Abstract
Background:
α-Amylases are starch-degrading enzymes and used widely, the study on thermostability of α-amylase is a central requirement for its application in life science and biotech-nology.
Objective:
In this article, our motivation is to study how the effect of Ca2+ ions on the structure and thermal characterization of α-amylase (AGXA) from thermophilic Anoxybacillus sp.GXS-BL.
Methods:
α-Amylase activity was assayed with soluble starch as the substrate, and the amount of sugar released was determined by DNS method. For AGXA with calcium ions and without calcium ions, optimum temperature (Topt), half-inactivation temperature (T50) and thermal inactivation (half-life, t1/2) was evaluated. The thermal denaturation of the enzymes was determined by DSC and CD methods. 3D structure of AGXA was homology modeled with α-amylase (5A2A) as the template.
Results:
With calcium ions, the values of Topt, T50, t1/2, Tm and ΔH in AGXA were significantly high-er than those of AGXA without calcium ions, showing calcium ions had stabilizing effects on α-amylase structure with the increased temperature. Based on DSC measurements AGXA underwent thermal denaturation by adopting two-state irreversible unfolding processes. Based on the CD spectra, AGXA without calcium ions exhibited two transition states upon unfolding, including α-helical contents increasing, and the transition from α-helices to β-sheet structures, which was obviously dif-ferent in AGXA with Ca2+ ions, and up to 4 Ca2+ ions were located on the inter-domain or intra-domain regions according to the modeling structure.
Conclusion:
These results reveal that Ca2+ ions have pronounced influences on the thermostability of AGXA structure.
Keywords: α-amylase, calcium ions, circular dichroism, differential scanning calorimetry, homology modeling, thermostability
1. INTRODUCTION
Thermophilic bacteria are extremophiles adapted to life at high temperatures (optimum growth is observed at temperatures above 45°C) [1]. They inhabit separate permanently hot ecological niches, such as areas with geothermal and volcanic activity [2]. Their enzymes (thermozymes) have unique characteristics that include increased temperature, chemical, and pH stability [3]. Among these interesting characteristics thermostability is an important consideration for proteins, particularly for those which are involved in biological processes at elevated temperatures [4].
α-Amylases (E.C. 3.2.1.1.) are starch-degrading enzymes that catalyze the hydrolysis of internal a-1,4-O-glycosidic bonds in polysaccharides while retaining the α-anomeric configuration of the products [5]. They mostly belong to family 13 (GH-13) of the glycoside hydrolase group of enzymes [6, 7]. α-Amylases can be obtained from a variety of organisms, including plants, animals and microbes [8]. Bacterial α-amylases, especially those from the bacillus species, are used widely in the food, pharmaceutics, textile, paper, detergent and bioenergy industries [9]. Thermostable α-amylases are especially available from certain thermophilic bacillus species of bacteria, such as Geobacillus stearothermophilus, Anoxybacillus, Bacillus subtilis, Bacillus licheniformis, Bacillus amyloliquefaciens, Bacillus alvei, Bacillus cereus, and Bacillus globisporus [10]. Compared to the well-studied genera Geobacillus or Bacillus, Anoxybacillus was a new genus first proposed by Pikuta in the year 2000 [11]. To date, a total of 22 species and two subspecies of Anoxybacillus with validly published names have been reported (http://www.bacterio.net). Most reported data have revealed that members of Anoxybacillus produce biotechnologically important enzymes that are thermostable and tolerant of alkaline pH conditions [12].
Calcium ions often have crucial roles in structure, function, and stability of α-amylases and are especially crucial in thermophilic α-amylases [13, 14]. They are considered to be important for maintaining protein structures in their correct conformations and for resisting thermal inactivation of enzymes [15, 16]. It has been shown that the removal of calcium ions from barley α-amylase irreversibly inactivate the enzyme, whereas the bacterial α-amylases restored its activity after the addition of calcium ions [17]. Some reports indicate that the role of calcium ions in α-amylases is mainly structural, as their catalytic sites are far from the calcium-binding sites [18, 19]. Although most α-amylases are Ca2+-dependent, there are reports of Ca2+-independent α-amylases [20-22], and there are also some α-amylases that are inhibited by Ca2+ ions [23, 24]. The study of the effect of calcium ions on the activity and stability of α-amylases from thermophiles may be helpful to determine a mechanism of Ca2+-binding protein in the thermal environment, furthermore to wider application range of the enzymes. We have previously reported that alkalitolerant α-amylase (AGXA) from thermophilic Anoxybacillus sp.GXS-BL was moderately thermostable and did not require Ca2+ for its activity [25]. In the present study, we investigate the impact of calcium ions on the thermal characterization of the α-amylase.
It is reported that the α-amylases from thermophilic Anoxybacillus species (ASKA and ADTA) and Geobacillus thermoleovorans (Pizzo, GTA, and Gtamy II), and halophilic Bacillus aquimaris (BaqA) have been proposed as a novel subfamily of the α-amylase family, which exhibiting (1) high maltose production; (2) the ability to degrade raw starch; (3) a long C-terminal sequence containing five conserved aromatic residues; (4) dual tryptophan residues between CSR-V and CSR-II; and (5) a unique stretch of amino acids (LPDIx) in CSR-V [26]. The crystal structure of GTA (PDB ID: 4E2O) provided the first insight into the overall structure and Ca2+ binding sites of this new GH13 subfamily of α-amylases [27]. To increase understanding of the unique GH13 subfamily, the homologous structures of ASKA (PDB ID: 5A2A, 5A2B and 5A2C) from Anoxybacillus in the novel GH13 subfamily were published recently [28]. The results show that there are four Ca2+ binding sites in all three ASKA structures. And sequence alignment suggests that all the residues that interact with the four Ca2+ ions are conserved in most α-amylases from Anoxybacillus spp. In this article, we just want to study how the effect of Ca2+ ions on the structure and thermal characterization of AGXA using homology modeling, Differential Scanning Calorimetry (DSC) and Circular Dichroism (CD) techniques. To our knowledge, this is the first report on the calorimetric and spectroscopic characteristics of the novel GH13 subfamily. As revealed in this article, the roles of Ca2+ ions on the structure, activity and thermostability of AGXA are interesting and profound. We think these results may be helpful to deep insight into the mechanisms of Ca2+-binding α-amylase adapting to the thermal environment.
2. MATERIALS AND METHODS
2.1. Materials
Anoxybacillus sp.GXS-BL was utilized as the α-amylase gene donor strain, which was isolated from the Tengchong hot spring of Yunnan province of China (GenBank Accession No. JF831203). Escherichia coli DH5α and E. coli M15 [pREP4] from Novagen (WI, USA) were used as cloning and expression hosts, respectively. The plasmid vector used for cloning and expression was pQE30 (Novagen, USA). Isopropyl-β-D-thiogalactopyranoside (IPTG), ampicillin and kanamycin were prepared from Sangon (Shanghai, China). Nickel-nitrilotriacetic acid (Ni-NTA) metal-affinity chromatography matrices were obtained from Qiagen (CA, USA). HiTrap desalting G-25 and Superdex 75 10/300GL were procured from GE Health (Uppsala, Sweden). Amicon Ultra15 Centrifugal Filter Units (MWCO, 10 kDa) and D-tube dialyzers (MWCO, 12 kDa) were procured from Millipore (MA, USA). Soluble potato starch, calcium chloride and other used chemicals were analytical-grade and obtained from Sigma-Aldrich (St. Louis, USA). All solutions were prepared in Milli-Q (Millipore, USA) water. After treatment with EDTA purified α-amylase samples in 50 mM Tris-HCl buffer (pH 8.0) were dialyzed extensively in the same buffer to remove EDTA. All Ca2+-removal and following steps were performed in plastic vessels to avoid any Ca2+ contamination from any glass. For Ca2+-added proteins, small aliquots of a 1 M Ca2+ solution was added to Ca2+-free proteins to achieve a final Ca2+ concentration of 5 mM.
2.2. Protein Estimation
The purified protein samples were concentrated by an Amicon centrifugal filter unit (MWCO, 10 kDa), enzyme protein concentration was measured using a Bradford assay using Bovine Serum Albumin (BSA) as the standard [29].
2.3. α-Amylase Activity
α-Amylase activity was assayed with soluble starch as the substrate, and the amount of sugar released was determined by using the 3,5-dinitrosalicylic acid (DNS) method [30]. Ten μl of diluted purified Ca2+-added and Ca2+-free enzyme (0.1 mg/ml) were added to 490 μl of Tris–HCl buffer (50 mM, pH 8.0) at 1% (w/v) soluble-starch. After a 10-min incubation at the specified temperature, the reaction was stopped by adding 500 μl of DNS reagent, the absorbance was recorded at 540 nm using a DU800 UV-Visible spectrophotometer (BeckmanCoulter, Germany), and the reducing sugar equivalent was determined using a maltose standard. All enzyme activity measurements were performed in triplicate.
2.4. Homology Modeling
Three-dimensional structure of the α-amylase was homology modeled using BIOVIA Discovery Studio 2016 software. The crystal structure of α-amylase (5A2A, at 1.9 Å resolution) from Anoxybacillus sp. SK3-4 was used as the template [28]. After aligning the model sequence to the template, a 3D model was built using Modeller 9.15 [31]. Prior to the modeling, the ligand glucose and crystallographic water molecules were removed from the template structure. Ten AGXA models were generated. Models with the lowest energy value (DOPE score) [32] were selected and evaluated by the Root-Mean Square Deviation (RMSD), Ramachandran plot [33] and Profiles-3D protocols [34]. A model with the best scores was selected for refinement and validation.
2.5. Thermal Stabilities
Optimum temperature (Topt) for activity was determined in a range of 40°C to 80°C for 10 min in 50 mM Tris-HCl buffer (pH 8.0). The highest activity at a specified temperature was designated 100%. Half-inactivation temperature (T50) was the temperature where the enzyme retains 50% of its initial activity after 30 min of incubation. The α-amylase was incubated at temperatures between 50-80°C for 30 min, followed by cooling at 4°C for 10 min, and residual activities were then measured at standard conditions. Half-life (t1/2) was the time at which the enzyme was reduced to 50% of its initial activity at a set temperature. Thermal inactivation of the α-amylase (0.1 mg/ml)was determined at 70°C by taking samples at regular intervals which were then cooled on ice, and residual activity was measured as described above. All reactions were performed in triplicate.
2.6. DSC Measurements
The thermal denaturation was determined by differential scanning calorimeter (Microcal VP Capillary DSC system, Malvern). Ca2+-added and Ca2+-free protein samples (0.80 mg/ml) in 50 mM Tris-HCl buffer (pH 8.0) were degassed with a Microcal thermovac instrument (GE Healthcare). Then, 400 μl of protein and buffer were loaded into the sample and reference cells, respectively. Measurements were performed by increasing the temperature from 10°C to 120°C at 1°C/min scan rate. Protein sample scans were corrected by subtraction of the respective buffer from the baseline values. Molar excess heat capacities (Cp) were obtained by normalizing to the sample concentration. The data were analyzed according to a non-two state model using Origin 7.0 software (Microcal). The apparent Tm was determined as the temperature corresponding to maximum Cp, the calorimetric enthalpy ΔH corresponded to the area under the peak of the heat capacity versus temperature graph, and the van’t Hoff enthalpy ΔHv was determined by the shape of the transition peak. The test was repeated three times.
2.7. CD Spectroscopy Measurements
The Chirascan instrument (Applied Photophysics) was also utilized to conduct the thermal denaturation experiments. The spectrometer was equipped with a Peltier-type temperature controller attached to a water bath. Ca2+-added and Ca2+-free protein samples (0.28 mg/ml) in 5 mM Tris-HCl buffer (pH 8.0) were degassed as described above. Next, 300 μl of a protein sample was pipetted into Hellma quartz cuvettes with a path length of 0.1 cm. The CD spectra were observed in the 201-260 nm range in 1 nm steps, and the time-per-point was 0.7 s. The bandwidth was set to 0.8 nm, and the temperature was varied from 35°C-85°C in step with a ramp rate of 1°C/min. The sample temperature was measured directly using an external probe immersed in the sample. The data was processed by Global3 software.
3. RESULTS AND DISCUSSION
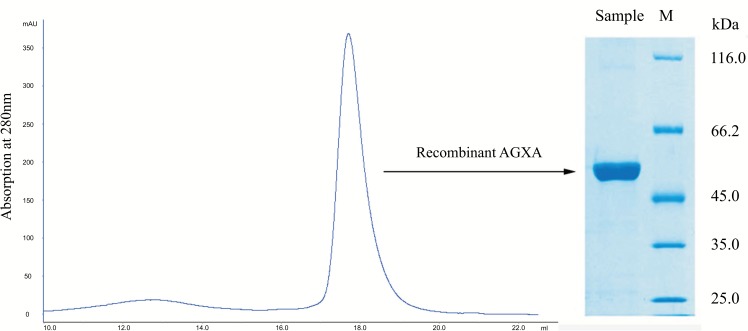
In this work, the recombinant α-amylase AGXA was overexpressed in E. coli M15 [pREP4] and purified by Ni-NTA and size-exclusion chromatography. The AGXA purity was showed by SDS-PAGE and the molecular mass was about 55 kDa (Figure 1).
Figure 1.
Recombinant α-amylase AGXA purification by gel filtration. Size-exclusion chromatography (left) showed a single narrow peak, and the SDS-PAGE analysis (right) also showed a single band, indicating sample homogeneity.
3.1. Homology Modeling
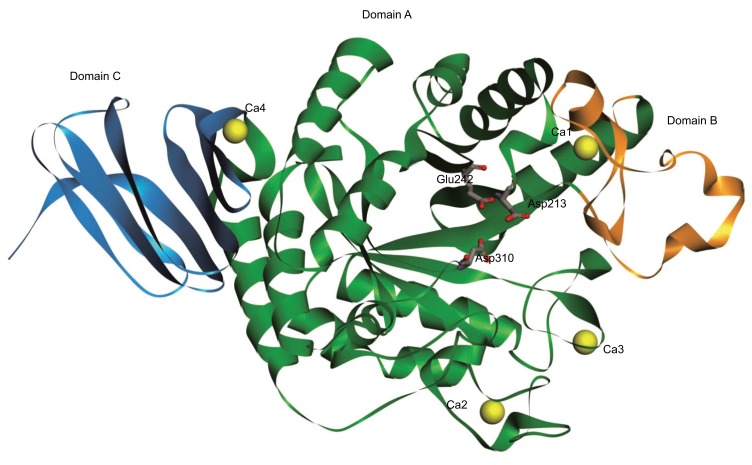
Searching sequences by similarity, results showed that the highest similarity with AGXA was observed in 5A2A (97%), followed by 4E2O (72%), with the others showing less than 35% similarity. A 3D model of AGXA was constructed using Discovery Studio 2016 software, with 5A2A used as the template. As shown in Figure 2, the model structure consists of three domains containing characteristic of GH13 α-amylase [35]: central domain A (residues 26–139 / 187–393 and the three catalytic residues of D213, E242 and D310) made up of a (α/β)8 TIM barrel; domain B (residues 140–186); and domain C (residues 394–475). The overall structure of the AGXA model was compact and similar to Geobacillus thermoleovorans CCB US3 UF5 α-amylase (GTA) [27], Bacillus KSM-1378 α-amylase (LAMY) [36], and a truncated Bacillus sp. strain TS-23 α-amylase (BACΔNC) [37]. There were four Ca2+ ions (Ca1–4) bound to the model structure. Ca1 was in the region between domain A and domain B, coordinated by the N139, D182, H217, and E173 residues and three water molecules. Ca2 and Ca3 were both located in domain A, with Ca2 coordinated by the N44, N46, N49, D50, G63, and D65 residues and a water molecule, and Ca3 coordinated by the N92, E109, and E110 residues and three water molecules. Ca4 was in the region between domain A and domain C, coordinated by the E400 residue and five water molecules. All the identified Ca2+-ion locations were consistent with what was been observed in 5A2A [28].
Figure 2.
Model structure of AGXA. The model structure of AGXA predicted by homology modeling based on the crystal structure of Anoxybacillus sp. SK3-4 α-amylase (PDB code: 5A2A). The three conserved active site residues are labeled by sticks. Domains A, B, and C are shown in green, orange, and blue, respectively. The calcium ions are shown in yellow. (The color version of the figure is available in the electronic copy of the article).
3.2. Effects of Calcium Ions on the Thermal Stability of Recombinant AGXA
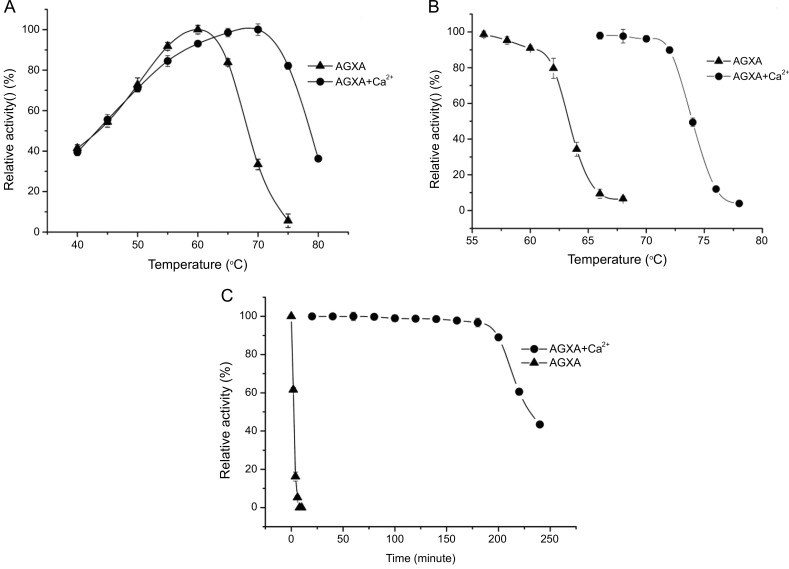
Optimum temperature of maximal enzymatic activity (Topt) was an important parameter in characterizing the thermal adaptation process [38]. Topt of the recombinant AGXA with calcium ions and without calcium ions were determined by evaluating their activity at different temperatures (40–80 °C). As the recombinant AGXA was stable at pH 6.0-9.5(above 70% of its highest activity) [25], and so in the above temperature range, the change in pH of buffer Tris-HCl was small and negligible. The results showed that the recombinant AGXA without calcium ions was active from 40°C to 70°C, with the Topt observed at 60°C, which was similar to the values previously reported for the alkaliphilic α-amylases from Bacillus sp. KSM-K38 [39] and in agreement with the 60°C and pH 8.0 reported for Anoxybacillus sp.SK3-4 and DT3-1 α-amylases (ASKA and ADTA, respectively) [40], which were the closest α-amylases to AGXA by sequence comparisons. The recombinant AGXA was active over a wide range of temperatures with calcium ions from 40 to 80°C and maintained more than 80% of its maximum activity between 55 and 75°C, with the maximal enzymatic activity observed at 70.0°C (Figure 3A). This value was similar to what has been reported for α-amylase from A. beppuensis TSSC-1(70°C and pH 5.5) [41] and GTA (70 °C and pH 6.0) [27].
Figure 3.
The optimal temperature curves (A), and stability curves (B), the inactivation curves at 70 °C of AGXA with Ca2+ and without Ca2+ ions (C). The optimal temperature curves were determined by measuring the enzymatic activities at a temperature range of 40–80 °C with 5 °C intervals. The stability curves were determined by measuring the enzymatic activity after treatment from 60 to 80 °C (with 2 °C steps) for 30 min following a 10-min incubation on ice. Inactivation curves were determined by incubating enzymes at 70 °C and enzymatic activities were measured at various time points. Error bars were obtained from the standard deviation of triplicates.
Half-inactivation temperature (T50) of the recombinant AGXA was individually assessed at different incubation temperatures in the absence and presence of Ca2+. After heat treatment from 60 to 80°C (with 2 °C steps) for 30 min, the residual activity of the α-amylase was assessed. As showed in Figure 3B, the T50 value of AGXA without calcium ions was 63.5°C, while 73.8°C with calcium ions, which was 10.3°C higher than that observed in AGXA without calcium ions. As T50 approaches or reaches the critical denaturation temperature, using the thermostable curve, we could speculate that the denaturation temperatures of the α-amylase with and without Ca2+ were likely to be approximately 65°C, and 75°C, respectively. These values were slightly higher than those reported for α-amylase in Aspergillus oryzae (TAKA) [42].
Thermal inactivation of the recombinant AGXA with calcium ions and without calcium ions was evaluated by incubating the enzymes at 70°C and residual activity was measured at various incubation times. As shown in Figure 3C, without calcium ions, recombinant AGXA lost its activity completely at 70°C after 4 min of incubation, while with Ca2+, more than 90% of the activity was retained even after 200 min of incubation at the same temperature. It was indicated that, with Ca2+, the α-amylase was 50-fold more thermostable than without calcium ions at 70°C. Similar results were shown for GTA [27], ASKA and ADTA [40]. GTA without Ca2+ lost its activity after 15 min, whereas GTA with 2.0 mM Ca2+ retained 85% of its activity even after 72 h of incubation at 70°C [34]. With Ca2+, ASKA and ADTA could remain stable for up to 48 h at 70°C [40]. In comparison, the thermostability in ASKA and ADTA were greater than in AGXA.
3.3. DSC Measurements
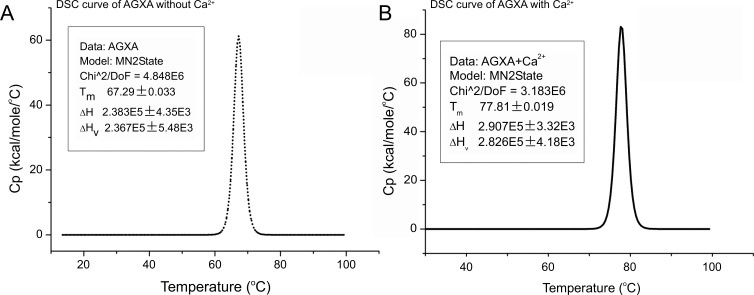
Thermal denaturation of recombinant AGXA was investigated using DSC. DSC is a tool used to study thermodynamic properties of macromolecules and protein unfolding processes that gives the unfolding temperature (Tm) and enthalpy (ΔH) of a protein simultaneously [43]. After data processing, including buffer correction, normalization, and baseline subtraction, the results are shown in Figure 4. The transition curves of recombinant AGXA with Ca2+ and without Ca2+ were fitted using a non-two-state model, and the values of ΔH/ΔHv were approximately equal to 1.0, showing the α-amylases underwent thermal denaturation following a two-state irreversible model. In the absence of calcium ions, the Tm value of recombinant AGXA was 67.3°C, and in the presence of calcium ions, the thermal denaturation curve was found to be shifted to a higher temperature range, with the Tm value at 77.8°C, where the increased temperature was 10.5°C. Tm was the midpoint temperature of transition, where the folded and the unfolded states of the protein were in equilibrium. The higher the Tm of a protein, the higher its thermal stability [44]. It has been reported that, with saturated Ca2+ and without Ca2+, differences in unfolding temperatures (ΔTm) were large for α-amylase from Bacillus licheniformis (BLA) and α-amylase from Bacillus amyloliquefaciens (BAA), for which the values were 50°C, and 48°C, respectively [45]. However, for the Alteromonas haloplanctis α-amylase (AHA), Bacillus halmapalus α-amylase (BHA), Aspergillus oryzae α-amylase (TAKA) and pig pancreatic α-amylase (PPA), the differences were relatively smaller, showing ΔTm values of 0°C, 5°C, 14°C, and 17°C, respectively [42, 46, 47], as shown in Table 1.
Figure 4.
DSC curves of AGXA without Ca2+ (A) and with Ca2+ (B) were fitted using a non-two-state model. The Tm, ΔH and ΔHv for AGXA without Ca2+ were 67.3 °C, 238.3 kcal.mol-1 and 236.7 kcal.mol-1, respectively. The Tm, ΔH and ΔHv for AGXA with Ca2+ were 77.8 °C, 290.7 kcal.mol-1 and 282.6 kcal.mol-1, respectively. Ca2+ ions caused a thermal increase (∼10 °C and ∼52 kcal.mol-1) in Tm and ΔH, and the values of ΔH/ΔHv were approximately equal to 1.0 for AGXA both with Ca2+ and without Ca2+ ions.
Table 1.
The effect of calcium ions on thermostability in terms of melting temperatures of various α-amylases.
The thermal stability of the α-amylase AGXA with calcium ions, also reflected in a higher ΔH, reached 290.7 k cal.mol-1 in the presence of calcium ions and reached 238.3 k cal.mol-1 in the absence of calcium ions. The ΔH value for AGXA without Ca2+ was similar to the reported ΔH for AHA without Ca2+ (238 kcal.mol-1) [47]. The ΔH value for AGXA with Ca2+ was larger but still less than the values reported for BHA (581.7 kcal.mol-1) [46], TAKA (535.9 kcal.mol-1) [42], BAA(336.2 kcal.mol-1) and BLA (362.1 kcal.mol-1) [45]. ΔH was the energy required to denature proteins, and a higher ΔH demonstrated that more energy was required to unfold the protein.
Generally, irreversible denaturation of a protein has been suggested to occur according to the Lumry and Eyring model [48]:
| (1) |
where N is the native state, U is the unfolded state and F is the final state [48, 49]. When a native, functional protein underwent irreversible alteration processes (aggregation, proteolysis, strong interactions with other macromolecules, and other similar events), it would eventually end up in the nonfunctional, final state [50]. When most of the unfolded molecules were converted to final state, the thermal denaturation could be regarded as a one-step process by a first-order irreversible process:
| (2) |
in which the conversion from N to F was determined by the temperature-dependent, first-order rate constant (k) [50, 51]. This model has been used successfully to describe the irreversible unfolding processes of various proteins [52-55]. In the present study, the ratio of calorimetric and van’t Hoff enthalpies (ΔH/ΔHv) was near 1.0, suggesting that AGXA with Ca2+ and without Ca2+ adopts a two-state thermal denaturation process. Similarly, it has been reported that thermal denaturation of Bacillus α-amylases has been characterized in terms of a two-state irreversible denaturation model [49, 56-58]. Nazmi et al. [59] reported on the apparent thermodynamic and kinetic properties of BLA renatured with CaCl2 using DSC, CD and DLS studies, indicating that partially-renatured BLA could be represented well through superposition of two irreversible processes, each of which following the two-state irreversible model.
3.4. CD Measurements
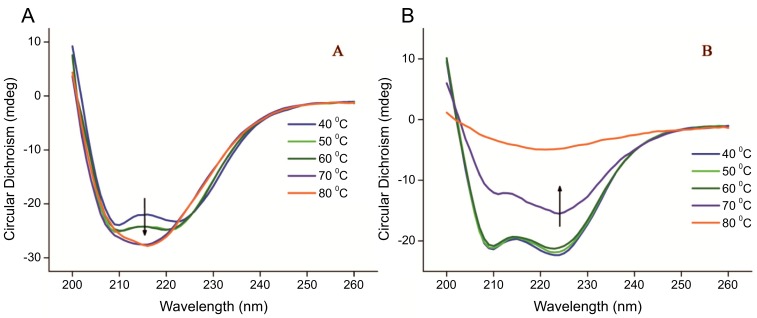
The effect of calcium ions on the thermal unfolding of recombinant AGXA has also been studied by recording CD spectra as a function of temperature. As shown in Figure 5, at low temperature (<40°C), the CD spectra of recombinant AGXA with Ca2+ and without Ca2+ were equivalent minima near 208 and 222 nm, which is the characteristic in α-amylase that suggests a predominant α-helix structure [60]. As temperature increased α-amylases with Ca2+ and without Ca2+ underwent different denaturation processes. For α-amylase without Ca2+ there were two significant changes: the first change was between 40 and 50°C, with the CD spectra of two deeper negative ellipticities, indicating the α-helical contents of the structure increase; the second change was observed above 60°C, with the emergence of a new peak with a minima ellipticity at approximately 215 nm, demonstrating the structural transition from an α-helix to a β-sheet structure, and the skeleton structure became loose. For the α-amylase with Ca2+, when the temperature was between 40°C and 60°C, there were few changes in the spectra; when the temperature increased to 70°C and 80°C, the spectra showed a gradual diminution of a decrease in α-helix structures. With temperatures over 80°C, the ellipticity of the spectra tended to have a value of 0, indicating aggregation and even preci-pitation of the α-amylase. As an excellent method for rapidly evaluating the secondary structure and folding properties of proteins [61], CD has been widely used to study enzyme structure stability, thermodynamics, thermal unfolding and its mechanism [62, 63]. CD spectra have examined thermal stability and catalytic activity of α-amylase in the presence of different concentrations of CaCl2, and the results show that the enzyme displays optimum catalytic activity in the presence of 1.0-2.0 mM CaCl2, while further addition of CaCl2 leads to enzyme inhibition [64]. Recently, the stability and changes in the secondary structure of α-amylase from thermophilic Bacillus sp. TSSC-3 had been reported and discovered to have a reduced α-helix content and increased amount of β-sheets after denaturation [65]. Far-UV CD studies revealed α-helical structures were shifted to 215 nm in porcine pancreatic α-amylase upon AgNP binding, with structural conformation changes with peak shifts confirmed through FTIR spectroscopy [66].
Figure 5.
The far UV-CD spectra of AGXA without Ca2+ (A) and with Ca2+ (B) from 40 °C to 80 °C. As temperature increased, AGXA without Ca2+ and with Ca2+ underwent different denaturation processes. (The color version of the figure is available in the electronic copy of the article).
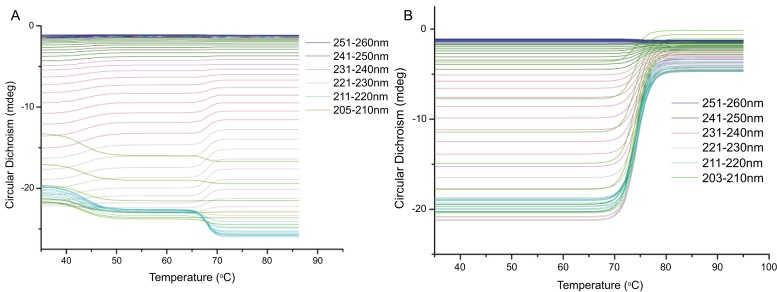
Global 3 was a package software to calculated structural and thermodynamic data developed by Applied Photophysics. After processing with Global 3 software the calculated spectra are shown in Figure 6, and the unfolding temperatures (Tm) were 67.5°C and 77.5°C for AGXA without Ca2+ and with Ca2+, respectively, which were consistent with the values from DSC measurements. From the CD-temperature profiles, there were two transition states (the low temperature changes were minor and the high temperature changes were obvious) for the α-amylase without Ca2+, whereas only one transition was observed in α-amylase with Ca2+, which were similar with the CD spectra shown above. From homology modeling, this α-amylase was identified to have four calcium ions binding to its structure, one in the region between domain A and B, another in the region between domain A and C, and the other two in domain A. These calcium ions could form extra bridges between the different domains of α-amylase to enhance enzyme stability [45]. For AGXA without Ca2+, the spectra changes could be speculated to be due to temperature increases cause higher thermal-induced mobility of domain C with respect to central domain A, which might be related to the first subtransition. As partially unfolded or completely unfolded proteins were much more susceptible to proteolytic degradation [59], β-sheet-like structures of the TIM on domain A lead to a higher probability of forming random inter-domain and intermolecular hydrogen bonds, which increase the chance of conformational scrambling or of protein aggregation, and in particular the formation of amyloid-like fibrils [67]. Ca2+ ions, however, are possibly causing irreversibility of thermal unfolding due to high temperature chemical modifications, and the Ca2+ ions act as intermolecular cross-links to adjacent anionic molecules by forming protein–Ca2+– protein complexes, exhibiting intramolecular electrostatic shielding of negative charges on the protein, and inducing ion-induced conformational changes leading to altered hydrophobic interactions [68-70]. The increased aggregation tendency then results from hydrophobic residues that became exposed to the solvent and interact preferably with hydrophobic residues from other unfolded protein molecules to minimize their exposure to the solvent [71, 72]. Recently, Uzma et al. [73] reported a unique thermostable dimer α-amylase Tp-AmyS, which with a Ca2+ ion in each monomer and showing catalytic cooperativity within the dimer. Just as the sequence similarity between AGXA and Tp-AmyS was only 20%, how the role of dimerization in the thermostability of the α-amylase was not involved in AGXA.
Figure 6.
The CD-temperature profiles of AGXA without Ca2+ (A) and with Ca2+ (B) from 35 °C to 85 °C were calculated with Global 3 software. The spectra with absorbance above 2.0AU were excluded. The results showed that the unfolding temperatures (Tm) were 67.5 °C and 77.5 °C for AGXA without Ca2+ and with Ca2+, respectively. (The color version of the figure is available in the electronic copy of the article).
CONCLUSION
In summary, the thermostability of Anoxybacillus sp. GXS-BL α-amylase with calcium ions and without calcium ions has been studied. It was found that, in the presence of calcium ions, the values of Topt, T50, t1/2, Tm and ΔH in AGXA were significantly higher than those observed in the absence of calcium ions, showing calcium ions had stabilizing effects on α-amylase structure with the increased temperature. Based on DSC measurements AGXA underwent thermal denaturation by adopting two-state irreversible unfolding processes for the ΔH/ΔHvh to be close to 1.0. Based on the CD spectra, AGXA without calcium ions exhibits two transition states upon unfolding, including α-helical contents increasing, and the transition from α-helices to β-sheet structures, which was obviously different with AGXA added Ca2+ ions. According to the modeled structure up to 4 Ca2+ ions were located on the inter-domain or intra-domain regions. These results reveal that Ca2+ ions have pronounced influences on the structural features accountable for the thermostability of α-amylase from Anoxybacillus sp GXS-BL.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundations of China (31560251), the Natural Science Foundations of Guangxi (2015GXNSFAA139053 and 2017GXNSFBA198008), the Key Project of Guangxi Science &Technology for Research and Development (Guangxi Science and Technology Cooperation Program 15104001-6), and the basic research program of the Guangxi Academy of Sciences (15YJ22SW02, 2017YJJ23004).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Zeldes B.M., Keller M.W., Loder A.J., Straub C.T., Adams M.W.W., Kelly R.M. Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels and industrial chemicals. Front. Microbiol. 2015;6:1209. doi: 10.3389/fmicb.2015.01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rezanka T., Kambourova M., Derekova A., Kolouchova I., Sigler K. LC-ESI-MS/MS identification of polar lipids of two thermophilic anoxybacillus bacteria containing a unique lipid pattern. Lipids. 2012;47(7):729–739. doi: 10.1007/s11745-012-3675-0. [DOI] [PubMed] [Google Scholar]
- 3.Bruins M.E., Janssen A.E.M., Boom R.M. Thermozymes and their applications. A review of recent literature and patents. Appl. Biochem. Biotechnol. 2001;90(2):155–186. doi: 10.1385/abab:90:2:155. [DOI] [PubMed] [Google Scholar]
- 4.Singh B., Bulusu G., Mitre A. Understanding the thermostability and activity of Bacillus subtilis lipase mutants: Insights from molecular dynamics simulations. J. Phys. Chem. B. 2015;119(2):392–409. doi: 10.1021/jp5079554. [DOI] [PubMed] [Google Scholar]
- 5.Janecek S. Alpha-amylase family: Molecular biology and evolution. Prog. Biophys. Mol. Biol. 1997;67(1):67–97. doi: 10.1016/s0079-6107(97)00015-1. [DOI] [PubMed] [Google Scholar]
- 6.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991;280(Pt 2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. The Carbohydrate-active enzymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Maarel M., van der Veen B., Uitdehaag J.C.M., Leemhuis H., Dijkhuizen L. Properties and applications of starch-converting enzymes of the alpha-amylase family. J. Biotechnol. 2002;94(2):137–155. doi: 10.1016/s0168-1656(01)00407-2. [DOI] [PubMed] [Google Scholar]
- 9.Gupta R., Gigras P., Mohapatra H., Goswami V.K., Chauhan B. Microbial alpha-amylases: A biotechnological perspective. Process Biochem. 2003;38(11):1599–1616. [Google Scholar]
- 10.Aguloglu S., Ensari N.Y., Uyar F., Otludil B. The effects of amino acids on production and transport of alpha-amylase through bacterial membranes. Starke. 2000;52(8-9):290–295. [Google Scholar]
- 11.Pikuta E., Lysenko A., Chuvilskaya N., Mendrock U., Hippe H., Suzina N., Nikitin D., Osipov G., Laurinavichius K. Anoxybacillus pushchinensis gen. nov., sp nov., a novel anaerobic, alkaliphilic, moderately thermophilic bacterium from manure, and description of Anoxybacillus falvithermus comb. nov. Int. J. Syst. Evol. Microbiol. 2000;50:2109–2117. doi: 10.1099/00207713-50-6-2109. [DOI] [PubMed] [Google Scholar]
- 12.Goh K.M., Kahar U.M., Chai Y.Y., Chong C.S., Chai K.P., Ranjani V., Illias R.M., Chan K.G. Recent discoveries and applications of Anoxybacillus. Appl. Microbiol. Biotechnol. 2013;97(4):1475–1488. doi: 10.1007/s00253-012-4663-2. [DOI] [PubMed] [Google Scholar]
- 13.Declerck N., Machius M., Joyet P., Wiegand G., Huber R., Gaillardin C. Hyperthermostabilization of Bacillus licheniformis alpha-amylase and modulation of its stability over a 50 degrees C temperature range. Protein Eng. 2003;16(4):287–293. doi: 10.1093/proeng/gzg032. [DOI] [PubMed] [Google Scholar]
- 14.Torrance J.W., MacArthur M.W., Thornton J.M. Evolution of binding sites for zinc and calcium ions playing structural roles. Proteins. 2008;71(2):813–830. doi: 10.1002/prot.21741. [DOI] [PubMed] [Google Scholar]
- 15.Goyal N., Gupta J.K., Soni S.K. A novel raw starch digesting thermostable alpha-amylase from Bacillus sp I-3 and its use in the direct hydrolysis of raw potato starch. Enzyme Microb. Technol. 2005;37(7):723–734. [Google Scholar]
- 16.Khajeh K., Ranjbar B., Naderi-Manesh H., Habibi A.E., Nemat-Gorgani M. Chemical modification of bacterial alpha-amylases: Changes in tertiary structures and the effect of additional calcium. Bba-Protein Struct. M. 2001;1548(2):229–237. doi: 10.1016/s0167-4838(01)00236-9. [DOI] [PubMed] [Google Scholar]
- 17.Bush D.S., Sticher L., Vanhuystee R., Wagner D., Jones R.L. The calcium requirement for stability and enzymatic-activity of 2 isoforms of barley aleurone alpha-amylase. J. Biol. Chem. 1989;264(32):19392–19398. [PubMed] [Google Scholar]
- 18.Larson S.B., Greenwood A., Cascio D., Day J., McPherson A. Refined molecular-structure of pig pancreatic alpha-amylase at 2-center-dot-1 angstrom resolution. J. Mol. Biol. 1994;235(5):1560–1584. doi: 10.1006/jmbi.1994.1107. [DOI] [PubMed] [Google Scholar]
- 19.Buisson G., Duee E., Haser R., Payan F. 3 dimensional structure of porcine pancreatic alpha-amylase at 2.9 A resolution-role of calcium in structure and activity. EMBO J. 1987;6(13):3909–3916. doi: 10.1002/j.1460-2075.1987.tb02731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hmidet N., Bayoudh A., Berrin J.G., Kanoun S., Juge N., Nasri M. Purification and biochemical characterization of a novel alpha-amylase from Bacillus licheniformis NH1 - Cloning, nucleotide sequence and expression of amyN gene in Escherichia coli. Process Biochem. 2008;43(5):499–510. [Google Scholar]
- 21.Asoodeh A., Chamani J., Lagzian M. A novel thermostable, acidophilic alpha-amylase from a new thermophilic “Bacillus sp Ferdowsicous” isolated from Ferdows hot mineral spring in Iran: Purification and biochemical characterization. Int. J. Biol. Macromol. 2010;46(3):289–297. doi: 10.1016/j.ijbiomac.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Sharma A., Satyanarayana T. High maltose-forming, Ca2+-independent and acid stable alpha-amylase from a novel acidophilic bacterium, Bacillus acidicola. Biotechnol. Lett. 2010;32(10):1503–1507. doi: 10.1007/s10529-010-0322-9. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka A., Hoshino E. Secondary calcium-binding parameter of Bacillus amyloliquefaciens alpha-amylase obtained from inhibition kinetics. J. Biosci. Bioeng. 2003;96(3):262–267. doi: 10.1016/s1389-1723(03)80191-3. [DOI] [PubMed] [Google Scholar]
- 24.Mehta D., Satyanarayana T. Biochemical and molecular characterization of recombinant acidic and thermostable raw-starch hydrolysing alpha-amylase from an extreme thermophile Geobacillus thermoleovorans. J. Mol. Catal., B Enzym. 2013;85-86:229–238. [Google Scholar]
- 25.Liao S.M., Sun L., Wang Q.Y., Shen N.K., Zhu J., Huang G.Y., Huang J.M., Chen D., Huang R.B. Screening of thermostable α-amylase producing strain and cloning, expression and characterization of the gene AmyGX. Guangxi Sci. 2017;2017(1):92–99. [Google Scholar]
- 26.Janecek S., Kuchtova A., Petrovicova S. A novel GH13 subfamily of alpha-amylases with a pair of tryptophans in the helix alpha 3 of the catalytic TIM-barrel, the LPDlx signature in the conserved sequence region V and a conserved aromatic motif at the C-terminus. Biologia. 2015;70(10):1284–1294. [Google Scholar]
- 27.Mok S.C., Teh A.H., Saito J.A., Najimudin N., Alam M. Crystal structure of a compact alpha-amylase from Geobacillus thermoleovorans. Enzyme Microb. Technol. 2013;53(1):46–54. doi: 10.1016/j.enzmictec.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Chai K.P., Othman N.F.B., Teh A.H., Ho K.L., Chan K.G., Shamsir M.S., Goh K.M., Ng C.L. Crystal structure of anoxybacillus alpha-amylase provides insights into maltose binding of a new glycosyl hydrolase subclass. Sci. Rep. UK. 2016;6:23126. doi: 10.1038/srep23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 30.Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31(3):426–428. [Google Scholar]
- 31.Sali A., Potterton L., Yuan F., van Vlijmen H., Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23(3):318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 32.Shen M.Y., Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15(11):2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovell S.C., Davis I.W., Adrendall W.B., de Bakker P.I.W., Word J.M., Prisant M.G., Richardson J.S., Richardson D.C. Structure validation by C alpha geometry: Phi, psi and C beta deviation. Proteins. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 34.Eisenberg D., Luthy R., Bowie J.U. In: VERIFY3D: Assessment of protein models with three-dimensional profiles, in macromolecular crystallography. Pt B., Carter C.W., Sweet R.M., editors. 1997. pp. 396–404. [DOI] [PubMed] [Google Scholar]
- 35.MacGregor E.A., Janecek S., Svensson B. Relationship of sequence and structure to specificity in the alpha-amylase family of enzymes. BBA-Protein Struct. M. 2001;1546(1):1–20. doi: 10.1016/s0167-4838(00)00302-2. [DOI] [PubMed] [Google Scholar]
- 36.Igarashi K., Hatada Y., Ikawa K., Araki H., Ozawa T., Kobayashi T., Ozaki K., Ito S. Improved thermostability of a Bacillus alpha-amylase by deletion of an arginine-glycine residue is caused by enhanced calcium binding. Biochem. Biophys. Res. Commun. 1998;248(2):372–377. doi: 10.1006/bbrc.1998.8970. [DOI] [PubMed] [Google Scholar]
- 37.Lin L.L., Huang C.C., Lo H.F. Impact of Arg210-Ser211 deletion on thermostability of a truncated Bacillus sp strain TS-23 alpha-amylase. Process Biochem. 2008;43(5):559–565. [Google Scholar]
- 38.D’Amico S., Marx J.C., Gerday C., Feller G. Activity-stability relationships in extremophilic enzymes. J. Biol. Chem. 2003;278(10):7891–7896. doi: 10.1074/jbc.M212508200. [DOI] [PubMed] [Google Scholar]
- 39.Hagihara H., Igarashi K., Hayashi Y., Endo K., Ikawa-Kitayama K., Ozaki K., Kawai S., Ito S. Novel alpha-amylase that is highly resistant to chelating reagents and chemical oxidants from the alkaliphilic Bacillus isolate KSM-K38. Appl. Environ. Microbiol. 2001;67(4):1744–1750. doi: 10.1128/AEM.67.4.1744-1750.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chai Y.Y., Abd Rahman R.N.Z.R., Illias R.M., Goh K.M. Cloning and characterization of two new thermostable and alkalitolerant alpha-amylases from the Anoxybacillus species that produce high levels of maltose. J. Ind. Microbiol. Biotechnol. 2012;39(5):731–741. doi: 10.1007/s10295-011-1074-9. [DOI] [PubMed] [Google Scholar]
- 41.Kikani B.A., Singh S.P. The stability and thermodynamic parameters of a very thermostable and calcium-independent alpha-amylase from a newly isolated bacterium, Anoxybacillus beppuensis TSSC-1. Process Biochem. 2012;47(12):1791–1798. [Google Scholar]
- 42.Fukada H., Takahashi K., Sturtevant J.M. Differential scanning calorimetric study of the thermal unfolding of Taka-amylase A from Aspergillus oryzae. Biochemistry. 1987;26(13):4063–4068. doi: 10.1021/bi00387a048. [DOI] [PubMed] [Google Scholar]
- 43.Johnson C.M. Differential scanning calorimetry as a tool for protein folding and stability. Arch. Biochem. Biophys. 2013;531(1-2):100–109. doi: 10.1016/j.abb.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Durowoju I.B., Bhandal K.S., Hu J., Carpick B., Kirkitadze M. Differential scanning calorimetry. A method for assessing the thermal stability and conformation of protein antigen. J. Vis. Exp. 2017;2017(121) doi: 10.3791/55262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fitter J., Herrmann R., Dencher N.A., Blume A., Hauss T. Activity and stability of a thermostable alpha-amylase compared to its mesophilic homologue: Mechanisms of thermal adaptation. Biochemistry. 2001;40(35):10723–10731. doi: 10.1021/bi010808b. [DOI] [PubMed] [Google Scholar]
- 46.Nielsen A.D., Pusey M.L., Fuglsang C.C., Westh P. A proposed mechanism for the thermal denaturation of a recombinant Bacillus halmapalus alpha-amylase - the effect of calcium ions. BBA-Proteins Proteom. 2003;1652(1):52–63. doi: 10.1016/j.bbapap.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Feller G., d’Amico D., Gerday C. Thermodynamic stability of a cold-active alpha-amylase from the Antarctic bacterium Alteromonas haloplanctis. Biochemistry. 1999;38(14):4613–4619. doi: 10.1021/bi982650+. [DOI] [PubMed] [Google Scholar]
- 48.Lumry R., Eyring H. Conformation changes of proteins. J. Phys. Chem. 1954;58(2):110–120. [Google Scholar]
- 49.Sanchezruiz J.M. Theoretical-analysis of lumry-eyring models in differential scanning calorimetry. Biophys. J. 1992;61(4):921–935. doi: 10.1016/S0006-3495(92)81899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.del Pino I.M.P., Ibarra-Molero B., Sanchez-Ruiz J.M. Lower kinetic limit to protein thermal stability: A proposal regarding protein stability in vivo and its relation with misfolding diseases. Proteins. 2000;40(1):58–70. doi: 10.1002/(sici)1097-0134(20000701)40:1<58::aid-prot80>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez V.B., Alameda E.J., Gallegos J.F.M., Requena A.R., Lopez A.I.G. Enzymatic hydrolysis of soluble starch with an alpha-amylase from Bacillus licheniformis. Biotechnol. Prog. 2006;22(3):718–722. doi: 10.1021/bp060057a. [DOI] [PubMed] [Google Scholar]
- 52.Sanchezruiz J.M., Lopezlacomba J.L., Cortijo M., Mateo P.L. Differential scanning calorimetry of the irreversible thermal-denaturation of thermolysin. Biochemistry. 1988;27(5):1648–1652. doi: 10.1021/bi00405a039. [DOI] [PubMed] [Google Scholar]
- 53.Kurganov B.I., Lyubarev A.E., Sanchez-Ruiz J.M., Shnyrov V.L. Analysis of differential scanning calorimetry data for proteins. Criteria of validity of one-step mechanism of irreversible protein denaturation. Biophys. Chem. 1997;69(2-3):125–135. doi: 10.1016/s0301-4622(97)80552-2. [DOI] [PubMed] [Google Scholar]
- 54.Vogl T., Jatzke C., Hinz H.J., Benz J., Huber R. Thermodynamic stability of annexin V E17G: Equilibrium parameters from an irreversible unfolding reaction. Biochemistry. 1997;36(7):1657–1668. doi: 10.1021/bi962163z. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez A., Pina D.G., Yelamos B., Leon J.J.C., Zhadan G.G., Villar E., Gavilanes F., Roig M.G., Sakharov I.Y., Shnyrov V.L. Thermal stability of peroxidase from the african oil palm tree Elaeis guineensis. Eur. J. Biochem. 2002;269(10):2584–2590. doi: 10.1046/j.1432-1033.2002.02930.x. [DOI] [PubMed] [Google Scholar]
- 56.Tomazic S.J., Klibanov A.M. Mechanisms of irreversible thermal inactivation of Bacillus alpha-amylases. J. Biol. Chem. 1988;263(7):3086–3091. [PubMed] [Google Scholar]
- 57.Sanchez-Ruiz J.M. Differential scanning calorimetry of proteins. Subcell. Biochem. 1995;24:133–176. doi: 10.1007/978-1-4899-1727-0_6. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka A., Hoshino E. Calcium-binding parameter of Bacillus amyloliquefaciens alpha-amylase determined by inactivation kinetics. Biochem. J. 2002;364:635–639. doi: 10.1042/BJ20011436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nazmi A.R., Reinisch T., Hinz H.J. Calorimetric studies on renaturation by CaCl2 addition of metal-free alpha-amylase from Bacillus licheniformis (BLA). J. Therm. Anal. Calorim. 2008;91(1):141–149. [Google Scholar]
- 60.Fazili N.A., Bhat W.F., Naeem A. Induction of amyloidogenicity in wild type HEWL by a dialdehyde: Analysis involving multi dimensional approach. Int. J. Biol. Macromol. 2014;64:36–44. doi: 10.1016/j.ijbiomac.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 61.Greenfield N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006;1(6):2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hegde K., Dasu V.V. Structural stability and unfolding properties of cutinases from Thermobifida fusca. Appl. Biochem. Biotechnol. 2014;174(2):803–819. doi: 10.1007/s12010-014-1037-5. [DOI] [PubMed] [Google Scholar]
- 63.Ropiak H.M., Lachmann P., Ramsay A., Green R.J., Mueller-Harvey I. Identification of structural features of condensed tannins that affect protein aggregation. PLoS One. 2017;12(1):e0170768. doi: 10.1371/journal.pone.0170768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yadav J.K. A differential behavior of alpha-amylase, in terms of catalytic activity and thermal stability, in response to higher concentration CaCl2. Int. J. Biol. Macromol. 2012;51(1-2):146–152. doi: 10.1016/j.ijbiomac.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 65.Kikani B.A., Singh S.P. Enzyme stability, thermodynamics and secondary structures of alpha-amylase as probed by the CD spectroscopy. Int. J. Biol. Macromol. 2015;81:450–460. doi: 10.1016/j.ijbiomac.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 66.Ernest V., Sekar G., Mukherjee A., Chandrasekaran N. Studies on the effect of AgNP binding on alpha-amylase structure of porcine pancreas and Bacillus subtilis by multi-spectroscopic methods. J. Lumin. 2014;146:263–268. [Google Scholar]
- 67.Fitter J. The perspectives of studying multi-domain protein folding. Cell. Mol. Life Sci. 2009;66(10):1672–1681. doi: 10.1007/s00018-009-8771-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Privalov P.L., Khechinashvili N.N. A thermodynamic approach to the problem of stabilization of globular protein structure: A calorimetric study. J. Mol. Biol. 1974;86(3):665–684. doi: 10.1016/0022-2836(74)90188-0. [DOI] [PubMed] [Google Scholar]
- 69.Segawa T., Sugai S. Interactions of divalent metal ions with bovine, human, and goat alpha-lactalbumins. J. Biochem. 1983;93(5):1321–1328. doi: 10.1093/oxfordjournals.jbchem.a134266. [DOI] [PubMed] [Google Scholar]
- 70.Simons J., Kosters H.A., Visschers R.W., de Jongh H.H.J. Role of calcium as trigger in thermal beta-lactoglobulin aggregation. Arch. Biochem. Biophys. 2002;406(2):143–152. doi: 10.1016/s0003-9861(02)00429-0. [DOI] [PubMed] [Google Scholar]
- 71.Violet M., Meunier J.C. Kinetic-study of the irreversible thermal-denaturation of bacillus-licheniformis alpha-amylase. Biochem. J. 1989;263(3):665–670. doi: 10.1042/bj2630665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vieille C., Zeikus G.J. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 2001;65(1):1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hameed U., Price I., Ikram Ul H., Ke A.L., Wilson D.B., Mirza O. Functional characterization and crystal structure of thermostable amylase from Thermotoga petrophila, reveals high thermostability and an unusual form of dimerization. BBA-Proteins Proteom. 2017;1865(10):1237–1245. doi: 10.1016/j.bbapap.2017.06.015. [DOI] [PubMed] [Google Scholar]