Abstract
Several protein kinases, including protein kinase C, Ca2+/calmodulin-dependent protein kinase II, and extracellular signal–regulated kinase, play key roles in the regulation of dopamine transporter (DAT) functions. These functions include surface expression, internalization, and forward and reverse transport, with phosphorylation sites for these kinases being linked to distinct regions of the DAT N terminus. Protein phosphatases (PPs) also regulate DAT activity, but the specific residues associated with their activities have not yet been elucidated. In this study, using co-immunoprecipitation followed by MS and immunoblotting analyses, we demonstrate the association of DAT with PP1 and PP2A in the mouse brain and heterologous cell systems. By applying MS in conjunction with a metabolic labeling method, we defined a PP1/2A-sensitive phosphorylation site at Thr-48 in human DAT, a residue that has not been previously reported to be involved in DAT phosphorylation. Site-directed mutagenesis of Thr-48 to Ala (T48A) to prevent phosphorylation enhanced dopamine transport kinetics, supporting a role for this residue in regulating DAT activity. Moreover, T48A-DAT displayed increased palmitoylation, suggesting that phosphorylation/dephosphorylation at this site has an additional regulatory role and reinforcing a previously reported reciprocal relationship between C-terminal palmitoylation and N-terminal phosphorylation.
Keywords: dopamine transporter, protein phosphatase, phosphorylation, mass spectrometry (MS), protein-protein interaction, post-translational modification (PTM), dephosphorylation, neurotransmission, okadaic acid, palmitoylation, synaptic transmission
Introduction
The plasma membrane dopamine transporter (DAT)3 belongs to the Na+/Cl−-dependent neurotransmitter transporter (neurotransmitter:sodium symporter; SLC6) family and contributes to dopamine (DA) homeostasis by reuptake of DA from the synaptic cleft into presynaptic neurons (1). DAT also mediates the reverse transport of DA from the presynaptic terminals to the synaptic cleft induced by amphetamine (2). DAT consists of 12 transmembrane domains and cytosolic N and C termini harboring functional domains for protein–protein and protein–lipid interactions as well as for post-translational modifications, such as palmitoylation and phosphorylation (3, 4). Multiple kinase pathways regulate DAT activity and surface expression (reviewed in Ref. 5). Ca2+/calmodulin-dependent protein kinase IIα (CaMKIIα) interacts with the C terminus of DAT and modulates amphetamine-induced DA efflux via N-terminal phosphorylation sites (6, 7). Proline-directed kinases, including extracellular signal–regulated kinase 1, phosphorylate the DAT N terminus (8). Activation of cAMP-dependent protein kinase (PKA) increases the efficacy of DA uptake in rat striatal synaptosomes (9, 10) but not in Sf9 cells (11). However, neither the PKA activator forskolin nor 8-bromo-cAMP increased the metabolic incorporation of [32P]orthophosphate in DAT, indicating an indirect effect of PKA on DAT (12). Activation of protein kinase C (PKC) enhances DAT internalization and results in a decrease of DAT-mediated DA uptake (13). In addition, PKC is involved in amphetamine-mediated DA efflux (14). PKC stimulates phosphorylation of the distal N terminus, which includes a cluster of serines at positions 2, 4, 7, 12, and 13, but the deletion of this region does not eliminate PKC-mediated internalization (15). Hence, this is suggestive of transporter modulation by other interacting proteins or other PKC-responsive residues in DAT.
There are many other serine and threonine residues in DAT intracellular domains that represent potential phosphorylation sites that may contribute to kinase-mediated functions. To search for additional phosphorylation sites, we have utilized MS previously to identify phosphorylation of rat DAT (rDAT) at PKC site Ser-7 and extracellular signal–regulated kinase site Thr-53 (16, 17). In a series of studies, we demonstrated that these residues mediate complex kinase- and phosphorylation-dependent functions, including regulation of cocaine analog affinity and amphetamine-induced DA efflux (16, 17). A further picture emerging from these studies is that regulation of DAT function occurs through integration of multiple post-translational inputs. For example, transport kinetic capacity is established by reciprocal phosphorylation of Ser-7 and palmitoylation of Cys-580 (18). Interaction of these effects could occur at the level of the enzyme pathways for phosphorylation (kinases and phosphatases) and palmitoylation (protein acyltransferases and acyl protein thioesterases) (19), which could be regulated by distinct signaling cascades. Alternatively, modification status could be regulated through accessibility of the sites, which may be affected by transporter activity, interactome alterations, or N-terminal conformation. Identification of enzymes that catalyze modification of these sites and determining how they are regulated or have access to DAT domains is thus crucial for elucidating DAT regulatory mechanisms.
Reversible protein phosphorylation is mediated by coordinated control of protein kinases and protein phosphatases (PPs). Most studies on DAT regulation by phosphorylation have focused on kinase inputs, with less known regarding dephosphorylation mechanisms. It has previously been shown that okadaic acid (OA), an inhibitor of PP1/2A, down-regulates DAT activity and increases DAT phosphorylation level by reducing dephosphorylation (8, 10, 12, 20, 21). Studies with specific PP1/2A peptide and small molecule inhibitors support dephosphorylation of DAT in rat striatum by PP1 and to a lesser extent PP2A (21), and the catalytic subunit of PP2A (PP2Ac) has been identified in co-immunoprecipitation complexes with DAT in mouse striatal synaptosomes (22). Interactions between DAT and PP2Ac in the rat caudate putamen were increased by cocaine self-administration followed by 3 weeks of abstinence (23), supporting a role for this enzyme in regulating DAT responses to drug exposure. However, the phosphorylation sites regulated by these enzymes remain unknown.
To investigate these issues, we aimed at identifying DAT-associated protein phosphatases and characterize their respective phosphorylation sites. Here, we demonstrate that PP1 and PP2A were complexed with both mouse striatal DAT (mDAT) and heterologously expressed human DAT (hDAT) and that pharmacological inhibition of PP1/2A increased phosphorylation of hDAT at Thr-48, a previously unknown DAT phosphorylation site. Functional analysis of this site via site-directed mutagenesis revealed a role for its involvement in DA uptake and reciprocal palmitoylation events in hDAT.
Results
PP1 and PP2A associate with DAT
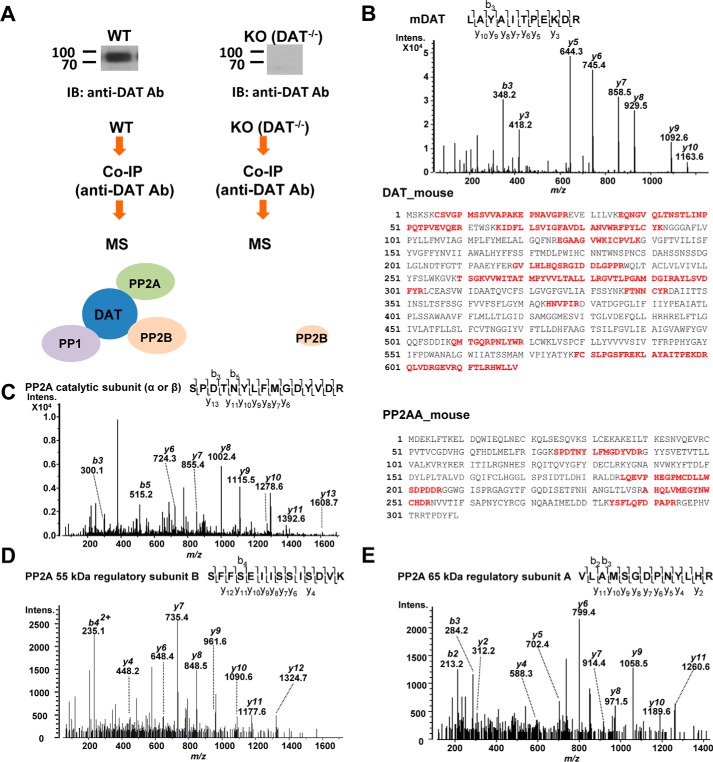
To screen for DAT association with PPs, we applied co-immunoprecipitation (co-IP) followed by LC-coupled tandem MS (LC-MS/MS) to confirm the association between DAT and PP2Ac, as well as to screen for association of any other protein phosphatases with DAT. DAT protein complexes were isolated from striatal synaptosomes prepared from WT and DAT−/− knockout (KO) mice by immunoprecipitation with anti-DAT antibody. Proteins were separated by SDS-PAGE with colloidal blue staining. Each lane was excised and divided into 12 pieces each for in-gel tryptic digestion, and the resulting tryptic peptides were subjected to LC-MS/MS. Specific associations were determined by comparison of proteins from the WT and KO samples (Fig. 1A).
Figure 1.
Identification of protein phosphatases in the mDAT complex by LC-MS/MS. A, flow diagram of co-IP and MS for the identification of DAT interacting PPs from WT and DAT−/− KO mouse striata. Immunoblot images show DAT protein expression exclusively in the WT striatal lysate. B (top), the double-charged (2+) peptide obtained at m/z 638.85 was fragmented to produce an MS/MS spectrum with y- and b-ion series that identified the sequence LAYAITPEKDR (amino acids 591–601) from mouse DAT. B (bottom), sequence coverage of mouse DAT with identified peptides (boldface type) by MS/MS in mouse striata. C (left), the MS/MS spectrum, obtained at m/z 897.41 (2+), for SPDTNYLFMGDYVDR (amino acids 75–89) of PP2A catalytic subunit (α or β isoforms). C (right), sequence coverage of mouse PP2Aα (Uniprot ID: PP2AA_mouse) with peptides identified by MS/MS in the DAT complex from the WT mouse striatum. D, the MS/MS spectrum of ion at m/z 779.90 (2+) for the sequence SFFSEIISSISDVK (amino acids 279–292) of PP2A subunit B isoform B55 identified in the DAT complex. E, the MS/MS spectrum representing VLAMSGDPNYLHR (amino acids 486–498) of co-purified PP2A 65-kDa regulatory subunit Aα isoform with DAT.
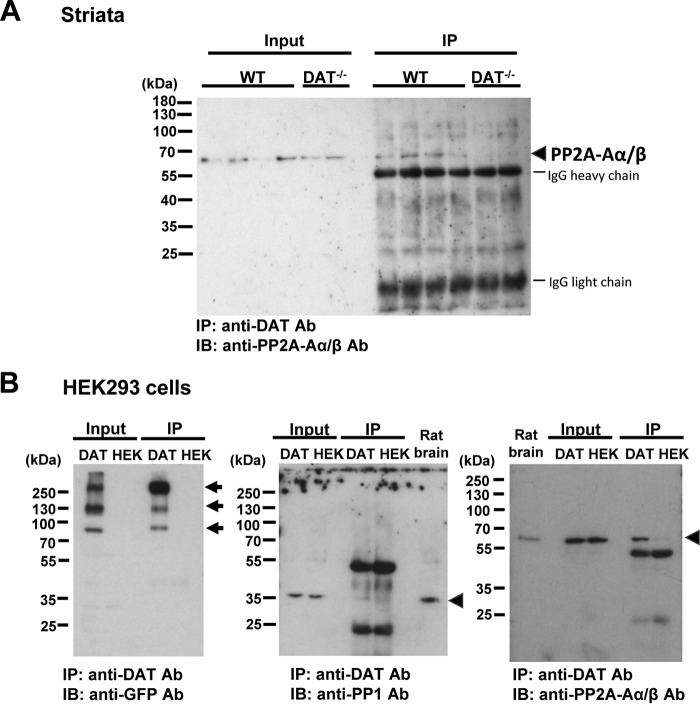
MS identified 39 representative DAT peptides in the immunopurified protein complex from the WT mouse striatum, which covered 34.89% of the mouse DAT sequence (Fig. 1B and Table S1). In the same protein complex from the WT striatum, MS identified three peptides for PP1 α, β, and/or γ catalytic subunits (Table 1). In addition, PP2A regulatory subunits A and B as well as PP2A catalytic subunit α/β were identified as co-immunoprecipitated proteins with DAT exclusively in the WT striatum (Table 1 and Fig. 1 (C–E)). The association between DAT and PP2A regulatory subunit Aα was confirmed by co-IP coupled with immunoblotting for mouse striata and HEK293 cells stably expressing YFP-tagged hDAT (YhDAT-HEK cells) (Fig. 2). Immunoreactivities of PP2A regulatory subunit Aα were observed in immunopurified DAT complexes from WT striata and YhDAT-HEK cells but not KO mouse striata and parent HEK293 cells, which do not express DAT (Fig. 2). PP1α catalytic subunit was also co-purified with DAT in YhDAT-HEK cells (Fig. 2B). MS identified co-purified PP2B with 24 peptides in the mouse WT striata, but parallel MS analysis also detected five peptides in the KO mouse striata (Table S2). Moreover, this association was not verified in a heterologous cell system examined in parallel (Fig. S1), and thus, we excluded this protein for further studies.
Table 1.
Identification of PP1 and PP2A in mouse DAT complex by MS
| Uniprot ID | Protein name | Residues | Peptide sequence | Mascot ions scorea (for each peptide) |
|
|---|---|---|---|---|---|
| WT | DAT−/− KO | ||||
| PP1 | |||||
| PP1A_MOUSE | Serine/threonine-protein phosphatase PP1-α catalytic subunit (PP1α catalytic subunit) | 44–60 | EIFLSQPILLELEAPLKb | 27 | – |
| 247–260 | AHQVVEDGYEFFAKb | 44 | – | ||
| PP1B_MOUSE or PP1G_MOUSE | Serine/threonine-protein phosphatase PP1-β or γ catalytic subunit (PP1β or -γ catalytic subunit) | 43–59 | EIFLSQPILLELEAPLKb | 27 | – |
| 111–121 | IKYPENFFLLR | 13 | – | ||
| 246–259 | AHQVVEDGYEFFAKb | 44 | – | ||
| PP2A | |||||
| 2ABA_MOUSE, 2ABB_MOUSE or 2ABD_MOUSE | Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B α, β, or δ isoform (PP2A subunit B isoform B55-α, -β, or -δ) | 279–292 | SFFSEIISSISDVK | 40 | – |
| 2AAA_MOUSE | Serine/threonine-protein phosphatase 2A 65-kDa regulatory subunit A α isoform (PP2A subunit A isoform PR65-α) | 106–113 | DKAVESLR | 10, 10 | – |
| 332–342 | ELVSDANQHVK | 11 | – | ||
| 486–498 | VLAMSGDPNYLHR | 21 | – | ||
| PP2AA_MOUSE and/or PP2AB_MOUSE | Serine/threonine-protein phosphatase 2A catalytic subunit α or β isoform (PP2A-α or -β) | 75–89 | SPDTNYLFMGDYVDR | 35 | – |
| 186–206 | LQEVPHEGPMoxcCDLLWSDPDDR.G | 17 | – | ||
| 240–254 | AHQLVMEGYNWCHDR | 32 | – | ||
| 240–254 | AHQLVMoxEGYNWCHDR | 31 | – | ||
| 284–294 | YSFLQFDPAPR | 50 | – | ||
a The ions score for an MS/MS match is based on the calculated probability, p, that the observed match between the experimental data and the database sequence is a random event. The reported score is −10log(p).
b Assigned to both PP1α and PP1β/γ.
c Mox, oxidation at methionine.
Figure 2.
Co-purified PPs with DAT in WT mouse striata and heterologous cells. A, PP2A regulatory subunit Aα protein (PP2A-Aα, Uniprot ID: 2AAA_Mouse) was detected in DAT immunoprecipitates from WT but not DAT−/− KO striatal lysates. Striatal synaptosomes were solubilized (Input) and immunoprecipitated (IP) by an affinity-purified polyclonal anti-DAT antibody (Ab). PP2A proteins (arrowhead) were detected by a monoclonal PP2A-Aα/β antibody. The numbers refer to the positions of prestained molecular weight markers. B, representative immunoblot images for co-purified PP1α (Uniprot ID: PP1A_Mouse) and PP2A-Aα with YFP-tagged hDAT stably expressed in HEK293 cells. Cell lysates (Input) were obtained from YhDAT-HEK (DAT) and parent HEK293 (HEK) cells and immunoprecipitated (IP) by an anti-DAT antibody. Immunoprecipitated DAT (arrows) was detected by using an anti-GFP antibody, and co-purified PP1α and PP2A-Aα were immunoblotted (arrowheads). Rat brain synaptosomal membrane fraction (Rat brain) was subjected to SDS-PAGE to compare the expression of endogenous PP1α and PP2A-Aα in HEK293 cells. The images represent three experiments with the same results.
Identification of phosphorylation in DAT
We detected phosphorylation of DAT residue Thr-53 in the mouse striatum by LC-MS/MS (Fig. S2), confirming previous findings obtained by immunoblotting with a Thr-53–phosphospecific antibody (16). However, because this identification could not provide any evidence for PP1/2A-responsive phosphorylation sites, we stably expressed hDAT in LLC-PK1 cells in the presence or absence of a PP1/2A inhibitor OA to determine which phosphorylation sites were affected.
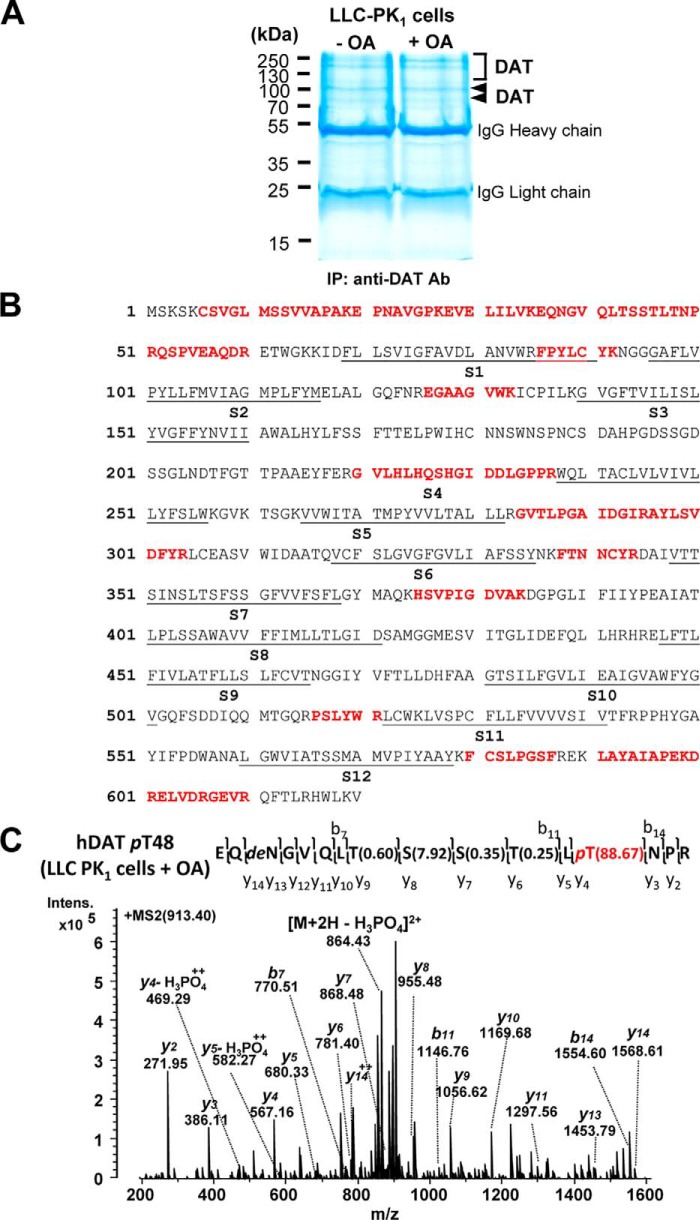
After immunopurification of hDAT from cell lysates, we repeated the MS analyses as applied for mouse DAT proteins. MS identified DAT from three different gel regions (Fig. 3, A and B), which corresponded to three immunoreactive YFP-DAT bands (Fig. 2B, left). MS identified a few phosphopeptides in hDAT under basal conditions, but treatment with various concentrations of OA increased the MS identification rate for hDAT phosphopeptides, especially the sequence 36EQNGVQLTSSTLTNPR51 (Table 2). We manually confirmed these phosphopeptides with a peak representing neutral loss of phosphoric acid, a signature of phosphopeptides, in MS/MS spectra. This tryptic peptide contains two serine (Ser-44 and -45) and three threonine (Thr-43, -46, and -48) residues that could represent potential phosphoacceptor sites, and the Mascot database search also indicated probabilities for phosphorylation at Ser-45, Thr-46, or Thr-48.
Figure 3.
Identification of PP1/2A-responsive DAT phosphorylation at Thr-48 by LC-MS/MS. A, representative colloidal blue–stained SDS-polyacrylamide gel image with proteins immunopurified by an anti-DAT antibody (Ab) from LLC-PK1 cells heterologously expressing hDAT in the absence (−) and presence (+) of OA. Arrowheads and square bracket, bands and region containing DAT protein identified by LC-MS/MS. The numbers refer to the positions of molecular weight markers. B, sequence coverage of human DAT with identified peptides (boldface type) by MS/MS from transfected LLC-PK1 cells. The putative transmembrane segments (S1–S12) of hDAT are underlined. C, MS/MS-identified hDAT phosphorylation site at Thr-48 (pT48) from transfected LLC-PK1 cells upon OA treatment. Shown is the MS/MS spectrum from the precursor ion at m/z 913.40 (2+) assigned to hDAT EQNGVQLTSSTLpTNPR (amino acids 36–51) with phosphorylation at Thr-48, which was verified by β-eliminated y-ions and neutral loss of H3PO4 from the precursor. The phosphosite localization probabilities determined by the Mascot delta score (38) are shown in parentheses. pT, phosphorylated threonine; deN, deamidated asparagine.
Table 2.
Increased number of phosphopeptides containing Thr-48 by inhibition of PP1/2A
| Peptide sequence | No treatment | Okadaic acid | |
|---|---|---|---|
| hDAT WT (LLC-PK1) | 28EVELILVKEQNGVQLTSSTLTNPR51 | 0:19a | 2:11 (0.25 μm, 2 h) |
| 36EQNGVQLTSSTLTNPR51 | 0:28 | 11:29 (0.5 μm, 2 h) | |
| 3:38 | 6:12 (1.0 μm, 2 h) |
a The values in boldface type denote the sum of the two different recovered phosphopeptide sequences displayed in the second column versus the sum of the two corresponding unphosphorylated peptides recovered in three individual experiments using three different concentrations of OA.
To determine the identity of the phosphorylation site, we mutated Thr-48 to Ala to prevent phosphorylation and expressed the T48A hDAT mutant in heterologous cells under OA treatment. MS analysis showed no phosphorylation of this peptide in either basal or OA-stimulated conditions (Tables S3 and S4). Therefore, we concluded that Thr-48 is a PP1/2A-responsive phosphorylation site in hDAT (Fig. 3C). As an additional control, we analyzed the phosphorylation status of this peptide derived from rDAT, which contains isoleucine rather than threonine at position 48 (Fig. S3), although several other Ser and Thr residues are present in this sequence. The resulting tryptic peptide also showed no phosphorylation, providing further support for the specific regulation of Thr-48 phosphorylation status by PP1/2A inhibition.
Inhibition of PP1/2A increases phosphorylation of DAT at Thr-48
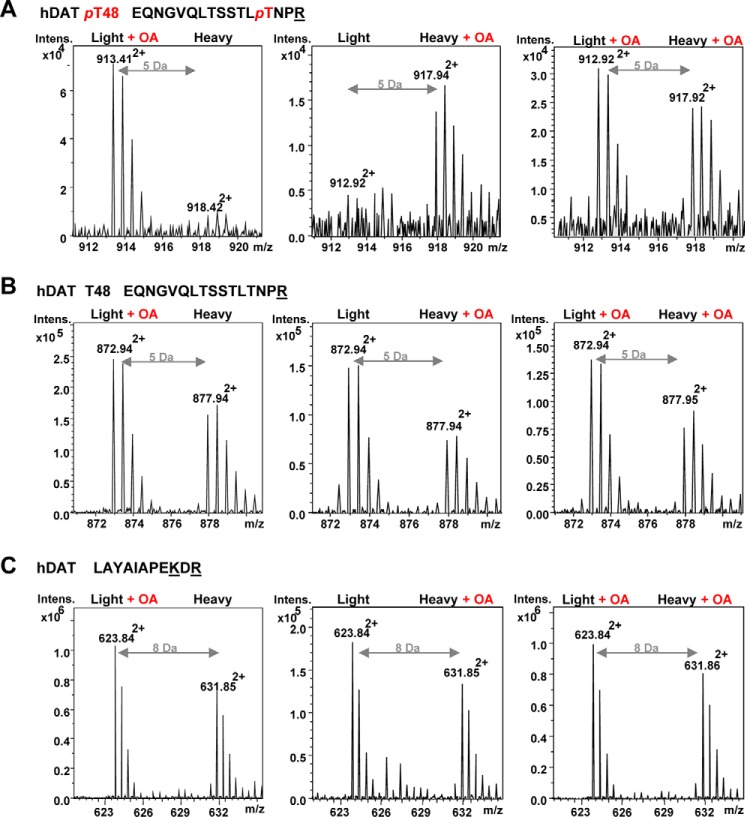
OA-inhibited hDAT dephosphorylation was indicated by the increased number of phosphopeptides isolated (Table 2). To determine the relative increase in Thr-48 phosphorylation mediated by OA, we applied “stable isotope labeling with amino acids in cell culture” (SILAC), which distinguishes peptides derived from different conditions by the stable isotope-introduced mass tag (24, 25). The LLC-PK1 cells stably expressing hDAT were cultured in media containing either Lys-0/Arg-0 (“Light”) or Lys-6/Arg-10 (“Heavy”) in the presence or absence of OA (0.5 μm for 1 h). Because the doubly charged (2+)-peptide 36EQNGVQLTSSTLTNPR51 contains one Arg and Lys-6/Arg-10 labeling would increase its mass by 5 Da (m/z = 10 Da/2+), we could compare the identified phosphopeptide peak with the peak that is located at +5 or −5 Da according to the labeling of the identified phosphopeptides. SILAC certainly indicated increased levels of phosphopeptide for Thr-48 in the presence of OA (Fig. 4A, left and middle). When treated with OA in both Light and Heavy culture media, MS identified comparable phosphopeptides for Thr-48 in the Light- and Heavy-labeled DAT at a different m/z of 5 Da (Fig. 4A, right). However, the increase of tryptic peptide peaks by OA was not observed in the corresponding unphosphorylated peptide 36EQNGVQLTSSTLTNPR51 (Fig. 4B) and the hDAT 591LAYAIAPEKDR601 peptide (Fig. 4C).
Figure 4.
Relative quantification of OA-induced DAT phosphorylation at Thr-48 by SILAC. A, the doubly charged (2+) phosphopeptide (EQNGVQLTSSTLpTNPR) presenting phosphorylated hDAT at Thr-48 (pT48) was abundant in the samples from hDAT-LLCPK1 cells upon OA treatment (0.5 μm for 1 h) but limited in the untreated samples. hDAT-LLCPK1 cells were cultured in metabolically different culture media with either Light medium containing normal lysine (12C6, 14N2) and arginine (12C6, 14N4) or Heavy medium containing isotopic lysine (13C6, 14N2) and arginine (13C6, 15N4) in the absence and presence of OA, which provides a difference of 5 Da between unlabeled and labeled doubly charged EQNGVQLTSSTLTNPR peptides. B, the corresponding unphosphorylated peptide (EQNGVQLTSSTLTNPR, 2+) was not affected by OA treatment in both Light and Heavy culture conditions. C, a representative hDAT peptide (LAYAIAPEKDR, 2+) demonstrates the overall ratio of hDAT and no effect of OA in hDAT expression between two different culture media. SILAC labeling at one Lys and one Arg provided the 8-Da (m/z = 6 + 10 Da/2+) difference of the labeling.
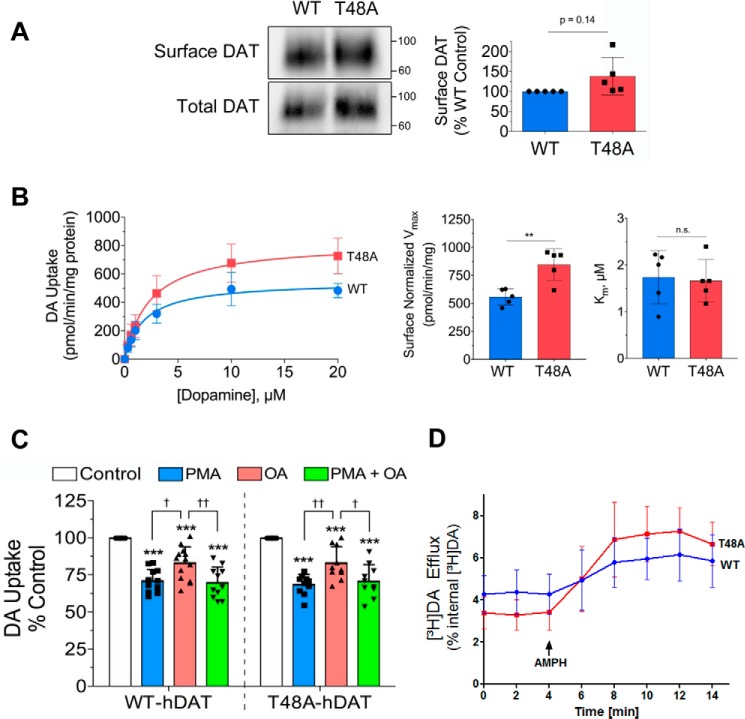
Enhanced DA uptake in the dephosphomimetic T48A mutant
To investigate the functional effect of Thr-48 dephosphorylation, we examined DA uptake mediated by the dephosphomimetic T48A mutant. The construct was transiently expressed in LLC-PK1 cells and showed plasma membrane expression comparable with that of the WT DAT by surface biotinylation analysis (p > 0.05; Fig. 5A). To validate that our cell surface analysis was within the linear range of detection and not saturated, we determined a NeutrAvidin binding curve using increasing amounts of cell lysate (25–200 μg of protein) from our cell surface biotinylation assay (Fig. S4). We found that our assessment of DAT surface levels was within the linear range of our assay. In saturation analyses performed in parallel and normalized to DAT surface levels, there was no difference in Km (WT, 1.7 ± 0.6 μm; T48A, 1.7 ± 0.5 μm; p > 0.05), but Vmax was significantly increased in T48A-DAT (WT, 558 ± 72 pmol/min/mg; T48A, 847 ± 142 pmol/min/mg; p < 0.01, n = 5) (Fig. 5B). Enhanced activity in the dephosphomimetic mutant suggests that uptake is reduced by phosphorylation of the site and returns to basal levels by PP1/2A-mediated dephosphorylation. The lower Vmax in the WT protein than in the T48A form is presumably driven by basal phosphorylation of this site.
Figure 5.
Expression and kinetic analysis of the dephosphomimetic T48A-DAT. A, representative immunoblots showing comparative amounts of surface and total DAT levels in transfected LLC-PK1 cells (left). Surface and total expression of WT and T48A-DATs were analyzed in each experiment in parallel for DA uptake saturation analysis (B). B, saturation analysis (left) of [3H]DA uptake by WT and T48A-DATs in LLC-PK1 cells. Uptake values shown are the mean ± S.D. of five independent experiments performed in triplicate. The histograms show significantly increased Vmax values for the T48A mutant (middle) and no change in the Km (right) compared with the WT DAT (means ± S.D. (error bars); **, p < 0.01). C, LLC-PK1 cells transfected and expressing WT or T48A-DATs were treated with 0.4 μm PMA, 1 μm OA, or both for 30 min followed by a DA uptake assay. The bar graph shows the significantly decreased DA uptake by both WT and T48A-DATs with each treatment (means ± S.D.; ***, p < 0.001 versus control with the appropriate group; †, p < 0.05; ††, p < 0.01 for indicated pairs, n ≥ 10; ANOVA with Tukey post hoc test). D, amphetamine (AMPH)-stimulated DA efflux by WT and T48A-DATs in LLC-PK1 cells. The addition of AMPH at t = 4 min is indicated by an arrow. Data are means ± S.D. of three experiments performed in sextuplicate, p > 0.05, ANOVA with Tukey post hoc test).
Other phosphorylation sites, including Ser-7, are regulated by PKC and PP1/2A (17) and could contribute to outcomes in WT and T48A-DATs. Hence, in order to determine whether Thr-48 mediates transport regulation by PP1/2A or PKC, we performed DA uptake analyses in the absence or presence of OA, the PKC activator phorbol 12-myristate 13-acetate (PMA), or both compounds together. DA uptake activity in WT and T48A-DATs was significantly decreased in all three conditions (Fig. 5C). In the WT protein, levels of down-regulation induced by OA and PMA (17 ± 11% and 29 ± 8%, respectively) were similar to that seen in previous studies (12, 18, 20), and there was no additivity with the combined PMA/OA treatment (Fig. 5C, 30 ± 10%), suggesting a shared mechanism of action. In addition, there was no difference in down-regulation responses between WT and T48A-DAT for each treatment (p > 0.05; Fig. 5C), suggesting that the inability of Thr-48 to undergo phosphorylation had no influence on either PKC or PP1/2A responses.
We further examined amphetamine-stimulated DA efflux by the dephosphomimetic T48A mutant. LLC-PK1 cells transiently expressing WT or T48A-DAT were preloaded with [3H]DA and then treated with amphetamine (3 μm) to stimulate DA efflux. In superfusion experiments, no significant differences of efflux were observed at each fraction between WT and T48A-DAT (p > 0.05) (Fig. 5D).
Increased palmitoylation in the dephosphomimetic T48A-DAT
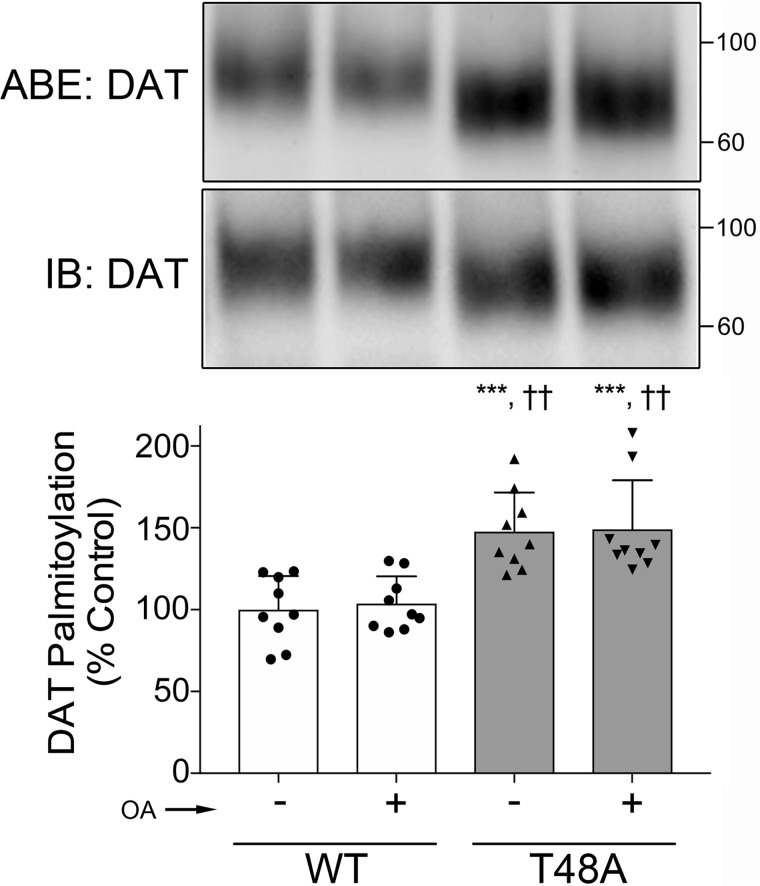
Previously, we reported a reciprocal mechanism between DAT phosphorylation at Ser-7 and palmitoylation at Cys-580 that acted in concert to set levels of DA uptake capacity (18). Therefore, we examined whether DAT palmitoylation could also be affected by Thr-48 phosphorylation status. We transiently expressed WT and T48A-DAT in LLC-PK1 cells in the absence and presence of OA and assessed transporter palmitoylation levels by an acyl-biotinyl exchange (ABE) assay. In this procedure, free thiols in the protein are blocked using methyl methanethiosulfonate (MMTS), endogenous palmitate groups are cleaved with hydroxylamine (NH2OH), and the liberated free SH groups are tagged using sulfhydryl-reactive (N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide (HPDP biotin). Biotinylated proteins are extracted, identified by immunoblotting, and modification levels are expressed relative to total transporter protein.
The results show that in basal conditions, palmitoylation of T48A-DAT was significantly enhanced relative to the WT protein (54 ± 14%, p < 0.05, versus WT, n = 9) (Fig. 6), supporting a role for Thr-48 phosphorylation in regulating this modification. However, in contrast to our findings that palmitoylation was reduced by PMA (18), we found no effect of OA on this modification (Fig. 6), indicating mechanistic differences between functions regulated by PKC versus PP1/2A.
Figure 6.
Enhanced palmitoylation in the dephosphomimetic T48A-DAT mutant. LLC-PK1 cells expressing WT or T48A mutant DAT were treated with either vehicle (control) or 1 μm OA for 30 min, followed by assessment of DAT palmitoylation by ABE. Shown are representative immunoblots (top panel) with an anti-DAT antibody for NH2OH-treated ABE samples (top blots) and total cell lysates (bottom blots). The histogram (bottom panel) shows quantification of DAT palmitoylation (***, p < 0.001 versus WT control; ††, p < 0.01 versus WT plus OA; ANOVA with Tukey post hoc test). Error bars, S.D.
Discussion
In this study, we found associations of DAT with PP1 and PP2A by unbiased MS-based proteomics and verified the findings by parallel analysis of striatal samples from DAT WT and KO mice that eliminated nonspecific and false-positive proteins. PP2A is characterized as having a heterodimeric core enzyme and a heterotrimeric holoenzyme, where the former is composed of scaffold and catalytic subunits and interacts with one of four regulatory subunits to form a holoenzyme (26). Our MS analysis not only confirmed the association with the previously immunoblotting-detected PP2A catalytic subunit (22) but also identified a scaffold subunit, PP2A PR65-α, and a regulatory subunit, B55, in the immunopurified DAT protein complex from mouse striata (Table 1). MS also presented PP1 association with DAT, which we previously showed is a major contributor to rat striatal DAT dephosphorylation (8, 21).
Previous MS-based proteomics studies have identified DAT-interacting proteins including PKCs and CaMKIIs, but not PP2A, in mouse and rat brains (7, 27, 28). In other SLC sodium carrier transporters, MS approaches have shown the interaction of PP2A with norepinephrine transporter and organic cation/carnitine transporter OCTN2 (29, 30), but PP2A-responsible dephosphorylation sites were not determined in these transporter proteins. Although MS is a very powerful and accurate tool to identify phosphorylation sites (31), some additional procedures are required for MS sample preparation to identify protein kinase/phosphatase-specific sites by MS. For this purpose, a target protein kinase/protein phosphatase needs to be identified, and experimental methods are determined accordingly. Because we detected a few phosphopeptides for DAT purified from LLC-PK1 cells under basal culture conditions, we presumed that the association of PP1/2A maintains the dephosphorylated state of DAT. Therefore, we pharmacologically inhibited PP1/2A using OA in LLC-PK1 cells expressing hDAT to prevent dephosphorylation. Then MS with stable isotope labeling manifested the increased phosphorylation at Thr-48 in hDAT, indicating PP1/2A-directed dephosphorylation at this site in hDAT (Fig. 4).
We observed a significant effect of the dephosphomimetic T48A mutant on DA uptake (Fig. 5B) but not on amphetamine-stimulated DA efflux (Fig. 5D), indicating the specific role of dephosphorylation at Thr-48 in the forward transport mechanism. The enhanced DAT-mediated DA uptake rate in the T48A mutant strongly supports a distinctive role of this PP1/2A-directed residue in DAT function in suppressing transport activity (Fig. 5B). However, the T48A mutant still displayed transport down-regulation with OA treatment (Fig. 5C). This could implicate additional PP1/2A-responsive phosphorylation sites not detected by our MS system, probably due to difficulty of ionization and/or unsuitable tryptic peptide lengths for MS analysis. Previous studies also support the existence of other PP1/2A-responsive phosphorylation sites besides Thr-48 in DAT by means of the OA-induced increase of 32PO4 incorporation in rDAT (12) and the OA-mediated decrease in DA uptake in rodent DAT, which lacks Thr-48 (12, 20). Therefore, the persistent decrease in DA uptake induced by OA in the hDAT T48A mutant (Fig. 5C) could be attributable to the effect of other OA-responsible sites or from other events such as transporter internalization. This raises the question of the identities of these additional sites.
In our previous studies, the OA treatment of rat striatal slices caused increased phosphorylation primarily on distal N-terminal serines (21, 32) and Thr-53 (equivalent to hDAT Ser-53) (16). In addition, our previous MS study pinpointed phosphorylation at hDAT Ser-7 in HEK293 cells upon OA treatment (17). In the present study, however, we could not identify OA-responsible phosphorylation at Ser-7 in LLC-PK1 cells, probably due to different profiles of protein kinases and protein phosphatases between experimental cell lines. This also may be related to different PP response to OA and against OA-induced cell toxicity between the two cell lines (data not shown). We also identified phosphorylation at hDAT Ser-53 in LLC-PK1 cells in the presence and absence of OA by chymotryptic in-gel digestion (48TNPRQSPVEAQDRETW63; data not shown). However, we could not determine its phosphorylation status upon OA treatment by SILAC because MS did not identify phosphorylation at Ser-53 in tryptic peptide (52QSPVEAQDR60 or 52QSPVEAQDRETWGK65), probably due to different properties for MS analysis between chymotryptic and tryptic Ser-53 phosphopeptides. The lack of sequence coverage for Ser-2, Ser-4, Thr-62, and Thr-613 in human DAT by our MS approach (Fig. 3B) needs to be mentioned. Thus, if functional relevance of OA might be achieved by increased phosphorylation of multiple sites, these unidentified residues together with Ser-7 and Ser/Thr-53 will be considered for PP1/2A-regulated DAT function. When phosphorylation was prevented at each of these potential PP1/2A sites by individual single-point mutations, DA uptake was increased in the hDAT T48A mutant (Fig. 5B) and the rDAT S7A mutant (18) but decreased in rDAT T53A (16). This indicates that PP1/2A-mediated dephosphorylation does not simply drive DAT for unidirectional transport function but rather tunes DAT functions through different PP1/2A-sensitive residues. In addition, the phosphorylation status of these multiple residues is achieved via coordinated activity of different protein kinases and phosphatases, providing more systematic DAT regulation upon cellular responses. This may also explain why OA treatment could not always achieve statistical significance for down-regulation of DAT activity. Previously, OA treatment significantly reduced the dopamine transport Vmax by 13–14% in mouse and rat striatal synaptosomes (10, 12, 20). Additionally, in human DAT expressed in heterologous cells, only a low concentration (0.3 μm) of OA treatment slightly decreased DA uptake (33), which obscured the effect of PP1/2A on DAT function. In the present study, we showed that OA treatment significantly decreased DA uptake by 17 ± 3% in heterologous cells expressing the WT hDAT (Fig. 5C). As mentioned above, this inconsistency may be caused by different OA sensitivity to several phosphoacceptor sites in DAT according to the extent of protein kinase and PP1/2A activity under certain conditions in different experimental subjects.
PKC-stimulated phosphorylation at Ser-7 is reciprocally regulated by palmitoylation (18), and thus we assumed that palmitoylation may also be related to Thr-48 phosphorylation. Indeed, we detected increased palmitoylated DAT in the T48A mutant, but OA did not affect the palmitoylation status in both the WT and T48A mutant (Fig. 6), although we did observe effects of OA on DA uptake in the T48A mutant. This may be due to a comparatively small extent of OA-induced phosphorylation in the total phosphorylation level of DAT from a heterologous cell system. Although we still need to determine the interplay between phosphorylation at Thr-48 and palmitoylation at Cys-580 in more detail, we propose that PP1/2A-mediated dephosphorylation at Thr-48 may regulate DAT palmitoylation and, furthermore, DA reuptake.
Here, we showed PP1/2A-mediated dephosphorylation at Thr-48 in DAT by MS and its functional implication in DA uptake by utilizing the dephosphorylation-mimetic mutant of this residue. In conjunction with our effort to pinpoint kinase-specific phosphorylation sites with their functional roles (16, 17), this study demonstrated the significance of the N terminus in DAT regulation by not only phosphorylation but also dephosphorylation. As we have found up- or down-regulation of DA activity in DAT dephosphomimetic mutants of potential PP1/2A-directed sites, a comprehensive study needs to be performed for phosphorylation/dephosphorylation effects on DAT functions, and for this purpose, identification of specific protein kinases/phosphatases as well as their specific sites of action is a prerequisite. This is the first time a protein phosphatase–directed site in SLC6 neurotransmitter transporters with functional involvement has been identified, representing progress toward establishing a mechanism for the modulation of SLC6 transporters by coordinated phosphorylation/dephosphorylation and, in the case of DAT, additionally palmitoylation.
Experimental procedures
Animals
Functional DAT knockout mice were generated by crossing heterozygous mice expressing the CRE recombinase under the DAT promoter (34). The mice have a C57BL/6J background, and the genotype was confirmed twice for each mouse. The mice were bred at the Core Unit of Biomedical Research, Division of Laboratory Animal Science and Genetics, Medical University of Vienna (Vienna, Austria). All animal care procedures were in accordance with the ARRIVE guidelines, the Austrian animal protection law, and the Austrian animal experiment by-laws that implement European law (directive 2010/63/EU) into Austrian law; they were approved by the local animal ethics committee at the Medical University of Vienna and authorized by the Austrian Federal Ministry of Science and Research (GZ: BMWFW-66.009/0016-WF/V/3b/2015).
Preparation of mouse striatal synaptosomes
Mice were dispatched by cervical dislocation, and subsequently the brain was removed immediately to dissect the dorsal striata. The right and left striata of one mouse were homogenized in ice-cold buffer containing 0.32 m sucrose, 5 mm sodium phosphate (pH 7.4), 0.1 m sodium fluoride, and protease inhibitors (cOmplete®, Roche Applied Science). The suspension was centrifuged at 800 × g for 10 min, and subsequently the supernatant was centrifuged at 38,000 × g for 90 min. The resulting crude synaptosomal membrane pellet was suspended in the homogenization buffer.
cDNA constructs
The hDAT pcDNA3.0 vector was created by excising the hDAT coding region from a His6-hDAT vector (35) using endonucleases Acc65I and EcoRI and ligating the excised fragment into pcDNA3.0 using these same restriction endonuclease sites. The hDAT pEYFP vector was created by excising the hDAT coding region from the hDAT pcDNA3.0 vector using endonucleases Acc65I and XbaI and ligating the excised fragment into pEYFP-C1 using these same restriction endonuclease sites. Mutagenesis to create DAT T48A was performed using hDAT pcDNA 3.0 as the template with the Stratagene QuikChange® kit, and codon substitution was verified by sequencing (Eurofins MWG, Louisville, KY). For production of stable transformants, LLC-PK1 cells (ATCC, Manassas, VA) were transfected (X-tremeGENE HP, Roche Applied Science) and after 48 h maintained under selection with 600 μg/ml G418 (8). HEK293 and tsA201 cells were transiently transfected with the indicated vector using the Turbofect (Thermo Scientific) according to the manufacturer's protocol.
Cell culture and transient transfection
Parental LLC-PK1 cells and LLC-PK1 cells stably expressing hDAT (hDAT-LLCPK1), T48A hDAT (T48A-LLCPK1), and rat DAT were maintained in α-minimum essential medium supplemented with 5% FBS, 2 mm l-glutamine, 200 μg/ml G418, 25 IU/ml penicillin, and 25 μg/ml streptomycin. Parental HEK293 cells and HEK293 cells stably expressing YFP-tagged human DAT (YhDAT-HEK293) as well as tsA201 cells were cultured in Dulbecco's modified Eagle's medium with 10% FBS, 4.5 mg/ml glucose, 25 IU/ml penicillin, and 25 μg/ml streptomycin. Additionally, 250 μg/ml G418 was added to the medium for stable cell lines to maintain DAT protein expression. Cells were maintained in a humidified atmosphere of 5% CO2 at 37 °C.
IP and immunoblotting (IB)
Mouse striata membranes and cell lysates from heterologous cells expressing DAT proteins were prepared as described previously (16). For IP, tissue and cell lysates were solubilized in 0.5 and 1% Triton X-100 lysis buffer, respectively, and incubated overnight with a goat anti-DAT (sc-1433, Santa Cruz Biotechnology, Inc.) or rabbit anti-GFP polyclonal antibodies (A6455, Life Technologies, Inc.). Antigen–antibody complexes were then incubated with protein G beads (GE Healthcare) for 4 h at 4 °C. After six washes, bound proteins were eluted in SDS sample buffer at 95 °C for 3 min. Eluted proteins were size-fractionated on SDS-polyacrylamide gels and visualized by a colloidal blue staining (Invitrogen).
For IB, proteins were separated on SDS-polyacrylamide gels and transferred to polyvinylidene fluoride membranes (Waters, Milford, MA), which were then blocked and immunostained with anti-PP1 (sc-7482, Santa Cruz), PP2A-Aα/β (sc-74580, Santa Cruz), or PP2B-Aα (sc-17808, Santa Cruz) antibody.
In-gel digestion
The protein bands were directly excised from SDS-polyacrylamide gels, destained with 50% acetonitrile in 50 mm ammonium bicarbonate, and dried in a speed-vacuum concentrator. After reduction and alkylation of Cys, gel pieces were washed and dehydrated. Dried gel pieces were swollen with 25 mm ammonium bicarbonate (pH 8.0) containing 10 ng/μl trypsin (Promega) or chymotrypsin (Promega) and incubated at 37 °C for 2–18 h. Digested peptides were extracted with 50% acetonitrile in 5% formic acid (FA) and concentrated in a speed-vacuum concentrator (16).
LC-MS/MS
An electrospray ionization (ESI)- ion trap (IT) mass spectrometer (HCT and Amazon speed ETD, Bruker) and an ESI-quadrupole-TOF (QTOF; Compact, Bruker) coupled with an Ultimate 3000 nano-HPLC system (Dionex) were used for LC-MS/MS data acquisition. A PepMap100 C-18 trap column (300 μm × 5 mm) and PepMap100 C-18 analytic column (75 μm × 150 mm) were used for reverse-phase chromatographic separation with a flow rate of 300 nl/min. The two buffers used for the reverse-phase chromatography were 0.1% FA and 80% acetonitrile in 0.08% FA with variable gradient conditions. Eluted peptides were then directly sprayed into the mass spectrometer, and the MS/MS spectra were interpreted with the Mascot search engine (version 2.4.1, Matrix Science, London, UK) against Swissprot database (556,196 sequences, released in November 2017), and the taxonomy was restricted to Homo sapiens (human; 20,244 sequences) for human DAT from cell lines as well as to Rodentia (rodents; 26,656 sequences) for mouse proteins and rat DAT purified from the mouse striata and heterologous cells, respectively.
The search parameters were used with a mass tolerance of 20 ppm (QTOF) or 0.5 Da (IT) and an MS/MS tolerance of 0.1 Da (QTOF) or 0.5 Da (IT). Carbamidomethylation on Cys, oxidation on Met, phosphorylation on Ser/Thr, and deamidation on Asn/Gln were allowed with two missing cleavage sites. The Mascot cut-off score was set to 10, and each filtered MS/MS spectrum exhibiting possible phosphorylation was carefully examined based on the existence of a neutral loss of H3PO4 (97.977 Da) (16, 36).
SILAC
hDAT-LLCPK1 cells were grown for at least five cell divisions in either normal lysine (12C6, 14N2; “Lys-0”) and arginine (12C6, 14N4; “Arg-0”) media or in isotopic Lys (13C6, 14N2; “Lys-6”) and Arg (13C6, 15N4; “Arg-10”) labeling media in the absence and presence of okadaic acid. After labeling, cells were solubilized in lysis buffer containing 1% Triton X-100, 20 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1 mm EDTA, 1 mm sodium orthovanadate, 5 mm NaF, 5 mm sodium pyrophosphate, and a protease inhibitor mixture. The cell lysates were mixed at a 1:1 ratio based on protein quantification for IP with an anti-GFP antibody. After size fractionation by SDS-PAGE, hDAT proteins were in-gel trypsin-digested and analyzed by ESI-QTOF as described above. After Mascot database searches, relative spectral intensities were examined using Compass DataAanlysis (version 4.2, Bruker) software.
DA uptake and cell surface biotinylation assays
Cells were grown in 24-well plates to ∼80% confluence and then transfected (XtremeGene, Sigma) with WT hDAT or T48A hDAT cDNA (pcDNA3.0). After 36 h post-transfection, cells were rinsed twice with 0.5 ml of 37 °C Krebs-Ringer/HEPES (KRH) buffer (25 mm HEPES, 125 mm NaCl, 4.8 mm KCl, 1.2 mm KH2PO4, 1.3 mm CaCl2, 1.2 mm MgSO4, 5.6 mm glucose, pH 7.4) followed by incubation at 37 °C for 30 min with vehicle (MeSO4), 1 μm OA, 400 nm PMA, or both OA and PMA followed by a [3H]DA transport assay. Uptake was performed in triplicate for 8 min and initiated by the addition of 10 nm [3H]DA plus 3 μm unlabeled DA. For saturation analysis, unlabeled DA concentration varied from 0.3 to 20 μm, with nonspecific uptake determined in the presence of 100 μm (−)-cocaine. Cells were rapidly washed two times with ice-cold KRH buffer and solubilized in 1% Triton X-100, and radioactivity contained in lysates was assessed by liquid scintillation counting.
For determination of cell surface expression, cells were plated, transfected, and treated in parallel with those analyzed for DA transport. These cells were washed three times with ice-cold PBS/Ca-Mg (138 mm NaCl, 2.7 mm KCl, 1.5 mm KH2PO4, 9.6 mm Na2HPO4, 1 mm MgCl2, 0.1 mm CaCl2, pH 7.4) and incubated twice with a 0.5 mg/ml concentration of the membrane-impermeable reagent sulfo-NHS-SS-biotin for 25 min at 4 °C in PBS/Ca-Mg. The biotinylating reagent was removed, and the reaction was quenched by two sequential incubations with 100 mm glycine in PBS/Ca-Mg for 20 min at 4 °C. Cells were washed with PBS/Ca-Mg and lysed at 4 °C with 250 μl of radioimmunoprecipitation assay buffer (10 mm sodium phosphate, pH 7.3, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing protease inhibitor. Total cell lysates (50 μg of protein) were incubated with NeutrAvidin beads overnight at 4 °C. The beads were washed three times with radioimmunoprecipitation assay buffer, and bound proteins were eluted with 40 μl of Laemmli buffer (62.5 mm Tris-HCl, pH 6.8, 20% glycerol, 2% SDS, 5% β-mercaptoethanol, and 0.01% bromophenyl blue), followed by immunoblotting with a DAT-specific polyclonal antibody (C-20, Santa Cruz, Inc., sc-1433).
DA uptake values from the saturation analysis were normalized to DAT surface expression by dividing WT and T48A uptake values by relative surface abundance for each, where the WT surface abundance was set to 1 and the T48A was either less than or greater than 1, as determined by cell surface biotinylation with equal amounts of total protein for WT and T48A samples loaded on the high-capacity NeutraAvidin resin. Vmax and Km values were determined by nonlinear regression analysis of the normalized saturation analysis uptake values.
Superfusion experiments
Substrate efflux assays were performed as described previously (16) with the following modification. The LLC-PK1 cells transiently expressing WT or T48A-DAT were preincubated with 0.4 μm [3H]DA (55 Ci/mmol; American Radiolabeled Chemicals, St. Louis, MO) for 60 min at 37 °C in a final volume of 0.1 ml of KRH buffer/well and subsequently transferred into superfusion chambers. Immediately, superfusion was initiated with KRH buffer at 25 °C at a perfusion rate of 0.7 ml/min. After 45 min, a stable efflux of radioactivity was achieved, and the experiment was started with the collection of 2-min fractions. After three fractions, amphetamine (3 μm) was added to stimulate DA efflux. Finally, the cells were lysed with 1% SDS. Afterward, the amount of tritiated substrate present in each vial was determined by β-scintillation counting (PerkinElmer Life Sciences). Efflux of tritium was expressed as a fractional rate (i.e. the radioactivity released during a fraction was expressed as the percentage of the total radioactivity present in the cells at the beginning of that fraction).
Analysis of palmitoylation by ABE
Analysis of palmitoylation by ABE was performed as described previously (37). Briefly, cells were treated with vehicle or 1 μm OA for 30 min, followed by cell membrane preparation as described previously (18). Membrane pellets were solubilized in lysis buffer (50 mm HEPES, pH 7.0, 2% SDS, 1 mm EDTA) containing protease inhibitors and 25 mm N-ethylmaleimide to block free thiols. Lysates were incubated at room temperature for 1 h with mixing followed by acetone precipitation and resuspension in lysis buffer containing MMTS and incubation at room temperature overnight with end-over-end rotation. Excess MMTS was removed by three sequential acetone precipitations, followed by resuspension of the precipitated proteins in 300 μl of a buffer containing 4% (w/v) SDS (4SB: 4% SDS, 50 mm Tris, 5 mm EDTA, pH 7.4). Each sample was divided into two equal portions that were treated for 2 h at room temperature with 50 mm Tris-HCl, pH 7.4, as control or 0.7 m NH2OH, pH 7.4, to cleave the thioester bonds. NH2OH was removed by three sequential acetone precipitations followed by resuspension of the precipitated proteins in 240 μl of 4SB buffer. Samples were diluted with 900 μl of 50 mm Tris-HCl, pH 7.4, containing 0.4 mm HPDP biotin and incubated at room temperature for 1 h with end-over-end mixing. Unreacted HPDP biotin was removed by three sequential acetone precipitations followed by resuspension of the final pellet in 75 μl of lysis buffer without MMTS. Samples were diluted with 50 mm Tris-HCl, pH 7.4, to contain 0.1% SDS, and biotinylated proteins were extracted using NeutrAvidin resin. Proteins bound to the resin were eluted with sample buffer (60 mm Tris, pH 6.8, 2% SDS, 10% glycerol) containing 100 mm DTT plus 3% β-mercaptoethanol and subjected to SDS-PAGE. DAT was identified by immunoblotting using polyclonal antibody (C-20, sc-1433, Santa Cruz Biotechnology).
Statistics
Palmitoylated and cell surface DAT band intensities were quantified by densitometry using Quantity One (Bio-Rad) software. The palmitoylated DAT signal was normalized to the amount of total DAT protein loaded on the NeutrAvidin resin. Signals for treatment groups were expressed relative to control samples set to 100%. All experiments were performed a minimum of three times with similar results, and values were analyzed for statistical significance using Student's t test or one-way ANOVA as indicated.
Author contributions
J.-W. Y., H. H. S., and J. D. F. conceptualization; J.-W. Y., H. H. S., and J. D. F. data curation; J.-W. Y., H. H. S., and J. D. F. supervision; J.-W. Y., R. A. V., H. H. S., and J. D. F. funding acquisition; J.-W. Y., G. Larson, L. K., M. S., M. H., K. J., M. K., S. H., and F. A. E. investigation; J.-W. Y., R. A. V., H. H. S., and J. D. F. writing-original draft; J.-W. Y., R. A. V., H. H. S., and J. D. F. writing-review and editing; G. Lubec resources; G. Lubec and J. D. F. methodology; H. H. S. project administration.
Supplementary Material
Acknowledgment
We are grateful to Dr. Cornelius Gross (EMBL, Monterotondo, Italy) for providing the DAT Cre mice.
This work was supported by Austrian Science Fund (FWF) Grants F3506 (to H. H. S.) and P23670 (to J.-W. Y.) and National Institutes of Health Grants R15 DA031991 (to J. D. F.), R01 DA13147 (to R. A. V.), and P20 GM104360 (to U. N. D.) from the COBRE program and Grant P20 GM103442 (to U. N. D.) from the INBRE program of the NIGMS. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S4 and Figs. S1–S4.
- DAT
- dopamine transporter
- ABE
- acyl-biotinyl exchange
- CaMKIIα
- Ca2+/calmodulin-dependent protein kinase II α
- IP
- immunoprecipitation
- DA
- dopamine
- hDAT
- human DAT
- YhDAT-HEK
- HEK293 cells stably expressing YFP-tagged hDAT
- HPDP biotin
- sulfhydryl-reactive (N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide
- mDAT
- mouse DAT
- MMTS
- methyl methanethiosulfonate
- MS/MS
- tandem MS
- OA
- okadaic acid
- PKA
- cAMP-dependent protein kinase
- PKC
- protein kinase C
- PMA
- phorbol 12-myristate 13-acetate
- PP
- protein phosphatase
- PP2Ac
- the catalytic subunit of PP2A
- rDAT
- rat DAT
- SILAC
- stable isotope labeling with amino acids in cell culture
- SLC6
- Na+/Cl−-dependent neurotransmitter transporter
- KO
- knockout
- IB
- immunoblot
- FA
- formic acid
- ESI
- electrospray ionization
- IT
- ion trap
- QTOF
- quadrupole TOF
- KRH
- Krebs-Ringer/HEPES
- ANOVA
- analysis of variance.
References
- 1. Mortensen O. V., and Amara S. G. (2003) Dynamic regulation of the dopamine transporter. Eur. J. Pharmacol. 479, 159–170 10.1016/j.ejphar.2003.08.066 [DOI] [PubMed] [Google Scholar]
- 2. Sulzer D., Chen T. K., Lau Y. Y., Kristensen H., Rayport S., and Ewing A. (1995) Amphetamine redistributes dopamine from synaptic vesicles to the cytosol and promotes reverse transport. J. Neurosci. 15, 4102–4108 10.1523/JNEUROSCI.15-05-04102.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hamilton P. J., Belovich A. N., Khelashvili G., Saunders C., Erreger K., Javitch J. A., Sitte H. H., Weinstein H., Matthies H. J. G., and Galli A. (2014) PIP2 regulates psychostimulant behaviors through its interaction with a membrane protein. Nat. Chem. Biol. 10, 582–589 10.1038/nchembio.1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vaughan R. A., and Foster J. D. (2013) Mechanisms of dopamine transporter regulation in normal and disease states. Trends Pharmacol. Sci. 34, 489–496 10.1016/j.tips.2013.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ramamoorthy S., Shippenberg T. S., and Jayanthi L. D. (2011) Regulation of monoamine transporters: role of transporter phosphorylation. Pharmacol. Ther. 129, 220–238 10.1016/j.pharmthera.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fog J. U., Khoshbouei H., Holy M., Owens W. A., Vaegter C. B., Sen N., Nikandrova Y., Bowton E., McMahon D. G., Colbran R. J., Daws L. C., Sitte H. H., Javitch J. A., Galli A., and Gether U. (2006) Calmodulin kinase II interacts with the dopamine transporter C terminus to regulate amphetamine-induced reverse transport. Neuron 51, 417–429 10.1016/j.neuron.2006.06.028 [DOI] [PubMed] [Google Scholar]
- 7. Steinkellner T., Yang J. W., Montgomery T. R., Chen W. Q., Winkler M. T., Sucic S., Lubec G., Freissmuth M., Elgersma Y., Sitte H. H., and Kudlacek O. (2012) Ca2+/calmodulin-dependent protein kinase IIα (αCaMKII) controls the activity of the dopamine transporter: implications for Angelman syndrome. J. Biol. Chem. 287, 29627–29635 10.1074/jbc.M112.367219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gorentla B. K., Moritz A. E., Foster J. D., and Vaughan R. A. (2009) Proline-directed phosphorylation of the dopamine transporter N-terminal domain. Biochemistry 48, 1067–1076 10.1021/bi801696n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Batchelor M., and Schenk J. O. (1998) Protein kinase A activity may kinetically upregulate the striatal transporter for dopamine. J. Neurosci. 18, 10304–10309 10.1523/JNEUROSCI.18-24-10304.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Page G., Barc-Pain S., Pontcharraud R., Cante A., Piriou A., and Barrier L. (2004) The up-regulation of the striatal dopamine transporter's activity by cAMP is PKA-, CaMK II- and phosphatase-dependent. Neurochem. Int. 45, 627–632 10.1016/j.neuint.2004.04.002 [DOI] [PubMed] [Google Scholar]
- 11. Pristupa Z. B., McConkey F., Liu F., Man H. Y., Lee F. J., Wang Y. T., and Niznik H. B. (1998) Protein kinase-mediated bidirectional trafficking and functional regulation of the human dopamine transporter. Synapse 30, 79–87 [DOI] [PubMed] [Google Scholar]
- 12. Vaughan R. A., Huff R. A., Uhl G. R., and Kuhar M. J. (1997) Protein kinase C-mediated phosphorylation and functional regulation of dopamine transporters in striatal synaptosomes. J. Biol. Chem. 272, 15541–15546 10.1074/jbc.272.24.15541 [DOI] [PubMed] [Google Scholar]
- 13. Melikian H. E., and Buckley K. M. (1999) Membrane trafficking regulates the activity of the human dopamine transporter. J. Neurosci. 19, 7699–7710 10.1523/JNEUROSCI.19-18-07699.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Q., Bubula N., Brown J., Wang Y., Kondev V., and Vezina P. (2016) PKC phosphorylates residues in the N-terminal of the DA transporter to regulate amphetamine-induced DA efflux. Neurosci. Lett. 622, 78–82 10.1016/j.neulet.2016.04.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Granas C., Ferrer J., Loland C. J., Javitch J. A., and Gether U. (2003) N-terminal truncation of the dopamine transporter abolishes phorbol ester- and substance P receptor-stimulated phosphorylation without impairing transporter internalization. J. Biol. Chem. 278, 4990–5000 10.1074/jbc.M205058200 [DOI] [PubMed] [Google Scholar]
- 16. Foster J. D., Yang J. W., Moritz A. E., Challasivakanaka S., Smith M. A., Holy M., Wilebski K., Sitte H. H., and Vaughan R. A. (2012) Dopamine transporter phosphorylation site threonine 53 regulates substrate reuptake and amphetamine-stimulated efflux. J. Biol. Chem. 287, 29702–29712 10.1074/jbc.M112.367706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moritz A. E., Foster J. D., Gorentla B. K., Mazei-Robison M. S., Yang J. W., Sitte H. H., Blakely R. D., and Vaughan R. A. (2013) Phosphorylation of dopamine transporter serine 7 modulates cocaine analog binding. J. Biol. Chem. 288, 20–32 10.1074/jbc.M112.407874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moritz A. E., Rastedt D. E., Stanislowski D. J., Shetty M., Smith M. A., Vaughan R. A., and Foster J. D. (2015) Reciprocal phosphorylation and palmitoylation control dopamine transporter kinetics. J. Biol. Chem. 290, 29095–29105 10.1074/jbc.M115.667055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rastedt D. E., Vaughan R. A., and Foster J. D. (2017) Palmitoylation mechanisms in dopamine transporter regulation. J. Chem. Neuroanat. 83, 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Copeland B. J., Vogelsberg V., Neff N. H., and Hadjiconstantinou M. (1996) Protein kinase C activators decrease dopamine uptake into striatal synaptosomes. J. Pharmacol. Exp. Ther. 277, 1527–1532 [PubMed] [Google Scholar]
- 21. Foster J. D., Pananusorn B., Cervinski M. A., Holden H. E., and Vaughan R. A. (2003) Dopamine transporters are dephosphorylated in striatal homogenates and in vitro by protein phosphatase 1. Brain Res. Mol. Brain Res. 110, 100–108 10.1016/S0169-328X(02)00645-9 [DOI] [PubMed] [Google Scholar]
- 22. Bauman A. L., Apparsundaram S., Ramamoorthy S., Wadzinski B. E., Vaughan R. A., and Blakely R. D. (2000) Cocaine and antidepressant-sensitive biogenic amine transporters exist in regulated complexes with protein phosphatase 2A. J. Neurosci. 20, 7571–7578 10.1523/JNEUROSCI.20-20-07571.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Samuvel D. J., Jayanthi L. D., Manohar S., Kaliyaperumal K., See R. E., and Ramamoorthy S. (2008) Dysregulation of dopamine transporter trafficking and function after abstinence from cocaine self-administration in rats: evidence for differential regulation in caudate putamen and nucleus accumbens. J. Pharmacol. Exp. Ther. 325, 293–301 10.1124/jpet.107.130534 [DOI] [PubMed] [Google Scholar]
- 24. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 10.1074/mcp.M200025-MCP200 [DOI] [PubMed] [Google Scholar]
- 25. Yang J. W., Vacher H., Park K. S., Clark E., and Trimmer J. S. (2007) Trafficking-dependent phosphorylation of Kv1.2 regulates voltage-gated potassium channel cell surface expression. Proc. Natl. Acad. Sci. U.S.A. 104, 20055–20060 10.1073/pnas.0708574104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484 10.1016/j.cell.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 27. Hadlock G. C., Nelson C. C., Baucum A. J. 2nd, Hanson G. R., and Fleckenstein A. E. (2011) Ex vivo identification of protein-protein interactions involving the dopamine transporter. J. Neurosci. Methods 196, 303–307 10.1016/j.jneumeth.2011.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maiya R., Ponomarev I., Linse K. D., Harris R. A., and Mayfield R. D. (2007) Defining the dopamine transporter proteome by convergent biochemical and in silico analyses. Genes Brain Behav. 6, 97–106 10.1111/j.1601-183X.2006.00236.x [DOI] [PubMed] [Google Scholar]
- 29. Juraszek B., and Nałęcz K. A. (2016) Protein phosphatase PP2A: a novel interacting partner of carnitine transporter OCTN2 (SLC22A5) in rat astrocytes. J. Neurochem. 139, 537–551 10.1111/jnc.13777 [DOI] [PubMed] [Google Scholar]
- 30. Sung U., Jennings J. L., Link A. J., and Blakely R. D. (2005) Proteomic analysis of human norepinephrine transporter complexes reveals associations with protein phosphatase 2A anchoring subunit and 14-3-3 proteins. Biochem. Biophys. Res. Commun. 333, 671–678 10.1016/j.bbrc.2005.05.165 [DOI] [PubMed] [Google Scholar]
- 31. Olsen J. V., and Mann M. (2013) Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteomics 12, 3444–3452 10.1074/mcp.O113.034181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Foster J. D., Pananusorn B., and Vaughan R. A. (2002) Dopamine transporters are phosphorylated on N-terminal serines in rat striatum. J. Biol. Chem. 277, 25178–25186 10.1074/jbc.M200294200 [DOI] [PubMed] [Google Scholar]
- 33. Zhang L., Coffey L. L., and Reith M. E. (1997) Regulation of the functional activity of the human dopamine transporter by protein kinase C. Biochem. Pharmacol. 53, 677–688 10.1016/S0006-2952(96)00898-2 [DOI] [PubMed] [Google Scholar]
- 34. Zhuang X., Masson J., Gingrich J. A., Rayport S., and Hen R. (2005) Targeted gene expression in dopamine and serotonin neurons of the mouse brain. J. Neurosci. Methods 143, 27–32 10.1016/j.jneumeth.2004.09.020 [DOI] [PubMed] [Google Scholar]
- 35. Vaughan R. A., Parnas M. L., Gaffaney J. D., Lowe M. J., Wirtz S., Pham A., Reed B., Dutta S. M., Murray K. K., and Justice J. B. (2005) Affinity labeling the dopamine transporter ligand binding site. J. Neurosci. Methods 143, 33–40 10.1016/j.jneumeth.2004.09.022 [DOI] [PubMed] [Google Scholar]
- 36. Salzer I., Erdem F. A., Chen W. Q., Heo S., Koenig X., Schicker K. W., Kubista H., Lubec G., Boehm S., and Yang J. W. (2017) Phosphorylation regulates the sensitivity of voltage-gated Kv7.2 channels towards phosphatidylinositol-4,5-bisphosphate. J. Physiol. 595, 759–776 10.1113/JP273274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Foster J. D., and Vaughan R. A. (2011) Palmitoylation controls dopamine transporter kinetics, degradation, and protein kinase C-dependent regulation. J. Biol. Chem. 286, 5175–5186 10.1074/jbc.M110.187872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Savitski M. M., Lemeer S., Boesche M., Lang M., Mathieson T., Bantscheff M., and Kuster B. (2011) Confident phosphorylation site localization using the Mascot Delta Score. Mol. Cell. Proteomics 10, M110.003830 10.1074/mcp.M110.003830 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.