Abstract
Introduction
Alzheimer's disease (AD) diagnosis requires invasive CSF analysis or expensive brain imaging. Therefore, a minimal-invasive reliable and cost-effective blood test is requested to power large clinical AD trials at reduced screening failure.
Methods
We applied an immuno-infrared sensor to measure the amyloid-β (Aβ) and tau secondary structure distribution in plasma and CSF as structure-based biomarkers for AD (61 disease controls, 39 AD cases).
Results
Within a first diagnostic screening step, the structure-based Aβ blood biomarker supports AD identification with a sensitivity of 90%. In a second diagnostic validation step, the combined use of the structure-based CSF biomarkers Aβ and tau excluded false-positive cases which offers an overall specificity of 97%.
Discussion
The primary Aβ-based blood biomarker funnels individuals with suspected AD for subsequent validation of the diagnosis by structure-based combined analysis of the CSF biomarkers Aβ and tau. Our novel two-step recruitment strategy substantiates the diagnosis of AD with a likelihood of 29.
Keywords: Alzheimer's disease, Amyloid-beta, Tau, Protein misfolding, Diagnostics
1. Introduction
Alzheimer's disease (AD) pathology is accompanied by misfolding of amyloid-β (Aβ) and tau from monomeric into β-sheet–enriched pathogenic species. This process is suggested to precede about 10-15 years before clinical onset of the disease [1], [2]. The aggregated Aβ and tau isoforms result in macroscopic plaques and neurofibrillary tangles in the brain of patients with AD [3], [4]. So far, guidelines and actual recommendations for AD diagnosis intend the quantitative analysis of CSF biomarkers and additional imaging methods such as amyloid positron emission tomography (Amyloid-PET) to detect and correlate Aβ burden in the brain with AD pathology [5], [6], [7], [8], [9], [10]. These techniques require either invasive lumbar punctures or expensive PET analyses, which rely on the use of radioactive compounds. Thus, there is a general agreement that a minimal-invasive, reliable, and cost-effective blood test is requested as a first screening funnel to identify individuals with high risk for AD [11], [12], [13], [14], [15]. To date, only few blood tests present the potential to detect AD [16], [17], [18], [19]. Already in 2016, we have shown that the secondary structure distribution of Aβ in blood plasma, measured by an immuno-infrared sensor, is an excellent biomarker for AD, reflecting the Aβ burden in the brain [20], [21]. In contrast to quantitative ELISA assays, the immuno-infrared sensor does not detect the concentration decline of Aβ42, which is a secondary event associated with Aβ42 deposition in the brain, but directly monitors the misfolding of Aβ, which is proposed to be a primary event in AD pathology occurring 15-20 years before clinical onset. We propose that our structure-based biomarker offers a unique additional molecular feature to the use of Aβ peptide ratios as promising AD biomarkers. Because β-sheet–enriched misfolding of Aβ peptides is thought to be an initial event in the pathophysiological cascade of AD, our immuno-infrared sensor is promising to identify patients at risk for AD at an early preclinical stage [22]. Recently, we have validated the diagnostic performance of our structure-based biomarker for mild to severe AD, prodromal AD, and for preclinical AD in three independent clinical studies [20], [22]. We also demonstrated in the latter studies that the Aβ secondary structure distribution as structure-based AD biomarker correlates significantly with already well established standard CSF core biomarkers (Aβ42/40 ratio, total-tau (ttau), phospho-tau181 [ptau]) and neuroimaging (Amyloid-PET) biomarkers of AD. Most remarkable, in a 15-year longitudinal aging study comprising 10,000 participants, we identified in a subcohort of 890 healthy participants, 48 of 65 participants who subsequently developed AD. We predicted AD progression in participants without any cognitive symptoms in an average of 8 years before their clinical onset with a sensitivity of 71% and specificity of 91% [22].
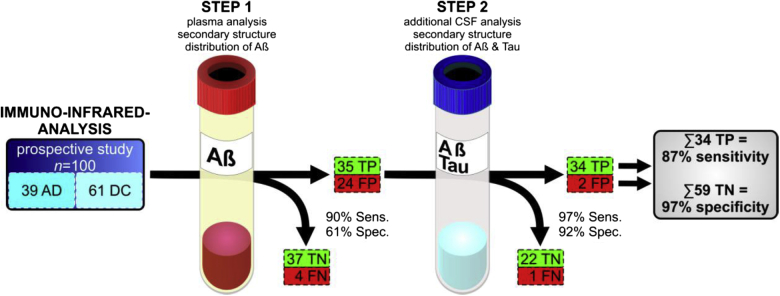
To increase the sensitivity of the structure-based blood biomarker for screening applications, we shift the diagnostic threshold within our novel two-step recruitment strategy to higher wavenumbers which yielded more false-positive cases at reduced specificity. Within a second validation test, the specificity could be increased by immuno-infrared sensor–based CSF analysis to exclude false-positive cases. A majority vote classifier was applied to categorize all CSF samples and to exclude false-positive cases. Here, we present the performance of the novel two-step AD recruitment strategy which preselects subjects at risk for AD by the blood-based immuno-infrared sensor screening assay. Individuals to be at risk for AD according to the blood-based immuno-infrared sensor screen underwent subsequently CSF-based analyses of the Aβ peptide and tau conformation. Here, in addition to Aβ, tau also was used as a structure-based biomarker. This two-step AD recruitment strategy provides an overall sensitivity of 87% and specificity of 97%. In principle, the structure-based blood screen is promising to identify preclinical and prodromal AD, which subsequently can be studied by CSF biomarkers and/or Amyloid-PET to validate the screen diagnosis. However, the blood screen test preselected 59 of 100 subjects, whereas 34 of 39 cases were correctly classified as AD according to the clinical diagnosis. Only two cases were misdiagnosed as false-positive cases.
2. Methods
2.1. Participants and clinical phenotyping
The prospective study fulfills the international standards for studies of diagnostic test accuracy in dementia [23].
For the present study, 61 disease control (DC) subjects and 39 AD cases were acquired from a prospective study designed and initiated by the gerontopsychiatric unit of the department of psychiatry and psychotherapy at the LVR Clinics, University of Duisburg-Essen, between 2009 and 2013 (PI J.W.). The study was approved by the ethical board of the University of Duisburg-Essen (ID 12 5160 BO) and the research use of the samples and data was in accord with the terms of the informed consents. Sample acquisition and dementia diagnostics were made according to the criteria of the National Institute for Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association and included the 2011 recommendations from the National Institute on Aging. The clinical diagnosis of DC was performed according to the International Classification of Diseases (ICD-10). Patients were investigated by psychometric testing (Mini-Mental State Examination and/or extended neuropsychological evaluation) and CSF-guided neurochemical dementia diagnostics. A detailed description of the clinical cohort can be found in the study by Nabers et al. [20].
Importantly, for all subjects, CSF levels of Aβ40, Aβ42, ttau, ptau, the Aβ42/40 ratio, and demographic data were available (Supplementary Table 1). Gerontopsychiatrists and neuropsychologists had access to all available clinical, neuroimaging, psychometric, and conventional CSF dementia biomarker data but were blinded for the immuno-infrared analysis. The DC group comprised patients with dementia of other origin and nondemented patients with heterogonous neurological or psychiatric diseases but without memory complaints. Most patients with AD presented with early AD.
All CSF samples were assessed with the Meso Scale Discovery (MSD) V-Plex Aβ peptide panel multiplex kit using monoclonal antibody 6E10 for detection (MSD, Rockville, MD). The Aβ peptides Aβ38, 40, and 42 were determined according to the manufacturer's instructions after 16-fold dilution of the CSF samples with “Diluent 35” (MSD). CSF concentrations of ttau and ptau were measured in duplicate by commercially available ELISA in an accredited expert laboratory for CSF-guided neurochemical dementia diagnostics (P. Lewczuk, University of Erlangen-Nürnberg, Germany) [20], [24].
2.2. Immuno-infrared sensor workflow
Preparation of the immuno-infrared sensor was described in detail previously [20], [22]. Briefly, the sensor surface was functionalized with monoclonal antibody A8978 (Sigma-Aldrich, Germany) for Aβ detection in blood plasma and CSF and monoclonal antibody TAU-5 for tau detection in CSF, respectively. After surface functionalization, the sensor was saturated to prevent unspecific binding. Finally, Aβ and tau was separately extracted from plasma (200 μL) and/or CSF (50 μL) for 1 hour followed by excessive PBS rinsing for 30 minutes. The recorded amide I absorbance band represented the biomarker secondary structure distribution in the respective body fluid by its maximum frequency. The immuno-infrared sensor technology is detailed in the study by Nabers et al. [20], [21], [22].
2.3. Bioinformatics
Before data interpretation, infrared-difference spectra were corrected for water vapor contributions by scaled subtraction as described in detail previously [20], [22]. Afterward, the amide I maximum frequency was determined by in-house procedures programmed with MATLAB 2015A (Mathworks). Statistical tests were performed using Origin 2016 (Origin Laboratories). Data distribution and group differences were analyzed with nonparametric Kruskal–Wallis analysis of variance. Thereby, statistical tests were conducted two-sided at a significance level of 0.05. Significance levels are denoted as follows: *P < .05, **P < .01, ***P < .001.
For the first diagnostic step based on blood plasma, the decisive threshold for AD and DC differentiation was set to <1647 cm−1 indicative for AD to reveal a sensitivity of ≈90%. For the second diagnostic step, the decisive threshold for all biomarkers was set to <1643 cm−1 determined by receiver operator characteristics (ROC)-curve analysis to reveal the highest diagnostic accuracy for each marker. These ROC-curve analyses were also described in the study by Nabers et al. [20]. Using a simple majority vote classifier, AD diagnosis was confirmed in step-2, as well as most false-positive cases were excluded. Therefore, each of the three biomarkers, Aβ from plasma, Aβ from CSF, and tau from CSF, equally contributed to the decision process of the majority vote classifier. In case the amide I maximum frequency of two biomarkers was below the decisive threshold at 1643 cm−1, the final diagnosis was AD. On the other hand, if two biomarkers had a maximum above or equal to 1643 cm−1, the classifier decision was non-AD.
3. Results
The Aβ secondary structure distribution in blood plasma was determined by the immuno-infrared assay as described previously [20], [21], [22]. Briefly, the sensor element is functionalized with highly specific antibodies recognizing all structural isoforms of Aβ or tau (Supplementary Fig. 1A), including helical or disordered monomers and β-sheet–enriched oligomers, prefibrillar, and fibrillar species. The recorded so-called amide I band represents the C=O stretching vibration of the protein backbone. The wavenumber or frequency of the corresponding absorbance band as shown in Supplementary Fig. 1B reflects the secondary structure distribution of Aβ or tau, respectively. Monomeric alpha-helical or disordered isoforms absorb between 1653 and 1649 wavenumbers (cm−1), whereas β-sheet isoforms absorb around 1626 cm−1. Increased β-sheet content, which is characteristic for Aβ and tau in AD pathology, shift the amide I band to lower wavenumbers. The more β-sheet isoforms are present in the blood, the larger is the spectral downshift to lower wavenumbers (Supplementary Fig. 1B). In step 1 of our two-step AD recruitment strategy, total-Aβ was extracted from blood plasma. The amide I frequency indicates the content of β-sheet–enriched, pathogenic Aβ in the total-Aβ fraction.
In the first step, we analyzed blood plasma of 61 DCs and 39 AD cases from a prospective study (Essen, Germany). The study design with SOPs for sample acquisition preanalytical sampling handling, sample storage, as well as diagnostic criteria for clinical phenotyping (including inclusion or exclusion criteria) is summarized in detail elsewhere [20]. AD and DCs differed significantly in the plasma Aβ amide I band frequency at maximum position (Fig. 1, P < .0001, two-sided nonparametric Kruskal–Wallis analysis of variance test). Importantly, our structure-based plasma biomarker significantly correlates (Spearman rank correlation with a significance value p for a niveau of α = 0.05) with well-established neurochemical CSF biomarkers such as Aβ42/40 ratio (rS = 0.47, P value = 6 × 10−7, determined by MSD), ttau (rS = -0.41, P value = 3 × 10−5, determined by ELISA), and ptau (rS = -0.48, P value = 7 × 10−7, determined by ELISA). We have detailed the analysis of the latter CSF biomarkers elsewhere [20]. However, in our former studies, we established 1643 cm−1 as decisive threshold (by ROC curve and Youden's cutoff) to reveal highest test accuracy. But the first diagnostic step, which serves as funnel for subsequent more invasive CSF analyses, should reveal high test sensitivity. To obtain 90% sensitivity, the decisive threshold had to be up-shifted from 1643 cm−1 used in former studies [20] to 1647 cm−1. The latter threshold up-shift increased the diagnostic sensitivity on the cost of specificity, which dropped to 61% (Fig. 2; step 1) as compared with the clinical diagnosis. In total numbers, we identified 35 of 39 AD cases with the first diagnostic step based on blood plasma; the number of false-positives (FP) was 24 of 61 DC subjects. Four AD cases could not be identified in the first step and thus remained false-negatives. Interestingly, three of the four AD cases that were misclassified in our blood test—as compared with the clinical diagnosis—were also indicated as non AD by Aβ or tau in CSF (see Supplementary Table 1B). In addition, two of this three clinical cases even had normal CSF Aβ42 levels (≥600 pg/mL) as determined by MSD and one of them was cognitively unimpaired (MMSE ≥ 27). Thus, these three cases might be actually misdiagnosed as AD. Prospective follow-up examination of these subjects is intended in the future to further validate the clinical diagnoses.
Fig. 1.
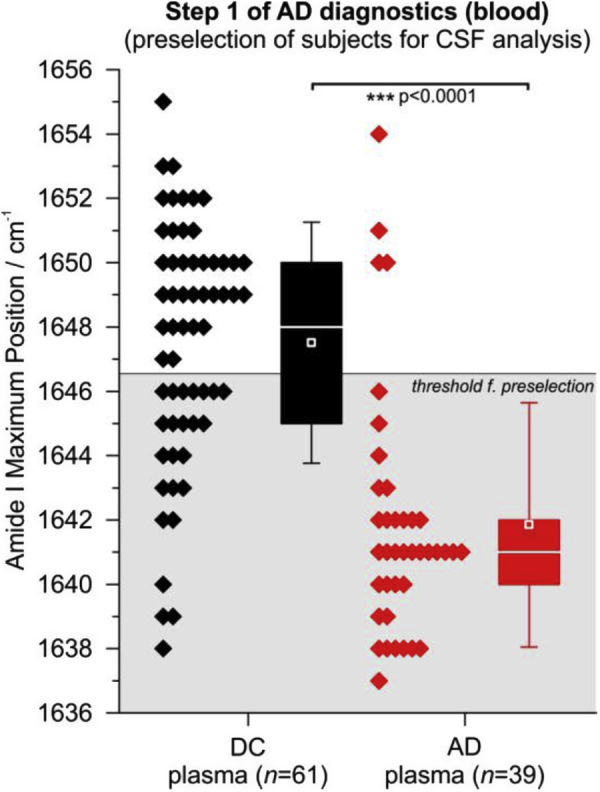
The Aβ amide I band frequencies of 61 disease controls and 39 AD cases (prospective study, Essen) are shown as diamonds. Aβ was extracted by mAb A8978 from blood plasma. The frequency of the amide I band is the measure of the structure-based biomarker, indicating the Aβ secondary structure distribution in blood plasma. The threshold was shifted to 1647 cm−1 (solid horizontal line) to increase the sensitivity to 90%. In former studies, 1643 cm−1 was used as decisive threshold for highest test accuracy. By up-shifting the threshold to 1647 cm−1, the specificity dropped from 88% to 61%. The 24 false-positives (FP) and 35 true-positives (TP) (gray background) were further examined in step 2 (Fig. 2, Supplementary Fig. 3) to exclude FP and validate TP. In box plots, 25/50/75% quantiles are shown as horizontal lines, the average amide I band position as square, and ±standard deviation as whiskers. Significant group differences are indicated by P value (two-sided nonparametric Kruskal–Wallis analysis of variance test) and by asterisks: ∗∗∗P < .001.
Fig. 2.
Procedure of the two-step AD recruitment strategy. In step 1, the structure-based biomarker was analyzed in blood plasma of 61 DC and 39 AD patients by the immuno-infrared sensor. As a result, 37 true-negatives (TN), 4 false-negatives (FN), 35 true-positives (TP), and 24 false-positives (FP) were identified in step 1 (sensitivity 90%, specificity 61%). In step 2, CSF of positive-tested individuals was analyzed regarding the Aβ and tau secondary structure distribution. Therewith, AD diagnosis could be confirmed in 34 of 35 cases. On the other hand, FP could be excluded in 22 of 24 cases, thus only two FP remained (sensitivity 97%, specificity 92%). This yielded in an overall sensitivity of 87% and a specificity of 97%.
In the second diagnostic step, we analyzed CSF samples of the preselected 35 AD cases and 24 FP. Analog to the Aβ analysis from blood plasma, the immuno-infrared assay was now used to separately extract and analyze the Aβ peptide and tau protein secondary structure distribution in CSF (Supplementary Fig. 1). The amide I band frequency of Aβ and tau in CSF of each person are presented in Supplementary Fig. 2 and Supplementary Fig. 4. Especially the tau secondary structure distribution is not suitable as a stand-alone biomarker for AD classification. But the combination of the plasma Aβ data obtained in step one and CSF Aβ and tau values provides a panel of three data sets that were used for validation of the AD diagnosis. For all three biomarker data sets—plasma Aβ, CSF Aβ, and CSF tau—the decisive threshold was set to <1643 cm−1 for AD identification as used in our former study [20]. In the second diagnostic step, 1643 cm−1 was used as decisive threshold, which was experimentally determined as decisive threshold for highest test accuracy in our former studies, for each biomarker Aβ in plasma, Aβ in CSF, and tau in CSF. Thus, each biomarker was set to provide the maximum accuracy within the majority vote classifier, respectively. Using a simple majority vote classifier, we confirmed the AD diagnosis suggested in step 1 in 34 of 35 (97%) preselected AD cases. Interestingly, the blood test amide I value of the one misclassified participant was directly at the threshold of 1643 cm−1 of the assay as used in the study by Nabers et al. [20]. By contrast, the Aβ amide I value in CSF was actually below the threshold indicating AD while tau was above the threshold in CSF (see Supplementary Fig. 4). The clinical diagnosis was early dementia due to AD. This finding might indicate that participants who are just at the border to switch to clinical Alzheimer's showed opposing biomarker states. This observation will be followed up in a more detailed study. However, the second diagnostic step excluded false-positive cases in 22 of 24 subjects (92%) (Fig. 2; step 2). In sum based on both diagnostic steps, 34 AD cases (of 39) and 59 controls (of 61) were classified correctly. Thus, the two-step AD recruitment strategy yielded in an overall sensitivity of 87% and a specificity of 97% (Fig. 2). For the majority vote classifier, all three biomarker values were equally weighted in the diagnostic decision process. Frequencies below the decisive threshold at 1643 cm−1 voted for AD. As soon as two biomarkers were below 1643 cm−1, the final diagnostic decision of the classifier was AD.
In summary, step 1 of our two-step AD recruitment strategy identified individuals with a largely increased likelihood for AD based on blood plasma analyses. The second diagnostic step, based on CSF analyses, confirmed AD and excluded FP suggested by step 1. Thus, an overall diagnostic accuracy of 93% and a likelihood ratio (LR+) of 29 were observed for AD/non-AD differentiation relative to the CSF biomarker cross-validated clinical diagnosis.
4. Discussion
Already in 2016, we have discovered that Aβ can be used as a structure-based biomarker in a blood test [20]. In the following, the diagnostic performance of our structure-based biomarker was validated for mild to severe AD, prodromal AD, and even for preclinical AD 8 years before clinical symptoms occurred [20], [22] in three independent clinical studies. Furthermore, the structure-based biomarker correlates with the Aβ42/40 ratio in CSF and with PET scanning [20], [22]. However, the sensitivity and specificity of our blood test had to be increased for clinical application. In general, immuno-infrared analyses based on CSF showed a better diagnostic performance than blood-based analyses (see also Supplementary Fig. 5). This difference might be explained by different secondary structure stabilizing effects, Aβ concentrations, and the origin of Aβ generation in blood plasma. However, here we show that the immuno-infrared sensor–based two-step AD recruitment strategy can be used to identify AD with an overall sensitivity of 87% and specificity of 97%. Therefore, in addition to Aβ, tau also was used as a structure-based biomarker. This shows nicely that the specificity of the test is increased by use of other structure-based biomarkers. Hence, we will use the structure-based tau biomarker also in blood, as soon as we establish an antibody for tau detection in blood in our assay. The blood test can be applied to identify subjects with high probability for AD in a blood-based first step. The second step application of the immuno-infrared sensor substantially increases the diagnostic specificity, thus validating the final neurochemical diagnosis of AD. In the present study, most cases presented assured AD and the blood plasma–based immuno-infrared sensor offered in the first diagnostic step a sensitivity of 90%. Hence, the assay is suitable to identify patients with dementia due to AD. Subjects that were preselected in this first step entered a second step, where CSF was analyzed by the immuno-infrared sensor. Therewith, an overall specificity of 97% was obtained with only 2 FP of 61 DCs. Regarding the patient cohort, our study is limited by a challenge bias (STARDdem criteria) because only patients with cognitive decline have been investigated, and only patients with probable AD were included (possible AD excluded). At this disease stage, neuropsychiatrists may directly recommend a lumbar puncture to substantiate the AD diagnosis. But our previous studies clearly demonstrated that our immuno-infrared assay is already able to detect preclinical disease stages. Thus, we have evidence that the two-step diagnostic process can also be applied to recruit individuals in asymptomatic disease stages. The two-step procedure presents a promising recruitment strategy for the preselection of individuals for clinical prevention trials focusing on the Aβ and/or tau as a therapeutic target. We could demonstrate that our structure-based biomarker (Aβ) significantly correlates with Amyloid-PET scanning on prodromal AD cases and with neurochemical CSF biomarkers in previous studies [20], [22]. In clinical practice, the two-step diagnostic process funnels individuals that may undergo comprehensive and expensive examinations such as lumbar puncture or brain imaging. However, for large prevention studies, the initial sensitive blood test will also be a reliable tool to preselect individuals with high risk for AD progression with a positive predictive value of 8 for study participation. This will also reduce the screening failure, that is, the number of individuals and costs that may undergo unnecessary comprehensive and expensive examinations such as lumbar puncture or brain imaging as recommended by FDA guidelines for clinical prevention studies.
Nevertheless, individuals who were preselected by our first blood plasma test may also be considered for conventional CSF-based neurochemical dementia diagnostics (measurement of Aβ40, Aβ42, ttau, and ptau levels) or for amyloid- or glucose-based PET to image Aβ burden or glucose dysmetabolism in the brain. Furthermore, the blood-based immuno-infrared sensor can also be combined with other promising novel blood tests for a two-step AD recruitment strategy [16], [17], [18]. For instance, Nakamura et al. [18] and Ovod et al. [17] previously reported mass-spectrometric approaches for the quantification of Aβ42/40 and Aβ669-711 levels in blood plasma. In clinical studies focusing on Aβ-PET–positive severe AD cases, both approaches revealed an accuracy of 85-90% with the Aβ-PET status. Most recently, Verberk et al. [16] focused on the identification of cognitively normal individuals with subjective cognitive decline, which converted to MCI or AD over years, by measuring the A42/40 ratio in blood plasma using the Single Molecule Array technology. This approach resulted in the Amsterdam study in a diagnostic accuracy of 76% [16]. All technologies may be applied in combination to receive a powerful solely blood-based AD recruitment platform.
Research in context.
-
1.
Systematic review: There is an urgent need for a minimal-invasive blood test to identify patients with Alzheimer's disease (AD) for clinical studies/trials focusing on amyloid-β (Aβ). Such test would also reduce the number of individuals for invasive or expensive examinations. To date, guidelines recommend only CSF and PET analyses because of the lack of reliable blood tests.
-
2.
Interpretation: The Aβ secondary structure distribution in plasma provides a sensitive biomarker for the preselection of individuals with increased probability for AD. Subsequent analyses of the Aβ and tau secondary structure distribution in CSF were efficient to confirm AD diagnoses and exclude false-positives with high diagnostic precision.
-
3.
Future directions: Our two-step diagnostic process provides an alternative in AD diagnosis. Especially, the plasma analysis presents a minimal-invasive test for routine use to select individuals with increased probability for AD. Subsequent comprehensive examinations, e.g., our CSF-based procedure, standard ELISAs, and PET, can finally be arranged to confirm the diagnosis.
Acknowledgments
Prof. Jens Wiltfang is supported by an Ilídio Pinho professorship and iBiMED (UID/BIM/04501/2013), and FCT project PTDC/DTP_PIC/5587/2014 at the University of Aveiro, Portugal.
Footnotes
Conflict of interest: The authors declare no competing financial interest.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.dadm.2019.01.008.
Contributor Information
Jens Wiltfang, Email: jens.wiltfang@med.uni-goettingen.de.
Klaus Gerwert, Email: klaus.gerwert@bph.rub.de.
Supplementary Data
References
- 1.Bateman R.J., Xiong C., Benzinger T.L.S., Fagan A.M., Goate A., Fox N.C. Clinical and Biomarker Changes in Dominantly Inherited Alzheimer's Disease. N Engl J Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowe C.C., Bourgeat P., Ellis K.A., Brown B., Lim Y.Y., Mulligan R. Predicting Alzheimer disease with β-amyloid imaging: Results from the Australian imaging, biomarkers, and lifestyle study of ageing. Ann Neurol. 2013;74:905–913. doi: 10.1002/ana.24040. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y., Mandelkow E. Tau in physiology and pathology. Nat Rev Neurosci. 2016;17:5–21. doi: 10.1038/nrn.2015.1. [DOI] [PubMed] [Google Scholar]
- 4.Blennow K., de Leon M.J., Zetterberg H. Alzheimer's disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 5.Paraskevaidi M., Martin-Hirsch P.L., Martin F.L. Progress and Challenges in the Diagnosis of Dementia: A Critical Review. ACS Chem Neurosci. 2018;9:446–461. doi: 10.1021/acschemneuro.8b00007. [DOI] [PubMed] [Google Scholar]
- 6.Simonsen A.H., Herukka S.K., Andreasen N., Baldeiras I., Bjerke M., Blennow K. Recommendations for CSF AD biomarkers in the diagnostic evaluation of dementia. Alzheimer's Dement. 2017;13:274–284. doi: 10.1016/j.jalz.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Hendrix J.A., Finger B., Weiner M.W., Frisoni G.B., Iwatsubo T., Rowe C.C. The Worldwide Alzheimer's Disease Neuroimaging Initiative: An update. Alzheimer's Dement. 2015;11:850–859. doi: 10.1016/j.jalz.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 8.McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Kawas C.H. The diagnosis of dementia due to Alzheimer's disease: Recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubois B., Feldman H.H., Jacova C., Hampel H., Molinuevo J.L., Blennow K. Advancing research diagnostic criteria for Alzheimer's disease: The IWG-2 criteria. Lancet Neurol. 2014;13:614–629. doi: 10.1016/S1474-4422(14)70090-0. [DOI] [PubMed] [Google Scholar]
- 10.Jack C.R., Therneau T.M., Wiste H.J., Weigand S.D., Knopman D.S., Lowe V.J. Transition rates between amyloid and neurodegeneration biomarker states and to dementia: A population-based, longitudinal cohort study. Lancet Neurol. 2016;15:56–64. doi: 10.1016/S1474-4422(15)00323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laske C., Sohrabi H.R., Frost S.M., López-De-Ipiña K., Garrard P., Buscema M. Innovative diagnostic tools for early detection of Alzheimer's disease. Alzheimer's Dement. 2015;11:561–578. doi: 10.1016/j.jalz.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Zhao X., Lejnine S., Spond J., Zhang C., Ramaraj T.C., Holder D.J. A candidate plasma protein classifier to identify Alzheimer's disease. J Alzheimer's Dis. 2015;43:549–563. doi: 10.3233/JAD-141149. [DOI] [PubMed] [Google Scholar]
- 13.Mapstone M., Cheema A.K., Fiandaca M.S., Zhong X., Mhyre T.R., Macarthur L.H. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014;20:415–418. doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snyder H.M., Carrillo M.C., Grodstein F., Henriksen K., Jeromin A., Lovestone S. Developing novel blood-based biomarkers for Alzheimer's disease. Alzheimer's Dement. 2014;10:109–114. doi: 10.1016/j.jalz.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olsson B., Lautner R., Andreasson U., Öhrfelt A., Portelius E., Bjerke M. CSF and blood biomarkers for the diagnosis of Alzheimer's disease: a systematic review and meta-analysis. Lancet Neurol. 2016;15:673–684. doi: 10.1016/S1474-4422(16)00070-3. [DOI] [PubMed] [Google Scholar]
- 16.Verberk I.M.W., Slot R.E., Verfaillie S.C.J., Heijst H., Prins N.D., van Berckel B.N.M. Plasma amyloid as pre-screener for the earliest Alzheimer's pathological changes. Ann Neurol. 2018;138:1328–1336. doi: 10.1002/ana.25334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ovod V., Ramsey K.N., Mawuenyega K.G., Bollinger J.G., Hicks T., Schneider T. Amyloid β concentrations and stable isotope labeling kinetics of human plasma specific to central nervous system amyloidosis. Alzheimer's Dement. 2017;13:841–849. doi: 10.1016/j.jalz.2017.06.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura A., Kaneko N., Villemagne V.L., Kato T., Doecke J., Doré V. High performance plasma amyloid-β biomarkers for Alzheimer's disease. Nature. 2018;554:249–254. doi: 10.1038/nature25456. [DOI] [PubMed] [Google Scholar]
- 19.Fandos N., Pérez-Grijalba V., Pesini P., Olmos S., Bossa M., Villemagne V.L. Plasma amyloid β 42/40 ratios as biomarkers for amyloid β cerebral deposition in cognitively normal individuals. Alzheimer's Dement Diagnosis, Assess Dis Monit. 2017;8:179–187. doi: 10.1016/j.dadm.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nabers A., Ollesch J., Schartner J., Kötting C., Genius J., Hafermann H. Amyloid-β-Secondary Structure Distribution in Cerebrospinal Fluid and Blood Measured by an Immuno-Infrared-Sensor: A Biomarker Candidate for Alzheimer's Disease. Anal Chem. 2016;88:2755–2762. doi: 10.1021/acs.analchem.5b04286. [DOI] [PubMed] [Google Scholar]
- 21.Nabers A., Ollesch J., Schartner J., Kötting C., Genius J., Haußmann U. An infrared sensor analysing label-free the secondary structure of the Abeta peptide in presence of complex fluids. J Biophotonics. 2016;9:224–234. doi: 10.1002/jbio.201400145. [DOI] [PubMed] [Google Scholar]
- 22.Nabers A., Perna L., Lange J., Mons U., Schartner J., Güldenhaupt J. Amyloid blood biomarker detects Alzheimer's disease. EMBO Mol Med. 2018;10:e8763. doi: 10.15252/emmm.201708763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noel-Storr A.H., McCleery J.M., Richard E., Ritchie C.W., Flicker L., Cullum S.J. Reporting standards for studies of diagnostic test accuracy in dementia: The STARDdem Initiative. Neurology. 2014;83:364–373. doi: 10.1212/WNL.0000000000000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewczuk P., Zimmermann R., Wiltfang J., Kornhuber J. Neurochemical dementia diagnostics: A simple algorithm for interpretation of the CSF biomarkers. J Neural Transm. 2009;116:1163–1167. doi: 10.1007/s00702-009-0277-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.