Abstract
Characterization of the epitopes on antigen recognized by monoclonal antibodies (mAb) is useful for the development of therapeutic antibodies, diagnostic tools, and vaccines. Epitope mapping also provides functional information for sequence-based repertoire analysis of antibody response to pathogen infection and/or vaccination. However, development of mapping strategies has lagged behind mAb discovery. We have developed a site-directed mutagenesis approach that can be used in conjunction with bio-layer interferometry (BLI) biosensors to map mAb epitopes. By generating a panel of single point mutants in the recombinant hemagglutinin (HA) and neuraminidase (NA) proteins of influenza A viruses, we have characterized the epitopes of hundreds of mAbs targeting the H1 and H3 subtypes of HA and the N9 subtype of NA.
1. Introduction
Antibodies are an important mechanism of protection against many pathogens, especially after vaccination. Individual antibodies recognize pathogen antigen (Ag) through contact of the antibody paratope with its epitope, a cluster of amino acids on the Ag surface (Cohn, 2002). The degree of antibody mediated protection from lethal infection is largely dependent on the precise targeting on pathogen Ag(s), typically those surface associated. After the pathogen is tagged by the antibody, it can be cleared or otherwise neutralized by other components of the immune system. Binding of the antibody may also directly neutralize the pathogen if the epitope is located on a functional domain essential for invasion or replication of the pathogen. The polyclonal antibody repertoire produced during a humoral immune response has the potential to recognize many different epitopes, whereas epitope specificity is a distinguishing characteristic for each monoclonal antibody (mAb) within this polyclonal response (Wilson and Cox, 1990). Understanding the fine specificity of mAb contained within the overall polyclonal response can greatly aid our understanding of an immune response. Further, mAb are widely used for many purposes, including research applications, diagnostic tools and therapeutics. Therapeutic mAbs are rapidly being developed as potential tools to treat major diseases and several anti-influenza mAbs are in clinical trials (Sparrow et al., 2016).
Although mAb can be used without understanding the precise area to which they bind, mapping their epitopes offers a molecular understanding of the mAb function. Current experimental methods for epitope mapping can be categorized into structural or functional in approach (Potocnakova et al., 2016). Structural approaches such as X-ray crystallography (Abbott et al., 2014), nuclear magnetic resonance (NMR) (Blech et al., 2013), and cryo-electron microscopy (Liu et al., 2017) can provide a complete high-resolution map of an epitope. Functional approaches such as peptide scanning (Reineke, 2009), phage/yeast display (Rojas et al., 2014; Najar et al., 2017), mutagenesis (Greenspan and Di Cera, 1999), and the generation of escape mutants (Knossow et al., 1984) can reveal the contribution of particular residues to the binding strength of the antibody-antigen complex.
The ability to obtain large numbers of mAbs has been greatly improved by recent technological advancements. For example, human mAbs against the hemagglutinin (HA) of 1918 A(H1N1) influenza pandemic virus were generated from limited numbers of donor cells by using optimized human hybridoma approaches (Yu et al., 2008). More than 500 mAbs specific for the HIV envelope glycoprotein were generated from single B cells by single cell RT-PCR and expression vector cloning (Scheid et al., 2009). At the same time, next-generation sequencing techniques have made it practical to directly sequence the antibody variable region repertoire amplified from primary cells, and generate large sequence databases that are valuable for mAb discovery and immune profiling (Finn and Crowe, 2013; Parola et al., 2017). In contrast, high-throughput fine mapping of mAb epitope remains technically challenging and new screening strategies are therefore needed to meet the growing demands of this rapidly advancing field.
Influenza A viruses (IAV) cause major respiratory disease each year and the immune response to IAV infection has been extensively studied (Couch and Kasel, 1983; Wilson and Cox, 1990). The major membrane glycoproteins of IAV, HA and NA, are the primary targets for the humoral immune response initiated by IAV infection or vaccination (Steinhauer and Skehel, 2002; Fiore et al., 2009). Many antigenically distinct strains and subtypes of IAV exist in nature. Currently, in humans, the A(H1N1) strain arising from the 2009 pandemic (“A(H1N1)pdm09”) and an A(H3N2) strain cause seasonal epidemics each year. Zoonotic IAV present risks of further human pandemics, with avian A(H7N9) representing the highest concern for a pandemic at the moment (Jernigan and Cox, 2015). This strain causes severe illness in sporadically-infected humans with a mortality rate approaching 40%. Neuraminidase inhibitors (NAIs) have been the main drugs available for the treatment of A(H7N9)-infected patients (Hai et al., 2013; Itoh et al., 2015). Alarmingly, NAI-resistant strains have been isolated from A(H7N9)-infected patients that have been treated with NAIs, confirming the need for new antiviral strategies.
The HA and NA surface proteins each carry out functions critical to the virion life cycle and anti-HA and -NA antibodies play a dominant role in protection against influenza virus infection (Steinhauer and Skehel, 2002). HA facilitates viral entry into a host cell through binding to terminal sialic acids of glycoproteins on the cell surface, and mediates fusion between endosomal and viral membranes that is triggered by the low pH of the endosomal compartment. NA catalyzes the hydrolysis of terminal sialic acid residues from cell receptors as well as newly formed virions, and therefore help release virus from cells for the spread of infection and to prevent the aggregation of viral particles. HA exists as a homotrimer. The ectodomain of each monomer has a globular head containing the receptor binding site and the major antigenic sites, and a stem region that possesses the membrane fusion peptide. NA is a homotetramer with the ectodomain of each monomer consisting of a stalk region and a box-like head where the enzyme active site and calcium binding domain are located.
Classically defined antigenic sites on the HA head have been mapped by tracking patterns of amino acid changes in isolated viruses and by the generation of mAb escape mutants (Wiley et al., 1981; Caton et al., 1982). These sites typically include multiple overlapping epitopes and represent regions on the protein that are particularly immunogenic. Five such sites (Sa, Sb, Ca1, Ca2 and Cb) have been identified on the H1 subtype of HA and five (A through E) on the H3 subtype (Webster and Laver, 1980; Wiley et al., 1981; Caton et al., 1982). These sites are highly variable between strains of virus, and have been considered as hot-spots for the antigenic drift that causes seasonal influenza epidemics. Recent structural characterization of broadly neutralizing antibodies (bnAbs)-HA complexes has identified two new epitope locations with less sequence variability: the receptor binding site (RBS) and the stem region (Lee and Wilson, 2015). Binding of antibody to the RBS or the stem region may block attachment of virus to the host cell or prevent fusion of viral and endosomal membranes, respectively. Because these regions are more conserved than the classical antigenic sites, antibody responses to them are more likely to protect against multiple IAV strains.
As in the case of HA, epitopes of antibodies targeting NA have been partially mapped by sequence analysis of viral escape mutants. The positions of these epitopes appear to be similar in different NA subtypes, with the majority of those identified surrounding the enzyme active site (Air, 2012). Although a few high-resolution epitope maps of the NA subtype N1, N2 and N9 have been obtained through structural analysis of NA-antibody complexes by X-ray crystallography (Tulip et al., 1992a; Malby et al., 1994; Venkatramani et al., 2006; Wan et al., 2015), the full antigenic nature of NA remains to be determined.
2. A site-directed mutagenesis epitope mapping system coupled with the use of BLI biosensor
Site-directed mutagenesis is a powerful approach for rapidly identifying key residues in mAb epitopes (Benjamin and Perdue, 1996). During the public health responses to the 2009 influenza A pandemic caused by influenza A(H1N1)pdm09 viruses and the A(H7N9) outbreaks, we have developed a site-directed mutagenesis method to determine the epitopes of mAbs targeting HA and NA (Wilson et al., 2015; Wilson et al., 2016). Briefly, this approach involves constructing many recombinant proteins, each of which contains a single point mutation in a solvent exposed residue that could affect mAb binding, based on previously identified epitopes of the IAV HA and NA. The binding affinity as well as the association/disassociation pattern of the mAb of interest is then measured against each recombinant protein individually using biolayer interferometry (BLI).
To generate correctly folded antigens for screening, we have taken advantage of well-defined recombinant HA/NA expression systems (Stevens et al., 2004; Xu et al., 2008). For HA expression, a codon-optimized cDNA encoding the entire ectodomain of the mature HA is sub-cloned into a pIEx-4 vector (EMD Millipore, MA) using the in-Fusion HD cloning system (Clontech, CA). The expressed recombinant HA (recHA) contains a thrombin cleavage site at the C-terminus followed by a trimerizing sequence (foldon) from the bacteriophage T4 fibritin for generating functional trimers, and a His-tag to aid with subsequent assays and detection (Stevens et al., 2004). Similarly, a codon-optimized cDNA encoding the entire ectodomain of the NA is cloned into the pIEx-4 vector. The produced recombinant NA (recNA) possesses a His-tag at the N-terminus followed by a tetramerization domain from human vasodilator-stimulated phosphoprotein, and a thrombin site (Xu et al., 2008).
HA or NA mutants containing a single point mutation within or near known antigenic sites are generated from the wild type pIEx-4-HA or NA clone using the QuickChange Lightning Site-Directed Mutagenesis kit (Agilent, CA). The point mutations are designed to induce significant size and/or charge change in surface-accessible residues. To express wild type and mutated HA or NA, the constructs are individually transiently transfected into suspension sf9 cells using the Cellfectin II transfection reagent (Life Technologies, NY) and the transfected cells are maintained at 27 °C in an orbital shaker/incubator. The expressed HA/NAs are secreted in the culture supernatant and harvested 5 days post transfection. Quantitative western blot using anti-His antibody is performed to assess the expression of each construct and to normalize the amount of recombinant protein used for epitope mapping. This is required to ensure equal loading of recombinant protein onto each biosensor so that the difference in binding affinities observed is caused by the mutated residue and not by variations of Ag quantity in Ab-Ag reactions. The supernatants containing the respective recombinant protein can be aliquoted and frozen at −80 °C for later use. It is not necessary to purify the antigens used in the mapping, allowing a new panel to be quickly generated; the turnaround time is about 11 days for production of a panel consisting of the wild type and 20 to 30 recHA/NA variants (Fig. 1). Similarly, mAbs in crude supernatants can be screened without being purified.
Figure 1. A timeline for generation of recombinant HA/NA panels for epitope mapping.
BLI-based platform has been routinely used to screen large mAb libraries by epitope binning (Abdiche et al., 2012; Abdiche et al., 2014). Unlike ELISA and Luminex, this system detects changes of thickness at the tip of biosensor caused by Ab-Ag binding, and therefore, it does not require labeling of reagent and provides real-time kinetics data during the Ab-Ag binding. The use of BLI biosensors minimizes the possibility of false negative/positive results caused by non-specific coating of the antigen to a solid phase such as ELISA plates (Dekker et al., 1989; Gan and Patel, 2013). The BLI-based platforms, such as the Octet Systems (ForteBio) can measure kinetic constants for bindings between purified molecules, whereas end-point and kinetic profile can be determined for crude sample interactions. We use end-point as a measurement of the binding affinity of an unpurified mAb for each recHA/NA of a panel. Previous studies have shown that mutations enabling escape from antibody neutralization were associated with a reduction in binding of 50% or more (Throsby et al., 2008); 50% reduction in binding is therefore used as a cutoff for considering a particular residue to be part of an epitope. In addition, kinetic profiles of the Ab bindings to the panel are plotted against that of the wild type to identify residues that cause altered association and/or disassociation patterns without significantly affecting the end-point response. These residues are considered to be involved but not critical in the Ab-Ag interactions.
To perform epitope mapping, the supernatants of recHA/NA are normalized and directly added into wells of a 96-well plate. The anti-His biosensor tips (ForteBio) are then immersed in the solutions and are labelled with the HA/NA through the binding of His-tag in the recombinant proteins. The HA/NA-loaded biosensors are washed in binding buffer to remove excessive antigen and subjected to epitope mapping using Octet Red 96 (ForteBio). The globular head of HA is extremely tolerant of point mutations (Doud and Bloom, 2016). To validate the structural integrity of expressed recHAs or NAs, monoclonal or polyclonal antibodies known to bind conformational epitopes of the target proteins are tested for their ability to bind to the HA or NA panel by BLI. In addition, correct folding of recNAs can be confirmed by analyzing neuraminidase activities of recNAs using the 2′-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid (MUNANA) assay (Yang et al., 2015). Assays based on this method can be performed in high- (or at least moderate-) throughput format with the use of the Octet HTX system (ForteBio), which has the capacity to read up to 96 wells in parallel. Screening of two mAbs with a panel consisting of 24 recHAs/NAs can be completed in about 10 mins when the 48 biosensor mode is used.
3. Epitope mapping of mAbs against the H1 and H3 HA of influenza A virus
The development of next-generation sequencing technology make it possible to assess antibody response to influenza vaccination and infection using B-cell receptor sequence-based repertoire analysis. To fully understand the antibody repertoire, analyses need to correlate the variable region sequence signatures with antibody binding activities, including epitope mapping. We cloned sequences for immunoglobulin heavy (IgH) and light (IgL) from memory B cells of mice immunized by influenza A(H1N1)pdm09 virus, and multiple recombinant mAbs (rmAbs) with high affinity to the A(H1N1)pdm09 HA were generated (Wilson et al., 2015). A panel of 20 recHAs of A/California/7/2009 A(H1N1)pdm09 was produced, with each containing a single point mutation within or near a previously defined epitope of H1 (Fig. 2A), and the relative binding affinities of rmAbs to each HA mutant were determined by BLI (Supplementary Fig. 1). To validate correct folding of the recHAs, two control mAbs, 26-D11 (Immune Technology Corp., New York) and Y2_50132_1C04 (provided by Patrick Wilson, University of Chicago) that have been known to bind to the head (antigenic site Sa) or stem of the H1pdm HA, were also tested against the HA panel. As expected, 26-D11 and Y2_50132_1C04 bound with the same affinity to the wild-type recHA and to 19 of the 20 mutants; binding was reduced only by mutations at site Sa (S121K) or the stem region (I372K), respectively, indicating that all of the HA mutants were correctly folded (Fig. 2B). Therefore, reduced binding of a mAb to particular mutant(s) in the panel would suggest the mutated residue(s) are components of the epitope.
Figure 2. Epitope mapping of recombinant mAbs targeting the HA of influenza A(H1N1)pdm09 virus.
A panel of 20 recHAs of A(H1N1)pdm09 virus was produced with each containing a single point mutation. (A) The sequence change and location for each mutant are shown on the 3D structure of trimeric A(H1N1)pdm09 HA (PDB ID 3M6S); mutation G155K is shown twice, once on each monomer. Classical H1 antigenic sites are indicated by different colors on the structure surface: Sa (red), Sb (magenta), Ca (cyan), and Cb (blue). Mutations not overlapping with the classical antigenic sites are indicated in golden yellow color; (B) Mutations that caused significant reductions (≥50%) in mAb binding are indicated on the HA 3D structure. For simplicity, only control antibodies with their corresponding epitopes are signified.
We found that all of the nine tested mAbs bound to the A(H1N1)pdm09 HA head with high affinities (Wilson et al., 2014), displaying seven distinct binding footprints covering antigenic sites Sa, Sb, Ca2 and Cb (Fig. 2B). We also identified a number of HA mutations associated with faster antibody off-rate in spite of showing less than 50% reduction in binding were also identified; their significance in Ag-Ab interactions is not known, but the presence of residues associated with faster off-rates in close proximity to those associated with greater than 50% reduction in binding suggests that faster off-rates may also help indicate residues associated with, but not critical for, antibody binding (Supplementary Fig. 1). We then used natural A(H1N1)pdm09 isolates that carried variant residues within the predicted epitope sites, and showed that binding of the appropriate mAb to these mutants was generally altered, based on a reduction in ELISA titer of at least 8-fold (Wilson et al., 2015). This confirmed that the ectodomain structure of recHA used in the mapping is similar to that of HA on virus surface, and the identified epitopes are applicable to natural viral HA, demonstrating the validity of this general approach.
In order to increase the resolution of the approach, we generated a panel of 68 recHAs of A(H3N2) A/Hong Kong/4801/2014 (HK4801) with each containing a single point mutation within or near the classical H3 antigenic sites. For structural validation of the H3 panel, a control mAb (F045–092), which has been characterized as a broadly neutralizing antibody against HAs from all H3 strains, was recombinantly expressed, purified and tested against the panel (Supplementary Fig. 2). X-ray crystallography of the complex of F045–092 Fab with the A/Victoria/3/1975 (H3N2) (Vic75) HA has shown that the Ab blocks virus entry by contacting a minimal epitope in the H3 RBS (Lee et al., 2014), with residues at 131, 133 through 137, 145, 153, 155 through 158, 193, 194 and 226 being considered contact residues in the F045–092/Vic75 H3 complex. Our epitope mapping, based on at least a 50% reduction in affinity, also identified the RBS of HK4801 H3 as the region bound by F045–092, with T131, T135, S145 and Y159 acting key residues for binding (Fig. 3A and Supplementary Figure 2). Three residues implicated in binding of F045–092 to Vic75 (137, 158, and 193) were mutated in our panel but did not lead to at least a 50% reduction in binding to HK4801 (Supplementary Figure 2). This may be because the contact site between F045–092 and HK4801 (as opposed to Vic75) may be slightly different due to the extensive sequence changes between these viruses. BLI also identified a number of residues adjacent to these key residues (Q132K, T167K, K189E, D190K, S219K, and R222E) as causing faster off-rates of the bound antibody even though affinity was not reduced by 50%, suggesting these residues may also be partially involved in the binding footprint (Fig. 3B).
Figure 3. Epitope mapping of a broadly neutralizing antibody targeting the H3 HA of influenza A virus.
The mAb F045–092 was produced by co-transfection of its heavy and light chain expression vectors in 293T cells, purified and tested against the H3 HA panel. (A) The sequence changes and locations of mutations as part of the epitope are shown on the 3D structure of trimeric H3 HA (adapted from PDB ID 4WE8). Mutations that caused significant reductions (≥50%) in mAb binding are highlighted in red. Percentage of the Ab binding to the mutants compared to wild type is indicated in the brackets. The overlapped H3 antigenic sites are indicated in different colors: Site A (red), Site B (blue), and Site D (cyan). (B) Association/disassociation curves of BLI for the F045–092 binding to the wild type and mutants with increased off-rate. The biosensors loaded with recHAs were incubated with the Ab for 300 sec at the association step followed by incubation in buffer for 180 sec at the disassociation step as shown on the X-axis. The changes of thickness at the tip of biosensors caused by Ab-Ag binding are shown by the Y-axis. The binding curve for each recHA is labeled with a unique color.
Having confirmed the correctly folded structures of the H3 panel and applicability of the panel for epitope mapping studies, we tested a series of mAb generated from H3-vaccinated mice. As with anti-H1 recombinant mAb, epitopes fell predominately within known antigenic sites, with most mAb recognizing epitopes within sites B and C (unpublished results).
4. Epitope mapping of mAbs against the N9 NA of H7N9 IAV
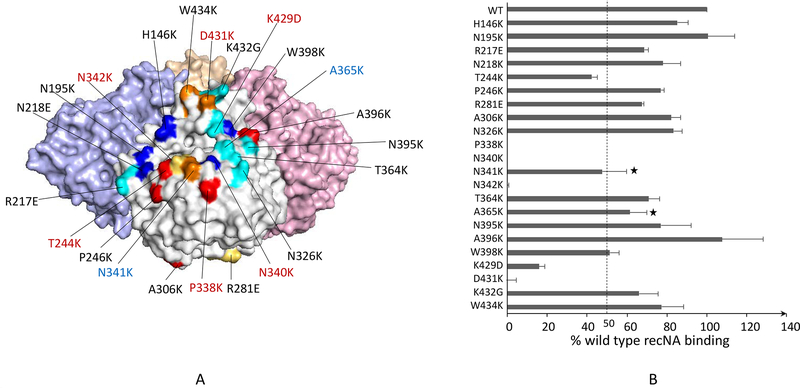
To analyze Ab response to the N9 and characterize anti-N9 mAbs with therapeutic potential, we generated a panel of 22 recNAs of A(H7N9) A/Shanghai/2/2013, each containing a single point mutation within or near the previously known NA epitopes identified by generation of escape mutants (Fig 4a) (Wilson et al., 2016). We confirmed that each recNA was correctly folded by testing neuraminidase activity using a MUNANA assay (Supplementary Fig. 3) (Yang et al., 2015). mAbs generated from mice vaccinated with inactivated A(H7N9) virus or recNA, were epitope mapped using the panel. We determined that the mAbs typically targeted residues near the enzyme active site with 13 distinct footprints (unpublished results). One of the mAbs (“3c10–3”) was found to bind with high affinity to residues spanning the enzyme active site, such that the antibody is expected to completely block access to the active site (Fig 4A). As expected from this binding footprint, 3c10–3 was able to inhibit N9 enzymatic activity at low concentrations (Wilson et al., 2016), making it a candidate therapeutic antibody. Studies in mice demonstrated that 3c10–3 does indeed have therapeutic and prophylactic activity against A(H7N9) infection (Wilson et al., 2017). Importantly, the binding affinity of 3c10–3 to a N9 mutant carrying a R289K change known to confer NAI resistance was not reduced, making this mAb an option for treating NAI-resistant H7N9 infections (Supplementary Table 1).
Figure 4. Epitope mapping of a mAb with therapeutic potential targeting the N9 NA of influenza A(H7N9) virus.
(A) The sequence changes and locations of the N9 panel mutants are shown on the 3D structure of tetrameric N9 NA (PDB ID 4WMJ). Mutations that caused ≥50% reduction in the mAb 3c10–3 binding are highlighted in red, and increased off-rate in blue. The panel is primarily based on previously identified epitopes of NA as indicated: N1 (red) (Wan et al., 2013), N2 (blue) (Lentz et al., 1984; Webster et al., 1984; Gulati et al., 2002), N8 (yellow) (Saito et al., 1994), and N9 (cyan) (Air et al., 1990; Tulip et al., 1992a; Tulip et al., 1992b). Unique sites selected by us are indicated in golden yellow. (B) Percent response of 3c10–3 binding to each mutant NA compared to the wild type was determined. A ≥50% reduction in binding activity was the cutoff for significance. “*” indicates mutations that caused increased off-rate but less than 50% reduction in binding.
5. Evaluation of epitope mapping methods
Other mutagenesis methods have been developed for epitope mapping of IAV. Wan et al (Wan et al., 2015) analyzed the epitopes of a N1 NA antibody by expressing the NA wild type and single point mutants on the cell surface followed by ELISA. This cell-based ELISA allows the expression of antigens in their native conformations, and is rapid and cost-effective. However, it is not convenient to normalize the expression levels of antigens with this method and the expressed antigens can only be used once. A similar method with large-scale shotgun mutagenesis has been described for characterizing mAbs against several viruses and cellular proteins in a high-throughput format (Davidson and Doranz, 2014). However, construction of plasmid libraries containing several hundreds or even thousands of vectors is a daunting task making this method less flexible in change of antigen target.
As previously described, generation of escape mutants has been useful for determining epitopes of the HA/NA, but this approach is time-consuming and involves growing virus which may require special lab facilities and permissions for highly pathogenic strains of IAV and potential gain-of-function mutations (Davis et al., 2014; Schultz-Cherry et al., 2014). Phage display has been used to analyze the antibody repertoires of sera from individuals exposed to A(H5N1) or A(H7N7) IAVs but with low mapping resolution (Khurana et al., 2009; Khurana et al., 2016). X-ray crystallography has been used as another strategy to obtain high-resolution epitope maps of the HA and NA. This method is also time-consuming, expensive and is unsuitable for high-throughput analysis. For other antigens, hydrogen-deuterium exchange approaches have also produced low-resolution epitope maps (Pandit et al., 2012).
One of the major limitations of our method is the lack of full coverage of potential epitopes. Of the 9 tested mAbs against the A(H1N1)pdm09, 065-D01 was the only mAb that failed to show any significant reductions in binding to the HA panel. This could be due to novel epitopes outside the classical antigenic sites recognized by 065-D01, or the mutated residues in the panel were parts of the epitope but the amino acid change was not significant enough to disrupt the Ab-Ag interaction, or the binding was the net result of multiple weak simultaneous interactions. For the last scenario, a combinational mutagenesis approach using recHAs that contain multiple point mutations in discrete antigenic sites on one single protein might be required (Infante et al., 2014). The other main limitation is the cost of biosensors used in the system which may be reduced if regenerable biosensors can be used. Despite the above limitations, our mapping method is rapid, flexible with high-throughput capacity, and can provide epitope maps with high resolution. Our method can be used as a pre-screening method to identify mAbs with potential novel epitopes which can be further defined by escape mutation or X-ray crystallography.
6. Conclusion
We have developed a site-directed mutagenesis method to determine epitopes of isolated mAbs against the HA or NA of influenza A viruses. Data from mapping the epitopes of mAbs against H1, H3, and N9, as well as testing with mAb with known epitopes, have demonstrated the feasibility of this approach for characterizing IAV HA/NA mAbs. A similar strategy can be applied to determine epitopes of mAbs against other antigens. Assays based on this method can be performed in a high-throughput format with the use of the Octet HTX system (ForteBio) and are therefore compatible with antibody repertoire analysis. Compared to other approaches to epitope mapping, this method offers high resolution, ease of use, and quick results.
Supplementary Material
Supplementary Figure 1. Epitope maps of monoclonal antibodies on A(H1N1)pdm09 HA (H1pdm09). Binding affinities of mAbs for wild type (WT) and each mutant of the H1pdm09 panel were determined by BLI and are presented as endpoint responses. “*” indicates mutations with ≥50% reduction in binding activity. “▲” indicates mutations with faster off-rate and <50% reduction in binding activity.
Supplementary Figure 2. Epitope map of mAb F045–092 on A(H3N2) A/Hong Kong/4801/2014 HA (HK4801 H3). Binding affinities of mAb F045–092 for WT and each mutant of the HK4801 H3 panel were determined by BLI and presented as endpoint response. “*” indicates mutations with ≥50% reduction in binding activity. “▲” indicates mutations with faster off-rate and <50% reduction in binding activity.
Supplementary Figure 3. Neuraminidase activities of recNAs of the A(H7N9) A/Shanghai/2/2013 (SH2 N9) panel. Neuraminidase activities of WT and mutants of the SH2 N9 panel were determined by MUNANA assay following procedures as described previously (Yang et al., 2015). The results were presented as percentage of the wild type N9 neuraminidase activity.
Supplementary Table 1 Kinetic data for mAb 3c10–3 for A(H7N9) A/Shanghai/2/2013 NA (SH2 N9) and its R289K mutant as determined by BLI.
Acknowledgments
This work was supported by the Centers for Disease Control and Prevention. The findings and conclusions are those of the authors and do not necessarily reflect the views of the funding agency.
References
- Abbott WM, Damschroder MM and Lowe DC, 2014, Current approaches to fine mapping of antigen-antibody interactions. Immunology 142, 526–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdiche YN, Lindquist KC, Stone DM, Rajpal A and Pons J, 2012, Label-free epitope binning assays of monoclonal antibodies enable the identification of antigen heterogeneity. J Immunol Methods 382, 101–16. [DOI] [PubMed] [Google Scholar]
- Abdiche YN, Miles A, Eckman J, Foletti D, Van Blarcom TJ, Yeung YA, Pons J and Rajpal A, 2014, High-throughput epitope binning assays on label-free array-based biosensors can yield exquisite epitope discrimination that facilitates the selection of monoclonal antibodies with functional activity. PLoS One 9, e92451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Air GM, 2012, Influenza neuraminidase. Influenza Other Respir Viruses 6, 245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Air GM, Laver WG and Webster RG, 1990, Mechanism of antigenic variation in an individual epitope on influenza virus N9 neuraminidase. Journal of virology 64, 5797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DC and Perdue SS, 1996, Site-Directed Mutagenesis in Epitope Mapping. Methods 9, 508–15. [DOI] [PubMed] [Google Scholar]
- Blech M, Peter D, Fischer P, Bauer MM, Hafner M, Zeeb M and Nar H, 2013, One target-two different binding modes: structural insights into gevokizumab and canakinumab interactions to interleukin-1beta. J Mol Biol 425, 94–111. [DOI] [PubMed] [Google Scholar]
- Caton AJ, Brownlee GG, Yewdell JW and Gerhard W, 1982, The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31, 417–27. [DOI] [PubMed] [Google Scholar]
- Cohn M, 2002, The immune system: a weapon of mass destruction invented by evolution to even the odds during the war of the DNAs. Immunol Rev 185, 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch RB and Kasel JA, 1983, Immunity to influenza in man. Annu Rev Microbiol 37, 529–49. [DOI] [PubMed] [Google Scholar]
- Davidson E and Doranz BJ, 2014, A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology 143, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, Buhay C, Kang H, Kim SC, Fahey CC, Hacker KE, Bhanot G, Gordenin DA, Chu A, Gunaratne PH, Biehl M, Seth S, Kaipparettu BA, Bristow CA, Donehower LA, Wallen EM, Smith AB, Tickoo SK, Tamboli P, Reuter V, Schmidt LS, Hsieh JJ, Choueiri TK, Hakimi AA, Cancer Genome Atlas Research, N., Chin L, Meyerson M, Kucherlapati R, Park WY, Robertson AG, Laird PW, Henske EP, Kwiatkowski DJ, Park PJ, Morgan M, Shuch B, Muzny D, Wheeler DA, Linehan WM, Gibbs RA, Rathmell WK and Creighton CJ, 2014, The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 26, 319–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker EL, Porta C and Van Regenmortel MH, 1989, Limitations of different ELISA procedures for localizing epitopes in viral coat protein subunits. Arch Virol 105, 269–86. [DOI] [PubMed] [Google Scholar]
- Doud MB and Bloom JD, 2016, Accurate Measurement of the Effects of All Amino-Acid Mutations on Influenza Hemagglutinin. Viruses 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn JA and Crowe JE Jr., 2013, Impact of new sequencing technologies on studies of the human B cell repertoire. Curr Opin Immunol 25, 613–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore AE, Shay DK, Broder K, Iskander JK, Uyeki TM, Mootrey G, Bresee JS, Cox NJ, Centers for Disease C. and Prevention. 2009, Prevention and control of seasonal influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2009. MMWR Recomm Rep 58, 1–52. [PubMed] [Google Scholar]
- Gan SD and Patel KR, 2013, Enzyme immunoassay and enzyme-linked immunosorbent assay. J Invest Dermatol 133, e12. [DOI] [PubMed] [Google Scholar]
- Greenspan NS and Di Cera E, 1999, Defining epitopes: It’s not as easy as it seems. Nat Biotechnol 17, 936–7. [DOI] [PubMed] [Google Scholar]
- Gulati U, Hwang CC, Venkatramani L, Gulati S, Stray SJ, Lee JT, Laver WG, Bochkarev A, Zlotnick A and Air GM, 2002, Antibody epitopes on the neuraminidase of a recent H3N2 influenza. Journal of virology 76, 12274–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai R, Schmolke M, Leyva-Grado VH, Thangavel RR, Margine I, Jaffe EL, Krammer F, Solorzano A, Garcia-Sastre A, Palese P and Bouvier NM, 2013, Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat Commun 4, 2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante YC, Pupo A and Rojas G, 2014, A combinatorial mutagenesis approach for functional epitope mapping on phage-displayed target antigen: application to antibodies against epidermal growth factor. MAbs 6, 637–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Shichinohe S, Nakayama M, Igarashi M, Ishii A, Ishigaki H, Ishida H, Kitagawa N, Sasamura T, Shiohara M, Doi M, Tsuchiya H, Nakamura S, Okamatsu M, Sakoda Y, Kida H and Ogasawara K, 2015, Emergence of H7N9 Influenza A Virus Resistant to Neuraminidase Inhibitors in Nonhuman Primates. Antimicrobial agents and chemotherapy 59, 4962–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernigan DB and Cox NJ, 2015, H7N9: preparing for the unexpected in influenza. Annu Rev Med 66, 361–71. [DOI] [PubMed] [Google Scholar]
- Khurana S, Chung KY, Coyle EM, Meijer A and Golding H, 2016, Antigenic Fingerprinting of Antibody Response in Humans following Exposure to Highly Pathogenic H7N7 Avian Influenza Virus: Evidence for Anti-PA-X Antibodies. J Virol 90, 9383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Suguitan AL Jr., Rivera Y, Simmons CP, Lanzavecchia A, Sallusto F, Manischewitz J, King LR, Subbarao K and Golding H, 2009, Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med 6, e1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knossow M, Daniels RS, Douglas AR, Skehel JJ and Wiley DC, 1984, Three-dimensional structure of an antigenic mutant of the influenza virus haemagglutinin. Nature 311, 678–80. [DOI] [PubMed] [Google Scholar]
- Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y and Wilson IA, 2014, Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nature communications 5, 3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS and Wilson IA, 2015, Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr Top Microbiol Immunol 386, 323–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz MR, Air GM, Laver WG and Webster RG, 1984, Sequence of the neuraminidase gene of influenza virus A/Tokyo/3/67 and previously uncharacterized monoclonal variants. Virology 135, 257–65. [DOI] [PubMed] [Google Scholar]
- Liu Y, Pan J, Jenni S, Raymond DD, Caradonna T, Do KT, Schmidt AG, Harrison SC and Grigorieff N, 2017, CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface. J Mol Biol 429, 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malby RL, Tulip WR, Harley VR, McKimm-Breschkin JL, Laver WG, Webster RG and Colman PM, 1994, The structure of a complex between the NC10 antibody and influenza virus neuraminidase and comparison with the overlapping binding site of the NC41 antibody. Structure 2, 733–46. [DOI] [PubMed] [Google Scholar]
- Najar TA, Khare S, Pandey R, Gupta SK and Varadarajan R, 2017, Mapping Protein Binding Sites and Conformational Epitopes Using Cysteine Labeling and Yeast Surface Display. Structure 25, 395–406. [DOI] [PubMed] [Google Scholar]
- Pandit D, Tuske SJ, Coales SJ, E SY, Liu A, Lee JE, Morrow JA, Nemeth JF and Hamuro Y, 2012, Mapping of discontinuous conformational epitopes by amide hydrogen/deuterium exchange mass spectrometry and computational docking. J Mol Recognit 25, 114–24. [DOI] [PubMed] [Google Scholar]
- Parola C, Neumeier D and Reddy ST, 2017, Integrating high-throughput screening and sequencing for monoclonal antibody discovery and engineering. Immunology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocnakova L, Bhide M and Pulzova LB, 2016, An Introduction to B-Cell Epitope Mapping and In Silico Epitope Prediction. J Immunol Res 2016, 6760830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke U, 2009, Antibody epitope mapping using de novo generated synthetic peptide libraries. Methods Mol Biol 524, 203–11. [DOI] [PubMed] [Google Scholar]
- Rojas G, Tundidor Y and Infante YC, 2014, High throughput functional epitope mapping: revisiting phage display platform to scan target antigen surface. MAbs 6, 1368–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Taylor G, Laver WG, Kawaoka Y and Webster RG, 1994, Antigenicity of the N8 influenza A virus neuraminidase: existence of an epitope at the subunit interface of the neuraminidase. Journal of virology 68, 1790–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV and Nussenzweig MC, 2009, Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–40. [DOI] [PubMed] [Google Scholar]
- Schultz-Cherry S, Webby RJ, Webster RG, Kelso A, Barr IG, McCauley JW, Daniels RS, Wang D, Shu Y, Nobusawa E, Itamura S, Tashiro M, Harada Y, Watanabe S, Odagiri T, Ye Z, Grohmann G, Harvey R, Engelhardt O, Smith D, Hamilton K, Claes F and Dauphin G, 2014, Influenza gain-of-function experiments: their role in vaccine virus recommendation and pandemic preparedness. MBio 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow E, Friede M, Sheikh M, Torvaldsen S and Newall AT, 2016, Passive immunization for influenza through antibody therapies, a review of the pipeline, challenges and potential applications. Vaccine 34, 5442–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer DA and Skehel JJ, 2002, Genetics of influenza viruses. Annu Rev Genet 36, 305–32. [DOI] [PubMed] [Google Scholar]
- Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P and Wilson IA, 2004, Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303, 1866–70. [DOI] [PubMed] [Google Scholar]
- Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J and Goudsmit J, 2008, Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PloS one 3, e3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulip WR, Varghese JN, Laver WG, Webster RG and Colman PM, 1992a, Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex. J Mol Biol 227, 122–48. [DOI] [PubMed] [Google Scholar]
- Tulip WR, Varghese JN, Webster RG, Laver WG and Colman PM, 1992b, Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface. J Mol Biol 227, 149–59. [DOI] [PubMed] [Google Scholar]
- Venkatramani L, Bochkareva E, Lee JT, Gulati U, Graeme Laver W, Bochkarev A and Air GM, 2006, An epidemiologically significant epitope of a 1998 human influenza virus neuraminidase forms a highly hydrated interface in the NA-antibody complex. J Mol Biol 356, 651–63. [DOI] [PubMed] [Google Scholar]
- Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, Easterbrook JD, Yang K, Zhong L, Rajabi M, Ye J, Sultana I, Wan XF, Liu X, Perez DR, Taubenberger JK and Eichelberger MC, 2013, Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. Journal of virology 87, 9290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Yang H, Shore DA, Garten RJ, Couzens L, Gao J, Jiang L, Carney PJ, Villanueva J, Stevens J and Eichelberger MC, 2015, Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers. Nature communications 6, 6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Brown LE and Laver WG, 1984, Antigenic and biological characterization of influenza virus neuraminidase (N2) with monoclonal antibodies. Virology 135, 30–42. [DOI] [PubMed] [Google Scholar]
- Webster RG and Laver WG, 1980, Determination of the number of nonoverlapping antigenic areas on Hong Kong (H3N2) influenza virus hemagglutinin with monoclonal antibodies and the selection of variants with potential epidemiological significance. Virology 104, 139–48. [DOI] [PubMed] [Google Scholar]
- Wiley DC, Wilson IA and Skehel JJ, 1981, Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289, 373–378. [DOI] [PubMed] [Google Scholar]
- Wilson IA and Cox NJ, 1990, Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol 8, 737–71. [DOI] [PubMed] [Google Scholar]
- Wilson JR, Belser JA, DaSilva J, Guo Z, Sun X, Gansebom S, Bai Y, Stark TJ, Chang J, Carney P, Levine MZ, Barnes J, Stevens J, Maines TR, Tumpey TM and York IA, 2017, An influenza A virus (H7N9) anti-neuraminidase monoclonal antibody protects mice from morbidity without interfering with the development of protective immunity to subsequent homologous challenge. Virology 511, 214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JR, Guo Z, Reber A, Kamal RP, Music N, Gansebom S, Bai Y, Levine M, Carney P, Tzeng WP, Stevens J and York IA, 2016, An influenza A virus (H7N9) anti-neuraminidase monoclonal antibody with prophylactic and therapeutic activity in vivo. Antiviral Res 135, 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JR, Guo Z, Tzeng WP, Garten RJ, Xiyan X, Blanchard EG, Blanchfield K, Stevens J, Katz JM and York IA, 2015, Diverse antigenic site targeting of influenza hemagglutinin in the murine antibody recall response to A(H1N1)pdm09 virus. Virology 485, 252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JR, Tzeng WP, Spesock A, Music N, Guo Z, Barrington R, Stevens J, Donis RO, Katz JM and York IA, 2014, Diversity of the murine antibody response targeting influenza A(H1N1pdm09) hemagglutinin. Virology 458–459, 114–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zhu X, Dwek RA, Stevens J and Wilson IA, 2008, Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J Virol 82, 10493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Nguyen HT, Carney PJ, Guo Z, Chang JC, Jones J, Davis CT, Villanueva JM, Gubareva LV and Stevens J, 2015, Structural and functional analysis of surface proteins from an A(H3N8) influenza virus isolated from New England harbor seals. J Virol 89, 2801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, Tumpey TM, Pappas C, Perrone LA, Martinez O, Stevens J, Wilson IA, Aguilar PV, Altschuler EL, Basler CF and Crowe JE Jr., 2008, Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 455, 532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Epitope maps of monoclonal antibodies on A(H1N1)pdm09 HA (H1pdm09). Binding affinities of mAbs for wild type (WT) and each mutant of the H1pdm09 panel were determined by BLI and are presented as endpoint responses. “*” indicates mutations with ≥50% reduction in binding activity. “▲” indicates mutations with faster off-rate and <50% reduction in binding activity.
Supplementary Figure 2. Epitope map of mAb F045–092 on A(H3N2) A/Hong Kong/4801/2014 HA (HK4801 H3). Binding affinities of mAb F045–092 for WT and each mutant of the HK4801 H3 panel were determined by BLI and presented as endpoint response. “*” indicates mutations with ≥50% reduction in binding activity. “▲” indicates mutations with faster off-rate and <50% reduction in binding activity.
Supplementary Figure 3. Neuraminidase activities of recNAs of the A(H7N9) A/Shanghai/2/2013 (SH2 N9) panel. Neuraminidase activities of WT and mutants of the SH2 N9 panel were determined by MUNANA assay following procedures as described previously (Yang et al., 2015). The results were presented as percentage of the wild type N9 neuraminidase activity.
Supplementary Table 1 Kinetic data for mAb 3c10–3 for A(H7N9) A/Shanghai/2/2013 NA (SH2 N9) and its R289K mutant as determined by BLI.