Abstract
During posttranslational translocation in Escherichia coli, polypeptide substrates are driven across the membrane through the SecYEG protein–conducting channel using the ATPase SecA, which binds to SecYEG and couples nucleotide hydrolysis to polypeptide movement. Recent studies suggest that SecA is a highly dynamic enzyme, able to repeatedly bind and dissociate from SecYEG during substrate translocation, but other studies indicate that these dynamics, here referred to as “SecA processivity,” are not a requirement for transport. We employ a SecA mutant (PrlD23) that associates more tightly to membranes than WT SecA, in addition to a SecA–SecYEG cross-linked complex, to demonstrate that SecA–SecYEG binding and dissociation events are important for efficient transport of the periplasmic protein proPhoA. Strikingly however, we find that transport of the precursor of the outer membrane protein proOmpA does not depend on SecA processivity. By exchanging signal sequence and protein domains of similar size between PhoA and OmpA, we find that SecA processivity is not influenced by the sequence of the protein substrate. In contrast, using an extended proOmpA variant and a truncated derivative of proPhoA, we show that SecA processivity is affected by substrate length. These findings underscore the importance of the dynamic nature of SecA–SecYEG interactions as a function of the preprotein substrate, features that have not yet been reported using other biophysical or in vivo methods.
Keywords: protein translocation, membrane transport, protein–protein interaction, protein dynamic, lipid–protein interaction, ATPase, Sec translocon
Introduction
In Escherichia coli, many periplasmic and extracellular proteins are transported across the inner membrane through the heterotrimeric SecYEG protein–conducting channel. Proteins can be transported either co-translationally or posttranslationally (1–4). During posttranslational translocation, the polypeptide substrate is synthesized by ribosomes before being directed toward SecYEG. The energy for protein translocation is then provided by the cytosolic ATPase SecA, which binds to the SecYEG channel and drives protein export through a series of ATP-dependent conformational changes (1, 2, 5–8). The proton motive force across the inner membrane also contributes to driving protein translocation, particularly at the later stages of the transport reaction (3, 9, 10).
A landmark previous study reported that ATP binding to SecA triggers a “power stroke,” resulting in forward movement of a polypeptide segment into the mouth of the SecYEG channel (11). Following ATP hydrolysis, when ADP is still bound to the enzyme, SecA adopts a conformation allowing the polypeptide to slide across the SecYEG channel until the next ATP-binding event (11, 12). In this model, SecA exists mainly in an ADP-bound state when translocation is taking place (11, 13). More recently, elegant FRET-based studies led to an alternative model in which protein translocation does not require a power stroke (14). Instead, SecA rather operates as a “Brownian ratchet,” allowing substrates to passively diffuse within the SecY channel while the enzyme is in an ADP-state. When a steric blockage is reached because of a stretch of bulky or aromatic residue within the substrate, ATP binding to SecA leads to temporary widening of the SecY channel, thus allowing the blockage to pass (14, 15). Both of these models, in which transport largely relies on passive sliding of substrates through the SecY channel without specific sequence recognition, explains why the translocon is able to handle such a wide variety of proteins, each with a different sequence (8, 11, 14, 15).
Despite these recent insights, some aspects of SecA–SecYEG interactions during the translocation reaction remain unclear. Studies by Morita et al. (5) and Mao et al. (16) suggest that SecA is a highly dynamic enzyme that must undergo successive cycles of binding and dissociation from the SecYEG complex for translocation to proceed efficiently. In both of these studies, SecA is hypothesized to dissociate from SecYEG while a polypeptide substrate is undergoing transit across the membrane (5, 16). Importantly, dissociation of SecA does not dislodge the substrate from SecYEG. Instead, a fresh copy of cytosolic SecA binds to SecYEG and completes translocation of the substrate (5, 16). Hereafter we refer to these cycles of SecA binding to and dissociating from SecYEG as “SecA processivity” (11). Other studies, however, have reported that SecA processivity is not essential for efficient protein translocation. Whitehouse et al. (17) and Gold et al. (18) showed that immobilization of SecA onto SecYEG via cysteine cross-linking results in the formation of a SecA–SecYEG complex that is still competent for translocation of proOmpA. Furthermore, a fusion construct of SecA–SecYEG also is able to catalyze translocation of proOmpA (19). Given the two differing views reported in the literature, the importance of SecA processivity warrants further investigation.
Here we used a SecA mutant, PrlD23 (SecA Y134S), that is less processive than WT SecA (20). The mutant exhibits elevated ATPase activity and associates more tightly with bacterial membranes and with SecYEG than WT SecA (20, 21). We reconstituted purified SecYEG with SecA or PrlD23 into proteoliposomes and assessed the in vitro translocation efficiency of proOmpA and proPhoA. As a complementary approach, we assessed translocation efficiency using a covalently cross-linked SecA–SecYEG complex. Interestingly, we observed an effect of SecA processivity on proPhoA translocation but not on proOmpA translocation. To determine which features of the two substrates lead to this difference, we generated a series of chimeric substrates by exchanging protein domains between proPhoA and proOmpA. Our data indicate that SecA processivity is mainly influenced by the length of the preprotein substrate.
Results
Effect of a SecA mutation that stabilizes SecA–SecYEG interactions
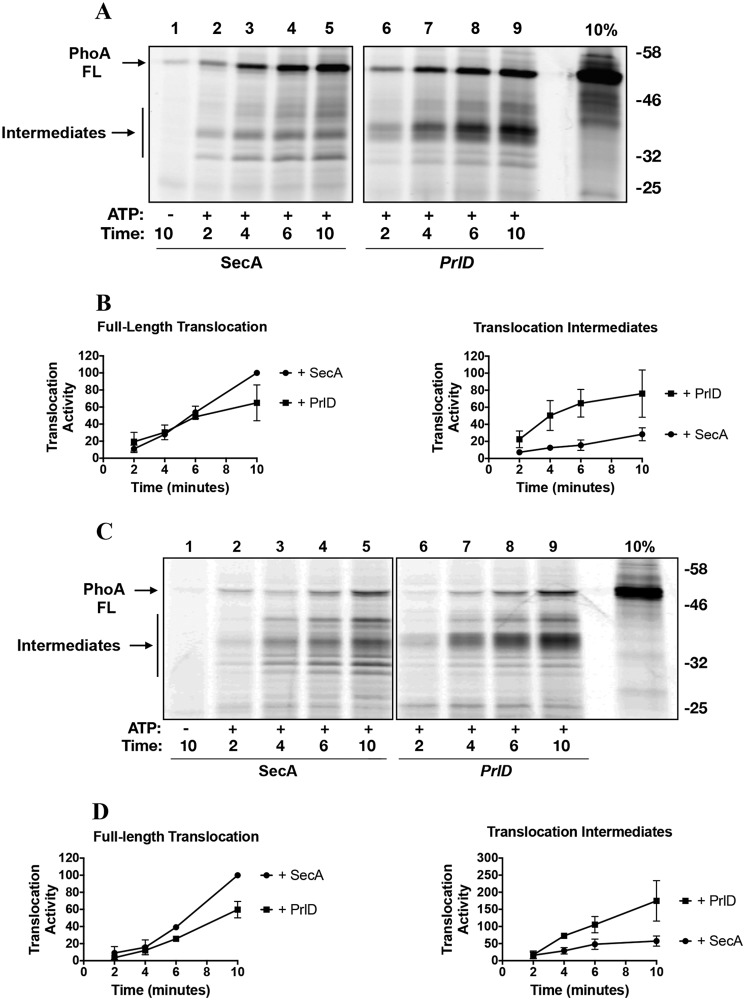
We reconstituted purified E. coli SecYEG into extruded 100-nm E. coli liposomes as described under “Experimental procedures.” Translocation assays were performed in the presence of ATP, fluorescently labeled proPhoA, and either WT or PrlD SecA. The data are shown in Fig. 1. With the PrlD mutant, a reproducible transport defect is observed; the amount of fully transported substrate measured in the presence of the PrlD mutant is only 60% of the amount observed with WT SecA. Additionally, there is a greater accumulation of translocation intermediates (partially transported substrate fragments) in the presence of the PrlD mutant. The apparent molecular mass of these intermediates is ∼35 kDa. Although some intermediates are formed in the presence of WT SecA, these are far less prominent and do not form a discrete band, in contrast to the intermediates seen with the PrlD mutant (Fig. 1A, quantified in Fig. 1B). Similar results were obtained when we performed translocation assays using IMVs2 containing overexpressed SecYEG instead of proteoliposomes (Fig. 1C, quantified in Fig. 1D). These data led us to hypothesize that the PrlD mutation stabilizes SecA–SecY interactions at the membrane surface, thereby reducing SecA processivity and, as a consequence, increasing formation of translocation intermediates while reducing full translocation.
Figure 1.
Translocation activity of PrlD23 with the substrate PhoA. A, translocation assays were performed using SecYEG proteoliposomes under conditions as described under “Experimental procedures.” Fluorescently labeled PhoA (10% of the total input for one reaction) was loaded onto the gel as a standard. Positions of fully translocated substrate (FL) and of the translocation intermediates (Intermediates) are indicated with arrows. B, densitometry analysis of A. Data were normalized to 100 units based on the densitometry obtained for fully translocated PhoA with WT SecA after 10 min at 30 °C (lane 5). The data represents the mean ± S.D. from three independent experiments. C, translocation assays were performed as in A but using IMVs containing overexpressed SecYEG. D, densitometry analysis of C.
PrlD is able to outcompete WT SecA at SecYEG
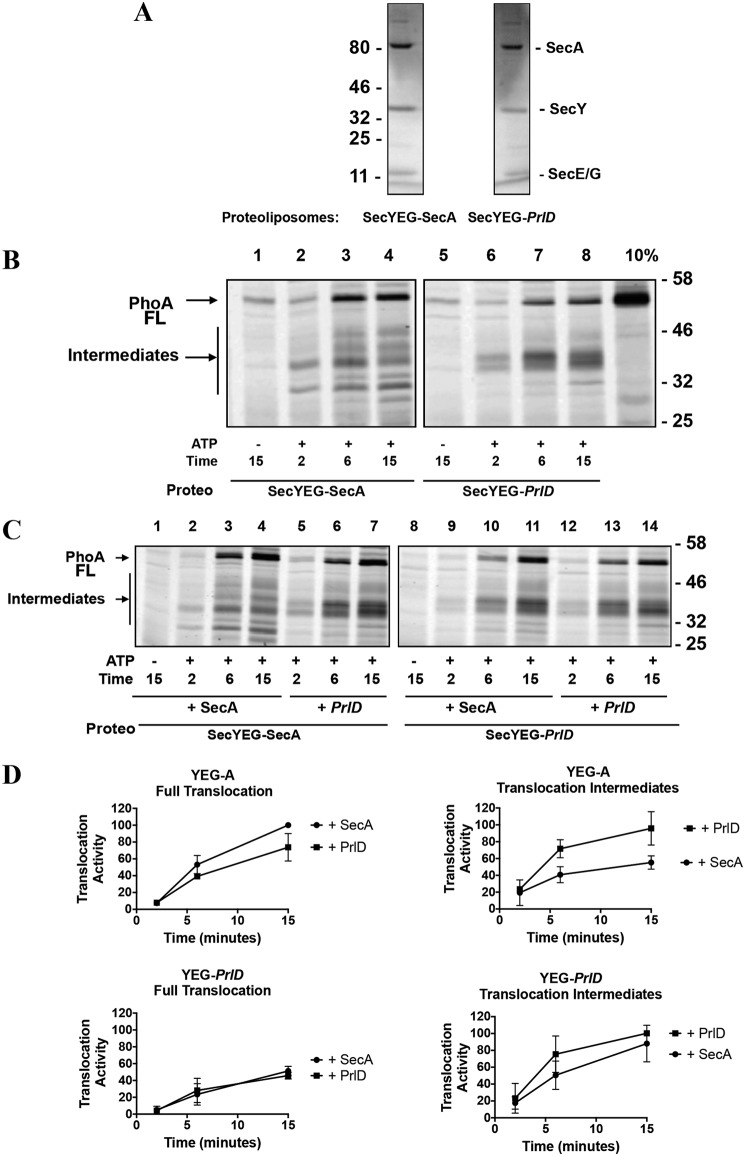
We next assessed the relative stabilities of SecYEG–SecA and SecYEG–PrlD complexes using a competition assay. We reconstituted preformed SecYEG–SecA and SecYEG–PrlD complexes into proteoliposomes as described under “Experimental procedures.” The reconstitution efficiency of both preparations was comparable, as assessed by SDS-PAGE (Fig. 2A). We also verified that SecA and SecYEG remained in complex during the co-reconstitution process by measuring the translocation activity without additional SecA. As seen earlier (Fig. 1), the amount of the 35-kDa translocation intermediates formed with PrlD SecA (Fig. 2B, lanes 5–8) is appreciably higher than the amount formed with the WT enzyme (Fig. 2B, lanes 1–4). In both cases, however, translocation does not require additional SecA, showing that SecA and SecYEG remain in a functional complex during the reconstitution process, in agreement with studies published previously (16, 22).
Figure 2.
Competition assay(s) between PrlD SecA and WT SecA. A, to verify the efficiency of our co-reconstitution procedure, 4 μg of co-reconstituted SecYEG-SecA and 4 μg of co-reconstituted SecYEG-PrlD were analyzed by 15% SDS-PAGE, followed by Coomassie Blue staining. B, translocation assays were performed with 4 μg of SecYEG co-reconstituted in liposomes with SecA (SecYEG–SecA) or with PrlD SecA (SecYEG–PrlD) without addition of extra SecA. FL, full-length. C, assays were performed as described in B in the presence of 1.5 μg of additional SecA or PrlD SecA, as indicated. D, densitometry analysis of C. The plotted data represent the mean ± S.D. for two independent experiments.
The co-reconstituted SecYEG–SecA complexes were subsequently incubated with a stoichiometric excess of WT or PrlD SecA (Fig. 2C). With WT SecA, the protein translocation profile is similar to that seen in Fig. 2B, left panel. In contrast, with PrlD, there is less proPhoA transported and more translocation intermediates (Fig. 2C, lanes 5–7; quantified in Fig. 2D). The reciprocal experiment, wherein a stoichiometric excess of SecA is added to the preformed SecYEG–PrlD complex, was performed in parallel (Fig. 2C, quantified in Fig. 2D). In that case, the protein translocation profile was unchanged with both SecA and PrlD. As expected, addition of PrlD SecA to the preformed SecYEG–PrlD complex had no effect (Fig. 2C, lanes 12–14).
These data indicate that PrlD can replace SecA during the translocation process but not the opposite, implying that the functional PrlD–SecYEG complex is more stable than SecA–SecYEG complexes. This is in agreement with an earlier study showing that PrlD SecA inserts stably into the membrane at SecYEG to a much greater extent than WT SecA (20). Importantly, the heightened stability of the SecYEG–PrlD complex is not due to an affinity difference between WT and PrlD SecA but, rather, due to the altered conformation of the PrlD mutant (20).
Covalent cross-linking of SecY–SecA mimics the effect of the PrlD mutation
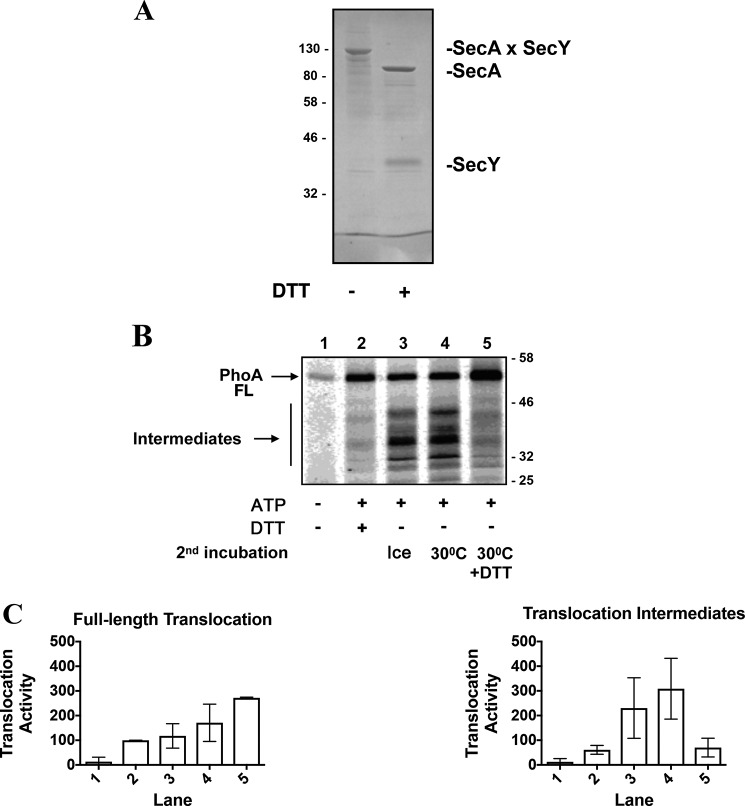
We measured translocation of proPhoA when SecA processivity is limited via a cysteine cross-link between SecA and SecY. The cysteine variants were cross-linked together and purified according to the protocol of Whitehouse et al. (17) before being reconstituted into proteoliposomes (Fig. 3A). As expected, the SecA–SecY cross-link is reducible with DTT (Fig. 3A). Translocation assays were then performed with the reconstituted cross-linked complex for 10 min at 30 °C. In that case, there was an accumulation of translocation intermediates, which are not detected in the presence of DTT (Fig. 3B, compare lane 2 with lane 3). These accumulated intermediates can, however, complete translocation upon addition of DTT (Fig. 3B, compare lane 4 with lane 5). Together, these results show that cleavage of the SecY–SecA covalent bond restores SecA processivity and promotes the forward movement of the jammed translocation intermediates. Quantification of the fully and partially transported species is shown in Fig. 3C.
Figure 3.
Translocation activity of a covalently linked SecA–SecYEG complex. A, the cross-linked SecYEG–SecA complex reconstituted into proteoliposomes (4 μg) was analyzed by 12% SDS-PAGE and Coomassie Blue staining with DTT (1 mm) as indicated. B, translocation assays were performed as described for Fig. 2. All incubations were for 10 min at 30 °C. The translocation intermediates formed in lane 3 were reincubated for an additional 10 min with or without DTT as indicated (lanes 4 and 5, respectively). FL, full-length. C, densitometry analysis of B. Data were normalized to 100 units based on the densitometry obtained for fully translocated PhoA in lane 2. The plotted data represent the mean ± S.D. for three independent experiments.
The importance of SecA–SecY processivity varies between preprotein substrates
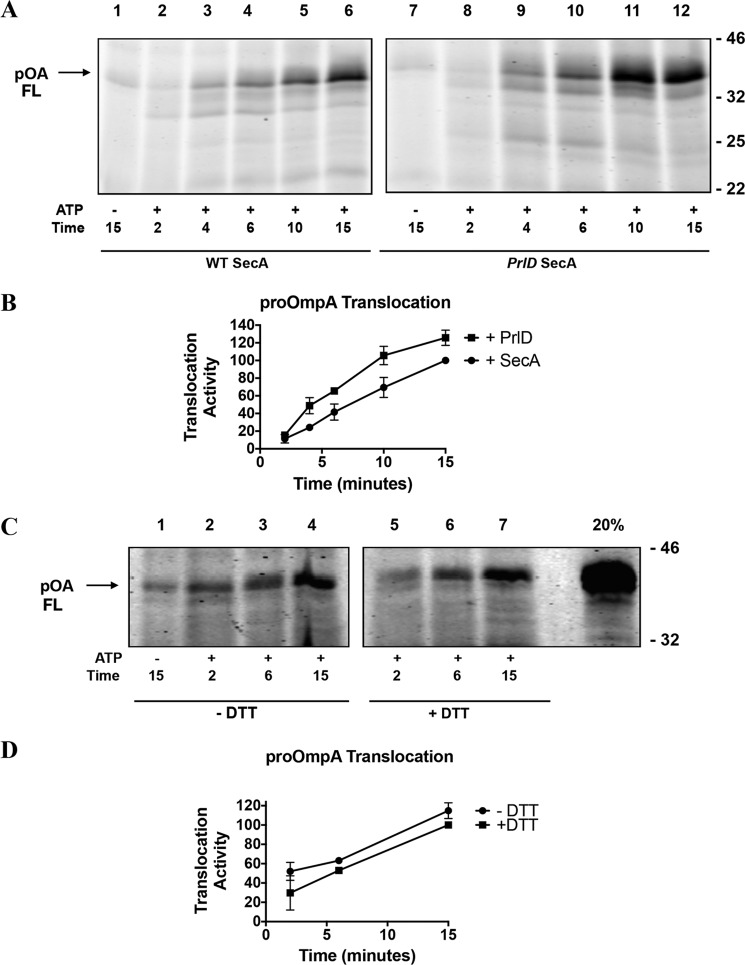
The results above indicate that SecA processivity is important for proPhoA translocation. To determine whether SecA processivity is required for translocation of a different substrate, we employed proOmpA. In contrast to proPhoA, translocation of proOmpA seems to not be influenced by SecA processivity. ProOmpA translocation proceeds modestly more efficiently under conditions where SecA processivity is restricted either by the PrlD23 mutant (Fig. 4A) or by the SecA–SecYEG cross-linked complex (Fig. 4C).
Figure 4.
Translocation activity of PrlD23 SecA with the substrate proOmpA. A, fluorescently labeled proOmpA was employed in translocation assays as in Fig. 1. FL, full-length. B, quantification of A. C, proOmpA was employed in translocation assays with the SecYEG–SecA cross-linked complex. Proteoliposomes, ATP, and labeled substrate were incubated at 30 °C for the indicated times in the presence or absence of 1 mm DTT. 50-μl aliquots were withdrawn at the indicated time points before being analyzed as described for Fig. 1. D, quantification of B. 100 units of translocation activity is defined as the level of fully translocated proOmpA observed after 15 min in the presence of ATP and 1 mm DTT (lane 7).
Influence of substrate leader peptide and mature domain on SecA
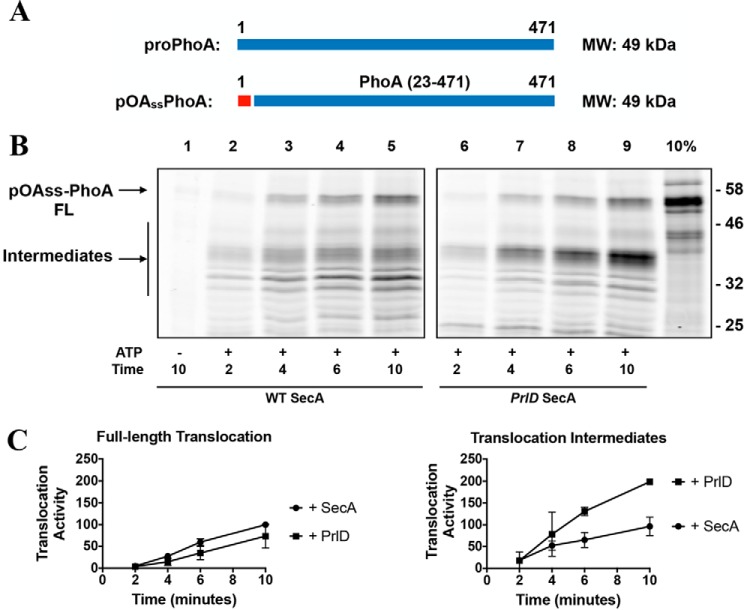
Why is an effect of SecA processivity apparent with the proPhoA substrate but not with proOmpA? We determined the influence of both the substrate leader peptide and the mature domain on SecA processivity. We attached the signal sequence of proOmpA to the PhoA mature domain (pOAss-PhoA, Fig. 5A). As seen with proPhoA, a significant amount of pOAss-PhoA translocation intermediates (size, ∼35 kDa) is formed with PrlD SecA. Thus, variations in the substrate signal sequence do not influence SecA processivity.
Figure 5.
Effect of the substrate leader peptide on SecA processivity. A, cartoon representations of proPhoA and the pOAss-PhoA fusion protein. The leader peptide of proOmpA (red) was fused to the mature sequence of PhoA (blue). The molecular masses of each protein are indicated on the right. B, pOAss-PhoA was fluorescently labeled and employed in translocation assays with SecYEG IMVs as described in Fig. 1C. FL, full-length. C, the amounts of fully translocated products and translocation intermediates were quantified as described for Fig. 1.
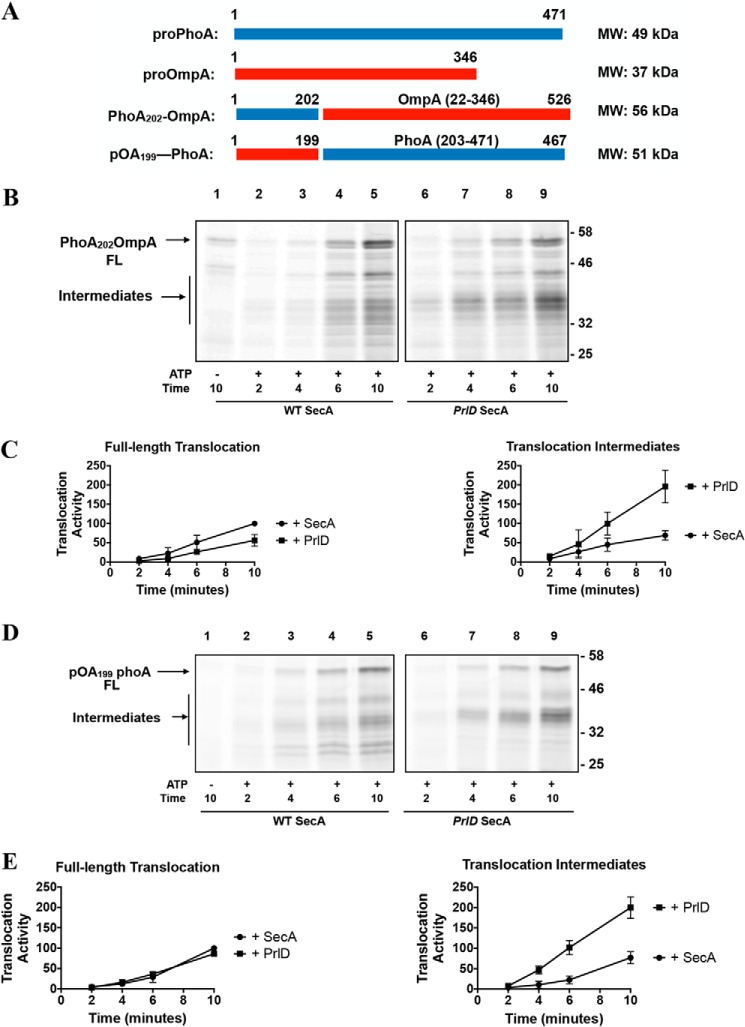
We next exchanged portions of the mature domains of proPhoA and proOmpA. We generated two constructs, PhoA202-OmpA and pOA199-PhoA, which have similar molecular weights as WT proPhoA (Fig. 6A). In the presence of the PrlD mutant, both of these substrates form the same, ∼35 kDa translocation intermediate observed with PhoA (Fig. 6, B and D; quantified in Fig. 6, C and E). This result suggests that the substrate sequence does not influence SecA processivity.
Figure 6.
Effect of substrate mature domain on SecA processivity. A, cartoon representations of proPhoA, proOmpA, and the two fusion substrates generated in this study. The molecular masses of each protein are indicated on the right. B, phoA202-OmpA, a fusion between the N-terminal 202 residues of proPhoA and the mature sequence of OmpA, was fluorescently labeled and employed in translocation assays as described in Fig. 6. FL, full-length. C, quantification of B. D, proOmpA199-PhoA, a fusion between the N-terminal 1–199 residues of proOmpA and residues 203–471 of PhoA, was fluorescently labeled and employed in translocation assays as described in Fig. 6. E, quantification of D.
Influence of substrate length on SecA processivity
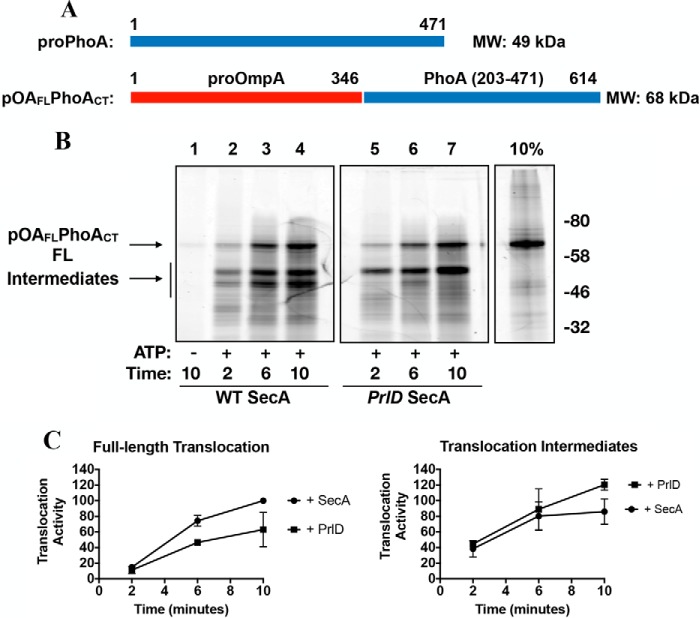
Given the difference in size between proOmpA (346 residues, 37 kDa) and proPhoA (471 residues, 49 kDa), we examined whether SecA processivity is influenced by the length of the substrate. We constructed an extended variant of proOmpA by fusing a C-terminal fragment of PhoA (PhoA residues 203–471) onto proOmpA. This generated the substrate pOAFLPhoACT (614 residues, 68 kDa, Fig. 7A). We have shown above that the 35-kDa translocation intermediates observed with proPhoA are not dependent on this fragment of PhoA (compare WT PhoA with PhoA202-OmpA). Thus, any new translocation intermediates we observe with this fusion protein can be attributed to its size as opposed to its sequence. In the presence of WT SecA, we observe time-dependent accumulation of both fully translocated material as well as an ∼50-kDa translocation intermediate. We observe less fully translocated material in the presence of the PrlD mutant along with more pronounced accumulation of the 50-kDa translocation intermediate (Fig. 7, B and C). Thus, it appears that substrate length does influence SecA processivity.
Figure 7.
Effect of substrate length on SecA processivity. A, cartoon representations of the recombinant substrate pOAFL PhoACT, using the same color scheme as Fig. 6A. A cartoon representation of the substrate PhoA is included here for reference. The molecular masses of each protein are indicated on the right. B, pOAFL PhoACT was fluorescently labeled and employed in translocation assays as described in Fig. 6. FL, full-length. C, quantification of B.
To accumulate more evidence for the possible role of substrate length, we also tested the effect of SecA processivity on translocation of an N-terminally truncated variant of proPhoA termed PhoA202 (202 residues, 22 kDa, Fig. 8A) (23). We expected that reducing the substrate length would decrease its dependence on SecA processivity. Consistent with this hypothesis, we found that translocation of PhoA202 was not influenced by SecA processivity. Indeed, as with proOmpA, we found that translocation of PhoA202 is modestly increased with the nonprocessive PrlD mutant (Fig. 8B, quantified in Fig. 8C).
Figure 8.
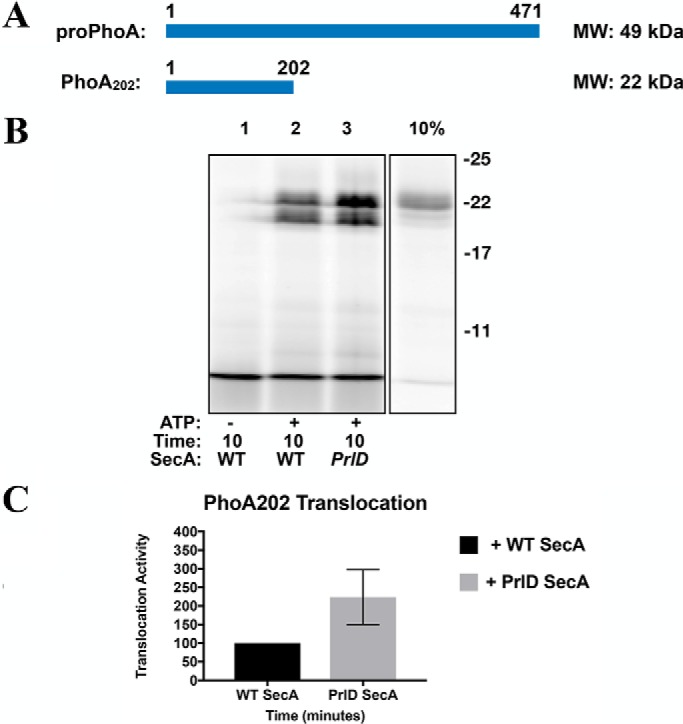
A proPhoA truncation mutant is no longer dependent on SecA processivity. A, cartoon representations of the substrate PhoA202. A cartoon representation of the substrate proPhoA is included here for reference. The molecular masses of each protein are indicated on the right. B, phoA202 was fluorescently labeled and employed in translocation assays as described in Fig. 6. C, quantification of B.
Discussion
We examined the importance of successive cycles of SecA binding and dissociation from SecYEG, termed SecA processivity, using a SecA mutant (PrlD23) that associates more tightly with bacterial membranes than WT SecA (20). Although previous work has shown that SecA undergoes cycles of binding and dissociation from SecYEG, this study was motivated by conflicting reports of whether this processivity is an absolute requirement for substrate translocation (5, 11, 16).
By monitoring the accumulation of translocation intermediates, we show that SecA processivity influences translocation of the substrate proPhoA but not of proOmpA. To unravel the reason for this difference between the two substrates, we examined the effect of the substrate signal sequence and mature domain on SecA processivity. We generated a series of chimeric substrates by exchanging substrate signal peptides as well as portions of the substrate mature domains. The sizes of the chimeric substrates are comparable with that of proPhoA (∼50 kDa), but these chimeric substrates display the same dependence on SecA processivity as WT proPhoA, leading us to conclude that substrate sequence is likely not the main factor determining SecA processivity.
When analyzing these results, we also considered the potential effect of substrate folding and secondary structure. A recent study by Tsirigotaki et al. (24) revealed important differences between the in-solution behavior of proOmpA and proPhoA. They show that proOmpA adopts some elements of secondary structure in solution whereas proPhoA does not (24). Additionally, the mature domains of both proOmpA and proPhoA adopt highly dynamic, loosely folded “folding intermediate” states. We consider it unlikely that the difference in the dependence on SecA processivity between proPhoA and proOmpA is caused by the potentially different folding dynamics of the two substrates because the exchange of portions of the substrate mature domains results in no apparent change in substrate dependence on SecA processivity.
We next tested the possible effect of substrate length. It has been speculated earlier that preprotein substrates of differing lengths and differing amino acid compositions may behave differently during the translocation reaction (25). However, this question has not been fully addressed, particularly in the context of SecA processivity. We extended the size of proOmpA beyond that of proPhoA, generating a substrate termed pOAFLPhoACT (68 kDa). In that case, we observed some accumulation of translocation intermediates when SecA was able to act in a processive manner. Accumulation of these intermediates is increased in the presence of the PrlD mutant, suggesting that increasing the length of proOmpA increases its dependence on SecA processivity.
We also tested whether decreasing the length of proPhoA would decrease its dependence on SecA processivity. As expected, translocation with the shorter PhoA202 is not dependent on SecA processivity, further supporting our conclusion regarding the importance of substrate length. We note that earlier studies by Tomkiewicz et al. (25) and Fessl et al. (15) examined the effect of substrate size using extended proOmpA derivatives, but these studies did not assess accumulation of translocation intermediates (15, 25). The authors employed either a fluorescent probe or an epitope tag located at the extreme C terminus of their substrates; thus, any intermediates that may have accumulated would not have been detectable (25). In our study, we suspect that the 35-kDa PrlD-dependent translocation intermediates observed with proPhoA as well as the ∼ 50-kDa intermediates seen with pOAFLPhoACT may represent rate-limiting steps in the translocation reaction. It could be, for instance, that ∼ 35 kDa is the maximum length of proPhoA that can be translocated by SecA before the enzyme needs to dissociate from the membrane and SecYEG. However, in the case of the longer substrate, pOAFLPhoACT, we no longer observe this ∼35 kDa intermediate. Instead, the size of the observed intermediates is shifted up to ∼50 kDa. This could be because the translocase is able to “sense” the length of the substrate and because the size of the rate-limiting step varies according to preprotein length.
Altogether, our findings are in line with the processive model of protein translocation that has been proposed previously, wherein SecA cycles on and off of the membrane and SecYEG during the translocation reaction (5, 16). We further report that the importance of this on/off cycling varies depending on the length of the translocating substrate. However, this is demonstrated using only two substrates. Therefore, a more comprehensive analysis testing multiple substrates on the scale of the recent studies by Bariya and Randall (22), Chatzi et al. (26), and Tsirigotaki et al. (24) will be needed to determine whether SecA processivity is a universal phenomenon.
Experimental procedures
Protein production and purification
Expression and purification of SecYEG were performed from the plasmid pBad22 his-EYG (pEYG) as described previously (23, 27–29). Inner membrane vesicles (IMVs) containing overexpressed SecYEG were prepared from the E. coli strain KM9 as described previously (23, 30). Expression and purification of WT and mutant variants of SecA were as described previously, using the plasmid pET23 SecAHis (23, 31). The coding region of alkaline phosphatase A (proPhoA) was cloned into the plasmid pBad33 and transformed into E. coli strain BL21. A 2-liter culture of pBad33-PhoA (C190S) was grown in Lysogeny Broth medium supplemented with 50 μg/ml chloramphenicol. At A600 0.4, 0.2% (w/v) l-arabinose was added to induce protein expression. After 3 h of induction, cells were harvested and lysed using a Microfluidizer (3 passes at 8000 p.s.i.). proPhoA was extracted from the resulting inclusion bodies in buffer A (50 mm Tris-HCl (pH 7.9) and 6 m urea). Expression and purification of proOmpA bearing a unique cysteine (mutant S108C) was carried out as described previously (27, 32). Cloning of all PhoA/OmpA recombinant translocation substrates used in this study was carried out using the polymerase incomplete primer extension method (33). All constructs were verified by DNA sequencing (Genewiz). Protein expression and purification of all derivatives were carried out as described above for proPhoA.
Fluorescent labeling
Dye labeling of translocation substrates was performed as described previously (23). Briefly, 100 μg of purified protein was incubated in the dark for 2 h with a molar excess of the cysteine-specific Alexa Fluor 680 labeling reagent (Invitrogen/Molecular Probes). Labeling was quenched by addition of 1 mm DTT. Excess dye was removed by gel filtration as described by Dalal et al. (23).
SecY–SecA cross-linking
The SecYEG–SecA cross-linked complex was prepared in accordance with protocols published previously with minor modifications (11, 17). The SecY cysteine mutation K268C was introduced into pEYG by site-directed mutagenesis. After transformation into BL21, membrane extracts bearing the overexpressed SecY complex were isolated from 6 liters of culture and resuspended in buffer B (50 mm Tris-HCl (pH 7.9), 100 mm NaCl, and 10% glycerol). Endogenous SecA was removed by incubating the membranes on ice for 1 h in buffer A. Urea was removed by ultracentrifugation (55,000 rpm, 4 °C, Beckman Ti70 rotor), and the washed membranes were resuspended in buffer B. Urea-treated membranes (50 mg) were incubated with purified N95 795C SecA (50 mg) on ice for 1 h in buffer B supplemented with 5 mm MgCl2. After the initial hour-long incubation, 0.1 mm copper phenanthroline was added to induce cross-link formation. After a further 1-h incubation, copper phenanthroline was removed by ultracentrifugation (55,000 rpm, 4 °C, Beckmann Ti60 rotor), and the membranes were resuspended in buffer B supplemented with 1% dodecyl maltoside. The cross-linked complex was then purified as described previously (11, 17).
Proteoliposome reconstitutions
Total E. coli lipid extract (100 mg, Avanti Polar Lipids) dissolved in chloroform was dried under a stream of nitrogen and desiccated overnight. The resulting lipid film was resuspended in buffer C (50 mm Tris-HCl (pH 7.9) and 50 mm NaCl) to a final concentration of 10 mg/ml and sonicated to homogeneity. Unilamellar vesicles were prepared by extrusion through a 100-nm filter at 300 p.s.i. using a Lipex 10/1.5 ml Thermobarrel and Regular Barrel Extruder (Northern Lipids Inc.). The size of the resulting liposomes was verified by dynamic light scattering using a Zetasizer (Malvern Technologies). A solution of Triton X-100 was titrated into a 0.5-mg aliquot of the extruded liposomes to determine the concentration of detergent required to swell but not disrupt the vesicles. Liposome swelling was assessed by monitoring the turbidity of the solution at A550 using a UV spectrophotometer. Optimal swelling was observed at a concentration of 0.1% Triton X-100. To form proteoliposomes, 0.5 mg of extruded liposomes was treated with 0.1% Triton X-100. The purified SecYEG complex (1 nmol) was added to this detergent–lipid mixture and incubated for 10 min on a rocking platform at 4 °C. Liposomes were incubated with a 1/10 volume of BioBeads overnight to facilitate detergent removal. Proteoliposomes were recovered by ultracentrifugation (55,000 rpm, Beckman TLA55 rotor) and resuspended in buffer C. Co-reconstitution of SecYEG with WT or PrlD23 SecA was performed as described previously with minor modifications (16, 22). Briefly, purified SecYEG (1 nmol) was mixed with either WT or PrlD mutant SecA (1 nmol each) for 30 min at 4 °C before being added to the lipids. The co-reconstituted preparations are termed SecYEG–SecA and SecYEG–PrlD throughout this paper.
Protein translocation assays
Translocation assays were performed essentially as described previously (23, 27). Briefly, SecYEG proteoliposomes (2 μg) or SecYEG-containing IMVs (5 μg) were mixed on ice with SecA (1.5 μg), fluorescently labeled substrate (2 ng), and ATP (2 mm) in 50 μl of TL buffer (50 mm Tris-HCl (pH 7.9), 50 mm NaCl, and 5 mm MgCl2). A negative control was performed in parallel, containing all reaction components except ATP. Reactions were incubated at 30 °C in a water bath for the indicated time before being chilled on ice. All untransported substrate was degraded by incubation with Proteinase K (5 μg, Bioshop) on ice for 10 min. The reactions were precipitated with ice-cold TCA (17% final, 30 min) followed by centrifugation (13,200 rpm, 10 min, 4 °C). The pellets were washed with ice-cold acetone before being dissolved in SDS-PAGE sample buffer and analyzed on 15% SDS-PAGE. Transported proteins were visualized by fluorescence scanning using a LI-COR Odyssey (LI-COR Biosciences).
Analysis of translocation data
Densitometry analysis of the translocation assays was performed using the LI-COR Odyssey software. Densitometry for full-length translocation was determined by selecting the area of each lane labeled full-length. The data were normalized to the densitometry obtained for fully translocated material with WT SecA at the final time point in each assay. The fluorescent intensity of this signal was arbitrarily assigned a value of 100. Densitometry for the translocation intermediates was obtained by selecting the area of each lane marked “intermediates.” As for full-length translocation, the fluorescence intensity in this area was plotted relative to the intensity obtained for fully translocated material with WT SecA at the final time point in each assay. Each assay was repeated at least twice to ensure reproducibility. The resulting data were plotted using GraphPad Prism 6 software (GraphPad, San Diego, CA). Error bars represent standard deviation.
Author contributions
J. Y. and F. D. conceptualization; J. Y. data curation; J. Y. and F. D. formal analysis; J. Y. investigation; J. Y. methodology; J. Y. and F. D. writing-original draft; J. Y. and F. D. writing-review and editing; F. D. resources; F. D. supervision; F. D. funding acquisition; F. D. project administration.
This work was supported by Canadian Institutes of Health Research Grant MOP 201309. The authors declare that they have no conflicts of interest with the contents of this article.
- IMV
- inner membrane vesicle.
References
- 1. Park E., and Rapoport T. A. (2012) Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41, 21–40 10.1146/annurev-biophys-050511-102312 [DOI] [PubMed] [Google Scholar]
- 2. Collinson I., Corey R. A., and Allen W. J. (2015) Channel crossing: how are proteins shipped across the bacterial plasma membrane? Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150025 10.1098/rstb.2015.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Crane J. M., and Randall L. L. (2017) The Sec system: Protein Export in Escherichia coli. EcoSal Plus 7 10.1128/ecosalplus.ESP-0002-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koch S., de Wit J. G., Vos I., Birkner J. P., Gordiichuk P., Herrmann A., van Oijen A. M., and Driessen A. J. (2016) Lipids activate SecA for high affinity binding to the SecYEG complex. J. Biol. Chem. 291, 22534–22543 10.1074/jbc.M116.743831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morita K., Tokuda H., and Nishiyama K. (2012) Multiple SecA molecules drive protein translocation across a single translocon with SecG inversion. J. Biol. Chem. 287, 455–464 10.1074/jbc.M111.301754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chatzi K. E., Sardis M. F., Economou A., and Karamanou S. (2014) SecA-mediated targeting and translocation of secretory proteins. Biochim. Biophys. Acta 1843, 1466–1474 10.1016/j.bbamcr.2014.02.014 [DOI] [PubMed] [Google Scholar]
- 7. Robson A., Booth A. E., Gold V. A., Clarke A. R., and Collinson I. (2007) A large conformational change couples the ATP binding site of SecA to the SecY protein channel. J. Mol. Biol. 374, 965–976 10.1016/j.jmb.2007.09.086 [DOI] [PubMed] [Google Scholar]
- 8. Cranford-Smith T., and Huber D. (2018) The way is the goal: how SecA transports proteins across the cytoplasmic membrane in bacteria. FEMS Microbiol. Lett. 365 10.1093/femsle/fny093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duong F., and Wickner W. (1997) The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 16, 4871–4879 10.1093/emboj/16.16.4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tsukazaki T., Mori H., Echizen Y., Ishitani R., Fukai S., Tanaka T., Perederina A., Vassylyev D. G., Kohno T., Maturana A. D., Ito K., and Nureki O. (2011) Structure and function of a membrane component SecDF that enhances protein export. Nature 474, 235–238 10.1038/nature09980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bauer B. W., Shemesh T., Chen Y., and Rapoport T. A. (2014) A “push and slide” mechanism allows sequence-insensitive translocation of secretory proteins by the SecA ATPase. Cell 157, 1416–1429 10.1016/j.cell.2014.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Erlandson K. J., Or E., Osborne A. R., and Rapoport T. A. (2008) Analysis of polypeptide movement in the SecY channel during SecA-mediated protein translocation. J. Biol. Chem. 283, 15709–15715 10.1074/jbc.M710356200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding H., Mukerji I., and Oliver D. (2003) Nucleotide and phospholipid-dependent control of PPXD and C-domain association for SecA ATPase. Biochemistry 42, 13468–13475 10.1021/bi035099b [DOI] [PubMed] [Google Scholar]
- 14. Allen W. J., Corey R. A., Oatley P., Sessions R. B., Baldwin S. A., Radford S. E., Tuma R., and Collinson I. (2016) Two-way communication between SecY and SecA suggests a Brownian ratchet mechanism for protein translocation. eLife 5, pii: e15598 10.7554/eLife.15598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fessl T., Watkins D., Oatley P., Allen W. J., Corey R. A., Horne J., Baldwin S. A., Radford S. E., Collinson I., and Tuma R. (2018) Dynamic action of the Sec machinery during initiation, protein translocation and termination. eLife 7, pii: e35112 10.7554/eLife.35112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mao C., Cheadle C. E., Hardy S. J., Lilly A. A., Suo Y., Sanganna Gari R. R., King G. M., and Randall L. L. (2013) Stoichiometry of SecYEG in the active translocase of Escherichia coli varies with precursor species. Proc. Natl. Acad. Sci. U.S.A. 110, 11815–11820 10.1073/pnas.1303289110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whitehouse S., Gold V. A., Robson A., Allen W. J., Sessions R. B., and Collinson I. (2012) Mobility of the SecA 2-helix-finger is not essential for polypeptide translocation via the SecYEG complex. J. Cell Biol. 199, 919–929 10.1083/jcb.201205191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gold V. A., Whitehouse S., Robson A., and Collinson I. (2013) The dynamic action of SecA during the initiation of protein translocation. Biochem. J. 449, 695–705 10.1042/BJ20121314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sugano Y., Furukawa A., Nureki O., Tanaka Y., and Tsukazaki T. (2017) SecY-SecA fusion protein retains the ability to mediate protein transport. PLoS ONE 12, e0183434 10.1371/journal.pone.0183434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gouridis G., Karamanou S., Sardis M. F., Schärer M. A., Capitani G., and Economou A. (2013) Quaternary dynamics of the SecA motor drive translocase catalysis. Mol. Cell 52, 655–666 10.1016/j.molcel.2013.10.036 [DOI] [PubMed] [Google Scholar]
- 21. Schmidt M., Ding H., Ramamurthy V., Mukerji I., and Oliver D. (2000) Nucleotide binding activity of SecA homodimer is conformationally regulated by temperature and altered by prlD and azi mutations. J. Biol. Chem. 275, 15440–15448 10.1074/jbc.M000605200 [DOI] [PubMed] [Google Scholar]
- 22. Bariya P., and Randall L. L. (2018) Co-assembly of SecYEG and SecA fully restores the properties of the native translocon. J. Bacteriol. 201, pii: e00493–18 10.1128/JB.00493-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dalal K., Chan C. S., Sligar S. G., and Duong F. (2012) Two copies of the SecY channel and acidic lipids are necessary to activate the SecA translocation ATPase. Proc. Natl. Acad. Sci. U.S.A. 109, 4104–4109 10.1073/pnas.1117783109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsirigotaki A., Chatzi K. E., Koukaki M., De Geyter J., Portaliou A. G., Orfanoudaki G., Sardis M. F., Trelle M. B., Jørgensen T. J. D., Karamanou S., and Economou A. (2018) Long-lived folding intermediates predominate the targeting-competent secretome. Structure 26, 695–707.e5 10.1016/j.str.2018.03.006 [DOI] [PubMed] [Google Scholar]
- 25. Tomkiewicz D., Nouwen N., van Leeuwen R., Tans S., and Driessen A. J. (2006) SecA supports a constant rate of preprotein translocation. J. Biol. Chem. 281, 15709–15713 10.1074/jbc.M600205200 [DOI] [PubMed] [Google Scholar]
- 26. Chatzi K. E., Sardis M. F., Tsirigotaki A., Koukaki M., Šoštarić N., Konijnenberg A., Sobott F., Kalodimos C. G., Karamanou S., and Economou A. (2017) Preprotein mature domains contain translocase targeting signals that are essential for secretion. J. Cell Biol. 216, 1357–1369 10.1083/jcb.201609022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dalal K., Nguyen N., Alami M., Tan J., Moraes T. F., Lee W. C., Maurus R., Sligar S. S., Brayer G. D., and Duong F. (2009) Structure, binding, and activity of Syd, a SecY-interacting protein. J. Biol. Chem. 284, 7897–7902 10.1074/jbc.M808305200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carlson M. L., Young J. W., Zhao Z., Fabre L., Jun D., Li J., Dhupar H. S., Wason I., Mills A. T., Beatty J. T., Klassen J. S., Rouiller I., and Duong F. (2018) The Peptidisc, a simple method for stabilizing membrane proteins in detergent-free solution. eLife 7, pii: e34085 10.7554/eLife.34085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alami M., Dalal K., Lelj-Garolla B., Sligar S. G., and Duong F. (2007) Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 26, 1995–2004 10.1038/sj.emboj.7601661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dalal K., and Duong F. (2009) The SecY complex forms a channel capable of ionic discrimination. EMBO Rep. 10, 762–768 10.1038/embor.2009.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duong F. (2003) Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 22, 4375–4384 10.1093/emboj/cdg418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Duong F., and Wickner W. (1998) Sec-dependent membrane protein biogenesis: SecYEG, preprotein hydrophobicity and translocation kinetics control the stop-transfer function. EMBO J. 17, 696–705 10.1093/emboj/17.3.696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klock H. E., and Lesley S. A. (2009) The polymerase incomplete primer extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol. Biol. 498, 91–103 10.1007/978-1-59745-196-3_6 [DOI] [PubMed] [Google Scholar]