Abstract
Membrane-associated RING-CH 8 (MARCH8) is one of 11 members of the MARCH family of RING finger E3 ubiquitin ligases and down-regulates several membrane proteins (e.g. major histocompatibility complex II [MHC-II], CD86, and transferrin receptor). We recently reported that MARCH8 also targets HIV-1 envelope glycoproteins and acts as an antiviral factor. However, it remains unclear whether other family members might have antiviral functions similar to those of MARCH8. Here we show that MARCH1 and MARCH2 are MARCH family members that reduce virion incorporation of envelope glycoproteins. Infectivity assays revealed that MARCH1 and MARCH2 dose-dependently suppress viral infection. Treatment with type I interferon enhanced endogenous expression levels of MARCH1 and MARCH2 in monocyte-derived macrophages. Expression of these proteins in virus-producing cells decreased the efficiency of viral entry and down-regulated HIV-1 envelope glycoproteins from the cell surface, resulting in reduced incorporation of envelope glycoproteins into virions, as observed in MARCH8 expression. With the demonstration that MARCH1 and MARCH2 are antiviral MARCH family members as presented here, these two proteins join a growing list of host factors that inhibit HIV-1 infection.
Keywords: HIV, membrane protein, glycoprotein, host–pathogen interaction, macrophage, antiviral factor, HIV-1, macrophage, MARCH family, membrane-associated RING-CH
Introduction
The membrane-associated RING-CH (MARCH)4 family of RING finger E3 ubiquitin ligases consists of 11 members that share a similar structure. MARCH family proteins have an N-terminal cytoplasmic tail containing a C4HC3 RING finger (RING-CH finger) motif and two or more transmembrane (TM) domains, except for MARCH7 and MARCH10, which have no predicted TM domains (Fig. 1A). The MARCH1 protein is mainly expressed in cells of the immune system, presumably playing a role in dendritic cell maturation (1, 2). MARCH10 and MARCH11 proteins are highly expressed in the testis and are involved in spermiogenesis (3, 4). Other MARCH members are widely expressed in various tissues. One of the family members, MARCH8, was originally identified as a cellular homolog of the viral RING-CH E3 ligases, such as K3 and K5 of Kaposi's sarcoma–associated herpesvirus, K3 of murine γ-2 herpesvirus 68, and the myxomavirus homolog M153R (5, 6). The viral RING-CH proteins mainly inhibit the cell surface expression of major histocompatibility complex (MHC)-I molecules (7–9). In contrast, MARCH family members down-regulate many cellular transmembrane proteins: MHC-II and CD86 are down-regulated by MARCH1 and MARCH8 (2, 6, 10–16), the NKG2D ligand Mult1 and CD166 by MARCH4 and MARCH9 (17, 18), CD81 and CD44 by MARCH4 and MARCH8 (19), ICAM-1 by MARCH9 (20), CD4 by MARCH11 (3), and TRAIL receptor 1, CD98, interleukin (IL)-1 receptor accessory protein, and transferrin receptor by MARCH8 (21–24). We recently reported that MARCH8 targets HIV-1 envelope glycoproteins (Env) by down-regulating them from the cell surface, resulting in reduced incorporation of Env into virions (25). Whether other MARCH family members might play a similar antiviral role as that observed in MARCH8 remains unknown. Here we show that MARCH1 and MARCH2 are such members and that they have similar functions as those observed for MARCH8.
Figure 1.
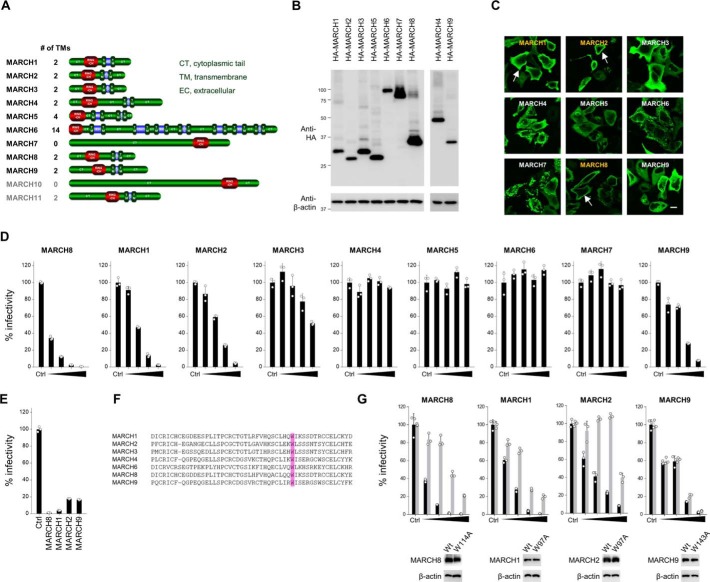
A, schematic of MARCH family members. Names in gray represent MARCH proteins that are highly expressed in the testis and are involved in spermiogenesis. B, Western blot analysis was performed by using extracts from 293T cells transfected with HA-tagged MARCH family expression plasmids. Antibodies specific for HA were used to detect MARCH proteins. C, to analyze the subcellular localization of MARCH proteins, immunofluorescence microscopy was performed using HeLa cells transfected with HA-tagged MARCH expression plasmids. An anti-HA mAb and Alexa 488–conjugated anti-mouse IgG were used as the primary and secondary antibodies, respectively. Arrows indicate plasma membrane localization. Bars, 10 μm. D, some MARCH family members inhibit HIV-1 infectivity. Viruses were prepared from 293T cells cotransfected with Env-defective HIV-1 luciferase (luc) reporter proviral DNA and either a control (Ctrl) or increasing amounts of the MARCH family plasmids together with the HIV-1 Env expression plasmid. Infectivity was determined by infecting MAGIC5 cells with HIV-1 Env-pseudotyped luc-reporter viruses and performing luc assays. Representative data from three independent experiments are shown as a percentage of the infectivity of control viruses (mean ± S.D., n = 3 technical replicates). E, similar experiments as described in D were performed, except that VSV-G–pseudotyped viruses were prepared from cells transfected with fixed amounts of MARCH family plasmids. Representative data from three independent experiments are shown as a percentage of the infectivity of control viruses (mean ± S.D., n = 3 technical replicates). F, amino acid sequence alignments of RING-CH domains of MARCH family members. A well-conserved tryptophan residue, which is essential for ubiquitin ligase activity, is boxed in violet. G, RING-CH domain–dependent inhibition of viral infectivity by expression of MARCH1 and MARCH2, but not MARCH9, in producer cells. Infectivity assays were performed as described in D (mean ± S.D., n = 3 technical replicates). Black and gray columns represent the WT and RING-CH-mutants, respectively. Immunoblot images of the expression of HA-tagged MARCH proteins (WT and RING-CH mutants) and β-actin in the producer cells are shown at the bottom.
Results and discussion
To examine whether other MARCH family members that are expressed in HIV-1 target cells would inhibit HIV-1 infection, we newly created plasmids expressing family members of MARCH (MARCH1, MARCH2, MARCH3, MARCH4, MARCH5, MARCH6, MARCH7, and MARCH9) by PCR-amplifying their complementary DNAs. The protein expression in cells transfected with each plasmid was confirmed by immunoblotting using an anti-hemagglutinin (HA) antibody (Fig. 1B). Using these plasmids, subcellular localization of the MARCH proteins was determined by immunofluorescence (Fig. 1C). MARCH1 and MARCH2 showed the same localization pattern as observed for MARCH8, which was clearly localized to the plasma membrane. In contrast, MARCH3, MARCH5, and MARCH9 proteins were diffused throughout the cytoplasm, whereas MARCH4, MARCH6, and MARCH7 showed punctate distribution in the cytoplasm. These observations implied that MARCH1 and MARCH2 might have similar antiviral activity as MARCH8.
Next we examined whether MARCH family members would be able to inhibit HIV-1 infectivity. We prepared HIV-1 luciferase reporter viruses from 293T cells transfected with incrementally increasing doses of MARCH family plasmids and compared their viral infectivity with that of a control virus. MARCH1, MARCH2, and MARCH9, as well as MARCH8, showed inhibitory activity in a dose-dependent manner, whereas the overexpression of MARCH3, MARCH4, MARCH5, MARCH6, and MARCH7 in virus-producing cells did not influence viral infectivity (Fig. 1D). A similar inhibitory activity of these proteins was observed in vesicular stomatitis virus G-glycoprotein (VSV-G)–pseudotyped viruses (Fig. 1E).
We also tested whether the RING-CH domains, which are essential for ubiquitin ligase activity, are important for the antiviral activity of MARCH1, MARCH2, and MARCH9, as observed for MARCH8. We introduced a point mutation into a well-conserved and critical tryptophan in the RING-CH domains of MARCH1, MARCH2, and MARCH9 proteins (26), as carried out previously for MARCH8 (25) (Fig. 1F). Infectivity assays showed that the RING-CH domain was indeed important for the inhibitory activity of MARCH1 and MARCH2. In the case of MARCH9, however, the RING-CH mutant also showed dose-dependent inhibition of HIV-1 infection, exactly as observed in the WT (Fig. 1G). We therefore assume that inhibition by MARCH9 could be RING-CH–independent or simply due to the overexpression artifact.
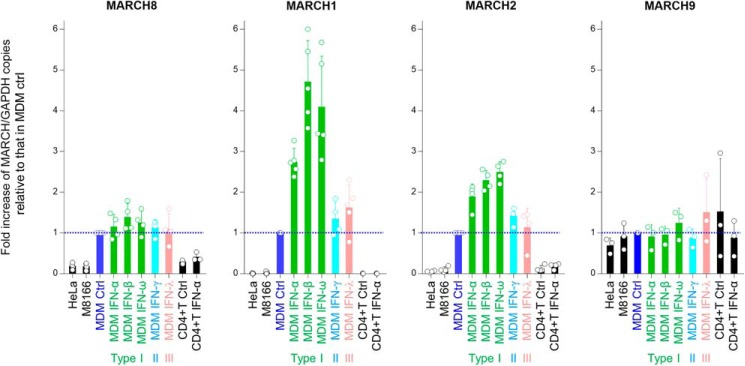
We next examined the endogenous expression levels of antiviral MARCH family members in different cell types by performing real-time RT-PCR (Fig. 2). As we reported previously, the expression of MARCH8 mRNA was higher in monocyte-derived macrophages (MDMs) and was only slightly up-regulated by interferons. MARCH1 and MARCH2 were also highly expressed in MDMs compared with transformed cell lines, and importantly, the expression of these two genes in MDMs was highly induced by type I interferons (α, β, and ω), especially in the case of MARCH1. In striking contrast, the levels of MARCH9 mRNA in MDMs with or without interferon treatment were nearly identical to those in cell lines, suggesting that expression of MARCH9 is not specific for primary cells and is not inducible by interferons. We therefore decided to focus on the function of MARCH1 and MARCH2, but not MARCH9, for the following experiments.
Figure 2.
Endogenous expression of MARCH genes in transformed human cell lines (HeLa cells and M8166 CD4-positive T cells) and human primary cells (stimulated CD4-positive T cells and MDMs in the presence or absence of IFN). The levels of MARCH mRNA were determined by real-time RT-PCR and normalized to those of GAPDH mRNA. Data are presented as the -fold increase of MARCH/GAPDH expression relative to that in control (Ctrl) MDMs (whose levels are shown by blue dotted lines) (mean ± S.D. from at least three independent experiments).
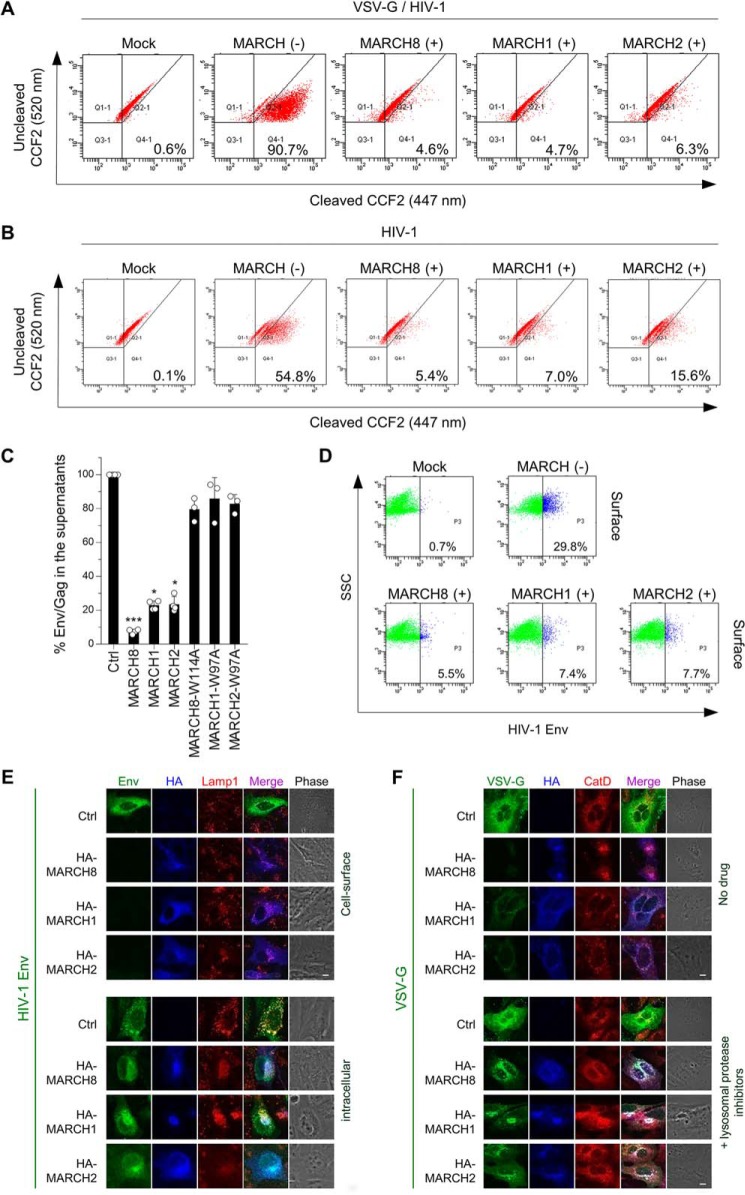
We next addressed whether the reduction in viral infectivity by MARCH1 and MARCH2 expression in the virus-producing cells was due to reduced viral entry, as observed previously in MARCH8 expression (25). β-lactamase (BlaM)–fused viral protein R (Vpr)–based entry assays indeed showed efficient inhibition of the entry of virions produced by cells expressing MARCH1 or MARCH2 proteins as well as MARCH8-expressing cells (Fig. 3, A and B). Furthermore, we pursued the possibility that the inhibition of viral entry by MARCH1 and MARCH2 expression might result from reduced virion incorporation of Env by these proteins, as observed previously in MARCH8 expression. An HIV-1 Env gp120 antigen capture ELISA showed that MARCH1 and MARCH2 decreased Env levels in viral supernatants whereas their RING-CH mutants did not (Fig. 3C).
Figure 3.
A and B, MARCH1, MARCH2, and MARCH8 reduce viral entry efficiency by blocking virion incorporation of envelope glycoproteins. Shown are BlaM-Vpr–based viral entry assays using VSV-G–pseudotyped viruses (A) or NL4–3 whole viruses (B) produced from cells expressing MARCH proteins or the vector control. Representative FACS dot plots from four independent experiments are shown. C, expression of MARCH1, MARCH2, and MARCH8 in producer cells decreases HIV-1 gp120 levels in viral supernatants. Shown are ELISA-based levels of Env gp120 in viral supernatants from 293T cells cotransfected with luc reporter proviral DNA and the NL-Env plasmid together with the MARCH expression plasmid. Data are shown as the percent of gp120 Env/p24Gag in the supernatants relative to that from control (Ctrl) cells (mean ± S.D. from at least three independent experiments). *, p < 0.05; ***, p < 0.005 compared with the control vector using one-way analysis of variance and Dunn's multiple comparisons test. D–F, MARCH1, MARCH2, and MARCH8 down-regulate envelope glycoproteins on the cell surface. Shown are FACS-based (D) and immunofluorescence-based (E and F) analyses of cell surface (D and E, top) and intracellular levels of HIV-Env (E, bottom) and VSV-G (F) with or without MARCH proteins in transfected HOS cells. Notably, for cell surface staining of HIV-1 Env (E, top), MARCH proteins were stained after permeabilization. VSV-G was rescued from MARCH8-induced degradation in the presence of lysosomal protease inhibitors. CatD, cathepsin D. Scale bars = 10 μm.
Flow cytometry experiments showed that the reduced virion incorporation of Env resulted from cell surface down-regulation of Env by MARCH1 and MARCH2 (Fig. 3D). The results were confirmed with immunofluorescence staining (Fig. 3, E and F); i.e. MARCH1 and MARCH2 blocked cell surface expression but not intracellular expression of HIV-1 Env, whereas both MARCH proteins blocked intracellular VSV-G expression. Treatment with lysosomal inhibitors rescued the expression of VSV-G from degradation by MARCH1 and MARCH2. Thus, we conclude that MARCH1 and MARCH2 down-regulate HIV-1 Env from the cell surface without degradation, whereas these host proteins lysosomally degrade VSV-G.
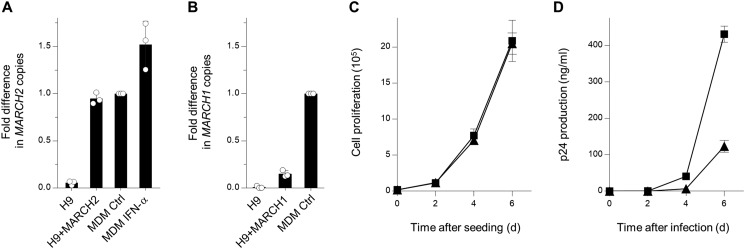
Finally, we performed replication assays to investigate whether MARCH1 and MARCH2 have an inhibitory effect on multiple rounds of HIV-1 replication. We first established H9 CD4-positive human T lymphoblast cell lines stably expressing MARCH1 or MARCH2. Real-time RT-PCR using the total RNA extracted from these cells showed that the levels of MARCH2 (Fig. 4A), but not MARCH1 (Fig. 4B), in the stable cell lines were nearly comparable with those in MDMs. Also, higher expression of MARCH2 did not affect cell proliferation (Fig. 4C). We therefore decided to use stable H9 cells expressing MARCH2 or control cells for infection by HIV-1 NL4-3 viruses. The supernatants were harvested every 2 days, and p24 antigen capture ELISA was performed to analyze the viral replication kinetics. As expected, viral growth in H9 cells stably expressing MARCH2 was slower than in control cells (Fig. 4D). Therefore, we speculated that interferon-induced levels of expression, of at least MARCH2, in MDMs contributed to the inhibition of HIV-1 replication.
Figure 4.
A, CD4-positive H9 cells were transfected with a MARCH2 expression plasmid carrying a neomycin-resistant gene and selected with G418; the resultant cells were cloned (H9+MARCH2). Cells were analyzed by real-time RT-PCR for MARCH2 expression. Note that the level of MARCH2 expressed in H9+MARCH2 is comparable with that endogenously expressed in MDMs. Data are shown as -fold difference in MARCH2 copies compared with those in the MDM control (mean ± S.D. from three independent experiments). B, MARCH1 stably expressed in H9 cells does not reach expression levels as those observed in MDMs. H9 cells transfected with MARCH1 expression plasmid were analyzed as shown in A. C, cell proliferation of control (squares) and MARCH2-expressing (triangles) H9 cells. The assay was started with 20,000 cells (mean ± S.D. from three independent experiments). D, multiple rounds of virus replication in H9 cells. Control H9 (squares) or H9+MARCH2 (triangles) cells (1 × 105) were infected with 0.1 ng of p24 antigen of NL4–3 viruses. Supernatants were harvested at the indicated times, and virus replication was monitored using p24 ELISA. The data shown are representative of three independent experiments (mean ± S.D. of three technical replicates). d, days.
In this study, we showed that the antiviral activity of the MARCH1 and MARCH2 proteins was similar to that observed previously for MARCH8 (25). Both MARCH1 and MARCH2 can down-regulate HIV-1 Env glycoproteins from the cell surface, thereby inhibiting virion incorporation of Env and leading to decreased entry efficiency, which results in the reduction of viral infectivity, exactly as MARCH8. The only difference is that expression of MARCH1 and MARCH2 is induced in MDMs by type I interferons, to which MARCH8 is insensitive. This caused experimental difficulty when proving the actual antiviral activity of MARCH1 and MARCH2 proteins in MDMs because treatment of MDMs with type I interferons up-regulates hundreds of different interferon-stimulated genes, making it impossible to distinguish specific protein activities. Instead, we established the CD4-positive T cell line H9, which stably expressed MARCH2 at levels similar to those in type I interferon–treated MDMs. Using these stable cells, we successfully observed that viral replication was indeed hampered by MARCH2 expression. Notably, MARCH1 stably expressed in H9 cells could not reach similar expression levels as those found in MDMs despite repeated attempts. This is probably because the highly AU-rich MARCH1 mRNA might be unstable when exogenously expressed in transformed cells, as reported for IL-8 mRNA (27, 28). Because our preliminary data suggest that MARCH8 can interact with both HIV-1 Env and VSV-G through TM interactions, we assume that especially MARCH1, whose TM domain is highly homologous to that of MARCH8, may also bind to these envelope glycoproteins, leading to cell surface down-regulation of these viral proteins. This needs to be elucidated with further experiments. Thus, we postulate that HIV-1 might rather take advantage of preferential expression of MARCH1 and MARCH2 as well as MARCH8 in macrophages that could serve as long-term viral reservoirs in vivo because cell surface down-regulation of HIV-1 Env by these MARCH proteins in macrophages could be beneficial for viruses in that they may have more chances to escape from immunosurveillance. Besides, HIV-1 is able to avoid stimulating type I interferon production by evading innate immune recognition (29) so that viruses could maintain suboptimal expression levels of interferon-induced proteins, including MARCH1 and MARCH2. Zhang et al. (30) recently reported that MARCH2 is up-regulated by HIV-1 infection and reduces viral production without being incorporated into virus particles. We were, however, unable to observe the same phenomenon (Fig. 5, A–C). This could be due to different experimental conditions. Overall, our present findings add both MARCH1 and MARCH2 to a growing list of host antiviral factors. Further investigations that probe whether the inhibitory activity of these MARCH family members against viral envelope glycoproteins could be linked to their ability to down-regulate cellular transmembrane proteins will help us to understand the molecular basis of the host defense mechanisms of these proteins.
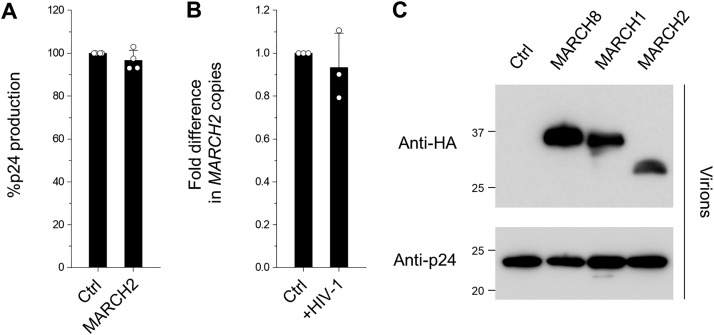
Figure 5.
A, MARCH2 does not reduce HIV-1 virion production. Data from four independent experiments are shown as a percentage of the production of viruses from 293T cells cotransfected with Env-defective HIV-1 luc reporter proviral DNA and either a control (Ctrl) or MARCH2 (+MARCH2) plasmid together with an HIV-1 Env plasmid. B, HIV-1 infection does not up-regulate MARCH2 expression. The levels of MARCH2 expression in the CD4-positive T cell line H9 (control and HIV-1 acutely infected) were determined by real-time RT-PCR and normalized to those of RPL27 mRNA. Data are presented as the -fold difference in MARCH2 copies (mean ± S.D. from three independent experiments). C, MARCH1, MARCH2, and MARCH8 are incorporated into HIV-1 particles. Virus particles were concentrated through a sucrose cushion by ultracentrifugation of the supernatants of virus-producing cells, and pelleted virions were treated with subtilisin. After the reaction was stopped with phenylmethylsulfonyl fluoride, virions were repelleted and subjected to immunoblot analyses using antibodies against HA (MARCH proteins) and p24.
Experimental procedures
DNA constructs
The Env-deficient HIV-1 proviral indicator construct pNL-Luc2-E(−), the HIV-1 Gag-Pol expression plasmid pC-GagPol-RRE, the HIV-1 Env expression vector pC-NLenv, the HIV-1 Rev expression plasmid pCa-Rev, the HIV-1 Tat expression plasmid pLTR-Tat, the lentiviral packaging vector pMDLg/pRRE, the GFP expression plasmid pCa-EGFP, the Vpr/BlaM expression plasmid pMM310, the MARCH8 expression plasmid pC-HA-MARCH8 and its RING-CH mutant pC-HA-MARCH8-W114A, and the neomycin-resistant version of the MARCH8 expression plasmid pCN-MARCH8 have been described previously (25). The N-terminally HA-tagged human MARCH family expression plasmids were created by inserting XhoI-NotI fragments of MARCH family members (except for an SalI-NotI fragment of MARCH6) into XhoI-NotI–digested pCAGGS-NHA. Each fragment was obtained as follows. MARCH1, MARCH2, MARCH3, MARCH4, MARCH5, and MARCH9 were PCR-amplified from MARCH1-Myc, MARCH2-Myc, MARCH3-Myc (22), pFLCI-MARCH4 (DNAFORM ID H023093F24), NKP66 pEGFP-N2 (31) (obtained from Eric Schirmer through Addgene, plasmid 62039), and pFLCIII-Fm-MARCH9 (DNAFORM ID H05D095M05) plasmids, respectively. MARCH6 and MARCH7 were RT-PCR–amplified from total RNA extracted from M8166 cells. The RING-CH mutants of MARCH1 and MARCH2 (W97A), in which tryptophan at position 97 was mutated to alanine, and of MARCH9 (W143A), in which tryptophan at position 143 was mutated to alanine, were created by inserting overlapping PCR fragments into XhoI/NotI-digested pCAGGS-NHA, designated pC-HA-MARCH1-W97A, pC-HA-MARCH2-W97A, and pC-HA-MARCH9-W143A, respectively. A neomycin-resistant version of the MARCH1 and MARCH2 expression plasmids (pCN-MARCH1 and pCN-MARCH2, respectively) were generated by replacing the MARCH8 gene of pCN-MARCH8 with KpnI/XhoI fragments of MARCH1 and MARCH2. All constructs were verified by the DNA sequencing service FASMAC.
Cell maintenance and stable cell line establishment
The cell lines 293T, M8166, MT4, HeLa, MAGIC5 (a HeLa derivative), and HOS were maintained under standard conditions. Cells were originally obtained from the ATCC (except for MAGIC5 cells) and routinely tested negative for mycoplasma contamination (PCR Mycoplasma Detection Set, Takara). To establish T cell lines stably expressing MARCH1 or MARCH2, H9 cells (1 × 106) were transfected with 1 μg of either pCN-MARCH1, pCN-MARCH2, or a control vector using the 4D-Nucleofector system (Lonza) according to the manufacturer's instructions. After 48 h, the cells were maintained under G418 selection (1 mg ml−1) for 14 days. The resultant G418-resistant H9 cells stably expressing MARCH1, MARCH2, or a vector control were subjected to RNA extraction, followed by real-time RT-PCR, as described below. Additionally, H9 cells were assayed every 2 days for cell proliferation by cell counting after trypan blue exclusion.
Virion production and infectivity assays
To prepare VSV-G–pseudotyped or HIV-1–Env–pseudotyped luciferase reporter viruses, 2.5 × 105 293T cells were cotransfected with 120 ng of the MARCH expression plasmid (WT or RING-CH mutants), either 20 ng of pC-VSVg or pC-NLenv, 500 ng of pNL-Luc2-E(−), and an empty vector up to 1 μg of total DNA using FuGENE 6. Sixteen hours later, the cells were washed with PBS, and 1 ml of fresh complete medium was added. After 24 h, the supernatants were treated as described above and then harvested. The p24 antigen levels in the viral supernatants were measured by HIV-1 p24 antigen capture ELISA. The transfection efficiencies were normalized with the luciferase activity. To determine viral infectivity, 1 × 104 MAGIC5 cells were incubated with 1 ng of p24 antigen from the HIV-1 supernatants. After 48 h, the cells were lysed in 100 μl of One-Glo luciferase assay reagent (Promega). Firefly luciferase activity was determined with a Centro LB960 (Berthold) luminometer.
Immunoblot assays
Cells transfected as descried above were lysed in 500 μl of lysis buffer (20 mm Tris (pH 8.0), 100 mm NaCl, 1 mm EDTA, 1.25% n-octyl-β-d-glucoside, and Complete protease inhibitor mixture (Roche Applied Science)). The cell extracts were then subjected to gel electrophoresis and transferred to a nitrocellulose membrane. The membranes were probed with an anti-HA mouse mAb (Sigma-Aldrich, H9658) or an anti-β-actin mouse mAb (Sigma-Aldrich, A5316). Reacted proteins were visualized by chemiluminescence using an ECL Western blotting detection system (GE Healthcare) and monitored using a LAS-3000 imaging system (FujiFilm).
Primary cell culture
All experiments using human samples were approved by the Medical Research Ethics Committee of the National Institute of Infectious Diseases, Japan (approval number 887), and the approved studies abided by the Declaration of Helsinki principles. Peripheral blood mononuclear cells (PBMCs) were obtained from healthy volunteer donors who signed an informed consent form. Briefly, PBMCs were isolated by Ficoll-Hypaque gradient centrifugation. The monocytes were then isolated from the PBMCs by positive selection for CD14 using CD14 microbeads (Miltenyi Biotec) and plated at 2.5 × 105 cells/well in 48-well tissue culture plates. Cells were then cultured in Dulbecco's modified Eagle's medium (Gibco BRL) supplemented with 10% FBS (Sigma-Aldrich) in the presence of 1,000 units ml−1 of macrophage colony-stimulating factor (R&D Systems) for 1 week to allow differentiation into MDMs. The cells were fed every 2 days by replacing half of the cell culture fluid with fresh medium. MDMs were then cultured in the presence or absence of 500 units ml−1 each of interferon (IFN)-α (Sigma-Aldrich), IFNβ (Wako), IFNγ (Wako), IFNλ (Peprotech), or IFNω (Wako) overnight. The total RNA was extracted from these MDMs and subjected to real-time RT-PCR as described below. The CD14-negative population was purified using a Dynabeads CD4-positive isolation kit (Invitrogen) to obtain CD4-positive T cells, and purified cells were cultured in the presence of 3 μg ml−1 phytohemagglutinin (Sigma-Aldrich) and 10 units ml−1 IL-2 (Peprotech) for 72 h.
Real-time RT-PCR
To measure the levels of MARCH gene expression, total RNA was extracted from transformed cells (HeLa, M8166, and stable H9 cells expressing either MARCH1 or MARCH2) and interferon-treated or nontreated primary cells (MDMs and stimulated CD4-positive T cells) derived from a healthy donor using a Reliaprep RNA cell Miniprep system (Promega). Real-time RT-PCR was performed with a Mx3005P (Stratagene) using the KAPA SYBR FAST One-Step qRT-PCR Kit (Kapa Biosystems) according to the manufacturer's instructions. The specific oligonucleotides used were as follows: MARCH8, 5′-CTCTCGCACTTCTATCACGCCA-3′/5′-AAGTGGAGGCTTCCTGTGCAGT-3′; MARCH1, 5′-GCAGCCACGTTTGTTGTAAT-3′/5′-GAGCTGTCCCTGTTGTTGG-3′; MARCH2, 5′-CTAACACCAGCTACTGCGAGCT-3′/5′-GGAAACACACCATGTCGCAGCA-3′; MARCH9, 5′-CTTCAGCCAAGTGGCAACGACA-3′/5′-GAAGATGCGGTAGACAGAGGAG-3′; glyceraldehyde-3-phosphate-dehydrogenase (GAPDH), 5′-ACCAGGTGGTCTCCTCTGAC-3′/5′-TGTAGCCAAATTCGTTGTCATACC-3′; and ribosomal protein L27 (RPL27), 5′-ATCGCCAAGAGATCAAAGATAA-3′/5′-TCTGAAGACATCCTTATTGACG-3′. The mRNA levels of MARCH8, MARCH1, MARCH2, and MARCH9 were normalized with either GAPDH or RPL27 mRNA levels.
Viral entry assays
HIV-1 particles incorporating a fusion protein between Vpr and the BlaM reporter protein (BlaM-Vpr) were produced by co-transfection of 293T cells with pNL4-3, pMM310 encoding BlaM-Vpr (provided by M. D. Miller), and either pC-MARCH8, pC-MARCH1, pC-MARCH2, or the control vector. Similarly, VSV-G–pseudotyped HIV-1 particles incorporating BlaM-Vpr were prepared by co-transfection with pNL-Luc-E(−), pC-VSVg, pMM310, and the MARCH expression plasmids or the control vector. The produced viruses were quantified by p24 antigen capture ELISA, and a CD4-positive T cell line MT4 (5 × 105 cells) was incubated with 100 ng of p24 antigen of the viruses at 37 °C for 4 h to allow viral entry. After washing three times with Hanks' balanced salt solution (HBSS, Invitrogen), the cells were resuspended and loaded with 1 μm CCF2-AM dye (Invitrogen), a fluorescent substrate for BlaM, in HBSS containing 1 mg ml−1 Pluronic F-127 surfactant (Invitrogen) and 0.001% acetic acid for 1 h at room temperature and then washed twice with HBSS. The BlaM reaction, which corresponds to the cleavage of intracellular CCF2 dye by BlaM-Vpr, was developed for 14 h at room temperature in HBSS supplemented with 10% fetal bovine serum (FBS). The cells were washed three times with PBS and fixed in a 1.2% solution of paraformaldehyde. The fluorescence was monitored at 520 and 447 nm (to detect uncleaved and cleaved CCF2, respectively) by flow cytometry using a BD FACS Canto II (BD Biosciences), and the data were collected and analyzed with BD FACS Diva Software (BD Biosciences).
Env incorporation assays
Viral supernatants containing NL4-3–Env–pseudotyped viruses were harvested from 293T cells transfected as described above and subjected to HIV-1 gp120 antigen capture ELISA using a commercially available kit (Advanced BioScience Laboratories), according to the manufacturer's instructions.
Flow cytometry
293T cells (5 × 105) were cotransfected with 1.8 μg of either the control vector and 200 ng of pCa-EGFP or a combination of 120 ng of the MARCH expression plasmids, 20 ng of either pC-VSVg or pC-NLenv, 500 ng of pNL-Luc2-E(−), and the control vector up to 1 μg. After 48 h, the transfected cells were incubated with either an anti-HIV-1 gp120 goat polyclonal antibody (Abcam, ab21179) or an anti-VSV-G mouse mAb, followed by staining for 30 min on ice with either a rabbit anti-goat IgG conjugated with Alexa 647 (Molecular Probes, A21446) or a goat anti-mouse IgG conjugated with R-phycoerythrin (Molecular Probes, P-852), respectively. The cells were then washed extensively with PBS with 4% FBS and fixed with 4% formaldehyde in PBS. To analyze intracellular expression, the transfected cells were fixed with 0.01% formaldehyde in PBS, permeabilized with 0.05% saponin for 10 min, and immunostained with an antibody against either gp120 or VSV-G, followed by incubation with secondary antibodies as described above. GFP-positive cells were sorted and analyzed for the expression of HIV-1 Env or VSV-G by flow cytometry using a BD FACS Canto II, and the data were collected and analyzed with BD FACS Diva Software.
Immunofluorescence microscopy
HeLa or HOS cells were plated on collagen-coated 13-mm glass coverslips, cotransfected with the indicated plasmids, and cultured for 24 h before fixation. HeLa cells were transfected with 0.5 μg of HA-tagged MARCH expression plasmids using FuGENE6. Cells were fixed with 4% paraformaldehyde at room temperature for 30 min, permeabilized with 0.05% saponin for 10 min, and immunostained with the anti-HA mAb followed by Alexa 488 goat anti-mouse IgG (Molecular Probes, A-11001). All immunofluorescence images were observed using Fluoview FV1000-IX81 (Olympus). Alternatively, HOS cells were cotransfected with 0.2 μg of either pC-NLenv or pC-VSVg-T7E, 0.2 μg of pC-Gag-Pol, 0.1 μg of pCa-Rev, and 0.5 μg of the MARCH expression plasmids using FuGENE6. For the cell surface staining of gp120 protein, cells were incubated with the anti-gp120 goat polyclonal antibody at 4 °C for 5 min and washed with PBS at 4 °C before fixation. For intracellular staining, cells were fixed and permeabilized as described above, and then immunostained with the indicated antibodies as follows. For triple-staining the gp120, lamp1, and HA-MARCH proteins, the primary antibodies (the anti-gp120 goat polyclonal antibody; an anti-mouse lamp1 mAb, clone H4A3, a gift from Koh Furuta; and an anti-HA rabbit polyclonal antibody, Sigma-Aldrich, H6908) were detected with Alexa 488 donkey anti-goat IgG (Molecular Probes, A-11055), Alexa 568 donkey anti-mouse IgG (Molecular Probes, A-10037), and Alexa 647 donkey anti-rabbit IgG (Molecular Probes, A-31573), respectively. For triple-staining the VSV-G–T7, cathepsin D, and HA-MARCH proteins, the primary antibodies (anti-T7 epitope mouse mAb, Novagen, 69522-4; an anti-cathepsin D rabbit polyclonal antibody, DAKO, A561; and an anti-HA goat polyclonal antibody, GenScript, A00168-40) were detected with Alexa 488 donkey anti-mouse IgG (Molecular Probes, A-21202), Alexa 568 donkey anti-rabbit IgG (Molecular Probes, A-10042), and Alexa 647 donkey anti-goat IgG (Molecular Probes, A-21447), respectively. To investigate the involvement of lysosomal degradation in MARCH-induced down-regulation of VSV-G, the transfected cells were cultured for 14 h in complete medium in the presence of lysosomal protease inhibitors (40 μm leupeptin and pepstatin A, Peptide Institute Inc.). Transfected cells treated with the drugs were fixed with 4% paraformaldehyde and subjected to intracellular staining. All immunofluorescence images were observed using a Fluoview FV10i (Olympus).
Virus replication assays
To investigate viral replication, either H9 cells or H9+MARCH2 cells (1 × 105) were infected for 3 h with NL4-3 viruses (0.1 ng of p24 antigen), washed extensively with serum-free medium, and then cultured in fresh complete medium. Supernatants were sampled every 2 days, and p24 antigen production was quantified by ELISA.
Virion purification
To examine whether MARCH proteins could be specifically incorporated into virions, subtilisin treatment was carried out as described by Perez-Caballero et al. (32). Briefly, viruses were prepared as described above, and pelleted virions were treated with or without subtilisin (1 mg ml−1) in buffer (10 mm Tris/HCl (pH 8.0), 1 mm CaCl2, 150 mm NaCl) for 45 min at room temperature. Subtilisin treatment was then stopped using DMEM containing 10% fetal calf serum, 5 mm phenylmethylsulfonyl fluoride, and 20 mm EGTA. Virions were then pelleted through sucrose and subjected to immunoblot analysis.
Author contributions
Y. Z., T. T., H. F., and K. T. data curation; Y. Z., T. T., S. O., W. Y., M. T., H. F., and K. T. formal analysis; Y. Z., T. T., H. F., and K. T. validation; Y. Z., T. T., S. O., W. Y., M. T., H. F., and K. T. investigation; M. T., H. F., and K. T. visualization; S. Y., S. K., and K. T. resources; H. F. and K. T. methodology; K. T. conceptualization; K. T. supervision; K. T. funding acquisition; K. T. writing-original draft; K. T. project administration; K. T. writing-review and editing.
Acknowledgments
We thank F. Barre-Sinoussi and P. Bieniasz for helpful discussions, M. D. Miller (Merck) for pMM310, Eric Schirmer (University of Edinburgh) for NKP66 pEGFP-N2, and K. Furuta (National Cancer Center Research Institute) for H4A3.
This work was supported by grants from the Japan Agency for Medical Research and Development (AMED, Research Program on HIV/AIDS, 17fk0410308j0503) and the Japan Society for the Promotion of Science (KAKENHI, 18K07156) to K. T. The authors declare that they have no conflicts of interest with the contents of this article.
- MARCH
- membrane-associated RING-CH
- TM
- transmembrane
- MHC
- major histocompatibility complex
- IL
- interleukin
- HA
- hemagglutinin
- VSV-G
- vesicular stomatitis virus G-glycoprotein
- MDM
- monocyte-derived macrophage
- BlaM
- β-lactamase
- Vpr
- viral protein R
- HBSS
- Hanks' balanced salt solution
- FBS
- fetal bovine serum
- PBMC
- peripheral blood mononuclear cell
- IFN
- interferon
- GAPDH
- glyceraldehyde-3-phosphate-dehydrogenase.
References
- 1. Matsuki Y., Ohmura-Hoshino M., Goto E., Aoki M., Mito-Yoshida M., Uematsu M., Hasegawa T., Koseki H., Ohara O., Nakayama M., Toyooka K., Matsuoka K., Hotta H., Yamamoto A., and Ishido S. (2007) Novel regulation of MHC class II function in B cells. EMBO J. 26, 846–854 10.1038/sj.emboj.7601556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baravalle G., Park H., McSweeney M., Ohmura-Hoshino M., Matsuki Y., Ishido S., and Shin J. S. (2011) Ubiquitination of CD86 is a key mechanism in regulating antigen presentation by dendritic cells. J. Immunol. 187, 2966–2973 10.4049/jimmunol.1101643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morokuma Y., Nakamura N., Kato A., Notoya M., Yamamoto Y., Sakai Y., Fukuda H., Yamashina S., Hirata Y., and Hirose S. (2007) MARCH-XI, a novel transmembrane ubiquitin ligase implicated in ubiquitin-dependent protein sorting in developing spermatids. J. Biol. Chem. 282, 24806–24815 10.1074/jbc.M700414200 [DOI] [PubMed] [Google Scholar]
- 4. Iyengar P. V., Hirota T., Hirose S., and Nakamura N. (2011) Membrane-associated RING-CH 10 (MARCH10 protein) is a microtubule-associated E3 ubiquitin ligase of the spermatid flagella. J. Biol. Chem. 286, 39082–39090 10.1074/jbc.M111.256875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartee E., Mansouri M., Hovey Nerenberg B. T., Gouveia K., and Früh K. (2004) Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J. Virol. 78, 1109–1120 10.1128/JVI.78.3.1109-1120.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goto E., Ishido S., Sato Y., Ohgimoto S., Ohgimoto K., Nagano-Fujii M., and Hotta H. (2003) c-MIR, a human E3 ubiquitin ligase, is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar activity. J. Biol. Chem. 278, 14657–14668 10.1074/jbc.M211285200 [DOI] [PubMed] [Google Scholar]
- 7. Coscoy L., and Ganem D. (2000) Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. U.S.A. 97, 8051–8056 10.1073/pnas.140129797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stevenson P. G., Efstathiou S., Doherty P. C., and Lehner P. J. (2000) Inhibition of MHC class I-restricted antigen presentation by γ 2-herpesviruses. Proc. Natl. Acad. Sci. U.S.A. 97, 8455–8460 10.1073/pnas.150240097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ishido S., Wang C., Lee B. S., Cohen G. B., and Jung J. U. (2000) Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 74, 5300–5309 10.1128/JVI.74.11.5300-5309.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corcoran K., Jabbour M., Bhagwandin C., Deymier M. J., Theisen D. L., and Lybarger L. (2011) Ubiquitin-mediated regulation of CD86 protein expression by the ubiquitin ligase membrane-associated RING-CH-1 (MARCH1). J. Biol. Chem. 286, 37168–37180 10.1074/jbc.M110.204040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Gassart A., Camosseto V., Thibodeau J., Ceppi M., Catalan N., Pierre P., and Gatti E. (2008) MHC class II stabilization at the surface of human dendritic cells is the result of maturation-dependent MARCH I down-regulation. Proc. Natl. Acad. Sci. U.S.A. 105, 3491–3496 10.1073/pnas.0708874105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohmura-Hoshino M., Matsuki Y., Aoki M., Goto E., Mito M., Uematsu M., Kakiuchi T., Hotta H., and Ishido S. (2006) Inhibition of MHC class II expression and immune responses by c-MIR. J. Immunol. 177, 341–354 10.4049/jimmunol.177.1.341 [DOI] [PubMed] [Google Scholar]
- 13. Young L. J., Wilson N. S., Schnorrer P., Proietto A., ten Broeke T., Matsuki Y., Mount A. M., Belz G. T., O'Keeffe M., Ohmura-Hoshino M., Ishido S., Stoorvogel W., Heath W. R., Shortman K., and Villadangos J. A. (2008) Differential MHC class II synthesis and ubiquitination confers distinct antigen-presenting properties on conventional and plasmacytoid dendritic cells. Nat. Immunol. 9, 1244–1252 10.1038/ni.1665 [DOI] [PubMed] [Google Scholar]
- 14. Jahnke M., Trowsdale J., and Kelly A. P. (2012) Structural requirements for recognition of major histocompatibility complex class II by membrane-associated RING-CH (MARCH) protein E3 ligases. J. Biol. Chem. 287, 28779–28789 10.1074/jbc.M112.381541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tze L. E., Horikawa K., Domaschenz H., Howard D. R., Roots C. M., Rigby R. J., Way D. A., Ohmura-Hoshino M., Ishido S., Andoniou C. E., Degli-Esposti M. A., and Goodnow C. C. (2011) CD83 increases MHC II and CD86 on dendritic cells by opposing IL-10-driven MARCH1-mediated ubiquitination and degradation. J. Exp. Med. 208, 149–165 10.1084/jem.20092203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lapaque N., Jahnke M., Trowsdale J., and Kelly A. P. (2009) The HLA-DRα chain is modified by polyubiquitination. J. Biol. Chem. 284, 7007–7016 10.1074/jbc.M805736200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bartee E., McCormack A., and Früh K. (2006) Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2, e107 10.1371/journal.ppat.0020107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nice T. J., Deng W., Coscoy L., and Raulet D. H. (2010) Stress-regulated targeting of the NKG2D ligand Mult1 by a membrane-associated RING-CH family E3 ligase. J. Immunol. 185, 5369–5376 10.4049/jimmunol.1000247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bartee E., Eyster C. A., Viswanathan K., Mansouri M., Donaldson J. G., and Früh K. (2010) Membrane-associated RING-CH proteins associate with Bap31 and target CD81 and CD44 to lysosomes. PLoS ONE 5, e15132 10.1371/journal.pone.0015132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoer S., Smith L., and Lehner P. J. (2007) MARCH-IX mediates ubiquitination and downregulation of ICAM-1. FEBS Lett. 581, 45–51 10.1016/j.febslet.2006.11.075 [DOI] [PubMed] [Google Scholar]
- 21. van de Kooij B., Verbrugge I., de Vries E., Gijsen M., Montserrat V., Maas C., Neefjes J., and Borst J. (2013) Ubiquitination by the membrane-associated RING-CH-8 (MARCH-8) ligase controls steady-state cell surface expression of tumor necrosis factor-related apoptosis inducing ligand (TRAIL) receptor 1. J. Biol. Chem. 288, 6617–6628 10.1074/jbc.M112.448209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujita H., Iwabu Y., Tokunaga K., and Tanaka Y. (2013) Membrane-associated RING-CH (MARCH) 8 mediates the ubiquitination and lysosomal degradation of the transferrin receptor. J. Cell Sci. 126, 2798–2809 10.1242/jcs.119909 [DOI] [PubMed] [Google Scholar]
- 23. Chen R., Li M., Zhang Y., Zhou Q., and Shu H. B. (2012) The E3 ubiquitin ligase MARCH8 negatively regulates IL-1β-induced NF-κB activation by targeting the IL1RAP coreceptor for ubiquitination and degradation. Proc. Natl. Acad. Sci. U.S.A. 109, 14128–14133 10.1073/pnas.1205246109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eyster C. A., Cole N. B., Petersen S., Viswanathan K., Früh K., and Donaldson J. G. (2011) MARCH ubiquitin ligases alter the itinerary of clathrin-independent cargo from recycling to degradation. Mol. Biol. Cell 22, 3218–3230 10.1091/mbc.e10-11-0874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tada T., Zhang Y., Koyama T., Tobiume M., Tsunetsugu-Yokota Y., Yamaoka S., Fujita H., and Tokunaga K. (2015) MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins. Nat. Med. 21, 1502–1507 10.1038/nm.3956 [DOI] [PubMed] [Google Scholar]
- 26. Dodd R. B., Allen M. D., Brown S. E., Sanderson C. M., Duncan L. M., Lehner P. J., Bycroft M., and Read R. J. (2004) Solution structure of the Kaposi's sarcoma-associated Herpesvirus K3 N-terminal domain reveals a novel E2-binding C4HC3-type RING domain. J. Biol. Chem. 279, 53840–53847 10.1074/jbc.M409662200 [DOI] [PubMed] [Google Scholar]
- 27. Mahmoud L., Al-Enezi F., Al-Saif M., Warsy A., Khabar K. S., and Hitti E. G. (2014) Sustained stabilization of interleukin-8 mRNA in human macrophages. RNA Biol. 11, 124–133 10.4161/rna.27863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Winzen R., Gowrishankar G., Bollig F., Redich N., Resch K., and Holtmann H. (2004) Distinct domains of AU-rich elements exert different functions in mRNA destabilization and stabilization by p38 mitogen-activated protein kinase or HuR. Mol. Cell. Biol. 24, 4835–4847 10.1128/MCB.24.11.4835-4847.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rasaiyaah J., Tan C. P., Fletcher A. J., Price A. J., Blondeau C., Hilditch L., Jacques D. A., Selwood D. L., James L. C., Noursadeghi M., and Towers G. J. (2013) HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503, 402–405 10.1038/nature12769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y., Lu J., and Liu X. (2018) MARCH2 is upregulated in HIV-1 infection and inhibits HIV-1 production through envelope protein translocation or degradation. Virology 518, 293–300 10.1016/j.virol.2018.02.003 [DOI] [PubMed] [Google Scholar]
- 31. Korfali N., Srsen V., Waterfall M., Batrakou D. G., Pekovic V., Hutchison C. J., and Schirmer E. C. (2011) A flow cytometry-based screen of nuclear envelope transmembrane proteins identifies NET4/Tmem53 as involved in stress-dependent cell cycle withdrawal. PLoS ONE 6, e18762 10.1371/journal.pone.0018762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perez-Caballero D., Zang T., Ebrahimi A., McNatt M. W., Gregory D. A., Johnson M. C., and Bieniasz P. D. (2009) Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139, 499–511 10.1016/j.cell.2009.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]