Abstract
Staphylococcus aureus is a Gram-positive bacterium that can cause both superficial and deep-seated infections. Histones released by neutrophils kill bacteria by binding to the bacterial cell surface and causing membrane damage. We postulated that cell wall–anchored proteins protect S. aureus from the bactericidal effects of histones by binding to and sequestering histones away from the cell envelope. Here, we focused on S. aureus strain LAC and by using an array of biochemical assays, including surface plasmon resonance and ELISA, discovered that fibronectin-binding protein B (FnBPB) is the main histone receptor. FnBPB bound all types of histones, but histone H3 displayed the highest affinity and bactericidal activity and was therefore investigated further. H3 bound specifically to the A domain of recombinant FnBPB with a KD of 86 nm, ∼20-fold lower than that for fibrinogen. Binding apparently occurred by the same mechanism by which FnBPB binds to fibrinogen, because FnBPB variants defective in fibrinogen binding also did not bind H3. An FnBPB-deletion mutant of S. aureus LAC bound less H3 and was more susceptible to its bactericidal activity and to neutrophil extracellular traps, whereas an FnBPB-overexpressing mutant bound more H3 and was more resistant than the WT. FnBPB bound simultaneously to H3 and plasminogen, which after activation by tissue plasminogen activator cleaved the bound histone. We conclude that FnBPB provides a dual immune-evasion function that captures histones and prevents them from reaching the bacterial membrane and simultaneously binds plasminogen, thereby promoting its conversion to plasmin to destroy the bound histone.
Keywords: Staphylococcus aureus (S. aureus), histone, plasminogen, innate immunity, cell surface protein, virulence factor, methicillin-resistant Staphylococcus aureus (MRSA), adhesin, extracellular trap, fibronectin-binding protein B
Introduction
Staphylococcus aureus is a leading cause of diverse infections ranging from mild skin diseases such as impetigo, cellulitis, and skin abscesses to serious invasive diseases, including sepsis, endocarditis, osteomyelitis, toxic shock syndrome, and necrotizing pneumoniae (1, 2). Strains that are resistant to multiple antibiotics are a major problem in healthcare settings in developed countries (3). These are referred to as hospital-associated methicillin-resistant S. aureus (MRSA)3 and occur in individuals with pre-disposing risk factors, such as surgical wounds and indwelling medical devices (4, 5). Recently, there has been a dramatic increase in the incidence of community-associated MRSA infections that occur in otherwise healthy individuals (6, 7). Community-associated MRSA strains, exemplified by the USA300 clone (8), express a low level of resistance to β-lactam antibiotics and cause serious skin and soft tissue infections (9–11).
S. aureus expresses a plethora of virulence factors, including both secreted and cell wall-anchored (CWA) proteins. The latter mediate adherence to the extracellular matrix, promote invasion of and survival within host cells, neutralize phagocytes, and modulate the immune response (12). CWA proteins are covalently anchored to peptidoglycan via a conserved C-terminal sorting signal mediated by the membrane-associated sortase A (13). Fibronectin-binding proteins FnBPA and FnBPB are CWA proteins belonging to the microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) family. The N-terminal A domain of FnBPs comprises three separately folded subdomains N1, N2, and N3 (14, 15). The minimum ligand-binding region of the A domain (N2–N3) comprises two IgG-like folded subdomains that bind fibrinogen (FBG) by the dock, lock, and latch mechanism (DLL) (15, 16) in a similar fashion to clumping factor A (ClfA) (17). Between the N2 and N3 subdomains lies a wide hydrophobic trench that accommodates the ligand, in the case of ClfA, FnBPA, and FnBPB the extreme C terminus of the γ-chain of FBG. According to this mechanism, the ligand first docks into the trench, and this is followed by a conformational change and redirection of the disordered C-terminal extension of the N3 subdomain resulting in its folding over the bound ligand to lock it in place. In the final latching step, the complex is stabilized by inserting the “latch” region in the N3 extension into the N2 subdomain through a β-strand complementation (18, 19). The N2–N3 subdomains of both FnBPA and FnBPB each comprise seven distinct isoforms (14, 20), also bind elastin (21) and plasminogen (PLG) (22), and promote biofilm formation by homophilic interactions (23). The C terminus of FnBPs comprises 10/11 tandemly repeated fibronectin-binding domains that bind type I modules of fibronectin by the tandem β-zipper mechanism (24, 25).
In eukaryotes, DNA is wrapped around a core complex of the histones H2A, H2B, H3, and H4 to form the nucleosome. Histones are the most abundant proteins in neutrophil extracellular traps (NETs), which are released by neutrophils as part of the innate defenses against infecting bacteria (26). In addition to histones, NETs contain nuclear DNA and proteases (e.g. elastase). Histones are also released into the bloodstream during severe sepsis (27). The potent antimicrobial activity of histones is due to characteristics akin to cationic antimicrobial peptides such as cathelicidins. Besides NETs, other myeloid cell lineages such as basophils (28), eosinophils (29), and macrophages (30) can deploy histones within DNA-based extracellular traps. In addition, histones can induce production of chemokines and elicit leukocyte recruitment (31).
Histones are classified as lysine-rich (H1, H2A, and H2B) and arginine-rich (H3 and H4) (32). H2A, H3, and H4 have been shown to have anti-staphylococcal activity (33), but the mechanistic basis differs. H3 and H4 cause membrane damage with blebbing and pore formation, whereas H2B disrupts the integrity of the cell without obvious morphological changes (33).
In Gram-positive (34, 35) and Gram-negative (36) bacteria, surface components can bind histones and provide protection from bactericidal effects. Lipoteichoic acid (37), the cell wall–anchored M1 protein (35), and the secreted protein SIC of Streptococcus pyogenes (38) bind to and promote resistance to histones. In the Gram-negative Klebsiella pneumoniae and Shigella flexneri, the polysaccharide O-antigen of lipopolysaccharide in the outer membrane can contribute to resistance to histones (36).
In this study, we have investigated the molecular basis of resistance to histones in S. aureus and found that FnBPB is the major surface component responsible. We studied the mechanism of histone binding by FnBPB and identified the most potent anti-staphylococcal histone as H3.
Results
Cell wall–anchored proteins bind histones
S. aureus is known to bind to histones, but the bacterial components responsible have not been characterized. To determine whether CWA proteins contribute to histone binding, the USA 300 strain LAC (8) and a sortase A-deficient mutant (srtA) (39) lacking CWA proteins were tested for their ability to adhere to a surface-coated with pooled calf thymus histones (CTH). WT LAC adhered about three times more strongly than the srtA mutant, which suggests that one or more CWA proteins are dominant histone binders (Fig. 1). The residual binding by the srtA mutant could be due to other surface components such as lipoteichoic acid (33, 37).
Figure 1.

Adherence of bacteria to CTH-coated microtiter wells. Microtiter wells coated with CTH were incubated with S. aureus USA 300 LAC cells. After washing with PBS, adherent cells were fixed and stained with crystal violet, and the absorbance at 595 nm was measured using an ELISA plate reader. Means and S.D. of the results of two independent experiments, each performed in triplicate, are indicated. A statistically significant difference is indicated (Student's t test; *, p < 0.05).
Recombinant FnBPB binds histones
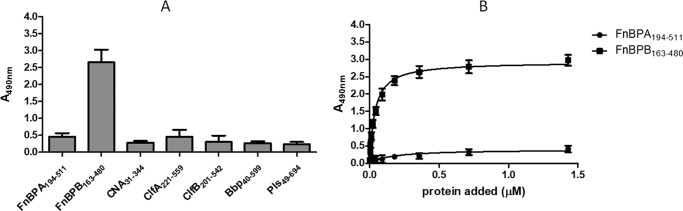
Purified recombinant ligand-binding domains of several CWA proteins were tested for binding to immobilized CTH in an ELISA-type assay. In particular, subdomains N2–N3 of isoform I of FnBPB, comprising residues 163–480 (FnBPB(163–480)), bound strongly to the histone mixture, whereas the N-terminal ligand-binding domains of isoform I of FnBPA, clumping factors A and B, the collagen-binding protein CNA, and also Bbp and Pls, did not bind detectably (Fig. 2A). Control experiments showed that the anti-His tag antibody used in these ELISA experiments recognize all recombinant proteins similarly.
Figure 2.
Binding of recombinant CWA proteins to immobilized CTH. A, microtiter wells coated with CTH were incubated with purified recombinant A domains of the indicated CWA proteins of S. aureus. Bound proteins were detected with mouse anti-hexahistidine mAb 7E8 followed by HRP-conjugated rabbit anti-mouse IgG. B, microtiter wells coated with CTH were incubated with increasing concentrations of purified recombinant N2N3 domains of FnBPA (FnBPA(194–511)) or FnBPB (FnBPB(163–480)). Bound proteins were detected as described above. The data points are the means (±S.D.) of two independent experiments, each performed in triplicate.
The specificity of FnBPB binding to histones was confirmed by performing dose-response binding assays with immobilized CTH. FnBPB(163–480) bound dose-dependently and saturably, whereas no binding of FnBPA(194–511) was observed (Fig. 2B).
The ability of recombinant FnBPB(163–480) to bind to individual histones was next measured by ELISA and far-Western blotting. FnBPB(163–480) bound H1 and H3 the strongest (Fig. 3, A and B). It was decided to focus on binding to H3 rather than H1 because the former exhibited stronger anti-bacterial activity (see below).
Figure 3.
FnBPB(163–480) binding to different histone subtypes. A, microtiter wells coated with different histone subtypes were incubated with increasing concentrations of recombinant FnBPB(163–480) domain. Bound protein was detected with rabbit FnBPB(163–480) polyclonal antibody followed by HRP-conjugated goat anti-rabbit IgG. Means and S.D. of results of two independent experiments, each performed in triplicate, are presented. B, different histone subtypes were subjected to SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was probed with FnBPB(163–480) followed by a rabbit anti-FnBPB(163–480) polyclonal IgG and HRP-conjugated goat anti-rabbit IgG. C, microtiter wells coated with histone H3 were incubated with increasing concentrations of recombinant variants of FnBPB A domain. Bound proteins were detected with mouse anti-hexahistidine mAb 7E8 followed by HRP-conjugated rabbit anti-mouse IgG. The data points are the means (±S.D.) of two independent experiments, each performed in triplicate.
The A domain of FnBPB binds to FBG by the dock, lock, and latch (DLL) mechanism (16). To investigate whether DLL is involved in histone binding, variants of the recombinant A domain lacking the ability to bind FBG were tested for histone binding. A truncate lacking the 17 residues involved in the locking and latching steps of DLL(464–480) and a mutant with substitutions of Asn-312 and Phe-314 located in the peptide binding trench, neither of which bound FBG, were also defective in binding to histone H3. Furthermore, neither single recombinant subdomains N2(163–308) nor N3(309–480) bound FBG or histone H3 (Fig. 3C). These data suggest that histones could bind FnBPB by the DLL mechanism.
Analysis of complex formation between FnBPB(163–480) and H3 by gel-filtration chromatography
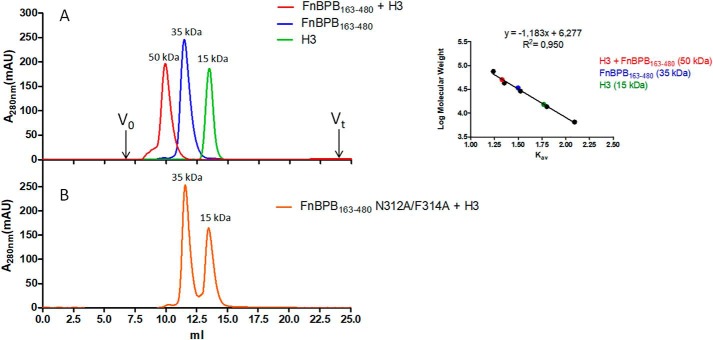
To determine whether FnBPB(163–480) forms a complex with H3 in solution, equimolar amounts of the two proteins were mixed and subjected to gel-filtration chromatography (Fig. 4). The two proteins alone eluted with distribution constants (Kav) corresponding to their molecular masses, i.e. ∼15 kDa for H3 and 35 kDa for FnBPB(163–480) (Fig. 4, inset), whereas co-incubation of the two proteins yielded a fast running peak of ∼50 kDa, in agreement with the predicted molecular mass of FnBPB(163–480)/H3 complex. In contrast, the nonhistone-binding trench mutant failed to form the 50-kDa complex. This shows that FnBPB(163–480) can form a stable complex with H3 in solution and that residues in the FBG-binding trench are crucially important.
Figure 4.
Size-exclusion chromatography analysis of the interaction of FnBPB(163–480) with histone H3. A, elution profiles of FnBPB(163–480), histone H3 alone, and a mixture of a co-incubated, equimolar concentrations of FnBPB and H3 loaded onto a Superdex 75 10/300 GL gel-filtration chromatography column. B, elution profile of a mixture of co-incubated, equimolar concentrations of FnBPB(163–480) trench mutant variant (N312A/F314A) and histone H3 loaded onto the gel-filtration chromatography column as indicated in A. In the inset, a calibration curve relating the Kav of standard proteins to their molecular mass is reported. The figure is representative of three independent experiments. mAU, milli-absorbance units.
Measurement of the affinity and ionic strength-dependence of H3–FnBPB(163–480) interaction
Surface plasmon resonance
The affinity of FnBPB for human H3 was measured by surface plasmon resonance. Recombinant FnBPB(163–480) was passed over human H3 immobilized on the surface of a dextran chip in concentrations ranging from 3.9 to 500 nm. The equilibrium dissociation constant, KD, for the interaction was estimated as 86.0 ± 12.5 nm, an affinity that is ∼20-fold higher than that reported for the interaction between FnBPB and FBG (Fig. 5A) (20). No signal was detected when the FnBPB(163–480) trench mutant was analyzed (Fig. 5B). These data show that FnBPB(163–480) has a high affinity for H3 in solution and that the interaction most likely occurs by the DLL mechanism.
Figure 5.
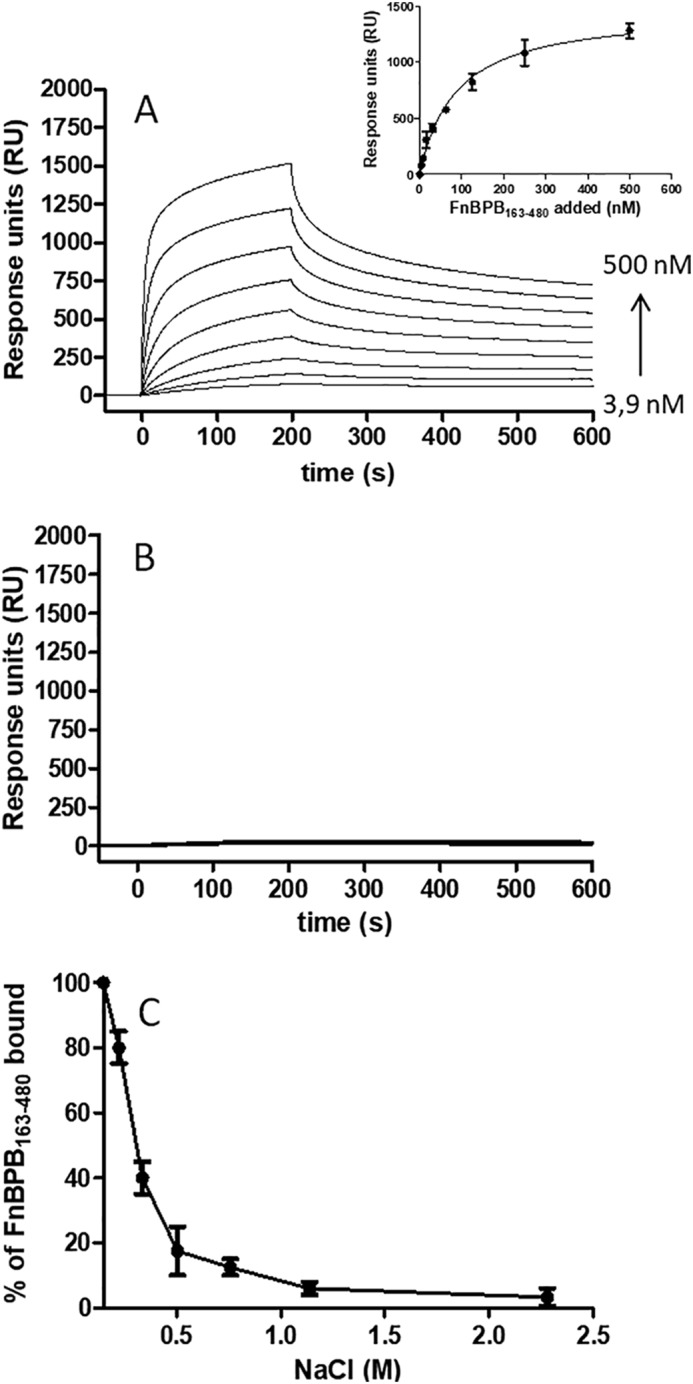
Affinity and ionic strength–dependence of interaction of the H3–FnBPB(163–480). A, representative sensorgrams display binding of FnBPB(163–480) to and dissociation from CM5 chip-coated histone H3. The affinity was calculated from curve fitting to a plot of the response unit values at the steady state (RUmax) against increasing concentrations of FnBPB(163–480) (inset). B, representative sensorgrams display binding of FnBPB(163–480) trench mutant variant (N312A/F314A) to and dissociation from H3. The figure shown is representative of three independent experiments. C, FnBPB(163–480) bound to H3 was analyzed under increasing concentrations of NaCl. FnBPB(163–480) bound to immobilized H3 was detected using a specific anti-FnBPB antibody.
Nature of the interaction between FnBPB(163–480) and H3
The FnBPB(163–480) protein has a hypothetical pI of 5.62 indicating that the protein is anionic at physiological pH, whereas H3 is highly positive (pI 12.5). To further investigate the nature of interaction of H3 with FnBPB(163–480), surface-coated H3 was incubated with FnBPB(163–480) in the presence of increasing amounts of NaCl. At a NaCl concentration of 500 mm, binding of FnBPB(163–480) was reduced by more than 80%. No FnBPB(163–480) binding was observed at higher NaCl concentrations, suggesting that hydrophobic forces are not involved in the interaction and that FnBPB(163–480) binding to H3 is an ionic strength-dependent process (Fig. 5C).
Interaction of H3 with FnBPB isoforms
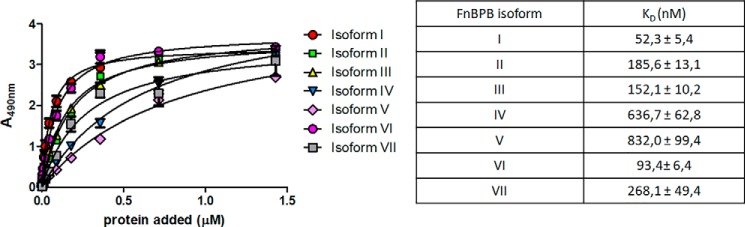
There are seven isoforms of FnBPB with amino acid sequence identities ranging from 61 to 85% (20). Each of the isoforms bind FBG, fibronectin, elastin, and PLG. To determine whether all isoforms bind H3, increasing concentrations of the recombinant N2–N3 subdomains were tested for their ability to bind surface-coated H3 in an ELISA-type assay (Fig. 6). Isoforms II, III, and VI bound similarly to isoform I with dissociation constants in the range of 50–185 nm, whereas the isoforms IV, V, and VII showed a lower binding affinity, with apparent KD values ranging from 270 to 830 nm (Fig. 6A, inset).
Figure 6.
Dose-dependent binding of FnBPB isoforms to surface-coated histone H3. Histone H3 was immobilized onto microtiter wells and tested for binding to recombinant N2–N3 domains of isoforms I–VII of FnBPB. Bound isoforms were detected with mouse anti-His mAb 7E8 followed by HRP-conjugated rabbit anti-mouse IgG. In the inset, the KD values of each isoform are reported. The data points are the means ± S.D. of three independent experiments each performed in triplicate.
FnBPB promotes binding of S. aureus to H3
To investigate whether FnBPB expressed on the surface of S. aureus mediates binding to H3, WT S. aureus LAC, a deletion mutant lacking the fnbA and fnbB genes (40), and the mutant expressing isoform I FnBPA or FnBPB from a multicopy plasmid (40) were tested for their ability to adhere to immobilized H3 and to capture soluble H3 onto the cell surface. The ΔfnbAfnbB mutant lacked the ability to adhere to immobilized H3, compared with the parental strain. In contrast, the mutant expressing FnBPB adhered more strongly than the WT strain, whereas the mutant expressing FnBPA did not adhere (Fig. 7A).
Figure 7.
Interaction of S. aureus USA 300 LAC with histone H3. A, microtiter wells coated with histone H3 were incubated with cells of the indicated bacteria. Wells were washed with PBS, fixed with formaldehyde, and stained with crystal violet, and the absorbance at 595 nm was measured in an ELISA plate reader. Means and S.D. of results of two independent experiments, each performed in triplicate, are presented. Statistically significant differences is indicated (Student's t test; *, p < 0.05). B, S. aureus strain USA 300 LAC, the double mutant ΔfnbAfnbB, or the mutant overexpressing FnBPB or FnBPA was incubated with histone H3. After several washings, proteins bound to the cell surface were released by extraction buffer, separated by SDS-PAGE under nonreducing conditions, and transferred to a nitrocellulose membrane. The membrane was sequentially probed with rabbit anti-histone IgG and HRP-conjugated goat anti-rabbit IgG. The figure is representative of two independent experiments. C, densitometric analysis of histone H3 binding to S. aureus LAC and its mutants as reported in B. The band intensity was quantified relative to a sample of pure histone H3 (5 μg, 100 intensity). The reported data are the mean values ± S.D. from two independent experiments. Statistically significant differences are indicated (Student's t test; *, p < 0.05).
Bacteria were incubated with soluble H3 and washed, and the bound H3 was eluted by incubating with extraction buffer containing SDS. Released H3 was detected by SDS-PAGE/Western immunoblotting and measured by ELISA. In both experiments, WT S. aureus LAC captured H3, whereas the ΔfnbAfnbB mutant did not bind H3 detectably. In contrast, the mutant expressing FnBPB captured ∼40% more H3 than the WT, most likely due to overexpression of FnBPB from the multicopy plasmid (Fig. 7, B and C). These data are consistent with results obtained above with recombinant proteins and show that FnBPB is the major surface protein involved in binding H3.
Histone bactericidal activity and resistance to killing promoted by FnBPB
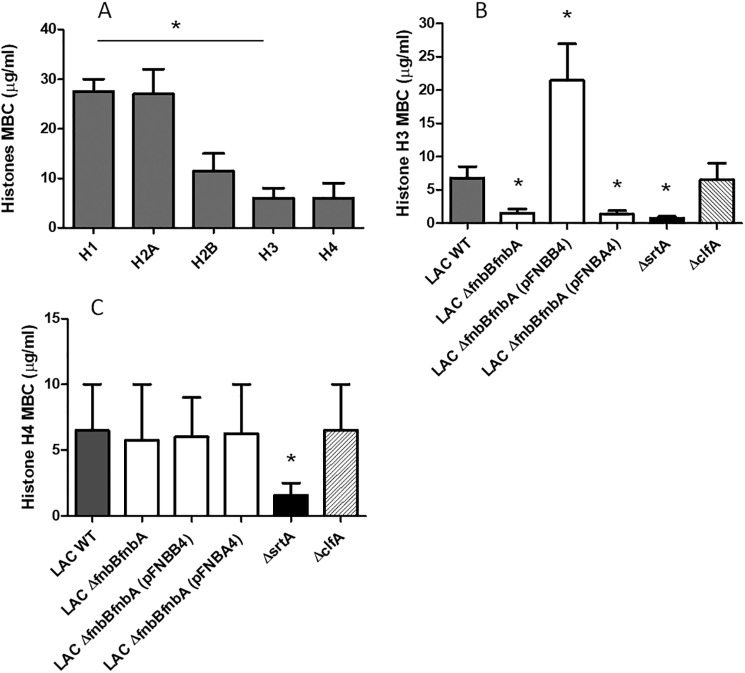
It has been widely reported that soluble histones kill many species of Gram-negative and Gram-positive bacteria and that they are often more potent than cationic antimicrobial defensin peptides. The minimum bactericidal concentration (MBC) required to kill >99.9% of a standard suspension of S. aureus cells was measured and used to compare the potency of different histones toward WT S. aureus LAC. Histones H3 and H4 had significantly lower MBC values compared with H1, H2A, and H2B indicating that they have more potent bactericidal activity (Fig. 8A).
Figure 8.
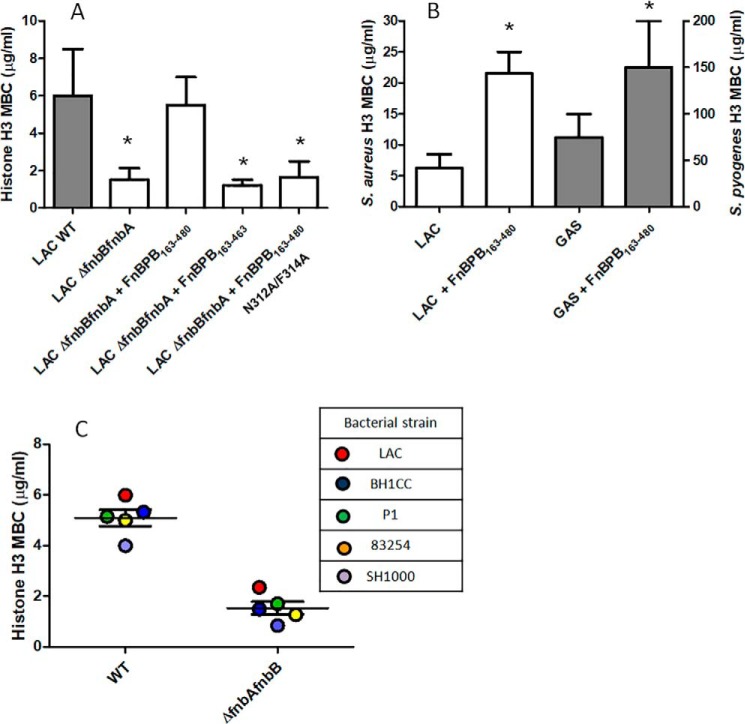
FnBPB expression on the surface of S. aureus LAC cells protects against the killing activity of histones. A, to determine the MBC of histones, cells of S. aureus LAC were incubated with the indicated increasing amounts of individual histones. After incubation, cell mixtures were plated on BHI agar, and colony counts were determined. Cells of S. aureus LAC, the ΔfnbAfnbB mutant, the mutant overexpressing FnBPA or FnBPB, and clfA and the srtA mutants were tested in the MBC assay with histone H3 in B or histone H4 in C. After incubation, serial dilutions of the cell mixtures were plated onto BHI agar and colony counts determined.
The MBC of histone H3 toward the variants of S. aureus LAC described above were measured. The MBC of H3 for the ΔfnbAfnbB mutant was ∼3-fold lower than the WT LAC, whereas the mutant overexpressing FnBPB from a multicopy plasmid had an ∼7-fold higher MBC than the mutant, which is consistent with the overexpressed FnBPB protein providing enhanced protection from the bactericidal activity of H3 (Fig. 8B). The MBC of H3 measured for the clfA mutant was similar to that for WT LAC, indicating that a mutation in a gene encoding a CWA protein other than fnbB does not affect the killing activity of H3 (Fig. 8B). Consistent with a significantly reduced binding to histones (Fig. 1), the srtA mutant showed a high susceptibility to H3 killing, as indicated by an ∼4-fold lower MBC value than the WT LAC (Fig. 8B). To further demonstrate the specificity of H3 binding to FnBPB, we also measured the MBC values of histone H4 for the LAC WT, the ΔfnbAfnbB mutant, the mutant overexpressing FnBPA or FnBPB, the srtA mutant, and the clfA mutant. As shown in Fig. 8C, the MBC values of H4 for the WT and the fnb mutants and complemented strains were very similar. In contrast, the MBC of H4 for srtA mutant was 4-fold lower than WT, indicating a higher susceptibility of the mutant to H4.
Soluble recombinant FnBPB(163–480) was added to a suspension of LAC ΔfnbAfnbB mutant cells to determine whether the protein protected susceptible bacteria from the bactericidal activity of H3. WT FnBPB(163–480) promoted survival of the mutant, whereas proteins defective in the ability to bind FBG and H3 failed to offer any protection. These data are consistent with FnBPB binding to soluble H3 and neutralizing its bactericidal activity (Fig. 9A).
Figure 9.
Comparison of MBC values of H3 for S. pyogenes and S. aureus strains. A, to evaluate the potential attenuation of microbicidal activity of histone H3 by soluble FnBPB(163–480), cells of S. aureus ΔfnbAfnbB mutant were tested in the MBC assay for killing by H3 in the presence of recombinant FnBPB(163–480) or its variants. After incubation, serial dilutions of the cell mixtures were plated on BHI agar and colonies counted. The MBC for WT is reported as control. Data are the mean values ± S.D. from three independent experiments. Statistically significant differences compared with the WT are indicated (Student's t test; *, p < 0.05). B, to determine the neutralizing effect of soluble FnBPB(163–480) on the bactericidal effect of H3 for S. pyogenes 71-695 and S. aureus LAC, bacteria were incubated in the presence/absence of FnBPB(163–480). Following incubation, serial dilutions of the mixtures were plated on BHI agar and colonies counted. Data shown are the mean values ± S.D. from three independent experiments. Statistically significant differences compared with the untreated WT are indicated (Student's t test; *, p < 0.05). C, protective effect of FnBPB expression against cytotoxicity of histone H3 was assessed by evaluating the MBC against different S. aureus strains and their fnb mutants.
S. pyogenes, also known as group A Streptococcus, is a Gram-positive bacterium for which the bactericidal action of histones has been previously demonstrated (34, 35). Thus, we asked whether soluble FnBPB(163–480) could competitively neutralize the bactericidal activity of H3 for this bacterium. The MBC value of H3 for the S. pyogenes serotype M1 strain 71-695 was ∼75 μg/ml, and exogenous addition of 5 μm FnBPB(163–480) to the MBC assay resulted in a 2-fold increase in resistance to the killing activity of H3. In a similar experimental approach, the MBC for S. aureus LAC was ∼6 μg/ml, and a 3-fold increase of MBC was produced when S. aureus WT LAC was incubated with H3 in the presence of 1 μm FnBPB(163–480) (Fig. 9B). Together, these data demonstrate that in both bacterial species soluble FnBPB(163–480) can confer resistance to H3-mediated killing.
To determine whether FnBPBs expressed by different strains of S. aureus also protected bacteria from the bactericidal effects of histones, the MBCs of H3 toward strains BH1CC (4), 8325-4 (42), and SH1000 (43) that express isoform I FnBPB and P1 (44) that expresses isoform IV FnBPB were very similar, although mutants lacking FnBPs (4, 21, 22, 45) were ∼3-fold more susceptible. This shows that both isoform I and isoform IV of FnBPB protect different strains of S. aureus to a similar degree despite isoform IV binding H3 less strongly than isoform I in vitro (Fig. 9C).
FnBPB expression protects S. aureus from NETs
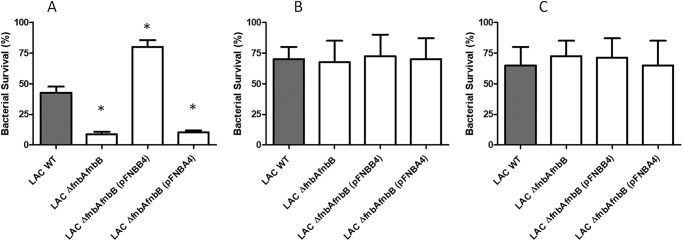
To determine whether histones released when neutrophils undergo NETosis exert bactericidal activity toward S. aureus, human neutrophils were stimulated to release NETs using a protein kinase C agonist and then incubated with the S. aureus LAC variants described previously. The ΔfnbAfnbB mutant survived less well than WT LAC, whereas the mutant overexpressing FnBPB survived much better than the mutant or indeed the WT. Strains expressing FnBPA were not protected (Fig. 10A). Treatment of the NETs with protease-free DNase allowed the ΔfnbAfnbB mutant and the mutant expressing FnBPA to survive to the same degree as WT LAC and the FnBPB-overexpressing strain (Fig. 10B). This indicates that bacteria must be entrapped within the DNA of the NETs for maximum bactericidal activity. Degradation of the NETs is likely to cause bactericidal proteins to be dissipated and diluted. Incubation of the NET with anti-histone antibody also restored survival of the ΔfnbAfnbB mutant and the mutant expressing FnBPA to the same level as WT and the mutant expressing FnBPB, which suggests that histones are the major bactericidal factors acting against S. aureus in NETs (Fig. 10C).
Figure 10.
FnBPB protein protects against histones released from neutrophils. A, neutrophils were stimulated with PMA to induce NETs and then incubated with S. aureus LAC, the ΔfnbAfnbB mutant, and the complemented mutant overexpressing FnBPA or FnBPB. After incubation, dilutions were plated on BHI agar and the colony counts determined. B, NETs treated with DNase I. C, NETs treated with anti-histone antibodies prior to infection. Surviving colony-forming units were calculated relative to initial inoculum incubated with untreated NETs.
Activated plasminogen captured by FnBPB cleaves H3 and enhances bacterial survival
We previously showed that FnBPs expressed on the surface of S. aureus can capture PLG, which can be activated by PLG activators to cleave FBG (22). Hence, we investigated whether recombinant FnBPB could simultaneously bind PLG and histone H3. FnBPB(163–480) was immobilized in microtiter plate wells and tested for binding of histone H3 in the presence of saturating amounts of PLG in an ELISA-type assay. The level of bound PLG remained the same (as detected with a specific antibody) in increasing concentrations of H3 (Fig. 11A). A similar result was obtained when increasing concentrations of PLG were added in the presence of a saturating concentration of H3 (Fig. 11B). Together, these data indicate that PLG and H3 bind to distinct sites on FnBPB consistent with H3 binding by the DLL mechanism.
Figure 11.

Binding of FnBPB to H3 in the presence of PLG. A, recombinant FnBPB(163–480) was immobilized on the surface of microtiter wells. Saturating concentrations of PLG were added along with increasing concentrations of H3. Bound PLG was detected with rabbit anti-PLG IgG followed by HRP-conjugated goat anti-rabbit IgG (solid squares). In the same panel, binding of increasing amounts of H3 to the wells is also reported (open circles). Bound H3 was detected with rabbit anti-histone antibodies followed by HRP-conjugated goat anti-rabbit IgG. B, ELISA-type assay with rFnBPB immobilized on the surface of microtiter wells. Saturating concentrations of H3 were added along with increasing amounts of PLG. Bound H3 was detected with rabbit anti-histones IgG followed by HRP-conjugated goat anti-rabbit IgG (solid squares). The panel shows binding of increasing amounts of PLG to the wells (open circles). Bound proteins were detected with specific antibodies as in A. The data points reported in the panels are the means ±S.D. of three independent experiments each performed in triplicate.
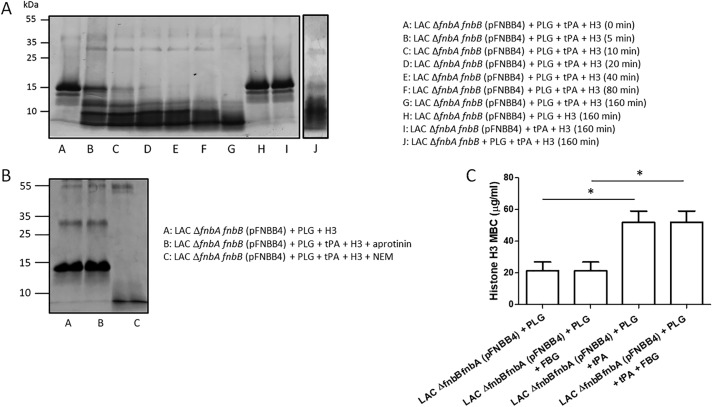
To determine whether bacteria-bound PLG can be activated by tissue plasminogen activator (t-PA) and can cleave H3, S. aureus LAC ΔfnbAfnbB cells expressing high levels of FnBPB from a multicopy plasmid were immobilized and incubated with PLG. After washing, both H3 and t-PA were added and incubated for different times. SDS-PAGE analysis revealed that H3 was rapidly degraded, whereas no cleavage of H3 was seen in controls incubated with PLG or t-PA alone. Interestingly, as reported previously (22), PLG bound to ΔfnbAfnbB mutant cells, and when activated to plasmin, it degraded H3 (Fig. 12A). In any case, it remains to be established whether activated PLG preferentially degrades bacteria-bound H3 rather than free H3 (or both).
Figure 12.
Specificity of H3 cleavage by bacteria-bound plasmin. A, cells of S. aureus LAC ΔfnbAfnbB mutant overexpressing FnBPB were immobilized on the surface of microtiter plates and then incubated with human PLG. After washing, t-PA and histone H3 were added to the wells and incubated for increasing periods of time. Supernatants were subjected to SDS-PAGE and the gels stained with Coomassie Blue. Controls made of mixtures without PLG or t-PA are reported. The effect of activated PLG captured by the ΔfnbAfnbB mutant cells on H3 cleavage is also reported. The figure is representative of two independent experiments. B, to demonstrate the specificity of proteolysis, immobilized bacteria were incubated with PLG, and t-PA and H3 were added and further incubated in the presence of aprotinin or NEM. Supernatants were then analyzed for H3 cleavage by SDS-PAGE. C, cells of the S. aureus LAC ΔfnbAfnbB mutant overexpressing FnBPB were mixed with human PLG, added to serial 1.3-fold diluted histone H3 in the presence/absence of t-PA, and incubated for additional periods of time. The MBC of histone H3 was determined by plating serial dilutions of bacteria on BHI agar. To analyze the effect of FBG on H3 cleavage by activated PLG, bacteria preincubated with PLG were mixed with equimolar concentrations of H3 and FBG and processed as above. The reported data are the mean values ± S.D. from three independent experiments. Statistically significant differences are indicated (Student's t test; *, p < 0.05).
In support of specific plasmin generation on the surface of bacterial cells, the serine protease inhibitor aprotinin was found to inhibit H3 degradation. In contrast, complete cleavage of H3 was observed when the incubation was carried out in the presence of the cysteine protease inhibitor N-ethylmaleimide (NEM) (Fig. 12B). To investigate the possibility that activated PLG also degraded surface-expressed FnBPB, immobilized cells of S. aureus LAC ΔfnbAfnbB expressing (p-FnBPB) were incubated with PLG, and the complex was activated with t-PA. No signal was detected when the supernatant obtained from the incubation mixture was analyzed for the presence of FnBPB digestion products by Western immunoblotting, suggesting that FnBPB is not cleaved by bacteria-bound PLG (data not shown).
Next, the ability of activated PLG to protect S. aureus from the bactericidal activity of histone H3 was investigated. S. aureus LAC ΔfnbAfnbB cells expressing high levels of FnBPB from a multicopy plasmid were incubated with PLG and t-PA, and the MBC of H3 was found to be ∼50 μg/ml. For bacteria incubated with PLG without t-PA, the MBC was ∼2-fold lower. An almost identical MBC value was obtained even in the presence of equimolar concentrations of FBG, suggesting that H3 preferentially binds to FnBPB and cleaved by PLG (Fig. 12C).
Discussion
Histones are constituents of the nucleosome in the nucleus of mammalian cells where they act to compact DNA and regulate gene expression. However, it is becoming apparent that histones also occur outside the nucleus, for example in granules of phagocytic cells or mucosal surfaces, including the human stomach, and are released into the bloodstream at elevated levels during sepsis. Histones are also secreted by sebocytes of the sebaceous gland in the skin and are thought to have antimicrobial activity against commensals living in the skin such S. aureus (46).
Several reports indicate that S. aureus can bind histones, but virtually nothing is known about the factors involved or the mechanism of bactericidal action (33). This study began by showing that a sortase mutant bound a lower level of histones than the WT, and we were able to show that a single CWA protein, FnBPB, was responsible for this interaction. The N2–N3 region of FnBPB bound specifically to CTH in vitro, although the most closely related surface proteins, such as FnBPA and ClfA, did not. When FnBPB binding to individual histones was tested, it was found that all types could bind but that H3 displayed the highest binding and bactericidal activity. We found that H3 binds to the N2N3 region of FnBPB, most likely by the dock, lock, and latch mechanism used to bind FBG (Fig. 13), because variants lacking the ability to bind FBG were also defective in H3 binding. To prove that the DLL mechanism is involved in the binding of FnBPB to H3, co-crystallization of FnBPB(163–480) in complex with H3 could be carried out. Notably, FnBPB has a 20-fold higher affinity for H3 than FBG suggesting that in plasma or other environments where FBG is present, FnBPB would bind H3 preferentially. Furthermore, increasing ionic strength had a dramatic effect on FnBPB binding to H3, indicating that H3/FnBPB complex formation is mainly driven by electrostatic interactions.
Figure 13.
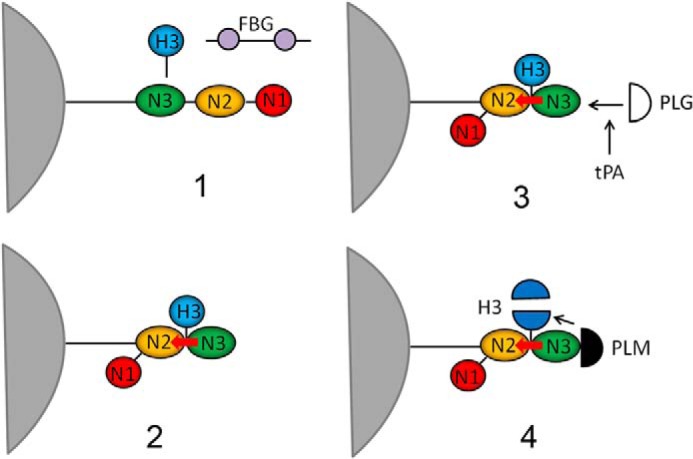
Proposed model of FnBPB in binding and inactivation of histone H3. The figure shows a sequence of events (numbered from 1 to 4) involving FnBPB expressed on the surface of S. aureus. At first, in preference to fibrinogen, H3 binds to the trench between the N2 and N3 subdomains of FnBPB, and once in place it induces a conformational change that enables C-terminal residues of the N3 extension (red arrow) to be inserted into the trench and to stabilize the complex (dock, lock, and latch mechanism). Then, plasminogen (PLG) (open semicircle) binds to a site on N3 subdomain and once activated by t-PA to plasmin (PLM) (solid semicircle) cleaves FnBPB-bound H3.
The specificity of the interaction between FnBPB(163–480) and H3 was demonstrated by capture ELISA, gel filtration, and surface plasmon resonance, which together indicate that the proteins form a complex even in the fluid phase. The almost identical susceptibility of S. aureus LAC and its fnbB mutants to the treatment with H4 suggests a specific role of FnBPB in the protection of bacteria by H3. The data obtained with srtA mutant also showed that H4 interacts with a bacterial surface component other than FnBPB and exhibits a toxicity for staphylococcal cells at a level equivalent to H3. This effect is consistent with the finding that histone H4 is an important component of the antimicrobial action of human sebocytes against S. aureus (46).
Seven different isoforms of FnBPB can bind to H3, but some have a higher affinity than others. This paper focused on S. aureus strain LAC, which expresses isoform I FnBPB. This isoform binds H3 strongly in vitro (KD of 50 nm). It is noteworthy that strain P1 that expresses the apparently weaker binding FnBPB isoform IV (apparent KD of 636.7 nm as estimated from ELISA) is protected to the same extent as LAC from the bactericidal activity. With respect with this point, it would be interesting to compare mutants of strains expressing other isoforms.
FnBPB can also bind to PLG at a site that does not overlap that of FBG. Indeed, FnBPB can bind both ligands independently (22). In a similar fashion, FnBPB can bind PLG to the same extent in the presence of H3 as in its absence. Indeed, bound PLG could be activated to plasmin by t-PA and rapidly degraded H3.
Previously, we showed that the double ΔfnbAfnbB mutant of S. aureus LAC still binds to PLG, which indicates that additional surface components can bind PLG (22). Consistent with this, we found that PLG captured by the ΔfnbAfnbB mutant efficiently digests H3 proving that, whatever the staphylococcal receptor, PLG retains its proteolytic potential. Importantly, cleavage of H3 by PLG was not affected by the presence of FBG, suggesting that even in closer physiological conditions H3 is degraded by PLG.
Summing up, we propose that FnBPB contributes to protection of S. aureus (i) by capturing/scavenging histones and preventing them from reaching the membrane and (ii) by promoting their degradation by binding PLG, which is activated by exogenously or endogenously expressed PLG activators (Fig. 13). It is unknown whether these mechanisms are operational in vivo. Thus, definition of the role of FnBPB and PLG as inhibitors of the bactericidal activity of H3 in animal models may be worth exploring.
In a study by Nitzsche et al. (34), S. pyogenes serotype M49 has been shown to protect itself from histone-killing through PLG acquisition and activation by streptokinase. However, the question was not addressed whether the M49 protein is directly involved in PLG and/or histone binding (34). Along this line, Finegoldia magna, a Gram-positive commensal of the skin and mucous membranes, binds histones extracted from human skin through the surface protein FAF (F. magna adhesion factor), and histones were found to be degraded by SufA (subtilase of F. magna), a subtilisin-like extracellular serine protease of F. magna (47). Thus, different bacterial species, including S. aureus, may use similar strategies to neutralize the antibacterial activity of histones.
It is clear that FnBPB is the major histone-binding cell wall–anchored surface protein expressed by S. aureus LAC under the in vitro growth conditions employed here. By using genetically manipulated strains, FnBPB expression was shown to be responsible for binding H3 and protecting S. aureus from the bactericidal effects of H3 or the total histones released by neutrophils when NETs were formed. Effects were severely reduced in the ΔfnbAfnbB mutant and, importantly, were enhanced to a greater extent than in the WT strain when FnBPB was expressed at enhanced levels from a multicopy plasmid. Loss of FnBPB reduced the bactericidal effect of H3 toward several other S. aureus strains. It could be argued that protecting S. aureus cells from traps extruded from the myeloid cell is the most important function of the A region of FnBPB given the high affinity of FnBPB for histones.
The WT LAC is efficiently killed by a relatively low dose of H3 (MBC: 5 μg/ml). Additionally, as reported previously, histones are expressed and extruded in NETs in abundant amounts, estimated at 2.5 μg/106 neutrophils, such that histones comprise more than two-thirds of the protein content within the NET structure (48). This information raises the question whether inhibition by FnBPB of histone-mediated bacterial killing is biologically significant. Notably, data reported here show that FnBPB protects staphylococci from killing by NETs, demonstrating that FnBPB-mediated resistance is important when H3 is present in a biologically relevant milieu. It is possible that other factors present in the NETs could down-regulate the bactericidal activity of histones. Among these, entrapment of histones by DNA might contribute to the reduction of bioavailability of histones in NETs and facilitate the histone-scavenging activity of FnBPB. Consideration should be also given to the fact that H3 is a fractional component (20%) of the total amount of histones in the NETs (48), and this could enhance the efficacy of the neutralizing activity of FnBPB.
Recently, it has been demonstrated that S. aureus extracellular protein Eap shows a neutrophil serine protease inhibitor activity in association with NET functions (49) and binds and aggregates DNA, thus blocking neutrophil extracellular trap formation (50). With this study, FnBPB is added to the list of potential factors that inhibit this important mechanism of host innate defense.
Other bacterial species block the killing activity of NETs through the neutralization of histones. For example, Dörhmann et al. (35) have shown the antimicrobial activity of histones against group A streptococci belonging to different serotypes. In particular, the surface-expressed M1 protein, a classical virulence factor of this pathogen, was required to bind and inactivate extracellular histones in NETs, deploying an FnBPB-like evasion strategy (35). Interestingly, soluble FnBPB(163–480) neutralized the ability of H3 to interact with and subsequently kill S. aureus as well as S. pyogenes. Hence, one can envisage the development of therapeutic derivatives of this protein.
In conclusion, we have shown that FnBPB is the dominant binding protein for capturing histone H3 by S. aureus and conferring resistance to the bacterium against the antimicrobial activity of histones. We have also investigated the molecular details of the histones binding to such an important virulence factor.
Experimental procedures
Bacterial strains and culture conditions
All strains are listed in Table 1. S. aureus was grown in Brain Heart Infusion broth (VWR International Srl, Milan, Italy) to mid-exponential phase (OD = 0.4) at 37 °C with shaking. The S. pyogenes serotype M1 strain 71-675 was received from Professor Bernd Kreikemeyer (Rostock, Germany). Bacteria were cultured in Todd-Hewitt broth supplemented with yeast extract at 37 °C with shaking.
Table 1.
Bacterial strains
Abbreviations used are as follows: Ermr, erythromycin resistance; Tcr, tetracycline resistance.
| Bacterial strains | Relevant properties | Refs. |
|---|---|---|
| S. aureus LAC | Community-associated MRSA of USA300 lineage | 8 |
| S. aureus LAC srtA | Constructed by the transduction of srtA::Ermr from Newman srtA (39) using bacteriophage 85 | This study |
| S. aureus LAC clfA | Constructed by the transduction of clfA::Ermr from Newman clfA (51) using bacteriophage 85 | This study |
| S. aureus LAC ΔfnbAfnbB | Deletion of fnbA and fnbB genes isolated by allelic exchange | 40 |
| S. aureus LAC ΔfnbAfnbB (pFNBA4) | Mutant transformed with plasmid-expressing FnBPA | 40 |
| S. aureus LAC ΔfnbAfnbB (pFNBB4) | Mutant transformed with plasmid-expressing FnBPB | 40 |
| S. aureus BH1CC | Hospital-associated MRSA | 4 |
| S. aureus BH1CC ΔfnbAfnbB | fnbA::Tcr fnbB::Ermr mutations transduced from 8325-4 fnbA fnbB | 4 |
| S. aureus P1 | Rabbit passaged strain derived from ATCC25923 | 44 |
| S. aureus P1 ΔfnbAfnbB | fnbA::Tcr fnbB::Er20mr mutations transduced from 8325-4 fnbAfnbB | 21 |
| S. aureus 8325-4 | NCTC8325 cured of three prophages | 42 |
| S. aureus 8325-4 ΔfnbAfnbB | fnbA::Tcr fnbB::Ermr mutations isolated by allelic exchange | 45 |
| S. aureus SH1000 | rbsU restored in 8325-4 | 43 |
| S. aureus SH1000 ΔfnbAfnbB | fnbA::Tcr fnbB::Ermrmutations transduced from 8325-4 fnbA fnbB | 22 |
In those experiments, where a defined number of cells were used, bacteria were harvested from the cultures by centrifugation, washed, suspended in PBS, and counted in a Petroff-Hausser chamber. Escherichia coli TOPP3 transformed with vector pQE30 (Stratagene, La Jolla, CA) or derivatives was grown in Luria agar and Luria broth (VWR International Srl, Milan, Italy) containing 100 μg/ml ampicillin. S. aureus LAC srtA was constructed by the transduction of srtA::Ermr from Newman srtA (39) using bacteriophage 85. S. aureus LAC clfA was constructed by the transduction of clfA::Ermr from Newman clfA using bacteriophage 85 (51).
DNA manipulation
DNA manipulation was performed as previously reported (22).
Expression and purification of recombinant proteins
Recombinant proteins FnBPB(163–308), FnBPB(309–480), and FnBPB(163–463) latch-truncated were expressed from pQE30 (Qiagen, Chatsworth, CA) in E. coli TOPP3 (Stratagene), as reported previously (22).
Recombinant FnBPB isotypes (20), FnBPB(163–480) N312A/F314A trench mutant (16), FnBPA(194–511) (15), CNA(31–344) (52), ClfA(221–559) (53), ClfB(201–542) (54), Bbp(40–599) (55), and Pls(49–694) (56), were each previously expressed with His6 N-terminal affinity tags using E. coli vectors and purified on a HiTrap chelating column (GE Healthcare, Buckinghamshire, UK) by Ni2+-chelate chromatography as described above. Protein purity was assessed to be 98% by SDS-PAGE, Coomassie Brilliant Blue staining, and densitometry analysis. A bicinchoninic acid protein assay (Pierce) was used to measure concentrations of purified proteins.
Proteins and reagents
Protease-free DNase I, BSA (BSA), skim milk, CTH, and human histones H1, H2A, H2B, H3, and H4 were purchased from Sigma.
Antibodies
Rabbit anti-mouse and goat anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Dako Cytomation (Glostrup, Denmark). Anti-human histone antibody was purchased from USBiological (Salem, MA). Polyclonal antiserum against FnBPB(163–480) was raised in a rabbit by routine immunization procedure using purified FnBPB(163–480) as antigen.
7E8 is an in-house–generated murine mAb recognizing recombinant His-tagged proteins. To validate ELISAs, where different His-tagged bacterial proteins were assessed for reactivity to antibody 7E8, increasing amounts of His-tagged CWA proteins, variants of FnBPB(163–480), or isoforms of FnBPB(163–480) were immobilized, and bound 7E8 was detected using HRP-conjugated rabbit anti-mouse IgG.
ELISA type solid-phase binding assays
The ability of soluble recombinant S. aureus proteins to bind to immobilized CTH or individual histones was determined using ELISA-type assays. Microtiter wells were coated overnight at 4 °C with 0.5 μg/well of CTH or individual histones in 0.1 m sodium carbonate, pH 9.5. The plates were washed with 0.5% (v/v) Tween 20 in PBS (PBST). To block additional protein-binding sites, the wells were treated for 1 h at 22 °C with bovine serum albumin (BSA) 2% (v/v) in PBS. The plates were then incubated for 1 h with 0.5 μmol of each ligand. After several washes with PBST, 0.5 μg of the specific mouse anti-hexahistidine tag mAb 7E8 in BSA (1% v/v) was added to the wells and incubated for 90 min. The plates were washed and incubated for 1 h with HRP-conjugated rabbit anti-mouse IgG diluted 1:1000. After washing, o-phenylenediamine dihydrochloride was added, and the absorbance at 490 nm was determined using an ELISA plate reader.
In some ELISA-type assays, a primary rabbit antibody binding was detected incubating the wells with HRP-conjugated goat anti-rabbit IgG secondary antibody. To calculate the relative affinity association constant (KA), values of each FnBPB isoform for H3 histone, the data were fitted using Equation 1,
| (Eq. 1) |
where [L] is the molar concentration of isoform. The reported dissociation constants (KD values) were calculated as reciprocals of the KA values.
To determine the effect of ionic strength on the H3–FnBPB interaction, microtiter wells coated with 500 ng of H3 were incubated with 1 μg/well of FnBPB(163–480) diluted in a phosphate buffer containing increasing concentrations of NaCl (0.15–2.3 m). Complex formation was detected by incubation of the wells with a rabbit anti-FnBPB antibody, followed by HRP-conjugated goat anti-rabbit secondary antibody.
Adherence of bacteria to surface-coated CTH or histone H3 was performed by incubating immobilized histones (2 μg/well) with log-phase S. aureus cells (108) for 2 h at 37 °C. After several washings with PBS, adherent cells were fixed with 2.5% formaldehyde for 30 min and stained with 1% crystal violet for 1 min. After washing with PBS, 100 μl of 10% acetic acid was added to the wells, and absorbance at 595 nm was recorded in an ELISA plate reader.
Size-exclusion chromatography analysis of the interaction of FnBPB(163–480) with histone H3
Samples of FnBPB(163–480), histone H3, and mixtures of co-incubated equimolar amounts of FnBPB(163–480) (or FnBPB(163–480) trench mutant variant (N312A/F314A)) and H3 were loaded onto a 75 10/300 GL gel-filtration chromatography column of Superdex (Vt = 24 ml). The column, equilibrated with PBS, was eluted with the same buffer at a flow rate of 0.75 ml/min. To calibrate the column, standard proteins (GE Healthcare) were allowed to pass through the column, and their partition coefficient Kav was plotted against the logarithm of the molecular weight.
Cleavage of histone H3 (or FnBPB) by S. aureus-associated activated plasminogen
5 × 107 cells of log-phase S. aureus LAC ΔfnbAfnbB (pFNBB4) or double ΔfnbAfnbB mutant were immobilized on the surface of microtiter plates and then incubated with human PLG (1 μg/well) for 1 h at 37 °C. After several washes, cell-coated wells were added with 27 nm t-PA and histone H3 (5 μg/well), and the mixtures were incubated at 22 °C for increasing periods of time (5–160 min). Supernatants containing cleaved histone H3 were subjected to 15% SDS-PAGE, and the gels were stained with Coomassie Brilliant Blue (see below).
To assess the specific activation of PLG to plasmin, surface-coated S. aureus LAC ΔfnbAfnbB (pFNBB4) was incubated with PLG, t-PA, and H3 in the presence of 5 μg/ml aprotinin or 100 μm NEM for 160 min, and the supernatants were subjected to 15% SDS-PAGE, as reported above.
To evaluate the possible cleavage of surface-expressed FnBPB by bacteria-bound plasmin, immobilized S. aureus cells were incubated with PLG and t-PA for 160 min, as reported above. Supernatant containing released material was subjected to 12.5% SDS-PAGE, and the proteins were transferred to a nitrocellulose membrane. The membrane was sequentially probed with a rabbit anti-FnBPB IgG and HRP-conjugated goat anti-rabbit IgG.
Capture of histone H3 by S. aureus cells
Log-phase S. aureus strain USA 300 LAC, the double mutant ΔfnbAfnbB, or the mutants transformed with plasmid overexpressing FnBPB or FnBPA (108 cells/ml) were mixed with 10 μg of histone H3 for 10 min. Bacteria were then harvested by centrifugation, washed with PBS, and treated with the extraction buffer (125 mm Tris-HCl, pH 7.0, containing 2% SDS) for 3 min at 95 °C and then centrifuged at 10,000 × g for 3 min. The supernatants were subjected to SDS-PAGE under nonreducing conditions, and the proteins were transferred to a nitrocellulose membrane. The membrane was sequentially probed with rabbit anti-histone IgG and HRP-conjugated goat anti-rabbit IgG (see below).
SDS-PAGE, Western immunoblotting, and far Western immunoblotting
Digestion products of histone H3 were boiled for 3 min in sample buffer (0.125 m Tris-HCl, 4% (w/v) SDS, 20% (v/v) glycerol, 10% (v/v) β-mercaptoethanol, 0.002% (w/v) bromphenol blue) and separated by 15% (w/v) SDS-PAGE. The gels were stained with Coomassie Brilliant Blue (Bio-Rad, Milan, Italy).
For Western immunoblotting, histone H3 captured and released from the bacterial cell surface was subjected to 15% SDS-PAGE and electroblotted onto a nitrocellulose membrane (GE Healthcare), and the membrane was blocked overnight at 4 °C with 5% (w/v) skim milk (Sigma) in PBS. Blotted proteins were probed with rabbit polyclonal anti-histone (1:5000) for 1 h at 22 °C. Following washes with PBST, the membrane was incubated for 1 h with HRP-conjugated goat anti-rabbit IgG (1:10,000). Finally, the blot was developed using the ECL Advance Western blotting detection kit (GE Healthcare), and an ImageQuantTM LAS 4000 mini-biomolecular imager (GE Healthcare) was used to capture images of the bands. The band intensities were quantified relative to the histone H3 (5 μg, 100% intensity) with the Quantity One software (Bio-Rad).
For far Western immunoblotting, the individual histones were subjected to 15% SDS-PAGE and electroblotted onto a nitrocellulose membrane (GE Healthcare), and the membrane was blocked overnight at 4 °C with 5% (w/v) skim milk in PBS. The membrane was probed with 1 μg/ml FnBPB(163–480) for 1 h at 22 °C followed by rabbit anti-FnBPB(163–480) polyclonal antibody (1:5000) and with HRP-conjugated goat anti-rabbit IgG (1:10,000), and the complexes were detected as reported above.
Surface plasmon resonance analysis of FnBPB binding to histone H3
To estimate the affinity of the interaction between H3 and FnBPB(163–480), surface plasmon resonance was conducted using a BIAcore X-100 instrument (GE Healthcare). Histone H3 was covalently immobilized on dextran matrix CM5 sensor chip surface by using a histone H3 solution (30 μg/ml in 50 mm sodium acetate buffer, pH 5) in a 1:1 dilution with N-hydroxysuccinimide and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride. The excess of active groups on the dextran matrix was blocked using 1 m ethanolamine, pH 8.5. On another flow cell, the dextran matrix was treated as described above but without any ligand to provide an uncoated reference flow cell. The running buffer used was PBS containing 0.005% (v/v) Tween 20. A 2-fold linear dilution series (3.9–500 nm) of FnBPB(163–480), in running buffer, was passed over the ligand at the flow rate of 10 μl/min, and all the sensorgrams were recorded at 22 °C. Assay channel data were subtracted from reference flow cell data. The response units at steady state were plotted as a function of FnBPB(163–480) concentration and fitted to the Langmuir equation to yield the KD values.
Isolation of neutrophils
Neutrophils were isolated from healthy donors using PolyMorphPrep solution (ThermoFisher Scientific) as described by Kristian et al. (57). Viability of neutrophils was assessed using 0.004% trypan blue staining by light microscopy.
NET-mediated killing of bacteria
Killing of bacteria by NETs was performed as reported previously (41). Briefly, 105 neutrophils were seeded into a 96-well plate and stimulated with 25 nm PMA for 4 h to induce NETosis. Log-phase bacteria were washed twice and added to neutrophils at a multiplicity of infection of 1 and incubated for 15 min at 37 °C, 5% CO2. To evaluate the role of DNA in NET-mediated killing of bacteria, NETs were degraded prior to infection with 2 units of DNase I for 10 min at 37 °C. The bactericidal activity of histones was blocked prior to infection with 2 μg/ml anti-human histone antibody for 30 min. In both cases, neutrophils were washed once prior to challenge. Colony-forming units were enumerated by plating onto brain heart infusion (BHI) the cells suspension. Bacterial survival was calculated as percentage of the initial inoculum.
Determination of histone minimal bactericidal concentrations
Resistance of S. aureus USA 300 LAC to individual histones was determined in a minimum bactericidal concentration (MBC) assay incubating bacteria (1 × 108) with 1.3-fold diluted histone starting from 60 to 0.93 μg/ml. The MBC value of H3 for S. pyogenes strain 71-675 was determined by using 1.3-fold dilution of histone starting from 400 to 29 μg/ml. After 2 h of incubation at 37 °C, serial dilutions of cell mixtures were plated on BHI agar, and bacterial colonies counts were determined the day after. The MBC was calculated by determining the lowest concentration of each histone that reduced the viability of the initial bacterial inoculum by ≥99.9%. The MBC of H4 for WT LAC and its mutants was evaluated as reported above. To determine the neutralizing effect of soluble FnBPB(163–480) and its mutated variants on killing by H3, the MBC of H3 for S. aureus ΔfnbAfnbB was determined in the presence of 1 μm FnBPB proteins. A similar experimental protocol was used to measure the effect of FnBPB(163–480) on MBC of H3 for S. pyogenes strain 71-675 in the presence of 5 μm FnBPB(163–480).
Determination of the protective effect of plasminogen on S. aureus from the bactericidal activity of histone H3
Log-phase cells of S. aureus LAC ΔfnbAfnbB (pFNBB4) (2 × 105/ml) were incubated with human PLG (10 μg) for 1 h. Cells were harvested by centrifugation, washed with PBS, and then incubated with serial 1.3-fold diluted histone H3 (from 100 to 0.78 μg/ml) in the presence/absence of t-PA (27 nm) for 2 h at 37 °C. In those experiments where the effect of fibrinogen on H3 cleavage by activated PLG captured by staphylococci was assessed, equimolar concentrations of H3 and fibrinogen were incubated with PLG captured by staphylococci, and the mixtures were processed as above.
To determine the MBC of histone H3, serial dilutions of the mixtures were plated on BHI agar and incubated at 37 °C overnight. Bacterial counts were determined the day after.
Statistical methods
Continuous data were expressed as means and standard deviations. Two group comparisons were performed by Student's t test. One-way analysis of variance, followed by Bonferroni's post hoc tests, was exploited for comparison of three or more groups. Analyses were performed using Prism 4.0 (GraphPad). Two-tailed p values of 0.05 were considered statistically significant.
Author contributions
G. P., T. J. F., J. A. G., V. D. F., and P. S. conceptualization; G. P. and P. S. resources; G. P. and P. S. data curation; G. P., G. N., M. J. A., and P. S. formal analysis; G. P. and P. S. supervision; G. P., G. N., and P. S. validation; G. P., M. J. A., and P. S. investigation; G. P. and P. S. visualization; G. P., G. N., M. J. A., T. J. F., and P. S. methodology; G. P., V. D. F., and P. S. writing-original draft; G. P., T. J. F., J. A. G., V. D. F., and P. S. writing-review and editing; V. D. F. and P. S. funding acquisition.
Acknowledgments
We thank Dara P. O'Halloran for constructing the LAC srtA mutant and Leanne M. Hays for constructing the LAC clfA mutant.
This work was supported by Fondazione CARIPLO Grant Vaccines 2009-3546 (to P. S.) and in part by PRAT-2015 Grant (to V. D. F.). The authors declare that they have no conflicts of interest with the contents of this article.
- MRSA
- methicillin-resistant S. aureus
- Bbp
- bone sialoprotein-binding protein
- ClfA
- clumping factor
- CNA
- collagen adhesin
- CTH
- calf thymus histone
- CWA
- cell wall–associated protein
- FBG
- fibrinogen
- FnBPA
- fibronectin-binding protein A
- FnBPB
- fibronectin-binding protein B
- MBC
- minimum bactericidal concentration
- NEM
- N-ethylmaleimide
- NET
- neutrophil extracellular trap
- MSCRAMM
- microbial surface component recognizing adhesive matrix molecule
- PLG
- plasminogen
- PBST
- PBS containing Tween 20
- Pls
- plasmin-sensitive protein
- t-PA
- tissue plasminogen activator
- PMA
- phorbol 12-myristate 13-acetate
- DLL
- dock, lock, and latch
- HRP
- horseradish peroxidase
- BHI
- brain heart infusion.
References
- 1. Chambers H. F. (2005) Community-associated MRSA-resistance and virulence converge. N. Engl. J. Med. 352, 1485–1487 10.1056/NEJMe058023 [DOI] [PubMed] [Google Scholar]
- 2. Lowy F. D. (1998) Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- 3. Noskin G. A., Rubin R. J., Schentag J. J., Kluytmans J., Hedblom E. C., Smulders M., Lapetina E., and Gemmen E. (2005) The burden of Staphylococcus aureus infections on hospitals in the United States: an analysis of the 2000 and 2001 Nationwide Inpatient Sample Database. Arch. Intern. Med. 165, 1756–1761 10.1001/archinte.165.15.1756 [DOI] [PubMed] [Google Scholar]
- 4. O'Neill E., Pozzi C., Houston P., Humphreys H., Robinson D. A., Loughman A., Foster T. J., and O'Gara J. P. (2008) A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190, 3835–3850 10.1128/JB.00167-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vergara-Irigaray M., Valle J., Merino N., Latasa C., García B., Ruiz de Los Mozos I., Solano C., Toledo-Arana A., Penadés J. R., and Lasa I. (2009) Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect. Immun. 77, 3978–3991 10.1128/IAI.00616-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fridkin S. K., Hageman J. C., Morrison M., Sanza L. T., Como-Sabetti K., Jernigan J. A., Harriman K., Harrison L. H., Lynfield R., Farley M. M., and Active Bacterial Core Surveillance Program of the Emerging Infections Program Network. (2005) Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352, 1436–1444 10.1056/NEJMoa043252 [DOI] [PubMed] [Google Scholar]
- 7. Moran G. J., Krishnadasan A., Gorwitz R. J., Fosheim G. E., McDougal L. K., Carey R. B., Talan D. A., and EMERGEncy ID Net Study Group. (2006) Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355, 666–674 10.1056/NEJMoa055356 [DOI] [PubMed] [Google Scholar]
- 8. Diep B. A., Gill S. R., Chang R. F., Phan T. H., Chen J. H., Davidson M. G., Lin F., Lin J., Carleton H. A., Mongodin E. F., Sensabaugh G. F., and Perdreau-Remington F. (2006) Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367, 731–739 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- 9. Otto M. (2013) Community-associated MRSA: what makes them special? Int. J. Med. Microbiol. 303, 324–330 10.1016/j.ijmm.2013.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Foster T. J. (2017) Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 41, 430–449 10.1093/femsre/fux007 [DOI] [PubMed] [Google Scholar]
- 11. Foster T. J. (2019) Can β-lactam antibiotics be resurrected to combat MRSA? Trends Microbiol. 27, 26–38 10.1016/j.tim.2018.06.005 [DOI] [PubMed] [Google Scholar]
- 12. Foster T. J., Geoghegan J. A., Ganesh V. K., and Höök M. (2014) Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62 10.1038/nrmicro3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marraffini L. A., Dedent A. C., and Schneewind O. (2006) Sortases and the art of anchoring proteins to the envelopes of Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70, 192–221 10.1128/MMBR.70.1.192-221.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loughman A., Sweeney T., Keane F. M., Pietrocola G., Speziale P., and Foster T. J. (2008) Sequence diversity in the A domain of Staphylococcus aureus fibronectin-binding protein A. BMC Microbiol. 8, 74 10.1186/1471-2180-8-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keane F. M., Loughman A., Valtulina V., Brennan M., Speziale P., and Foster T. J. (2007) Fibrinogen and elastin bind to the same region within the A domain of fibronectin binding protein A, an MSCRAMM of Staphylococcus aureus. Mol. Microbiol. 63, 711–723 [DOI] [PubMed] [Google Scholar]
- 16. Burke F. M., Di Poto A., Speziale P., and Foster T. J. (2011) The A domain of fibronectin-binding protein B of Staphylococcus aureus contains a novel fibronectin binding site. FEBS J. 278, 2359–2371 10.1111/j.1742-4658.2011.08159.x [DOI] [PubMed] [Google Scholar]
- 17. Deivanayagam C. C., Wann E. R., Chen W., Carson M., Rajashankar K. R., Höök M., and Narayana S. V. (2002) A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J. 21, 6660–6672 10.1093/emboj/cdf619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ponnuraj K., Bowden M. G., Davis S., Gurusiddappa S., Moore D., Choe D., Xu Y., Hook M., and Narayana S. V. (2003) A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115, 217–228 10.1016/S0092-8674(03)00809-2 [DOI] [PubMed] [Google Scholar]
- 19. Bowden M. G., Heuck A. P., Ponnuraj K., Kolosova E., Choe D., Gurusiddappa S., Narayana S. V., Johnson A. E., and Höök M. (2008) Evidence for the “dock, lock, and latch” ligand binding mechanism of the staphylococcal microbial surface component recognizing adhesive matrix molecules (MSCRAMM) SdrG. J. Biol. Chem. 283, 638–647 10.1074/jbc.M706252200 [DOI] [PubMed] [Google Scholar]
- 20. Burke F. M., McCormack N., Rindi S., Speziale P., and Foster T. J. (2010) Fibronectin-binding protein B variation in Staphylococcus aureus. BMC Microbiol. 10, 160 10.1186/1471-2180-10-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roche F. M., Downer R., Keane F., Speziale P., Park P. W., and Foster T. J. (2004) The N-terminal A domain of fibronectin-binding proteins A and B promotes adhesion of Staphylococcus aureus to elastin. J. Biol. Chem. 279, 38433–38440 10.1074/jbc.M402122200 [DOI] [PubMed] [Google Scholar]
- 22. Pietrocola G., Nobile G., Gianotti V., Zapotoczna M., Foster T. J., Geoghegan J. A., and Speziale P. (2016) Molecular interactions of human plasminogen with fibronectin-binding protein B (FnBPB), a fibrinogen/fibronectin-binding protein from Staphylococcus aureus. J. Biol. Chem. 291, 18148–18162 10.1074/jbc.M116.731125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Geoghegan J. A., Monk I. R., O'Gara J. P., and Foster T. J. (2013) Subdomains N2N3 of fibronectin binding protein A mediate Staphylococcus aureus biofilm formation and adherence to fibrinogen using distinct mechanisms. J. Bacteriol. 195, 2675–2683 10.1128/JB.02128-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwarz-Linek U., Werner J. M., Pickford A. R., Gurusiddappa S., Kim J. H., Pilka E. S., Briggs J. A., Gough T. S., Höök M., Campbell I. D., and Potts J. R. (2003) Pathogenic bacteria attach to human fibronectin through a tandem β-zipper. Nature 423, 177–181 10.1038/nature01589 [DOI] [PubMed] [Google Scholar]
- 25. Meenan N. A., Visai L., Valtulina V., Schwarz-Linek U., Norris N. C., Gurusiddappa S., Höök M., Speziale P., and Potts J. R. (2007) The tandem β-zipper model defines high affinity fibronectin-binding repeats within Staphylococcus aureus FnBPA. J. Biol. Chem. 282, 25893–25902 10.1074/jbc.M703063200 [DOI] [PubMed] [Google Scholar]
- 26. Sollberger G., Tilley D. O., and Zychlinsky A. (2018) Neutrophil extracellular traps: the biology of chromatin externalization. Dev. Cell 44, 542–553 10.1016/j.devcel.2018.01.019 [DOI] [PubMed] [Google Scholar]
- 27. Xu J., Zhang X., Pelayo R., Monestier M., Ammollo C. T., Semeraro F., Taylor F. B., Esmon N. L., Lupu F., and Esmon C. T. (2009) Extracellular histones are major mediators of death in sepsis. Nat. Med. 15, 1318–1321 10.1038/nm.2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schorn C., Janko C., Latzko M., Chaurio R., Schett G., and Herrmann M. (2012) Monosodium urate crystals induce extracellular DNA traps in neutrophils, eosinophils, and basophils but not in mononuclear cells. Front. Immunol. 3, 277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ueki S., Melo R. C., Ghiran I., Spencer L. A., Dvorak A. M., and Weller P. F. (2013) Eosinophil extracellular DNA trap cell death mediates lytic release of free secretion-competent eosinophil granules in humans. Blood 121, 2074–2083 10.1182/blood-2012-05-432088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chow O. A., von Köckritz-Blickwede M., Bright A. T., Hensler M. E., Zinkernagel A. S., Cogen A. L., Gallo R. L., Monestier M., Wang Y., Glass C. K., and Nizet V. (2010) Statins enhance formation of phagocyte extracellular traps. Cell Host Microbe 8, 445–454 10.1016/j.chom.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Westman J., Papareddy P., Dahlgren M. W., Chakrakodi B., Norrby-Teglund A., Smeds E., Linder A., Mörgelin M., Johansson-Lindbom B., Egesten A., and Herwald H. (2015) Extracellular histones Induce chemokine production in whole blood ex vivo and leukocyte recruitment in vivo. PLoS Pathog. 11, e1005319 10.1371/journal.ppat.1005319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DeLange R. J., and Smith E. L. (1971) Histones: structure and function. Annu. Rev. Biochem. 40, 279–314 10.1146/annurev.bi.40.070171.001431 [DOI] [PubMed] [Google Scholar]
- 33. Morita S., Tagai C., Shiraishi T., Miyaji K., and Iwamuro S. (2013) Differential mode of antimicrobial actions of arginine-rich and lysine-rich histones against Gram-positive Staphylococcus aureus. Peptides 48, 75–82 10.1016/j.peptides.2013.07.025 [DOI] [PubMed] [Google Scholar]
- 34. Nitzsche R., Köhler J., Kreikemeyer B., and Oehmcke-Hecht S. (2016) Streptococcus pyogenes escapes killing from extracellular histones through plasminogen binding and activation by streptokinase. J. Innate Immun. 8, 589–600 10.1159/000448039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Döhrmann S., LaRock C. N., Anderson E. L., Cole J. N., Ryali B., Stewart C., Nonejuie P., Pogliano J., Corriden R., Ghosh P., and Nizet V. (2017) Group A streptococcal M1 protein provides resistance against the antimicrobial activity of histones. Sci. Rep. 7, 43039 10.1038/srep43039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chaput C., Spindler E., Gill R. T., and Zychlinsky A. (2013) O-antigen protects Gram-negative bacteria from histone killing. PLoS ONE 8, e71097 10.1371/journal.pone.0071097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rose-Martel M., and Hincke M. T. (2014) Antimicrobial histones from chicken erythrocytes bind bacterial cell wall lipopolysaccharides and lipoteichoic acids. Int. J. Antimicrob. Agents 44, 470–472 10.1016/j.ijantimicag.2014.07.008 [DOI] [PubMed] [Google Scholar]
- 38. Westman J., Chakrakodi B., Snäll J., Mörgelin M., Bruun Madsen M., Hyldegaard O., Neumann A., Frick I. M., Norrby-Teglund A., Björck L., and Herwald H. (2018) Protein SIC secreted from Streptococcus pyogenes forms complexes with extracellular histones that boost cytokine production. Front. Immunol. 9, 236 10.3389/fimmu.2018.00236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mazmanian S. K., Liu G., Ton-That H., and Schneewind O. (1999) Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285, 760–763 10.1126/science.285.5428.760 [DOI] [PubMed] [Google Scholar]
- 40. McCourt J., O'Halloran D. P., McCarthy H., O'Gara J. P., and Geoghegan J. A. (2014) Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol. Lett. 353, 157–164 10.1111/1574-6968.12424 [DOI] [PubMed] [Google Scholar]
- 41. Corriden R., Hollands A., Olson J., Derieux J., Lopez J., Chang J. T., Gonzalez D. J., and Nizet V. (2015) Tamoxifen augments the innate immune function of neutrophils through modulation of intracellular ceramide. Nat. Commun. 6, 8369 10.1038/ncomms9369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Novick R. (1967) Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33, 155–166 10.1016/0042-6822(67)90105-5 [DOI] [PubMed] [Google Scholar]
- 43. Horsburgh M. J., Aish J. L., White I. J., Shaw L., Lithgow J. K., and Foster S. J. (2002) σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184, 5457–5467 10.1128/JB.184.19.5457-5467.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sherertz R. J., Carruth W. A., Hampton A. A., Byron M. P., and Solomon D. D. (1993) Efficacy of antibiotic-coated catheters in preventing subcutaneous Staphylococcus aureus infection in rabbits. J. Infect. Dis. 167, 98–106 10.1093/infdis/167.1.98 [DOI] [PubMed] [Google Scholar]
- 45. Greene C., McDevitt D., Francois P., Vaudaux P. E., Lew D. P., and Foster T. J. (1995) Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17, 1143–1152 10.1111/j.1365-2958.1995.mmi_17061143.x [DOI] [PubMed] [Google Scholar]
- 46. Lee D. Y., Huang C. M., Nakatsuji T., Thiboutot D., Kang S. A., Monestier M., and Gallo R. L. (2009) Histone H4 is a major component of the antimicrobial action of human sebocytes. J. Invest. Dermatol. 129, 2489–2496 10.1038/jid.2009.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Murphy E. C., Mohanty T., and Frick I. M. (2014) FAF and SufA: proteins of Finegoldia magna that modulate the antibacterial activity of histones. J. Innate Immun. 6, 394–404 10.1159/000356432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Urban C. F., Ermert D., Schmid M., Abu-Abed U., Goosmann C., Nacken W., Brinkmann V., Jungblut P. R., and Zychlinsky A. (2009) Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 5, e1000639 10.1371/journal.ppat.1000639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stapels D. A., Ramyar K. X., Bischoff M., von Köckritz-Blickwede M., Milder F. J., Ruyken M., Eisenbeis J., McWhorter W. J., Herrmann M., van Kessel K. P., Geisbrecht B. V., and Rooijakkers S. H. (2014) Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors. Proc. Natl. Acad. Sci. U.S.A. 111, 13187–13192 10.1073/pnas.1407616111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eisenbeis J., Saffarzadeh M., Peisker H., Jung P., Thewes N., Preissner K. T., Herrmann M., Molle V., Geisbrecht B. V., Jacobs K., and Bischoff M. (2018) The Staphylococcus aureus extracellular adherence protein Eap is a DNA binding protein capable of blocking neutrophil extracellular trap formation. Front. Cell. Infect. Microbiol. 8, 235 10.3389/fcimb.2018.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McDevitt D., Francois P., Vaudaux P., and Foster T. J. (1994) Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11, 237–248 10.1111/j.1365-2958.1994.tb00304.x [DOI] [PubMed] [Google Scholar]
- 52. Zong Y., Xu Y., Liang X., Keene D. R., Höök A., Gurusiddappa S., Höök M., and Narayana S. V. (2005) A “Collagen Hug” model for Staphylococcus aureus CNA binding to collagen. EMBO J. 24, 4224–4236 10.1038/sj.emboj.7600888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. O'Connell D. P., Nanavaty T., McDevitt D., Gurusiddappa S., Höök M., and Foster T. J. (1998) The fibrinogen-binding MSCRAMM (clumping factor) of Staphylococcus aureus has a Ca2+-dependent inhibitory site. J. Biol. Chem. 273, 6821–6829 10.1074/jbc.273.12.6821 [DOI] [PubMed] [Google Scholar]
- 54. Mulcahy M. E., Geoghegan J. A., Monk I. R., O'Keeffe K. M., Walsh E. J., Foster T. J., and McLoughlin R. M. (2012) Nasal colonization by Staphylococcus aureus depends upon clumping factor B binding to the squamous epithelial cell envelope protein loricrin. PLoS Pathog. 8, e1003092 10.1371/journal.ppat.1003092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vazquez V., Liang X., Horndahl J. K., Ganesh V. K., Smeds E., Foster T. J., and Hook M. (2011) Fibrinogen is a ligand for the Staphylococcus aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMM) bone sialoprotein-binding protein (Bbp). J. Biol. Chem. 286, 29797–29805 10.1074/jbc.M110.214981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roche F. M., Meehan M., and Foster T. J. (2003) The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology 149, 2759–2767 10.1099/mic.0.26412-0 [DOI] [PubMed] [Google Scholar]
- 57. Kristian S. A., Datta V., Weidenmaier C., Kansal R., Fedtke I., Peschel A., Gallo R. L., and Nizet V. (2005) d-Alanylation of teichoic acids promotes group a streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 187, 6719–6725 10.1128/JB.187.19.6719-6725.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]