Abstract
The bacterial type III secretion system (T3SS) delivers virulence proteins, called effectors, into eukaryotic cells. T3SS comprises a transmembrane secretion apparatus and a complex network of specialized chaperones that target protein substrates to this secretion apparatus. However, the regulation of secretion switching from early (needle and inner rod) to middle (tip/filament and translocators) substrates is incompletely understood. Here, we investigated chaperone-mediated secretion switching from early to middle substrates in the T3SS encoded by Salmonella pathogenicity island 2 (SPI2), essential for systemic infection. Our findings revealed that the protein encoded by ssaH regulates the secretion of an inner rod and early substrate, SsaI. Structural modeling revealed that SsaH is structurally similar to class III chaperones, known to associate with proteins in various pathogenic bacteria. The SPI2 protein SsaE was identified as a class V chaperone homolog and partner of SsaH. A pulldown analysis disclosed that SsaH and SsaE form a heterodimer, which interacted with another early substrate, the needle protein SsaG. Moreover, SsaE also helped stabilize SsaH and a middle substrate, SseB. We also found that SsaE regulates cellular SsaH levels to translocate the early substrates SsaG and SsaI and then promotes the translocation of SseB by stabilizing it. In summary, our results indicate that the class III chaperone SsaH facilitates SsaI secretion, and a heterodimer of SsaH and the type V chaperone SsaE then switches secretion to SsaG. This is the first report of a chaperone system that regulates both early and middle substrates during substrate switching for T3SS assembly.
Keywords: chaperone, type III secretion system (T3SS), protein secretion, protein assembly, protein folding, Salmonella enterica, virulence factor, effector protein, protein secretion switching, Salmonella pathogenicity island 2, SsaE, SsaH
Introduction
Many Gram-negative pathogens use type III secretion systems (T3SSs)2 to deliver virulence proteins, termed effectors, directly into the eukaryotic cytoplasm (1). T3SSs comprise several structures, including a sorting platform containing an ATPase complex and a cytoplasmic ring, an inner membrane export apparatus, a basal body with a needle and an inner rod, and a translocation pore. For T3SSs to fully assemble and function correctly, numerous proteins must be secreted in a predefined order. On the basis of the T3SS secretion hierarchy, secreted proteins are divided into early (needle and inner rod), middle (tip/filament and translocators), and late (effectors) substrates.
The components of the T3SS have been uniformly named (1, 2). The needle (SctF) and inner rod (SctI) proteins are secreted through the basal body. When the needle reaches a defined length, it signals the completion of the functional type III secretion apparatus, and the system switches to secretion of the middle substrates (3). A needle-length ruler (SctP) is a key regulator responsible for controlling the needle length and for secretion switching. The loss of SctP results in long, unregulated T3SS needles (4, 5). The secretion of SctF and SctI is significantly increased upon deletion of SctP (6–8). SctU, a member of the export apparatus, is also involved in controlling needle length and secretion switching (3, 7, 9, 10).
In some bacterial species, specific chaperones are necessary for SctF secretion. Chaperones in T3SSs bind to newly synthesized substrates and keep them partially unfolded in a secretion-competent state (1, 11). Chaperones are classified on the basis of substrate specificity into classes IA/IB, II, III, IV, and V (12). Class III chaperones are known to bind to needle proteins. In Pseudomonas aeruginosa and Yersinia spp., SctF (PscF and YscF) forms a complex between the class III chaperone (PscG and YscG) and a partner protein known as the class V chaperone (PscE and YscE). Structural studies have revealed that class III chaperones bind to the C termini of needle proteins to maintain their soluble state and prevent self-aggregation (13, 14). However, the homologs of class III and V chaperones have not been identified in most T3SSs. Thus, it is still unclear whether chaperone-mediated regulation of early substrate secretion contributes to the control of the needle length and substrate switching in other T3SSs.
Salmonella enterica serovar Typhimurium is an enteropathogenic bacterium that causes gastroenteritis in humans and typhoid-like fever in mice. Salmonella produces two T3SSs encoded chromosomally by the Salmonella pathogenicity islands (SPIs) 1 and 2 (15). The SPI1-T3SS plays an important role in gastrointestinal disease but seems to be largely dispensable for systemic infection in mice (16). In contrast, the SPI2-T3SS is required for growth within host cells such as macrophages to establish systemic infection in mice (17, 18). After the phagocytosis of Salmonella by macrophages, SPI2 expression is induced in bacteria growing intracellularly in response to phagosomal conditions such as acidic pH and nutrient limitations, whereas SPI1 expression is repressed (19, 20). Thirty-one genes in SPI2 are organized into two operons encoding components of the secretion apparatus, ssaBCDE and ssaG–U; one transcriptional unit encodes effectors (sse) and chaperones (ssc), and the other encodes the two-component regulatory system, SsrAB (21) (Fig. S1). To date, two subgroups of substrate proteins transported by the SPI2-T3SS have been identified: translocators and effectors. SseBCD function as translocators of the SPI2-T3SS and show a certain similarity to the translocon proteins EspABD of enteropathogenic Escherichia coli (EPEC) (17, 22, 23). SseB is a protein with sequence similarity to EspA and composes a filament, whereas SseC and SseD are similar to EspD and EspB, respectively, and compose the pore-forming translocon. In vitro secretion analysis has shown that all three proteins are secreted by the SPI2-T3SS and are loosely attached to the bacterial surface (22, 24). Although SseB secretion is independent of SseC and SseD (22), efficient secretion of the latter two proteins occurs only in the presence of SseB (23). After assembly of the translocation pores in phagosomal membranes by SseC and SseD, effectors are translocated into the host cytoplasm upon sensing of the host cytosolic nutrient pH (25). Although many studies have focused on the regulation of secretion and switching from middle (translocators) to late (effectors) substrates, the secretion of early substrates by the SPI2-T3SS is not well understood.
In this study, we aimed to investigate chaperone-mediated secretion switching from early to middle substrates in the SPI2-T3SS. Our findings show that the class V chaperone SsaE regulates the cellular level of the class III chaperone SsaH to translocate early substrates and subsequently promotes secretion of the middle substrate, SseB, through its stabilization. The class V chaperone that regulates both early and middle substrates affecting substrate switching, described here, is expected to provide insight into the mechanism of secretion switching from early to middle substrates in the T3SSs of pathogenic bacteria.
Results
SsaI is an early substrate of SPI2-T3SS
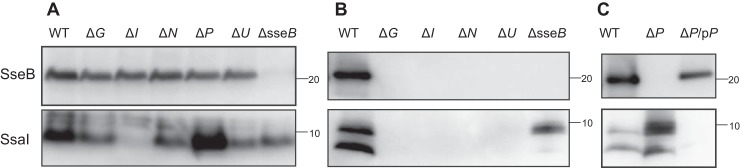
SsaI is a homolog of the inner rod, commonly known as SctI (1). The SsaI ORF is 246 bp long and encodes a peptide of 82 amino acid residues with a predicted molecular mass of 8.96 kDa. SsaI seems to have low levels of identity with other inner rod SctI proteins (Fig. S2A). SctI proteins are known to be secreted as early T3SS substrates. To determine whether SsaI is also secreted by the SPI2-T3SS, we established a rabbit anti-SsaI antibody (Fig. 1). The antibody showed a band corresponding to ∼9 kDa in the cytoplasm of the wildtype (WT) strain (Fig. 1A), whereas the protein was not detected in the ssaI-disrupted mutant (ΔssaI) strain. However, the corresponding protein was detected in the ΔssaI strain carrying plasmid pTKY1245, capable of overexpressing ssaI following the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) under the regulation of the PA1lacO−1 promoter system (Fig. 2A), indicating that the anti-SsaI antibody established in this study has the ability to specifically detect SsaI produced in Salmonella cells. The anti-SsaI antibody detected the protein in the supernatant fraction prepared from WT cells (Fig. 1B), demonstrating the secretion of the SsaI protein.
Figure 1.
SsaI is an early substrate of SPI2-T3SS. Effects of disruption of several SPI2 genes on the production (A) and secretion (B and C) of SsaI and SseB are shown. SsaI and SseB were determined via immunoblotting using anti-SsaI and anti-SseB antibodies. Bacterial cells of strains χ3306 (WT), CS3491 (ΔsseB), CS4269 (ΔssaI; ΔI), CS4270 (ΔssaP; ΔP), CS10072 (ΔP/pP), CS10092 (ΔssaU; ΔU), CS10107 (ΔssaN; ΔN), and CS10135 (ΔssaG; ΔG) were used.
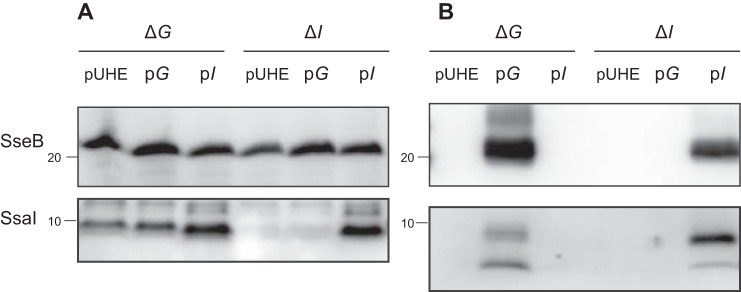
Figure 2.
SsaG is required for SsaI secretion. Salmonella strains CS10157 (ΔG/pUHE), CS10155 (ΔG/pG), CS10156 (ΔG/pI), CS10068 (ΔI/pUHE), CS10159 (ΔI/pG), and CS10069 (ΔI/pI) were grown at 37 °C for 8 h in low-Mg2+ minimal medium with 0.1 mm IPTG for the induction of ssaI. Each protein was detected in cell lysates (A) and supernatants (B) by immunoblotting using anti-SseB and anti-SsaI antibodies.
SsaG, SsaP, and SsaU are homologs of the needle protein SctF, the needle-length ruler SctP, and the inner membrane protein SctU, respectively (1). SsaN is an ATPase associated with SPI2 (26, 27). SseB is the filament protein secreted by the SPI2-T3SS (23, 28). We also detected SseB in the supernatant fraction by immunoblotting with a rabbit anti-SseB antibody used previously (23). To examine whether the disruption of the SPI2 apparatus influenced the production of SsaI and SseB, the cellular levels of SsaI and SseB in the disrupted mutants of ssaG, ssaN, ssaP, ssaU, and sseB were examined (Fig. 1A). Levels of SsaI in ΔssaG, ΔssaN, ΔssaU, and ΔsseB strains were detectable but slightly lower than that in the WT strain. In contrast, SseB levels in ΔssaG, ΔssaI, ΔssaN, and ΔssaU strains were similar to that in the WT strain. In the ΔssaP strain, the amount of SsaI was increased, but the production of SseB was not affected. These results suggest that the disruption of the SPI2 apparatus may not reduce the production of SsaI and SseB.
To determine whether SsaI is secreted extracellularly via the SPI2-T3SS, we examined the amounts of SsaI and SseB in the supernatants of WT and SPI2 mutant strains (Fig. 1B). Both SsaI and SseB were detected in the supernatants of the WT strain. The anti-SsaI antibody detected two bands in the supernatant fraction. On the basis of molecular size, the upper band appeared to correspond to the intact SsaI protein, which can be detected in the cell lysate, whereas the lower band was assumed to be truncated SsaI. In the supernatant of the ΔsseB strain, SsaI was detected at a reduced level compared with that in the WT strain. In contrast, neither SsaI nor SseB was detected in the supernatants of the ΔssaN and ΔssaU strains (Fig. 1B). These results suggest that SsaI may be secreted via the SPI2-T3SS prior to secretion of the middle substrate SseB.
SsaI and SsaG coregulate SseB secretion
The ΔssaI strain was not capable of secreting SseB (Fig. 1B). Moreover, the ΔssaG strain did not secrete SsaI or SseB (Fig. 1B). To confirm that the loss of SseB secretion in both ΔssaG and ΔssaI strains was due to disruption of ssaG and ssaI, respectively, plasmids pTKY1243 and pTKY1245, capable of overexpressing ssaG and ssaI, respectively, under the regulation of the PA1lacO−1 promoter system, were introduced into the corresponding strains. The resultant strains were tested for complementation of SseB secretion via detection of SseB protein in cell lysates and supernatants (Fig. 2). The cellular level of SseB in the ΔssaI strain containing pTKY1245, which enables inducible expression of ssaI, was increased compared with that in ΔssaI strains carrying the empty vector pUHE21-2Δfd12 and pTKY1243 (Fig. 2A). In the supernatant of the ΔssaI strain carrying pTKY1245, SseB was detected (Fig. 2B), indicating that ssaI gene on pTKY1245 was translated to the functional SsaI. In the ΔssaG strain, ssaG expression by the introduced pTKY1243 recovered the secretion of both SseB and SsaI (Fig. 2), indicating that ssaG gene on pTKY1243 was also translated to the functional SsaG. In contrast, the expression of ssaI by pTKY1245 in the ΔssaG strain recovered the secretion of neither SseB nor SsaI, whereas the cellular level of SsaI in this strain was increased compared with those in the ΔssaG strain carrying pUHE21-2Δfd12 and pTKY1243. Furthermore, ssaG expression in the ΔssaI strain did not recover the secretion of SseB. SsaG is the needle protein (Fig. S2B), which is secreted as the early substrate. Therefore, the results suggest that the secretion of both SsaG and SsaI may be involved in SseB secretion.
SsaI secretion is negatively regulated by SsaP
In the supernatant of the ΔssaP strain, an increased level of SsaI was detected compared with that in the WT strain, but no SseB was detected (Fig. 1C). The increased secretion of SsaI was suppressed by the introduction of plasmid pTKY1251, which expresses a functional ssaP gene under the regulation of the lac promoter in a low-copy plasmid. Furthermore, SseB secretion was recovered in the ssaP-complemented strain. The secretion of SPI2 effectors is activated by the exposure of bacteria to pH 7.2 following growth at pH 5.0 (25). SseB was not secreted even when the ΔssaP strain was exposed to the medium at pH 7.2 after growth at pH 5.0 (Fig. S3). These results suggest that the repression of SsaI secretion by SsaP may be necessary for substrate switching to SseB in the SPI2-T3SS.
Cytosolic protein SsaH is required for secretion of SsaI
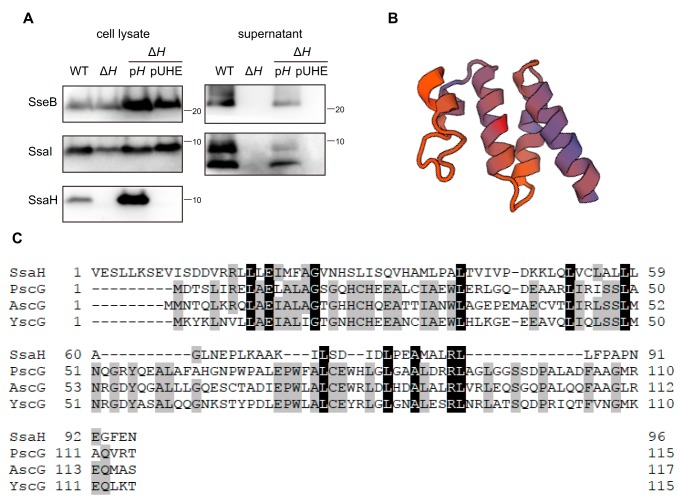
The ssaH gene is located between ssaG and ssaI in the ssaG–U operon in SPI2 (Fig. S1). The ORF of ssaH is 288 bp long and encodes a peptide of 96 amino acid residues with a predicted molecular mass of 10.5 kDa and a predicted pI of 4.83. The homolog of SsaH has not been identified in other T3SSs. To examine the function of SsaH in the SPI2-T3SS, we established a rabbit anti-SsaH antibody and characterized an ssaH-disrupted mutant (Fig. 3A). In the cell lysate prepared from the WT strain, a band corresponding to ∼10 kDa was detected by immunoblotting using the anti-SsaH antibody. The protein was not detected in the cell lysate of the ΔssaH strain. The corresponding protein was also detected in the ΔssaH strain carrying plasmid pTKY1246, which overexpressed ssaH under the regulation of the PA1lacO−1 promoter upon the addition of IPTG (Fig. 3A), indicating the ability of the established anti-SsaH antibody to detect endogenous SsaH. In the ΔssaH strain, SsaI protein, encoded by a gene located downstream of ssaH, was produced at the same level as that in the WT strain (Fig. 3A). Additionally, the amount of SseB was similar to that in the WT strain. However, neither SsaI nor SseB was detected in the supernatant of the ΔssaH strain (Fig. 3A). The loss of SPI2 secretion was complemented by ssaH expression in the ΔssaH strain. In addition, SsaH was not detected in the supernatants of WT or ssaH-overexpressing strains (data not shown). These results suggest that SsaH in the bacterial cytoplasm is required for the secretion of SsaI via the SPI2-T3SS.
Figure 3.
SsaH is required for SsaI secretion. A, effect of disruption of ssaH on secretion and production of SseB, SsaI, and SsaH. Each protein was detected in cell lysates and supernatants via immunoblotting using anti-SseB, anti-SsaI, and anti-SsaH antibodies. Bacterial cells of strains χ3306 (WT), CS10091 (ΔssaH; ΔH), CS10095 (ΔH/pH), and CS10096 (ΔH/pUHE) were used. B, ribbon representation of SsaH structure predicted by the SWISS-MODEL expert system. C, amino acid alignment of SsaH and class III chaperones PscG, AscG, and YscG.
SsaH forms a heteromolecular complex with SsaE
Structural modeling of SsaH was performed with the SWISS-MODEL expert system for protein structure modeling (https://swissmodel.expasy.org) to evaluate the biological function of SsaH (29). The structure of SsaH was based on the class III chaperone PscG in P. aeruginosa (SWISS-MODEL Template Library (SMTL) ID 2uwj.1.C) (Fig. 3B), although the sequence identity between SsaH and PscG was only 12.5% (Fig. 3C). AscG, in Aeromonas hydrophila, and YscG, in Yersinia spp., are also known as class III chaperones. Sequence alignment of SsaH showed low identities of 10.4 and 14.9% with AscG and YscG, respectively (Fig. 3C).
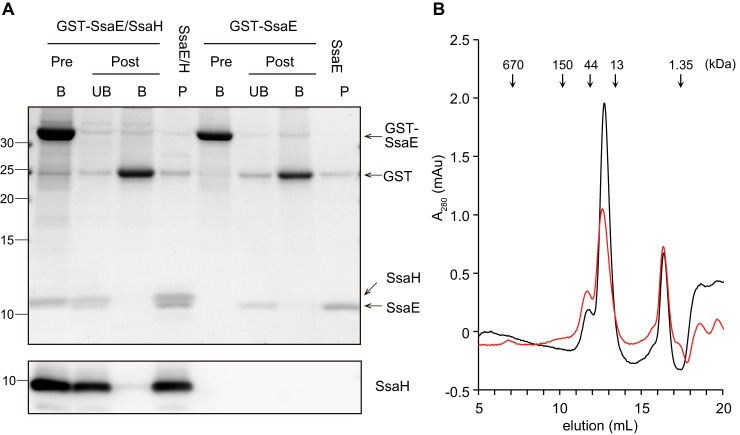
PscG interacts with PscE, a class V chaperone, to form the heteromonomeric chaperone PscGE (14). AscG and YscG also interact with AscE and YscE, respectively, which are class V chaperones (30, 31). Another homolog of a class V chaperone in the SPI2-T3SS is SsaE (28), encoded by a gene located within the ssaBCDE operon in the SPI2 locus (Fig. S1). Recently, functionality of the SWISS-MODEL system has been extended to modeling heteromeric complexes (32). Thus, a structural model of the complex of SsaH with SsaE was predicted using SWISS-MODEL. Three heteromolecular complexes of SsaH and SsaE were feasible based on a PscGE hetero-2–2-mer, a YscGE heterodimer, and an AscGE heterodimer (Fig. S4, A–C). This suggested that SsaH interacts with SsaE. To test this possibility, GSH S-transferase (GST)-tagged SsaE (GST-SsaE) and SsaH were coexpressed in E. coli and subsequently used to assess the interaction between SsaH and SsaE via a GST pulldown assay (Fig. 4A). When GST-SsaE and SsaH were expressed together in E. coli, both bound to the GSH particles. Following cleavage of the GST tag from SsaE by the addition of PreScission protease, SsaH could be coeluted with SsaE but not with GST, indicating that SsaH directly may interact with SsaE. The gel-filtration profile of the SsaH–SsaE complex showed that the complex was eluted at a volume corresponding to its expected molecular weight as a heterodimer (Fig. 4B). As shown in Fig. 4A, when proteins in the SsaH–SsaE complex were eluted by gel filtration and separated by a Tricine gel system, the resultant band intensity corresponding to the SsaH protein was similar to that of SsaE. These results indicated that SsaH forms a 1:1 heteromolecular complex with SsaE. The SsaH–SsaE heterodimer model predicted that the N-terminal region of SsaH may be involved in the interaction with SsaE (Fig. S4, A–C). Coexpression of SsaH lacking the 21 N-terminal residues and GST-SsaE demonstrated that SsaH lacking the 21 N-terminal residues could not bind to GST-SsaE (Fig. S4D). This result suggests that the N terminus of SsaH is involved in the direct interaction with SsaE in the complex.
Figure 4.
SsaH directly interacts with SsaE. A, E. coli strains CS7141 (GST-SsaE) and CS7145 (GST-SsaE and SsaH) were grown to exponential phase followed by the induction of GST-SsaE and SsaH by the addition of 1 mm IPTG for 3 h. GST-SsaE in each cell lysate was bound to MagneGST glutathione particles for 1 h at 4 °C. After washing, the particles were divided into two groups. One part was incubated with 50 mm GSH to elute GST-SsaE and bound proteins (Pre, B). The other was incubated with PreScission protease for 16 h at 4 °C to cleave the GST tag from the SsaE protein. Next, cleaved proteins were collected (Post, UB), and the proteins bound to particles were eluted (Post, B). P indicates the peak fraction separated using gel-filtration chromatography. Proteins separated via 16% Tricine gel were visualized with Oriole fluorescent gel stain (upper panel) and immunostained with anti-SsaH antibody (lower panel). B, stoichiometric analysis of SsaE (molecular mass, 9.65 kDa) and SsaE-SsaH (molecular mass, 10.3 kDa for SsaH and 20 kDa for the complex) via gel filtration. A total of 12 μg of each of SsaE–SsaH (red) and SsaE (black) protein was subjected to size-exclusion chromatography using Superose 12. Absorbance at A280 was monitored to determine the elution profile. mAU, milliabsorbance units.
Additionally, the gel-filtration profile of SsaE showed that SsaE exists as a homodimer in solution (Fig. 4B). The crystal structures of YscE from Yersinia pestis and AscE from A. hydrophila have been reported as that of dimeric helical proteins (30, 33). These findings suggested that SsaE may exist as a homodimer as well as form a heterodimer with SsaH.
SsaE regulates the cellular levels of SsaH and SseB in the bacterial cytoplasm
Although SsaE has been shown to recognize SseB as a middle substrate to regulate its secretion via the SPI2-T3SS (28), the involvement of SsaE in the secretion of early substrates, including SsaI, is not clear. Thus, we constructed an ssaE-disrupted mutant and assessed its ability to secrete SsaI (Fig. 5A). Immunoblotting of the supernatant indicated that neither SsaI nor SseB was secreted by the ΔssaE strain. To confirm that the loss of secretion of these proteins in the ΔssaE strain was due to the ssaE disruption, plasmid pTKY1261, expressing ssaE under the regulation of the ssaB promoter, was introduced into the ΔssaE strain. The amount of SsaI secreted by the ΔssaE strain carrying pTKY1261 was significantly increased compared with that in the ΔssaE strain, whereas the amount of SseB was only slightly increased. This result indicated that SsaE may also be involved in the secretion of SsaI. The cellular level of SsaI was not affected by the ssaE disruption (Fig. 5A). Interestingly, cellular levels of SsaH and SseB in the ΔssaE strain were significantly reduced compared with that in the WT strain. This reduction in the ΔssaE strain was complemented by introducing the functional ssaE gene into pTKY1261, suggesting that SsaE may contribute to the stabilization of SsaH and SseB. In contrast, ssaH disruption did not affect the cellular level of SseB (Fig. 3A). These results suggest that SsaE may regulate the production of SseB in an SsaH-independent manner.
Figure 5.
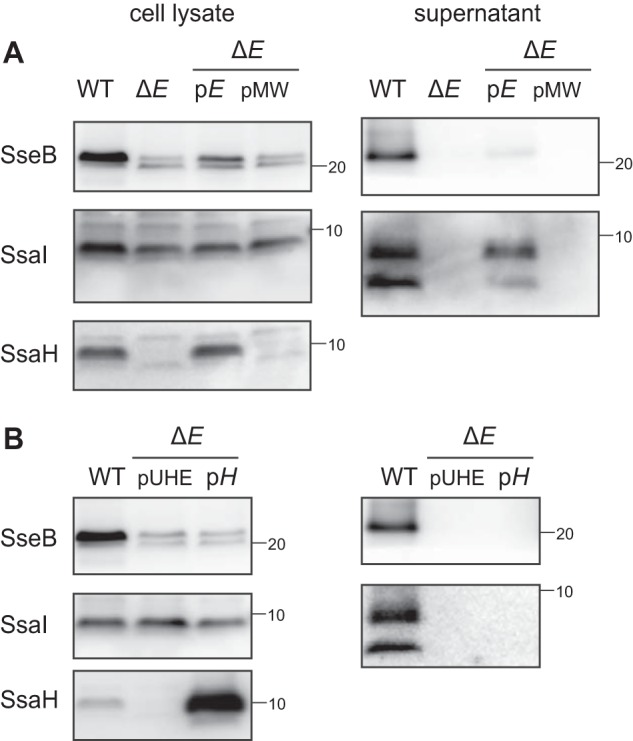
SsaE controls the cellular level of SsaH and SseB. A, effect of disruption of ssaE on secretion and production of SseB, SsaI, and SsaH. Each protein was detected in supernatants and cell lysates via immunoblotting using anti-SseB, anti-SsaI, and anti-SsaH antibodies. Bacterial cells of strains χ3306 (WT), CS10106 (ΔssaE; ΔE), CS10150 (ΔE/pE), and CS10151 (ΔE/pMW) were used. B, overproduction of SsaH in ssaE-disrupted cells did not complement the secretion of SsaI. Bacterial strains χ3306 (WT), CS10153 (ΔE/pUHE), and CS10165 (ΔE/pH) were grown at 37 °C for 8 h in low-Mg2+ minimal medium with 0.1 mm IPTG for the induction of ssaH. Each protein in supernatants and cell lysates was detected via immunoblotting using anti-SseB, anti-SsaI, and anti-SsaH antibodies.
To examine the possibility that impaired SsaH production by the ssaE-disrupted strain could result in the loss of SsaI secretion, ssaH was expressed in the ΔssaE strain by introducing pTKY1246 (Fig. 5B). The enhanced production of SsaH protein in the ssaE-disrupted background did not recover SsaI secretion. These results suggest that both SsaH and SsaE may function as a complex to regulate SsaI secretion.
SsaH–SsaE complex interacts with SsaG but not SsaI
Most T3SS chaperones directly interact with their substrates in the bacterial cytoplasm to stabilize them and recruit them to the sorting platform, resulting in the translocation of the substrates into target host cells (34). Although SsaH and SsaE were involved in the secretion of SsaI, neither appeared to be involved in the stabilization of SsaI (Figs. 3A and 5A). We examined the binding of SsaI to SsaH–SsaE in E. coli using the GST pulldown assay (Fig. 6). When GST-SsaE, SsaH, and SsaI were coexpressed in E. coli and proteins bound to SsaE were collected, SsaH but not SsaI was detected in the eluted fraction of SsaE by immunoblotting analysis (Fig. 6). We also performed the GST pulldown assay in the ssaE-disrupted strain of Salmonella (Fig. S5). Cellular SsaH and SseB were detected in the GST-SsaE–expressing strain but not in the GST-expressing strain. Moreover, the GST pulldown assay demonstrated that GST-SsaE interacted with SsaH and SseB but not SsaI on a chromosomal level, suggesting that GST-SsaE may bind to both SsaH and SseB (Fig. S5). These results indicated that the SsaH–SsaE complex recognizes other substrates involved in SsaI secretion.
Figure 6.
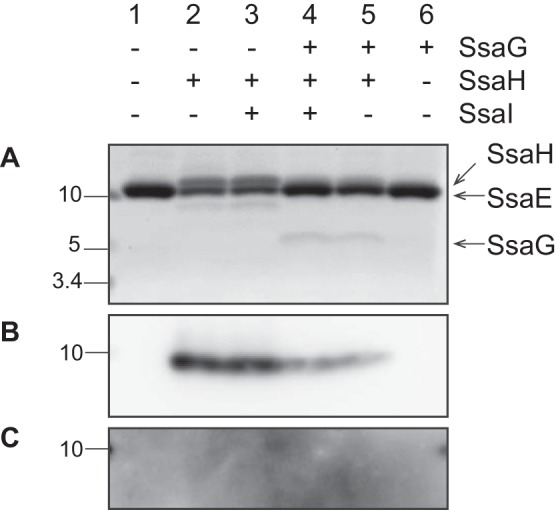
SsaG binds to SsaE in the presence of SsaH. E. coli strains CS7141 (GST-SsaE), CS7142 (GST-SsaE, SsaG, SsaH, and SsaI), CS7143 (GST-SsaE, SsaG, and SsaH), CS7144 (GST-SsaE, SsaH, and SsaI), CS7145 (GST-SsaE and SsaH), and CS7168 (GST-SsaE and SsaG) were grown to exponential phase followed by the induction of gene expression by the addition of 1 mm IPTG for 3 h. GST-SsaE in each cell lysate was bound to MagneGST glutathione particles for 1 h at 4 °C. The bound proteins were incubated with PreScission protease to cleave the GST tag from the SsaE protein. Proteins separated by 16% Tricine gel were visualized with Oriole fluorescent gel stain (A) and immunostained with anti-SsaH antibody (B).
The PscGE, YscGE, and AscGE complexes form ternary structures with needle proteins PscF, YscF, and AscF, respectively, to avoid premature assembly in the cytoplasm (14, 30, 35). Furthermore, the needle proteins PscF and YscF are necessary for the secretion of inner rod proteins PscI and YscI, respectively (10, 36). In the SPI2-T3SS, the needle protein SsaG is also involved in SsaI secretion (Figs. 1 and 2). Therefore, we hypothesized that SsaH–SsaE complex may act as a chaperone of SsaG to regulate SsaI secretion. Because the establishment of an anti-SsaG antibody was unsuccessful in the present study, the GST pulldown assay in E. coli was performed to investigate the possibility of interaction between the SsaH–SsaE complex and SsaG (Fig. 6). When SsaG was coexpressed with GST-SsaE and SsaH in E. coli, a band corresponding to ∼8 kDa was observed after collecting proteins bound to SsaE. This protein was also detected upon coexpression of SsaG, SsaH, and SsaI. In contrast, it was not detected in the absence of ssaH expression. The ORF for SsaG is 213 bp long and encodes a peptide of 71 amino acid residues with a predicted molecular mass of 7.9 kDa. Thus, the observed protein corresponds to the predicted size of SsaG. These findings indicated that SsaG may be a substrate of the SsaH–SsaE complex in the SPI2-T3SS.
Discussion
Class III chaperones, which form heterodimeric complexes with partner proteins known as class V chaperones, bind and stabilize the monomeric needle protein to prevent self-aggregation in the bacterial cytosol. Complexes of class III and V chaperones and needle proteins have been identified in P. aeruginosa (PscG–PscE–PscF) (14), Yersinia spp. (YscG–YscE–YscF) (35), A. hydrophila (AscG–AscE–AscF) (30), EPEC (EscG–EscE–EscF) (37), and Chlamydia trachomatis (CdsG–CdsE–CdsF) (38). In this study, we found that SsaH functions as a class III chaperone in S. enterica Typhimurium SPI2-T3SS. S. enterica Typhimurium strains with disruptions in ssaG, ssaH, or ssaE showed no secretion of the inner rod, SsaI, suggesting that SsaG, SsaH, and SsaE may be required for secretion of SsaI, an early substrate of SPI2-T3SS. Prediction of the structures of SsaH and SsaE revealed that their tertiary structures could be aligned and superimposed over the known structures of PscG–PscE, YscG–YscE, and AscG–AscE (Fig. S3). Biological analysis showed that SsaH forms a heterodimer with the class V chaperone SsaE (Fig. 4). The binding of the needle protein SsaG to SsaE could be detected only in the presence of SsaH (Fig. 6). Therefore, it is suggested that SsaH may interact directly with SsaG, promoting the secretion of both SsaG and SsaI. Previous studies of PscG–PscE–PscF, YscG–YscE–YscF, and AscG–AscE–AscF complexes have shown that the chaperone-binding region of the needle protein is located in the C-terminal region (13, 30, 39). In the PscG–PscE–PscF complex, the C terminus of PscF is contained within the hydrophobic groove of PscG (39). Site-directed mutagenesis revealed that the hydrophobic residues Ile-83 and Leu-84, located within the last five C-terminal residues (“ILQKI”) of AscF, are essential for AscG binding (30). The C-terminal sequence “IIAKI” of SsaG is also hydrophobic, and the Ile residue is conserved (Fig. S2B). Another study demonstrated that the last five C-terminal residues of MxiH and PrgI, which are needle proteins in Shigella and SPI1, respectively, are essential for oligomerization. When these five residues were removed, these proteins could be expressed as soluble proteins but were unable to assemble (40). Therefore, the hydrophobic C-terminal region of SsaG is likely protected by the binding of SsaH to prevent oligomerization of the premature SsaG inside the cell, contributing to SPI2-T3SS assembly.
The amount of SsaH in presence of SsaG was decreased compared with that in the absence of SsaG (Fig. 6). The structure of the YscG–YscE–YscF complex (13) revealed that the N terminus of YscG is involved in the interaction between both YscG/YscE and YscG/YscF. The 21 N-terminal residues of SsaH may be involved in the interaction with SsaE (Fig. S4D). The hydrophobic C-terminal region of SsaG likely binds to the N-terminal region of SsaH. The binding activity of SsaH may change in the presence of SsaG because a portion of the region of SsaH that binds to SsaG and SsaE is overlapped.
The present results suggest that SsaE may be involved in the stabilization of SsaH via a direct interaction (Figs. 4 and 5A). In P. aeruginosa, PscE and PscG appear to costabilize each other (14). Reportedly, SsaE interacts directly with SseB and regulates its secretion via SPI2-T3SS (28). The secretion of SsaI and SseB by the ssaP-disrupted mutant strain indicates that SsaI secretion is negatively regulated by SsaP prior to SseB secretion (Fig. 1). This suggests that SsaE may function only as a chaperone for the early substrate SsaG through SsaH stabilization but not as a chaperone for the middle substrate SseB. Interestingly, overexpression of GST-SsaE in the ssaE-disrupted mutant restored the production of both SsaH and SseB (Fig. S5), but neither SsaI nor SseB was secreted (data not shown). The chaperone–substrate complex is recruited to the sorting platform (34). Therefore, SsaE may recruit SsaG and SseB to the secretion apparatus of SPI2-T3SS, facilitating the secretion of early and middle substrates.
Recently, Souza et al. (41) showed that YscE of Y. pestis may play a direct role in the secretion of YscF by mediating contact with the T3SS apparatus. SsaE interacts with SsaK, SsaN, and SsaQ in SPI2, and binding of SseB to SsaE is abrogated by the ATPase SsaN (27, 28). SsaK and SsaQ are homologs of OrgB and SpaO, respectively, in SPI1-T3SS (1), and SpaO, OrgB, and OrgA comprise the sorting platform in SPI1-T3SS. The SpaO–OrgA–OrgB complex ensures secretion of translocators before effectors (34). This platform may also contribute to substrate selection during needle assembly as well as to the substructure of the needle complex (42). In this process, the binding of the chaperone to its substrate is required for targeting of type III secreted proteins to the SpaO–OrgA–OrgB sorting platform (34). The hierarchy of secretion may reflect different affinities of the different substrate–chaperone complexes for the sorting platform. Thus, this suggests a possible hierarchy of secretion from the early substrates SsaG and SsaI to the middle substrate SseB in the SPI2-T3SS as the affinity of SsaE in the SsaE–SsaH–SsaG and SsaE–SseB complexes to the sorting platform differed even though the common chaperone SsaE interacted directly with the sorting platform in both complexes.
In addition to the sorting platform, substrate switching from early to middle substrates in the T3SS assembly depends on the termination of the needle complex assembly, regulated by the needle-length ruler SctP (5, 9, 43–45). SsaP, the homolog of SctP, is also required for substrate switching from SsaI to SseB secretion (Fig. 1). Although SctP secretion in other T3SSs is known to be involved in regulating needle length, no SsaP was detected in the fraction of secreted proteins from the S. enterica Typhimurium WT strain (data not shown). Interestingly, SsaP interacted with GST-SsaE (Fig. S5). SsaH production and SsaI secretion were complemented in the ssaE-disrupted mutant when ssaE was expressed from a plasmid, although only some SseB was secreted (Fig. 5). These results raise the possibility of SsaE involvement in the titration of SsaP in bacterial cells to regulate substrate switching. EscP in EPEC has been reported to interact with the multieffector chaperone CesT, suggesting that EscP may prevent effector secretion until the translocation pore has been formed in the host cell membrane (6).
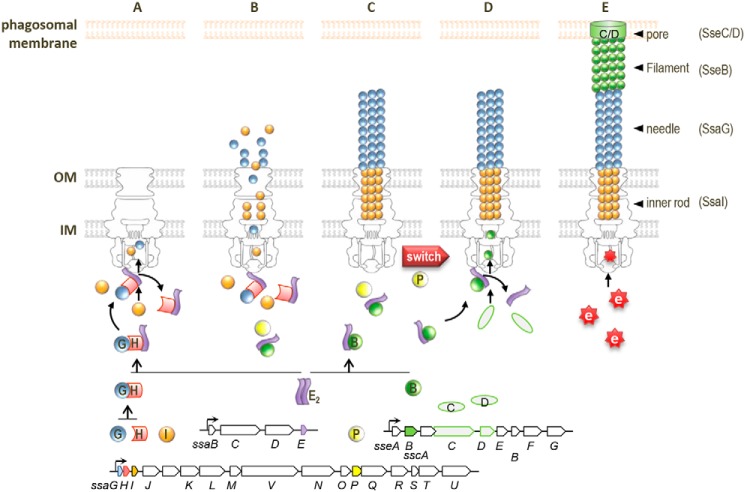
Considering our findings and previous studies together, we propose a model for chaperone function in the substrate switching of SPI2 (Fig. 7). Until the assembly of outer and inner membrane rings and export apparatus, including the sorting platform (46–48), SsaE binds to the SsaG–SsaH complex to stabilize SsaG and SsaH in the bacterial cytosol. SsaE also forms a complex with SseB for stabilization. Following the assembly of the sorting platform, SsaG is recognized by the sorting platform via an interaction with SsaE in the SsaE–SsaG–SsaH complex, and secretion is initiated (Fig. 7A). Simultaneously, SsaI is also recognized and secreted regardless of binding to SsaE and SsaH. During the assembly of the needle and the inner rod, the binding of SsaP to SsaE likely occurs to regulate recognition of the SsaE–SseB complex by the sorting platform (Fig. 7B). Once SsaG and SsaI polymerization is complete, SsaP promotes substrate switching to the translocators (Fig. 7C). This initiates interaction between SsaE in the SsaE–SseB complex and the sorting platform, resulting in the secretion of translocators (Fig. 7D). Finally, following the formation of the translocation pore, the membrane-bound regulatory complex consisting of SsaL, SsaM, and SsaB is degraded upon sensing change in host cytosolic pH, which initiates the switch from translocators to effectors (Fig. 7E) (25). To our knowledge, this is the first report of a chaperone that regulates both early and middle substrates affecting substrate switching in the T3SS assembly. Class V chaperones such as SsaE have been identified in other T3SSs of pathogenic bacteria. Thus, the regulation of both early and middle substrates by class V chaperones affecting substrate switching demonstrated here may provide insight into the mechanism of secretion switching from early to middle substrates in other T3SSs of pathogenic bacteria. However, it is unclear how the inner rod protein is secreted at the same time as the needle protein. The inner rod controls switching from early to middle substrates and needle length (3, 9, 49). Cao et al. (36) revealed that interaction between the inner rod protein YscI and the needle protein YscF is required to assemble the needle structure of the Yersinia T3SS. YscI is necessary for the needle tip protein LcrV. They demonstrated that direct interaction between YscF and YscI is critical for this process. Similarly, it is assumed that the secretion of SseB may require an interaction between SsaG and SsaI. Additional studies regarding the function and regulation of the inner rod are felt to be required for a better understanding of substrate switching.
Figure 7.
Schematic of chaperone-mediated mechanism of substrate stabilization and switching in SPI2-T3SS. A, SsaH (H) interacts with SsaG (G), and the SsaG–SsaH complex interacts with SsaE (E) for stabilization. The SsaE–SsaH–SsaG complex interacts with the export apparatus through SsaE, leading to initiation of the secretion of SsaG and SsaI (I). B, SseB (B) in the cytosol also interacts with SsaE for stabilization. Secretion of SseB is prevented during SsaG and SsaI secretion. C, SsaP (P) promotes substrate switching (switch) upon completion of SsaG and SsaI polymerization. D, the SsaE–SseB complex associates with the export apparatus to guide the secretion of SseB, SseC (C), and SseD (D). E, following assembly of the translocation pore (C/D) in the phagosomal membrane, effectors (e) are translocated upon sensing neutral pH of the host cytosol environment. IM, inner membrane; OM, outer membrane.
Experimental procedures
Bacterial strains, plasmids, and DNA oligonucleotides
All Salmonella strains were derivatives of S. enterica serovar Typhimurium χ3306 (50). Bacteria were routinely grown in L broth (1% Bacto tryptone (Difco), 0.5% Bacto yeast extract (Difco), 0.5% sodium chloride, pH 7.4) and L agar. When necessary, the medium was supplemented with chloramphenicol (20 μg/ml), ampicillin (50 μg/ml), kanamycin (25 μg/ml), spectinomycin (25 μg/ml), and/or nalidixic acid (25 μg/ml). For SPI2 expression, bacterial strains were grown in low-Mg2+ minimal medium at pH 5.0 (23) for 8 h in aerobic condition. Bacterial strains, plasmids, and DNA oligonucleotides used for construction of mutant strains and plasmids are detailed in Tables S1–S3.
Construction of plasmids
Plasmids pTKY941, pTKY942, and pTKY943, encoding N-terminally His-tagged ssaG, N-terminally His-tagged ssaH64–291, and N-terminally His-tagged ssaI, respectively, were constructed via PCR amplification of BamHI-HindIII fragments carrying ssaG, ssaH64–291, and ssaI with the primer sets pUHE-ssaG-BamHI-F and pUHE-ssaG-HindIII-R, pUHE-ssaH-BamHI-F and pUHE-ssaH-HindIII-R, and pUHE-ssaI-BamHI-F and pUHE-ssaI-HindIII-R, respectively, followed by cloning the fragment into pUHE212-1 (51).
Plasmids pTKY1243 and pTKY1245, which allow IPTG-controlled induction of ssaG and ssaI, respectively, were constructed by subcloning the BamHI-HindIII fragments carrying ssaG and ssaI of plasmids pTKY941 and pTKY943, respectively, into pUHE21-2Δfd12 (51). Plasmid pTKY1246, which allows IPTG-controlled induction of ssaH, was recombined with the BamHI-HindIII fragment carrying the ssaH gene amplified with the primer set pUHE-ssaH-new-BamHI-F and pUHE-ssaH-HindIII-R.
Prior to construction of the plasmids pTKY1251 and pTKY1261 encoding ssaP and ssaE, respectively, pTKY1231 encoding ssaB and its endogenous promoter was constructed by PCR amplification of the KpnI-SphI fragment carrying ssaB, including the 359 bp upstream, with the primer set pMW119-ssaB-KpnI-F and pMW119-ssaB-SphI-R. The fragment was then cloned into pMW119. Using the resultant plasmid, pTKY1231, as a template, a vector including ssaB promoter was amplified with the primer set pMW119-ssaB-XbaI-F and pMW119-ssaB-BamHI-R. To construct pTKY1251, the BamHI-XbaI fragment of ssaP, amplified with the primer set ssaP-BamHI-F and ssaP-XbaI-R, was ligated into the vector. The EcoRI-HindIII fragment of ssaP with ssaB promoter was digested from the resultant plasmid and then ligated into pMW118. To construct pTKY1261, the BglII-SphI fragment of ssaE, amplified with the primer set BglII-SD-ssaE-F and ssaE-SphI-R, was ligated into the vector. To construct plasmid pTKY1263 for expression of SsaE protein fused to GST, ssaE was amplified via PCR with the primer set GST-SsaE-BglII-F and GST-SsaE-EcoRI-R, and the BglII-EcoRI fragment was cloned into BamHI-EcoRI–digested pGEX6p-1.
Plasmids pTKY1265, pTKY1266, pTKY1267, pTKY1268, and pTKY1269, for coexpression of ssaG, ssaGHI operon, ssaGH operon, ssaHI operon, and ssaH gene with GST-tagged SsaE, respectively, were constructed via PCR of EcoRI-SalI fragments carrying ssaG, ssaGHI operon, ssaGH operon, ssaHI operon, and ssaH gene with the primer sets EcoRI-SD-SsaG-F and ssaG-SalI-R, EcoRI-SD-SsaG-F and ssaI-SalI-R, EcoRI-SD-SsaG-F and ssaH-SalI-R, and EcoRI-SD-SsaH-F and ssaH-SalI-R, respectively, followed by cloning the fragment into pTKY1263.
Construction of Salmonella mutant strains
Strains CS3491 (ΔsseB::FRT), CS4269 (ΔssaI::FRT), CS4270 (ΔssaP::FRT), CS10091 (ΔssaH::FRT), CS10092 (ΔssaU::FRT), CS10106 (ΔssaE::FRT), CS10107 (ΔssaN::FRT), and CS10135 (ΔssaG::FRT) were constructed as follows by λRed and FLP-mediated recombination essentially as described by Datsenko and Wanner (52). PCR products used to construct gene replacements were generated with template plasmid pKD3 and the primer sets ssaH-P1-F and ssaH-P2-R for CS10091, ssaU-P1-F and ssaU-P2-R for CS10092, ssaE-P1-F and ssaE-P2-R for CS10106, and ssaN-P1-F and ssaN-P2-R for CS10107 and template plasmid pKD4 and the primer sets sseB-P1-F and sseB-P2-R for CS3491, ssaI-P1-F and ssaI-P2-R for CS4269, ssaP-P1-F and ssaP-P2-R for CS4270, and ssaG-P1-F and ssaG-P2-R for CS10135. Each 1.1- and 1.4-kbp fragment generated by pKD3 and pKD4, respectively, was introduced into strain χ3306 carrying pKD46, encoding the λRed recombinase, by transformation. Insertion of Cm- or Km-resistance genes into target genes was confirmed by PCR amplification of the chromosomal DNA with each primer set. To remove the Cm- or Km-resistance gene, pCP20 encoding Flp recombinase (53) was introduced into each Cm- or Km-resistance strain via transformation. The FRT insertion in the target gene was checked via PCR amplification of the chromosomal DNA with each primer set and DNA sequencing. The primer sets used for the PCR amplification of chromosomal DNA were as follows: sseB-check-F and sseB-check-R for CS3491, ssaP-check-F and ssaP-check-R for CS4270, ssaU-check-F and ssaU-check-R for CS10092, ssaE-check-F and ssaE-check-R for CS10106, ssaN-check-F and ssaN-check-R for CS10107, and ssaG-check-F and ssaI-check-R for CS4269, CS10091, and CS10135.
Purification of N-terminally His-tagged proteins and generation of anti-SsaH and anti-SsaI antibodies
For purification of N-terminally His-tagged SsaHΔN and SsaI, 3-liter cultures of E. coli strains CS6234 and CS6235 were incubated at 37 °C until the cell density reached an A600 of 1.0. Next, IPTG was added to a final concentration of 1 mm for 3 h followed by collection of cells via centrifugation. Wet cell pastes were resuspended in buffer (100 mm sodium phosphate buffer, pH 8.0, 0.1 m NaCl, 8 m urea) and sonicated. After further centrifugation, the supernatant was loaded onto a Ni2+-nitrilotriacetic acid Superflow column (10 ml; Millipore). After washing with buffer (100 mm sodium phosphate buffer, pH 8.0, 0.1 m NaCl, 8 m urea, 20 mm imidazole), the protein was eluted with 50, 75, 100, 150, 250, and 500 mm imidazole. A 5.0-ml portion of the peak fraction was electrophoresed by 15% SDS-PAGE, and the eluent corresponding to SsaHΔN and SsaI bands was used to immunize a rabbit. The resultant anti-SsaH and -SsaI antibodies were purified from antisera using a MelonTM Gel IgG spin purification kit (Thermo Fisher Scientific).
Preparation of cell lysates and proteins secreted into the medium
To prepare cell lysates, bacterial cells were harvested by centrifuging 1 ml of the culture and suspended in SDS sample buffer. The suspension was incubated at 95 °C for 5 min, sonicated, and recentrifuged. A portion of the supernatant was used as cell lysates. To prepare the proteins secreted into the medium, the same culture was centrifuged to remove cells. The filtrated supernatant was mixed with prechilled TCA (final concentration, 10%), chilled on ice, and centrifuged. The resultant pellet was washed once with acetone and then solubilized in SDS sample buffer.
GST pulldown assay
GST pulldown assay was performed using the MagneGSTTM Protein Purification System (Promega) according to the manufacturer's instructions.
Preparation of SsaE–SsaH complex and SsaE protein
E. coli strains CS7141 and CS7145 were grown at 37 °C to an A600 of 0.5 in L broth containing 50 μg/ml ampicillin and 0.5% glucose before adding IPTG to 1 mm to induce GST-SsaE and SsaH. Following a 3-h incubation at 37 °C, cells were pelleted and lysed in B-PER Bacterial Protein Extraction Reagent buffer (Thermo Fisher Scientific) containing DNase I. Following centrifugation, the supernatant was added to MagneGST particles (Promega) equilibrated with reaction buffer (50 mm Tris-HCl, pH7.0, 150 mm NaCl, 1 mm EDTA, 1 mm DTT) and then rotated at 4 °C for 1 h. After washing with reaction buffer, the particles were incubated for 16 h at 4 °C in reaction buffer containing PreScission protease (GE Healthcare) to cleave the GST tag from the SsaE protein. The proteins were separated by gel chromatography (SuperoseTM 12 10/300, GE Healthcare) with reaction buffer containing 10% glycerol. The eluents were analyzed by gel electrophoresis (Fig. S6). The peak fractions containing SsaE at 14.5 ml and SsaE/SsaH at 14 ml, respectively, were concentrated using VivaSpin 500 with 3000 molecular weight cutoff (GE Healthcare). For stoichiometric analysis, a total of 12 μg of each of SsaE/SsaH and SsaE was subjected to size-exclusion chromatography.
Separation of SPI2 proteins by gel electrophoresis and immunoblotting
Separation of low-molecular-weight SPI2 proteins was carried out using the Invitrogen NovexTM Tricine gel systems (Thermo Fisher Scientific). Protein samples were mixed with Tricine SDS sample buffer (Thermo Fisher Scientific), and proteins were electrophoresed using Novex 16% Tricine protein gels (Thermo Fisher Scientific). Precision Plus ProteinTM WesternCTM Standards (Bio-Rad) or PageRuler Unstained Low Range Protein Ladder (Thermo Fisher Scientific) was used as protein standards. Separated proteins were stained with OrioleTM fluorescent gel stain (Bio-Rad) and visualized using a Gel DocTM Plus system (Bio-Rad). For immunoblotting, separated proteins were transferred onto Immun-Blot polyvinylidene difluoride membranes (Bio-Rad) and incubated with rabbit anti-SsaI, anti-SsaH, anti-SsaP (22), and anti-SseB (22) antibodies followed by HRP-conjugated anti-rabbit IgG (Jackson ImmunoResearch Laboratories). Precision ProteinTM Streptactin–HRP complex (Bio-Rad) was used for detection of protein standards. Enzymatic reactions were carried out in the presence of ECL Prime (GE Healthcare), and the resultant reaction was visualized via LAS4000mini (GE Healthcare). All results were based on at least three independent experiments.
Author contributions
A. T. and T. Y. conceptualization; A. T. and H. T. resources; A. T., H. T., and S. T. data curation; A. T., H. T., and S. T. formal analysis; A. T. funding acquisition; A. T. and T. Y. validation; A. T. investigation; A. T. writing-original draft; A. T. and T. Y. project administration; H. K. and T. Y. writing-review and editing; T. Y. supervision.
Supplementary Material
Acknowledgments
We thank Dr. Isogai for generation of anti-SsaH and SsaI antibodies and M. Ohya, K. Yamada, R. Suzuki, N. Kimura, and K. Harufuku for technical assistance. We also thank M. Hensel for SseB and SsaP antibodies.
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grants 16K15272 and 18K07102 (to A. T.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6 and Tables S1–S3.
- T3SS
- type III secretion system
- SPI
- Salmonella pathogenicity island
- EPEC
- enteropathogenic E. coli
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- GST
- GSH S-transferase
- Cm
- chloramphenicol
- Km
- kanamycin
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- HRP
- horseradish peroxidase
- FRT
- Flp recombination target.
References
- 1. Deng W., Marshall N. C., Rowland J. L., McCoy J. M., Worrall L. J., Santos A. S., Strynadka N. C. J., and Finlay B. B. (2017) Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337 10.1038/nrmicro.2017.20 [DOI] [PubMed] [Google Scholar]
- 2. Portaliou A. G., Tsolis K. C., Loos M. S., Zorzini V., and Economou A. (2016) Type III secretion: building and operating a remarkable nanomachine. Trends Biochem. Sci. 41, 175–189 10.1016/j.tibs.2015.09.005 [DOI] [PubMed] [Google Scholar]
- 3. Lefebre M. D., and Galán J. E. (2014) The inner rod protein controls substrate switching and needle length in a Salmonella type III secretion system. Proc. Natl. Acad. Sci. U.S.A. 111, 817–822 10.1073/pnas.1319698111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Agrain C., Callebaut I., Journet L., Sorg I., Paroz C., Mota L. J., and Cornelis G. R. (2005) Characterization of a type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica. Mol. Microbiol. 56, 54–67 10.1111/j.1365-2958.2005.04534.x [DOI] [PubMed] [Google Scholar]
- 5. Bergeron J. R., Fernández L., Wasney G. A., Vuckovic M., Reffuveille F., Hancock R. E., and Strynadka N. C. (2016) The structure of a type 3 secretion system (T3SS) ruler protein suggests a molecular mechanism for needle length sensing. J. Biol. Chem. 291, 1676–1691 10.1074/jbc.M115.684423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Monjarás Feria J., García-Gómez E., Espinosa N., Minamino T., Namba K., and González-Pedrajo B. (2012) Role of EscP (Orf16) in injectisome biogenesis and regulation of type III protein secretion in enteropathogenic Escherichia coli. J. Bacteriol. 194, 6029–6045 10.1128/JB.01215-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wood S. E., Jin J., and Lloyd S. A. (2008) YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J. Bacteriol. 190, 4252–4262 10.1128/JB.00328-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sukhan A., Kubori T., and Galán J. E. (2003) Synthesis and localization of the Salmonella SPI-1 type III secretion needle complex proteins PrgI and PrgJ. J. Bacteriol. 185, 3480–3483 10.1128/JB.185.11.3480-3483.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marlovits T. C., Kubori T., Lara-Tejero M., Thomas D., Unger V. M., and Galán J. E. (2006) Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441, 637–640 10.1038/nature04822 [DOI] [PubMed] [Google Scholar]
- 10. Monjarás Feria J. V., Lefebre M. D., Stierhof Y. D., Galán J. E., and Wagner S. (2015) Role of autocleavage in the function of a type III secretion specificity switch protein in Salmonella enterica serovar typhimurium. MBio 6, e01459–15 10.1128/mBio.01459-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Izoré T., Job V., and Dessen A. (2011) Biogenesis, regulation, and targeting of the type III secretion system. Structure 19, 603–612 10.1016/j.str.2011.03.015 [DOI] [PubMed] [Google Scholar]
- 12. Wilharm G., Dittmann S., Schmid A., and Heesemann J. (2007) On the role of specific chaperones, the specific ATPase, and the proton motive force in type III secretion. Int. J. Med. Microbiol. 297, 27–36 10.1016/j.ijmm.2006.10.003 [DOI] [PubMed] [Google Scholar]
- 13. Sun P., Tropea J. E., Austin B. P., Cherry S., and Waugh D. S. (2008) Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J. Mol. Biol. 377, 819–830 10.1016/j.jmb.2007.12.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quinaud M., Chabert J., Faudry E., Neumann E., Lemaire D., Pastor A., Elsen S., Dessen A., and Attree I. (2005) The PscE-PscF-PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa. J. Biol. Chem. 280, 36293–36300 10.1074/jbc.M508089200 [DOI] [PubMed] [Google Scholar]
- 15. Jennings E., Thurston T. L. M., and Holden D. W. (2017) Salmonella SPI-2 type III secretion system effectors: molecular mechanisms and physiological consequences. Cell Host Microbe 22, 217–231 10.1016/j.chom.2017.07.009 [DOI] [PubMed] [Google Scholar]
- 16. Galán J. E., and Curtiss R. 3rd (1989) Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. U.S.A. 86, 6383–6387 10.1073/pnas.86.16.6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hensel M., Shea J. E., Waterman S. R., Mundy R., Nikolaus T., Banks G., Vazquez-Torres A., Gleeson C., Fang F. C., and Holden D. W. (1998) Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30, 163–174 10.1046/j.1365-2958.1998.01047.x [DOI] [PubMed] [Google Scholar]
- 18. Ochman H., Soncini F. C., Solomon F., and Groisman E. A. (1996) Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. U.S.A. 93, 7800–7804 10.1073/pnas.93.15.7800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eriksson S., Lucchini S., Thompson A., Rhen M., and Hinton J. C. (2003) Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47, 103–118 [DOI] [PubMed] [Google Scholar]
- 20. Srikumar S., Kröger C., Hebrard M., Colgan A., Owen S. V., Sivasankaran S. K., Cameron A. D., Hokamp K., and Hinton J. C. (2015) RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog. 11, e1005262 10.1371/journal.ppat.1005262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuhle V., and Hensel M. (2004) Cellular microbiology of intracellular Salmonella enterica: functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell. Mol. Life Sci. 61, 2812–2826 10.1007/s00018-004-4248-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beuzón C. R., Banks G., Deiwick J., Hensel M., and Holden D. W. (1999) pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33, 806–816 10.1046/j.1365-2958.1999.01527.x [DOI] [PubMed] [Google Scholar]
- 23. Nikolaus T., Deiwick J., Rappl C., Freeman J. A., Schröder W., Miller S. I., and Hensel M. (2001) SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J. Bacteriol. 183, 6036–6045 10.1128/JB.183.20.6036-6045.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klein J. R., and Jones B. D. (2001) Salmonella pathogenicity island 2-encoded proteins SseC and SseD are essential for virulence and are substrates of the type III secretion system. Infect. Immun. 69, 737–743 10.1128/IAI.69.2.737-743.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu X. J., McGourty K., Liu M., Unsworth K. E., and Holden D. W. (2010) pH sensing by intracellular Salmonella induces effector translocation. Science 328, 1040–1043 10.1126/science.1189000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Allison S. E., Tuinema B. R., Everson E. S., Sugiman-Marangos S., Zhang K., Junop M. S., and Coombes B. K. (2014) Identification of the docking site between a type III secretion system ATPase and a chaperone for effector cargo. J. Biol. Chem. 289, 23734–23744 10.1074/jbc.M114.578476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoshida Y., Miki T., Ono S., Haneda T., Ito M., and Okada N. (2014) Functional characterization of the type III secretion ATPase SsaN encoded by Salmonella pathogenicity island 2. PLoS One 9, e94347 10.1371/journal.pone.0094347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miki T., Shibagaki Y., Danbara H., and Okada N. (2009) Functional characterization of SsaE, a novel chaperone protein of the type III secretion system encoded by Salmonella pathogenicity island 2. J. Bacteriol. 191, 6843–6854 10.1128/JB.00863-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tan Y. W., Yu H. B., Leung K. Y., Sivaraman J., and Mok Y. K. (2008) Structure of AscE and induced burial regions in AscE and AscG upon formation of the chaperone needle-subunit complex of type III secretion system in Aeromonas hydrophila. Protein Sci. 17, 1748–1760 10.1110/ps.036798.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Day J. B., Guller I., and Plano G. V. (2000) Yersinia pestis YscG protein is a Syc-like chaperone that directly binds yscE. Infect. Immun. 68, 6466–6471 10.1128/IAI.68.11.6466-6471.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F. T., de Beer T. A. P., Rempfer C., Bordoli L., Lepore R., and Schwede T. (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Phan J., Austin B. P., and Waugh D. S. (2005) Crystal structure of the Yersinia type III secretion protein YscE. Protein Sci. 14, 2759–2763 10.1110/ps.051706105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lara-Tejero M., Kato J., Wagner S., Liu X., and Galán J. E. (2011) A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331, 1188–1191 10.1126/science.1201476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chatterjee C., Kumar S., Chakraborty S., Tan Y. W., Leung K. Y., Sivaraman J., and Mok Y. K. (2011) Crystal structure of the heteromolecular chaperone, AscE-AscG, from the type III secretion system in Aeromonas hydrophila. PLoS One 6, e19208 10.1371/journal.pone.0019208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cao S. Y., Liu W. B., Tan Y. F., Yang H. Y., Zhang T. T., Wang T., Wang X. Y., Song Y. J., Yang R. F., and Du Z. M. (2017) An interaction between the inner rod protein YscI and the needle protein YscF is required to assemble the needle structure of the Yersinia type three secretion system. J. Biol. Chem. 292, 5488–5498 10.1074/jbc.M116.743591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sal-Man N., Setiaputra D., Scholz R., Deng W., Yu A. C., Strynadka N. C., and Finlay B. B. (2013) EscE and EscG are cochaperones for the type III needle protein EscF of enteropathogenic Escherichia coli. J. Bacteriol. 195, 2481–2489 10.1128/JB.00118-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Betts H. J., Twiggs L. E., Sal M. S., Wyrick P. B., and Fields K. A. (2008) Bioinformatic and biochemical evidence for the identification of the type III secretion system needle protein of Chlamydia trachomatis. J. Bacteriol. 190, 1680–1690 10.1128/JB.01671-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Quinaud M., Plé S., Job V., Contreras-Martel C., Simorre J. P., Attree I., and Dessen A. (2007) Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc. Natl. Acad. Sci. U.S.A. 104, 7803–7808 10.1073/pnas.0610098104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Darboe N., Kenjale R., Picking W. L., Picking W. D., and Middaugh C. R. (2006) Physical characterization of MxiH and PrgI, the needle component of the type III secretion apparatus from Shigella and Salmonella. Protein Sci. 15, 543–552 10.1110/ps.051733506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Souza C. A., Richards K. L., Park Y., Schwartz M., Torruellas Garcia J., Schesser Bartra S., and Plano G. V. (2018) The YscE/YscG chaperone and YscF N-terminal sequences target YscF to the Yersinia pestis type III secretion apparatus. Microbiology 164, 338–348 10.1099/mic.0.000610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sukhan A., Kubori T., Wilson J., and Galán J. E. (2001) Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J. Bacteriol. 183, 1159–1167 10.1128/JB.183.4.1159-1167.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Edqvist P. J., Olsson J., Lavander M., Sundberg L., Forsberg A., Wolf-Watz H., and Lloyd S. A. (2003) YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J. Bacteriol. 185, 2259–2266 10.1128/JB.185.7.2259-2266.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Journet L., Agrain C., Broz P., and Cornelis G. R. (2003) The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302, 1757–1760 10.1126/science.1091422 [DOI] [PubMed] [Google Scholar]
- 45. Agrain C., Sorg I., Paroz C., and Cornelis G. R. (2005) Secretion of YscP from Yersinia enterocolitica is essential to control the length of the injectisome needle but not to change the type III secretion substrate specificity. Mol. Microbiol. 57, 1415–1427 10.1111/j.1365-2958.2005.04758.x [DOI] [PubMed] [Google Scholar]
- 46. Diepold A., Amstutz M., Abel S., Sorg I., Jenal U., and Cornelis G. R. (2010) Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 29, 1928–1940 10.1038/emboj.2010.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Diepold A., Wiesand U., and Cornelis G. R. (2011) The assembly of the export apparatus (YscR,S,T,U,V) of the Yersinia type III secretion apparatus occurs independently of other structural components and involves the formation of an YscV oligomer. Mol. Microbiol. 82, 502–514 10.1111/j.1365-2958.2011.07830.x [DOI] [PubMed] [Google Scholar]
- 48. Wagner S., Königsmaier L., Lara-Tejero M., Lefebre M., Marlovits T. C., and Galán J. E. (2010) Organization and coordinated assembly of the type III secretion export apparatus. Proc. Natl. Acad. Sci. U.S.A. 107, 17745–17750 10.1073/pnas.1008053107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ho O., Rogne P., Edgren T., Wolf-Watz H., Login F. H., and Wolf-Watz M. (2017) Characterization of the ruler protein interaction interface on the substrate specificity switch protein in the Yersinia type III secretion system. J. Biol. Chem. 292, 3299–3311 10.1074/jbc.M116.770255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gulig P. A., and Curtiss R. 3rd (1987) Plasmid-associated virulence of Salmonella typhimurium. Infect. Immun. 55, 2891–28901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gamer J., Bujard H., and Bukau B. (1992) Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor σ 32. Cell 69, 833–842 10.1016/0092-8674(92)90294-M [DOI] [PubMed] [Google Scholar]
- 52. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cherepanov P. P., and Wackernagel W. (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14 10.1016/0378-1119(95)00193-A [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.