Abstract
Most clear cell renal cell carcinomas (ccRCCs) have inactivation of the von Hippel–Lindau tumor suppressor protein (pVHL), resulting in the accumulation of hypoxia-inducible factor α-subunits (HIF-α) and their downstream targets. HIF-2α expression is particularly high in ccRCC and is associated with increased ccRCC growth and aggressiveness. In the canonical HIF signaling pathway, HIF-prolyl hydroxylase 3 (PHD3) suppresses HIF-2α protein by post-translational hydroxylation under sufficient oxygen availability. Here, using immunoblotting and immunofluorescence staining, qRT-PCR, and siRNA-mediated gene silencing, we show that unlike in the canonical pathway, PHD3 silencing in ccRCC cells leads to down-regulation of HIF-2α protein and mRNA. Depletion of other PHD family members had no effect on HIF-2α expression, and PHD3 knockdown in non-RCC cells resulted in the expected increase in HIF-2α protein expression. Accordingly, PHD3 knockdown decreased HIF-2α target gene expression in ccRCC cells and expression was restored upon forced HIF-2α expression. The effect of PHD3 depletion was pinpointed to HIF2A mRNA stability. In line with these in vitro results, a strong positive correlation of PHD3 and HIF2A mRNA expression in ccRCC tumors was detected. Our results suggest that in contrast to the known negative regulation of HIF-2α in most cell types, high PHD3 expression in ccRCC cells maintains elevated HIF-2α expression and that of its target genes, which may enhance kidney cancer aggressiveness.
Keywords: cancer, cancer biology, hypoxia, hypoxia-inducible factor (HIF), mRNA decay, ccRCC, Egl-9 family hypoxia-inducible factor 3 (EGLN3), post-transcriptional regulation, prolyl hydroxylase PHD3, renal cell carcinoma
Introduction
Clear cell renal cell carcinoma (ccRCC)3 is the most common subtype of kidney cancer arising from renal tubular epithelial cells. Characteristic for ccRCC is the inactivation of von Hippel–Lindau tumor suppressor protein (pVHL) that occurs in the majority of both familial and sporadic ccRCC cases (1). pVHL functions as an ubiquitin E3 ligase in the proteasomal degradation pathway and the best-known targets of pVHL are the α-subunits of the hypoxia-inducible factors (HIFs) that are directed to degradation in oxygen-dependent manner. Loss of pVHL in ccRCC directly leads to a constant activation of HIF-α isoforms and HIF target genes despite the oxygen level within the tissue (2, 3).
HIFs are heterodimeric transcription factors that mediate the adaptive responses to low oxygen levels. HIFs comprise HIF-α and HIF-β. The two major HIF-α isoforms, HIF-1α and HIF-2α (also known as EPAS1) are strictly regulated in the presence of oxygen, whereas HIF-β subunit is considered to be stable. HIF–prolyl hydroxylases (PHD1–3, also known as EGLNs and HPHs) are a family of oxygen- and 2OG-dependent enzymes that hydroxylate HIF-α in the presence of oxygen, thus marking it to be recognized by pVHL and subsequently degraded (reviewed in Refs. 4, 5). It is generally thought that PHD2 is the main regulator of HIF-1α stability whereas HIF-2α isoform is mainly regulated by PHD3 (6, 7). HIF-1α and HIF-2α share structural and functional similarities, but their target genes are markedly different. Also, their function in ccRCC development and tumor growth is considerably different, as HIF-1α shows tumor suppressor functions and HIF-2α has been reported by several groups to promote tumorigenicity (reviewed in Ref. 3). In line, recent clinical phase 1 trial demonstrated clinical benefit for the inhibition of HIF-2α (8).
From the PHD isoforms, PHD3 (also known as EGLN3) shows the highest expression in ccRCC (9–12). However, the role of elevated PHD3 expression in ccRCC has not been thoroughly studied. Apart from its function in post-translational modification of HIF-α in the presence of oxygen, PHD3 has been suggested to have various targets that are independent of its hydroxylase activity (13). Recently, using proteome level analyses we have shown that PHD3 regulates a number of proteins involved in glucose metabolism, translational machinery, and proliferation in ccRCC cell lines. PHD3 depletion resulted in the down-regulation of most glycolytic enzymes together with decrease in lactate production. Importantly PHD3 depletion also caused deregulation of proteins acting in protein translation, mRNA processing and mTOR downstream signaling (14).
In addition to the oxygen-dependent regulation of HIF-α by PHDs, it has become evident that the HIF-α subunits are regulated and degraded by other mechanisms independent of the oxygen-sensing enzymes and pVHL activity (15–20). However, the transcriptional regulation of HIF2A is not well-characterized. Recently, it was shown that deubiquitylase Cezanne (also known as OTUD7B) regulates HIF2A expression via transcription factor E2F1 that directly affects the HIF2A gene expression (21). Moreover, poly (ADP-ribose) polymerase-1 (PARP-1) and JNK2 has been reported to regulate HIF2A expression level (22, 23). Also, insulin-like growth factor 2 (IGF2)–induced PI3K-mTORC2 signaling was shown to regulate HIF2A mRNA expression in neuroblastoma cells (24). These findings indicate a complex regulation of the HIF-signaling pathway.
Noticeably, besides PHD3, elevated expression also of HIF-2α in ccRCC has been reported in several studies (25–27). This is counterintuitive given the supposedly negative feedback of PHD3 on HIF-2α. Here we report that, in ccRCC, PHD3 rather surprisingly maintains elevated HIF-2α level whereas in other tested cell types PHD3 suppressed HIF-2α expression. We show that siRNA-mediated silencing of PHD3 leads to down-regulation of HIF-2α protein but also HIF2A mRNA expression and we pinpoint the regulation to mRNA stability. In addition, we demonstrate that PHD3 depletion leads to down-regulation of HIF-2α target genes VEGFA, OCT4, GLUT1, and LDHA. Importantly, PHD3 and HIF2A expression were found to correlate in clinical ccRCC data set. The findings suggest that high PHD3 expression is needed in ccRCC to maintain high HIF-2α expression.
Results
PHD3 silencing decreases LDHA and GLUT1 mRNA expression
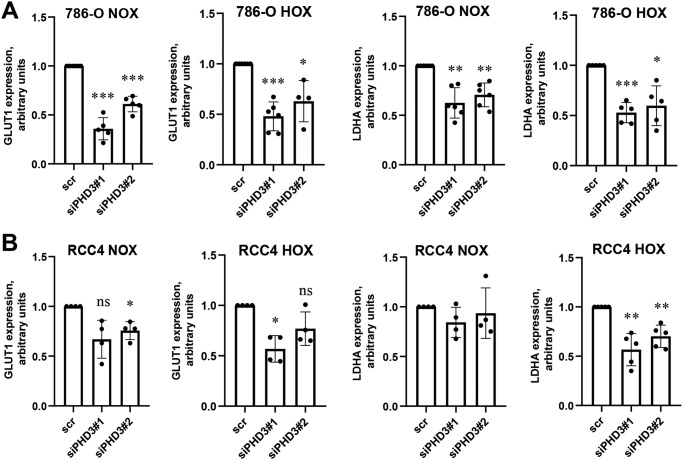
We have previously shown that PHD3 silencing down-regulates crucial glucose metabolism–related proteins such as lactate dehydrogenase A (LDHA) and glucose transporter 1 (GLUT1) in ccRCC cells (14). As the previous study was performed at the protein level we further studied the mRNA expression of these enzymes upon PHD3 knockdown. For PHD3 siRNA treatment we used two independent previously well-characterized siRNA sequences (7, 14, 28–30). Down-regulation of PHD3 expression was verified with both sequences in both cell lines used (Fig. S1, A and B) and the specificity of the PHD3 knockdown was also verified by studying PHD2 expression where no significant effect was observed (Fig. S1, C and D). The mRNA expression of both GLUT1 and LDHA was reduced in response to siRNA-mediated silencing of PHD3 in 786-O cells in both normoxic and hypoxic conditions (Fig. 1A). Similar reduction was also seen in RCC4 cells that was more pronounced under hypoxia (Fig. 1B).
Figure 1.
PHD3 knockdown results in decreased GLUT1 and LDHA mRNA expression in ccRCC cells. A, GLUT1 and LDHA expression in 786-O cells transfected with control (scr) or PHD3-targeted siRNA (siPHD3#1, siPHD3#2) followed by normoxic (NOX, 21% oxygen) or hypoxic (HOX, 1% oxygen) exposure. Quantification of five (GLUT1) or six (LDHA) individual experiments, -fold change to scr. Bar represents mean ± S.D.; ***, p <0.001; **, p <0.01; *, p <0.05. B, GLUT1 and LDHA expression in RCC4 cells transfected with control (scr) or siPHD3#1, siPHD3#2 followed by normoxic (NOX, 21% oxygen) or hypoxic (HOX, 1% oxygen) exposure. Quantification of four individual experiments, -fold change to scr. Bar represents mean ± S.D.; ns, not significant; **, p <0.01; *, p <0.05.
As the down-regulation of GLUT1 and LDHA occurred at mRNA level, it was likely to be mediated by a transcription factor. Several studies have shown that the glycolytic enzymes are regulated by HIF-α in ccRCC. Both of the used cell lines bear an inactivating mutation in VHL, leading to a constitutively up-regulated expression of HIF-α subunits. However, whereas RCC4 expresses both HIF-1α and HIF-2α, 786-O expresses a truncated form of HIF1A transcript and thus lacks a functional HIF-1α protein but expresses a high basal level of HIF-2α (31, 32). HIF-2α has previously been shown to transcriptionally regulate LDHA and GLUT1 (31, 33, 34). GLUT1 and LDHA expression responded similarly to PHD3 silencing in both cell lines suggesting that the responsible isoform may be HIF-2α.
Down-regulation of HIF-2α protein expression by PHD3 silencing
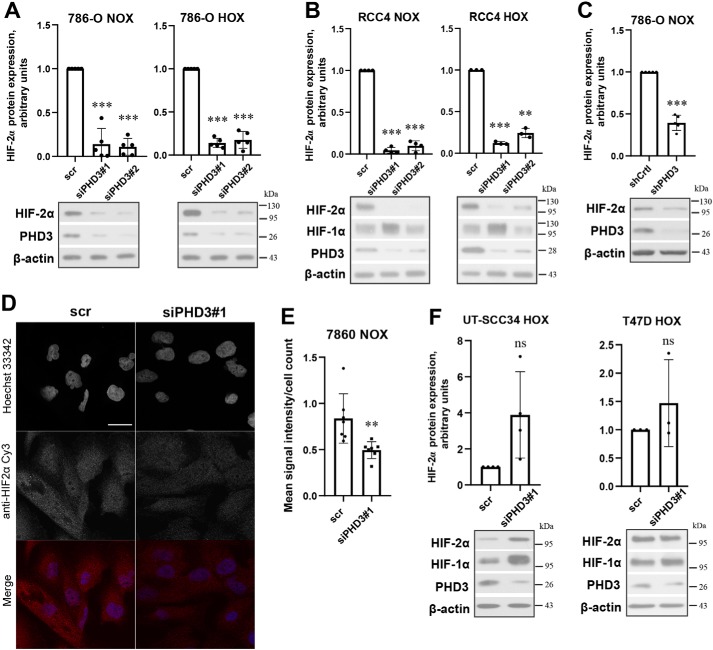
We studied whether PHD3 alters HIF-2α protein expression in ccRCC. Surprisingly, we observed a significant and reproducible down-regulation of HIF-2α protein levels under both normoxia and hypoxia by two independent PHD3 siRNAs. The effect was clear in both 786-O and RCC4 cells (Fig. 2, A and B). As expected, and in striking contrast to HIF-2α, HIF-1α protein level was not down-regulated, but rather up-regulated by PHD3 silencing in RCC4 cells (Fig. 2B). Furthermore, independent adenoviral shRNA-mediated knockdown of PHD3 resulted in similar decrease in HIF-2α protein, further verifying the effect (Fig. 2C). In addition, we used immunofluorescence staining to study HIF-2α protein expression and localization. In line with the western blot analyses, a marked reduction of HIF-2α protein in PHD3-depleted cells was detected compared with control, whereas the cellular localization of HIF-2α did not change (Fig. 2, D and E).
Figure 2.
PHD3 silencing leads to down-regulation of HIF-2α protein expression in ccRCC cells. A, HIF-2α protein expression in 786-O cells transfected with control (scr), siPHD3#1 or siPHD3#2 followed by normoxic (NOX, 21% oxygen) or hypoxic (HOX, 1% oxygen) exposure. Quantification of five independent experiments, -fold change to scr. Bar represents mean ± S.D.; ***, p <0.001. B, HIF-2α protein expression in RCC4 cells transfected with control (scr), siPHD3#1, siPHD3#2 followed by normoxic (NOX, 21% oxygen) or hypoxic (HOX, 1% oxygen) exposure. Quantification of at least three independent experiments, -fold change to scr. Bar represents mean ± S.D.; **, p <0.01; ***, p <0.001. C, 786-O cells infected with adenoviral control (shCtrl) or PHD3-targeted shRNA (shPHD3) in normoxia. Quantification of five independent experiments, -fold change to scr. Bar represents mean ± S.D.; ***, p < 0.001. D, immunostaining of HIF-2α in 786-O cells transfected with control (scr) or siPHD3. Representative images are shown, scale bar, 100 μm. E, quantification of the HIF-2α immunostainings, mean intensity normalized to cell count. Bar represents mean ± S.D.; n = 7 fields of views from two individual experiments; **, p <0.01. F, primary head and neck squamous cell carcinoma cells (UT-SCC-34) or breast cancer cells T47D cells were transfected with scr or siPHD3#1 followed by hypoxic (HOX, 1% oxygen) exposure. Quantification of four (UT-SCC34) or three (T47D) independent experiments, bar represents mean ± S.D.; -fold change to scr; ns = not significant.
Previous studies show that PHD3 depletion leads to an up-regulation of HIF-2α protein expression in some cell lines (28). Therefore, we next tested whether other cell lines respond similarly as ccRCC to the PHD3 depletion. Human primary head and neck squamous cell carcinoma cells (UT-SCC-34), exposed to hypoxia to induce PHD3 expression, showed increase in HIF-2α protein expression with siPHD3 treatment (Fig. 2F). In breast cancer cells (T47D), siPHD3 treatment did not show marked effect on HIF-2α protein expression under hypoxia (Fig. 2F) implying that the effect of PHD3 silencing on HIF-2α expression depends on the cell type studied. PHD3 has also been established as a target gene of HIF-2α, creating a feedback loop in the regulation of HIF-α (6, 7, 35–37). In line with this, we observed down-regulation of PHD3 with HIF-2α depletion both at protein and mRNA level in 786-O cells (Fig. S1, E and F).
We further used PHD1 and PHD2 silencing on 786-O cells but did not detect any significant decrease in HIF-2α protein expression (Fig. 3A), indicating that the effect on HIF-2α is specific to PHD3. Moreover, no effect on HIF-2α protein expression was detected with 8-h exposure to panhydroxylase inhibitor dimethyloxalylglysine (DMOG) in VHL mutated 786-O nor RCC4 cells, thus suggesting a hydroxylase-independent down-regulation of HIF-2α by PHD3 siRNA (Fig. 3B). RCC4 cells with restored functional VHL (RCC4+VHL) demonstrated an expected increase in both HIF-2α and HIF-1α protein levels in response to DMOG treatment (Fig. 3B). Similarly, exposure to cobalt chloride (CoCl2) in RCC4+VHL cells showed an expected increase but in RCC4 or 786-O cells CoCl2 treatment had no or minor effect on HIF-2α protein level (Fig. 3C).
Figure 3.
Down-regulation of HIF-2α protein is specific to PHD3 silencing and independent from hydroxylase activity. A, 786-O cells transfected with control (scr), siPHD1, siPHD2, or siPHD3. No decrease in HIF-2α protein expression was observed in response to PHD1 or PHD2 depletion. Quantification of three independent experiments, -fold change to scr. Bar represents mean ± S.D.; ns = not significant. B, 786-O, RCC4, and RCC4+VHL cells were treated with panhydroxylase inhibitor DMOG for 8 h under normoxia or hypoxia followed by western blot analysis of HIF-2α expression. In VHL mutated 786-O and RCC4 cells DMOG has no effect on HIF-2α protein expression. Representative blots from two individual experiments. C, 786-O, RCC4, and RCC4+VHL cells were treated with cobalt chloride (CoCl2) for 6 h under normoxia or hypoxia followed by western blot analysis of HIF-2α expression. Representative blots from four individual experiments. D, HIF-2α protein decay was studied using 4 h CHX treatment. HIF-2α protein decay remains unchanged with PHD3 depletion. Quantification of three individual experiments, individual data points are shown. -Fold change to untreated sample (0 h).
We next tested whether the stability of HIF-2α protein is affected by PHD3 depletion using protein translation inhibitor cycloheximide (CHX) to chase the post-translational degradation of HIF-2α protein. We did not see major change in HIF-2α protein decay with PHD3 depletion as compared with the control (Fig. 3D), indicating that the regulation of HIF-2α expression by PHD3 silencing does not occur at post-translational level.
Knockdown of PHD3 decreases HIF2A mRNA expression
Because the down-regulation of HIF-2α protein level upon PHD3 knockdown could not be explained by protein decay, we next examined whether the down-regulation of HIF-2α is seen at mRNA expression level. 786-O and RCC4 cells were transfected with two individual PHD3 siRNAs or PHD2 siRNA followed by quantitative RT-PCR for HIF2A mRNA. Noticeably, we observed significant down-regulation of HIF2A mRNA expression with PHD3 depletion in both 786-O and RCC4 cells whereas PHD2 depletion had no effect (Fig. 4, A and B). As expected based on the protein level studies, the expression of HIF1A in RCC4 cells was not affected by silencing of PHD3 (Fig. 4C). As we had observed decrease in GLUT1 and LDHA expression we were also interested if other known HIF-2α target genes react to PHD3 silencing in ccRCC cells. In line with the metabolic enzymes, a significant down-regulation in well-characterized HIF-2α target gene VEGFA mRNA level in both 786-O and RCC4 cell lines with PHD3 knockdown was seen (Fig. 4, D and E). In addition, 786-O cells showed decrease in OCT4, another established HIF-2α target gene (Fig. 4F). The data show that silencing of PHD3, can lead to down-regulation of HIF2A mRNA expression and to reduction of HIF-2α target gene expression in ccRCC cells.
Figure 4.
PHD3 silencing leads to down-regulation of HIF2A mRNA expression. A, qRT-PCR analysis of HIF2A expression in 786-O cells transfected with control (scr), siPHD3#1, siPHD3#2, or siPHD2 followed by normoxic (NOX, 21% oxygen) or hypoxic (HOX, 1% oxygen) exposure. Quantification of at least three independent experiments, -fold change to scr. Bar represents mean ± S.D.; ***, p <0.001; **, p <0.001; ns = not significant. B, HIF2A expression in RCC4 cells transfected with scr, siPHD3#1, siPHD3#2, or siPHD2 followed by normoxic or hypoxic exposure. Quantification of at least three independent experiments, -fold change to scr. Bar represents mean ± S.D.; *** p <0.001; ** p <0.01; ns = not significant. C, HIF1A expression in RCC4 cells remains unchained with PHD3 or PHD2 silencing. Quantification of three (siPHD3) or two (siPHD2) individual experiments, -fold change to scr. Bar represents mean ± S.D. D, VEGFA expression in 786-O cells transfected with scr, siPHD3#1, siPHD3#2 followed by normoxic or hypoxic exposure. Quantification of four individual experiments, -fold change to scr. Bar represents mean ± S.D.; ** p <0.01; * p <0.05. E, VEGFA expression in RCC4 cells transfected with scr, siPHD3#1, siPHD3#2 followed by normoxic or hypoxic exposure. Quantification of four individual experiments, -fold change to scr. Bar represents mean ± S.D.; *, p < 0.05. F, OCT4 expression in 786-O cells. Quantification of four individual experiments, -fold change to scr. Bar represents mean ± S.D.; **, p <0.01; *, p <0.05.
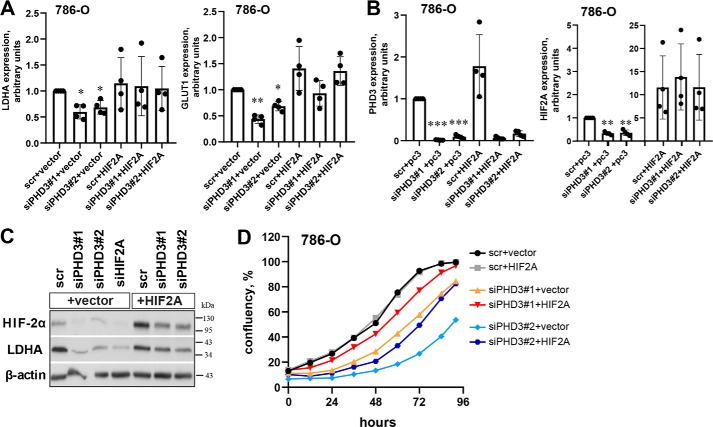
To further verify the effect of PHD3, we asked whether the decreased levels of HIF-2α target genes could be rescued by HIF2A overexpression under silenced PHD3. By using a transient HIF2A transfection in PHD3-depleted 786-O cells, expression of both GLUT1 and LDHA were rescued as compared with control (Fig. 5A). Also, PHD3 and HIF2A expression levels were verified by qRT-PCR (Fig. 5B) and by western blot analysis (Fig. 5C). In keeping with the mRNA expression, also the protein expression of LDHA was markedly rescued upon HIF2A overexpression (Fig. 5C). We have previously shown that PHD3 knockdown attenuated ccRCC cell proliferation on both 2D and 3D models (14). In line with the effect of PHD3 on HIF-2α, forced expression of HIF2A rescued the suppressed 96-h cell growth in PHD3-silenced cells when compared with the empty vector (Fig. 5D). In the control siRNA background HIF2A overexpression had no effect on cell proliferation, suggesting that the restored proliferation is specific to the effect of PHD3 silencing. Growth curves with PHD3 silencing and HIF2A overexpression treatment with their respective error bars are shown in Fig. S2.
Figure 5.
Forced expression of HIF2A in PHD3-silenced cells restore GLUT1 and LDHA expression. A, 786-O cells were transfected with scr or siPHD3 followed by forced expression of HIF2A or empty vector for 24 h. LDHA and GLUT1 expressions were determined with qRT-PCR. Quantification of four individual experiments, -fold change to scr+vector. Bar represents mean ± S.D. Statistics: one-way ANOVA; post hoc: Tukey, **, p <0.01; *, p <0.05. B, PHD3 and HIF2A expression in 786-O cells with PHD3 silencing and forced expression of HIF2A. Quantification of four individual experiments, -fold change to scr+vector. Bar represents mean ± S.D. Statistics: one-way ANOVA; post hoc: Tukey, ***, p <0.001; **, p <0.01. C, LDHA protein level is rescued by restoring expression of HIF2A in PHD3-depleted cells. A representative blot. D, Incucyte® Live Cell Analysis of 786-O cells treated with two distinct siRNA sequences targeting PHD3 and HIF2A overexpression or empty vector. Forced expression of HIF2A restores the proliferation of PHD3-silenced cells. Curves represent mean values of three individual biological experiments with eight replicate wells in each experiment.
PHD3 silencing leads to attenuated HIF2A mRNA stability
Previous reports suggest that inhibiting proteasome activity could decrease the expression of some mRNAs at transcriptional level (38). Thus, we next asked whether HIF2A transcription is regulated via proteasomal activity. However, HIF2A expression remained unchanged in siPHD3-treated 786-O cells after 4 h of proteasome inhibitor MG132 exposure (Fig. S3A), indicating that proteasomal activity has no role in the transcription of HIF2A. Furthermore, as mTOR pathway has been previously linked to HIF-2α regulation by several reports (24) and also we have previously shown suppression of mTOR signaling by PHD3 silencing (14), we also tested if mTOR inhibition results in decrease in HIF2A expression. By using 4-h exposure to mTOR inhibitors rapamycin or torin1, we did not see any major effect on HIF2A expression (Fig. S3B).
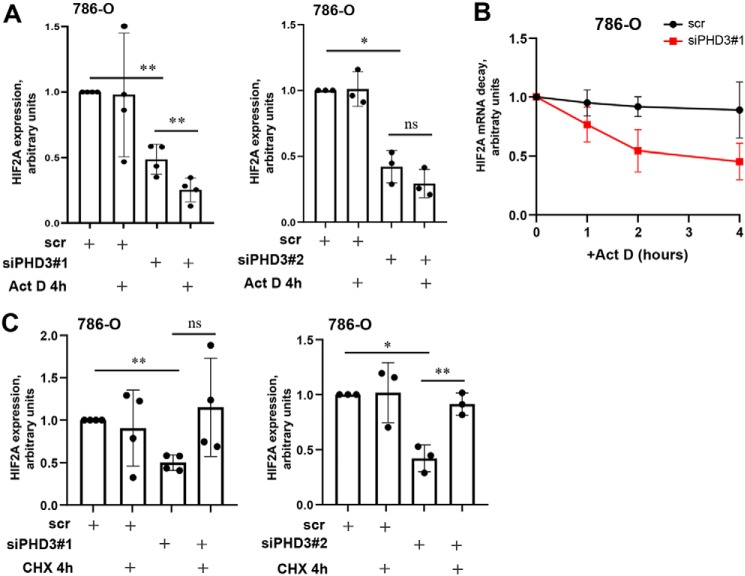
As we have previously shown that a number of mRNA regulating factors responded to PHD3 silencing in 786-O cells (14), we next asked whether the mRNA stability of HIF2A is altered upon PHD3 knockdown by using actinomycin D as an inhibitor of de novo mRNA synthesis. Indeed, we saw further reduction of HIF2A in PHD3-depleted cells by actinomycin D after 4 h (Fig. 6A). Furthermore, we scrutinized the dynamics of HIF2A mRNA decay in PHD3-silenced cells using an actinomycin D chase experiment. The data at four time points (0, 1, 2, and 4 h) show clearly enhanced decay of HIF2A mRNA in PHD3-silenced cells as compared with control (scr) cells, further implying that PHD3 silencing enhances HIF2A mRNA degradation (Fig. 6B). Moreover, we studied whether the effect of PHD3 silencing in enhancing mRNA degradation occurs more broadly. PHD3 silencing alone had no or increasing effect on the expression level of cyclin-dependent kinase inhibitors p27, p21 or an apoptosis regulator BAX genes. Most importantly, both the control and PHD3-silenced cells responded similarly to actinomycin D treatment. This implies that the effect of PHD3 in enhancing HIF2A mRNA degradation is not a generalized phenomenon (Fig. S3C).
Figure 6.
PHD3 silencing leads to attenuated HIF-2α mRNA stability. A, qRT-PCR analysis of HIF2A expression in 786-O cells transfected with control (scr), siPHD3#1, or siPHD3#2 and treated with transcription inhibitor actinomycin D (Act D) for 4 h. Act D treatment further reduces the HIF2A expression in PHD3-depleted cells. Quantification of at least three independent experiments, -fold change to untreated scr. Bar represents mean ± S.D.; Statistics: one-way ANOVA; post hoc: Tukey, **, p <0.01; *, p <0.05; ns = not significant. B, 786-O cells were transfected with scr or siPHD3#1 and treated with Act D for 4 h. Samples were collected at indicated time points and HIF2A decay was determined by qRT-PCR. GAPDH expression was used for normalization. Graph represents mean values of three individual experiments ± S.E., -fold change to untreated sample (0 h). C, 786-O cells were transfected with scr, siPHD3#1, siPHD3#2 and treated with protein translation inhibitor CHX for 4 h. Treatment with CHX restores the expression of HIF2A mRNA. Quantification of at least three independent experiments, -fold change to untreated scr. Bar represents mean ± S.D., Statistics: one-way ANOVA; post hoc: Tukey, **, p <0.01, *, p <0.05, ns = not significant.
To further study the mRNA stability in PHD3-depleted cells, we used CHX that is known to stabilize labile mRNAs via binding to the ribosomes (39–41). Surprisingly, we repeatedly detected almost complete rescue of HIF2A expression in PHD3-silenced cells in response to CHX treatment (Fig. 6C). Together, our data suggest that the post-transcriptional decay of HIF2A is enhanced upon PHD3 silencing.
PHD3 and HIF2A mRNA expression correlates in ccRCC tumor samples
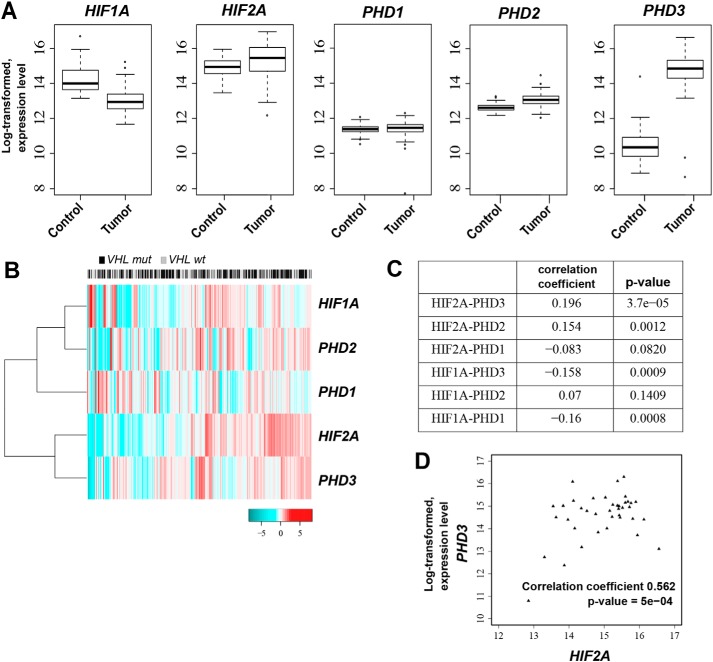
Finally, to investigate the relationship of PHD3 and HIF2A mRNA expression levels in clinical ccRCC tumor samples, we used the publicly available TCGA clear cell renal cell carcinoma data set (42). Out of 442 tumor samples, the original TCGA study provided normal matched samples for 71 patients. Our comparative analysis across the tumor and their corresponding normal adjacent tissues revealed a significant difference for HIF1A, HIF2A, PHD2, and PHD3 expression (p value <0.001) (Fig. 7A). This shows significant up-regulation of both HIF2A and PHD3 in tumor samples that is in line with previous studies (10, 27). Accordingly, we were able to divide the samples into two highly distinct groups of tumor and normal tissues based on the genes of interest using hierarchical clustering method (Fig. S4A). Similar clustering of all the VHL mutated (n = 217) and nonmutated (n = 194) tumor samples in the TCGA data set did not reveal any clear pattern in terms of the mutation status (Fig. 7B).
Figure 7.
PHD3 and HIF2A mRNA expression correlate in clinical ccRCC tumor samples. A, distribution of HIF1A, HIF2A, and PHD1–3 expression levels across tumor (n = 71) and their adjacent normal (n = 71) samples. Expression levels (normalized, log transformed) of both HIF1A and HIF2A in addition to PHD2 and PHD3 were found significantly different between tumor and normal samples (p <0.001). B, hierarchical clustering of TCGA ccRCC VHL mutated (n = 217) and nonmutated (n = 194) samples based on the expression levels of HIF1A, HIF2A and PHD1–3 genes. C, correlation coefficients (Pearson correlation) with corresponding p values for HIF1A or HIF2A and PHD1–3 in the whole ccRCC tumor sample data set (n = 442). Correlation of expression between HIF2A and PHD3 was higher than the correlation between other gene pairs (Pearson correlation 0.196, p <0.001). D, correlation of the expression levels (normalized, log transformed) between HIF2A and PHD3 for the patients with the poorest survival (n = 40). A significant correlation of 0.526 was observed (Pearson correlation, p <0.001).
Inspired by the strong signal for co-expression between HIF2A and PHD3 detected in RCC cell lines, we consequently explored the correlation between two HIFs and PHD1–3 expression for all 442 TCGA samples (Fig. 7C). In line with our data from cell lines, HIF2A and PHD3 presented the highest correlation value among the gene pairs (Pearson correlation 0.196, p value <0.001). Next, taking into account the known heterogeneity in ccRCC (3), we further narrowed down our investigations to a more homogeneous group of patients, focusing on 40 (∼10%) TCGA patients with the poorest prognosis (survival time <12 months) and reanalyzed the correlation of PHD3 and HIF2A expression. Using the poor prognosis group we observed a very strong correlation between HIF2A and PHD3 expression (Pearson correlation 0.526, p value <0.001) (Fig. 7D), whereas the expression of other PHD family members did not significantly correlate with HIF1A or HIF2A expression (Fig. S4B).
Discussion
The family of PHDs regulates the protein stability of HIF-α by post-translational hydroxylation that subsequently leads to ubiquitination and proteasomal degradation of HIF-α under sufficient oxygen availability (reviewed in Refs. 4, 5). This mechanism provides a rapid regulation of HIF-signaling activity as HIF-α can be induced or degraded within minutes in response to oxygen levels (43). A number of reports have demonstrated that silencing of either the expression or the activity of PHDs results in up-regulation of HIF-1α and/or HIF-2α protein expression. In line, PHD3 acts as a major regulator of HIF-2α protein in many cell types (6, 7, 28). However, in ccRCC, high expression of both HIF-2α and PHD3 because of the frequently occurring VHL mutation is well-established (10, 12).
Here we report a novel mechanism of HIF-2α regulation in ccRCC cells by PHD3 that may shed light on the simultaneous high expression of PHD3 and HIF-2α. Depletion of PHD3, but not other PHDs, resulted in significant decrease, rather than increase, in HIF-2α protein. Noticeably, HIF-2α protein degradation was not found to be affected, but instead we found a marked decrease in HIF2A mRNA level. Moreover, the decrease was pinpointed to HIF2A mRNA stability in PHD3-depleted cells. In line with this, we demonstrated that PHD3 and HIF2A expression positively correlated in a clinical ccRCC data set.
The regulation of HIF-2α expression by PHD3 seems to be cell type–specific, as previous studies show an up-regulation of HIF-2α protein in many widely studied cancer cells lines (28). Also, we showed that in squamous carcinoma and breast carcinoma cells silencing of PHD3 either up-regulated or did not have an effect on HIF-2α protein that is well in line with previous reports (28). Here we describe for the first time marked reduction of HIF-2α expression in response to PHD3 silencing, suggesting that in striking contrast to the canonical pathway, in ccRCC cells high PHD3 expression is needed to keep high expression levels of HIF-2α and its target genes. How widespread the effect is in ccRCC cells and whether it is associated with VHL status, remains to be investigated.
Recently, several regulation mechanisms independent from PHD and pVHL activity have been described for HIF-α isoforms. Besides proteasomal degradation, these include also lysosomal and autophagosomal degradation pathways (15–20). Together, the findings suggest more complex regulation of the HIF signaling pathway than previously known. Nonetheless, regulation of HIF-2α transcription level is not fully understood. Transcription factor E2F1 has been shown to directly regulate HIF2A expression, and deubiquitylase Cezanne was linked to the regulation of HIF2A by showing that Cezanne regulates the stability of E2F1 (21). The regulation was shown in several cell lines, including VHL mutated 786-O ccRCC cell line. Also PARP-1 has been suggested to directly enhance HIF2A transcription when studied in Parp knockout mouse embryonic fibroblasts, but not in RCC cells (22). In neuroblastoma cells, HIF2A expression has been shown to be regulated by mTORC2 complex (24). Furthermore, HIF-2α protein levels have been reported to be regulated by mTORC2 in ccRCC cells (44) highlighting the involvement of mTORC2 in regulation of HIF-2α across different cell types. These findings suggest that the transcriptional regulation of HIF-2α is complex and sensitive to many different stimuli and that tissue and cell type specificity is likely to occur. However, as actinomycin D treatment demonstrated decreased stability of HIF2A mRNA under PHD3 silencing, the above-listed known mechanisms are not likely to be responsible for HIF2A mRNA stability regulation by PHD3.
Although further studies are needed to determine the specific mechanism by which PHD3 regulates HIF2A mRNA stability, our recent data may give hints to this. We recently showed that PHD3 depletion leads to up-regulation of several mRNA processing factors, including hnRNPs D, H, L, and G that function in several steps of mRNA regulation, including mRNA stability, alternative splicing, and mRNA maturation as well as in down-regulation of a number of ribosomal proteins and other translational machinery components, including EIF4G (14). Thus, it is feasible that the up-regulation of mRNA processing factors by PHD3 depletion is responsible for the regulation of HIF2A mRNA stability. Supporting PHD3's function in regulating HIF2A mRNA stability, treatment with CHX that blocks the exit site at the ribosome and stabilizes labile mRNAs (39–41), completely restored the HIF2A mRNA level. Previously, HIF-1α regulation has been linked to RNA-binding proteins HuR and PTB, which are known regulators of mRNA stability. HuR and PTB have been shown to directly bind to and regulate the translation of HIF1A (45, 46). In fact, it has been proposed that the regulation of HIF-1α mRNA translation is an important regulatory step under hypoxic condition. It can be speculated that similar motifs for RNA-binding proteins occur in HIF2A sequence and that HIF2A could be post-transcriptionally regulated by certain mRNA processing factors.
We also observed that mRNA levels of glucose transporter GLUT1 and glycolytic enzyme LDHA were down-regulated by PHD3 silencing and the effect was restored by forced expression of HIF-2α. As ccRCC has been shown to rely on high glycolytic activity (42, 47, 48), we suggest that PHD3 participates in maintaining high glycolytic activity in ccRCC at least partially through HIF-2α. High expression of PHD3 may have other crucial roles in ccRCC via maintaining high HIF-2α expression and its tumor-promoting target genes, including VEGFA. On the other hand, given the range of metabolic and signaling pathways that have been shown to be regulated by PHD3, it is feasible that PHD3 has also independent functions in cancer progression, and the modified expression of several genes by PHD3 is not due only to the induction of HIF-2α.
In summary, we have demonstrated that PHD3 maintains high HIF-2α expression in post-transcriptional manner in ccRCC cells that is supported by a strong correlation of PHD3 and HIF2A expression in clinical ccRCC samples. In contrast to the common wisdom of a negative feedback loop between HIFs and PHDs, we suggest a positive feedback loop where high HIF-2α expression up-regulates PHD3, which then post-transcriptionally enhances HIF-2α expression. Together with the previous data this suggests that PHD3 enhances ccRCC progression and modulation of PHD3 could possibly be exploited as a therapeutic target in renal cell carcinomas.
Experimental procedures
Cell culture, reagents, and transient transfections
786-O (CRL-1932) cells were obtained from ATCC (Rockville, MD) and cultured in RPMI 1640 medium (Lonza). RCC4 and RCC4 cells with stably transfected VHL (RCC4+VHL) were acquired from Prof. Peppi Koivunen (University of Oulu, Finland) and cultured in DMEM (Sigma-Aldrich). Both media were supplied with 10% FBS (Biowest, Nuaillé, France), l-glutamine (Lonza, Basel, Switzerland), and penicillin/streptomycin (Lonza). When in culture, cells were regularly tested for mycoplasma contamination and all cell lines were tested negative. Cell lines were authenticated by ATCC Cell Authentication Service (STR profiling) in May 2018. After thawing, experiments were carried out after 2 passages and until 20 passages. Cells were cultured in a humidified atmosphere in 37 °C containing 5% CO2. For hypoxic experiments the cells were cultured in 1% O2 in a hypoxic work station (In vivo2, Ruskinn Technology Ltd, Bridgend, UK) with oxygen replaced with 99.5% pure N2 (AGA, Finland). Samples for Western blotting and RT-PCR were collected at indicated time points after normoxic (21% O2) or hypoxic incubation.
DMOG (Sigma-Aldrich) was used at 1 mm final concentration for 8 h, cobalt chloride (Sigma-Aldrich) was used at 100 μm final concentration for 6 h, actinomycin D (Sigma-Aldrich) was used at 5 μg/ml final concentration for up to 4 h, CHX (Sigma-Aldrich) was used at 10 μg/ml for 4 h, proteasome inhibitor MG132 (Sigma-Aldrich) at 10 μm final concentration for 4 h, rapamycin (LC Laboratories, Woburn, MA) was used at 100 nm final concentration for 4 h and torin1 (Selleckchem, Germany) was used at 100 nm final concentration for 4 h.
For siRNA transfections, two stranded oligonucleotides were used at final concentration of 10 nm. Reverse transfections were performed using Lipofectamine® RNAiMAX (Invitrogen) according to manufacturer's protocol. The siRNAs (MWG Biotech AG, Germany) used were nontarget (siScr) 5′-CCUACAUCCCGAUCGAUGAUG(dTdT)-3′, siHIF-2α 5′-GCGACA-GCUGGAGUAUGAAUU(dTdT)-3′, siHIF-1α 5′-AACUAACUGGACACAGUGUGU(dTdT)-3′, siPHD1 5′-ACAUUGCUGCAUGGUAGAA(dTdT)-3′, siPHD2 5′-GACGAAAGCCAUGGUUGCUUG(dTdT)-3′, siPHD3#1 5′-GUCUAAGGCAAUGGU-GGCUUG(dTdT)-3′, and siPHD3#2 5′-AGGAGAGGUCUAAGGCAAUG (dTdT)-3′.
For adenoviral shRNA delivery 786-O cells were transduced with either control (Ad-shScr) 5′-GACACGCGACTTGTACCACTTCAAGAGAGTGGTACAAGTCGCGTGTCTTTTTTACGCGT-3′ or with PHD3-targeting shRNA (Ad-shPHD3) 5′-CCGGCACCTGCATCTA-CTATCTGAACTCGAGTTCAGATAGTAGATGCAGGTGTTTTT-3′ (Vector BioLabs, Malvern, PA).
For plasmid transfections, cells were plated and allowed to attach for 24 h. 0.5 μg of plasmid DNA per well was used to achieve suitable overexpression of HIF-α protein. Empty pcDNA.3 plasmid as used as a vector control. Transfections were performed with FuGene® HD (Promega, Madison, WI) according to manufacturer's protocol.
Protein expression analysis
For protein expression analysis, cells were harvested in SDS-Triton lysis buffer with protease inhibitors (1 mm Na3VO4, 1 mm PMSF, and 10 mm NaF). Protein concentration was measured using Bio-Rad DC Protein assay followed by addition of 2-mercaptoethanol containing SDS loading buffer and boiling prior to loading. Equal amounts of protein were loaded and run on SDS-PAGE in a mini-gel chamber (Bio-Rad) and transferred to a PVDF membrane (Merck). Western blotting analyses with the following antibodies were performed: PHD3 (NB100-139, Novus Biologicals, Littleton, CO), PHD2 (NB100-137, Novus Biologicals), PHD1 (NB100-310, Novus Biologicals), HIF-1α (610959, BD Transduction Laboratories), HIF-2α/EPAS1 (NB100-122, Novus Biologicals), LDHA (no. 3582, Cell Signaling Technology, Danvers, MA), β-actin (AC-74, Sigma-Aldrich), GAPDH (5G4-6C5, HyTest), anti-mouse-HRP (DAKO), anti-rabbit-HRP (DAKO). Protein detection was performed using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific). β-Actin or GAPDH was used as a loading control. Blots were quantified using ImageJ (National Institutes of Health) and intensities of the protein of interest were normalized to β-actin.
Quantitative RT-PCR
Total RNA was extracted using NucleoSpin RNA II kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's protocol. Reverse transcription was performed using M-MuLV RNase H-reverse transcriptase (Finnzymes, Thermo Fisher Scientific) and random hexamer primers (Promega). qPCR reactions were run using QuantStudio 12K Flex (Thermo Fisher Scientific) and TaqMan Universal Master Mix II, no UNG (Applied Biosystems, Life Technologies). TaqMan primers (Oligomer, Finland) and probes (Universal Probe Library, Roche) used are listed in Table S1. Data analysis was performed with QuantStudio 12K Flex Software (Thermo Fisher Scientific) by using ΔΔCq method for quantification differences in gene expression. The expression of the genes of interest was normalized against the expression of GAPDH.
Immunocytochemistry and imaging
Cells were grown on coverslips, fixed with fresh 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Nuclei were stained with the nuclear stain Hoechst 33342 (Invitrogen). Antibody against HIF-2α/EPAS1 (NB100-122, Novus Biologicals) was used in 1:500 dilution and Cy3 conjugated anti-rabbit secondary antibody (Invitrogen) in 1:1000 dilution. Imaging was performed using LSM780 (Carl Zeiss, Germany) confocal microscope with C-Apochromat 40×/1.20 W Korr M27 objective. Experiments were performed as parallel treatments and repeated two times. The images were cropped for representation and the intensity was measured using ImageJ (National Institutes of Health).
Cell proliferation
To follow cell proliferation in 2D cell growth siRNA and HIF2A overexpression plasmid or control plasmid-treated cells were plated on 96-well plates, 8 wells for each treatment. After 24 h the well plates were placed into Incucyte® Live-Cell Analysis System (Essen BioScience) for 96 h. The wells were scanned every 12 h, and the proliferation rate was determined as confluency accordingly.
TCGA clear cell renal cell carcinoma data analysis
The ccRCC data set was published by TCGA (42) and is available through TCGA website under the name of The Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma (TCGA-KIRC). We utilized the raw mRNA expression files sequenced and preprocessed through IlluminaHiSeq_RNASeqV2 platform. A total of 442 patients had both clinical and expression data needed for current analysis.
The TCGA ccRCC analyses were performed using R version 3.4.3. The mRNA expression values were first normalized using the Trimmed Mean of M values (TMM) method (Bioconductor edgeR package version 3.18.1) and then the normalized values were log-2 transformed (49, 50). Unsupervised hierarchical clustering was performed based on euclidean distance and the average linkage method using the R package gplots (version 3.0.1).
Statistical analysis
Quantified data from the protein and mRNA expression analysis were reported as means together with their respective S.D.s. Statistical significance was analyzed using GraphPad Prism 8.00 software (La Jolla, CA). Significance was assessed with a Student's t test for comparisons of two groups or with one-way analysis of variance (ANOVA) for three or more groups and a post hoc Dunnett's multiple comparisons test unless otherwise specified. Nominal p values were reported; * indicates p value <0.05, ** indicates p value <0.01, *** indicates p value <0.001, and ns indicates not significant.
Author contributions
P. M., H. H., K. R., and P. M. J. conceptualization; P. M., H. H., and F. S. data curation; P. M. and F. S. formal analysis; P. M. and P. M. J. funding acquisition; P. M. and F. S. visualization; P. M., F. S., and P. M. J. writing-original draft; H. H., K. R., L. L. E., and P. M. J. supervision; H. H., L. L. E., and P. M. J. writing-review and editing; P. M. J. resources.
Supplementary Material
Acknowledgments
We thank Dr. Peppi Koivunen (University of Oulu, Finland) for kindly providing the RCC4 cell lines for our study. We acknowledge Taina Kalevo-Mattila for expert technical assistance. We acknowledge the Finnish Functional Genomics Centre and the Cell Imaging Core at the Turku Centre for Biotechnology for providing qPCR service and the microscopy imaging facilities.
This work was supported by Cancer Foundation Finland (to P.M. and P.M.J.), The Sigrid Juselius Foundation (to P.M.J.), Turku University Hospital (EVO1303) (to P.M., H.H. and P.M.J.), Helsinki University Hospital (to P.M. and P.M.J.), The University of Turku Graduate School (to P.M.), and Finnish Cultural Foundation Varsinais-Suomi Regional fund (to P.M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4 and Table S1.
- ccRCC
- clear cell renal cell carcinoma
- HIF
- hypoxia-inducible factor
- DMOG
- dimethyloxalylglycine
- CHX
- cycloheximide
- TCGA
- The Cancer Genome Atlas
- q
- quantitative.
References
- 1. Li L., and Kaelin W. G. (2011) New insights into the biology of renal cell carcinoma. Hematol. Oncol. Clin. North Am. 25, 667–686 10.1016/j.hoc.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ivan M., and Kaelin W. G. Jr. (2017) The EGLN-HIF O2-Sensing System: Multiple inputs and feedbacks. Mol. Cell 66, 772–779 10.1016/j.molcel.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schödel J., Grampp S., Maher E. R., Moch H., Ratcliffe P. J., Russo P., and Mole D. R. (2016) Hypoxia, hypoxia-inducible transcription factors, and renal cancer. Eur. Urol. 69, 646–657 10.1016/j.eururo.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaelin W. G. Jr., and Ratcliffe P. J. (2008) Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 10.1016/j.molcel.2008.04.009 [DOI] [PubMed] [Google Scholar]
- 5. Semenza G. L. (2012) Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408 10.1016/j.cell.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aprelikova O., Chandramouli G. V., Wood M., Vasselli J. R., Riss J., Maranchie J. K., Linehan W. M., and Barrett J. C. (2004) Regulation of HIF prolyl hydroxylases by hypoxia-inducible factors. J. Cell. Biochem. 92, 491–501 10.1002/jcb.20067 [DOI] [PubMed] [Google Scholar]
- 7. Berra E., Benizri E., Ginouvès A., Volmat V., Roux D., and Pouysségur J. (2003) HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 22, 4082–4090 10.1093/emboj/cdg392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Courtney K. D., Infante J. R., Lam E. T., Figlin R. A., Rini B. I., Brugarolas J., Zojwalla N. J., Lowe A. M., Wang K., Wallace E. M., Josey J. A., and Choueiri T. K. (2018) Phase I Dose-Escalation Trial of PT2385, a first-in-class hypoxia-inducible factor-2α antagonist in patients with previously treated advanced clear cell renal cell carcinoma. J. Clin. Oncol. 36, 867–874 10.1200/JCO.2017.74.2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sato E., Torigoe T., Hirohashi Y., Kitamura H., Tanaka T., Honma I., Asanuma H., Harada K., Takasu H., Masumori N., Ito N., Hasegawa T., Tsukamoto T., and Sato N. (2008) Identification of an immunogenic CTL epitope of HIFPH3 for immunotherapy of renal cell carcinoma. Clin. Cancer Res. 14, 6916–6923 10.1158/1078-0432.CCR-08-0466 [DOI] [PubMed] [Google Scholar]
- 10. Högel H., Rantanen K., Jokilehto T., Grenman R., and Jaakkola P. M. (2011) Prolyl hydroxylase PHD3 enhances the hypoxic survival and G1 to S transition of carcinoma cells. PLoS One 6, e27112 10.1371/journal.pone.0027112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tanaka T., Kitamura H., Torigoe T., Hirohashi Y., Sato E., Masumori N., Sato N., and Tsukamoto T. (2011) Autoantibody against hypoxia-inducible factor prolyl hydroxylase-3 is a potential serological marker for renal cell carcinoma. J. Cancer Res. Clin. Oncol. 137, 789–794 10.1007/s00432-010-0940-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fu L., Minton D. R., Zhang T., Nanus D. M., and Gudas L. J. (2015) Genome-wide profiling of TRACK kidneys shows similarity to the human ccRCC transcriptome. Mol. Cancer Res. 13, 870–878 10.1158/1541-7786.MCR-14-0423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jaakkola P. M., and Rantanen K. (2013) The regulation, localization, and functions of oxygen-sensing prolyl hydroxylase PHD3. Biol. Chem. 394, 449–457 10.1515/hsz-2012-0330 [DOI] [PubMed] [Google Scholar]
- 14. Miikkulainen P., Högel H., Rantanen K., Suomi T., Kouvonen P., Elo L. L., and Jaakkola P. M. (2017) HIF prolyl hydroxylase PHD3 regulates translational machinery and glucose metabolism in clear cell renal cell carcinoma. Cancer Metab. 5, 5 10.1186/s40170-017-0167-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bento C. F., Fernandes R., Ramalho J., Marques C., Shang F., Taylor A., and Pereira P. (2010) The chaperone-dependent ubiquitin ligase CHIP targets HIF-1α for degradation in the presence of methylglyoxal. PLoS One 5, e15062 10.1371/journal.pone.0015062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bremm A., Moniz S., Mader J., Rocha S., and Komander D. (2014) Cezanne (OTUD7B) regulates HIF-1α homeostasis in a proteasome-independent manner. EMBO Rep. 15, 1268–1277 10.15252/embr.201438850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Demidenko Z. N., Rapisarda A., Garayoa M., Giannakakou P., Melillo G., and Blagosklonny M. V. (2005) Accumulation of hypoxia-inducible factor-1α is limited by transcription-dependent depletion. Oncogene 24, 4829–4838 10.1038/sj.onc.1208636 [DOI] [PubMed] [Google Scholar]
- 18. Hubbi M. E., Hu H., Kshitiz, Ahmed I., Levchenko A., and Semenza G. L. (2013) Chaperone-mediated autophagy targets hypoxia-inducible factor-1α (HIF-1α) for lysosomal degradation. J. Biol. Chem. 288, 10703–10714 10.1074/jbc.M112.414771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kong X., Alvarez-Castelao B., Lin Z., Castaño J. G., and Caro J. (2007) Constitutive/hypoxic degradation of HIF-α proteins by the proteasome is independent of von Hippel Lindau protein ubiquitylation and the transactivation activity of the protein. J. Biol. Chem. 282, 15498–15505 10.1074/jbc.M700704200 [DOI] [PubMed] [Google Scholar]
- 20. Liu X. D., Yao J., Tripathi D. N., Ding Z., Xu Y., Sun M., Zhang J., Bai S., German P., Hoang A., Zhou L., Jonasch D., Zhang X., Conti C. J., Efstathiou E., et al. (2015) Autophagy mediates HIF2α degradation and suppresses renal tumorigenesis. Oncogene 34, 2450–2460 10.1038/onc.2014.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moniz S., Bandarra D., Biddlestone J., Campbell K. J., Komander D., Bremm A., and Rocha S. (2015) Cezanne regulates E2F1-dependent HIF2α expression. J. Cell Sci. 128, 3082–3093 10.1242/jcs.168864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gonzalez-Flores A., Aguilar-Quesada R., Siles E., Pozo S., Rodríguez-Lara M. I., López-Jiménez L., López-Rodríguez M., Peralta-Leal A., Villar D., Martin-Oliva D., del Peso L., Berra E., and Oliver F. J. (2014) Interaction between PARP-1 and HIF-2α in the hypoxic response. Oncogene 33, 891–898 10.1038/onc.2013.9 [DOI] [PubMed] [Google Scholar]
- 23. Sala M. A., Chen C., Zhang Q., Do-Umehara H. C., Wu W., Misharin A. V., Waypa G. B., Fang D., Budinger G. R. S., Liu S., Chandel N. S., Schumacker P. T., Sznajder J. I., and Liu J. (2018) JNK2 up-regulates hypoxia-inducible factors and contributes to hypoxia-induced erythropoiesis and pulmonary hypertension. J. Biol. Chem. 293, 271–284 10.1074/jbc.RA117.000440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mohlin S., Hamidian A., von Stedingk K., Bridges E., Wigerup C., Bexell D., and Påhlman S. (2015) PI3K-mTORC2 but not PI3K-mTORC1 regulates transcription of HIF2A/EPAS1 and vascularization in neuroblastoma. Cancer Res. 75, 4617–4628 10.1158/0008-5472.CAN-15-0708 [DOI] [PubMed] [Google Scholar]
- 25. Gordan J. D., Lal P., Dondeti V. R., Letrero R., Parekh K. N., Oquendo C. E., Greenberg R. A., Flaherty K. T., Rathmell W. K., Keith B., Simon M. C., and Nathanson K. L. (2008) HIF-α effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell 14, 435–446 10.1016/j.ccr.2008.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mandriota S. J., Turner K. J., Davies D. R., Murray P. G., Morgan N. V., Sowter H. M., Wykoff C. C., Maher E. R., Harris A. L., Ratcliffe P. J., and Maxwell P. H. (2002) HIF activation identifies early lesions in VHL kidneys: Evidence for site-specific tumor suppressor function in the nephron. Cancer Cell 1, 459–468 10.1016/S1535-6108(02)00071-5 [DOI] [PubMed] [Google Scholar]
- 27. Turner K. J., Moore J. W., Jones A., Taylor C. F., Cuthbert-Heavens D., Han C., Leek R. D., Gatter K. C., Maxwell P. H., Ratcliffe P. J., Cranston D., and Harris A. L. (2002) Expression of hypoxia-inducible factors in human renal cancer: Relationship to angiogenesis and to the von Hippel-Lindau gene mutation. Cancer Res. 62, 2957–2961 [PubMed] [Google Scholar]
- 28. Appelhoff R. J., Tian Y. M., Raval R. R., Turley H., Harris A. L., Pugh C. W., Ratcliffe P. J., and Gleadle J. M. (2004) Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 279, 38458–38465 10.1074/jbc.M406026200 [DOI] [PubMed] [Google Scholar]
- 29. Högel H., Miikkulainen P., Bino L., and Jaakkola P. M. (2015) Hypoxia inducible prolyl hydroxylase PHD3 maintains carcinoma cell growth by decreasing the stability of p27. Mol. Cancer 14, 143 10.1186/s12943-015-0410-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xue J., Li X., Jiao S., Wei Y., Wu G., and Fang J. (2010) Prolyl hydroxylase-3 is down-regulated in colorectal cancer cells and inhibits IKKβ independent of hydroxylase activity. Gastroenterology 138, 606–615 10.1053/j.gastro.2009.09.049 [DOI] [PubMed] [Google Scholar]
- 31. Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., and Ratcliffe P. J. (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399, 271–275 10.1038/20459 [DOI] [PubMed] [Google Scholar]
- 32. Shen C., Beroukhim R., Schumacher S. E., Zhou J., Chang M., Signoretti S., and Kaelin W. G. Jr. (2011) Genetic and functional studies implicate HIF1α as a 14q kidney cancer suppressor gene. Cancer Discov. 1, 222–235 10.1158/2159-8290.CD-11-0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cui X. G., Han Z. T., He S. H., Wu X. D., Chen T. R., Shao C. H., Chen D. L., Su N., Chen Y. M., Wang T., Wang J., Song D. W., Yan W. J., Yang X. H., Liu T., Wei H. F., and Xiao J. (2017) HIF1/2α mediates hypoxia-induced LDHA expression in human pancreatic cancer cells. Oncotarget 8, 24840–24852 10.18632/oncotarget.15266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raval R. R., Lau K. W., Tran M. G., Sowter H. M., Mandriota S. J., Li J. L., Pugh C. W., Maxwell P. H., Harris A. L., and Ratcliffe P. J. (2005) Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol. Cell. Biol. 25, 5675–5686 10.1128/MCB.25.13.5675-5686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. D'Angelo G., Duplan E., Boyer N., Vigne P., and Frelin C. (2003) Hypoxia up-regulates prolyl hydroxylase activity: A feedback mechanism that limits HIF-1 responses during reoxygenation. J. Biol. Chem. 278, 38183–38187 10.1074/jbc.M302244200 [DOI] [PubMed] [Google Scholar]
- 36. del Peso L., Castellanos M. C., Temes E., Martin-Puig S., Cuevas Y., Olmos G., and Landazuri M. O. (2003) The von Hippel Lindau/hypoxia-inducible factor (HIF) pathway regulates the transcription of the HIF-proline hydroxylase genes in response to low oxygen. J. Biol. Chem. 278, 48690–48695 10.1074/jbc.M308862200 [DOI] [PubMed] [Google Scholar]
- 37. Henze A. T., Riedel J., Diem T., Wenner J., Flamme I., Pouyseggur J., Plate K. H., and Acker T. (2010) Prolyl hydroxylases 2 and 3 act in gliomas as protective negative feedback regulators of hypoxia-inducible factors. Cancer Res. 70, 357–366 10.1158/0008-5472.CAN-09-1876 [DOI] [PubMed] [Google Scholar]
- 38. Yao T., and Ndoja A. (2012) Regulation of gene expression by the ubiquitin-proteasome system. Semin. Cell Dev. Biol. 23, 523–529 10.1016/j.semcdb.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koeller D. M., Horowitz J. A., Casey J. L., Klausner R. D., and Harford J. B. (1991) Translation and the stability of mRNAs encoding the transferrin receptor and c-fos. Proc. Natl. Acad. Sci. U.S.A. 88, 7778–7782 10.1073/pnas.88.17.7778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Newton R., Stevens D. A., Hart L. A., Lindsay M., Adcock I. M., and Barnes P. J. (1997) Superinduction of COX-2 mRNA by cycloheximide and interleukin-1β involves increased transcription and correlates with increased NF-κB and JNK activation. FEBS Lett. 418, 135–138 10.1016/S0014-5793(97)01362-8 [DOI] [PubMed] [Google Scholar]
- 41. Ohh M., and Takei F. (1995) Regulation of ICAM-1 mRNA stability by cycloheximide: Role of serine/threonine phosphorylation and protein synthesis. J. Cell. Biochem. 59, 202–213 10.1002/jcb.240590210 [DOI] [PubMed] [Google Scholar]
- 42. Cancer Genome Atlas Research Network. (2013) Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 10.1038/nature12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jewell U. R., Kvietikova I., Scheid A., Bauer C., Wenger R. H., and Gassmann M. (2001) Induction of HIF-1α in response to hypoxia is instantaneous. FASEB J. 15, 1312–1314 10.1096/fj.00-0732fje [DOI] [PubMed] [Google Scholar]
- 44. Nayak B. K., Feliers D., Sudarshan S., Friedrichs W. E., Day R. T., New D. D., Fitzgerald J. P., Eid A., Denapoli T., Parekh D. J., Gorin Y., and Block K. (2013) Stabilization of HIF-2α through redox regulation of mTORC2 activation and initiation of mRNA translation. Oncogene 32, 3147–3155 10.1038/onc.2012.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Galbán S., Kuwano Y., Pullmann R. Jr., Martindale J. L., Kim H. H., Lal A., Abdelmohsen K., Yang X., Dang Y., Liu J. O., Lewis S. M., Holcik M., and Gorospe M. (2008) RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1α. Mol. Cell. Biol. 28, 93–107 10.1128/MCB.00973-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schepens B., Tinton S. A., Bruynooghe Y., Beyaert R., and Cornelis S. (2005) The polypyrimidine tract-binding protein stimulates HIF-1α IRES-mediated translation during hypoxia. Nucleic Acids Res. 33, 6884–6894 10.1093/nar/gki1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Minton D. R., Fu L., Chen Q., Robinson B. D., Gross S. S., Nanus D. M., and Gudas L. J. (2015) Analyses of the transcriptome and metabolome demonstrate that HIF1α mediates altered tumor metabolism in clear cell renal cell carcinoma. PLoS One 10, e0120649 10.1371/journal.pone.0120649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Semenza G. L. (2007) HIF-1 mediates the Warburg effect in clear cell renal carcinoma. J. Bioenerg. Biomembr. 39, 231–234 10.1007/s10863-007-9081-2 [DOI] [PubMed] [Google Scholar]
- 49. Robinson M. D., and Oshlack A. (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Robinson M. D., McCarthy D. J., and Smyth G. K. (2010) edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.