Abstract
The development of ectodermal organs, such as teeth, requires epithelial–mesenchymal interactions. Basic helix–loop–helix (bHLH) transcription factors regulate various aspects of tissue development, and we have previously identified a bHLH transcription factor, AmeloD, from a tooth germ cDNA library. Here, we provide both in vitro and in vivo evidence that AmeloD is important in tooth development. We created AmeloD-knockout (KO) mice to identify the in vivo functions of AmeloD that are critical for tooth morphogenesis. We found that AmeloD-KO mice developed enamel hypoplasia and small teeth because of increased expression of E-cadherin in inner enamel epithelial (IEE) cells, and it may cause inhibition of the cell migration. We used the CLDE dental epithelial cell line to conduct further mechanistic analyses to determine whether AmeloD overexpression in CLDE cells suppresses E-cadherin expression and promotes cell migration. Knockout of epiprofin (Epfn), another transcription factor required for tooth morphogenesis and development, and analysis of AmeloD expression and deletion revealed that AmeloD also contributed to multiple tooth formation in Epfn-KO mice by promoting the invasion of dental epithelial cells into the mesenchymal region. Thus, AmeloD appears to play an important role in tooth morphogenesis by modulating E-cadherin and dental epithelial–mesenchymal interactions. These findings provide detailed insights into the mechanism of ectodermal organ development.
Keywords: basic helix-loop-helix transcription factor (bHLH), cadherin-1 (CDH1), epithelial cadherin (E-cadherin), cell migration, epithelial-mesenchymal transition (EMT), gene knockout, tooth development, inner enamel epithelium, AmeloD, epiprofin, ectodermal organ
Introduction
Ectodermal organs, such as teeth, all display a need for epithelial–mesenchymal interactions for their development. Tooth development is, in fact, a good model for understanding the mechanism of ectodermal development because it has well-defined stages and distinctive differentiated cell types (1). In the mouse, the morphogenesis of the molars is divided into four stages: the initiation stage (embryonic day 11.5 (E11.5)),2 bud stage (E13.5), cap stage (E15.5), and bell stage (E17.5). At the initiation stage, the dental epithelium starts to thicken and invades into the mesenchymal region. This invagination process forms the tooth bud, and the dental epithelium condenses at the bud stage. After the cap stage, the dental epithelial stem cells differentiate into various cell types to form the enamel organ: the inner enamel epithelium (IEE), outer enamel epithelium, stratum intermedium (SI), and stellate reticulum. The IEE cells are ameloblast progenitor cells, a unique cell population in the proliferation stage as they express proliferation markers but do not express E-cadherin, a negative regulator of cell division and migration (2, 3). The IEE cells actively proliferate and migrate to form a correctly sized enamel organ. After proliferation, the IEE cells differentiate into enamel-secreting ameloblasts. Proliferative IEE cells persist near the apical tip of the root, and these cells continue to invaginate to form roots. After postnatal day 7 (P7), these cells form the Hertwig's epithelial root sheath with outer enamel epithelium. Adjacent mesenchymal cells then receive signals from the IEE cells and differentiate into odontoblasts (4, 5).
Tooth morphogenesis is regulated by multiple genes; for example, epiprofin (Epfn/Sp6) has previously been identified as an essential transcription factor for tooth morphogenesis (6–8). During tooth development, Epfn is expressed in the dental epithelium at the initiation stage. Later, its expression is restricted to the ameloblast lineage (including IEE cells) and secretory and mature ameloblasts with increasing levels of its expression. Epfn is also expressed in mature odontoblasts. Epfn-deficient mice show severe enamel hypoplasia due to inhibition of IEE cell proliferation and ameloblast differentiation, but they also form supernumerary teeth due to random dental epithelial cell invagination (9). A more recent study demonstrated that Epfn regulates the balance between cell proliferation and cytodifferentiation in dental epithelial and mesenchymal cells during tooth development and morphogenesis (8). The spatiotemporal regulation of the epithelial–mesenchymal interaction during the developmental stages results in a proper tooth shape.
The cell type–specific bHLH transcription factors are key regulators of certain types of organ morphogenesis (10–12), and several of these bHLH transcription factors play important roles in tooth development (13–16). However, few studies have focused on tooth-specific bHLH transcription factors. Recently, we used a yeast two-hybrid system to identify a novel bHLH transcription factor from a tooth germ cDNA library, and we isolated a homologous sequence to Ascl5 (17). Ascl5 was previously predicted from a database to be a pseudogene, and we modified it as a protein-coding gene. Therefore, we speculated that Ascl5 was a novel gene in tooth development, and we named it AmeloD. The AmeloD sequence has been registered in GenBankTM under accession number MG575629. AmeloD forms a heterodimer with the protein E12 and binds to E-box cis-elements on the E-cadherin promoter to suppress its transcriptional activity (17). AmeloD is expressed in developing teeth and specifically in the IEE (17); however, its function in tooth development is unknown.
In the present study, we sought to identify the role of AmeloD in tooth development by creating AmeloD-KO mice. These KO mice showed enamel hypoplasia and a reduction in dental epithelial cell invagination, and the resulting molars had smaller crowns and shorter roots when compared with control wildtype (WT) molars. We found that AmeloD promotes proper tooth germ growth by the suppression of E-cadherin. We further analyzed the relationship between cell migration and tooth morphogenesis in AmeloD and Epfn double-knockout (AmeloD; Epfn-KO) mice. We found that AmeloD contributed to the multiple tooth formation observed in Epfn-KO mice by promoting the invagination of dental epithelial cells. Our results revealed that AmeloD acts as a suppressor of E-cadherin likely to promote dental epithelial cell migration and that it regulates the interactions between dental epithelial and mesenchymal cells to form proper tooth sizes.
Results
AmeloD-KO mice showed enamel hypoplasia and small teeth
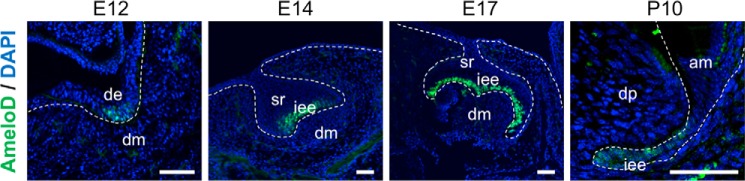
We first analyzed AmeloD expression during tooth development. We performed immunostaining of AmeloD in molars at E12, E14, and E17 and at P10 (Fig. 1). AmeloD was expressed in the thickening dental epithelium in E12 molars and in the invaginating epithelium in E14 molars. In P10 molars, AmeloD was not expressed in the differentiated ameloblasts, but it was expressed in the IEE cells in the apical root tip. These results indicate that AmeloD was expressed in the early stage of IEE. However, although AmeloD was strongly expressed in proliferating IEE cells, it was no longer expressed once the IEE cells differentiated into ameloblasts, in agreement with a previous observation by He et al. (17).
Figure 1.
AmeloD expression in developing molars. Immunofluorescence of AmeloD in developing molars is shown. Immunofluorescence staining analysis of AmeloD expression was performed using an anti-AmeloD antibody in E12, E14, E17, and P10 molars. Green, AmeloD; blue, DAPI. de, dental epithelium; dm, dental mesenchyme; iee, inner enamel epithelium; sr, stellate reticulum; am, ameloblast; dp, dental pulp. The dashed lines indicate the borders between the dental epithelium and mesenchyme. Scale bars, 50 μm.
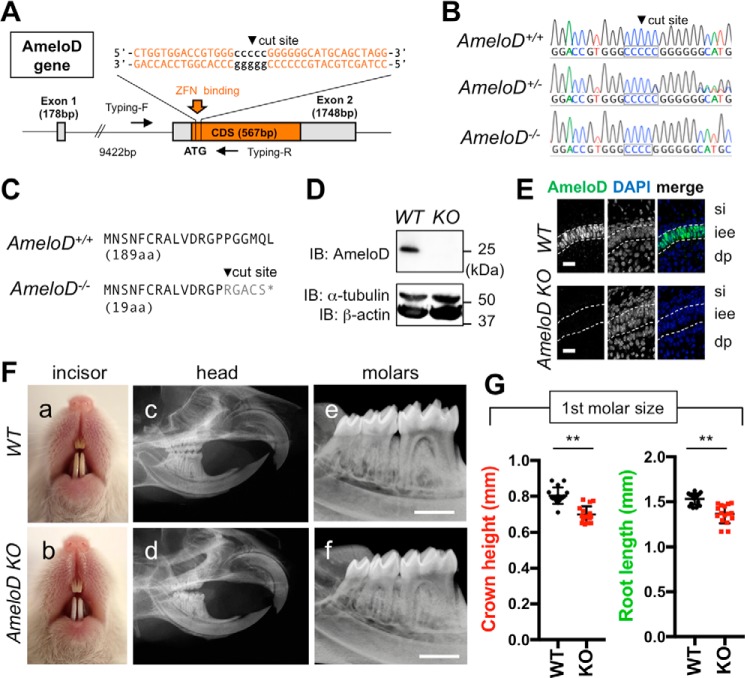
We also sought to identify the in vivo function of AmeloD by using zinc-finger nuclease technology to create AmeloD-KO mice (Fig. 2A). We analyzed the genotype of these mice by DNA sequencing (Fig. 2B) and obtained two mouse lines that had an identical deletion of 1 bp in the cut site; this deletion caused a frameshift and induced an early termination codon. In WT mice, the AmeloD protein consists of 188 amino acids (aa) and contains a bHLH domain. By contrast, the AmeloD-KO mice expressed a premature AmeloD protein consisting of only 19 aa (Fig. 2C). Consequently, Western blotting of protein extracts (Fig. 2D) and immunostaining using the anti-AmeloD antibody (Fig. 2E) did not detect the truncated AmeloD in P1 AmeloD-KO molars. The two AmeloD-KO mouse lines showed the same phenotype. The AmeloD-KO mice were fertile. However, the incisors of 6-week-old mice were chalky white in color, indicating enamel hypoplasia (Fig. 2F, a and b). Radiographic analysis of the heads of the 6-week-old AmeloD-KO mice revealed a length of the incisors similar to that determined in WT mice (Fig. 2F, c and d). Conversely, the roots were shorter in the AmeloD-KO molars than in the WT molars (Fig. 2F, e and f). We extracted WT and AmeloD-KO molars from 6-week-old mice; at this age, root formation was completed (Fig. 2F). Measurements of tooth crown height and root length revealed that AmeloD-KO teeth had smaller crowns and roots when compared with WT teeth (Fig. 2G).
Figure 2.
Generation of AmeloD-KO mice. A, targeting strategy for AmeloD-KO mice. ZFN binds near the cut site and deletes 1 bp in the cut site. Arrows indicate primer locations for genotyping. CDS, coding sequence. B, DNA sequences of the ZFN-binding site in the AmeloD locus. The cut site is shown in the boxed region. In AmeloD−/− (KO) mice, 1 bp was deleted from the cut site. In the AmeloD+/− (heterozygous) mice, both WT and KO alleles were detected. C, comparison of the AmeloD amino acid sequence of the WT and AmeloD-KO product. In AmeloD-KO mice, the frameshift caused an early termination codon, resulting in a short protein consisting of 19 aa. D, protein expression of AmeloD in WT and AmeloD-KO P1 molars. The AmeloD protein was stained with the AmeloD antibody (molecular mass, 25 kDa). α-Tubulin (molecular mass, 52 kDa) and β-actin (molecular mass, 42 kDa) are shown as internal controls. IB, immunoblotting. E, immunofluorescence staining analysis of AmeloD in WT and AmeloD-KO P1 molars. Green, AmeloD; blue, DAPI for nuclear staining. si, stratum intermedium; iee, inner enamel epithelium; dp, dental pulp. The dashed lines indicate the borders of IEE cells. Scale bars, 20 μm. F, photographic analyses (a and b) and radiographic analysis (c–f) of 6-week-old WT and AmeloD-KO heads. Skeletal bone abnormality was not observed between WT and AmeloD-KO mice. Scale bars, 1000 μm. G, the crown heights and root lengths of extracted first molars from 6-week-old WT and AmeloD-KO mice (n = 16). The mean is shown as lines. Error bars represent S.D. **, p < 0.01 with a two-tailed t test.
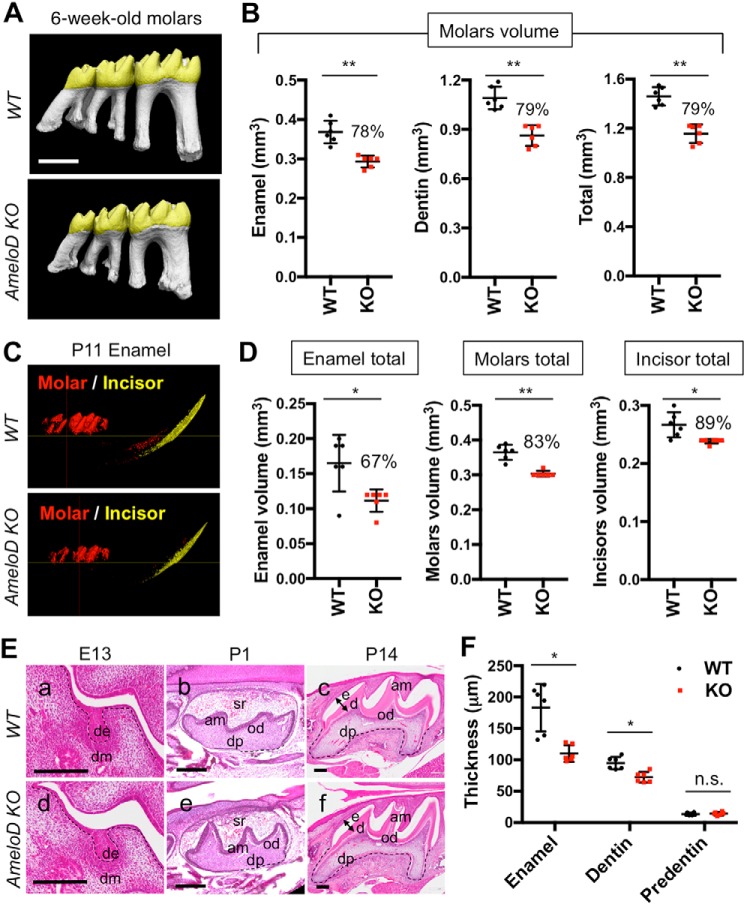
We also used micro-computed tomography (micro-CT) to analyze the phenotypes of the lower molars of 6-week-old WT and AmeloD-KO mice (Fig. 3A). The WT and AmeloD-KO teeth differed in size as the AmeloD-KO molars developed smaller cusps (Fig. 3A). The total enamel volume and dentin volume were also about 20% smaller in the AmeloD-KO molars than in the WT molars (Fig. 3B). Six-week-old WT and AmeloD-KO mice had similar incisor volumes. We examined the effects of deletion of AmeloD on incisor size by micro-CT analysis of the P11 mandible; at this age, the incisor had not yet erupted (Fig. 3C). This micro-CT analysis revealed that the size of the incisors and molars at the age of P11 and the total and combined volumes of enamel and dentin in the incisors and molars were smaller in AmeloD-KO mice than in WT mice (Fig. 3D). We also analyzed the developing molars to determine the histological details of the tooth abnormalities (Fig. 3E). We used hematoxylin-eosin (H-E) staining to analyze the histological differences between WT and AmeloD-KO molars. The bud stage (E13) molars showed no obvious histological differences. After the crown formation (P14), the enamel thickness and dentin thickness were thinner in AmeloD-KO mice than in WT mice (Fig. 3F), and this resulted in a smaller crown size in the AmeloD-KO mice. At this P14 stage, the developing root width was also smaller in the AmeloD-KO molars. These results indicate that deletion of AmeloD inhibited tooth germ growth.
Figure 3.
Tooth phenotype analysis of AmeloD-KO mice. A, micro-CT analysis of 6-week-old WT and AmeloD-KO molars. B, the total volume of enamel and dentin in 6-week-old WT and AmeloD-KO molars (n = 6). The number above the bar graph is the ratio of the volume (KO/WT). The mean is shown as lines. Error bars represent S.D. **, p < 0.01 with a two-tailed t test. Scale bars, 1000 μm. C, micro-CT analysis of P11 AmeloD-KO and WT incisor enamel and molar enamel. Enamel is defined by density as more than 1300 mg of hydroxyapatite/cm3. D, left panel, total volume of enamel in P11 WT and AmeloD-KO incisor and molars. Middle panel, total volume of enamel and dentin in molars. Right panel, total volume of enamel and dentin in incisors. The ratio of the volume (KO/WT) is shown as the number above the bar graph (n = 6). Red, molar enamel; yellow, incisor enamel. The mean is shown as lines. Error bars represent S.D. *, p < 0.05; **, p < 0.01 with a two-tailed t test. E, H-E staining of developing WT (a–c) and AmeloD-KO (d–f) molars. Arrows indicate the position used for quantification of thickness in F. de, dental epithelium; dm, dental mesenchyme; sr, stellate reticulum; am, ameloblasts; od, odontoblasts; dp, dental pulp; e, enamel; d, dentin. The dashed lines indicate the border between the dental epithelium and mesenchyme. Scale bars, 200 μm. F, quantification of enamel, dentin, and predentin thicknesses from the P14 distal cusps of the lower first molars (arrows in E indicate the region of measurement; n = 6). The mean is shown as lines. Error bars represent S.D. *, p < 0.05 with a two-tailed t test. n.s., not significant.
Deletion of AmeloD did not affect the proliferation of dental epithelium
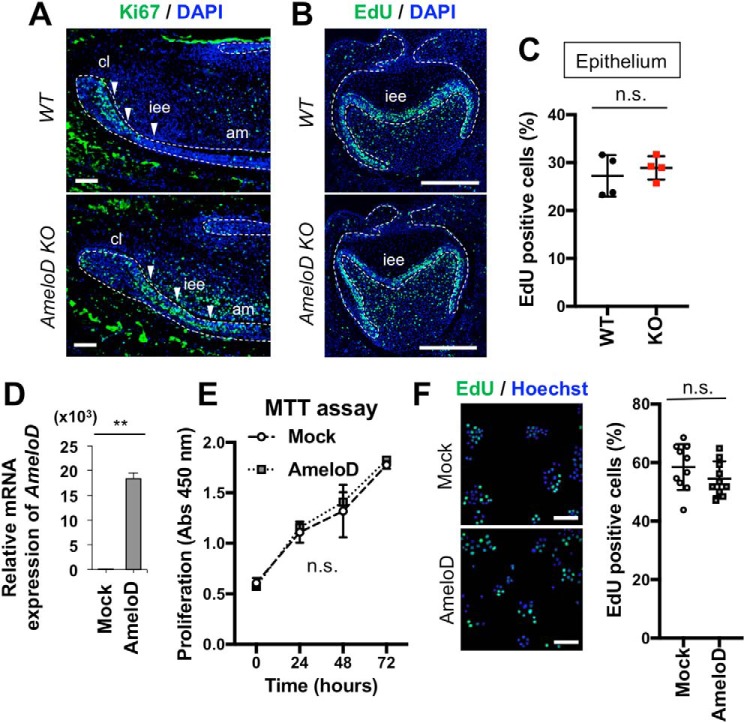
The AmeloD-KO mice had small teeth and enamel hypoplasia. Tooth size is related to dental epithelial proliferation and expansion of the tooth germ. We used immunofluorescence staining of Ki67, a proliferation marker, to determine the effect of AmeloD on the proliferation of the IEE cells in P1 WT and AmeloD-KO incisors (Fig. 4A). Ki67 was strongly expressed at an early stage in IEE cells in WT incisors (Fig. 4A). The Ki67-positive area was expanded in AmeloD-KO mice, indicating that deletion of AmeloD may affect the proliferation activity of the IEE cells or expansion of the area of Ki67-positive cells caused by the inhibition of IEE cell differentiation. We used an EdU labeling assay and the molar tooth germ to examine whether the deletion of AmeloD influences the proliferation of IEE cells (Fig. 4B). We found that the numbers of EdU-positive cells did not differ between WT and AmeloD-KO mice (Fig. 4C). We also examined the effect of AmeloD on dental epithelial proliferation in vitro. Overexpression of the AmeloD gene in the cervical loop–derived dental epithelial (CLDE) cell line resulted in high expression of AmeloD (Fig. 4D) without affecting the proliferation of these cells (Fig. 4E). Similar results were observed in EdU incorporation assays (Fig. 4F). These results suggested that IEE cell proliferation may not be affected by AmeloD expression.
Figure 4.
AmeloD has no effect on cell proliferation in IEE and in the CLDE dental epithelial cell line. A, immunostaining of Ki67 in P1 WT and AmeloD-KO incisors. Green, Ki67; blue, DAPI. Arrowheads indicate IEE cells. B, EdU staining of E18 WT and AmeloD-KO molars. C, quantification of EdU-positive cells. The EdU-positive ratio was calculated by counting the numbers of EdU-positive cells/stained nuclei in the epithelium. No significant difference was observed between WT and AmeloD-KO molars (n = 4). The mean is shown as lines. Error bars represent S.D. n.s., p > 0.05 with a two-tailed t test. D, AmeloD mRNA expression levels in AmeloD- and control mock vector–transfected CLDE cells (n = 3). Error bars represent S.D. **, p < 0.01 with a two-tailed t test. E, proliferation assay of AmeloD- and mock-transfected cells. The proliferation activity was detected as absorbance at 450 nm using a MMT kit (Dojindo; n = 3). Error bars represent S.D. n.s., p > 0.05 with two-way ANOVA. F, EdU incorporation and staining in AmeloD- and mock-transfected CLDE cells. Green, EdU; blue, Hoechst (n = 10). The mean is shown as lines. Error bars represent S.D. n.s., p > 0.05 with a two-tailed t test. Bars, 50 μm.
AmeloD inhibits E-cadherin expression and promotes migration of CLDE cells
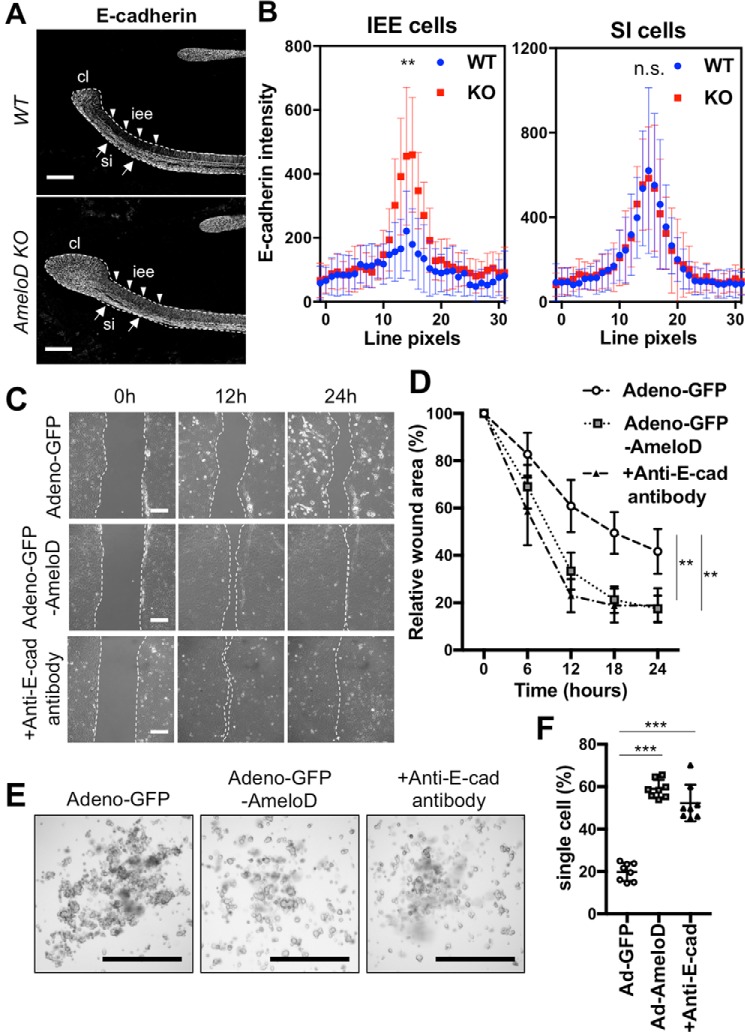
Tooth germ growth is regulated by the migration and proliferation of IEE cells, and E-cadherin negatively regulates cell migration in these cells (3, 18). We used immunofluorescence staining for E-cadherin to identify the mechanism of tooth growth inhibition in P1 WT and AmeloD-KO incisors (Fig. 5A). The early stage of IEE cells of WT incisors did not express E-cadherin. However, differentiation of the IEE cells into ameloblasts resulted in strong expression of E-cadherin by the ameloblasts to retain their polarity. By contrast, the IEE cells of the AmeloD-KO incisors expressed E-cadherin (Fig. 5A). We used fluorescence line intensity scans (30 pixels wide) across cell–cell junctions to quantify the expression levels of E-cadherin in the IEE cells (Fig. 5B). The IEE cells of the WT incisors showed no E-cadherin expression, whereas the IEE cells of the AmeloD-KO incisors showed significant E-cadherin expression. The SI cells of the WT and AmeloD-KO incisors showed no differences in E-cadherin expression levels.
Figure 5.
AmeloD inhibits E-cadherin expression in IEE and promotes cell migration in vitro. A, immunostaining of E-cadherin in P1 WT and AmeloD-KO incisors. White, E-cadherin. Arrowheads indicate IEE cells, and arrows indicate SI cells. cl, cervical loop; iee, inner enamel epithelium; si, stratum intermedium. The dashed lines indicate the border between the dental epithelium and mesenchyme. Scale bars, 200 μm. B, quantification of E-cadherin staining from 30-pixel-wide fluorescence line intensity scans across cell–cell junctions. SI cells were measured as an internal control (n = 20). The mean is shown as dots. Error bars represent S.D. **, p < 0.01 with one-way ANOVA. C, wound healing assays for the adeno-GFP– or the adeno-GFP-AmeloD–infected or anti-E-cadherin functional antibody–treated CLDE cells. Scale bars, 50 μm. D, cell migration and wound closure areas in the wound healing assay (n = 5). The mean is shown as dots. Error bars represent S.D. **, p < 0.01 with respect to the adeno-GFP as control by two-way ANOVA. E, hanging drop cultures of the adeno-GFP– or the adeno-AmeloD–infected or anti-E-cadherin (E-cad) functional antibody–treated CLDE cells. Cell spheroids in cultures 6 h after the cells were dropped. Scale bars, 1000 μm. F, single-cell ratio after passage through a cell strainer (n = 10). The mean is shown as lines. Error bars represent S.D. ***, p < 0.001 with respect to the adeno-GFP as a control by one-way ANOVA.
E-cadherin continued to be expressed in the proliferative IEE cells of the AmeloD-KO mice, suggesting an inhibition of migration activity of the IEE cells. We found that AmeloD suppressed the transcription of E-cadherin by binding to its promoter region (17), suggesting that AmeloD is required for E-cadherin modulation in the IEE cells. We used adeno-GFP-AmeloD–infected CLDE cells in wound healing assays to determine the activity of AmeloD during dental epithelial cell migration (Fig. 5C). Cell migration was significantly greater in CLDE cells overexpressing AmeloD than in control CLDE cells infected with adeno-GFP. Addition of the anti-E-cadherin antibody to CLDE cells without infection promoted cell migration, suggesting that the suppression of E-cadherin induces cell migration (Fig. 5D).
We then used a hanging drop culture to compare spheroid formation by the adeno-GFP-AmeloD–infected CLDE cells and the control adeno-GFP–infected cells (Fig. 5E). The control CLDE cells formed spheroids in 6 h, whereas the AmeloD-overexpressing CLDE cells and CLDE cells treated with anti-E-cadherin antibody failed to form spheroids. We quantified single cell numbers in the drops by counting cell numbers after passage through a cell strainer (Fig. 5F). We found significantly higher numbers of single cells in the drops containing AmeloD-overexpressing CLDE cells or the cells treated with anti-E-cadherin antibody than in the control infected with adeno-GFP cells (Fig. 5F). These results indicated that AmeloD expression inhibited spheroid formation through suppression of E-cadherin.
Deletion of AmeloD inhibits the growth of clipped incisors
The in vitro analysis revealed that AmeloD promoted migration of dental epithelial cells by inhibition of E-cadherin. During ectodermal organ development, the motility of epithelial cells affects organ morphogenesis (19). Therefore, we hypothesized that deletion of AmeloD in vivo inhibits tooth growth by the suppression of IEE cell motility. We examined whether the deletion of AmeloD affected incisor growth speed by performing incisor cutting and recovery assays in 6-week-old WT and AmeloD-KO mice (Fig. 6). At this age, the sizes of the incisors are similar in WT and AmeloD-KO mice as shown in Fig. 2F; this is because the continuous growth of the incisors might overcome the potential growth rate differences in the AmeloD-KO incisors.
Figure 6.
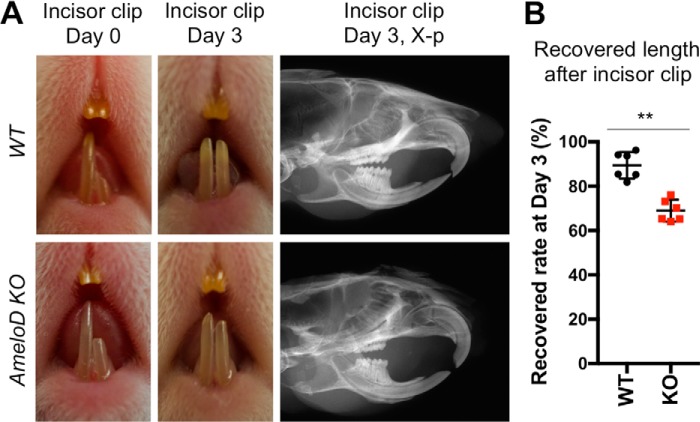
Deletion of AmeloD in adult mice inhibits incisor regeneration. A, photographic analysis of 6-week-old WT and AmeloD-KO mice after clipping the lower left incisor. First column, mice incisors at day 0 after clipping half of the left lower incisor. Second column, mice incisors at day 3 of recovery. Third column, radiographic analysis of clipped incisors after 3 days of recovery. B, quantified data of the recovered incisor length after cutting (n = 6). The mean is shown as lines. Error bars represent S.D. **, p < 0.01 with a two-tailed t test.
The lower left incisor was clipped to one-half its length on day 0, and we observed the rate of incisor growth by measuring incisor length at day 3 postcutting (Fig. 6A). At day 3, the WT incisors had recovered to almost the length of the uncut right lower incisor (Fig. 6A, middle panel). By contrast, the AmeloD-KO incisors did not recover their length. We quantified the growth speed of the recovered incisors by measuring their total length 3 days after cutting (Fig. 6B). The relatively slow recovery of the AmeloD-KO incisors suggests that a reduction in IEE cell motility had affected tooth growth speed; i.e. the deletion of AmeloD inhibits incisor growth, likely due to the inhibition of IEE cell motility.
AmeloD contributes to abnormal dental epithelium invasions in Epfn-KO teeth
Our data suggested an important role for AmeloD in IEE cell migration and invagination; therefore, we hypothesized that AmeloD could be involved in the random dental epithelium invasions into the mesenchyme region observed in Epfn-KO teeth. Epiprofin, an Sp zinc-finger family transcription factor, is essential for ameloblast development. Epfn is expressed in the IEE, ameloblasts, and odontoblasts (7, 8). Epfn has multiple functions in developing ameloblast lineages, including the regulation of IEE cell differentiation into ameloblasts and the promotion of IEE cell proliferation (6, 20, 21). The IEE cells in Epfn-KO teeth show drastically reduced proliferation activity (7, 9). In addition, Epfn-KO teeth lack enamel and develop a defect in dentin structure (7). The premature IEE cells of Epfn-KO teeth do not polarize, and they show abnormal invagination into the mesenchymal region (7, 9, 22). This random invasion of the dental epithelium results in multiple branching and the formation of supernumerary teeth in Epfn-KO mice (7, 9).
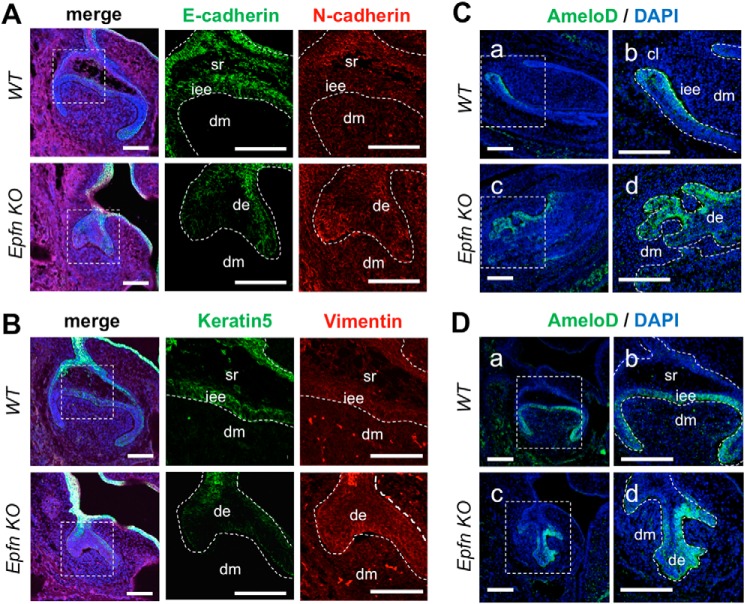
We characterized the dental epithelial cells in Epfn-KO teeth by immunofluorescence staining for E-cadherin in E17 molars (Fig. 7A). The invading epithelia showed a reduction in E-cadherin expression in the presumptive IEE region, indicating that random invasions were caused by an abnormal migration. Interestingly, the region showed a loss of keratin 5 and started to express N-cadherin and vimentin, which are markers of mesenchymal cells. These changes suggested the involvement of a partial epithelial–mesenchymal transition (EMT) in the invasions (Fig. 7, A and B).
Figure 7.
Epfn deficiency induces the partial epithelial-mesenchymal transition in the invading epithelia. A, immunofluorescence of E-cadherin and N-cadherin in E16 WT and Epfn-KO molars. Green, E-cadherin; red, N-cadherin; blue, DAPI. B, immunofluorescence of keratin 5 and vimentin in E16 WT and Epfn-KO molars. Green, keratin 5; red, vimentin; blue, DAPI. C, immunofluorescence of AmeloD in E17 WT and Epfn-KO incisors. b and d, enlargements of a and c. D, immunofluorescence of AmeloD in E17 WT and Epfn-KO molars. b and d, enlargements of a and c. Green, AmeloD; blue, DAPI. de, dental epithelium; dm, dental mesenchyme; iee, inner enamel epithelium; sr, stellate reticulum. Scale bars, 100 μm.
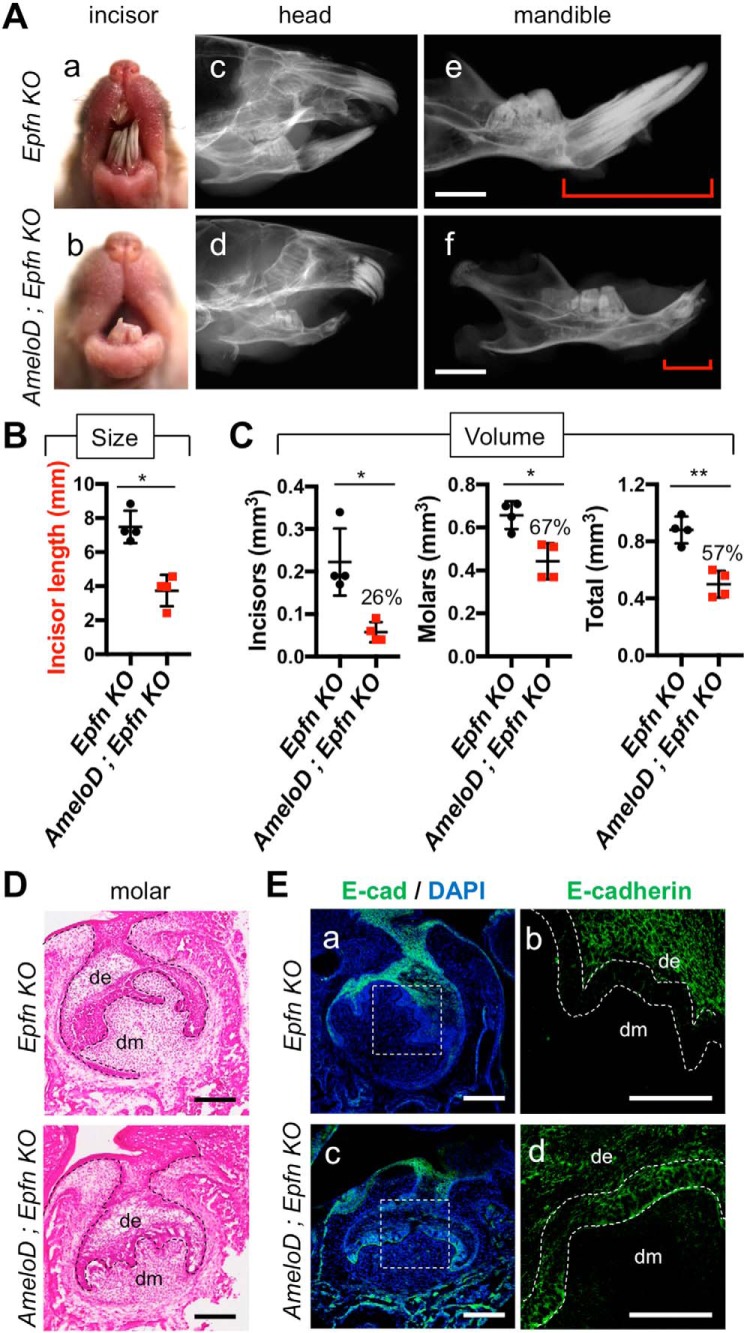
We also examined AmeloD expression in the developing incisors and molars of Epfn-KO mice. The E17 WT incisors and molars showed restriction of AmeloD expression to the IEE cells (Fig. 7, C and D). In the E17 Epfn-KO incisors and molars, the dental epithelium started to form multiple branches by random invasion into mesenchyme, and the Epfn-KO incisors developed an ectopic cervical loop–like structure to form multiple incisors. The AmeloD protein was expressed strongly in the invading epithelium regions in both Epfn-KO incisors and molars (Fig. 7, C and D). We further examined the relationship between multiple tooth formation and the AmeloD expression by creating AmeloD; Epfn-KO mice. The Epfn-KO mice developed more than 20 incisors at 3 months of age, whereas the AmeloD; Epfn-KO mice developed fewer incisors (Fig. 8A, a and b). The AmeloD; Epfn-KO mice erupted only one to two incisors at 3 months of age. Similar to the Epfn-KO mice, the AmeloD; Epfn-KO mice lacked enamel on their teeth. Radiographic analysis also showed a drastic reduction in the number and size of incisors (Fig. 8A, d and f). We confirmed an incisor length difference by measuring the length of extracted incisors (Fig. 8B). The incisors were significantly shorter in AmeloD; Epfn-KO mice than in Epfn-KO mice. The Epfn-KO molars failed to develop roots because of defects in the root-forming dental epithelium. Similarly, AmeloD; Epfn-KO molars lacked roots, suggesting a disturbance in root formation. We also performed micro-CT analysis of 6-week-old Epfn-KO and AmeloD; Epfn-KO teeth. The volume of the incisors and the molars was smaller for AmeloD; Epfn-KO mice than for Epfn-KO mice (Fig. 8C).
Figure 8.
AmeloD deficiency reduces the tooth size and number in Epfn-KO mice via inhibition of random dental epithelial invasions. A, photographic analyses (a and b) and radiographic analysis (c–f) of 3-month-old Epfn-KO and AmeloD; Epfn-KO heads. The red brackets indicate the length of the incisors. Scale bars, 1000 μm. B, incisor lengths of 3-month-old Epfn-KO and AmeloD; Epfn-KO mice (n = 5). The mean is shown as lines. Error bars represent S.D. *, p < 0.05 with a two-tailed t test. C, the total volume of dentin in 6-week-old Epfn-KO and AmeloD; Epfn-KO teeth by micro-CT analysis. Left panel, total volume of incisors. Middle panel, total volume of molars. Right panel, total volume of molars and total volume of incisor were quantified. The ratio of the volume (AmeloD; Epfn KO/Epfn KO) is shown as the number above the bar graph (n = 4). The mean is shown as lines. Error bars represent S.D. *, p < 0.05; **, p < 0.01 with a two-tailed t test. D, H-E staining of P1 Epfn-KO and AmeloD; Epfn-KO molars. The dashed lines indicate the border between the dental epithelium and mesenchyme. Scale bars, 100 μm. E, immunofluorescence staining analysis of E-cadherin (E-cad) in P1 Epfn-KO and AmeloD; Epfn-KO molars. b and d, enlargements of a and c. Green, E-cadherin; blue, DAPI. The dashed lines indicate the presumptive inner enamel epithelium. de, dental epithelium; dm, dental mesenchyme. Scale bars, 100 μm.
We performed a histological analysis of P1 Epfn-KO and AmeloD; Epfn-KO molars (Fig. 8, D and E). In Epfn-KO molars, the dental epithelium showed random invasions into the mesenchymal regions. In AmeloD; Epfn-KO molars, the dental epithelium also showed random invasions, but the elongation of these invasions was inhibited (Fig. 8D). We used immunofluorescence staining of E-cadherin in P1 Epfn-KO and AmeloD; Epfn-KO molars to analyze the mechanism of these differences in epithelium invasions (Fig. 8E). In Epfn-KO molars, the invading epithelial cells did not express E-cadherin. However, in the AmeloD; Epfn-KO molars, these cells strongly expressed E-cadherin, suggesting inhibition of the dental epithelium invasions. These results suggest that AmeloD promotes epithelium invasions by suppressing E-cadherin expression in the invading epithelial cells in Epfn-KO teeth.
Discussion
Tooth development requires the involvement of several bHLH transcription factors, such as dHand/Hand2, Math1, Twist1, Snail, and Id1 (13–15). The dHand2 factor shows restricted expression in lower incisors and functions in mesenchymal cell differentiation and apoptosis of the dental cells. Math1 is later expressed in differentiated ameloblasts and odontoblasts in P3 molars and is suggested to be involved in the differentiation of both ameloblasts and odontoblasts; however, its actual function remains unclear. Twist1 is expressed in the dental mesenchyme and is important for odontoblast differentiation. The deletion of Twist1 in the dental mesenchyme inhibits odontoblast differentiation by suppression of the fibroblast growth factor signaling pathway (23). Snail is expressed in the dental mesenchyme and is also regulated by fibroblast growth factor signaling. Id1 is expressed in both the dental mesenchyme and epithelium and is especially restricted to the enamel knot. We found that AmeloD shows a restricted expression pattern in proliferative IEE (Fig. 1). This expression pattern is unique among the reported bHLH factors. Therefore, we propose that AmeloD has a distinct role in tooth development.
Expression of AmeloD was restricted to IEE cells where it regulated E-cadherin expression in IEE cells. The AmeloD-deficient teeth of KO mice showed increased expression of E-cadherin in the IEE cells (Fig. 5A); consequently, the migration of those IEE cells might be inhibited. In vitro experiments showed that overexpression of AmeloD induced cell migration. A similar result was observed for the inhibition of E-cadherin using a blocking antibody. However, we did not confirm these findings in vivo. Further in vivo experiments are still needed to clarify the relationship between AmeloD and E-cadherin in the in vivo migration of IEE cells. The tooth germ size was similar in AmeloD-KO and WT mice in the bud stage (Fig. 3E). However, after the bell stage, the tooth germs were significantly smaller in the AmeloD-KO than in the WT mice. In addition, the IEE migration required for root formation was likely inhibited in the AmeloD-KO molars. Thus, AmeloD may regulate the whole tooth size by suppressing E-cadherin expression in IEE cells. This modulation of E-cadherin expression by AmeloD is consistent with our previous report that AmeloD binds to the E-box cis-regulatory element in the E-cadherin promoter region to suppress E-cadherin transcriptional activity in the dental epithelial CLDE cell line (17).
Modulation of E-cadherin expression is important for epithelial cell dynamics in organogenesis because E-cadherin acts as a negative regulator of cell division and migration (2). For example, Btbd7, a member of the BTB domain–containing protein family, is essential for cleft formation during in vivo branching morphogenesis, which occurs through down-regulation of E-cadherin (24, 25). A more recent report reveals that Btdb7 functions as a negative regulator of E-cadherin by promoting E-cadherin ubiquitination and degradation (19). Epithelium-specific conditional Btdb7-KO mice develop smaller salivary glands, lungs, and kidneys compared with WT mice due to inhibition of the division and migration of bud epithelial cells through the up-regulation of E-cadherin expression (19). Thus, regulation of epithelial cell division and migration through modulation of E-cadherin expression is critical for ectodermal organs to achieve an appropriate size. In fact, functional inhibition using a neutralizing antibody for E-cadherin promoted cell migration of the CLDE dental epithelial cell line (Fig. 5, C and D). However, inhibition of transcription using siRNA for E-cadherin did not rescue the AmeloD-deficient phenotype of CLDE cells (data not shown). Other factors regulated by AmeloD may be involved in this process.
Both cell migration and cell proliferation activities are important for tooth germ growth; however, AmeloD did not affect the cell proliferation activity of IEE cells (Fig. 4). IEE cell proliferation is most likely regulated by Epfn as suggested by several lines of evidence. For example, Epfn-KO teeth show a severe inhibition of IEE cell proliferation (7). In addition, a low level of exogenous Epfn expression promotes the proliferation of CLDE cells, whereas a high level of Epfn expression promotes CLDE differentiation into ameloblasts.3 The basal layer of the epidermis contains stem cells and proliferating transit-amplifying cells, which are equivalent to IEE cells in developing teeth. The Epfn-KO epidermis shows a severe reduction in transit-amplifying cell proliferation (26). In keratinocyte cell proliferation, Epfn functions as a cell cycle regulator by binding to Cdk4 and promoting the phosphorylation of Rb to activate the E2F transcription factor for cell cycle progression (26). The Epfn expression in AmeloD-KO teeth was similar to that found in the WT teeth (data not shown). Therefore, despite the inhibition of cell migration observed in the absence of AmeloD in the in vitro experiments, cell proliferation may not be suppressed in vivo in AmeloD-KO teeth because of the expression of Epfn in the IEE cells. We need further analysis using in vivo experimental models to understand the mechanism of cell proliferation and migration in IEE cells.
The shortened length of the early-stage incisors in P11 AmeloD-KO mice may arise due to the defect in IEE cell migration (Fig. 3, C and D). However, in 6-week-old AmeloD-KO mice, the lengths of the incisors were similar to those of the WT, which could reflect the continuous growth of adult mouse incisors (Fig. 2F). Indeed, incisor recovery speed was decreased by the deletion of AmeloD (Fig. 6), suggesting that incisor growth was inhibited in AmeloD-KO mice during tooth germ development. By contrast, the AmeloD-KO molars were smaller than the WT molars because IEE cells disappear after differentiation into ameloblasts or upon the completion of root formation. Interestingly, the AmeloD; Epfn-KO mice showed a reduced tooth size when compared with AmeloD-KO mice (Fig. 8). Previously, we reported that Epfn knockout results in inhibition of cell proliferation in the early stage of dental epithelium development (7). The reduced tooth size observed in the AmeloD; Epfn-KO mice may therefore result from the suppression of cell proliferation by the deletion of Epfn.
The EMT process contributes to organogenesis and cancer metastasis (27, 28). During the EMT, epithelial cells lose their polarity and cell–cell adhesion activity, thereby gaining migration capacity. Suppression of E-cadherin expression is commonly observed during the EMT (29), and various transcription factors that can suppress E-cadherin have been reported to act as EMT transcription factors (EMT-TFs). We previously suggested that AmeloD is an EMT-TF (17).
Twist1, a bHLH transcription factor, is a known EMT-TF (30). During tooth development, Twist1 is expressed in the mesenchyme, and mesenchymal-conditional Twist1-KO mice show an inhibition of odontoblast differentiation caused by the inhibition of epithelial–mesenchymal interactions (23). In Epfn-KO mice, the dental epithelial cells failed to polarize, and after the bell stage, the immature dental epithelium randomly invades into the mesenchymal region to form multiple teeth (7, 9, 31). In the present study, we demonstrated that the AmeloD expression in the invading dental epithelial cells of Epfn-KO teeth caused abnormal migration of epithelial cells through partial EMT processes (Fig. 7). These invasive dental epithelial cells did not express E-cadherin, but they expressed the mesenchymal markers N-cadherin and vimentin (Fig. 7). By contrast, in the AmeloD; Epfn-KO mice, dental epithelial cells expressed E-cadherin, and epithelial cell invasion was inhibited (Fig. 8), which, in turn, reduced the number and size of multiple teeth in the AmeloD; Epfn-KO mice.
These results suggest that AmeloD contributes to multiple tooth formation; however, some continuous formation of multiple teeth was still observed in the AmeloD; Epfn mice. We found that the invasive dental epithelium of Epfn-KO mice strongly expressed AmeloD as well as Twist1 (data not shown). The increased expression of Twist1 may contribute to the random invasion of dental epithelium observed in the Epfn-KO mice. Further analysis of the regulatory mechanism of AmeloD transcription and the relationship between AmeloD and Twist1 is required.
In summary, we identified a novel mechanism for the regulation of E-cadherin by AmeloD during tooth development. Our in vivo and in vitro results suggest that AmeloD is expressed in the IEE cells and that it suppresses E-cadherin expression. As a result, IEE cells might gain the migration capacity needed to increase tooth size. Taken together, our findings reveal that AmeloD is a novel factor that regulates tooth size during development, and this work provides new insights into the mechanism of ectodermal organ development.
Experimental procedures
Generation of AmeloD-KO mice
CompoZrTM Custom ZFN Service (Sigma) designed the zinc-finger nuclease (ZFN) construct for the AmeloD gene. The targeting construct was injected into embryonic stem cells. The AmeloD gene has two exons: noncoding exon 1 (1–178) and coding exon 2 (9599–11347). The AmeloD coding sequence is 567 bp long and encoded by exon 2 (10091–10658). We designed the targeting construct as shown in Fig. 2A. The 39-bp ZFN-binding sequence (10115–10154) is located near the transcriptional start site. Fertilized eggs were microinjected with the targeting construct and implanted into pseudopregnant mice. Two AmeloD-KO mouse lines were created. The following primers were used for genotyping: Forward, 5′-GAAGCCTTAGCAACTCGCAG-3′; Reverse, 5′-CCCAGTGTAGGCCGTCGTAGT-3′. A frameshift mutation was effected in both targeted AmeloD loci by deleting an identical single bp in the coding sequence of exon 2, resulting in a termination codon soon after the deletion. These two mouse lines showed a similar tooth phenotype. The AmeloD-KO mouse line was maintained by cross-mating with FVB/N mice. The following primers were used for Epfn-KO mouse genotyping: Epfn-Forward1, 5′-GCTTCCTCGTGCTTTACGGTATC-3′; Epfn-Forward2, 5′-GCTGGAGGCCGTGAAGGAAAGG-3′; Epfn-Reverse, 5′-GGGTTAGGGGTCATAAGGGATAGG-3′. The animal protocols for maintaining mice were approved by the National Institute of Dental and Craniofacial Research Animal Care and Use Committee. All animals were housed in an animal facility approved (Animal Protocol number 16-796) by the American Association for the Accreditation of Laboratory Animal Care.
Cell culture and transfection
The mouse CLDE cell line was established and maintained as described previously (32). Briefly, CLDE cells were maintained in a keratinocyte serum-free medium (K-SFM) supplemented with epidermal growth factor and bovine pituitary extract (Invitrogen) at 37 °C in 5% CO2. For the MTT assay and EdU staining, CLDE cells were transfected with the AmeloD expression vector (AmeloD-pCA1 vector) using Lipofectamine® LTX with PlusTM Reagent (Invitrogen). For the hanging drop culture and wound healing assay, CLDE cells were infected with a 100 multiplicity of infection of the adeno-associated virus expression vectors, adeno-GFP expression vector or adeno-GFP-AmeloD expression vector, in DMEM/F-12 without serum. After infection with the adeno-associated virus vectors, CLDE cells were maintained in DMEM/F-12 with 10% fetal bovine serum.
Western blotting
The P1 tooth germ was dissected, and the total protein was harvested with T-PER® Tissue Protein Extraction Reagent (Thermo Fisher Scientific). The protein amount was measured with Micro BCATM Protein Assay Reagent (Thermo Fisher Scientific), and 10 μg of protein was loaded onto a NuPAGE® Bis-Tris gel. Western blotting was performed as described previously (33).
Radiographic and micro-CT analysis
X-ray imaging was performed as described previously (34). For micro-CT analysis, the heads of 6-week-old mice were dissected and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS). Scanning was performed using a SCANCO μCT50 device. The micro-CT images were acquired with the X-ray source at 80 kV peak. The data were collected at a resolution of 2 μm. The 3D reconstruction and enamel and dentin volume quantification were conducted using AnalyzePro (AnalyzeDirect).
Histological analysis and immunofluorescence staining analysis
For histological analysis, several stages of developing mouse heads were dissected and processed as described previously (7). For H-E staining, sections were stained with hematoxylin 2 and eosin Y (Thermo Fisher Scientific). The widths of the enamel, dentin, and predentin layers were measured using a microscope (BioZero-8000) as described previously (35). Six mouse heads from each genotype were used for sectioning, and the thickest region in the distal cusp of the lower first molars was used for analysis. For immunostaining, antigen retrieval was performed with LAB solution (Cosmo Bio) for frozen sections and with citrate buffer (Sigma) for paraffin sections. After antigen retrieval, the sections were incubated in a power block reagent (Genetex) for 20 min prior to incubation with the primary antibody. The following primary antibodies were used in our study: keratin 5 (Covance; 1:400), vimentin (Abcam; 1:100), E-cadherin (Cell Signaling Technology; 1:200), and N-cadherin (Novus Biologicals; 1:200). Rabbit polyclonal antibody or the AmeloD peptide antibody was raised and purified on a peptide affinity column (17). Primary antibodies were detected by Alexa Fluor 488–conjugated antibody (Invitrogen; 1:400) and Cy-5–conjugated secondary antibody (Jackson ImmunoResearch Laboratories; 1:400). Nuclear staining was performed with DAPI (Sigma). Images were captured using a Nikon A1R confocal laser microscope system. Fluorescence line intensity scans were analyzed using ImageJ software as described previously (19).
MTT assay
The CLDE cells were plated on a 96-well plate at 4 × 103 cells/well. At days 0, 1, 2, and 3 after transfection, cell proliferation activity was measured using a Cell Counting Kit-8® (Dojindo). Two hours after incubation at 37 °C with reagent in K-SFM, the absorbance at 450 nm was measured using a Gen5TM microplate reader and Imager Software (BioTek).
EdU staining
EdU staining was performed using the Click-it® Plus reagent (Invitrogen) following the manufacturer's protocol. As in the in vitro experiment, 48 h prior to the assay, the CLDE cells were transfected with the AmeloD expression vector. The cells were incubated with 10 μm EdU containing K-SFM for 4 h. The EdU-positive ratio was measured using ImageJ. As in the in vivo experiment, pregnant mice were injected peritoneally with 1.5 μg of EdU in saline. After 4 h, the mice were euthanized in CO2, and E18 pups were immediately dissected for experiments. The EdU-positive ratio was calculated using ImageJ software.
Incisor cutting and recovery assay
Six-week-old WT and AmeloD-KO mice were used for this experiment. On day 0, the left lower incisor was clipped to half its length, and the length was recorded using a caliper as described previously (36). On day 3 after incisor clipping, the animals were sacrificed for analysis. The recovered incisor length was calculated as the total increase in length after the injury as follows: Recovered rate = Length difference at day 3/Length difference at day 0.
RT-PCR and real-time PCR
Total RNA was isolated using the RNeasy kit (Qiagen) following the manufacturer's protocol. The cDNA was synthesized from 500 ng of this total RNA using iScriptTM RT Supermix for RT-qPCR (Bio-Rad), and quantitative RT-PCR was performed using iQTM SYBR® Green Supermix (Bio-Rad) using a CFX384 Touch Real-Time PCR Detection System. The relative mRNA expression was determined using GAPDH as an internal control. The following primers were used; AmeloD-Forward, 5′-ACTACGACGCCTACACTGGG-3′; AmeloD-Reverse, 5′-ATGAAGGCAGGCTCGAACGG-3′; Gapdh-Forward, 5′-GGTGAAGGTCGGTGTGAACG-3′; Gapdh-Reverse, 5′-CTCGCTCCTGGAAGATGGTG-3′.
Hanging drop method
Forty-eight hours prior to the experiments, CLDE cells were infected with an adeno-GFP or adeno-GFP-AmeloD expression vector. Anti-E-cadherin functional antibody–treated CLDE cells were prepared by incubating the cells with antibody at 37 °C for an hour. The cells were dissociated from the plate using 0.25% trypsin, EDTA (Gibco). For the hanging drop culture, 3 × 106 cells were concentrated in a 20-μl drop. After a 6-h incubation, cell spheroids were imaged using an Advanced Microscopy Group (AMG) EVOS microscope. Single-cell numbers were counted after passage through a 70-μm cell strainer.
In vitro wound healing assay
CLDE cells were plated into an ibidi® 2-well cell culture insert at 4 × 104 cells/well. The next day, the cells were infected with the adeno-GFP or adeno-GFP-AmeloD expression vector. Anti-E-cadherin functional antibody–treated CLDE cells were incubated with antibody. After 24 h, the ibidi insert was removed and washed with PBS. Time-lapse CLDE cell images were captured by an Axiovert 135XT microscope after 24 h in DMEM/F-12 plus 1 ng/ml transforming growth factor-β1. The relative wound area was calculated using ImageJ.
Statistics
A two-tailed Student's t test was applied for statistical analysis of two independent variables. Fluorescence line intensity scan data and single-cell ratio in hanging drop culture were analyzed by one-way analysis of variance (ANOVA). Multiple analyses of the MTT assay and wound healing assay results were conducted using two-way ANOVA. A p value <0.05 was considered statistically significant.
Author contributions
Y. C., B. H., K. Y., M. I., C. K. B., E. S., and S. d. V. data curation; Y. C., B. H., M. I., C. K. B., E. S., and S. d. V. formal analysis; Y. C., B. H., S. F., and Y. Y. funding acquisition; Y. C., B. H., K. Y., M. I., C. K. B., E. S., and S. d. V. investigation; Y. C., B. H., K. Y., M. I., E. S., E. Y. C., and S. d. V. methodology; Y. C. writing-original draft; B. H., C. R., M. I., T. N., T. I., and K. S. resources; C. R., C. K. B., E. Y. C., T. N., T. I., K. S., S. F., and Y. Y. supervision; S. F. and Y. Y. writing-review and editing; Y. Y. conceptualization; Y. Y. project administration.
Acknowledgments
We thank Dr. William P. Daley for technical advice and Dr. Matthew Hoffman for various suggestions. We also thank Dr. Ashok Kulkarni, Glenn Longnecker, and the National Institute of Dental and Craniofacial Research Gene Targeting Core Facility for performing the embryonic stem cell injections to generate AmeloD-KO mice.
This work was supported in part by NIDCR, National Institutes of Health Intramural Research Program Grant 1ZIADE000720-07 (to Y. Y.). and NIDCR, National Institutes of Health Transfer Core Facility Grant ZIC DE000744-04. This work was also supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grants-in-aid 17H01606 (to S. F.) and 15J04116 (to Y. C.) and National Nature Science Foundation of China Grant NSFC81500811 (to B. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) MG575629.
Y. Chiba, B. He, K. Yoshizaki, C. Rhodes, M. Ishijima, C. K. E. Bleck, E. Stempinski, E. Y. Chu, T. Nakamura, T. Iwamoto, S. de Vega, K. Saito, S. Fukumoto, and Y. Yamada, unpublished data.
- E
- embryonic day
- bHLH
- basic helix–loop–helix
- EdU
- 5-ethynyl-2′-deoxyuridine
- EMT
- epithelial–mesenchymal transition
- Epfn
- epiprofin
- IEE
- inner enamel epithelium
- KO
- knockout
- P
- postnatal day
- SEM
- scanning electron microscopy
- SI
- stratum intermedium
- ZFN
- zinc-finger nuclease
- aa
- amino acids
- micro-CT
- micro-computed tomography
- H-E
- hematoxylin-eosin
- CLDE
- cervical loop–derived dental epithelial
- EMT-TF
- EMT transcription factor
- K-SFM
- keratinocyte serum-free medium
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- DMEM
- Dulbecco's modified Eagle's medium
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- DAPI
- 4′,6-diamidino-2-phenylindole
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ANOVA
- analysis of variance.
References
- 1. Pispa J., and Thesleff I. (2003) Mechanisms of ectodermal organogenesis. Dev. Biol. 262, 195–205 10.1016/S0012-1606(03)00325-7 [DOI] [PubMed] [Google Scholar]
- 2. Cai D., Chen S. C., Prasad M., He L., Wang X., Choesmel-Cadamuro V., Sawyer J. K., Danuser G., and Montell D. J. (2014) Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell 157, 1146–1159 10.1016/j.cell.2014.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li C. Y., Cha W., Luder H. U., Charles R. P., McMahon M., Mitsiadis T. A., and Klein O. D. (2012) E-cadherin regulates the behavior and fate of epithelial stem cells and their progeny in the mouse incisor. Dev. Biol. 366, 357–366 10.1016/j.ydbio.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li J., Parada C., and Chai Y. (2017) Cellular and molecular mechanisms of tooth root development. Development 144, 374–384 10.1242/dev.137216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sohn W. J., Choi M. A., Yamamoto H., Lee S., Lee Y., Jung J. K., Jin M. U., An C. H., Jung H. S., Suh J. Y., Shin H. I., and Kim J. Y. (2014) Contribution of mesenchymal proliferation in tooth root morphogenesis. J. Dent. Res. 93, 78–83 10.1177/0022034513511247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakamura T., Unda F., de-Vega S., Vilaxa A., Fukumoto S., Yamada K. M., and Yamada Y. (2004) The Krüppel-like factor epiprofin is expressed by epithelium of developing teeth, hair follicles, and limb buds and promotes cell proliferation. J. Biol. Chem. 279, 626–634 10.1074/jbc.M307502200 [DOI] [PubMed] [Google Scholar]
- 7. Nakamura T., de Vega S., Fukumoto S., Jimenez L., Unda F., and Yamada Y. (2008) Transcription factor epiprofin is essential for tooth morphogenesis by regulating epithelial cell fate and tooth number. J. Biol. Chem. 283, 4825–4833 10.1074/jbc.M708388200 [DOI] [PubMed] [Google Scholar]
- 8. Nakamura T., Jimenez-Rojo L., Koyama E., Pacifici M., de Vega S., Iwamoto M., Fukumoto S., Unda F., and Yamada Y. (2017) Epiprofin regulates enamel formation and tooth morphogenesis by controlling epithelial-mesenchymal interactions during tooth development. J. Bone Miner. Res. 32, 601–610 10.1002/jbmr.3024 [DOI] [PubMed] [Google Scholar]
- 9. Nakamura T., Fukumoto S., and Yamada Y. (2011) Diverse function of epiprofin in tooth development. J. Oral Biosci. 53, 22–30 10.1016/S1349-0079(11)80032-0 [DOI] [Google Scholar]
- 10. Jones S. (2004) An overview of the basic helix-loop-helix proteins. Genome Biol. 5, 226 10.1186/gb-2004-5-6-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poulin G., Turgeon B., and Drouin J. (1997) NeuroD1/β2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol. Cell. Biol. 17, 6673–6682 10.1128/MCB.17.11.6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rudnicki M. A., Schnegelsberg P. N., Stead R. H., Braun T., Arnold H.-H., and Jaenisch R. (1993) MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75, 1351–1359 10.1016/0092-8674(93)90621-V [DOI] [PubMed] [Google Scholar]
- 13. Abe M., Tamamura Y., Yamagishi H., Maeda T., Kato J., Tabata M. J., Srivastava D., Wakisaka S., and Kurisu K. (2002) Tooth-type specific expression of dHAND/Hand2: possible involvement in murine lower incisor morphogenesis. Cell Tissue Res. 310, 201–212 10.1007/s00441-002-0611-2 [DOI] [PubMed] [Google Scholar]
- 14. Borkosky S. S., Nagatsuka H., Orita Y., Tsujigiwa H., Yoshinobu J., Gunduz M., Rodriguez A. P., Missana L. R., Nishizaki K., and Nagai N. (2008) Sequential expressions of Notch1, Jagged2 and Math1 in molar tooth germ of mouse. Biocell 32, 251–258 [PubMed] [Google Scholar]
- 15. Rice R., Thesleff I., and Rice D. P. (2005) Regulation of Twist, Snail, and Id1 is conserved between the developing murine palate and tooth. Dev. Dyn. 234, 28–35 10.1002/dvdy.20501 [DOI] [PubMed] [Google Scholar]
- 16. Zhang Y., Blackwell E. L., McKnight M. T., Knutsen G. R., Vu W. T., and Ruest L. B. (2012) Specific inactivation of Twist1 in the mandibular arch neural crest cells affects the development of the ramus and reveals interactions with hand2. Dev. Dyn. 241, 924–940 10.1002/dvdy.23776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He B., Chiba Y., de Vega S., Tanaka K., Yoshizaki K., Ishijima M., Yuasa K., Ishikawa M., Rhodes C., Sakai K., Zhang P., Fukumoto S., Zhou X., and Yamada Y. (2018) Identification of the novel tooth-specific transcription factor AmeloD. J. Dent. Res., in press 10.1177/0022034518808254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Obara N., Suzuki Y., Nagai Y., and Takeda M. (1998) Expression of E- and P-cadherin during tooth morphogenesis and cytodifferentiation of ameloblasts. Anat. Embryol. 197, 469–475 10.1007/s004290050157 [DOI] [PubMed] [Google Scholar]
- 19. Daley W. P., Matsumoto K., Doyle A. D., Wang S., DuChez B. J., Holmbeck K., and Yamada K. M. (2017) Btbd7 is essential for region-specific epithelial cell dynamics and branching morphogenesis in vivo. Development 144, 2200–2211 10.1242/dev.146894 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Aurrekoetxea M., Irastorza I., García-Gallastegui P., Jiménez-Rojo L., Nakamura T., Yamada Y., Ibarretxe G., and Unda F. J. (2016) Wnt/β-catenin regulates the activity of epiprofin/Sp6, SHH, FGF, and BMP to coordinate the stages of odontogenesis. Front. Cell Dev. Biol. 4, 25 10.3389/fcell.2016.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jimenez-Rojo L., Ibarretxe G., Aurrekoetxea M., de Vega S., Nakamura T., Yamada Y., and Unda F. (2010) Epiprofin/Sp6: a new player in the regulation of tooth development. Histol. Histopathol. 25, 1621–1630 10.14670/HH-25.1621 [DOI] [PubMed] [Google Scholar]
- 22. Ibarretxe G., Aurrekoetxea M., Crende O., Badiola I., Jimenez-Rojo L., Nakamura T., Yamada Y., and Unda F. (2012) Epiprofin/Sp6 regulates Wnt-BMP signaling and the establishment of cellular junctions during the bell stage of tooth development. Cell Tissue Res. 350, 95–107 10.1007/s00441-012-1459-8 [DOI] [PubMed] [Google Scholar]
- 23. Meng T., Huang Y., Wang S., Zhang H., Dechow P. C., Wang X., Qin C., Shi B., D'Souza R. N., and Lu Y. (2015) Twist1 is essential for tooth morphogenesis and odontoblast differentiation. J. Biol. Chem. 290, 29593–29602 10.1074/jbc.M115.680546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Onodera T., Sakai T., Hsu J. C., Matsumoto K., Chiorini J. A., and Yamada K. M. (2010) Btbd7 regulates epithelial cell dynamics and branching morphogenesis. Science 329, 562–565 10.1126/science.1191880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sakai T., and Onodera T. (2013) Btbd7/Cleftin regulates cleft formation and branching morphogenesis of epithelial cells. J. Oral Biosci. 55, 73–75 10.1016/j.job.2013.03.002 [DOI] [Google Scholar]
- 26. Nakamura T., Yoshitomi Y., Sakai K., Patel V., Fukumoto S., and Yamada Y. (2014) Epiprofin orchestrates epidermal keratinocyte proliferation and differentiation. J. Cell Sci. 127, 5261–5272 10.1242/jcs.156778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kalluri R., and Weinberg R. A. (2009) The basics of epithelial-mesenchymal transition. J. Clin. Investig. 119, 1420–1428 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lim J., and Thiery J. P. (2012) Epithelial-mesenchymal transitions: insights from development. Development 139, 3471–3486 10.1242/dev.071209 [DOI] [PubMed] [Google Scholar]
- 29. Lamouille S., Xu J., and Derynck R. (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peinado H., Olmeda D., and Cano A. (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer 7, 415–428 10.1038/nrc2131 [DOI] [PubMed] [Google Scholar]
- 31. Li L., Tang Q., Nakamura T., Suh J. G., Ohshima H., and Jung H. S. (2016) Fine tuning of Rac1 and RhoA alters cuspal shapes by remolding the cellular geometry. Sci. Rep. 6, 37828 10.1038/srep37828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoshizaki K., Hu L., Nguyen T., Sakai K., He B., Fong C., Yamada Y., Bikle D. D., and Oda Y. (2014) Ablation of coactivator Med1 switches the cell fate of dental epithelia to that generating hair. PLoS One 9, e99991 10.1371/journal.pone.0099991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakamura T., Chiba Y., Naruse M., Saito K., Harada H., and Fukumoto S. (2016) Globoside accelerates the differentiation of dental epithelial cells into ameloblasts. Int. J. Oral Sci. 8, 205–212 10.1038/ijos.2016.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fukumoto S., Kiba T., Hall B., Iehara N., Nakamura T., Longenecker G., Krebsbach P. H., Nanci A., Kulkarni A. B., and Yamada Y. (2004) Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J. Cell Biol. 167, 973–983 10.1083/jcb.200409077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoshizaki K., Yamamoto S., Yamada A., Yuasa K., Iwamoto T., Fukumoto E., Harada H., Saito M., Nakasima A., Nonaka K., Yamada Y., and Fukumoto S. (2008) Neurotrophic factor neurotrophin-4 regulates ameloblastin expression via full-length TrkB. J. Biol. Chem. 283, 3385–3391 10.1074/jbc.M704913200 [DOI] [PubMed] [Google Scholar]
- 36. Sun Z., Yu W., Sanz Navarro M., Sweat M., Eliason S., Sharp T., Liu H., Seidel K., Zhang L., Moreno M., Lynch T., Holton N. E., Rogers L., Neff T., Goodheart M. J., et al. (2016) Sox2 and Lef-1 interact with Pitx2 to regulate incisor development and stem cell renewal. Development 143, 4115–4126 10.1242/dev.138883 [DOI] [PMC free article] [PubMed] [Google Scholar]