and a decrease being counted as a fusion event. The total number of fission events and fusion events per cell was estimated and divided by the total duration of the time lapse to obtain the fission and fusion frequencies, respectively.
Test for fit of mitochondrial partitioning during mitosis to binomial distribution
To test the fit of mitochondrial segregation during mitosis to a binomial distribution, the data were z-transformed as described previously (40). Briefly, given n mitochondria in the mother cell just prior to cell division, x and n − x mitochondria in the resulting daughter cells, z was given by 2x − n/√n to approximate the binomial distribution to a normal distribution of 0, 1. The z values obtained for each x and n were binned into k bins of equal sizes and subjected to χ2 test with k − 1 degrees of freedom. The z values are expected to be equally distributed among the k bins, with expected number of 1/k per bin.
Data analysis and plotting
Data analysis was performed in Matlab (Mathworks, Natick, MA). Box plots with the central line indicating the median and notches that represent the 95% confidence interval of the median were obtained by performing one-way ANOVA (“anova1” in Matlab) or Kruskal–Wallis test (“kruskalwallis” in Matlab). The former was used when data were found to be normally distributed, and the latter was used when data were nonnormally distributed (tested using “chi2gof” in Matlab). Following this, significant difference (p < 0.05) was tested using Tukey's honestly significant difference procedure (“multcompare” in Matlab). All the plots were generated using Matlab.
Author contributions
K. M., L. A. C., S. J., and V. A. formal analysis; K. M., L. A. C., and V. A. validation; K. M., L. A. C., M. K. C., S. J., and V. A. investigation; K. M., L. A. C., and V. A. visualization; K. M., L. A. C., M. K. C., and V. A. methodology; V. A. conceptualization; V. A. resources; V. A. data curation; V. A. software; V. A. supervision; V. A. funding acquisition; V. A. writing-original draft; V. A. project administration; V. A. writing-review and editing.
Supplementary Material
Acknowledgments
We thank J. M. Thankachan, S. S. Nuthalapati, and M. Ayushman for pilot experiments; High Content Imaging Facility at Centre for BioSystems Science and Engineering, Indian Institute of Science, and P. I. Rajyaguru for the use of the InCell 6000 and Deltavision RT microscopes, respectively; P. Delivani, I. Jourdain, R. C. Salas, M. Takaine, I. Tolic, Y. Gachet, P. Tran, and National BioResource Project Japan for yeast strains and constructs; and S. Jain for CellROX reagent.
This work was supported by the Department of Science and Technology (India)–Innovation of Science Pursuit for Inspired Research (INSPIRE) Faculty Award, the Department of Biotechnology (India) Innovative Young Biotechnologist Award, and the Science and Engineering Research Board (SERB; India) Early Career Research Award (to V. A.) and by the SERB Early Career Research Award (to S. J.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supporting Experimental Methods, Figs. S1–S6, Tables S1 and S2, and Movies S1–S13.
- Dnm1
- dynamin-like protein 1
- MBC
- methyl benzimidazol-2-yl-carbamate
- Klp
- kinesin-like protein
- MT
- microtubule bundles
- EMM
- Edinburg minimal medium
- ANOVA
- analysis of variance.
References
- 1. Smirnova E., Griparic L., Shurland D. L., and van der Bliek A. M. (2001) Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 12, 2245–2256 10.1091/mbc.12.8.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bleazard W., McCaffery J. M., King E. J., Bale S., Mozdy A., Tieu Q., Nunnari J., and Shaw J. M. (1999) The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298–304 10.1038/13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jourdain I., Gachet Y., and Hyams J. S. (2009) The dynamin related protein Dnm1 fragments mitochondria in a microtubule-dependent manner during the fission yeast cell cycle. Cell Motil. Cytoskeleton 66, 509–523 10.1002/cm.20351 [DOI] [PubMed] [Google Scholar]
- 4. Ingerman E., Perkins E. M., Marino M., Mears J. A., McCaffery J. M., Hinshaw J. E., and Nunnari J. (2005) Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170, 1021–1027 10.1083/jcb.200506078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Basu K., Lajoie D., Aumentado-Armstrong T., Chen J., Koning R. I., Bossy B., Bostina M., Sik A., Bossy-Wetzel E., and Rouiller I. (2017) Molecular mechanism of DRP1 assembly studied in vitro by cryo-electron microscopy. PLoS One 12, e0179397 10.1371/journal.pone.0179397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Delettre C., Lenaers G., Griffoin J.-M., Gigarel N., Lorenzo C., Belenguer P., Pelloquin L., Grosgeorge J., Turc-Carel C., Perret E., Astarie-Dequeker C., Lasquellec L., Arnaud B., Ducommun B., Kaplan J., et al. (2000) Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26, 207–210 10.1038/79936 [DOI] [PubMed] [Google Scholar]
- 7. Sesaki H., Southard S. M., Yaffe M. P., and Jensen R. E. (2003) Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol. Biol. Cell. 14, 2342–2356 10.1091/mbc.e02-12-0788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eura Y., Ishihara N., Yokota S., and Mihara K. (2003) Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J. Biochem. 134, 333–344 10.1093/jb/mvg150 [DOI] [PubMed] [Google Scholar]
- 9. Hermann G. J., Thatcher J. W., Mills J. P., Hales K. G., Fuller M. T., Nunnari J., and Shaw J. M. (1998) Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143, 359–373 10.1083/jcb.143.2.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mishra P., and Chan D. C. (2016) Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 212, 379–387 10.1083/jcb.201511036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen H., and Chan D. C. (2009) Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum. Mol. Genet. 18, R169–R176 10.1093/hmg/ddp326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hirai K., Aliev G., Nunomura A., Fujioka H., Russell R. L., Atwood C. S., Johnson A. B., Kress Y., Vinters H. V., Tabaton M., Shimohama S., Cash A. D., Siedlak S. L., Harris P. L., Jones P. K., et al. (2001) Mitochondrial abnormalities in Alzheimer's disease. J. Neurosci. 21, 3017–3023 10.1523/JNEUROSCI.21-09-03017.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deng H., Dodson M. W., Huang H., and Guo M. (2008) The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 14503–14508 10.1073/pnas.0803998105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Graves J. A., Wang Y., Sims-Lucas S., Cherok E., Rothermund K., Branca M. F., Elster J., Beer-Stolz D., Van Houten B., Vockley J., and Prochownik E. V. (2012) Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PLoS One 7, e37699 10.1371/journal.pone.0037699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ashrafian H., Docherty L., Leo V., Towlson C., Neilan M., Steeples V., Lygate C. A., Hough T., Townsend S., Williams D., Wells S., Norris D., Glyn-Jones S., Land J., Barbaric I., et al. (2010) A mutation in the mitochondrial fission gene Dnm1l leads to cardiomyopathy. PLoS Genet. 6, e1001000 10.1371/journal.pgen.1001000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pilling A. D., Horiuchi D., Lively C. M., and Saxton W. M. (2006) Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell. 17, 2057–2068 10.1091/mbc.e05-06-0526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stowers R. S., Megeath L. J., Górska-Andrzejak J., Meinertzhagen I. A., and Schwarz T. L. (2002) Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron 36, 1063–1077 10.1016/S0896-6273(02)01094-2 [DOI] [PubMed] [Google Scholar]
- 18. Glater E. E., Megeath L. J., Stowers R. S., and Schwarz T. L. (2006) Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol. 173, 545–557 10.1083/jcb.200601067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Spronsen M., Mikhaylova M., Lipka J., Schlager M. A., van den Heuvel D. J., Kuijpers M., Wulf P. S., Keijzer N., Demmers J., Kapitein L. C., Jaarsma D., Gerritsen H. C., Akhmanova A., and Hoogenraad C. C. (2013) TRAK/milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron 77, 485–502 10.1016/j.neuron.2012.11.027 [DOI] [PubMed] [Google Scholar]
- 20. Chen Y., and Sheng Z.-H. (2013) Kinesin-1-syntaphilin coupling mediates activity-dependent regulation of axonal mitochondrial transport. J. Cell Biol. 202, 351–364 10.1083/jcb.201302040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kang J.-S., Tian J.-H., Pan P.-Y., Zald P., Li C., Deng C., and Sheng Z.-H. (2008) Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell 132, 137–148 10.1016/j.cell.2007.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yaffe M. P., Stuurman N., and Vale R. D. (2003) Mitochondrial positioning in fission yeast is driven by association with dynamic microtubules and mitotic spindle poles. Proc. Natl. Acad. Sci. U.S.A. 100, 11424–11428 10.1073/pnas.1534703100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chiron S., Bobkova A., Zhou H., and Yaffe M. P. (2008) CLASP regulates mitochondrial distribution in Schizosaccharomyces pombe. J. Cell Biol. 182, 41–49 10.1083/jcb.200712147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fu C., Jain D., Costa J., Velve-Casquillas G., and Tran P. T. (2011) Mmb1p binds mitochondria to dynamic microtubules. Curr. Biol. 21, 1431–1439 10.1016/j.cub.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li T., Zheng F., Cheung M., Wang F., and Fu C. (2015) Fission yeast mitochondria are distributed by dynamic microtubules in a motor-independent manner. Sci. Rep. 5, 11023 10.1038/srep11023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fehrenbacher K. L., Yang H. C., Gay A. C., Huckaba T. M., and Pon L. A. (2004) Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr. Biol. 14, 1996–2004 10.1016/j.cub.2004.11.004 [DOI] [PubMed] [Google Scholar]
- 27. Huh D., and Paulsson J. (2011) Random partitioning of molecules at cell division. Proc. Natl. Acad. Sci. U.S.A. 108, 15004–15009 10.1073/pnas.1013171108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lawrence E., and Mandato C. (2013) Mitochondrial inheritance is mediated by microtubules in mammalian cell division. Commun. Integr. Biol. 6, e27557 10.4161/cib.27557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krüger N., and Tolić-Nørrelykke I. M. (2008) Association of mitochondria with spindle poles facilitates spindle alignment. Curr. Biol. 18, R646–R647 10.1016/j.cub.2008.06.069 [DOI] [PubMed] [Google Scholar]
- 30. Jajoo R., Jung Y., Huh D., Viana M. P., Rafelski S. M., Springer M., and Paulsson J. (2016) Accurate concentration control of mitochondria and nucleoids. Science 351, 169–172 10.1126/science.aaa8714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bergeland T., Widerberg J., Bakke O., and Nordeng T. W. (2001) Mitotic partitioning of endosomes and lysosomes. Curr. Biol. 11, 644–651 10.1016/S0960-9822(01)00177-4 [DOI] [PubMed] [Google Scholar]
- 32. Shima D. T., Cabrera-Poch N., Pepperkok R., and Warren G. (1998) An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J. Cell Biol. 141, 955–966 10.1083/jcb.141.4.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tischer C., Brunner D., and Dogterom M. (2009) Force- and kinesin-8-dependent effects in the spatial regulation of fission yeast microtubule dynamics. Mol. Syst. Biol. 5, 250 10.1038/msb.2009.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. West R. R., Malmstrom T., Troxell C. L., and McIntosh J. R. (2001) Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol. Biol. Cell. 12, 3919–3932 10.1091/mbc.12.12.3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Browning H., Hayles J., Mata J., Aveline L., Nurse P., and McIntosh J. R. (2000) Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J. Cell Biol. 151, 15–28 10.1083/jcb.151.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Busch K. E., Hayles J., Nurse P., and Brunner D. (2004) Tea2p kinesin is involved in spatial microtubule organization by transporting Tip1p on microtubules. Dev. Cell. 6, 831–843 10.1016/j.devcel.2004.05.008 [DOI] [PubMed] [Google Scholar]
- 37. Pletjushkina O. Y., Lyamzaev K. G., Popova E. N., Nepryakhina O. K., Ivanova O. Y., Domnina L. V., Chernyak B. V., and Skulachev V. P. (2006) Effect of oxidative stress on dynamics of mitochondrial reticulum. Biochim. Biophys. Acta 1757, 518–524 10.1016/j.bbabio.2006.03.018 [DOI] [PubMed] [Google Scholar]
- 38. Ding R., West R. R., Morphew D. M., Oakley B. R., and McIntosh J. R. (1997) The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell. 8, 1461–1479 10.1091/mbc.8.8.1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Birky C. W. (1983) The partitioning of cytoplasmic organelles at cell division. Int. Rev. Cytol. Suppl. 15, 49–89 [DOI] [PubMed] [Google Scholar]
- 40. Hennis A. S., and Birky C. W. (1984) Stochastic partitioning of chloroplasts at cell division in the alga Olisthodiscus, and compensating control of chloroplast replication. J. Cell Sci. 70, 1–15 [DOI] [PubMed] [Google Scholar]
- 41. Guillou E., Bousquet C., Daloyau M., Emorine L. J., and Belenguer P. (2005) Msp1p is an intermembrane space dynamin-related protein that mediates mitochondrial fusion in a Dnm1p-dependent manner in S. pombe. FEBS Lett. 579, 1109–1116 10.1016/j.febslet.2004.12.083 [DOI] [PubMed] [Google Scholar]
- 42. Höög J. L., Schwartz C., Noon A. T., O'Toole E. T., Mastronarde D. N., McIntosh J. R., and Antony C. (2007) Organization of interphase microtubules in fission yeast analyzed by electron tomography. Dev. Cell 12, 349–361 10.1016/j.devcel.2007.01.020 [DOI] [PubMed] [Google Scholar]
- 43. Mears J. A., Lackner L. L., Fang S., Ingerman E., Nunnari J., and Hinshaw J. E. (2011) Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 18, 20–26 10.1038/nsmb.1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brazer S. C., Williams H. P., Chappell T. G., and Cande W. Z. (2000) A fission yeast kinesin affects Golgi membrane recycling. Yeast 16, 149–166 [DOI] [PubMed] [Google Scholar]
- 45. Varadi A., Johnson-Cadwell L. I., Cirulli V., Yoon Y., Allan V. J., and Rutter G. A. (2004) Cytoplasmic dynein regulates the subcellular distribution of mitochondria by controlling the recruitment of the fission factor dynamin-related protein-1. J. Cell Sci. 117, 4389–4400 10.1242/jcs.01299 [DOI] [PubMed] [Google Scholar]
- 46. Liu X., Weaver D., Shirihai O., and Hajnóczky G. (2009) Mitochondrial “kiss-and-run”: interplay between mitochondrial motility and fusion-fission dynamics. EMBO J. 28, 3074–3089 10.1038/emboj.2009.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Taguchi N., Ishihara N., Jofuku A., Oka T., and Mihara K. (2007) Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 282, 11521–11529 10.1074/jbc.M607279200 [DOI] [PubMed] [Google Scholar]
- 48. Mitra K., Wunder C., Roysam B., Lin G., and Lippincott-Schwartz J. (2009) A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. U.S.A. 106, 11960–11965 10.1073/pnas.0904875106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S. O., Masuda K., Otera H., Nakanishi Y., Nonaka I., Goto Y., Taguchi N., Morinaga H., Maeda M., Takayanagi R., Yokota S., et al. (2009) Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11, 958–966 10.1038/ncb1907 [DOI] [PubMed] [Google Scholar]
- 50. Mishra P., and Chan D. C. (2014) Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 15, 634–646 10.1038/nrm3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ferreira-da-Silva A., Valacca C., Rios E., Pópulo H., Soares P., Sobrinho-Simões M., Scorrano L., Máximo V., and Campello S. (2015) Mitochondrial dynamics protein Drp1 is overexpressed in oncocytic thyroid tumors and regulates cancer cell migration. PLoS One 10, e0122308 10.1371/journal.pone.0122308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Itoh K., Nakamura K., Iijima M., and Sesaki H. (2013) Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 23, 64–71 10.1016/j.tcb.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chang C.-R., and Blackstone C. (2007) Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem. 282, 21583–21587 10.1074/jbc.C700083200 [DOI] [PubMed] [Google Scholar]
- 54. Han X.-J., Lu Y.-F., Li S.-A., Kaitsuka T., Sato Y., Tomizawa K., Nairn A. C., Takei K., Matsui H., and Matsushita M. (2008) CaM kinase Iα–induced phosphorylation of Drp1 regulates mitochondrial morphology. J. Cell Biol. 182, 573–585 10.1083/jcb.200802164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Forsburg S. L., and Rhind N. (2006) Basic methods for fission yeast. Yeast 23, 173–183 10.1002/yea.1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kanter-Smoler G., Dahlkvist A., and Sunnerhagen P. (1994) Improved method for rapid transformation of intact Schizosaccharomyces pombe cells. BioTechniques 16, 798–800 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.