Abstract
RNA granules consist of membrane-less RNA–protein assemblies and contain dynamic liquid-like shells and stable solid-like cores, which are thought to function in numerous processes in mRNA sorting and translational regulation. However, how these distinct substructures are formed, whether they are assembled by different scaffolds, and whether different RNA granule scaffolds induce these different substructures remains unknown. Here, using fluorescence microscopy–based morphological and molecular-dynamics analyses, we demonstrate that RNA granule scaffold proteins (scaffolds) can be largely classified into two groups, liquid and solid types, which induce the formation of liquid-like and solid-like granules, respectively, when expressed separately in cultured cells. We found that when co-expressed, the liquid-type and solid-type scaffolds combine and form liquid- and solid-like substructures in the same granules, respectively. The combination of the different types of scaffolds reduced the immobile fractions of the solid-type scaffolds and their dose-dependent ability to decrease nascent polypeptides in granules, but had little effect on the dynamics of the liquid-type scaffolds or their dose-dependent ability to increase nascent polypeptides in granules. These results suggest that solid- and liquid-type scaffolds form different substructures in RNA granules and differentially affect each other. Our findings provide detailed insight into the assembly mechanism and distinct dynamics and functions of core and shell substructures in RNA granules.
Keywords: RNA binding protein, scaffold protein, molecular dynamics, fluorescence recovery after photobleaching (FRAP), cell permeabilization, core-shell, liquid, liquid–liquid phase separation (LLPS), RNA granule, solid
Introduction
RNA granules are membrane-less assemblies consisting of mRNAs, ribosomes, and RNA-binding proteins (1–3). Several kinds of RNA granules have been identified, e.g. stress granules, which are transiently formed in response to cellular stress and sequester untranslated mRNAs and signaling proteins, and neuronal RNA granules, which are constantly formed to sequester mRNAs and transport them from the soma to dendrites for local translation (2, 4). In addition to the sequestration of untranslated mRNAs, RNA granules function in the selective translation of specific mRNAs and rapid translational reactivation of mRNAs released from the granules. Thus, RNA granules have both stable and dynamic characteristics (5–7). Abnormalities in RNA granule dynamics are associated with degenerative diseases, such as amyotrophic lateral sclerosis and frontotemporal lobar degeneration, in which aggregates of RNA granule components are formed in neurons (8, 9).
Many components of RNA granules possess intrinsically disordered regions (IDRs),2 which are involved in weak multivalent molecular interactions. These interactions promote liquid–liquid phase separation (LLPS) to form dynamic RNA granules (3, 10–13). Parker and co-workers (14) recently reported that RNA granules are more than simply structures for LLPS; they contain stable core substructures surrounded by dynamic shells. The cores are densely concentrated structures, and the shells are less concentrated liquid-like structures (14). This uneven distribution of materials in RNA granules was also observed by EM (15). As the core substructures are not disassembled and are purified as small foci even after cell lysis, they are thought to be solid-like structures rather than liquid droplets. It has been demonstrated that the assembly of cores and shells consists of distinct processes, i.e. core formation precedes shell formation after the induction of stress granule formation (16). However, the mechanism by which the distinct substructures are formed, e.g. whether the substructures simply differ in concentration or are assembled by different scaffolds, remains unknown. This question can be refined to a more specific question of whether different RNA granule scaffolds induce different types, i.e. core-type or shell-type, of granules in cells.
Here, we expressed RNA granule scaffolds in cultured cells and analyzed the morphology of the granules formed and the dynamics of the scaffolds in the granules. As a result, the scaffolds were largely classified into two types: scaffolds that assembled liquid-like smooth (S) granules and those that assembled solid-like rough (R) granules. Furthermore, co-expression of sets of S- and R-granule scaffolds in cells promoted the formation of RNA granules with S- and R-substructures. The two types of substructures had different influences on each other such that S-substructures increased the mobility of R-substructures, although R-substructures had little effect on the dynamics of S-substructures. These results suggest that combinations of RNA granule scaffolds have the ability to form substructures in granules, providing insight into the formation and interaction of dynamic shell-like and stable core-like substructures in RNA granules.
Results
S- and R-granules assembled with distinct scaffolds
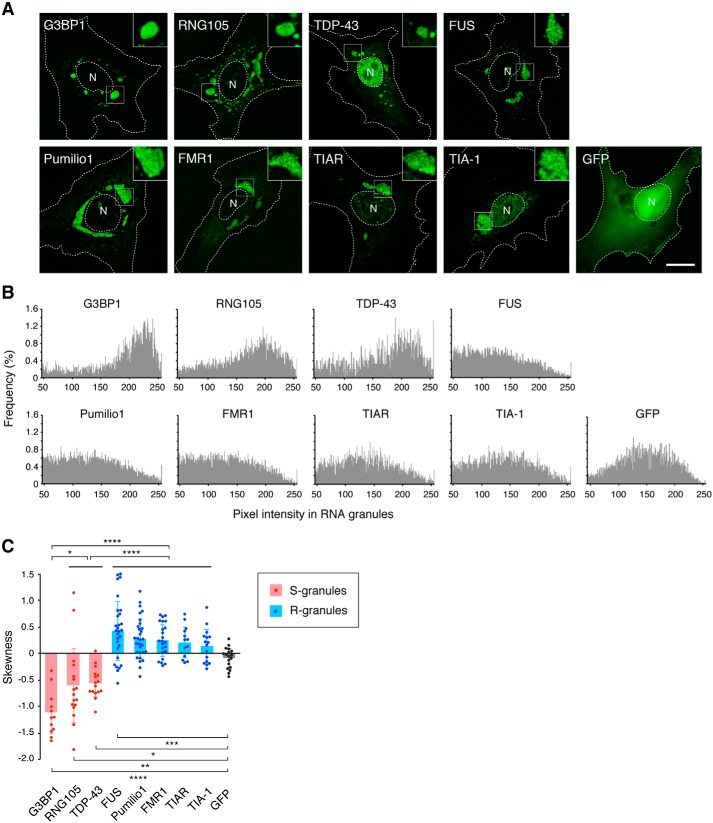
Many RNA granule-associated proteins have been identified, among which several proteins are known to induce RNA granule assembly when expressed in cells and are designated as scaffolds (2, 17). We expressed the following scaffolds as GFP-tagged proteins in cultured A6 cells: T-cell intracellular antigen 1 (TIA-1); TIA-1–related protein (TIAR); RNA granule protein 105 (RNG105)/caprin1; Ras-GTPase–activating protein SH3 domain-binding protein 1 (G3BP1); TAR DNA–binding protein 43 (TDP-43); fused in sarcoma (FUS); fragile X mental retardation 1 (FMR1); and Pumilio1 (18–24). When expressed separately, each scaffold formed cytoplasmic granules (Fig. 1A). These granules contained a higher concentration of mRNA than the cytoplasm (Fig. S1), which was consistent with the previous reports that these scaffolds induce the formation of RNA granules containing mRNA.
Figure 1.
Each RNA granule scaffold preferentially assembles into S- or R-granules. A, granules formed by GFP-tagged RNA granule scaffolds in A6 cells. Insets show magnified images of the boxed areas. N, nucleus. The outlines of the cell and the nucleus are indicated by dotted lines. Scale bar, 10 μm. B, histograms of the pixel intensity of GFP-tagged scaffolds in granules and control GFP in the cytoplasm. C, skewness of the histograms in B. Data are represented by scatter plots of the individual data with the mean ± S.D. n = 12 (G3BP1), 17 (RNG105), 16 (TDP-43), 29 (FUS), 30 (Pumilio1), 23 (FMR1), 13 (TIAR), 16 (TIA-1), and 18 (GFP) cells. *, p < 0.05; **, p < 0.01; ***, p < 0.005; ****, p < 0.001, one-way ANOVA (F(8,165) = 25.6, p < 2.2e-16) followed by the Tukey-Kramer test.
The granules formed with the scaffolds were morphologically classified into two types: granules with smooth surfaces and those with rough surfaces. G3BP1, RNG105, and TDP-43 induced the formation of granules with smooth surfaces, in which the scaffolds were evenly distributed (Fig. 1A). We designated these granules as S-granules. In contrast, TIA-1, TIAR, FUS, Pumilio1, and FMR1 induced the formation of granules consisting of numerous small foci (Fig. 1A). The size of the foci in the granules was 252 ± 55 nm (FUS), 340 ± 104 nm (Pumilio1), 281 ± 54 nm (FMR1), 227 ± 52 nm (TIAR), and 235 ± 53 nm (TIA-1) in diameter (mean ± S.D., n = 20 each), which was similar to stress granule cores whose sizes are 200–300 nm in diameter (14, 16). As the small foci gathered, the overall images of the granules had a rugged texture and rough surfaces. Thus, we designated these granules as R-granules.
To more objectively distinguish the two types of granules, the texture of the granules was analyzed quantitatively. It is known that the skewness of a profile can be used to evaluate surface roughness (25). We applied this method to RNA granules because if granules have a rough texture, they have many “fluorescence valleys,” and their fluorescence histogram will have a higher frequency of small values than the histogram of smooth granules. As a result, the histograms of rough and smooth granules exhibit different skew patterns. We measured the pixel intensity of GFP fluorescence in the granules and generated histograms (Fig. 1B). The histograms demonstrated asymmetrical shapes that were different between S- and R-granules: S-granules had a left-skewed distribution, and R-granules had a right-skewed distribution. As a result, the skewness of S-granules was negative, but that of R-granules was positive (Fig. 1C). These results quantitatively indicated that S- and R-granule scaffolds induced the formation of different types of RNA granules.
S- and R-granules exhibit different shape-change behavior after cell permeabilization
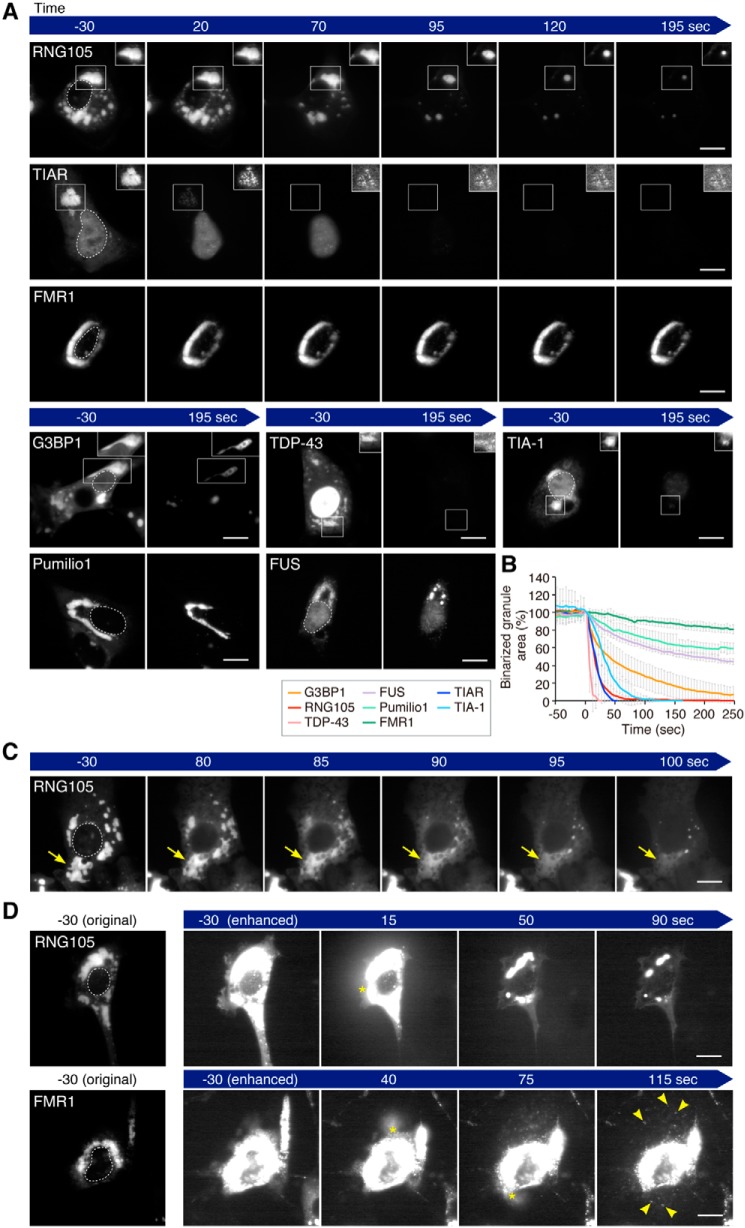
One different property between the core and the shell of RNA granules is that the cores can be purified as foci, but the shells cannot, after cell lysis (14). We permeabilized the cells expressing the GFP-tagged scaffolds with digitonin and analyzed the changes in the shape and fluorescence intensity of the granules by time-lapse imaging (Fig. 2, A–D). A series of time-lapse images was binarized and measured for the area of the binarized granules (Fig. 2B). FMR1- and Pumilio1-induced R-granules were the most stable, and their overall shapes and fluorescence intensity changed less than those of the other scaffold-induced granules (Fig. 2, A and B, and Movie S1). In some cells, FMR1 and Pumilio1 foci blew out of the permeabilized cells and were deposited on the dish (Fig. 2D, Movie S2). These results suggested that FMR1- and Pumilio1-induced R-granules were resistant to cell lysis and were purifiable, like cores. The fluorescence intensity of FUS-induced granules also did not decrease much after cell permeabilization, but the shape changed significantly. They were converted from R-type to S-type after cell permeabilization (Fig. 2A and Movie S3).
Figure 2.
S- and R-granules exhibit different shape-change behavior after cell permeabilization. A, time-lapse images of scaffold-induced granules in the cell permeabilization analysis. Digitonin was added to the cells at time 0. Fluorescence levels in the boxed areas were adjusted using the automatic window/level tool of ImageJ to visualize the shapes of the granules regardless of the decrease in fluorescence in the granules after permeabilization, and the adjusted images are shown in the insets. The outlines of the nuclei are indicated by dotted lines in the left panels. Scale bars, 10 μm. B, area of binarized granules in the cell permeabilization analysis. Note that the time just before the cells became permeable was set to time 0, and the area of the binarized granules at time 0 was adjusted to 100%. n = 7 (G3BP1), 9 (RNG105), 8 (TDP-43), 7 (FUS), 8 (Pumilio1), 8 (FMR1), 8 (TIAR), and 8 (TIA-1) cells. C, representative image series of RNG105-induced granules to dissolve rapidly after cell permeabilization. Arrows indicate dissolution of a granule. D, representative image series of RNG105 and FMR1 blowing out from permeabilized cells. Asterisks indicate the positions of the blowout. RNG105-induced granules were dissolved in the medium, but FMR1-induced granules blew out from the cells as foci (arrowheads). Scale bars, 10 μm. See also Movies S1–S7.
In contrast to the R-granules, RNG105-, G3BP1-, and TDP-43-induced S-granules underwent rapid decreases in the binarized granule areas (Fig. 2B). These decreases were due to shrinkage and dissolution of the granules. For example, RNG105-induced granules rapidly shrank (Fig. 2A and Movie S4) or dissolved (Fig. 2C and Movie S5) after cell permeabilization. Shrinkage and dissolution were also observed in G3BP1-induced granules and TDP-43-induced granules, respectively (Fig. 2A). These types of granule deformation were reminiscent of the shrinkage and dissolution of liquid–liquid phase-separated droplets triggered by perturbing the equilibrium conditions in vitro (26). In some cells, RNG105, G3BP1, and TDP-43 blew out of the permeabilized cells, but they were not observed as foci (Fig. 2D and Movie S6), suggesting that the granules formed by the S-scaffolds were not purifiable, which contrasted with the R-granules.
TIAR- and TIA-1-induced R-granules also underwent rapid decreases in the binarized granule areas (Fig. 2, A and B). However, these R-granules were different from the S-granules in that they appeared to not shrink or dissolve after cell permeabilization. Instead, the fluorescence intensity of foci in the granules was rapidly decreased without greatly changing the overall shape of the granules (Fig. 2A and Movie S7), suggesting that the fluorescence decrease was due to the rapid dissociation of the scaffolds from the foci embedded in the granules.
Taken together, R-granules induced by FMR1 and Pumilio1 were stable in shape, resistant to cell permeabilization, and purifiable, similar to the properties of solid-like cores. In contrast, S-granules were similar to liquid-like droplets in that they were not purifiable and underwent shrinkage and dissolution after cell permeabilization. Although R-granules induced by TIAR and TIA-1 were not resistant to cell permeabilization, they were not similar to liquid-like droplets because TIAR and TIA-1 were localized to foci in granules that did not shrink or dissolve after cell permeabilization. An exception was FUS-induced granules, which converted from R-type to S-type granules after cell permeabilization.
R-granules with slow τ½ have large immobile fractions, whereas S-granules with fast τ½ have high deformability
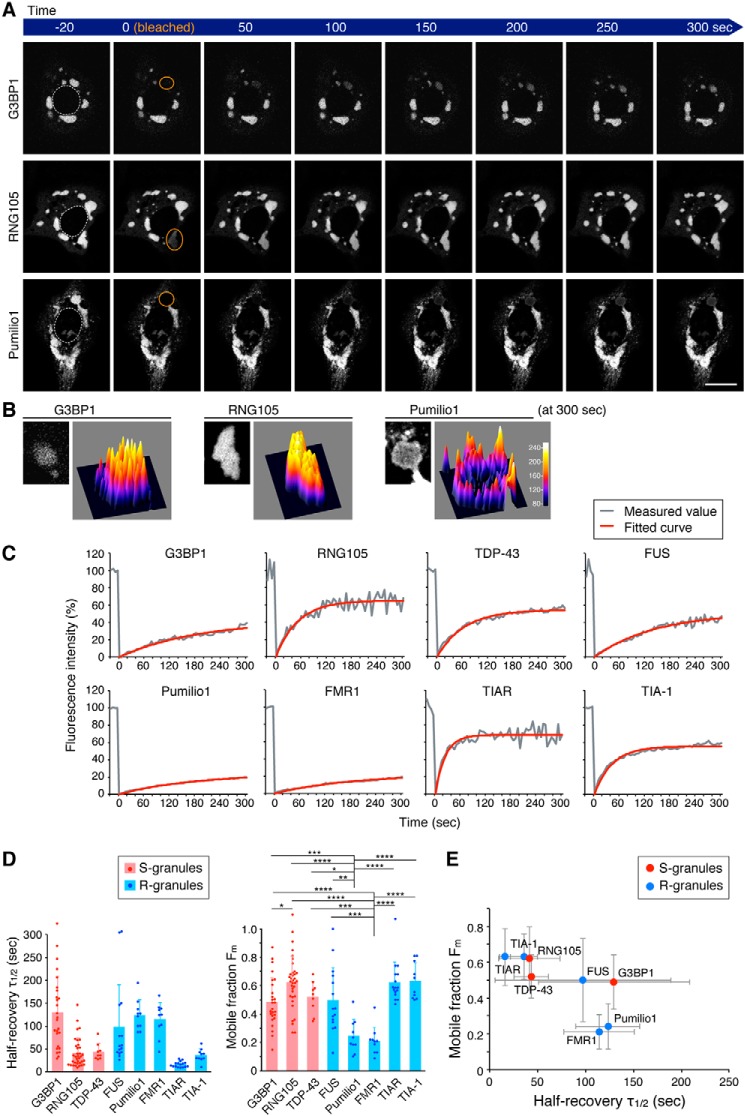
To examine the dynamics of the scaffolds in granules in living cells, we conducted fluorescence recovery after photobleaching (FRAP) analysis (Fig. 3 and Movies S8–S10). Each FRAP curve was fitted with an exponential equation, and the halftime of recovery (τ½) and the mobile fraction (Fm) were extracted (Fig. 3, C and D). Comparison of τ½ among the scaffolds revealed that they had distinct τ½ values, and the τ½ was not dependent on which type of granule the scaffold formed (Fig. 3D, left).
Figure 3.
R-scaffolds, but not S-scaffolds, with slower exchange rates have less mobile fractions in RNA granules. A, representative time-lapse images of RNA granules in FRAP analysis. Orange circles, regions bleached just before 0 s. Dotted lines in the left panels, nuclei. Scale bar, 10 μm. B, surface plots of the fluorescence intensity of the scaffolds in the bleached RNA granules 300 s after photobleaching. The left panel of each scaffold shows a magnified image of the granule in the ROI at 300 s in A; right panel of each scaffold, surface plot. C, representative FRAP curves and fitted curves with exponential equations. D, halftime of recovery (τ½) and mobile fraction (Fm) extracted from the fitted equations. n = 26 (G3BP1), 35 (RNG105), 9 (TDP-43), 15 (FUS), 10 (Pumilio1), 10 (FMR1), 16 (TIAR), and 10 (TIA-1). p = 3.49e-12 (F(7,123) = 12.7) for τ½ and p = 5.01e-12 (F(7,123) = 12.5) for Fm, one-way ANOVA. *, p < 0.05; **, p < 0.01; ***, p < 0.005; ****, p < 0.001, Tukey-Kramer test. E, relationship between the τ½ and Fm of the scaffolds. See also Movies S8–S10.
In contrast, comparison of the Fm among the scaffolds revealed that only R-granule scaffolds, FMR1 and Pumilio1, had low Fm values (Fig. 3D, right). Plotting the relationship between τ½ and the Fm of the scaffolds demonstrated that the Fm of R-granule scaffolds was smaller when the scaffolds had a slower τ½, e.g. FMR1 and Pumilio1 (Fig. 3E). In contrast, the Fm of S-granule scaffolds was not small even when the scaffold had a slow τ½, e.g. G3BP1 (Fig. 3E). Thus, the τ½–Fm relationship differed between R- and S-granules, which may be related to the different fluorescence recovery of R- and S-granule scaffolds with a slow τ½ (Pumilio1 and G3BP1) at the center of the bleached granules: fluorescence recovery of Pumilio1 at the center was low, whereas that of G3BP1 was high (Fig. 3B). This low and high mobility of the R- and S-scaffolds may be due to low and high miscibility of the R- and S-scaffolds in granules, respectively, as discussed under the “Discussion.”
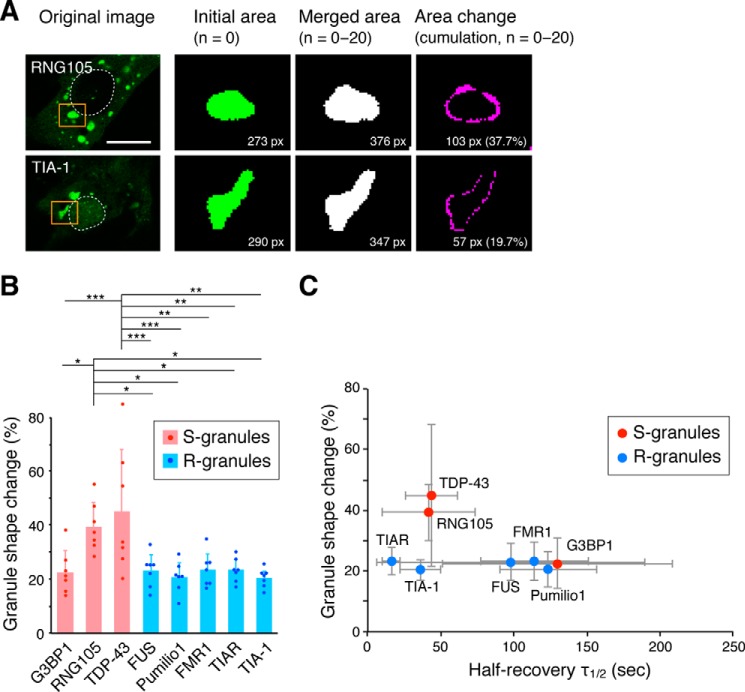
Next, the time-dependent shape change of granules was analyzed. The deformation rate over a set period of time (100 s) was measured, and it was significantly larger for RNG105- and TDP-43–induced S-granules than the other granules (Fig. 4, A and B, and Movies S11 and S12). Plotting the relationship between the τ½ and the shape change rate demonstrated that the shape change rate was almost the same among the R-granule scaffolds regardless of the τ½ (Fig. 4C). In contrast, among the S-granule scaffolds, the shape change rate was significantly higher for RNG105 and TDP-43, which had a fast τ½ (Fig. 4C). Thus, the τ½-shape change rate relationship differed between R- and S-granules, which may be due to different abilities to resist deformation forces between solid and liquid structures. In addition, the τ½-dependent shape change rate of S-granules may be related to the property of liquid droplets to become spherical by surface tension, as discussed under the “Discussion.”
Figure 4.
RNA granules formed by S-scaffolds, but not R-scaffolds, with faster exchange rates dynamically change their shapes. A, methods for the quantification of granule shape change. Granules of interest (orange squares) were converted into binary images. Twenty one images (n = 0–20) taken over 100 s were merged, and the area of the merged granule was compared with the average area of the granule. Dotted lines, nuclei. Scale bar, 10 μm. B, granule shape change rates over 100 s. n = 7 cells. *, p < 0.05; **, p < 0.01; ***, p < 0.005, one-way ANOVA (F(7,48) = 5.99, p = 4.82e-5) followed by the Tukey-Kramer test. C, relationship between τ½ in the FRAP analysis and the granule shape change rates of the scaffolds. See also Movies S11 and S12.
Taken together, these results suggested that R-granules are solid-like structures that contain larger immobile fractions when the R-granule scaffolds have a slower τ½. In contrast, S-granules had higher fluidity in terms of miscibility and deformability, and this property was clearer when the S-granule scaffolds had a faster τ½.
S- and R-substructure formation with S- and R-scaffolds
The formation of different types of RNA granules by distinct scaffolds raised the question of whether the scaffolds constitute the same granules. Therefore, we next co-transfected A6 cells with sets of two scaffolds as GFP- and mRFP1-tagged proteins. RNG105, G3BP1, and TIAR were tagged with mRFP1 and co-expressed with the other scaffolds tagged with GFP (Fig. 5, A–C). Any combination of the two scaffolds was combined into the same granules, indicating that the scaffolds constituted the same granules. Sets of S-granule scaffolds were highly co-localized in S-granules with smooth surfaces (Fig. 5A). Sets of R-granule scaffolds were also well co-localized in the small foci embedded in the R-granules (Fig. 5B). Thus, although the turnover of the scaffolds varied from each other (Figs. 2, A and B, and 3, C–E), the S-scaffolds and R-scaffolds were co-localized to the same S-granules and to the same foci in the R-granules, respectively. When scaffolds of different types were co-expressed, they were combined into the same granules but formed different subgranular structures, except for well co-localized combinations FMR1–RNG105 and TIAR–TDP-43 (Fig. 5, C and D). In general, S-scaffolds formed smooth granules, whereas R-scaffolds formed punctate substructures therein.
Figure 5.
S- and R-subgranular structures are formed by co-expressing sets of S- and R-scaffolds. A–C, granules formed by GFP- and mRFP1-tagged scaffolds co-expressed in A6 cells. Cells co-expressing two S-granule scaffolds (A), two R-granule scaffolds (B), or S- and R-granule scaffolds (C). Attached images on the right are magnified GFP, mRFP1, and merged images of the boxed areas. Dotted lines, nuclei. Scale bars, 1 μm. D, Pearson's r values for the localization correlation of the two scaffolds in the granules. The mean, S.D., and n are indicated. G3BP versus the other scaffolds, p < 2.2e-16 (F(6,141) = 25.6); RNG105 versus the other scaffolds, p = 3.78e-10 (F(6, 122) = 11.5); TIAR versus the other scaffolds, p = 4.30e-8 (F(6,111) = 9.10), one-way ANOVA.
By calculating Pearson's correlation coefficient, co-localization of the two scaffolds in the granules was quantified (Fig. 5D). Pearson's correlation coefficient between S-granule scaffolds was significantly high (>0.9) for any combination. The correlation coefficient between R-granule scaffolds was also significantly high (>0.9), except between TIAR and FMR1. These results indicated that scaffolds of the same type were concentrated in the same subgranular structures. However, the correlation coefficient between scaffolds of different types was relatively low. Among the combinations, the correlation coefficients between G3BP1 and FMR1, G3BP1 and FUS, and G3BP1 and Pumilio1 were particularly low. In contrast, the correlation coefficients between RNG105 and FMR1 and TIAR and TDP-43 were significantly high, although they are different scaffold types (Fig. 5D). One of the reasons for this may be direct binding of the scaffolds (27). Taken together, these results supported the observation that S- and R-scaffolds formed distinct substructures in granules and suggested that the scaffolds were not randomly mixed but localized in a moderate order in the granules.
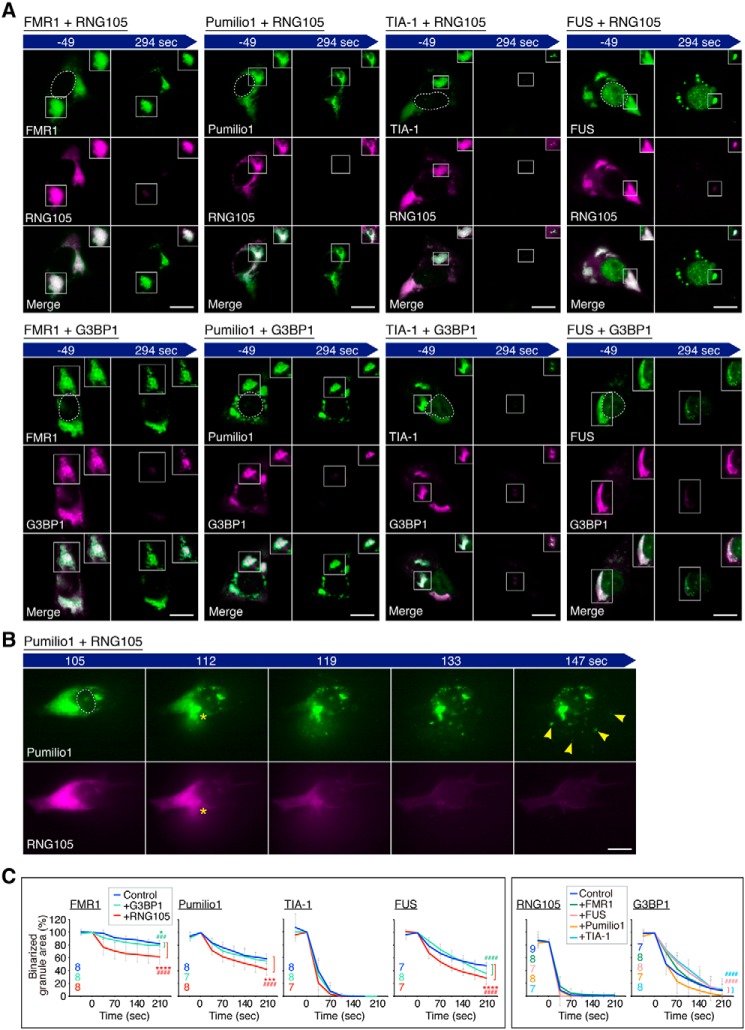
Next, we examined whether S- and R-substructures that combined into the same granules retained the respective properties of S- and R-granules. Cells were co-transfected with sets of R- and S-scaffolds and subjected to the cell permeabilization experiment with digitonin (Fig. 6A). R-scaffolds retained the properties observed when expressed alone, even in the presence of S-scaffolds as follows: FMR1- and Pumilio1-induced R-substructures were resistant to cell permeabilization and retained their R-texture (Fig. 6A and Movie S13). In some cells, the R-scaffolds blew out of the permeabilized cells as foci, whereas S-scaffolds blew out of the same cells without forming any granular structures (Fig. 6B and Movie S14). TIA-1 rapidly dissociated from the granules, but foci of the substructures remained after cell permeabilization (Fig. 6A). FUS-induced R-substructures were converted from R-type to S-type after cell permeabilization, although the conversion was partially inhibited by the co-expression of G3BP1 (Fig. 6A).
Figure 6.
Differences in resistance to cell permeabilization between S- and R-granules are retained after convergence into the same granules. A, time-lapse images of R- and S-substructures in the same cells. The cells were treated with digitonin at time 0. The fluorescence levels of the boxed areas were adjusted as in Fig. 2A, and the adjusted images are shown in the insets. Dotted lines in the left GFP panels, nuclei. Scale bars, 10 μm. B, representative image series in which Pumilio1 and RNG105 co-expressed in the same cell blew out from the cell after permeabilization. Asterisks indicate the position of the blowout. Pumilio1-induced substructures blew out from the cell as foci (arrowheads), whereas RNG105-induced substructures were dissolved in the medium. Scale bar, 10 μm. C, area of binarized granules was measured as in Fig. 2B. The numbers of cells analyzed are indicated in the graphs. *, p < 0.05; ***, p < 0.005; ****, p < 0.001, main effect; ###, p < 0.005; ####, p < 0.001, interaction effect in two-way repeated measures ANOVA. * and # are colored corresponding to the co-expressed scaffolds. See also Movies S13 and S14.
S-scaffolds also retained their properties even in the presence of R-scaffolds: RNG105 and G3BP1 rapidly disappeared from the granules, whereas co-expressed R-scaffolds remained in the granules after cell permeabilization (Fig. 6, A and B, and Movies S13 and S14). One exception was that the S-substructures did not exhibit clear spherical shapes during shrinking, which may have been because a fraction of S-scaffolds was associated with R-substructures formed by R-scaffolds (Fig. 6A, insets). Thus, the properties of R- and S-granules were retained in R- and S-substructures, respectively, even when they were combined with the other granule types.
However, the dynamics of the scaffolds were not unaffected by the co-expression of the other types of scaffolds (Fig. 6C). In particular, the reduction in the binarized granule area of R-scaffolds was significantly accelerated by RNG105, suggesting that R-granules were converted to more dynamic structures via association with RNG105. To further investigate the effects of the co-expression on the scaffold dynamics in living cells, we next conducted FRAP analysis.
Immobility of R-granules is reduced by combination with S-granules
We examined the effects of co-expression of S- and R-scaffolds on their dynamics in granules in living cells using FRAP. For example, when RNG105 and FMR1 were co-expressed, neither the τ½ nor Fm of RNG105 was affected by FMR1 (Fig. 7, A–C). In contrast, although the τ½ of FMR1 was not affected, the Fm of FMR1 was significantly increased by RNG105 co-expression (Fig. 7C). Likewise, when other combinations were co-expressed (Fig. 7, D and E), neither the τ½ nor Fm of the S-scaffolds, i.e. RNG105 or G3BP1, was affected by R-scaffolds (Fig. 7E). In contrast, although the τ½ of R-scaffolds was not significantly changed, the Fm of the R-scaffolds was significantly increased by the co-expression of S-scaffolds in most of the combinations (Fig. 7D). RNG105 more effectively increased the Fm of R-scaffolds than G3BP1, which was consistent with the results of the cell permeabilization experiments (Fig. 6C). These results suggested that the dynamics of S-scaffolds were minimally influenced by R-scaffolds; however, the Fm values of R-scaffolds were significantly increased by S-scaffolds, suggesting that the immobile fraction of R-substructures was reduced by the association with S-substructures (Fig. 7F).
Figure 7.
Mobile fraction of R-granule scaffolds is increased by being combined with S-granule scaffolds. A, representative time-lapse FRAP images of R- and S-scaffolds (FMR1 and RNG105) co-expressed in the same cell. Orange circles indicate the region bleached just before 0 s. Dotted line in the left panel, nucleus. Scale bar, 10 μm. B, representative FRAP curves fitted with exponential equations. C, halftime of recovery (τ½) and the mobile fraction (Fm) extracted from the fitted equations. + and − indicate with and without co-expression, respectively. n = 8. **, p < 0.01, Student's t test. D, τ½ and Fm of R-scaffolds (FMR1, Pumilio1, FUS, and TIA-1) with and without the co-expression of S-scaffolds (RNG105 and G3BP1). E, τ½ and Fm of S-scaffolds (RNG105 and G3BP1) with and without the co-expression of R-scaffolds (FMR1, Pumilio1, FUS, and TIA-1). D and E, top tables, the mean ± S.D. Bottom tables, statistical analysis. ↑, p < 0.05; ↑↑, p < 0.01; ↑↑↑, p < 0.005; ↑↑↑↑, p < 0.001, significant increase; ↓, p < 0.01, significant decrease; −, no significance, Student's t test with control. n = 8. F, model of the decrease in the immobile fraction of R-scaffolds by being combined with S-scaffolds.
Conversion of R-scaffold dose-dependent decrease in the amount of nascent polypeptides in RNA granules to a dose-dependent increase by S-scaffolds
As changes in the dynamics of RNA granules may influence physiological roles of RNA granules, we next conducted ribopuromycilation analysis to examine the effects of the presence of S-scaffolds on protein synthesis, i.e. the amount of nascent polypeptides, in R-scaffold–expressing cells. Labeling of nascent polypeptides with puromycin is largely used for two purposes: one is to label translating polysomes in which nascent polypeptides maintain their association with ribosomes (28, 29), and the other is to produce and label defective ribosomal products (DRiPs) that are truncated and released from ribosomes (30, 31). The difference in the protocol between them is the existence of cycloheximide during the labeling only for the former purpose, which we applied to our experiments.
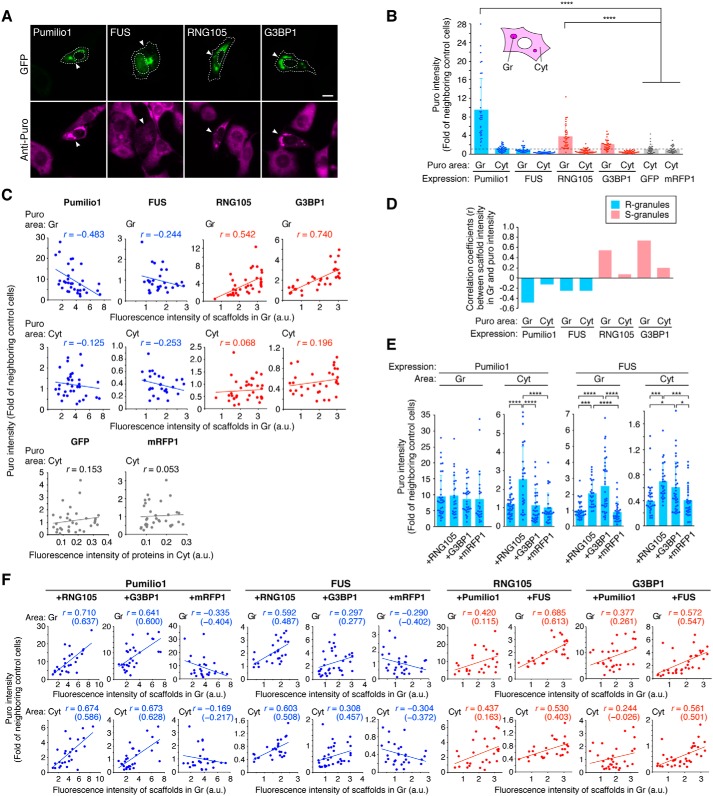
Before analyzing the effects of co-expression of the scaffolds, we first measured the amount of nascent polypeptides (puro intensity) in cells expressing Pumilio1, FUS, RNG105, or G3BP1 alone using the ribopuromycilation analysis (Fig. 8, A and B). Puro intensity was higher in and/or near the granules than in the cytoplasm in all cells expressing the scaffolds (Fig. 8B), as reported previously for G3BP1 (29), which may be due to the accumulation of mRNA in/near RNA granules. The puro intensity was not dependent on which type of scaffold, S or R, was expressed. For example, although both Pumilio1 and FUS formed R-granules, Pumilio1-expressing cells had higher puro intensity than control cells in/near the granules, whereas FUS-expressing cells had lower puro intensity than control cells in both the granules and cytoplasm, suggesting their intrinsically opposite effects on translation.
Figure 8.
Effects of R-scaffolds on nascent polypeptide levels are converted from a dose-dependent decrease to an increase following convergence with S-scaffolds. A, nascent polypeptides in cells were stained using the ribopuromycilation method. Arrowheads indicate cells expressing GFP-tagged scaffolds. The outlines of the cells and the nuclei are indicated by dotted lines. Scale bar, 10 μm. B, Puro intensity in the granules (Gr) and cytoplasm (Cyt) of cells expressing the scaffolds or control fluorescent proteins was measured and then normalized by that in the cytoplasm of neighboring untransfected cells. n = 34 (Pumilio1), 28 (FUS), 36 (RNG105), 32 (G3BP), 32 (GFP), and 33 (mRFP1). ****, p < 0.001, one-way ANOVA (F(9,315) = 44.7, p < 2.2e-16) followed by the Tukey-Kramer test between scaffold-expressing cells and control GFP- or mRFP1-expressing cells. C, relationship between the scaffold concentration in the granules and puro intensity in the granules or cytoplasm. Regression lines and Pearson's correlation coefficients are indicated. D, Pearson's correlation coefficients in C are represented as a bar graph. E, Puro intensity in cells expressing GFP-tagged Pumilio1 or FUS with or without co-expression of mRFP1-tagged RNG105, G3BP1, or mRFP1. n = 34 (Pumilio1), 26 (Pumilio1 + RNG105), 30 (Pumilio1 + G3BP1), 26 (Pumilio1 + mRFP1), 28 (FUS), 26 (FUS + RNG105), 35 (FUS + G3BP1), and 28 (FUS + mRFP1). p = 0.879 (F(3,112) = 0.225) for Pumilio1 (Gr), p = 6.21e-7 (F(3,112) = 13.7) for Pumilio1 (Cyt), p = 6.52e-9 (F(3,113) = 16.4) for FUS (Gr), and p = 0.000122 (F(3,113) = 7.54) for FUS (Cyt), one-way ANOVA. *, p < 0.05; ***, p < 0.005; ****, p < 0.001, Tukey-Kramer test. F, effects of the co-expression of the scaffolds on the relationship between the scaffold concentration and puro intensity. Regression lines and Pearson's correlation coefficients are indicated. Numbers in parentheses indicate partial correlation coefficients.
However, the dose-dependent effects of Pumilio1 and FUS on puro intensity were similar; both Pumilio1 and FUS caused dose-dependent decreases in puro intensity, particularly in the granules (Fig. 8, C and D). In contrast to the R-scaffolds, S-scaffolds, i.e. RNG105 and G3BP1, dose-dependently increased puro intensity in the granules, whereas the cytoplasmic puro intensity remained lower than that in control cells (Fig. 8, C and D). These results suggested that increasing concentrations of R-scaffolds in granules gradually decrease nascent polypeptides in the granules, but in contrast, increasing concentrations of S-scaffolds in granules gradually increase nascent polypeptides in the granules, regardless of their intrinsic activity on translation.
Large G3BP1-induced granules, but not small granules, were reported to induce phosphorylation of eIF2α and decrease puro intensity (29), which is inconsistent with our result that G3BP1 increased puro intensity in granules in a dose-dependent manner (Fig. 8C). Thus, we examined the eIF2α phosphorylation level in G3BP1-expressing cells. eIF2α phosphorylation was not significantly increased in the granules or in the cytoplasm of G3BP1-expressing cells (Fig. S2, A and B), and eIF2α phosphorylation was not increased in a G3BP1 dose-dependent manner (Fig. S2C). These results did not contradict our ribopuromycilation results but were different from the previous report (29). This difference may be explained by the puro intensity in G3BP1-expressing cells being biphasic, ending with a decrease if G3BP1 expression increases more than in our experiments. However, cells with very high expression of G3BP1 underwent cell death until the 3rd day of transfection of the cells with G3BP1, at which time the cells were subjected to the experiments in our study.
eIF2α phosphorylation levels in cells expressing the other scaffolds were also evaluated. eIF2α phosphorylation levels were higher in and/or near the granules than in the cytoplasm in all cells expressing the scaffolds (Fig. S2B), but they were not likely to be sufficient to shut off all translation, as the phosphorylation levels were significantly less than in arsenite-stressed cells (Fig. S2B). In stress granules induced transiently by stress, such as arsenite, eIF2α is phosphorylated, and translation preinitiation complexes are stalled. However, granules in this study were not induced by stress but by expression of scaffolds, which may be the reason for the low phosphorylation levels of eIF2α. Furthermore, eIF2α phosphorylation levels did not depend on scaffold dose, except for Pumilio1 (Fig. S2C), suggesting that eIF2α phosphorylation is not a major cause of the scaffold dose-dependent decrease or increase in the puro intensity in granules (Fig. 8C).
Next, we analyzed the effects of the presence of S-scaffolds on the puro intensity in R-scaffold–expressing cells (Fig. 8E). Puro intensity in Pumilio1-expressing cells were increased by RNG105, but not by G3BP1, in the cytoplasm (Fig. 8E), which coincided with the increase in the Fm values of Pumilio1 by RNG105 but not by G3BP1 (Fig. 7D). In contrast, puro intensity in FUS-expressing cells was increased by both RNG105 and G3BP1 in both the granules and the cytoplasm (Fig. 8E), which coincided with the significant increase in the Fm value of FUS by both RNG105 and G3BP1 (Fig. 7D).
We further analyzed the influence of the S-scaffolds on the R-scaffold dose-dependent effects on puro intensity. In contrast to the Pumilio1 and FUS dose-dependent decreases in puro intensity in cells expressing them alone (Fig. 8C), co-expression of RNG105 and G3BP1 in cells reversed the dose dependence for Pumilio1 and FUS (Fig. 8F). As the expression levels of the R-scaffolds and the S-scaffolds in the co-expressed cells were slightly positively correlated, the R-scaffold dose-dependent increases in puro intensity may reflect the S-scaffold dose-dependent increases in puro intensity. However, partial correlation coefficients between the R-scaffolds and the puro intensity were positive, indicating R-scaffold dose-dependent increases in puro intensity in the S-scaffold co-expressing cells (Fig. 8F). This change in the R-scaffold dose dependence by S-scaffolds may underlie the increase in the puro intensity in R-scaffold–expressing cells by co-expression of S-scaffolds. Although G3BP1 co-expression resulted in a Pumilio1 dose-dependent increase in puro intensity, the overall puro intensity was not altered (Fig. 8E), which may have been because the degree of reversal was not as large as with RNG105 co-expression (Fig. 8F). In contrast to the change in the R-scaffold dose dependence, the dose-dependent increase in puro intensity in granules by S-scaffolds was not reversed by the co-expression of R-scaffolds (Fig. 8F). These results were reminiscent of the limited effects of R-scaffold co-expression on the dynamics of S-scaffolds (Fig. 7E). Taken together, the effects of R-scaffolds on puro intensity were altered by S-scaffolds, suggesting that the effects of R-scaffolds on translation were altered by being combined with S-scaffolds in RNA granules, which may not exclude other possibilities such as alteration in the effects of R-scaffolds on the accumulation of DRiPs by combination with S-scaffolds (30, 31). Thus, these results suggested that the combination with S-granules influenced the physiological properties as well as dynamics of R-granules.
Discussion
RNA granules were recently revealed not to be simply uniform structures but instead to consist of stable solid-like cores and less concentrated liquid-like shells (14, 16). However, it was unknown whether these two substructures differed solely in concentration or whether they had different constituents and properties. In this study, by expressing RNA granule scaffolds in cells, we demonstrated that solid core-like R-structures and liquid shell-like S-structures were formed by distinct scaffolds, as judged by morphological analysis and dynamics analysis. R-granules were induced by TIA-1, TIAR, FMR1, FUS, and Pumilio1, whereas S-granules were induced by G3BP1, RNG105, and TDP-43. The R- and S-scaffolds were combined into the same granules and formed distinct subgranular structures therein. The R- and S-substructures retained their solid-like and liquid-like properties, respectively, after the convergence but were also influenced by the other substructure. In particular, large immobile fractions of R-scaffolds were reduced by combination with S-scaffolds. The decrease in the immobile fractions coincided with changes in physiological properties of the scaffolds, e.g. the R-scaffold dose-dependent decrease in the amount of nascent polypeptides was reversed by the combination with S-scaffolds. These results suggested that R- and S-substructures are formed by distinct scaffolds and have different dynamics, different influences on each other, and different effects on translation, thereby providing novel insight into the assembly, dynamics, and physiological roles of core-shell substructures in RNA granules.
In this study, four experiments were conducted to evaluate the liquid-like and solid-like properties of granules. First, the surface texture of granules was analyzed. S-granules had smooth surfaces, whereas R-granules had rough surfaces. If granules are liquid droplets and have surface tension, their surfaces should be smooth due to surface area minimization. In contrast, solid granules do not need to be smooth to maintain their assembled state. Therefore, the surface textures of S- and R-granules supported them being liquid-like and solid-like granules, respectively.
Second, the cell permeabilization experiment was conducted. This experiment evaluated the characteristic behavior of liquid droplets and the purifiability of solid structures. S-granules underwent shrinkage and/or dissolution after cell permeabilization, which may have been triggered by the decrease in the scaffold concentration in the cytoplasm, i.e. perturbed equilibrium between the granules and the cytoplasm. These changes in the granule shape were similar to those observed for liquid–liquid phase-separated droplets in vitro (26). In contrast, R-granules exhibited little shape change after cell permeabilization. In addition, they were purifiable as foci outside the permeabilized cells when they had slow exchange rates (τ½). Although TIAR and TIA-1 foci were not purifiable, this was not due to the shrinkage or dissolution of the R-granules that they formed. As TIAR was localized to the same foci with the other R-scaffolds that formed solid-like structures, the rapid disappearance of TIAR and TIA-1 from the foci was likely due to their rapid dissociation from the foci embedded in the solid-like R-granules. These results indicated that S- and R-granules have properties of liquid droplets and solid-like cores, respectively. As for FUS, the granules were converted from R-type to S-type in this experiment. FUS was able to undergo LLPS in the equilibrium where the FUS concentration was high in granules and very low outside the granules, although it is not known whether such conditions can be established in living cells where abrupt concentration changes do not occur.
Third, granule dynamics were measured by FRAP. The most notable difference between S- and R-granules was the mobile fraction of scaffolds that had a slow τ½. If solid-type scaffolds have a slow τ½, they will not change position in the granules over time, thereby increasing the probability that the scaffolds at the center of the granules are not exchanged with the cytoplasmic pool. However, τ½ may have little influence on liquid-type scaffolds because they can be mixed within the granules and come to the surface of the granules to be exchanged with the cytoplasmic pool even if their τ½ is slow. Thus, the difference in the mobile fractions supported the notion that S- and R-granules had properties of liquid and solid phases, respectively.
Fourth, granule shape change was measured. The deformation rate was different between S- and R-scaffolds with a fast τ½. In liquid droplets, if scaffolds have a fast τ½, the attraction of the scaffolds to each other within the droplets will be weak, resulting in a low surface tension in the droplets. The lower surface tension will make the shape of the droplets easier to deform because spheronization of droplets is driven by higher surface tension. In contrast to liquid droplets, the shape of solid granules is considered to be stable regardless of the τ½ of the scaffolds because of the high resistance of solids to deformation forces. Taken together, the four experiments demonstrated that the S- and R-granules have properties of liquid-like shells and solid-like cores, respectively.
The components in the shell were hypothesized to exchange rapidly with the cytoplasmic pool, whereas those in the cores are likely less dynamic. However, our results indicated that the exchange rate (τ½) of the scaffolds is not related to which type of granule the scaffolds formed. These results suggested that cores and shells are not be characterized by the exchange rate of the scaffolds in the structures. Nevertheless, the τ½ of the scaffolds likely influenced the solid-like and liquid-like properties of the scaffolds. For example, R-scaffolds with a slower τ½ (FMR1 and Pumilio1) more clearly exhibited solid-like properties, such as large immobile fractions and resistance to cell permeabilization, than R-scaffolds with a faster τ½ (TIAR and TIA-1). In contrast, S-scaffolds with a faster τ½ (RNG105 and TDP-43) more clearly exhibited liquid-like properties, such as a high shape change rate, than S-scaffolds with a slower τ½ (G3BP1) (Fig. S3).
Contrary to the τ½, the Fm of the scaffolds was related to which type of granule the scaffolds formed. Large immobile fractions were formed only in the R-granules depending on the slow exchange rate of the R-scaffolds. Furthermore, the immobile fraction of the R-scaffolds was reduced by combination with S-scaffolds, although their exchange rates were minimally affected. Taken together, the difference between R- and S-substructures may be characterized not by the exchange rate of the components, but by the ability to make a large immobile fraction in the R-substructures compared with the S-substructures. This difference in dynamics may be related to the dose-dependent decrease and increase in the amount of nascent polypeptides in RNA granules by R- and S-scaffolds, respectively.
The core-shell structure is not limited to RNA granules and is also observed in other RNA–protein (RNP) complexes such as nucleoli (32, 33). The core-shell structure in the nucleolus was reconstructed by phase-separated liquid droplets of distinct components of the nucleolus, which were immiscible with each other because of differences in the surface tension of the different kinds of liquid droplets (33). The core-shell structures of RNA granules and the nucleoli are similar in that the core and shell (R and S) substructures were formed by specific scaffolds. However, they differ in that the core (R-substructure) of RNA granules exhibited solid-like properties, whereas that of the nucleoli had liquid-like properties. Although the cores (R-substructures) of RNA granules changed their dynamics toward a fluid state by combination with the shells (S-substructures), they were not likely liquid droplets because their shapes were rough with small foci and resistant to cell permeabilization. As the core (R) scaffolds of the RNA granules and the nucleoli formed solid-like granules and liquid droplets by themselves, respectively, whether the core is solid-like or liquid-like may depend on which type of granule the scaffolds form by themselves.
R- and S-scaffolds facilitated convergence of R- and S-granules instead of forming R- and S-granules separately. This convergence suggested that there was interaction between R- and S-granules. This interaction may be mediated by direct binding between specific R- and S-scaffolds, e.g. FMR1 and RNG105 (27). However, such relatively strong binding will inhibit separation of R- and S-substructures if the scaffolds were abundantly expressed, as observed in the cells expressing FMR1 and RNG105 (Fig. 5C). Another possible interaction mechanism is weak interactions between IDRs of R-scaffolds and IDRs of S-scaffolds (16). As R-granules contain scaffolds that have IDRs, R-granules may provide a high concentration of IDRs. These IDRs may also weakly interact with S-scaffolds that also have IDRs, allowing R-granules to be surrounded by S-granules.
RNA granules have functions to sequester untranslated mRNAs while selectively translating specific mRNAs. RNA granules also function to sort other specific mRNAs for decay in processing bodies (5, 6). Our results suggested that R- and S-substructures enable this multifunctionality of RNA granules. R-substructures can sequester mRNAs and decrease their translation in large immobile fractions, whereas S-substructures can increase the mobility of granules and facilitate the translation of mRNAs. The difference in the molecular constituents between R- and S-substructures may confer a different preference of the substructures for specific mRNAs. Some mRNAs may be preferentially stored stably in R-substructures, whereas others may be preferentially associated with S-substructures and translated effectively in the liquid phase and/or dynamically sent back to the cytoplasm for translation. Although sorting of mRNAs for decay was not examined here, our assay system will be useful to assess which of the R- and S-substructures function in mRNA decay and stability. Taken together, the differences in the subgranular structures and their molecular constituents impact the coordination of mRNA sorting and translational regulation by RNA granules.
RNA granule scaffolds play key roles, especially in the brain. Their functions are involved in learning and memory (34), and abnormalities in their dynamics are associated with neurodegenerative diseases (8, 9). Therefore, it is important to understand the relationship between the dynamics of RNA granules and neural functions. Our study provided insight into the assembly, dynamics, and effects on translation of the core- and shell-like substructures in RNA granules, which will be useful for studies on the brain regarding health and disease.
Experimental procedures
Cell culture and transfection
A6 cells were cultured as described previously (35). Briefly, the cells were cultured on glass-bottomed dishes in 10% fetal bovine serum (FBS) and 50% Leibovitz's L-15 medium (ThermoFisher Scientific, Waltham, MA) at 23 °C without a CO2 atmosphere. Transfection was performed using Lipofectin (ThermoFisher Scientific) in accordance with the manufacturer's protocol, and the transfected cells were observed 3 days after transfection.
Plasmid construction
The pEGFP-N1 vector (Clontech) was used to construct plasmids for GFP-tagged proteins. To construct plasmids for mRFP1-tagged proteins, the GFP-coding sequence in the pEGFP-N1 vector was replaced with the mRFP1-coding sequence (19). Plasmids for RNG105, G3BP1, and FUS were constructed previously (19, 35, 36). cDNAs for TDP-43, Pumilio1, FMR1, TIAR, and TIA-1 were obtained by RT-PCR from mouse brain RNA with the following primers: 5′-caagcttatgtctgaatatattcgggtaacaga-3′ and 5′-gcgtcgacccaccccctccccccattccccagccagaagact-3′ for TDP-43; 5′-gcgtcgacatgagcgttgcatgtgtcttg-3′ and 5′-tccccgcggtccgccccctcctccgatgataccattagggggaccac-3′ for Pumilio1; 5′-ggaattcatggaggagctggtggtgg-3′ and 5′-gcgtcgacccgccgcctcctccgggtactccattcaccagcg-3′ for FMR1; 5′-ccctcgagatgatggaagacgacggaca-3′ and 5′-cgggatccccacccccacctccctgtgtcgggaagcttgc-3′ for TIAR; and 5′-ggaattcatggaggacgagatgcccaa-3′ and 5′-gcgtcgacccaccccctcctccctgggtttcatacccggcc-3′ for TIA-1. The cDNAs were cloned into the HindIII/SalI sites (TDP-43), SalI/SacII sites (Pumilio1), EcoRI/SalI sites (FMR1 and TIA-1), and XhoI/BamHI sites (TIAR) of the vectors. These plasmids encoded proteins tagged with GFP or mRFP1 at their C termini through five-glycine linkers.
Fluorescence imaging of fixed cells
Cells expressing GFP- and mRFP1-tagged proteins were fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) for 10 min. After washing with PBS, the cells were treated with 0.5% Triton X-100 in PBS for 10 min to allow a mounting agent to permeate into the cells in the later procedure. After washing with PBS, the specimens were mounted in Mowiol (Merck Millipore, Billerica, MA). Fluorescence images were acquired using a DeltaVision optical sectioning microscope (GE Healthcare) equipped with an IX70 inverted microscope (Olympus, Tokyo, Japan) with a PlanApo ×60 oil objective lens. The diameter of the foci in R-granules was calculated by measuring the full width at half-maximum of the fluorescence profiles of the foci.
Analysis of the texture of S- and R-granules
Fluorescence images of GFP-tagged proteins were acquired as described above. Each RNA granule with surrounding cytoplasm was selected as a region of interest (ROI). The fluorescence intensity of each pixel in the ROI was measured and divided into 256 gradations, with the brightest signal being 256. Gradations 1–47 were eliminated from the data as background (cytoplasmic) levels. In the case of GFP-only expressing cells, the cytoplasm and an adjacent extracellular region (background) were selected as a ROI and measured in the same way. The normalized pixel intensity was plotted on a histogram. Skewness of the histogram was calculated as shown in Equations 1 and 2,
| (Eq. 1) |
where
| (Eq. 2) |
and x̄ is the mean intensity of the pixels.
In situ hybridization
Cells expressing the GFP-tagged scaffolds were fixed with 3.7% formaldehyde in PBS containing 5 mm MgCl2 (PBSM) for 10 min. After washing with PBSM, the cells were treated with 0.5% Triton X-100 in PBSM for 10 min. The cells were washed with PBSM, equilibrated with 15% formamide, 2× saline-sodium citrate (SSC), and 10 mm sodium phosphate (pH 7.0) for 10 min and then probed with 0.25 μg/ml 3′-digoxigenin–labeled poly(dT) probe (55-mer) in 0.5 mg/ml yeast t-RNA (Roche Diagnostics, Basel, Switzerland), 0.5 mg/ml salmon sperm DNA (FUJIFILM Wako Pure Chemical, Osaka, Japan), 10% dextran sulfate, 0.1% BSA, 15% formamide, 2× SSC, and 10 mm sodium phosphate (pH 7.0) at 37 °C for 1.5 h. The cells were washed in 15% formamide and 2× SSC at 37 °C for 20 min, and then in 1× SSC for 30 min. After blocking with 10% FBS in Dulbecco's modified Eagle's medium, the probe was labeled with an anti-digoxigenin antibody (Roche Diagnostics) and a Cy3-conjugated anti-goat IgG antibody (1:400, 705-165-147, Jackson ImmunoResearch, West Grove, PA). Fluorescence images were acquired as described under “Fluorescence imaging of fixed cells.” The fluorescence intensity of the immunolabeled mRNA was measured in the granule areas and the other cytoplasmic areas of the scaffold-expressing cells and was normalized by the fluorescence intensity in the cytoplasm of neighboring untransfected cells.
Cell permeabilization analysis
Time-lapse fluorescence images of cells were acquired at 5-s intervals for cells expressing only a GFP-tagged protein or at 7-s intervals for cells expressing both GFP- and mRFP1-tagged proteins using an IX83 inverted microscope (Olympus) with a UPlanFL N ×40 objective lens and an ORCA-R2 digital CCD camera (Hamamatsu Photonics, Hamamatsu, Japan). After 10 images (for GFP-expressing cells) or 7 images (for both GFP- and mRFP1-expressing cells) were taken, 0.5 ml of 0.06% digitonin dissolved in medium was added to the cells cultured in 2 ml of the same medium. Fluorescence images were taken for ∼300 s after the addition of digitonin.
The first image of the time-lapse data was binarized using the MaxEntropy threshold algorithm in ImageJ software, and the other images of the same series were also binarized using the same threshold value. The binarized granule areas were measured using ImageJ software. As the duration between the digitonin addition and cell permeabilization varied from cell to cell, the time just before the cells became permeable was adjusted to time 0 to calculate the average of the binarized granule areas (Figs. 2B and 6C). Note that the time 0 of the fluorescence images (Figs. 2, A, C, and D, and 6, A and B) was not adjusted and indicates the time at which digitonin was added.
FRAP analysis
Time-lapse images of live cells expressing GFP- and mRFP1-tagged proteins were acquired at 5-s intervals using an A1 confocal laser microscope equipped with a Ti-E inverted microscope (Nikon, Tokyo, Japan) with a PlanApo VC ×60 oil objective lens. Four control images were taken before bleaching, and then a ROI, an ellipse covering the whole area of an RNA granule, was bleached using the 488-nm laser (40 milliwatts) and 561-nm laser (10 milliwatts) at 60% power and 8 s/frame (Figs. 3A and 7A). The fluorescence intensity at each time point was measured for the RNA granule in the ROI (F), the other granules in the same cell (Fg), and the cytoplasm of the cell (Fc) using ImageJ. The fluorescence intensity of the target RNA granule at each time point (F(t)) was normalized as shown in Equation 3,
| (Eq. 3) |
where t = 0 was just after photobleaching. Then fluorescence intensity (% of before bleaching) was calculated as F(t)/Fcont, where Fcont is the average of the F(t) of the four control images. A FRAP curve of the measured value (F(t)/Fcont, t ≥ 0) was fitted with Equation 4,
| (Eq. 4) |
where Fm is the mobile fraction (maximum recovery), and τ½ is the halftime of recovery. The least-squares method with Excel Solver was used for fitting. A surface plot of RNA granules (Fig. 3B) was created using the interactive 3D surface plot plugin in ImageJ.
Granule shape change
Time-lapse images taken at 5-s intervals were acquired as in the FRAP analysis. Deformation of each granule over 100 s (21 images) was analyzed. A granule in the 21 time-lapse images was selected and converted into binary images using the default mode in ImageJ. If the granule was displaced by intracellular transport or cell migration, the place was corrected such that the differential area of the granule throughout the time course reached a minimum. The 21 binary images were merged, and the merged area was compared with the average area of the granule using Equation 5,
| (Eq. 5) |
where S is the area of the granule.
Co-localization analysis
Fluorescence images of GFP- and mRFP1-tagged scaffolds in the same cells were acquired as described under “Fluorescence imaging of fixed cells.” GFP and mRFP1 images were merged, and then the granule area was selected and converted into binary images using the default mode in ImageJ. The binary image was used to select a ROI, and an RNA granule in the ROI in the original images was analyzed for co-localization of the scaffolds. Pearson's r value for the two scaffolds in the ROI was calculated using the Coloc 2 plugin in Fiji (37).
Ribopuromycilation analysis
Ribopuromycilation analysis was conducted as described previously with a modification (35). Cells were pulse-labeled with 50 μg/ml puromycin and 100 μg/ml cycloheximide at 23 °C for 10 min to puromycilate nascent polypeptide chains in the cells. After washing with 100 μg/ml cycloheximide in PBS on ice for 3 min, the cells were permeabilized and fixed in 0.015% digitonin, 3.7% formaldehyde, 100 μg/ml cycloheximide, 5 mm MgCl2, 25 mm KCl, and 50 mm Tris-HCl (pH 7.5) on ice for 5 min. After post-fixation with 3.7% formaldehyde in PBS for 10 min, the puromycilated nascent polypeptide chains were labeled with an anti-puromycin antibody (1:125, 3RH11, Kerafast, Boston, MA) and a DyLight 649-conjugated anti-mouse IgG antibody (1:400, 715-495-151, Jackson ImmunoResearch). After washing with PBS, the specimens were mounted in Mowiol. Fluorescence images were acquired using an IX83 inverted microscope (Olympus) with an UPlanFL N ×40 objective lens and a 4.2 Mpixel sCMOS camera (Photometrics, Tucson, AZ). The fluorescence intensity of the immunolabeled polypeptides was measured in the granule areas and the other cytoplasmic areas of the cells and was normalized by the fluorescence intensity in the cytoplasm of neighboring untransfected cells.
Immunostaining
Cells were stressed with or without 0.5 mm arsenite for 30 min. The cells were fixed and permeabilized as described under “Fluorescence imaging of fixed cells.” After blocking with 10% FBS in PBS, the specimens were labeled with an anti-phospho-eIF2α (Ser-51) antibody (1:100, 119A11, Cell Signaling Technology, Danvers, MA) and a cyanin 3–conjugated anti-rabbit IgG (1:400, Jackson ImmunoResearch). The specimens were mounted and visualized as described under “Ribopuromycilation analysis.”
Statistical analysis
Data are represented as scatter plots that show all the individual data points and the mean ± S.D. Sample numbers are indicated in the figure legends. Statistical analysis was performed using R. Significance was determined using the Student's t test, one-way analysis of variance (ANOVA), one-way repeated measures ANOVA, and post hoc Tukey-Kramer test as indicated in the figure legends.
Author contributions
N. S. conceptualization; N. S. data curation; N. S. formal analysis; N. S. funding acquisition; N. S. investigation; N. S. visualization; N. S. methodology; N. S. writing-original draft; N. S. project administration; N. S. writing-review and editing.
Supplementary Material
Acknowledgments
I thank C. Matsuda for technical assistance and the Spectrography and Bioimaging Facility and Functional Genomic Facility, NIBB Core Research Facilities, for technical support.
This work was supported by the Takeda Science Foundation, the Astellas Foundation for Research on Metabolic Disorders, and the Orion Project of Okazaki Institute for Integrative Bioscience to N. S. The author declares that he has no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3 and Movies S1–S14.
- IDR
- intrinsically disordered region
- LLPS
- liquid–liquid phase separation
- S
- smooth
- R
- rough
- TIAR
- TIA-1–related protein
- FUS
- fused in sarcoma
- FMR1
- fragile X mental retardation 1
- FRAP
- fluorescence recovery after photobleaching
- ANOVA
- analysis of variance
- ROI
- region of interest
- DRiP
- defective ribosomal product
- FBS
- fetal bovine serum.
References
- 1. Anderson P., and Kedersha N. (2006) RNA granules. J. Cell Biol. 172, 803–808 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kedersha N., Ivanov P., and Anderson P. (2013) Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38, 494–506 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weber S. C., and Brangwynne C. P. (2012) Getting RNA and protein in phase. Cell 149, 1188–1191 10.1016/j.cell.2012.05.022 [DOI] [PubMed] [Google Scholar]
- 4. Bramham C. R., and Wells D. G. (2007) Dendritic mRNA: transport, translation and function. Nat. Rev. Neurosci. 8, 776–789 10.1038/nrn2150 [DOI] [PubMed] [Google Scholar]
- 5. Buchan J. R., and Parker R. (2009) Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson P., and Kedersha N. (2008) Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141–150 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- 7. Krichevsky A. M., and Kosik K. S. (2001) Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32, 683–696 10.1016/S0896-6273(01)00508-6 [DOI] [PubMed] [Google Scholar]
- 8. Ling S. C., Polymenidou M., and Cleveland D. W. (2013) Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438 10.1016/j.neuron.2013.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramaswami M., Taylor J. P., and Parker R. (2013) Altered ribostasis: RNA–protein granules in degenerative disorders. Cell 154, 727–736 10.1016/j.cell.2013.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee K. H., Zhang P., Kim H. J., Mitrea D. M., Sarkar M., Freibaum B. D., Cika J., Coughlin M., Messing J., Molliex A., Maxwell B. A., Kim N. C., Temirov J., Moore J., Kolaitis R. M., et al. (2016) C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788.e17 10.1016/j.cell.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shin Y., Berry J., Pannucci N., Haataja M. P., Toettcher J. E., Clifford P., and Brangwynne C. P. (2017) Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168, 159–171.e14 10.1016/j.cell.2016.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hyman A. A., Weber C. A., and Jülicher F. (2014) Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- 13. Uversky V. N. (2017) Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 44, 18–30 [DOI] [PubMed] [Google Scholar]
- 14. Jain S., Wheeler J. R., Walters R. W., Agrawal A., Barsic A., and Parker R. (2016) ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Souquere S., Mollet S., Kress M., Dautry F., Pierron G., and Weil D. (2009) Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci. 122, 3619–3626 10.1242/jcs.054437 [DOI] [PubMed] [Google Scholar]
- 16. Wheeler J. R., Matheny T., Jain S., Abrisch R., and Parker R. (2016) Distinct stages in stress granule assembly and disassembly. Elife 5, e18413 10.7554/eLife.18413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Banani S. F., Rice A. M., Peeples W. B., Lin Y., Jain S., Parker R., and Rosen M. K. (2016) Compositional control of phase-separated cellular bodies. Cell 166, 651–663 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kedersha N. L., Gupta M., Li W., Miller I., and Anderson P. (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431–1442 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shiina N., Shinkura K., and Tokunaga M. (2005) A novel RNA-binding protein in neuronal RNA granules: regulatory machinery for local translation. J. Neurosci. 25, 4420–4434 10.1523/JNEUROSCI.0382-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J. M., Bertrand E., and Tazi J. (2003) The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Liu-Yesucevitz L., Bilgutay A., Zhang Y. J., Vanderweyde T., Vanderwyde T, Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M., Petrucelli L., and Wolozin B. (2010) Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS ONE 5, e13250 10.1371/journal.pone.0013250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujii R., Okabe S., Urushido T., Inoue K., Yoshimura A., Tachibana T., Nishikawa T., Hicks G. G., and Takumi T. (2005) The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr. Biol. 15, 587–593 10.1016/j.cub.2005.01.058 [DOI] [PubMed] [Google Scholar]
- 23. Mazroui R., Huot M. E., Tremblay S., Filion C., Labelle Y., and Khandjian E. W. (2002) Trapping of messenger RNA by fragile X mental retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 11, 3007–3017 10.1093/hmg/11.24.3007 [DOI] [PubMed] [Google Scholar]
- 24. Vessey J. P., Vaccani A., Xie Y., Dahm R., Karra D., Kiebler M. A., and Macchi P. (2006) Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J. Neurosci. 26, 6496–6508 10.1523/JNEUROSCI.0649-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gadelmawla E. S., Koura M. M., Maksoud T. M., Elewa I. M., and Soliman H. H. (2002) Roughness parameters. J. Mater. Process Technol. 123, 133–145 10.1016/S0924-0136(02)00060-2 [DOI] [Google Scholar]
- 26. Moon B. U., Hwang D. K., and Tsai S. S. (2016) Shrinking, growing, and bursting: microfluidic equilibrium control of water-in-water droplets. Lab. Chip. 16, 2601–2608 10.1039/C6LC00576D [DOI] [PubMed] [Google Scholar]
- 27. Wu Y., Zhu J., Huang X., and Du Z. (2016) Crystal structure of a dimerization domain of human Caprin-1: insights into the assembly of an evolutionarily conserved ribonucleoprotein complex consisting of Caprin-1, FMRP and G3BP1. Acta Crystallogr. D Struct. Biol. 72, 718–727 10.1107/S2059798316004903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. David A., Netzer N., Strader M. B., Das S. R., Chen C. Y., Gibbs J., Pierre P., Bennink J. R., and Yewdell J. W. (2011) RNA binding targets aminoacyl-tRNA synthetases to translating ribosomes. J. Biol. Chem. 286, 20688–20700 10.1074/jbc.M110.209452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reineke L. C., Dougherty J. D., Pierre P., and Lloyd R. E. (2012) Large G3BP-induced granules trigger eIF2α phosphorylation. Mol. Biol. Cell 23, 3499–3510 10.1091/mbc.e12-05-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ganassi M., Mateju D., Bigi I., Mediani L., Poser I., Lee H. O., Seguin S. J., Morelli F. F., Vinet J., Leo G., Pansarasa O., Cereda C., Poletti A., Alberti S., and Carra S. (2016) A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol. Cell 63, 796–810 10.1016/j.molcel.2016.07.021 [DOI] [PubMed] [Google Scholar]
- 31. Mateju D., Franzmann T. M., Patel A., Kopach A., Boczek E. E., Maharana S., Lee H. O., Carra S., Hyman A. A., and Alberti S. (2017) An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 36, 1669–1687 10.15252/embj.201695957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brangwynne C. P., Mitchison T. J., and Hyman A. A. (2011) Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U.S.A. 108, 4334–4339 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feric M., Vaidya N., Harmon T. S., Mitrea D. M., Zhu L., Richardson T. M., Kriwacki R. W., Pappu R. V., and Brangwynne C. P. (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakayama K., Ohashi R., Shinoda Y., Yamazaki M., Abe M., Fujikawa A., Shigenobu S., Futatsugi A., Noda M., Mikoshiba K., Furuichi T., Sakimura K., and Shiina N. (2017) RNG105/caprin1, an RNA granule protein for dendritic mRNA localization, is essential for long-term memory formation. Elife 6, e29677 10.7554/eLife.29677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shiina N., and Nakayama K. (2014) RNA granule assembly and disassembly modulated by nuclear factor associated with double-stranded RNA 2 and nuclear factor 45. J. Biol. Chem. 289, 21163–21180 10.1074/jbc.M114.556365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shiina N., Yamaguchi K., and Tokunaga M. (2010) RNG105 deficiency impairs the dendritic localization of mRNAs for Na+/K+ ATPase subunit isoforms and leads to the degeneration of neuronal networks. J. Neurosci. 30, 12816–12830 10.1523/JNEUROSCI.6386-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.