Abstract
With the unprecedented rise of drug-resistant pathogens, particularly antibiotic-resistant bacteria, and no new antibiotics in the pipeline over the last three decades, the issue of antimicrobial resistance has emerged as a critical public health threat. Antimicrobial Peptides (AMP) have garnered interest as a viable solution to this grave issue and are being explored for their potential antimicrobial applications. Given their low bioavailability in nature, tailoring new AMPs or strategizing approaches for increasing the yield of AMPs, therefore, becomes pertinent.
The present review focuses on biotechnological interventions directed towards enhanced AMP syn-thesis and revisits existing genetic engineering and synthetic biology strategies for production of AMPs. This review further underscores the importance and potential applications of advanced gene editing technologies for the synthesis of novel AMPs in future.
Keywords: Antimicrobial peptides, antimicrobial resistance, magnifection, genetic engineering, gene editing, CRISPR-Cas9
1. INTRODUCTION
Given the unprecedented rise of drug-resistant pathogens, mainly antibiotic-resistant bacteria, and no new antibiotics in the pipeline over the last three decades, the issue of antimicrobial resistance has emerged as a critical public health threat. In this context, Antimicrobial Peptides (AMPs) have garnered interest and are being explored for their potential role in antimicrobial and therapeutic applications [1-3]. Being one of the important and naturally occurring anti-infectives with potent antimicrobial properties, AMPs find application as promising alternatives to current antibiotics for treatment of disease-resistant infections. AMPs are known to be produced by almost all living species and form an essential component of their innate immunity. Additionally, AMPs have also been found to influence the inflammatory responses of the host during infection. Hence these are also referred to as host-defense peptides. A large number of AMPs with antimicrobial properties against bacteria, virus or even parasites have been documented [4, 5].
At the molecular level, AMPs are small sized peptides and largely cationic in nature owing to the presence of large number lysine or arginine residues in the amino acid primary backbone. The positive nature of the AMPs facilitates interaction with the microbial membranes which are negatively charged on their respective surfaces. However, anionic AMPs have also been reported [5]. The mechanism of
antimicrobial action of AMPs involves interaction with the microbial membrane, followed by their internalization, and further interference with cellular metabolic processes. According to the literature, the effect of AMPs have been described as “disruptive,” when linked to membrane lysis and “undisruptive,” when AMPs cross the membrane barrier, internalize into cells and inhibit normal cellular functions [6, 7]. Their structure, as well as functions, classify these peptides. Regarding structure, AMPs can be categorized into four major groups namely, amphipathic α-helical antimicrobial peptides, β-sheet antimicrobial peptides, extended antimicrobial peptides, and hairpin or loop antimicrobial peptides. Functionally, they are categorized as antiviral, antibacterial, antifungal and antiparasitic peptides [4, 5].
The characteristic properties in AMPs that make them potential candidates for therapeutic application include their distinct mechanism of action, broad-spectrum antibacterial activity, higher efficacy at low concentrations, target specificity, low propensity towards drug resistance, biodegradability, smaller size, and synergistic action with classical antibiotics [7, 8]. The demand for AMPs has therefore seen a surge over the last couple of years owing to the dearth of effective antibiotics and the concomitant rise of antimicrobial-resistant bacteria, which have stopped responding to conventional antibiotics. Furthermore, with possible applications of AMPs in human health, agriculture and food, generation of antimicrobial surfaces being reported, which indicates a promising future for extensive application of these peptides.
While such diverse applications of AMPs call for their continuous and large-scale production, there are several associated limitations with AMPs. The synthesis rates of AMPs that occur naturally are quite low and readily susceptible to proteolytic degradation, owing to the presence of L-amino acids in them which are easy targets for proteases [7]. This often leads to low bioavailability of the AMPs. Once produced, the downstream processing of the AMP involves a tedious process along with inputs of high cost and time. Considering these factors, it is crucial to tailor new AMPs or strategize approaches for increasing yield of high-quality AMPs, which have high rates of biosynthesis, reduced production costs, and enhanced antimicrobial properties.
Genetic engineering has been one important strategy that has been adopted for higher yields or higher quality of AMPs. The use of recombinant microbial platforms such as bacteria and yeast, as well as transgenic plants or animals for expression of AMPs has been explored. However, production of such genetically tailored AMPs is often impacted by low yield and poor quality of the end product. As such, only limited numbers of recombinant AMPs have reached stages of clinical trial, leading to fewer commercial AMPs in the market [9, 10]. Nonetheless, newer approaches to design and synthesize new AMPs with unique properties are also finding their way. The present review focuses on biotechnological approaches for enhanced AMP production and revisits existing genetic engineering and synthetic biology strategies employed for recombinant or transgenic expression of AMPs in recent years. The review further underscores the importance and potential applications of advanced gene editing technologies for synthesis or generation of novel next-generation AMPs.
2. BIOTECHNOLOGICAL APPROACHES FOR ENHANCED ANTIMICROBIAL PEPTIDE PRODUCTION
2.1. Heterologous Expression of Antimicrobial Peptides
Recombinant DNA technology has often been considered as the most effective method for enhanced production of proteins, peptides or enzymes. The technology proves to be advantageous not just regarding reduced time and well-established protocols, but also reduced production costs and easy scale up. Genes expressing the target protein/peptide of interest are cloned into specific vectors for expression in host cellular expression systems. Bacteria and yeast are the most widely used host systems for the expression of recombinant products [11]. In case of AMPs as well, these two expression hosts have been reported to produce more than 95% of the heterologously expressed AMPs [12].
Among bacteria selected for AMP expression, E. coli, particularly strain E. coli BL2l (DE3) has been most popularly used. The choice of E. coli as a suitable host is attributed to its faster growth rate, higher yields, established expression protocols, large commercial availability of expression vectors and cost-effectiveness. Other bacterial systems such as B. subtilis have also been used as hosts for expression of AMPs, but not to the same extent as E. coli. Among yeast, Pichia pastoris has been employed as a potential host. Table 1 shows several AMPs produced recombinantly from engineered microorganisms in the recent years [13-32].
Table 1. Recent evidence on production of recombinant antimicrobial peptides from engineered microorganisms.
| Antimicrobial Peptide | Isolated From | Expression System | Fusion Partner | References |
|---|---|---|---|---|
| LL-37 | Human | E. coli strain JM109 | Thioredoxin–SUMO dual-tag | [13] |
| Cecropin XJ | Bombyx mori | E. coli BL2l (DE3) | Thioredoxin | [14] |
| Ranalexin |
Rana catesbeiana (American bullfrog) |
E. coli BL21 (DE3) | Thioredoxin | [15] |
| Protegrin-1 dimer, LL-37-linker-histatin-5 hybrid peptide | - | E. coli BL21 (DE3) pLysS | Biotin carboxyl carrier protein | [16] |
| Cathelicidin-BF | Bungarus fasciatus (snake) venom | B. subtilis WB800N | SUMO technology | [17] |
| Piscidin 1 and piscidin 3 | Mast cells of hybrid striped sea bass (fish) | E. coli | TrpLE-piscidin fusion partners | [18] |
| β-defensin 2 and LL-37 | Human | E. coli | Self cleaving tag ∆I-CM mini-intein | [19] |
| NZ17074 gene | Arenicola marina (lugworm) | P. pastoris | SUMO | [20] |
| ORBK (cyclic cationic peptide) | Derivative of ORB1 | E. coli | Maltose Binding Protein | [21] |
| Gibberellin Stimulated-Like (GSL) or Snakin peptides | Solanum tuberosum L. (potato) | E. coli | His6-thioredoxin | [22] |
| Snakin-2 (SN2), | Solanum lycopersicum | E. coli | Thioredoxin | [23] |
| a-defensin 5 (HD5) and Mytilin-1 | Human | E. coli | Thioredoxin | [24] |
| Snakin-1 | Plant | P. pastoris | - | [25] |
| Fowlicidin-2 | Chicken | P. pastoris | - | [26] |
| Persea americana var. drymifolia defensin (PaDef) | Avocado fruit | P. pastoris | - | [27] |
| LsGRP1C | Defense-related LsGRP1 protein of Lilium | E. coli | SUMO-based | [28] |
| Dermcidin-derived DCD-1L | Human | E. coli | SUMO-based | [29] |
| Radiolabelled peptide UBI18-35 | Fragment of ubiquicidine, a human natural cationic peptide |
E. coli | Ketosteroid isomerase | [30] |
| Magainin II-cecropin B | Hybrid AMP constructed | Cordyceps militaris | - | [31] |
| Apidaecin | Apis mellifera (honeybee) | P. pastoris | Human serum albumin | [32] |
It becomes clear that AMPs from numerous and different sources have further been expressed recombinantly. Interestingly, the target AMP in most cases is expressed as a fusion protein (i.e., in combination with a carrier protein) and later cleaved from the same [33]. This is primarily to avoid the toxicity of the AMP to the host strain. Carrier proteins have specific anionic properties, which when fused with an AMP, neutralize the overall cationic charge on the AMP. Thus toxicity of the AMP to the host cells is reduced. Also known as fusion partners, these carrier proteins reportedly also increase the solubility of target AMPs being expressed [33]. Common fusion partners that have been used to express and purify AMPs include thioredoxin, Small Ubiquitin-like Modifier (SUMO), Glutathione S-transferase (GST), a Biotin Carboxyl Carrier Protein (BCCP), Green Fluorescent Protein (GFP), etc.
Cleaving the carrier protein from the target AMP requires the use of chemicals or enzymes. While proteolytic cleavage of recombinant AMP from fusion partner has been reported in case of the SN2 peptide from tomato [23], cyanogen bromide induced chemical-cleavage was explored in case of piscidin 1 and 3 [18]. Chemical cleavage is said to be more efficient than enzymatic cleavage, but it is less specific and can bring about modifications in the side chain [34]. Both types of cleavages have been found to leave behind one or two residues at the N-terminus of the cleaved AMP, which is non-native to the AMP. To ease the detachment of the fusion protein from the AMP, conjugation of affinity tags to the fusion partner has also been attempted. This facilitates easy purification by affinity chromatography methods. For example, His6-thioredoxin tagged GSL1 fusion protein expressed in E. coli could be easily purified by affinity chromatography [22]. Sometimes, the fusion partner, such as in the case of glutathione S-transferase, may itself have affinity properties and thus any additional affinity tag is not required [35]. Use of an affinity tag can, however, be an expensive proposition, given the costly resins and buffers required for its purification by chromatography. Another particular group of fusion tags is self-cleaving tags that possess inducible proteolytic activity. When combined with the appropriate affinity tags, they enable easy separation of the fusion tag at less time, effort and cost [36]. Self-cleaving tag ∆I-CM mini-intein was used for recombinant production of AMPs, β-defensin 2 and LL-37 in E. coli [19].
Among the fusion proteins which are used for expression of recombinant AMPs, thioredoxin and SUMO appear to be the most preferred. Insect AMP cecropin XJ was highly expressed in E. coli, as a fusion peptide along with thioredoxin [14]. The purified AMP cecropin XJ not only showed strong antibacterial and antifungal activity but also exhibited toxicity to human cancer cells. Similar antibacterial and anticancer activities have also been reported for the recombinant AMP, ranalexin, expressed in E. coli using thioredoxin tag as a fusion partner [15]. On the other hand, production of recombinant LsGRP1C protein was assisted with the help of yeast SUMO tag, in E. coli host system, which led to a high yield of SUMO-LsGRP1C soluble fusion protein [28]. Likewise, SUMO-based DCD-1L production in E. coli was also reported recently [29].
Interestingly, when Li [13] tried to express AMP, LL37 as a SUMO-based fusion protein, the expression levels were lower than that obtained from the thioredoxin based fusion protein. To increase production levels, novel thioredoxin–SUMO dual-tag was used to express LL37, which was later cleaved by SUMO-protease and purified by size-exclusion chromatography. In this case, the amino acid sequence of the cleaved recombinant AMP LL37 was identical to that of the native peptide and cleavage did not leave behind any additional amino acid residue on the target AMP sequence. Biotin Carboxyl Carrier Protein (BCCP) is another fusion protein that has reportedly been used for expression of AMP [16]. B. subtilis has been used for recombinant expression of cathelicidin-BF sourced from Bungarus fasciatus (snake) venom [17].
The importance of yeasts in genetic engineering and recombinant protein production has increased owing to the ease of their genetic manipulation, the capability of complex post-translational modifications, and rapid growth in the inexpensive medium [37]. Methylotrophic yeast Pichia pastoris has evolved as a popular platform for heterologous expression of recombinant AMPs [38]. Some of the critical features that confer advantages in P. pastoris as a suitable expression host over E. coli include the presence of methanol-induced alcohol oxidase promoter, lack of endotoxins, correct folding ability, large-scale production ability [38]. NZ17074 gene was synthesized and fused with SUMO3 in P. pastoris X-33, following which the carrier protein was cleaved by formic acid [20]. Other AMPs sourced from plants, fruits, chicken have also been expressed in P. pastoris, but have not made use of fusion protein [25-27]. In a recently published interesting study, AMP expression was attempted in a fungus. A recombinant hybrid magainin II-cecropin B AMP was expressed in the mycelium of the medicinal fungus Cordyceps militaris [31]. The purified recombinant AMPs and C. militaris mycelium producing AMPs displayed antibacterial and immunomodulatory effects in mice.
2.2. Transgenic Expression of Antimicrobial Peptides in Plants
Recombinant expression of AMPs in bacteria and yeasts is associated with limitations such as AMP-mediated inhibition of growth in the host, instability of the AMP, and the inability to carry out the correct post-translational modifications [39]. While the use of fusion proteins, as discussed earlier, solves the limitations to an extent, new arenas in genetic engineering such as transgenic expression have garnered significant interest over the last decade.
Transgenes are external DNA sequences which have been introduced into the genome of an organism. Upon insertion of this external DNA, the resulting plant, animal, or microorganism is said to have become transformed. Advances in genetic engineering have enabled the creation of transgenic plants with the help of different transformation techniques like Agrobacterium-mediated transformation etc. These transgenic plants have been explored for expression of desired products such as AMPs or other metabolites from bacterial sources [40]. The development of transgenic plants as a source of industrially relevant and valuable products has been termed as “molecular farming”. Use of higher plants is preferred owing to lesser production costs involved, easier handling and faster scale-up, lesser contamination, and greater stability as compared to transgenic animals [41]. Moreover, higher plants generally synthesize proteins with correct folding, and post-translational modifications, leading to products which are biologically active.
Tobacco plant (Nicotiana tabacum) has been most popularly used as a transgenic expression system. An important advantage of using tobacco as plant production systems is the high volume of biomass that can be produced with only a few processing steps. This expression system is also said to be highly amenable to genetic manipulation, coupled with well-established transformation and regeneration protocols [40, 41]. Other expression systems have also been identified among vegetables (e.g., potato, tomato, cabbage), fruits (e.g., citrus fruits, banana) and cereals (e.g., rice, wheat, barley). Some of the recent evidence on transgenic expression of AMPs are showed in Table 2 [42-54].
Table 2. Antimicrobial peptides expressed transgenically in recent years.
| Antimicrobial Peptide | Source |
Transgenic
Expression Platform |
Property of Transgenic Line or Antimicrobial Peptide | References |
|---|---|---|---|---|
| Floral defensins | Petunia hybrida |
Musa spp. (banana) |
Transgenic lines showed significant resistance against infection of filamentous fungi Fusarium oxysporum f. sp. cubense | [42] |
| Snakin-2 (SN2) | - |
Solanum lycopersicum (tomato) |
Transgenic lines showed enhanced tolerance to Clavibacter michiganensis subsp. michiganensis (Cmm) | [43] |
| Lactoferricin B | Bovine | Nicotiana tabacum (tobacco) | Transgenic plant showed enhanced resistance to bacterial and fungal diseases | [44] |
| PmAMP1 | Pinus monticola (western white pine) | Brassica napus (canola) | In planta expression conferred greater protection against Alternaria brassicae, Leptosphaeria maculans and Sclerotinia sclerotiorum | [45] |
| Cathelicidin anti-microbial peptide (hCAP18/LL-37) | Human | Brassica rapa (Chinese cabbage) | Transgenic plant exhibited varying levels of resistance to bacterial and fungal pathogens | [46] |
| Antimicrobial peptide SN1 | Solanum tuberosum (potato) | Triticum aestivum (wheat) | Transgenic wheat showed increased resistance to Gaeumannomyces graminis var. tritici | [47] |
| Thanatin(S) | Podisus maculiventris (insect) |
Arabidopsis thaliana (flowering plant) |
Transgenic plant acquired resistance to phytopathogenic fungi and bacteria Thanatin(S) has in-vitro antifungal and antibacterial activity |
[48] |
| SP1-1 | de-novo designed | Nicotiana benthamiana (tobacco) | Antimicrobial activity in transgenically produced SP1-1 was only slightly higher than that for the synthetic SP1-1 | [49] |
| Protegrin 1(PG-1) | Porcine leukocytes | Low-alkaloid N. tabacum (tobacco) | Growth of several bacterial and fungal human pathogens inhibited by PG-1 | [50] |
| Cecropin A | Synthetic peptide | Oryza sativa (rice) | Transgenic cecropin A seeds exhibited resistance to fungal and bacterial pathogens | [51] |
| AMP from pro-SmAMP2 gene | Chickweed (Stellaria media L.) |
Solanum tuberosum (potato) | AMP showed enhanced resistance against phytopathogens only in the resistant potato cultivar and not in susceptible potato cultivar | [52] |
| D2A21 | Synthetic peptide |
Carrizo citrange
(citrus fruit) |
Transgenic Carrizo expressing D2A21 showed significant resistance to canker as compared to control plant | [53] |
| LFchimera | Bovine | Nicotiana tabacum (tobacco) | Total protein extracts showed an inhibitory effect on the growth of clinical and phytopathogen indicator bacteria | [39] |
| LL‐37 | Human |
Hordeum vulgare L. (Barley) |
Antimicrobial activity displayed by recombinant LL-37 | [54] |
2.2.1. Antimicrobial Activity of Transgenically Expressed Antimicrobial Peptides
Transgenically expressed AMPs in tobacco plants have been reported for their antibacterial and antifungal activities [55]. In a study by Patiño-Rodríguez et al., [50], a low-alkaloid N. tabacum was used for the expression of Protegrin 1(PG-1), a broad-spectrum AMP, using a transient expression system. The expressed PG-1, showed antimicrobial activity against K. pneumoniae, S. aureus, E. coli, M. bovis BCG and C. albicans. The transient expression refers to the temporary expression of genes in an expression host that are not expressed later on in the development stages of the host. The transient expression was achieved in this case by using magnifection, a platform technology developed by Icon Genetics for the production of recombinant proteins in a plant-based expression system. In magnifection process, a suspension of transgenic Agrobacterium tumefaciens carrying AMP coding genes (introduced into viral vectors such as Tobacco Mosaic Virus (TMV) or Potato Mosaic Virus (PMV)), is infiltrated into the whole of the tobacco plants. The bacterium, once infiltrated, spreads throughout the plant and delivers the vectors for transient expression in the plant. Da Cunha et al., [7] discusses that one of the key advantages of magnifection is the speed at which recombinant peptides can be produced, thereby allowing for easy scale-up and reduced costs. Amphipathic AMP, SP1-1 was transiently expressed in Nicotiana benthamiana using a TMV based transient production system [49]. In this case, the AMP was produced as a fused protein with the viral coat protein of the virus, which was later cleaved using bromocyanide. Furthermore, the possibility of expressing AMPs in whole tobacco plant has also been investigated. Chahardoli and group have reported the expression of Lfchimera, a chimerical peptide, in tobacco hairy roots as well as an in vitro plant culture system [56]. In a following study, they could also successfully express Lfchimera in tobacco leaf [39]. The expressed AMP displayed notable antibacterial activity against clinical and phytopathogenic bacteria. Rajasekaran et al., [57] used computational and synthetic biology approaches to rationally design an tachyplesin1-derived synthetic peptide AGM182, which was transgenically expressed in maize plants. Transgenic maize lines expressing the synthetic peptide AGM182 not only showed 72% reduction in growth of fungus Aspergillus flavus, but also significantly brought down levels of aflatoxin, which are toxic secondary metabolites produced when A. flavus infects maize.
2.2.2. Disease Resistance Conferred by Transgenically Expressed Antimicrobial Peptides
Transgenically expressed AMPs have not just displayed antimicrobial activities, but have also been investigated for their ability to resist bacterial and fungal diseases in other plants. Transgenic expression of plant defensin gene from Jatropha curcas (JcDef) in tobacco showed enhanced resistance against sheath blight disease caused by R. solani [58]. Similarly, overexpression of an antimicrobial protein, Psc-AFP, in transgenic tobacco showed significant enhancement in the disease resistance of tobacco, including complete tolerance to Ralstonia solanacearum and A. alternata exhibited by some of the transgenic lines [59]. However, transgenic potato has been commonly studied for this purpose [60, 61]. Goyal et al., [62] found that transgenic potato expressing msrA3 was not only able to confer resistance against potato pathogen Fusarium solani, but also slowed down floral bud development and extended vegetative phase of the plant. msrA3, expressed in potato, was earlier reported to exhibit resistance against selected fungal and bacterial pathogens [63]. Hevein-like peptides with in-vitro antimicrobial activity are encoded by the chickweed-SmAMP2 gene. Upon introducing this gene into two different cultivars of potato plants (that differed in their resistance to a plant pathogen species; a resistant and a susceptible cultivar), it was found that the expression of AMP from pro-SmAMP2 gene enhanced the resistance against phytopathogens only in case of the resistant cultivar in comparison to the susceptible cultivar [52]. In an interesting study by Hao et al., [53], the author explored the transgenic expression of D2A21 peptide to achieve citrus fruits resistant to canker (a disease caused by P. syringae pv. tabaci and X. citri) and citrus Huanglongbing (HLB; a disease caused by Candidatus Liberibacter asiaticus) was investigated. Firstly, transgenic tobacco expressing D2A21 was obtained, and its applicability in conferring disease resistance to the plant was confirmed. This was then followed by similarly transforming the citrus plant, Carrizo citrange. The transgenic Carrizo citrange expressing D2A21 showed significant resistance to canker as compared to control plants with notable disease symptoms. Sarcotoxin IA, an AMP isolated from the flesh fly (Sarcophaga peregrina), is known to efficiently control different plant pathogenic bacteria. Agrobacterium mediated transformation of mature sweet orange (Citrus sinensis) with gene corresponding for sacrotoxin IA (STX IA), imparted resistance to the plant against Xanthomonas citri subp. Citri infection, which causes citrus canker [64]. STX IA was expressed as a fusion peptide with the PRIa signal peptide.
Apart from expression of AMPs, other mechanisms of conferring resistance in transgenic plants include expression of genes for inducing bacterial toxin tolerance, expression of gene products that directly inhibit pathogen virulence products, activation of general plant defense responses or those that are involved in response or interactions with avirulence factors. Saharan et al., [65] in their chapter has critically reviewed the several such approaches and advances in developing transgenic crop plants resistant to viral, fungal or bacterial diseases.
2.3. Chloroplast Engineering
Chloroplast in plants is known to have their genetic systems and genomes. Therefore, chloroplasts have also been explored as bioreactors for expression of AMPs, as well as resistance to herbicides, insects, or disease [66]. Chloroplast engineering involves alteration of the chloroplast genome (plastome) to introduce target AMP encoding gene sequence. Chloroplast transformation provides a high expression level of AMPs, as compared to that obtained by nuclear transformation. This is because a single plant cell has numerous chloroplasts, thereby yielding a higher copy number of the gene being carried within the chloroplast genome and eventually higher expression levels [66].
The gene encoding for MSI-99, an AMP known to confer resistance against bacterial and fungal diseases, was introduced into the genome of the tobacco plant chloroplast using particle bombardment technique that delivers foreign DNA to a cell culture [67]. Transgenic tobacco plants, so developed, displayed enhanced resistance to fungal disease and also showed inhibitory effects on the mycelial growth of rice blast pathogens. Other examples of AMPs production by chloroplast engineering in tobacco plants include expression of AMPs retrocyclin-101 (RC101) and Protegrin-1 (PG1) [68]. Both RC101 and PG1 were biologically active when expressed in chloroplasts, and prevented bacterial infection caused by E. carotovora to the tobacco plant. RC101 also additionally displayed anti-viral properties. Engineering chloroplast genome has also recently allowed the expression of fused AMPs, interlinked by linkers, to enable the production of multifunctional molecules, while at the same time, reducing possibilities of protease-mediated degradation of the individual AMPs [69].
3. FUTURE PERSPECTIVES
While new frontiers in genetic engineering are paving ways for synthesis and enhanced production of a variety of AMPs, there lies immense future potential in this area which is yet untapped. For example, the applications of gene editing tools and technologies for manipulating genes that code of AMPs is an area that remains inadequately understood and therefore provides tremendous scope for future research and investigations.
Advanced gene editing tools such as Zinc-Finger Nucleases (ZFNs), Transcription Activator-Like Effector Nucleases (TALENS) and Clustered Regularly Interspaced Short Palindromic Repeat-CRISPR-associated protein (CRISPR-Cas) are opening newer opportunities in gene editing space [70]. With such modern tools being applied, manipulation of expression hosts has become relatively easier and can be used for effectively editing target genes towards achieving specific goals. The genome of host cells can, therefore, be easily manoeuvred such that potential and cost-effective recombinant cellular products could be obtained. Gene editing technology can further revolutionize AMP production, especially when there is a greater demand for industrially and therapeutically relevant AMPs.
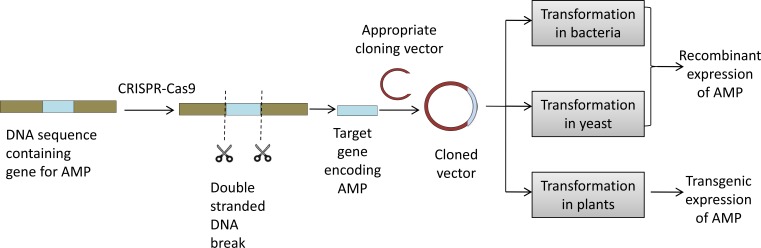
One of the most effective gene editing technology being recently exploited is the CRISPR-Cas, which controls pathogens based on the natural defense mechanism in bacteria [70]. With the help of Cas9 endonuclease, CRISPR RNA (crRNA) can introduce double-stranded DNA breaks at particular sequences of the target DNA. CRISPR-Cas mediated gene repair, disruption, insertion or deletion has been finding applications in several areas of biomedical research, medicine, agriculture, and biotechnology. It will therefore not be wrong to predict that such tools will prove to be extremely relevant and useful for editing AMP coding gene sequences before their insertion in a suitable host. This can, therefore, enable expression of customized AMPs or AMPs with specific properties. Figure. 1 shows how modern gene editing tools such as this can be integrated with existing genetic engineering approaches for easier, faster and enhanced AMP production. Furthermore, CRISPR-Cas9 mediated gene silencing, gene knock out, or manipulation of particular DNA sequences can also facilitate enhanced heterologous AMP production, their correct/faster folding or appropriate post-translational modifications.
Figure 1.
Integrating CRISPR-Cas9 based gene editing with existing genetic engineering approaches for production of novel antimicrobial peptides.
Apart from gene editing, there has also been simultaneous progress in the development of protein engineering techniques or computational tools to tweak proteins or peptides, to be able to meet the growing industrial demands [71, 72]. Such approaches are being recently exploited for the screening of new peptide sequences with antimicrobial properties, or for the computer-aided discovery of AMPs [73-75].
CONCLUSION
Antimicrobial peptides present significant potential as antimicrobial agents for addressing the growing burden of drug-resistant infections. As we near the post-antibiotic area, these peptides are emerging as suitable alternatives to conventional antibiotics. The present review looks at the genetic engineering approaches for the synthesis of AMPs and recent progress which have been made in this direction. While gene repair, disruption, insertion or deletion, using advanced gene editing tools, are making inroads into several areas of biomedical research, medicine, agriculture and biotechnology, their ability to revolutionize antimicrobial therapeutic approaches through the generation of novel AMPs remains yet unexplored. The review sheds light on the tremendous opportunity that CRISPR-Cas9 presents, for development of efficient and effective AMPs, poised to be next generation antimicrobial alternatives.
ACKNOWLEDGEMENTS
The authors acknowledge Maharshi Dayanand University, Rohtak, India for providing infrastructure facility. PS acknowledges the infrastructural support from Department of Science and Technology, New Delhi, Govt. of India, through FIST grant (Grant No. 1196 SR/FST/LS-I/ 2017/4). PS acknowledges Department of Microbiology, Barkatullah University, Bhopal, India for their infrastructural support for D. Sc. Work.
LIST OF ABBREVIATIONS
- AMPs
Antimicrobial Peptides
- BCCP
Biotin Carboxyl Carrier Protein
- BCG
Bacillus Calmette Guerin
- CRISPR-Cas
Clustered Regularly Interspaced Short Palindromic Repeat-CRISPR-associated protein
- DNA
Deoxyribonucleic Acid
- GFP
Green Fluorescent Protein
- GST
Glutathione S-Transferase
- HLB
Huanglongbong
- PMV
Potato Mosaic Virus
- SUMO
Small Ubiquitin like Modifier
- TALENS
Transcription Activator Like Effector Nucleases
- TMV
Tobacco Mosaic Virus
- ZFNs
Zinc-Finger Nucleases
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Seo M.D., Won H.S., Kim J.H., Mishig-Ochir T., Lee B.J. Antimicrobial peptides for therapeutic applications: A review. Molecules. 2012;17:12276–12286. doi: 10.3390/molecules171012276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang H.K., Kim C., Seo C.H., Park Y. The therapeutic applications of antimicrobial peptides (AMPs): A patent review. J. Microbiol. 2017;55:1–2. doi: 10.1007/s12275-017-6452-1. [DOI] [PubMed] [Google Scholar]
- 3.Strempel N., Strehmel J., Overhage J. Potential application of antimicrobial peptides in the treatment of bacterial biofilm infections. Curr. Pharm. Des. 2015;21:67–84. doi: 10.2174/1381612820666140905124312. [DOI] [PubMed] [Google Scholar]
- 4.Bahar A.A., Ren D. Antimicrobial peptides. Pharmaceuticals. 2013;6:1543–1575. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narayana J.L., Chen J.Y. Antimicrobial peptides: Possible anti-infective agents. Peptides. 2015;72:88–94. doi: 10.1016/j.peptides.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Mahlapuu M., Håkansson J., Ringstad L., Björn C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016;6:194. doi: 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.da Cunha N.B., Cobacho N.B., Viana J.F., Lima L.A., Sampaio K.B., Dohms S.S., Ferreira A.C.R., de la Fuente-Núñez C., Costa F.F., Franco O.L., Dias S.C. The next generation of Antimicrobial Peptides (AMPs) as molecular therapeutic tools for the treatment of diseases with social and economic impacts. Drug Discov. Today. 2017;22:234–248. doi: 10.1016/j.drudis.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huerta-Cantillo J., Navarro-García F. Properties and design of antimicrobial peptides as potential tools against pathogens and malignant cells. Investigación en Discapacidad. 2016;5:96–115. [Google Scholar]
- 9.Vlieghe P., Lisowski V., Martinez J., Khrestchatisky M. Synthetic therapeutic peptides: Science and market. Drug Discov. Today. 2010;15:40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Kosikowska P., Lesner A. Antimicrobial Peptides (AMPs) as drug candidates: A patent review (2003-2015). Expert Opin. Ther. Pat. 2016;26:689–702. doi: 10.1080/13543776.2016.1176149. [DOI] [PubMed] [Google Scholar]
- 11.Gupta S.K., Shukla P. Sophisticated cloning, fermentation, and purification technologies for an enhanced therapeutic protein production: A review. Front. Pharmacol. 2017;8:419. doi: 10.3389/fphar.2017.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y.F., Chen Z.X. RAPD: A database of recombinantly-produced antimicrobial peptides. FEMS Microbiol. Lett. 2008;289:126–129. doi: 10.1111/j.1574-6968.2008.01357.x. [DOI] [PubMed] [Google Scholar]
- 13.Li Y. Production of human antimicrobial peptide LL-37 in Escherichia coli using a thioredoxin-SUMO dual fusion system. Protein Expr. Purif. 2013;87:72–78. doi: 10.1016/j.pep.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Xia L., Zhang F., Liu Z., Ma J.I., Yang J. Expression and characterization of cecropinXJ, a bioactive antimicrobial peptide from Bombyx mori (Bombycidae, Lepidoptera) in Escherichia coli. Exp. Ther. Med. 2013;5:1745–1751. doi: 10.3892/etm.2013.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aleinein R.A., Hamoud R., Schäfer H., Wink M. Molecular cloning and expression of ranalexin, a bioactive antimicrobial peptide from Rana catesbeiana in Escherichia coli and assessments of its biological activities. Appl. Microbiol. Biotechnol. 2013;97:3535–3543. doi: 10.1007/s00253-012-4441-1. [DOI] [PubMed] [Google Scholar]
- 16.Orrapin S., Intorasoot S. Recombinant expression of novel protegrin-1 dimer and LL-37-linker–histatin-5 hybrid peptide mediated biotin carboxyl carrier protein fusion partner. Protein Expr. Purif. 2014;93:46–53. doi: 10.1016/j.pep.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Luan C., Zhang H.W., Song D.G., Xie Y.G., Feng J., Wang Y.Z. Expressing antimicrobial peptide cathelicidin-BF in Bacillus subtilis using SUMO technology. Appl. Microbiol. Biotechnol. 2014;98:3651–3658. doi: 10.1007/s00253-013-5246-6. [DOI] [PubMed] [Google Scholar]
- 18.Chen W., Cotton M.L. Expression, purification, and micelle reconstitution of antimicrobial piscidin 1 and piscidin 3 for NMR studies. Protein Expr. Purif. 2014;102:63–68. doi: 10.1016/j.pep.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Luan C., Xie Y.G., Pu Y.T., Zhang H.W., Han F.F., Feng J., Wang Y.Z. Recombinant expression of antimicrobial peptides using a novel self-cleaving aggregation tag in Escherichia coli. Can. J. Microbiol. 2014;60:113–120. doi: 10.1139/cjm-2013-0652. [DOI] [PubMed] [Google Scholar]
- 20.Wang X.J., Wang X.M., Teng D., Zhang Y., Mao R.Y., Wang J.H. Recombinant production of the antimicrobial peptide NZ 17074 in Pichia pastoris using SUMO 3 as a fusion partner. Lett. Appl. Microbiol. 2014;59:71–78. doi: 10.1111/lam.12246. [DOI] [PubMed] [Google Scholar]
- 21.Li Y., Wang J., Yang J., Wan C., Wang X., Sun H. Recombinant expression, purification and characterization of antimicrobial peptide ORBK in Escherichia coli. Protein Expr. Purif. 2014;95:182–187. doi: 10.1016/j.pep.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Meiyalaghan S., Latimer J.M., Kralicek A.V., Shaw M.L., Lewis J.G., Conner A.J., Barrell P.J. Expression and purification of the antimicrobial peptide GSL1 in bacteria for raising antibodies. BMC Res. Notes. 2014;7:777. doi: 10.1186/1756-0500-7-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbel V., Schäfer H., Wink M. Recombinant production of snakin-2 (an antimicrobial peptide from tomato) in E. coli and analysis of its bioactivity. Molecules. 2015;20:14889–14901. doi: 10.3390/molecules200814889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shan Y., Dong Y., Jiang D. Recombinant expression of a novel antimicrobial peptide consisting of human α-defensin 5 and Mytiluscoruscus mytilin-1 in Escherichia coli. J. Korean Soc. Appl. Biol. Chem. 2015;58:807–812. [Google Scholar]
- 25.Kuddus M.R., Rumi F., Tsutsumi M., Takahashi R., Yamano M., Kamiya M., Kikukawa T., Demura M., Aizawa T. Expression, purification and characterization of the recombinant cysteine-rich antimicrobial peptide snakin-1 in Pichia pastoris. Protein Expr. Purif. 2016;122:15–22. doi: 10.1016/j.pep.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Xing L.W., Tian S.X., Gao W., Yang N., Qu P., Liu D., Jiao J., Wang J., Feng X.J. Recombinant expression and biological characterization of the antimicrobial peptide fowlicidin-2 in Pichia pastoris. Exp. Ther. Med. 2016;12:2324–2330. doi: 10.3892/etm.2016.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng D.M., Zhao J.F., Ling X., Dai H.X., Guo Y.J., Gao X.F., et al. Recombinant expression, purification and antimicrobial activity of a novel antimicrobial peptide PaDef in Pichia pastoris. Protein Expr. Purif. 2017;130:90–99. doi: 10.1016/j.pep.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Lin C.H., Pan Y.C., Liu F.W., Chen C.Y. Prokaryotic expression and action mechanism of antimicrobial LsGRP1 C recombinant protein containing a fusion partner of small ubiquitin-like modifier. Appl. Microbiol. Biotechnol. 2017;101:8129–8138. doi: 10.1007/s00253-017-8530-z. [DOI] [PubMed] [Google Scholar]
- 29.Mohanraj U., Kinnunen O., Kaya M.E., Aranko A.S., Viskari H., Linder M. Escherichia coli. bioRxiv. 2018. SUMO-based expression and purification of dermcidin-derived DCD-1L, a human antimicrobial peptide; p. 343418. [Google Scholar]
- 30.Ashcheulova D.O., Efimova L.V., Lushchyk A.Y., Yantsevich A.V., Baikov A.N., Pershina A.G. Production of the recombinant antimicrobial peptide UBI18-35 in Escherichia coli. Protein Expr. Purif. 2018;143:38–44. doi: 10.1016/j.pep.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 31.Zhang M., Shan Y., Gao H., Wang B., Liu X., Dong Y., Liu X., Yao N., Zhou Y., Li X., Li H. Expression of a recombinant hybrid antimicrobial peptide magainin II-cecropin B in the mycelium of the medicinal fungus Cordyceps militaris and its validation in mice. Microb. Cell Fact. 2018;17:18. doi: 10.1186/s12934-018-0865-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao J., de la Fuente-Nunez C., Ou R.W., Torres M.D., Pande S.G., Sinskey A.J., Lu T.K. Yeast-based synthetic biology platform for antimicrobial peptide production. ACS Synth. Biol. 2018;7:896–902. doi: 10.1021/acssynbio.7b00396. [DOI] [PubMed] [Google Scholar]
- 33.Li Y. Carrier proteins for fusion expression of antimicrobial peptides in Escherichia coli. Biotechnol. Appl. Biochem. 2009;54:1–9. doi: 10.1042/BA20090087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y. Recombinant production of antimicrobial peptides in Escherichia coli: A review. Protein Expr. Purif. 2011;80:260–267. doi: 10.1016/j.pep.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Schäfer F., Seip N., Maertens B., Block H., Kubicek J. Purification of GST-tagged proteins. In: Lorsch J.R., editor. Methods in Enzymology (Laboratory Methods in Enzymology: Protein Part D). Cambridge, MA: Academic Press; 2015. pp. 127–139. [DOI] [PubMed] [Google Scholar]
- 36.Li Y. Self-cleaving fusion tags for recombinant protein production. Biotechnol. Lett. 2011;33:869–881. doi: 10.1007/s10529-011-0533-8. [DOI] [PubMed] [Google Scholar]
- 37.Kim H., Yoo S.J., Kang H.A. Yeast synthetic biology for the production of recombinant therapeutic proteins. FEMS Yeast Res. 2015;15:1–6. doi: 10.1111/1567-1364.12195. [DOI] [PubMed] [Google Scholar]
- 38.Ahmad M., Hirz M., Pichler H., Schwab H. Protein expression in Pichia pastoris: Recent achievements and perspectives for heterologous protein production. Appl. Microbiol. Biotechnol. 2014;98:5301–5317. doi: 10.1007/s00253-014-5732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chahardoli M., Fazeli A., Niazi A., Ghabooli M. Recombinant expression of LFchimera antimicrobial peptide in a plant-based expression system and its antimicrobial activity against clinical and phytopathogenic bacteria. Biotechnol. Biotechnol. Equip. 2018;32:714–723. [Google Scholar]
- 40.Yevtushenko D.P., Misra S. Transgenic expression of antimicrobial peptides in plants: Strategies for enhanced disease resistance, improved productivity, and production of therapeutics. In: Rajasekaran K., Cary J.W., Jaynes J.M., Montesinos E., editors. Small Wonders: Peptides for Disease Control. Vol. 1095. Washington, D.C: American Chemical Society; 2012. pp. 445–458. [Google Scholar]
- 41.Wani S.H., Sah S.K. Transgenic plants as expression factories for bio pharmaceuticals. Research and Reviews: J. Bot. Sci. 2015 Phytopathology/ Genes & Diseases- S1. [Google Scholar]
- 42.Ghag S.B., Shekhawat U.K., Ganapathi T.R. Petunia floral defensins with unique prodomains as novel candidates for development of Fusarium wilt resistance in transgenic banana plants. PLoS One. 2012;7:e39557. doi: 10.1371/journal.pone.0039557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balaji V., Smart C.D. Over-expression of snakin-2 and extensin-like protein genes restricts pathogen invasiveness and enhances tolerance to Clavibacter michiganensis subsp. michiganensis in transgenic tomato (Solanumly copersicum). Transgenic Res. 2012;21:23–37. doi: 10.1007/s11248-011-9506-x. [DOI] [PubMed] [Google Scholar]
- 44.Fukuta S., Kawamoto K.I., Mizukami Y., Yoshimura Y., Ueda J.I., Kanbe M. Transgenic tobacco plants expressing antimicrobial peptide bovine lactoferricin show enhanced resistance to phytopathogens. Plant Biotechnol. 2012;29:383–389. [Google Scholar]
- 45.Verma S.S., Yajima W.R., Rahman M.H., Shah S., Liu J.J., Ekramoddoullah A.K., Kav N.N. A cysteine-rich antimicrobial peptide from Pinus monticola (PmAMP1) confers resistance to multiple fungal pathogens in canola (Brassica napus). Plant Mol. Biol. 2012;79:61–74. doi: 10.1007/s11103-012-9895-0. [DOI] [PubMed] [Google Scholar]
- 46.Jung Y.J., Lee S.Y., Moon Y.S., Kang K.K. Enhanced resistance to bacterial and fungal pathogens by overexpression of a human cathelicidin antimicrobial peptide (hCAP18/LL-37) in Chinese cabbage. Plant Biotechnol. Rep. 2012;6:39–46. doi: 10.1007/s11816-011-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rong W., Qi L., Wang J., Du L., Xu H., Wang A., Zhang Z. Expression of a potato antimicrobial peptide SN1 increases resistance to take-all pathogen Gaeumannomyces graminis var. tritici in transgenic wheat. Funct. Integr. Genomics. 2013;13:403–409. doi: 10.1007/s10142-013-0332-5. [DOI] [PubMed] [Google Scholar]
- 48.Wu T., Tang D., Chen W., Huang H., Wang R., Chen Y. Expression of antimicrobial peptides thanatin (S) in transgenic Arabidopsis enhanced resistance to phytopathogenic fungi and bacteria. Gene. 2013;527:235–242. doi: 10.1016/j.gene.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 49.Zeitler B., Bernhard A., Meyer H., Sattler M., Koop H.U., Lindermayr C. Production of a de-novo designed antimicrobial peptide in Nicotiana benthamiana. Plant Mol. Biol. 2013;81:259–272. doi: 10.1007/s11103-012-9996-9. [DOI] [PubMed] [Google Scholar]
- 50.Patiño-Rodríguez O., Ortega-Berlanga B., Llamas-González Y.Y., Flores-Valdez M.A., Herrera-Díaz A., Montes-de-Oca-Luna R., Korban S.S., Alpuche-Solís Á.G. Transient expression and characterization of the antimicrobial peptide protegrin-1 in Nicotiana tabacum for control of bacterial and fungal mammalian pathogens. Plant Cell Tissue Organ Cult. 2013;115:99–106. [Google Scholar]
- 51.Bundó M., Montesinos L., Izquierdo E., Campo S., Mieulet D., Guiderdoni E., Rossignol M., Badosa E., Montesinos E., San Segundo B., Coca M. Production of cecropin A antimicrobial peptide in rice seed endosperm. BMC Plant Biol. 2014;14:102. doi: 10.1186/1471-2229-14-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vetchinkina E.M., Komakhina V.V., Vysotskii D.A., Zaitsev D.V., Smirnov A.N., Babakov A.V., Komakhin R.A. Expression of plant antimicrobial peptide pro-SmAMP2 gene increases resistance of transgenic potato plants to Alternaria and Fusarium pathogens. Russ. J. Genet. 2016;52:939–951. [PubMed] [Google Scholar]
- 53.Hao G., Zhang S., Stover E. Transgenic expression of antimicrobial peptide D2A21 confers resistance to diseases incited by Pseudomonas syringae pv. tabaci and Xanthomonas citri, but not Candidatus Liberibacter asiaticus. PLoS One. 2017;12:e0186810. doi: 10.1371/journal.pone.0186810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holásková E., Galuszka P., Mičúchová A., Šebela M., Öz M.T., Frébort I. Molecular farming in barley: Development of a novel production platform to produce human antimicrobial peptide LL‐37. Biotechnol. J. 2018;13:e1700628. doi: 10.1002/biot.201700628. [DOI] [PubMed] [Google Scholar]
- 55.Cary J.W., Rajasekaran K., Jaynes J.M., Cleveland T.E. Transgenic expression of a gene encoding a synthetic antimicrobial peptide results in inhibition of fungal growth in vitro and in planta. Plant Sci. 2000;154:171–181. doi: 10.1016/s0168-9452(00)00189-8. [DOI] [PubMed] [Google Scholar]
- 56.Chahardoli M., Fazeli A., Ghabooli M. Recombinant production of bovine Lactoferrin-derived antimicrobial peptide in tobacco hairy roots expression system. Plant Physiol. Biochem. 2018;123:414–421. doi: 10.1016/j.plaphy.2017.12.037. [DOI] [PubMed] [Google Scholar]
- 57.Rajasekaran K., Sayler R.J., Sickler C.M., Majumdar R., Jaynes J.M., Cary J.W. Control of Aspergillus flavus growth and aflatoxin production in transgenic maize kernels expressing a tachyplesin-derived synthetic peptide, AGM182. Plant Sci. 2018;270:150–156. doi: 10.1016/j.plantsci.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 58.Wang Q., Zhu S., Liu Y., Li R., Tan S., Wang S., Tang L., Chen F. Overexpression of Jatropha curcas defensin (JcDef) enhances sheath blight disease resistance in tobacco. J. Phytopathol. 2017;165:15–21. [Google Scholar]
- 59.Luo X.M., Xie C.J., Wang D., Wei Y.M., Cai J., Cheng S.S., Yang X.Y., Sui A.P. Psc-AFP from Psoralea corylifolia L. overexpressed in Pichia pastoris increases antimicrobial activity and enhances disease resistance of transgenic tobacco. Appl. Microbiol. Biotechnol. 2017;101:1073–1084. doi: 10.1007/s00253-016-7768-1. [DOI] [PubMed] [Google Scholar]
- 60.Almasia N.I., Bazzini A.A., Hopp H.E., Vazquez‐Rovere C.E. Overexpression of snakin‐1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol. Plant Pathol. 2008;9:329–338. doi: 10.1111/j.1364-3703.2008.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rivero M., Furman N., Mencacci N., Picca P., Toum L., Lentz E., Bravo-Almonacid F., Mentaberry A. Stacking of antimicrobial genes in potato transgenic plants confers increased resistance to bacterial and fungal pathogens. J. Biotechnol. 2012;15:334–343. doi: 10.1016/j.jbiotec.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 62.Goyal R.K., Hancock R.E., Mattoo A.K., Misra S. Expression of an engineered heterologous antimicrobial peptide in potato alters plant development and mitigates normal abiotic and biotic responses. PLoS One. 2013;8:e77505. doi: 10.1371/journal.pone.0077505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Osusky M., Osuska L., Hancock R.E., Kay W.W., Misra S. Transgenic potatoes expressing a novel cationic peptide are resistant to late blight and pink rot. Transgenic Res. 2004;13:181–190. doi: 10.1023/b:trag.0000026076.72779.60. [DOI] [PubMed] [Google Scholar]
- 64.Kobayashi A.K., Vieira L.G.E. BespalhokFilho, J.C.; Leite, R.P.; Pereira, L.F.P.; Molinari, H.B.C.; Marques, V.V. Enhanced resistance to citrus canker in transgenic sweet orange expressing the sarcotoxin IA gene. Eur. J. Plant Pathol. 2017;149:865–873. [Google Scholar]
- 65.Saharan V., Jain D., Pareek S., Pal A., Kumaraswamy R.V., Jakhar S.K., Singh M. Viral, fungal and bacterial disease resistance in transgenic plants. In: Al-Khayri J.M., Jain S.M., Johnson D.V., editors. Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits. Vol. 2. Berlin: Springer; 2016. pp. 627–656. [Google Scholar]
- 66.Khan M.S. Plastid genome engineering in plants: Present status and future trends. Mol. Plant Breed. 2012;3:91–102. [Google Scholar]
- 67.Wang Y.P., Wei Z.Y., Zhang Y.Y., Lin C.J., Zhong X.F., Wang Y.L., Ma J.Y. M, J.; Xing, S.-C. Chloroplast-expressed MSI-99 in tobacco improves disease resistance and displays inhibitory effect against rice blast fungus. Int. J. Mol. Sci. 2015;16:4628–4641. doi: 10.3390/ijms16034628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee S.B., Li B., Jin S., Daniell H. Expression and characterization of antimicrobial peptides Retrocyclin‐101 and Protegrin‐1 in chloroplasts to control viral and bacterial infections. Plant Biotechnol. J. 2011;9:100–115. doi: 10.1111/j.1467-7652.2010.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoelscher M., Forner J., Bock R. Chloroplast produced antimicrobial peptide fusions for pharma and plant protection.; Helsinki Congress Paasitorni; Finland. 2018. p. 196. [Google Scholar]
- 70.Dangi A.K., Sinha R., Dwivedi S., Gupta S.K., Shukla P.S. Cell line techniques and gene editing tools for antibody production: A review. Front. Pharmacol. 2018;9:630. doi: 10.3389/fphar.2018.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baweja M., Nain L., Kawarabayasi Y., Shukla P. Current technological improvements in enzymes toward their biotechnological applications. Front. Microbiol. 2016;7:965. doi: 10.3389/fmicb.2016.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumar V., Baweja M., Liu H., Shukla P. Microbial enzyme engineering: Applications and perspectives. In: Shukla P., editor. Recent Advances in Applied Microbiology. Singapore: Springer; 2017. pp. 259–273. [Google Scholar]
- 73.Tucker A.T., Leonard S.P., DuBois C.D., Knauf G.A., Cunningham A.L., Wilke C.O., Trent M.S., Davies B.W. Discovery of next-generation antimicrobials through bacterial self-screening of surface-displayed peptide libraries. Cell. 2018;172:618–628. doi: 10.1016/j.cell.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haney E.F., Brito-Sánchez Y., Trimble M.J., Mansour S.C., Cherkasov A., Hancock R.E. Computer-aided discovery of peptides that specifically attack bacterial biofilms. Sci. Rep. 2018;8:1871. doi: 10.1038/s41598-018-19669-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guralp S.A., Murgha Y.E., Rouillard J.M., Gulari E. From design to screening: A new antimicrobial peptide discovery pipeline. PLoS One. 2013;8:e59305. doi: 10.1371/journal.pone.0059305. [DOI] [PMC free article] [PubMed] [Google Scholar]