Abstract
Visual analytics and visualisation can leverage the human perceptual system to interpret and uncover hidden patterns in big data. The advent of next-generation sequencing technologies has allowed the rapid production of massive amounts of genomic data and created a corresponding need for new tools and methods for visualising and interpreting these data. Visualising genomic data requires not only simply plotting of data but should also offer a decision or a choice about what the message should be conveyed in the particular plot; which methodologies should be used to represent the results must provide an easy, clear, and accurate way to the clinicians, experts, or researchers to interact with the data. Genomic data visual analytics is rapidly evolving in parallel with advances in high-throughput technologies such as artificial intelligence (AI) and virtual reality (VR). Personalised medicine requires new genomic visualisation tools, which can efficiently extract knowledge from the genomic data and speed up expert decisions about the best treatment of individual patient’s needs. However, meaningful visual analytics of such large genomic data remains a serious challenge. This article provides a comprehensive systematic review and discussion on the tools, methods, and trends for visual analytics of cancer-related genomic data. We reviewed methods for genomic data visualisation including traditional approaches such as scatter plots, heatmaps, coordinates, and networks, as well as emerging technologies using AI and VR. We also demonstrate the development of genomic data visualisation tools over time and analyse the evolution of visualising genomic data.
Keywords: multidimensional data, genomic data, analytics, visualisation, virtual reality, augmented reality, immersive, artificial intelligence, machine learning, personalised medicine
Introduction
Visual analytics of genomic data is widely used in biology to help understand the data and communicate its contents, generate ideas, and to gain insight into biological processes. Visualisation plays an essential role in genomics research by making it possible to observe correlations and trends in large datasets as well as communicate findings to others. Visual analytics combine visualisation with analysis tools to enable seamless use of both approaches for scientific enquiring and offer a powerful method for performing complex genomic analyses.
Genomic is the convergence of many sciences including genetics, molecular biology, biochemistry, statistics, and computer sciences.1 Since Gregor Mendel discovered the basic principles of heredity, which became the foundation of modern genetics with the study of heredity,2 huge amounts of genomic data have now been collected around the world by different organisations. For example, one of the world’s largest pharmaceutical companies, AstraZeneca, launched a massive effort to compile genome sequences and health records from 2 million people. The company and its collaborators hoped to unearth rare genetic sequences that are associated with diseases and with responses to treatment.3 By the end of 2003, the Human Genome Project4 had successfully completed the ambitious goal of collecting sequence code covering 3 billion base pairs in the human genome, 2 years ahead of the previous projects.5 Sequencing is becoming the most popular high-throughput technology including the study of various genetic diseases as well as drug design and discovery for the diseases. With the development of computer technologies, genomic data can be collected at a faster pace and at a lower cost. The significance of this has launched the age of individual genome sequencing which supports an era of personalised medicine.6 Personalised cancer medicine based on the molecular characteristics of a tumour from an individual patient has great potential in the therapy of any type of cancer.7 DNA sequencing capacities continue to grow rapidly. If the growth continues at the current rate by doubling every 7 months, then we should reach more than 1 exabyte (1018) of sequence per year in the next 5 years and the approach 1 zettabyte (1021) of sequence per year by 2025.8
In human health, the major need driven by the vast amount of genomic data is how to interpret genomic sequences and how to find patterns9 over the large collections in high dimensions. Data visualisation is a way to convey meaningful concepts in a universal manner that is rapid and efficient and can allow humans to find potential value in big data. How we visualise complex data is becoming an increasingly significant part of the cognitive system and can provide the highest bandwidth channel from the computer to the human. The term visualisation, in the past, meant constructing a visual image in the mind. Now, it means a physical or graphical representation of data or concepts that clinicians or researchers graph with genomic big data. Visualisation, as a cognitive tool, has the following advantages: provides an ability to comprehend huge amounts of data; allows the perception of emergent properties that were not anticipated; enables problems with the data to become immediately apparent; facilitates understanding of both large- and small-scale features of the data; and facilitates hypothesis formation.10-12 Some intuitive visualisation tools are used to visualise multidimensional cancer genomic data, and they integrate different types of alterations with clinical data to extract useful knowledge from the vast amount of data which is generated by high-throughput technologies.13,14
New technologies are starting to be used in visual analytics of genomics such as artificial intelligence (AI) and virtual reality (VR) or augmented reality (AR). Artificial intelligence is already a part of our everyday lives and has been heralded as the key to our civilisation’s brightest future.15 Machine learning, as an approach to achieve AI, is the practice of using algorithms to parse data, learning from it, and then making a determination or prediction about something in the world.16 Machine learning boosts the next generation of visualisation which is named as intelligent visualisation. Intelligent visualisation assists a human user to handle tedious or repetitive tasks by learning from previous sessions and input data. Intelligent visualisation combines machine-learning algorithms to make high-level, goal-oriented decisions, which makes data visualisation technology directly accessible to a wide range of application scientists.17,18
Intelligent data visualisation can be used to find the relationship between genomic data and diseases and aid in the process of targeted personalised therapy.19 In the analysis of genomic data, the current statistical analysis methods are not enough for achieving data insight from the data-analysed applications. Meanwhile, applications of machine learning and data visualisation have become more attractive. Intelligent visualisation combined with machine-learning algorithms for genomic data is a big challenge and is becoming a new trend in the genomic visualisation evolution. Some modern data visualisation tools use AI technology, modern three-dimensional (3D) plots, mobile devices, and VR or AR techniques to tell the full story of genomic data. Three-dimensional and VR/AR techniques immerse the user into a digitally created space and simulate movement in three dimensions to greatly increase the bandwidth of data available to our brains.20-22 All the tools allow users to interact with the data in a way that is more natural to human cognition and movement. This includes reaching out to manipulate virtual objects constructed from the data with our hands, moving around them to view them from a clearer perspective and highlighting objects of interest with a point of the finger.
In this article, we focus on selected intelligent visual analytic tools for genomic and cancer data that are essential to support the effective disease and patient assessment. We provide a comprehensive comparison of the tools in both aspects: (1) the visualisation methods in genomic and cancer data fields and (2) the trends of visualisation in genomic analytic fields from 2000 to now. We reviewed the situation of current genomic and cancer data, the potential application to personalised medicine, and methods for genomic data visualisation. Here, we assess the units of traditional approaches such as scatter plots, heatmaps, coordinates, networks, and clustering, as well as emerging technologies involving AI and VR. We also review the evolution of genomic data visualisation tools from the speed of technology development, effective interactions, current tool status, tool integrations, and new features.
Review Strategy
Methods
This systematic review was conducted in accordance with the guidelines provided in the PRISMA statement. ‘Computational methods and resources for the interpretation of genomic variants in cancer’23 was reviewed in 2015, and ‘Expanding the computational toolbox for mining cancer genomes’24 was reviewed in 2014. In this article, we focus on tools, methods, and trends for visual analytics of genomic data, particularly cancer data. This study has no direct involvement of the handling or inclusion of personal data, so ethical approval was not necessary.
Search strategy
We commenced with a general search on a search engine, such as Google, and then in several databases, namely, BMC Genomics, Nature, Genome Research, IEEE, and ACM. We also searched through the relevant reports such as Scientific Report. In addition, a forward search of authors mentioned and the website of a tool in selected articles was also conducted. The search terms included ‘Genomic visualisation’, ‘Genomic visual analytics’, ‘Cancer data visualisation’, and ‘Genomic data visualisation tools’. These words were used for all the other database searches. Only studies published in English language from year 2000 onwards were included for review. The main reviewer extracted and analysed data from all articles in consultation with the other authors.
Bias assessment
In this article, we focused on reviewing the methods and trends of all the selected genomic data visualisation tools. There is no specific data collection process and no specific source of data, so this systematic review has no bias related to data. There is no meta-analysis in this systematic review either to avoid statistical procedure bias. We classified the tools in a tabular form and we discussed both positive and negative aspects in the main document. We aimed to minimise the bias in the discussion by referring to details that were presented in the previous publications or respectable sources.
Outcomes
Related work
Massive genomic datasets are generated by different projects, stored and shared with the different group of professionals. To help downstream analysts to access and manipulate the massive sequencing datasets in a programmatic way, new feature-rich, efficient, and robust analysis tools have been developed to process data to answer specific scientific questions.25,26 Through this, knowledge about associations between genomic factors and diseases have rapidly accumulated. Genomic analyses have provided new biologic insights into the pathogenesis and classification of diseases and insights into determinants of success and failure of therapies, which lead to develop analytic approaches that use multidimensional datasets and embrace the complexity of genomic data for personalised medicine.27,28
Personalised medicine is the tailoring of medical treatment to the individual characteristics, needs, and preferences of each patient. Personalised medicine presents the unique challenge for new tools that can efficiently extract knowledge from the data, explore the multiple relationships between the data, and speed up experts’ decisions about individual patients. Then, patients can be treated and monitored in specific ways to meet their individual needs.29-32
Personal health data are soaring with increasing number of mobile health applications. Mobile health has grown exponentially over the last several years and is expected to worth about $20.7 billion by 2018, with nearly 96 million users.33 Thousands of applications are being developed and used to collect personal health and lifestyle data, which make personalised health more personal than ever imagined. Data analytical tools can be used to visualise data from the population level to a more personalised approach, from the reactive method to proactive method, to focus on prevention, wellness, and most importantly – the individual.34,35
In the following two sections, we provide a comprehensive comparison of the tools in both aspects: (1) the visualisation methods in genomic and cancer data fields (section ‘Comparison of traditional and new methods for genomic data visualisation’) and (2) the trend of visualisation in genomic analytic fields from 2000 to 2018 (section ‘The trend of genomic data visual analytics’).
Comparison of traditional and new methods for genomic data visualisation
Along with personalised cancer medicine development, cancer genomic data visualisation in the clinical setting is becoming a key topic. Using computational and statistical methodologies, effective visualisation is crucial to successful extraction of knowledge from oncogenomic data by experts. High-throughput technologies allow the comparison of the genomic sequences, epigenomics profiles, and transcriptomes of tumour cells with those of normal cells. Visualisation techniques and tools can integrate different type of alterations with clinical experience to show vast amount of multidimensional oncogenomic data in different types of plots such as heatmaps, genomic coordinates, and networks.13,36,37 Efficient tools, that support the visual stratification of a tumour genomic profiles and that highlight their relationships to know drugs or treatments, will be more useful than the existing research-oriented tools.13,38
Researchers and doctors usually combine different visualisation methods in a typical analysis procedure to assist their work. For example, they need first to normalise experimental and batch differences between samples and then to identify differentially regulated genes based on a fold-change level when comparing across samples, such as between a healthy and a non-healthy tissue. In this procedure, principal component analysis or partitioned clustering algorithms39,40 can be used to group together genes with similar behaviour patterns, then scatter-plotting is the typical visualisation to represent such groupings. Then, categorising genes with similar behaviour patterns across time, hierarchical clustering based on expression correlation can be performed with clustering heatmaps which can allow data from distant genome loci to be grouped and visualised together for comparison.41,42
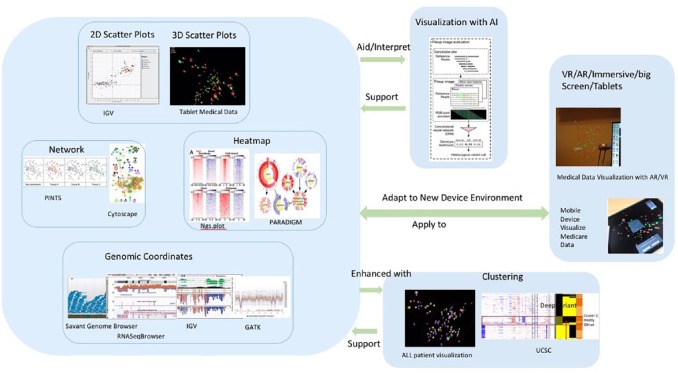
Nowadays, new visualisation tools and methods such as cluster analysis, AI, and VR are introduced by different groups of people including designers, software developers, and scientists. They try to combine existing visualisation tools with new technological opportunities, especially AI and VR, to maximise human knowledge and intuition.43-45 Figure 1 shows the genomic visualisation methods used in recent years: scatter plots, cluster, matrix heatmaps, genomic coordinates, networks, AI, and VR from screenshots of tools that are frequently used in cancer genomics research distributed according to their visualisation principles. Two-dimensional and 3D scatter plots, networks, heatmaps, and coordinates are four traditional statistical visualisation methods for genomic data which are still key methods in current popular visual analytic tools, and clustering could support all the four methods to enhance the classification of these methods. Clustering is also an AI technique that involves the grouping of data points to classify each data point into a specific group. Artificial intelligence algorithms support visualisation by automatically identifying patterns and making highly accurate prediction, meanwhile visualisation methods can interpret AI by framing predictive modelling problem and evaluating the outcome. Interactive visualisation work has been extended to emerging environments such as VR, AR, large, and high-resolution displays as well as mobile devices. Virtual reality, augmented reality, immersive, and mobile are the new environments for data visualisation to make the interactions with data in a more natural or easier way. Genomic and cancer visualisation tools have supported new environments to enhance human’s perception in such environments.46 The tools usually include multiple visualisation methods, for example, Integrative Genomics Viewer (IGV) uses both scatter plot and genomic coordinate and UCSC uses scatter plot, clustering, and genomic coordinate. We provide a summary of popular visualisation methods, their description, and the tools in Table 1. We also illustrate the popular genomic data visualisation methods and the environments in Figure 1, including scatter plots, cluster, heatmap, networks, genomic coordinates, AI, and VR.
Figure 1.
Genomic data visualisation methods and environments: scatter plots, cluster, heatmap, networks, genomic coordinates, AI, and VR for visualisation. Two-dimensional and 3D scatter plots, networks, heatmaps, and coordinates are four traditional statistical visualisation methods for genomic data which are still main methods in current popular visual analytic tools and clustering could support all the four methods to enhance the classification of these methods. Artificial intelligence algorithms support visualisation by automatically identifying patterns and making highly accurate prediction, while visualisation methods can aid or interpret AI by framing predictive modelling problem and evaluating model. Virtual reality/augmented reality/immersive/big screen/tablets are new environments for data visualisation to make the interactions with data in a more natural or easier way.
Table 1.
Summary of popular visualisation methods, their description, and the tools.
| Description | Example vVisualisation tools | |
|---|---|---|
| Two-dimensional scatter plot | The scatter diagram graphs pairs of numerical data, with one variable on each axis, to look for a relationship between them. If the variables are correlated, the points will fall along a line or curve. The better the correlation, the tighter the points will hug the line.47 | IGV48
UCSC49 |
| Three-dimensional scatter plot | Three-dimensional scatter plots are used to plot data points on three axes in the attempt to show the relationship between three variables. Each row in the data table is represented by a marker whose position depends on its values in the columns set on the X, Y, and Z axes. The fourth variable can be set to correspond to the colour or size of the markers, thus adding yet another dimension to the plot.50 | Medical Data Visualisation51 |
| Heatmap | A heatmap is a graphical representation of data that uses a system of colour-coding to represent different values. A common method of visualising gene expression data is to display it as a heatmap. In heatmaps, the data are displayed in a grid where each row represents a gene and each column represents a sample. The colour and intensity of the boxes are used to represent changes in gene expression.52 | ngs.plot53
Gitools54 PARADIGM55 |
| Clustering | A cluster is a group of similar elements. Each cluster can be represented by a profile, either a summary measure such as a cluster means or one of the elements itself, which is called a medoid or centroid.56 | Medical Data Visualisation51
UCSC57 |
| Network | A network graph uses information from both the link and the node datasets to generate a graphical depiction of the network. The nodes and links in a network graph can be arranged in a variety of layout patterns.58 | Cytoscape59 |
| Genomic coordinate | Genomic coordinate can visualise single-nucleotide polymorphism (SNP) including their physical location relative to their host gene and the structure of the relevant transcripts to provide intuitive supplements to the understanding of their functions.60 | UCSC57
IGV48 RNASeqBrowser61 GATK25 Savant Genome62 |
| Artificial intelligence (AI) | Artificial intelligence is a term of cognitive technologies and a big forest of academic and commercial work around the science and engineering intelligent machines. Artificial intelligence has many branches with many significant connections and commonalities among them, in which machine-learning is one of the branches.15 | DeepVariant63
GDC DAVE64 |
| Virtual reality (VR) | Virtual reality is by immersing the user in a digitally created space and simulated movement in three dimensions, it should be possible to greatly increase the bandwidth of data available to our brains.65 | UWS Microsoft HoloLens Visualisation |
We now explain and evaluate each visualisation method with example tools in the following paragraphs. We also analyse the combinations between these methods and how to use them in research and clinical fields.
Scatter plots
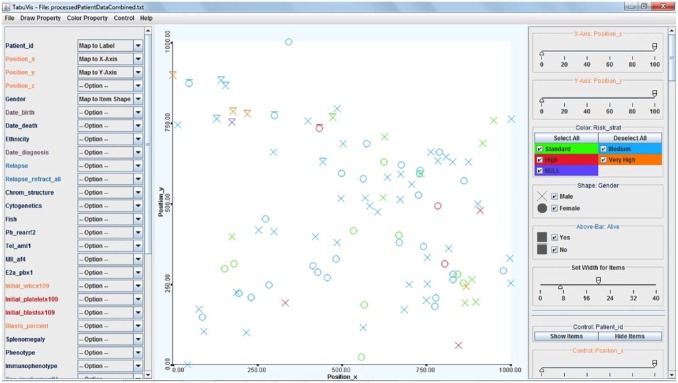
The scatter plots use horizontal and vertical axes to plot data points and display how much one variable is affected by another. The diagram graphs pairs of numerical data, with one variable on each axis, to look for a relationship between them.47 A scatter plot is a simple way to visualise genetic similarity of the patients. For example, Figure 2 shows a scatter plot of 100 acute lymphoblastic leukaemia patients with a 2D scatter plot that shows their genetic similarity. Patients’ locations are decided by their genetic properties. Two patients are close together if their genes are similar, while they are located far from each other if their genetic properties are different. The visual mapping includes the following: (1) colour → risk stratification (red, very high risk; orange, high risk; blue, medium risk; green, normal; and purple, unknown), (2) shape → gender (O, female; X, male), and (3) bar → status (top-bar, deceived; no-bar, survived). We can see from it that most of the deceased patients are located in the top-left area.51
Figure 2.
A scatter plot of 100 acute lymphoblastic leukaemia patients. Two-dimensional scatter plot showing their genetic similarity. The visual mapping includes the following: (1) colour → risk stratification (red, very high risk; orange, high risk; blue, medium risk; green, normal; and purple, unknown), (2) shape → gender (O, female; X, male), and (3) bar → status (top-bar, deceived; no-bar, survived). It shows that most of the deceased patients are located in the top-left area.51
UCSC Cancer Genomics Browser is a web-based application for hosting, visualising, and analysing cancer genomic datasets with multidimensional visualisations.57 The UCSC scatter plots are used to quickly and easily see the relationship between any two variables or columns of data such as glioblastoma multiforme (GBM) and lower grade glioma (LGG) samples.49
Three-dimensional scatter plot is used to discover relationships between three variables at the same time and is boosted by the recent widespread use of VR devices. Even though VR has been in development for decades, only recently are into producing compelling experiences. Virtual reality reveals spatially complex structures behind 3D data and 3D scatter plots and can solve the problematic issues on common 2D scatter plots such as overlapping of data and the absence of depth perception.66 Some genomic and cancer data visualisation tools such as Medical Data Visualisation started to use 3D scatter plots and supported mixed reality devices such as Microsoft HoloLens.
Heatmaps
Heatmap is a 2D graphical false-colour image representation of data which makes use of a predefined colour scheme, and different colours display different values and variations in a data matrix. Heatmap plot is a fundamental method in genomic data visualisation and is broadly used to unravel patterns hidden in genomic data, especially popular used for gene expression analysis and methylation profiling.67 Many genomic visualisation tools provide heatmap plots, such as ngs.plot, Gitools, and PARADIGM. Figure 3 shows a heatmap for comparing gene of interests between four patients: ALL92, ALL129, ALL321, and ALL323 which were chosen by users.
Figure 3.
Heatmap for comparing gene between different patients: ALL92, ALL129, ALL321, and ALL323.51
Heatmaps are very handy for large, multidimensional dataset visualisation. High-throughput gene expression data are often displayed using heat maps: data are displayed in a grid where each row represents a gene and each column represents a sample. Colour and intensity of each box represent variations of gene expression. Scientists often use green-black-red heat maps to visualise gene expression data from microarrays.68
Most heatmap representations are also combined with clustering methods to group genes or samples based on their expression patterns. Each gene is represented as a row and is colour-coded to represent the intensity of its variation, such as positive or negative, relative to a reference value, and biological samples are represented as columns in the grid.69
Genomic coordinates
Genomic coordinate plot is a common way to visualise oncogenomic data to show alterations tied to their genomic loci. UCSC, IGV, RNASeqBrowser, GATK, and Savant Genome provide genomic coordinates. The different tool may have the different focus but most of them can display genomic topography of alterations in each tumour samples as genomic tracks to inspect particular genome loci.
Integrative Genomics Viewer is a lightweight visualisation tool for interactive exploration of integrated genomic datasets and it makes use of efficient, multi-resolution file formats to enable intuitive real-time exploration of diverse, large-scale genomic datasets on standard desktop computers. Integrative Genomics Viewer can handle large heterogeneous dataset to provide a smooth and intuitive user experience at all levels of genome resolution. It uses special data tiling technique which is a pyramidal data structure to support interactive exploration of large-scale genomic datasets on standard desktop computers.70
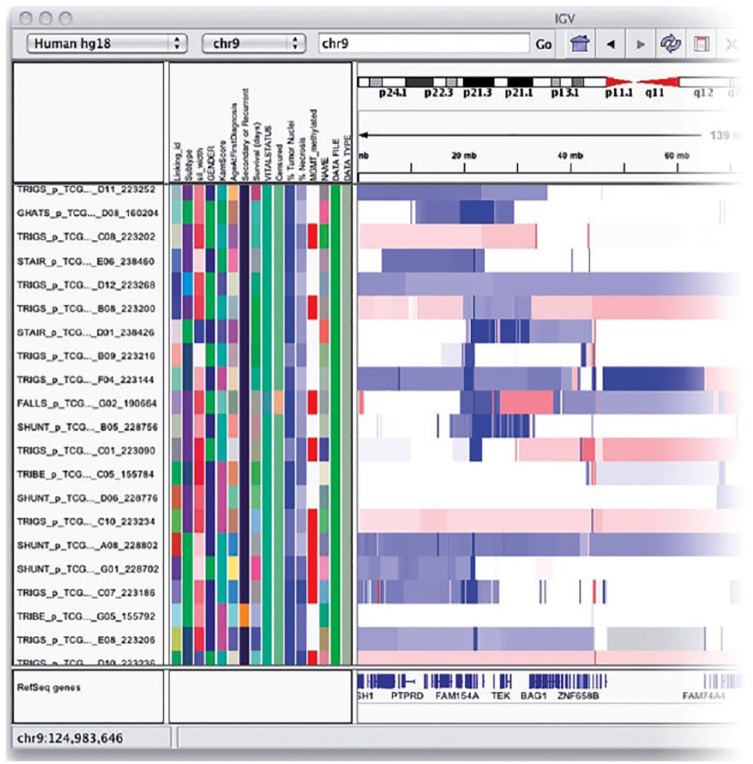
In IGV, all tracks can be annotated with a coordinate application colour-coded sample and clinical information. Genomic regions can be annotated with text labels.71 Figure 4 shows an IGV attribute panel that displays a colour-coded matrix of phenotypic and clinical data. Just below the command bar is a header panel with an ideogram representation of the currently viewed chromosome, along with a genome coordinate ruler that indicates the size of the region in view. The remainder of the window is divided into one or more data panels and an attribute panel. Data are mapped to the genomic coordinates of the reference genome and are displayed in the data panels as horizontal rows called ‘tracks’. Each track typically represents one sample, experiment, or genomic annotation. If any sample or track attributes have been loaded, they are displayed as a colour-coded matrix in the attribute panel. Each column in the matrix corresponds to an attribute, and a track’s attribute values are displayed as a row of coloured cells adjacent to the track.70
Figure 4.
Integrative Genomics Viewer’s genomic coordinates show a colour-coded matrix of phenotypic and clinical data. Just below the command bar is a header panel with an ideogram representation of the currently viewed chromosome, along with a genome coordinate ruler that indicates the size of the region in view. Data are mapped to the genomic coordinates of the reference genome and are displayed in the data panels as horizontal rows called ‘tracks’. Each track typically represents one sample, experiment, or genomic annotation.70
Networks
Networks can show functional relationships between different genomic entities to allow the researchers to explore visually clusters of nodes representing highly interconnected altered genes that can constitute driver pathways or subnetworks. Cytoscape provides network visualisation in genomic research.
Cytoscape is an open-source software for visualising complex networks and integrating these with any type of attribute networks desktop data such as genomic data and clinical patient information. Cytoscape is most powerful when used in conjunction with large databases of protein-protein, protein-DNA, and genetic interactions that are increasingly available for humans and model organisms. The software is extensible through a straightforward plug-in architecture, allowing rapid development of additional computational analyses and features.59
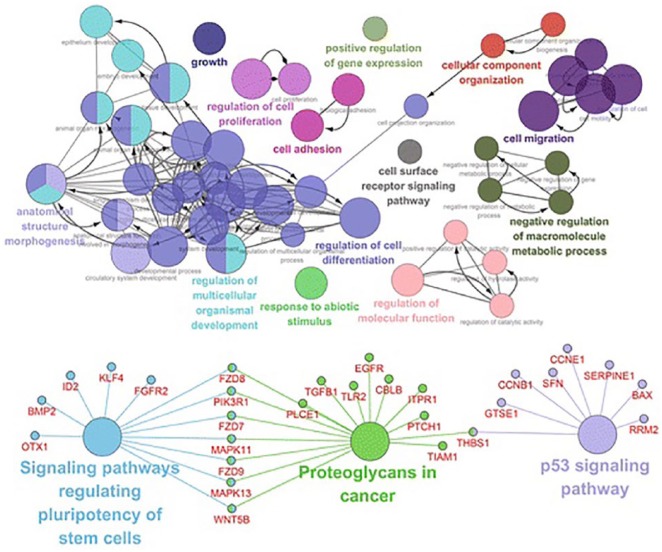
Figure 5 shows breast cancer genomic data visualisation with network method from Cytoscape v3.4.0. The upper network shows the gene ontology (GO) analysis based on the biological process of the 513 differentially expressed genes (DEGs), and the bottom network shows the KEGG pathway analysis of the 513 DEGs.72
Figure 5.
Network visualisation from Cytoscape v3.4.0. The upper network shows the GO analysis based on the biological process of the 513 DEGs, and the bottom network shows the KEGG pathway analysis of the 513 DEGs.72
Cluster
Cluster is a strategy that is used to combine other visualisation methods such as scatter plots, heatmaps, and networks. For example, Medical Data Visualisation uses scatter plot cluster, while UCSC uses heatmap cluster. A cluster is usually a group of similar elements that can be represented by a profile, either a summary measure such as a cluster means or one of the elements itself.
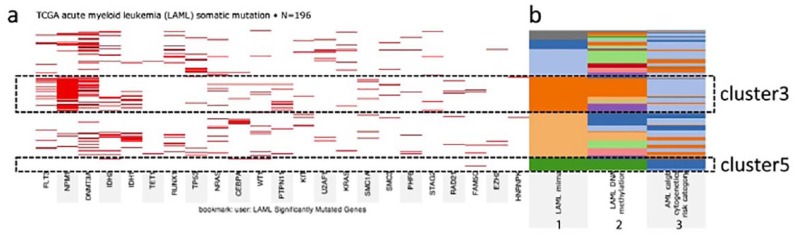
Clustering combined with heatmaps enable grouping of genes or samples which can be obtained through high-throughput sequencing methods such as RNA sequencing or DNA microarray studies together. Clustering is useful in visualising similarity of gene expression pattern.68 Figure 6 shows a clustering heatmap to explore relationships between somatic mutation profiles, genomic subtypes, and survival. It illustrates the somatic mutation profile of the significantly mutated genes in The Cancer Genome Atlas (TCGA) project acute myeloid leukaemia (AML) cohort, as well as the corresponding AML subtype designations for these samples.73
Figure 6.
UCSC shows clustering heatmaps to explore relationships between somatic mutation profiles, genomic subtypes, and survival. (A) Somatic mutations for the most significantly mutated genes in The Cancer Genome Atlas (TCGA) project AML tumour samples. Samples are arranged in rows and genes in columns. A strong concordance is observed between miRNA cluster 3 (orange), DNA methylation cluster 3 (also orange), and intermediate cytogenetic risk (light blue); and between miRNA cluster 5 (green), DNA methylation cluster 5 (also green), and favourable cytogenetic risk (dark blue). (B) Column 1 represents the miRNA expression clusters, Column 2 represents the DNA methylation clusters, and Column 3 represents cytogenetic risk category for the AML cohort.
Clustering method can combine scatter plots, network, and genomic coordinate methods to show a group of similar elements. Clustering data can identify a subset of representative examples to process sensory signals and detect patterns in data. Clustering data based on a measure of similarity is a critical step in scientific data analysis and in engineering systems. A common approach is to use data to learn a set of centres such as the sum of squared error between data points and their nearest centres is small.74
Artificial intelligence for genomic data visualisation
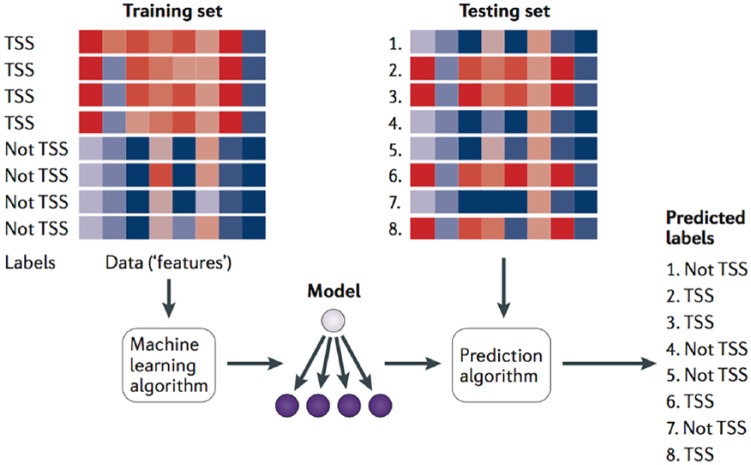
In recent years, AI has started to be used in big data visualisations, including multivariate genomic data for development of quicker hardware.75 Machine learning is one branch of the field of AI, and it is a way of solving problems without explicitly codifying the solution and a way of building systems that improve themselves over time. Machine-learning goals are typically used to build predictive or descriptive models from characteristic features within a dataset and then use those features to draw conclusions from other similar datasets. For example, in cancer detection, diagnosis, and management, machine learning helps identify significant factors in high-dimensional datasets of genomic, proteomic, chemical, or clinical data which can be used to understand the predicate underlying diseases, as well as to provide possible insights into effective disease-management strategies. Machine learning combined with data visualisation should have three stages: developing an algorithm, applying genomic data to the algorithm, and predicting new unlabelled data.76 Figure 7 shows a canonical example of a machine-learning application with these three stages. A training set of DNA sequences is provided as input to a learning procedure, along with binary labels indicating whether each sequence is centred on a transcription start site (TSS) or not. The learning algorithm produces a model that can then be subsequently used, in conjunction with a prediction algorithm, to assign predicted labels (such as ‘TSS’ or ‘not TSS’) to unlabelled test sequences. In Figure 7, the red-blue gradient might represent, for example, the scores of various motif models (one per column) against the DNA sequence.
Figure 7.
A canonical example of a machine-learning application for DNA sequences, A training set of DNA sequences is provided as input to a learning procedure, along with binary labels indicating whether each sequence is centred on a transcription start site (TSS) or not. The learning algorithm produces a model that can then be subsequently used, in conjunction with a prediction algorithm, to assign predicted labels (such as ‘TSS’ or ‘not TSS’) to unlabelled test sequences.76
DeepVariant is a tool that uses the latest AI techniques to build a more accurate picture of a person’s genome from sequencing data. The tool fed the data from millions of high-throughput reads and fully sequenced genomes from the Genome in a Bottle (GIAB) project, a public-private effort to promote genomic sequencing tools and techniques, to a deep-learning system and painstakingly tweaked the parameters of the model until it learned to interpret sequenced data with a high level of accuracy.77 DeepVariant is a genomic variant caller which uses deep neural networks to call genetic variants in germline genomes. It is originally developed by Google Brain and Verily Life Science and it won the 2016 PrecisionFDA Truth Challenge award for Highest SNP Performance.63
The future of big data visual exploration will involve the tight integration of visualisation tools with traditional techniques from such disciplines as statistics, machine learning, operations research, and simulation. Visual exploration also needs to combine fast automatic data mining algorithms with the intuitive power of the human mind which can improve the quality and speed of the data exploration process.78
Virtual reality and augmented reality
Virtual reality enables the psychophysical immersive experience in an artificially computer-generated virtual environment.79 Augmented reality, usually, is built upon VR in integrating and overlaying the virtual environment into the user’s real world and allowing the user to interact with the virtual objects in the context of his or her actual surroundings.80,81 Special equipment such as a head-mounted display (HMD) or cave automatic virtual environment (CAVE) system is required for the use of VR/AR technologies. The sensor and camera on the equipment will help the system to determine and track the user moment and move the point of view accordingly.
Shan et al80 developed an AR visualisation which runs on the mobile platform to deliver real-time 3D brain tumour volume rendering. It allows the clinician to visualise and communicate with the patients on their tumour sizes and locations. The visualisation uses the facial features of the patient as the tracking point to project the reconstructed brain tumour model onto the same location as the subject’s actual anatomy. Chang et al81 have created a 3D AR visualisation for archaeological purposes. It uses the ARToolKit in rendering the objects. The purpose of the visualisation is to create a platform for underground cultural heritage protection and research.
Some analysts even think the application of AI to VR enables important possibilities such as AI-based continuous image recognition reporting results in a VR display.82 One of the biggest challenges of big data is extracting information in a way that enables clinicians to quickly use the vast amount of data to analyse the purpose of making better decisions in a timely manner. Immersive environments such as AR and VR can measure people’s reactions of large datasets to understand the subconscious process of the human brain to determine the optimum amount of information. Virtual reality either simplifies the visualisations so as to reduce the cognitive load, thus keeping the user less stressed and more able to focus, or it will guide the person to the areas of the data representation that are not as heavy in information.83,84
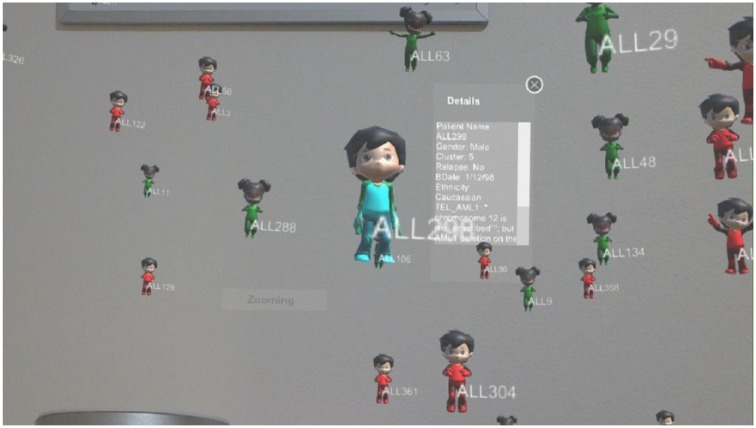
Children Cancer Data Visualisation tool can show the whole group of patients’ data with a 3D scatter plot and check a single patient’s details. It can also zoom and rotate the visualisation plot, compare gene among several patients, and interact with users and shows the comparison visualisation between selected patients.22 The tool supports different mobile operating systems such as iOS and Android, and VR devices. Figure 8 shows a 3D scatter plot from the tool running on Microsoft HoloLens, which is a pair of mixed reality smart glasses developed and manufactured by Microsoft. HoloLens gained popularity for being one of the first computers running the Windows Mixed Reality platform under the Windows 10 operating system and it can trace its lineage to Kinect, an add-on for Microsoft’s Xbox gaming console that was introduced in 2010.85
Figure 8.
Children Cancer Data Visualisation tool running in Microsoft HoloLens. It shows a 3D scatter plot and checks individual patient’s details.
The trend of genomic data visual analytics
We compared genomic data visualisation tools via the timeline since 2000s. We evaluated the trend of visual analytics and the current status of these tools. Particularly, the usefulness of the software and how the tools assist with genomic analysis are evaluated.
Rapidly evolving genomic and cancer data, and intelligent visualisations
‘A picture is worth a thousand words’ – this is an adage especially for life science which is one of the biggest generators of enormous datasets because of recent and rapid technological advances. The complexity of genomic data makes these datasets incomprehensible without effective visualisation methods. Genomic data visualisation is a rapidly evolving field and great progress has been achieved in many areas including hardware acceleration, standardised exchangeable file formats, dimensionality reduction, visual feature selection, multivariate data analyses, interoperability, 3D rendering, and visualisation of complex data at different resolutions, especially the area of image processing combined with AI-based pattern recognition.86
Interactive visualisation of complex genomic data is an effective way to bring the insight of information and to discover the relationships, non-trivial structures, and irregularities that may pertain to the disease course of the patient. Basic statistics and visualisations without effective interaction and capabilities to control the visual data mining process are often insufficient for the analysis and exploration process. Intelligent visualisation can focus on patient-to-patient comparisons through the biological data and then display the multidimensional data in cooperation with the automated analysis.51 Intelligent genomic visualisation can support experts in the process of hypotheses generation concerning the roles of genes in diseases and find the complex interdependencies between genes by bringing gene expressions into context with pathways.87
The evolution of genomic data visual analytics
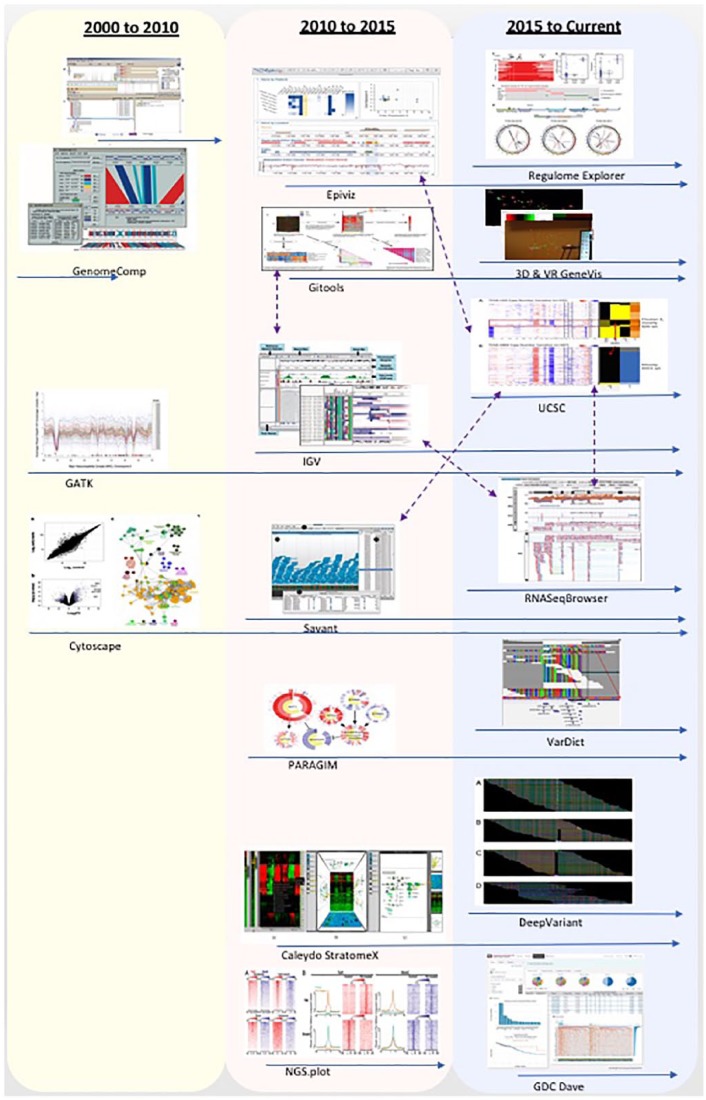
Figure 9 shows the tools for visual analytics of genomic and cancer data grouped in the years they started to be developed or extracted from papers written during those years. We can see that between 2000 and 2015, most genomic data visualisation tools only use some traditional methods such as scatter plots, heatmaps, genomic coordinates, networks, and clustering. From 2016, new visualisation techniques started to be used such as machine-learning algorithms for predictions and personalised medicine. Some visualisation tools can be ran on environments such as mobile devices and VR/AR/immersive big screen. Some tools were used for a short time such as X:Map and GenomeComp, while some tools were developed very early before 2010, but kept being updated and added new features until now such as GATK and Cytoscape, which are still very popular genomic data visualisation tools now. Integration among tools is also a key to keep a tool lasting for a longer time. For example, Epiviz can obtain annotation data from the UCSC, Gitools can get heatmaps from IGV, and RNASeqBrowser is compatible with UCSC as shown in Figure 9 with purple arrows.
Figure 9.
Timeline and integration of tools; the blue arrows stand for timeline and the purple arrows stand for integration. Between 2000 and 2015, most genomic data visualisation tools only used some traditional methods such as scatter plots, heatmaps, genomic coordinates, networks, and clustering. From 2016, new visualisation methods started to be used such as machine-learning algorithms for predictions and personalised medicine.
Table 2 shows the tool list for visual analytics of genomic and cancer data. Some tools have not been updated recently such as GenomeComp, X:Map, PARADIGM, and ngs.plot, while most tools are still being maintained very well or upgraded with new technologies such as IGV and other tools as shown in Figure 9. Some non-updated tools are still used and can be downloaded from online. GenomeComp is a visualisation tool which is implemented as a stand-alone programme that can compare, parse, and visualise large genomic sequences, especially closely related genomes such as interspecies or interstrain.90 It was developed by Laboratory of Bioinformatics, Institute of Biophysics, Beijing, and use Perl/TK, and can run on Linux, Unix, Mac OS X, and Microsoft Windows operating systems. The last version update happened in 2004.98 X:Map is a genome annotation database browser developed by the University of Manchester, UK, around 2008. It is a tool designed for annotation and visualisation of genome structure for Affymetrix exon array analysis.89 PARADIGM is a tool which focuses on inferring patient-specific genetic activities incorporating curated pathway interactions among genes and can predict the degree to which a pathway’s activities are altered in the patient using probabilistic inference.55
Table 2.
Tools for visual analytics of genomic and cancer data.
| Tool name/website | Description | Visualisation methods | Developer/year | Tool type |
|---|---|---|---|---|
| Genome Analysis Toolkit (GATK) https://software.broadinstitute.org/gatk/ |
Genome Analysis Toolkit (GATK) is designed to process exomes and whole genomes generated with illumine sequencing technology and can also be adapted to handle a variety of other technologies and experimental designs. This toolkit focuses on the variant discovery and also includes many utilities to perform related tasks such as processing and quality control of high-throughput sequencing data.88 | Genomic coordinates, cluster, 2D scatter plot, AI | Broad Institute of MIT and Harvard 2004 to current |
Structured Java programming framework |
| X:Map http://xmap.picr.man.ac.uk |
X:Map is a tool which is designed specifically for high-density microarrays that are required to show for each gene, transcript, and exon the probe sets that match it, their specificity, and for each probe, their locations of potential hybridisation and for each individual exon, its sequence.89 | Heatmap, genomic coordinates | University of Manchester, UK 2008 |
Genome annotation database browser |
| GenomeComp http://www.mgc.ac.cn/GenomeComp/ |
GenomeComp is a visualisation tool which is implemented as a stand-alone programme that can compare, parse, and visualise large genomic sequences, especially closely related genomes such as interspecies or interstrain.90 | Genomic coordinates | Laboratory of Bioinformatics, Institute of Biophysics,
Beijing 2002-2004 |
Use Perl/TK, run in Linux, Unix, Mac OS X, and Microsoft Windows |
| Epiviz http://epiviz.cbcb.umd.edu/ |
Epiviz is a genomic information visualisation tool which can quickly and easily visualise and compare large amounts of genomic information resulting from high-throughput sequencing experiments. It is the first system to provide tight integration between a state-of-the-art analytics platform and a modern, powerful, integrative visualisation system for functional genomics.91 | Heatmaps, 2D scatter plot, genomic coordinates | University of Maryland 2014 to now |
Web-based genome browsing application |
| Gitools http://www.gitools.org/ |
Gitools is a desktop application for analysis and visualisation of matrices using interactive heatmaps which contain multiple dimensions. It has interactive capabilities to allow the user to filter, sort, move, and hide rows and columns in the heatmaps. Gitools is especially useful for cancer genomic analysis as it includes all the methods implemented for some integrative sources and can import data directly from some other tools.54 | Heatmaps | Biomedical Genomics Group located in Barcelona at the
Biomedical Research Park in Barcelona 2011 to current |
Desktop application |
| UCSC https://genome-cancer.ucsc.edu/ |
UCSC Cancer Genomics Browser is a web-based application for hosting, visualising, and analysing cancer genomic datasets. The browser provides interactive views of data from genomic regions to annotated biological pathways and user-contributed collections of genes.57 | Heatmap, cluster | UCSC in the University of California system 2015 to current |
Web-based application |
| Integrative Genomics Viewer (IGV) http://software.broadinstitute.org/software/igv/ |
Integrative Genomics Viewer (IGV) is a lightweight visualisation tool for interactive exploration of integrated genomic datasets and it supports a wide range of genomic data including aligned sequence reads, mutations, copy number, RNAi screen, gene expression, methylation, and genomic annotations.71 | Heatmap, genomic coordinates, cluster, 2D scatter plot | Broad Institute, the University of
California 2013 to current |
Visualisation tool for integrated genomic datasets |
| Savant Genome Browser http://www.genomesavant.com/p/home/index/ |
Savant Genome Browser is a sequence annotation, desktop visualisation, and analysis browser for genomic data. This tool was primarily developed for the effective visualisation of large sets of high-throughput sequencing data. Multiple visualisation modes enable the exploration of genome-based sequence, points, intervals, or continuous datasets. Plug-ins are available, among which is the WikiPathways plug-in, which aids the navigation of the data by the integration of pathways.62 | Genomic coordinates, heatmap, cluster | The Computational Biology Lab at the University of Toronto.92
2010 to current |
Desktop visualisation and analysis browser for genomic data |
| PARADIGM http://sbenz.github.io/Paradigm/ |
PARADIGM is a tool which focuses on inferring patient-specific genetic activities incorporating curated pathway interactions among genes and can predict the degree to which a pathway’s activities are altered in the patient using probabilistic inference. CircleMap is one of the PARADIGM visualisation methods that produce heatmaps with a circular layout.55 | Heatmap | Charles Vaske, Steve Benz, University of California,
Santa Cruz 2010 |
A factor graph framework for pathway inference on high-throughput genomic data |
| CaleydoStratomeX http://caleydo.org/tools/stratomex/ |
CaleydoStratomeX is a visual analytic framework prepared for the visualisation of interdependencies between multiple datasets. It allows exploration of relationships between multiple groupings and different datasets. It can cluster genomic data of different alterations and represents them as matrix heatmaps. The different groupings are connected by ribbons whose width corresponds to the number of samples shared by the connected clusters. Clinical data and pathway maps can be integrated to characterise the clusters.93 | Heatmap, cluster | Marc Streit, Linz, Alexander Lex, Nils Gehlenborg,
Christian Partl, Samuel Gratzl, Hanspeterpfister, Dieter
Schmalstieg, and Peter J. Park.94
2012 to current |
StratomeX is a visual analytic framework for the analysis of multiple stratified datasets |
| Regulome Explorer http://explorer.cancerregulome.org |
Regulome Explorer is a tool for the visualisation options that includes circular and linear genomic coordinates and networks.95 The Cancer Genome Atlas takes an integrated approach towards a systems-level understanding of regulatory disruptions in cancer which are intertwined within complex dynamical networks through a multitude of interactions among different types of molecules.96 | Heatmap, genomic coordinates |
Institute for Systems Biology and MD Anderson Cancer
Centre 2016 to current |
A tool for the integrative exploration of associations between clinical and molecular features of data |
| Cytoscape http://www.cytoscape.org |
Cytoscape is an open-source software for visualising complex networks and integrating these with any type of attribute Networks Desktop data such as genomic data and clinical patient information.59 | Networks | US National Institute of General Medical Sciences
(NIGMS) and National Resource for Network Biology
(NRNB). 2003 to current |
An open-source software platform for visualising complex networks |
| ngs.plot https://code.google.com/p/ngsplot |
ngs.plot is a tool to help understand the relationship between the millions of functional DNA elements and their protein regulators and demonstrate how they work in conjunction to manifest diverse phenotypes. ngs.plot uses two steps to quickly mine and visualise genome samples: the first step is to define a region of interest and the second step is to plot something meaningful.53 | Heatmap | Peter Briggs from the University of
Manchester supported by the Friedman Brain Institute; and the National Institutes of Health 2014 |
A quick mining and visualisation tool for NGS
data Programming language is R and Python |
| GDC DAVE (Genomic Data Commons Data Analysis,
Visualisation, and Exploration) https://gdc.cancer.gov/analyse-data/gdc-dave-tools |
GDC DAVE Tools allow users to interact intuitively with GDC data and promote the development of a true cancer genomics knowledge base, which including the following key features: view most frequently mutated genes, plot high-impact mutations using oncoGrid, perform survival analysis, visualise mutations for protein-coding regions, view cancer distribution, view top mutated genes across projects, view genes annotated by COCMIC, build and compare custom cohorts, and perform set operations.64 | Heatmap, 2D scatter plot, cluster | The National Cancer Institute (NCI) Centre for Cancer
Genomics (CCG) from Maryland, USA 2016 to current |
GDC Data Portal |
| VarDict https://github.com/AstraZeneca-NGS/VarDict |
VarDict is a novel and versatile variant caller for both DNA- and RNA-sequencing data and it simultaneously calls SNA, MNV, InDels, complex and structural variants, expanding the detected genetic driver landscape of tumours.97 | Heatmap, genomic coordinates | AstraZeneca which is in the United States. 2016 to current |
VarDict is implemented in Perl |
| DeepVariant https://github.com/google/deepvariant |
DeepVariant is a tool that uses the latest AI techniques to build a more accurate picture of a person’s genome from sequencing data. The tool fed the data from millions of high-throughput reads and fully sequenced genomes to a deep-learning system and painstakingly tweaked the parameters of the model until it learned to interpret sequenced data with a high level of accuracy.77 | Artificial intelligence, genomic coordinates, heatmap | Google Brain and Verily Life Science. 2016 to current |
Deep neural networks to call genetic variants in germline genomes |
| RNASeqBrowser | RNASeqBrowser is a visualisation tool that incorporates and extends the function of the UCSC genome browser and NGS visualisation tools such as IGV.61 | Genomic coordinates, cluster | JA, Australian Government Department of
Health 2015 to current |
A visualisation tool that incorporates and extends the function of UCSC and IGV |
| Children Cancer Data Visualisation | Children Cancer Data Visualisation tool can show the whole group of patients’ data with a 3D scatter plot and check a single patient’s details, zoom and rotate the visualisation plot, compare gene among several patients, and interact with users and shows the comparison visualisation between selected patients22 | 3D scatter plot, heatmap, cluster, VR | Western Sydney University 2016 to current |
Developed by Java, Unity 3D |
CircleMap is one of the PARADIGM visualisation methods that produce heatmaps with a circular layout. Different datasets coming from the same samples can be plotted as different layered circles that form a node. The data layers are plotted application maintaining the sample order, which can be adjusted by the user. CircleMap visualisation can be used to display multiple datasets centred around each gene in a pathway.55 The tool is a factor graph framework for pathway inference on high-throughput genomic data and was developed by Charles Vaske and Steve Benz from the Regents of the University of California, Santa Cruz in around 2010.
ngs.plot is a tool to quick mining and visualisation of next-generation sequencing data by integrating genomic databases. The tool visualises massive datasets and genomic information based on big sequencing data and it can produce 1 billion sequencing reads in a few days. ngs.plot uses two steps to quickly mine and visualise genome samples. The first step is to define a region of interest and the second step is to plot something meaningful.53 It is platform independent, and the programming languages are R and Python. It was produced by Peter Briggs from the University of Manchester, supported by the Friedman Brain Institute and the National Institutes of Health, and was developed in around 2014.
New visualisation techniques are applied to tools
More and more modern visualisation methods are applied to popular genomic visualisation tools. For example, Genome Analysis Toolkit (GATK) now has features for deep learning with AI technology using variants and annotations encoded as tensors, which carry the precise read and reference sequences, read flags, as well as base and mapping qualities.99 Genome Analysis Toolkit is a structured programming framework designed to process exomes and whole genomes generated with illumine sequencing technology and can also be adapted to handle a variety of other technologies and experimental designs. This toolkit focuses on the variant discovery and also includes many utilities to perform related tasks such as processing and quality control of high-throughput sequencing data.88 The GATK provides a small but rich set of data access patterns that encompass the majority of analysis tool needs and it can separate specific analysis calculations from common data management infrastructure for correctness, stability, and efficiency.25
DeepVariant is also a visualisation tool that uses machine-learning technique to identify all the mutations that an individual inherits from their parents and modelled loosely on the networks of neurons in the human brain.100 DeepVariant helps turn high-throughput sequencing readouts into a picture of a full genome. The tool developers are the researchers from the Google Brain team, who fed the data to a deep-learning system to interpret sequenced data with a high level of accuracy.77
VarDict is a tool that uses polymerase chain reaction (PCR) technology to amplify genes before submitting them to sequencing. VarDict’s abilities to detect PCR artefacts, such as amplicon bias and mispaired primers, together with the linear scalability to depth, make it desirable in such studies to reduce both false positives and false negatives. VarDict is a novel and versatile variant caller for both DNA- and RNA-sequencing data and it simultaneously calls special nucleic acids (SNAs), murine norovirus (MNV), insertion and deletion (InDels), complex and structural variants, and expanding the detected genetic driver landscape of tumours. VarDict has three main features: (1) performing scales linearly to sequencing depth, enabling ultra-deep sequencing used to explore tumour evolution or detect tumour DNA circulating in blood; (2) performing amplicon-aware variant calling for PCR-based targeted sequencing which is often used in diagnostic setting; and (3) detecting differences in somatic and loss of heterozygosity variants between paired samples. VarDict uses data from TCGA Lung Adenocarcinoma dataset to call known driver mutations in KRAS, EGFR, BRAF, PIK3CA, and MET in 16% more patients than previously published variant calls.97
Some visualisation tools start to be available on VR/AR/immersive big screen and mobile devices such as Children Cancer Data Visualisation tools. It can show the whole group of patients’ data with a 3D scatter plot and check a single patient’s details, zoom and rotate the visualisation plot, compare gene among several patients, and interact with users and shows the comparison visualisation between selected patients.22 The tool now supports mobile devices, VR devices, and other immersive environments.
Tools are integrated with each other
Some visualisation tools can be integrated to do the tool-to-tool communication. For example, Epiviz can obtain annotation data from the UCSC57 genome browser.91 Epiviz is a genomic information visualisation tool that can quickly and easily visualise and compare large amounts of genomic information resulted from high-throughput sequencing experiments. As the first system to provide tight integration between a state-of-the-art analytic platform and a modern, powerful, integrative visualisation system for functional genomics, Epiviz can interactively support a number of widely used, state-of-the-art methods for (1) ChIP-seq where iterative visualisation of data and results of peak-calling algorithms is necessary; (2) RNA-seq analysis where both location-based coverage and feature-based expression levels are required; and (3) methylation analyses using location-based analysis at multiple genomic scales.91
Gitools can get heatmaps from IGV71 through load command and then send locate commands for selected rows in the heatmaps to IGV via IGV logo in the Gitools toolbar, which makes it easy to spot and compare genes of interest within IGV.101 Gitools is a desktop application for analysis and visualisation of matrices using interactive heatmaps which contain multiple dimensions. It has interactive capabilities to allow the user to filter, sort, move, and hide rows and columns in the heatmaps. Gitools is especially useful for cancer genomic analysis as it includes all the methods implemented for some integrative sources and can import data directly from some other tools. Gitools can be used by researchers without advanced knowledge on bioinformatics as well as more experienced users who need to perform many of the operations available using the command line.54
Savant Genome Browser is a sequence annotation, desktop visualisation, and analysis browser for genomic data. This tool was primarily developed for the effective visualisation of large sets of high-throughput sequencing data. Multiple visualisation modes enable the exploration of genome-based sequence, points, intervals, or continuous datasets. Plug-ins are available, among which is the WikiPathways plug-in, which aids the navigation of the data by the integration of pathways.62 Savant also planned to expand by allowing users to automatically download annotation tracks from various public resources such as the UCSC Genome Browser.62
RNASeqBrowser is another tool that can be compatible with UCSC files and extend the functionality over IGV. RNASeqBrowser is a visualisation tool that adds several new types of tracks to show NGS data such as individual raw reads, SNPs, and InDel; it can dynamically generate RNA secondary structure which is useful for identifying non-coding RNA such as miRNA, and it overlays NGS wiggle data to display differential expression. Paired reads are also connected in the browser to enable easier identification of novel exon/intron borders and chimaeric transcripts. Strand-specific RNA-seq data are also supported by RNASeqBrowser that displays reads above (positive strand transcript) or below (negative strand transcripts) the central line.61
Tools allow more interactions and more visual analytical methods
The active tools usually allow users to interact intuitively with data and choose multi-visualisation methods to support different research purpose. For example, the Genomic Data Commons Data Analysis, Visualisation, and Exploration (GDC DAVE)Visualisation tools use scatter plot to visualise mutations and their frequency across cases mapped to a graphical visualisation of protein-coding regions and use heatmap to visualise the top mutated genes across projects and the number of cases affected. GDC DAVE Tools’ web interface can analyse cancer genomic data, in real time, online, without the need to download or process the data. Users can navigate from project cohorts to individual patients, to specific genes and mutations of interest. DAVE uses specialised graphs to visualise genomic signatures of cancer and identify potential drivers of disease and also visualise patient survival curves and identify the molecular consequence of a mutation on resultant protein.102 DAVE Tools allow users to interact intuitively with GDC data and promote the development of a true cancer genomic knowledge base, which includes the following key features: view most frequently mutated genes, plot high-impact mutations using oncoGrid, perform survival analysis, visualise mutations for protein-coding regions, view cancer distribution, view top mutated genes across projects, view genes annotated by COCMIC, build and compare custom cohorts, and perform set operations.64
Discussion
Genomic research is critical to progress against cancer. Through the study of cancer genomes, abnormalities in genes have been revealed to drive the development and growth of many types of cancer. Genomic and cancer data visualisation tools can assist in improving our understanding of the biology of cancer and lead to new methods of diagnosing and treating the disease. Over the past decade, large-scale research projects have begun to survey and catalogue the genomic changes associated with a number of types of cancer which have revealed unexpected genetic similarities across different types of tumours. For instance, mutations in the HER2 gene, distinct from amplifications of this gene, for which therapies have been developed for breast, esophageal, and gastric cancers, have been found in a number of cancers, including breast, bladder, pancreatic, and ovarian.103
Personalised medicine refers to diagnosis and treatment based on a person’s entire DNA sequence. Variants in the DNA sequence determine the differences between individuals and differences between types of cells such as tumour cells and non-tumour cells. Targeted genomic cancer medicine uses the latest genome sequencing to look at the genetics of cancer rather than treating it based on location to allow us to understand the inherited cancer risk and find more effective treatments for people with cancer.104
The cancer genomic research field is rapidly evolving in parallel with advances in high-throughput genomic technologies. This evolution of the field requires continuous advancement in visualisation techniques and tools. As this rapid scientific evolution continues, cancer researchers are highly dependent on computers to manage, analyse, and visualise data. The conventional genomic and cancer data visualisation tools are two-dimensional and present data by enhancing with the creative use of colour and size, combination of space and time, and advanced computer graphics. Most visualisation tools have four visualisation methods: two-dimensional scatter plot, networks, heatmaps, and genomic coordinates. These traditional visualisation methods are used to graph genomic and cancer data, for example, IGV supports all the four visualisation methods.
Genomic and cancer data visualisation is entering a new era with emerging sources of AI and new visual environment equipment such as VR/AR/immersive big screen and mobile devices. New technologies and evolving cognitive framework are opening new horizons to enable more accurate and contextual data visualisation.
Artificial intelligence is playing an integral role in the evolution of the field of genomics. Genomics is closely related to precision medicine whose market size projected to reach $87 billion by 2023.105 The field of personalised medicine is an approach to patient care that encompasses genetics, behaviours, and environment with a goal of implementing a patient- or population-specific treatment method in contrast to a one-size-fits-all approach. Artificial intelligence and machine learning have been applied in genomics for analysing genome sequencing, gene editing, clinical workflow, and direct-to-consumer genomics. Future applications of machine learning in the field of genomics are diverse and may potentially contribute to the development of patient or population-specific pharmaceutical drugs to look at the role of genetics in the context of how an individual responds to drugs.106 While the field is still quite new, there is already some evidences of research involving machine learning. For example, what is regarded as the first study to apply machine-learning models to determine a stable dose of Tacrolimus in renal transplant patients was published in February 2017. Tacrolimus is commonly administered to patients following a solid organ transplantation to prevent ‘acute rejection’ of the new organ.107
Virtual reality and related technologies have been adopted in health care industry. Medical researchers have been exploring ways to create 3D models of patients’ internal organs using VR since the 1990s. Recently, VR and related technologies are used to plan complex operations, reduce anxiety in cancer patients, and help patients overcome balance and mobility problems resulting from stroke or head injury. Virtual reality environment is expected to bring a revolution in genomic data visualisation as one could integrate meta-genomic data in virtual worlds. Approaching the problem from a different angle, mixed reality devices such as Google Glass, HoloLens, and Magic Leap offer an AR experience which can facilitate the learning process of the biological systems because it builds on exploratory learning.
In summary, genomic and cancer data visualisation tools are essential to facilitate decision-making for the treatment methods or targeted medicine. New technologies have been used in recent years to create visualisation tools that can explore complex genomic data. Further efforts are needed to develop new tools to meet the changing needs of the field.
Acknowledgments
The authors would like to thank Hien Dang and Jesse Tran for their invaluable comments and proof reads.
Footnotes
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research has been partially supported by the Impact Funding at Western Sydney University and Big Data, Big Impact Grant-Stage 2 from Cancer Institute of NSW, Australia.
Author Contributions: ZQ led the writing of the manuscript and did the pilot group study with the end-users of genomic visualisation tools who are cancer researchers and medical doctors. CWL contributed partially to the manuscript. QVN and YZ provided guidance and revision on the article, particularly on the technologies and methodology. DRC gave general direction on genomics and cancer research perspective as well as revision on the manuscript. DRC and QVN provide oversight and leadership to the team and initiated the projects.
ORCID iD: Zhonglin Qu  https://orcid.org/0000-0003-4500-004X
https://orcid.org/0000-0003-4500-004X
References
- 1. Dubey RC. Advanced Biotechnology. New Delhi, India: S. Chand & Company Pvt. Ltd; 2014. [Google Scholar]
- 2. Biography: Gregor Mendel Biography.com. The Biography.com website; 2017. [Google Scholar]
- 3. Ledford H. AstraZeneca launches project to sequence 2 million genomes. Nature. 2016;532:427. [DOI] [PubMed] [Google Scholar]
- 4. Croce N. Science and Technology Behind the Human Genome Project. 1st ed. New York, NY: Britannica Educational Publishing; 2015. [Google Scholar]
- 5. Francis S, Collins AP, Jordan E, Chakravarti A, Gesteland R, Walters L. New goals for the U.S. human genome project: 1998-2003. Science. 2012;282:682–689. [DOI] [PubMed] [Google Scholar]
- 6. McClean P. A history of genetics and genomics. https://www.ndsu.edu/pubweb/~mcclean/plsc411/History-of-Genetics-and-Genomics-arrative-and-overheads.pdf. Up-dated 2011.
- 7. Wistuba II, Gelovani JG, Jacoby JJ, Davis SE, Herbst RS. Methodological and practical challenges for personalized cancer therapies. Nat Rev Clin Oncol. 2011;8:135–141. [DOI] [PubMed] [Google Scholar]
- 8. Stephens ZD, Lee SY, Faghri F, et al. Big Data: astronomical or genomical? PLoS Biol. 2015;13:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Colbran LL, Chen L, Capra JA. Short DNA sequence patterns accurately identify broadly active human enhancers. BMC Genomics. 2017;18:536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ware C. Information Visualization: Perception for Design. Burlington, MA: Morgan Kaufmann; 2013. [Google Scholar]
- 11. Keahey TA. Using visualization to understand big data (advanced visualization). https://dataconomy.com/wp-content/uploads/2014/06/IBM-WP_Using-vis-to-understand-big-data.pdf. Up-dated 2013.
- 12. Green TM, Ribarsky W, Fisher B. Visual analytics for complex concepts using a human cognition model. Paper presented at: 2008 IEEE Symposium on Visual Analytics Science and Technology; October 19-24, 2008; Atlantic City, NJ. [Google Scholar]
- 13. Schroeder MP, Gonzalez-Perez A, Lopez-Bigas N. Visualizing multidimensional cancer genomics data. Genome Med. 2013;5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nguyen QV, Qian Y, Huang ML, Zhang JW. TabuVis: A tool for visual analytics multidimensional datasets. Science China Informat Sci. 2013;56:1–12. [Google Scholar]
- 15. Mills M. Artificial Intelligence in law: the state of play 2016 Thomson Reuters. https://www.neotalogic.com/wp-content/uploads/2016/04/Artificial-Intelligence-in-Law-The-State-of-Play-2016.pdf. Up-dated 2016.
- 16. What’s the difference between artificial intelligence, machine learning, and deep learning? https://blogs.nvidia.com/blog/2016/07/29/whats-difference-artificial-intelligence-machine-learning-deep-learning-ai/.
- 17. Ma KL. Machine learning to boost the next generation of visualization technology. IEEE Comput Graph Appl. 2007;27:6–9. [DOI] [PubMed] [Google Scholar]
- 18. Fuchs R, Waser J, Groller ME. Visual human+machine learning. IEEE Trans Vis Comput Graph. 2009;15:1327–1334. [DOI] [PubMed] [Google Scholar]
- 19. Nguyen QV, Gleeson A, Ho N, Huang ML, Simoff S, Catchpoole D. Visual analytics of clinical and genetic datasets of acute lymphoblastic leukaemia. In: Lu B-L, Zhang L, Kwok J, eds. Neural Information Processing: 18th International Conference (ICONIP 2011), Shanghai, China, November 13–17, 2011, Proceedings, Part I. Berlin, Germany: Springer; 2011:113–120. [Google Scholar]
- 20. How augmented reality will change data visualization. http://blog.i2econsulting.com/how-augmented-reality-will-change-data-visualization/.
- 21. Leung MKK, Delong A, Alipanahi B, Frey BJ. Machine learning in genomic medicine: a review of computational problems and data sets. Proc IEEE. 2016;104:176–197. [Google Scholar]
- 22. Nguyen QV, Khalifa NH, Alzamora P, et al. Visual analytics of complex genomics data to guide effective treatment decisions. J Imaging. 2016;2:29. [Google Scholar]
- 23. Tian R, Basu M, Capriotti E. Computational methods and resources for the interpretation of genomic variants in cancer. BMC Genomics. 2015;16:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ding L, Wendl MC, McMichael JF, Raphael BJ. Expanding the computational toolbox for mining cancer genomes. Nat Rev Genet. 2014;15:556–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chittaro L. Visualization of Patient Data at Different Temporal Granularities on Mobile Devices. Udine, Italy: Department of Math and Computer Science, University of Udine; 2006. [Google Scholar]
- 27. Sikic BI, Tibshirani R, Lacayo NJ. Genomics of childhood leukemias: the virtue of complexity. J Clin Oncol. 2008;26:4367–4368. [DOI] [PubMed] [Google Scholar]
- 28. Procter JB, Thompson J, Letunic I, Creevey C, Jossinet F, Barton GJ. Visualization of multiple alignments, phylogenies and gene family evolution. Nat Methods. 2010;7:S16–S25. [DOI] [PubMed] [Google Scholar]
- 29. Margaret A, Hamburg MD. Paving the way for personalized medicine FDA’s role in a new era of medical product development FDA. https://www.fdanews.com/ext/resources/files/10/10-28-13-Personalized-Medicine.pdf. Up-dated 2013.
- 30. Vogenberg FR, Isaacson Barash C, Pursel M. Personalized medicine: part 1: evolution and development into theranostics. Pharm Therapeut. 2010;35:560–576. [PMC free article] [PubMed] [Google Scholar]
- 31. Savoia C, Volpe M, Grassi G, Borghi C, Agabiti Rosei E, Touyz RM. Personalized medicine-a modern approach for the diagnosis and management of hypertension. Clin Sci (Lond). 2017;131:2671–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cordeiro JV. Ethical and legal challenges of personalized medicine: paradigmatic examples of research, prevention, diagnosis and treatment. Rev Portuguesa Saúde Pública. 2014;32:164–180. [Google Scholar]
- 33. Juniper: digital health: vendor analysis, emerging technologies & market forecasts 2017–2022. https://www.juniperresearch.com/researchstore/iot-m2m/digital-health/subscription/vendor-analysis-emerging-technologies. Up-dated 2018.
- 34. Krisa D, Tailor SI. Data visualization in health care: optimizing the utility of claims data through visual analysis. https://support.sas.com/resources/papers/proceedings14/SAS176-2014.pdf. Up-dated 2014.
- 35. Boudreaux ED, Waring ME, Hayes RB, Sadasivam RS, Mullen S, Pagoto S. Evaluating and selecting mobile health apps: strategies for healthcare providers and healthcare organizations. Transl Behav Med. 2014;4:363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bhojwani D, Kang H, Menezes RX, et al. Gene expression signatures predictive of early response and outcome in high-risk childhood acute lymphoblastic leukemia: a children’s oncology group study. J Clin Oncol. 2008;26:4376–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rebeiz M, Posakony JW. GenePalette: a universal software tool for genome sequence visualization and analysis. Dev Biol. 2004;271:431–438. [DOI] [PubMed] [Google Scholar]
- 38. Albuquerque MA, Grande BM, Ritch EJ, et al. Enhancing knowledge discovery from cancer genomics data with Galaxy. Gigascience. 2017;6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pollard KS, van der Laan MJ. Cluster analysis of genomic data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, eds. Bioinformatics and Computational Biology Solutions Using R and Bioconductor (Statistics for Biology and Health). New York, NY: Springer; 2005:209–228. [Google Scholar]
- 40. Ciaramella A, Cocozza S, Iorio F, et al. Interactive data analysis and clustering of genomic data. Neural Netw. 2008;21:368–378. [DOI] [PubMed] [Google Scholar]
- 41. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Olshannikova E, Ometov A, Koucheryavy Y, Olsson T. Visualizing Big Data with augmented and virtual reality: challenges and research agenda. J Big Data. 2015;2:22. [Google Scholar]
- 44. García-Hernández RJ, Anthes C, Wiedemann M, Kranzlmüller D. Perspectives for using virtual reality to extend visual data mining in information visualization. Paper presented at: 2016 IEEE Aerospace Conference; March 5-12, 2016; Big Sky, MT. [Google Scholar]
- 45. Golestan Hashemi FS, Razi Ismail M, Rafii Yusop M, et al. Intelligent mining of large-scale bio-data: bioinformatics applications. Biotech Biotechnol Equip. 2017;32:10–29. [Google Scholar]
- 46. Matte-Tailliez O, Toffano-Nioche C, Ferey N, Kepes F, Gherbi R. Immersive visualization for genome exploration and analysis. Paper presented at: 2006 2nd International Conference on Information & Communication Technologies; April 24-28, 2006; Damascus, Syria. [Google Scholar]
- 47. Scatter diagram. http://asq.org/learn-about-quality/cause-analysis-tools/overview/scatter.html.
- 48. Scatter plots. http://software.broadinstitute.org/software/igv/ScatterPlots.
- 49. UCSC Xena: box plots & scatter plots. http://xena.ucsc.edu/bar-graph-scatter-plot/.
- 50. What is a 3D scatter plot? https://docs.tibco.com/pub/spotfire/6.5.1/doc/html/3d_scat/3d_scat_what_is_a_3d_scatter_plot.htm.
- 51. Nguyen QV, Nelmes G, Huang ML, Simoff S, Catchpoole D. Interactive visualization for patient-to-patient comparison. Genomics Inform. 2014;12:21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Biological interpretation of gene expression data. https://www.ebi.ac.uk/training/online/course/functional-genomics-ii-common-technologies-and-data-analysis-methods/biological-0.
- 53. Shen L, Shao NY, Liu XC, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Perez-Llamas C, Lopez-Bigas N. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS ONE. 2011;6:e19541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vaske CJ, Benz SC, Sanborn JZ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–i245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pollard KS. Cluster Analysis of Genomic Data. College Park, MD: Center for Bioinformatics and Computational Biology; 2003. [Google Scholar]
- 57. Goldman M, Craft B, Swatloski T, et al. The UCSC Cancer Genomics Browser: update 2015. Nucleic Acids Res. 2015;43:D812–D817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Network visualization Workshop2.1 user’s guide. http://support.sas.com/documentation/cdl/en/grnvwug/62918/HTML/default/viewer.htm#p0q343kxjyj36jn1e2z6lulkda3j.htm.
- 59. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang F, Xu Y, Cao H, et al. Mapsnp: an R package to plot a genomic map for single nucleotide polymorphisms. PLoS ONE. 2015;10:e0123609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. An J, Lai J, Wood DL, et al. RNASeqBrowser: a genome browser for simultaneous visualization of raw strand specific RNAseq reads and UCSC genome browser custom tracks. BMC Genomics. 2015;16:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fiume M, Williams V, Brook A, Brudno M. Savant: genome browser for high-throughput sequencing data. Bioinformatics. 2010;26:1938–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Running DeepVariant on Google Cloud Platform. https://cloud.google.com/genomics/deepvariant.
- 64. GDC Dave Tools. https://gdc.cancer.gov/analyze-data/gdc-dave-tools.
- 65. How VR will revolutionize big data visualizations. https://www.forbes.com/sites/bernardmarr/2016/05/04/how-vr-will-revolutionize-big-data-visualizations/#2f50d104e151.
- 66. Gray GE. Navigating 3D Scatter Plots in Immersive Virtual Reality. Seattle, WA: University of Washington; 2016. [Google Scholar]
- 67. Gu ZG, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. [DOI] [PubMed] [Google Scholar]
- 68. Why data visualization is so important in biology. https://www.fiosgenomics.com/data-visualization-and-data-analysis/.
- 69. Levin C. Your top 3 heatmap generation tools. Omic Tools Blog; 2017. [Google Scholar]
- 70. Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu MS, Liu Y, Deng L, et al. Transcriptional profiles of different states of cancer stem cells in triple-negative breast cancer. Molec Cancer. 2018;17:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cline MS, Craft B, Swatloski T, et al. Exploring TCGA pan-cancer data at the UCSC cancer genomics browser. Sci Rep. 2013;3:2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. [DOI] [PubMed] [Google Scholar]
- 75. Nilsson NJ. The Quest for Artificial Intelligence: A History of Ideas and Achievements. Burlington, MA: Morgan Kaufmann; 2009. [Google Scholar]
- 76. Libbrecht MW, Noble WS. Machine learning applications in genetics and genomics. Nat Rev Genet. 2015;16:321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Google has released an AI tool that makes sense of your genome. https://www.technologyreview.com/s/609647/google-has-released-an-ai-tool-that-makes-sense-of-your-genome/.
- 78. Keim DA. Visual exploration of large data sets. Comm ACM. 2001;44:38–44. [Google Scholar]
- 79. Simpson RM, LaViola JJ, Laidlaw DH, Forsberg AS, van Dam A. Immersive VR for scientific visualization: a progress report. IEEE Comp Graph Appl. 2000;20:26–52. [Google Scholar]
- 80. Shan Q, Doyle TE, Samavi R, Al-Rei M. Augmented reality based brain tumor 3D visualization. Procedia Comp Sci. 2017;113:400–407. [Google Scholar]
- 81. Chang Y, Peng Xu W, Wang L. Research on 3D visualization of underground antique tomb based on augmented reality. Appl Mech Mater. 2013;336–338:1434-1438. [Google Scholar]
- 82. Why AI with augmented and virtual reality will be the next big thing. https://tdwi.org/articles/2017/04/04/ai-with-augmented-and-virtual-reality-next-big-thing.aspx.
- 83. Verma P. When virtual reality meets big data; 2017. [Google Scholar]
- 84. Stolk B, Abdoelrahman F, Koning A, et al. Mining the human genome using virtual reality. Paper presented at: EGPGV’02 Proceedings of the Fourth Eurographics Workshop on Parallel Graphics and Visualization; September 9-10, 2002; Blaubeuren, Germany. [Google Scholar]
- 85. Microsoft HoloLens. https://www.microsoft.com/en-au/hololens.
- 86. Pavlopoulos GA, Malliarakis D, Papanikolaou N, Theodosiou T, Enright AJ, Iliopoulos I. Visualizing genome and systems biology: technologies, tools, implementation techniques and trends, past, present and future. Gigascience. 2015;4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lex A, Streit M, Kruijff E, Schmalstieg D. Caleydo: Design and evaluation of a visual analysis framework for gene expression data in its biological context. Paper presented at: 2010 IEEE Pacific Visualization Symposium (PacificVis); March 2-5, 2010; Taipei, Taiwan. [Google Scholar]
- 88. Genome Analysis Toolkit. https://software.broadinstitute.org/gatk/.
- 89. Yates T, Okoniewski MJ, Miller CJ. X:Map: annotation and visualization of genome structure for Affymetrix exon array analysis. Nucleic Acids Res. 2008;36:D780–D786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jian Yanga JW, Yaob ZJ, Jinc Q, Shenb Y, Chena R. GenomeComp: a visualization tool for microbial genome comparison. J Microbiol Methods. 2003;54:423–426. [DOI] [PubMed] [Google Scholar]
- 91. Chelaru F, Smith L, Goldstein N, Bravo HC. Epiviz: interactive visual analytics for functional genomics data. Nat Methods. 2014;11:938–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Genome Savant. http://www.genomesavant.com.
- 93. Lex A, Streit M, Schulz HJ, et al. StratomeX: visual analysis of large-scale heterogeneous genomics data for cancer subtype characterization. Comput Graph Forum. 2012;31:1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Integrative visualization of stratified heterogeneous data for disease subtype analysis. http://caleydo.org/tools/stratomex/.
- 95. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. TCGA Genome Data Analysis Center (GDAC) for systems analysis of the cancer regulome. http://www.cancerregulome.org.
- 97. Lai Z, Markovets A, Ahdesmaki M, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. GenomeComp: a whole genome comparison and visualization tool. http://www.mgc.ac.cn/GenomeComp/.
- 99. Samwell Deep Learning in GATK4. Cambridge, MA: Broad Institute; 2017. [Google Scholar]
- 100. Google is giving away AI that can build your genome sequence. https://www.wired.com/story/google-is-giving-away-ai-that-can-build-your-genome-sequence/.
- 101. Toot-to-tool communication. http://www.gitools.org/docs/UserGuide_ToolCommunication.html.
- 102. Introducing DAVE: online analysis tools for the genomic data commons. https://www.cancer.gov/news-events/cancer-currents-blog/2017/gdc-dave-tools.
- 103. Cancer genomic research. https://www.cancer.gov/research/areas/genomics.
- 104. Stevens EA, Rodriguez CP. Genomic medicine and targeted therapy for solid tumors. J Surg Oncol. 2015;111:38–42. [DOI] [PubMed] [Google Scholar]
- 105. Precision medicine market size to exceed $87 billion by 2023: Global Market Insights Inc. https://www.prnewswire.com/news-releases/precision-medicine-market-size-to-exceed-87-billion-by-2023-global-market-insights-inc-599454691.html.
- 106. Machine learning in genomics – current efforts and future applications. https://www.techemergence.com/machine-learning-in-genomics-applications/.
- 107. Tang J, Liu R, Zhang YL, et al. Application of machine-learning models to predict tacrolimus stable dose in renal transplant recipients. Sci Rep. 2017;7:42192. [DOI] [PMC free article] [PubMed] [Google Scholar]