Abstract
OBJECTIVE.
Antibiotic resistance is a major threat to public health. Resistance is largely driven by antibiotic usage, which in many cases is unnecessary and can be improved. The impact of decreasing overall antibiotic usage on resistance is unknown and difficult to assess using standard study designs. The objective of this study was to explore the potential impact of reducing antibiotic usage on the transmission of multidrug-resistant organisms (MDROs).
DESIGN.
We used agent-based modeling to simulate interactions between patients and healthcare workers (HCWs) using model inputs informed by the literature. We modeled the effect of antibiotic usage as (1) a microbiome effect, for which antibiotic usage decreases competing bacteria and increases the MDRO transmission probability between patients and HCWs and (2) a mutation effect that designates a proportion of patients who receive antibiotics to subsequently develop a MDRO via genetic mutation.
SETTING.
Intensive care unit.
INTERVENTIONS.
Absolute reduction in overall antibiotic usage by experimental values of 10% and 25%.
RESULTS.
Reducing antibiotic usage absolutely by 10% (from 75% to 65%) and 25% (from 75% to 50%) reduced acquisition rates of high-prevalence MDROs by 11.2% (P< .001) and 28.3% (P< .001), respectively. We observed similar effect sizes for low-prevalence MDROs.
CONCLUSIONS.
In a critical care setting, where up to 50% of antibiotic courses may be inappropriate, even a moderate reduction in antibiotic usage can reduce MDRO transmission.
Antibiotic resistance is a significant problem in health care. Multidrug-resistant organisms (MDROs) are increasingly prevalent and are associated with significant morbidity, mortality, and costs.1 The United States Centers for Disease Control and Prevention recognizes antibiotic usage as the most important factor driving resistance.1 Antibiotic exposure is known to contribute to resistance; it is estimated that the odds of developing a MDRO among patients who received antibiotic therapy is 1.8–5.1 times that of patients who did not receive antibiotics.2–4 Antibiotics are among the most common drugs prescribed in medicine; nearly 50% of all hospitalized patients and 75% of critically ill patients receive an antibiotic during a hospital stay.1 Furthermore, up to 50% of those antibiotics prescribed are considered inappropriate.5 Significant opportunities exist to optimize and reduce antibiotic usage. The impact of reducing overall antibiotic usage on antibiotic resistance is not known and would be difficult to assess using traditional study designs. Therefore, we applied mathematical modeling to estimate the effect of reducing antibiotic usage on antibiotic resistance.
In this study, we used agent-based modeling to explore the potential impact of antibiotic stewardship (ie, specifically reduced overall antibiotic usage) on acquisition rates of MDROs and the proportion of patients who acquired a MDRO during their hospital stay. Increased understanding of the potential effect of reducing unnecessary antibiotic usage is useful in promoting and implementing antimicrobial stewardship programs in healthcare settings.
MATERIALS AND METHODS
Agent-based modeling facilitates the explicit simulation of healthcare worker (HCW)–patient interactions that serve as the mechanism for MDRO transmission, and it allows for distinct representation of individual characteristics (ie, heterogeneity).6 We modified an existing agent-based model of MDRO transmission developed using NetLogo version 5.0.5 software7 that was used in a previous study.8 (A description of the model pseudocode is provided as an online supplement.) We modeled the intensive care unit (ICU), where MDROs are prevalent and antibiotic usage is high. In this model, we assumed that transmission among patients occurred via the contaminated hands of HCWs. We modeled nurses, physicians, and other HCWs separately, each with distinct, literature-based rates of hand-hygiene compliance on entry and exit to patient rooms.8 Clinically relevant MDROs were considered in the model; methicillin-resistant Staphylococcus aureus (MRSA) or vancomycin-resistant enterococci (VRE) were modeled as high-prevalence pathogens, while carbapenem-resistent Enterobacteriaceae (CRE), multidrug-resistant Acinetobacter baumannii (MDR-AB), and multidrug-resistant Pseudomonas aeruginosa (MDR-PA) were modeled as low-prevalence pathogens (definitions for MDR-AB and MDR-PA previously reported in the literature were considered).9 MDROs were considered as either high- or low-prevalence based on prevalence rates found predominately in the US literature. Colonization status and transmission with respect to each class of MDROs were modeled independently.
A conceptual diagram of the agent-based model is presented in Figure 1. A proportion of patients admitted to the ICU were colonized with each respective MDRO, modeled using an admission prevalence parameter. An independent proportion of all newly admitted patients was selected at random to receive antibiotics during their stay (ie, antibiotic usage). We assumed that the duration of antibiotic treatment was longer than the length of stay; therefore, we did not model the initiation and termination of antibiotic usage. During each patient’s hospital stay, there were HCW-patient interactions to provide care, and transmission of each MDRO depended on colonization of the patient and contamination of the HCW. We modeled transmission probability asymmetrically; that is, we assumed that a colonized patient was more likely to transmit an MDRO to a HCW than a HCW to a patient. In this model, if a HCW failed to comply with hand hygiene on room exit and again on entry to the subsequent patient’s room—or if the hand hygiene attempt was unsuccessful at removing the contamination—there was risk of transmission to the subsequent patient. The HCW’s hands remained contaminated until a successful hand-hygiene event was completed; therefore, multiple subsequent patients could have been at risk. Patients were discharged at the end of their predetermined length of stay and were immediately replaced with a new patient.
FIGURE 1.
Conceptual flow diagram for agent-based model of multidrug-resistant organism (MDRO) transmission. Patients are admitted to the intensive care unit with a parameterized probability of MDRO colonization and receiving antibiotics. Patients receiving antibiotics are more likely to acquire and transmit a MDRO (microbiome effect) and can also acquire a MDRO via genetic mutation. Healthcare workers periodically visit patients during their stay and can colonize patients if they transiently acquire an MDRO from a colonized patient and transmit it to a susceptible patient prior to successfully cleaning their hands.
As stated, a portion of the patients received antibiotics; we modeled the effect of this antibiotic exposure in 2 ways. First, the administration of antibiotics can lead to disruptions in the human microbiome, which may result in the overgrowth of MDROs that are already present at low levels or can leave patients susceptible to acquisition of new MRDOs. In turn, these patients are in turn more likely to shed and transmit these organisms to other patients via the contamination of HCW hands. For this scenario, we introduced a parameter, denoted the microbiome effect, which increased the transmission probability for relevant visits between patients who received antibiotics and their HCWs. Specifically, we increased the transmission probability from contaminated HCWs to susceptible patients who received antibiotics, and we increased the transmission probability from colonized patients who received antibiotics to susceptible visiting HCWs. We set the baseline microbiome effect at 1.0 (ie, no increase in transmission probability for visits involving patients who received antibiotics). Then, we varied the antibiotic usage and microbiome effect parameters from their respective baseline levels to explore the effect of decreasing overall antibiotic usage on transmission of MDROs in the ICU. Our goal was to identify the level at which the microbiome effect causes transmission to become statistically significantly higher, as well as the level at which an overall reduction in antibiotic usage causes a statistically significant decrease in transmission.
We also modeled the effect of antibiotic usage in a second manner. In some cases, the presence of antibiotics can lead to the development of antibiotic resistance via genetic mutation. Thus, we introduced another parameter, denoted the mutation effect, which designated a small proportion of patients who received antibiotics to develop a MDRO via this mechanism. The actual proportion of patients who received antibiotics and developed a resistant phenotype via mutation is unknown. We set the mutation effect conservatively at 1%, and we varied this parameter via sensitivity analysis to evaluate the effect of this assumption on our results, given the uncertainty surrounding the actual likelihood of the occurrence of a mutation event (see online supplement).
The model inputs for the simulation experiments are summarized in Table 1. Many nonexperimental parameters were configured using data from the research literature or observation data from a large, academic medical center.10–28 In the absence of other information, the remaining nonexperimental parameter values were selected to model a general hospital ICU. These nonexperimental parameters were generally held constant for all simulation experiments, with the exception of sensitivity analysis performed for the mutation effect and the hand hygiene of nurses. The key experimental parameters were the antibiotic usage and microbiome effect. At baseline, we assumed an antibiotic usage of 75%, that is, 75% of the patients in our model received antibiotics,26–29 and no microbiome effect (ie, equal to 1).
TABLE 1.
Agent-Based Model Input Parameters
| Parameter | Baseline Value | Experimental Values | Source, by reference no. |
|---|---|---|---|
| No. of patients | 18 | Selected | |
| No. of nurses | 9 | Selected | |
| No. of physicians | 2 | Selected | |
| No. of other HCWs | 6 | Selected | |
| Admission prevalence | 0.13 for high prevalence 0.02 for low prevalence |
6–12 | |
| Proportion of patients on contact precautions | 0.20 | 13–14 | |
| Mean (Median) length of stay | 3.5 (2) | 15–16 | |
| Proportion of visits to patients by nurses vs physicians and other HCWs | 0.5 | 17–18 | |
| Proportion of visits to patients by physicians vs other HCWs | 0.5 | 1–18 | |
| Patient visits per hour | 3 | 12, 17, 18 | |
| Transmission probabilitya | 0.025 from HCWto patient 0.075 from patient to HCW |
19, calibrated to fit observed acquisition rate data | |
| Hand-hygiene complianceb | 80% for nurses and 50% for physicians Equal on entry and exit |
13, 20, calibrated to fit observed data | |
| Hand-hygiene efficacy | 0.83 | 19,21 | |
| Gloves and gown compliance | 80% for all HCWs | 12 | |
| Gloves and gown effect | 0.60 | Calibrated to fit observed acquisition rate data | |
| Antibiotic usage | 0.75 | 0.50, 0.65 | 22–24 |
| Mutation effect | 0.01 | … | |
| Microbiome effect | 1 (no effect) | 1.1 to 2.0 in increments of 0.1 | … |
note. HCW, healthcare worker.
The probability of transmission given an interaction between a susceptible agent and a colonized agent.
For sensitivity analysis of the hand hygiene compliance of nurses and the mutation rate, please see the online supplement.
RESULTS
We simulated the transmission of the high- and low-prevalence MDROs for 1 year using the model inputs specified in Table 1. We performed 200 replications each for 33 parameter-based scenarios and compared the effects of antibiotic usage and the microbiome effect on prevalence-specific acquisition rates (acquisitions per 1,000 patient days) and the proportion of patients who acquired each class of MDRO during their stay. We first explored this effect using a baseline frequency of antibiotic usage of 75% and then explored the effect on transmission by reducing absolute antibiotic usage by 10% and 25%, respectively.
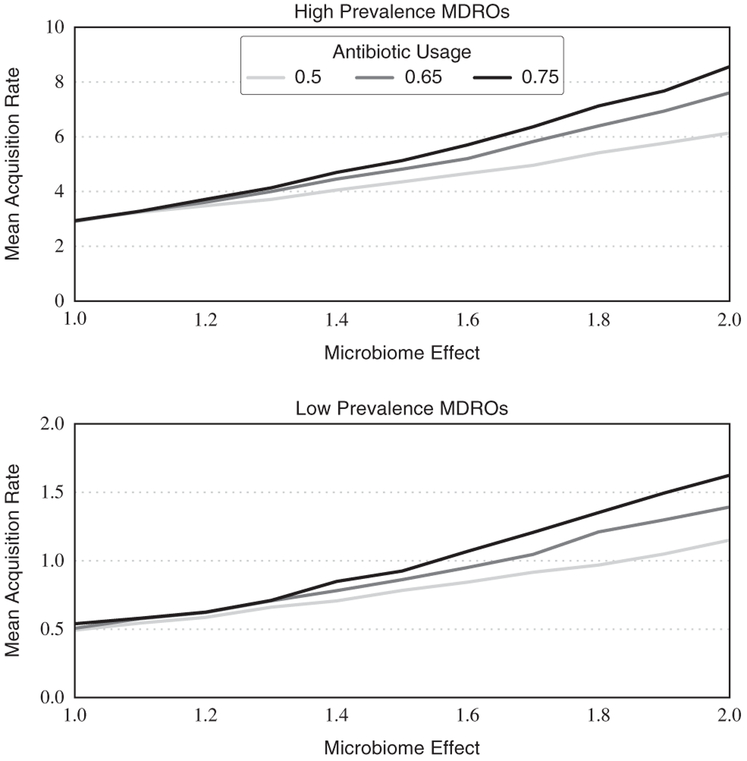
In Figure 2, we summarize the prevalence-specific acquisition rates (dependent outcome) as a function of the microbiome effect and antibiotic usage. When considering a microbiome effect of 2.0 (ie, that patients who received antibiotics were twice as likely to acquire or transmit a MDRO), reducing antibiotic usage by 10% (ie, from 75% to 65%) reduced acquisition rates of high-prevalence MDROs by 11.2% from 8.56 to 7.60 acquisitions per 1,000 patient days (P<.001). Reducing antibiotic usage by 25% (i.e., from 75% to 50%) reduced acquisition rates by 28.3% from 8.56 to 6.14 acquisitions per 1,000 patient days (P<.001). For low-prevalence MDROs, absolute reductions in antibiotic usage by 10% and 25% reduced acquisition rates by 14.3% (P<.001) and 29.8% (P<.001), respectively.
FIGURE 2.
Prevalence-specific multidrug-resistant organism (MDRO) acquisition rates as a function of microbiome effect and antibiotic usage levels. Acquisition rates for high- and low-prevalence MDROs grow exponentially with increasing microbiome effect, and statistically significant benefits of decreased antibiotic usage are observed at microbiome effect levels of ~ 1.2 and higher.
For the high-prevalence MDROs, when considering a microbiome effect of 1.1 (ie, a 10% increase in transmission probability between patients receiving antibiotics and HCWs) and higher, acquisition rates were statistically significantly higher than at baseline (ie, microbiome effect of 1.0) for all experimental levels of antibiotic usage. Statistically significant differences for low-prevalence MDROs were not consistently observed until a microbiome effect of 1.3 was used for all experimental levels of antibiotic usage. Using the same testing approach, we found that reducing the proportion of patients who received antibiotics from 75% to 65% significantly lowered acquisition rates at a microbiome effect of 1.5 for the high-prevalence MDROs and 1.7 for the low-prevalence MDROs, respectively. Reducing the proportion of patients who received antibiotics from 75% to 50% provided a consistent benefit at microbiome effects of 1.2 and 1.4 for high- and low-prevalence MDROs, respectively.
Figure 3 summarizes the proportion of patients who acquired an MDRO during their stay (dependent outcome). We observed similar trends with respect to the microbiome effect as for acquisition rates. In addition, among patients not receiving antibiotics, acquisition rates increased with the microbiome effect for both high- and low-prevalence MDROs.
FIGURE 3.
Proportion of patients who acquire each class of multidrug-resistant organism (MDRO) during their stay as a function of the microbiome effect and whether they are receiving antibiotics. Increasing the microbiome effects not only increases transmission for patients who are receiving antibiotics but also increases colonization pressure for all patients. Differences between the 2 groups of patients at a microbiome effect of 1.0 (no effect) are due to genetic mutation effects.
DISCUSSION
Antibiotic usage, and in particular unnecessary usage of antibiotics, is a major driver of antibiotic resistance. A primary goal of antimicrobial stewardship programs, therefore, is to reduce the incidence of resistant organisms by reducing antibiotic usage.30,31 We found measurable and significant benefits to be gained by reducing antibiotic usage in an ICU setting. Studies reporting on the outcomes of antimicrobial stewardship interventions suggest that a reduction in overall antibiotic usage by approximately 33% is feasible.5,32,33 Even a moderate absolute reduction of 10% in overall antibiotic usage has the potential to reduce acquisition rates of MDROs by 11.2%, and a 25% absolute reduction in antibiotic usage can reduce MDRO acquisition rates by 28.3%.
Mathematical modeling, such as the methods used in this paper, is an important tool to evaluate the impact of antibiotic reduction because large-scale clinical studies to evaluate the impact of reduced overall antibiotic usage on the spread of resistant organisms are complex and/or unavailable. Mathematical modeling of this problem to date has been limited; a time series analysis study by Willman et al34 only evaluated the potential effect of a specific antibiotic (meropenem) on resistance rates of a specific organism (multidrug-resistant Pseudomonas aeruginosa).34 In 2016, Pelat et al35 developed a stochastic, compartmental model of extended-spectrum β-lactamase–producing Enterobacteriaceae transmission in an ICU and explored the impact of reducing antibiotic prevalence at admission with or without reducing antibiotherapy duration. These researchers found that the reduction in antibiotic prevalence at admission by 50% reduced acquisition rates by as much as 33%, which is consistent with our results. Our study also focused on the effect of an overall reduction in antibiotic usage on transmission of multiple MDROs, but we utilized an agent-based model to simulate the potential effects of the intervention—including both microbiome and mutation effects—instead of a projection based on forecast estimates or compartmental modeling.
There are challenges in using traditional study methods and outcomes to examine the impact of antimicrobial stewardship. First, the use of the antibiogram does not capture changes in resistance patterns over time in a hospital unit, as it usually includes only the first isolate from each patient.36 Second, reliance on clinical cultures to assess changes in resistance patterns is challenging due to the infrequency of positive clinical cultures; therefore, few studies include development of antimicrobial resistance based on surveillance cultures as a primary outcome.37,38 The use of transmission dynamic modeling allows for the estimation of the impact of antimicrobial stewardship via experimentation with relevant parameters.
In this model, we considered 2 effects of antibiotic usage: the microbiome effect and the mutation effect. A few studies have examined the microbiome of individuals after antibiotic exposure.39,40 In one study, Malhotra et al39 examined the impact of macrolide versus placebo use on macrolide-resistant streptococci from pharyngeal swabs of healthy volunteers and found that exposure to azithromycin increased the proportion of streptococci that were macrolide resistant by 53.4%. However, the antibiotic effect on risk of transmission from one person to the next (ie, the microbiome effect) is not known. In our model, we examined microbiome effect over a spectrum and noted that benefits are statistically significant even at conservative estimates, particularly for high-prevalence MDROs (ie, ranging from 1.2 to 1.5, depending on the level of antibiotic usage). The benefits of antibiotic reduction were even larger at higher levels.
Furthermore, overall reduction of antibiotic usage lowers colonization pressure for the entire unit, including for patients who are not receiving antibiotics. In this scenario, fewer patients receive antibiotics and are therefore less likely to transmit (if they are colonized) or acquire a MDRO (either via a contaminated HCW or genetic mutation). As a result, other susceptible patients in the unit are at a decreased risk of acquisition because HCWs are not exposed as often to colonized patients.
The benefits observed in this model are proportionally similar for both high- and low-prevalence MDROs with similar relative risk reductions. For typical high-prevalence MDROs, such as MRSA or VRE, a significant reduction in antibiotic usage can lead to a reduction of multiple acquisitions per 1000 patient days. For low-prevalence MDROs, a reduction in antibiotic usage can have the same relative effect, although not as large of an absolute risk reduction (ie, in terms of acquisitions per 1,000 patient days). Such a reduction could be a key intervention to preventing further emergence and spread of these MDROs and to maintaining low prevalence.41 Notably, prevalence varies geographically, and the assumptions made regarding low- and high-prevalence MDROs may not hold true in specific areas.
For example, multidrug-resistant gram-negative bacteria are more common in regions such as India and Greece and may be considered high-prevalence pathogens in these locations.
This study has some limitations. First, we grouped the effects of all antibiotics together with respect to antibiotic usage and the microbiome effect. In reality, different antibiotics (or classes of antibiotics) may have differential effects on the transmission probabilities for various MDROs, but this level of detail is not currently supported by available data.41 In addition, we only considered a single category of antibiotic usage, which ignores differences in outcomes for patients who receive suboptimal dosing.42 This simplification limited the number of parameter assumptions required for the model and facilitated more direct experimentation regarding the primary interventional parameter (ie, antibiotic usage). Next, the results generated via simulation are sensitive to the input parameters and would likely change under different assumptions. For example, admission prevalence as well as compliance rates for hand hygiene and glove and gown usage are likely to vary by hospital; they are strong determinants for MDRO transmission in the model. In addition, antibiotic prescribing practices are also likely to vary. However, modifying these parameters tends to scale transmission proportionally and does not have a significant impact on the proportional effect of reducing antibiotic usage. For example, increasing the mutation rate from 1% to 5% of patients receiving antibiotics increased overall transmission; however, the percentage decrease in acquisition rates due to a reduction in antibiotic usage remained strongly significant. Additional details on the sensitivity analysis of hand hygiene and mutation effect parameters are available in an online supplement, which supports our results in the face of the variability and uncertainty associated with these parameters. In this study, we focused on setting the model parameters to evidence-based values, either informed by the literature or by calibrating those using observational data. Individual facilities could adapt the model parameters as needed to form more reasonable estimates of local prevalence and transmission levels, as well as the effect of reducing antibiotic usage on acquisition rates and other secondary outcomes.
In a critical-care setting—where nearly 75% of patients receive antibiotics and up to 50% of antibiotic courses may be considered inappropriate—even a moderate reduction in antibiotic usage is expected to cause a clinically significant reduction of MDRO transmission. In prior studies, antimicrobial stewardship programs have been shown to reduce overall antibiotic usage by nearly 33%, which in our model would result in at least a 25% decrease in MDRO acquisition. These reductions are statistically significant and proportionally similar for both high- and low-prevalence MDROs, and they can potentially decrease MDRO acquisition among patients who are receiving antibiotics as well as among patients who are not receiving antibiotics.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Gwen Robinson for her assistance with the preparation and submission of this manuscript.
Financial support: A.D.H. is supported by a grant from the National Institutes of Health (NIH; grant no. 5K24AI079040-05). K.A.T. is supported by an NIH Career Development Grant (grant no. 1K23AI08250-01A1). D.J.M. is supported by a grant from the Veterans Affairs Health Services Research and Development Department (grant no. CRE 12-307).
Footnotes
SUPPLEMENTARY MATERIAL
To view supplementary material for this article, please visit https://doi.org/10.1017/ice.2017.34
Potential conflicts of interest: All authors report no conflicts of interest relevant to this article.
REFERENCES
- 1.Antibiotic resistance threats in the United States. 2013. antibiotic/ antimicrobial resistance. Centers for Disease Control and Prevention website. https://www.cdc.gov/drugresistance/threat-report-2013/. Published 2013. Accessed January 19, 2017.
- 2.Tornieporth NG, Roberts RB, John J, Hafner A, Riley LW. Risk factors associated with vancomycin-resistant Enterococcus faecium infection or colonization in 145 matched case patients and control patients and control patients. Clin Infect Dis Off Publ Infect Dis Soc Am 1996;23:767–772. [DOI] [PubMed] [Google Scholar]
- 3.Ling ML, Tee YM, Tan SG, et al. Risk factors for acquisition of carbapenem resistant Enterobacteriaceae in an acute tertiary care hospital in Singapore. Antimicrob Resist Infect Control 2015;4:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Couderc C, Jolivet S, Thiébaut ACM, et al. Fluoroquinolone use is a risk factor for methicillin-resistant Staphylococcus aureus acquisition in long-term care facilities: a nested case-case-control study. Clin Infect Dis Off Publ Infect Dis Soc Am 2014;59:206–215. [DOI] [PubMed] [Google Scholar]
- 5.Dellit TH, Owens RC, McGowan JE, et al. Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clin Infect Dis Off Publ Infect Dis Soc Am 2007;44:159–177. [DOI] [PubMed] [Google Scholar]
- 6.Macal CM, North MJ. Tutorial on agent-based modelling and simulation. J Simul 2010;4:151–162. [Google Scholar]
- 7.Wilensky Uri. NetLogo home page. Northwestern University website. http://ccl.northwestern.edu/netlogo/. Published 2013. Accessed January 19, 2017. [Google Scholar]
- 8.Barnes SL, Morgan DJ, Harris AD, Carling PC, Thom KA. Preventing the transmission of multidrug-resistant organisms: modeling the relative importance of hand hygiene and environmental cleaning interventions. Infect Control Hosp Epidemiol 2014;35:1156–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magiorakos A-P, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis 2012;18:268–281. [DOI] [PubMed] [Google Scholar]
- 10.Climo MW, Sepkowitz KA, Zuccotti G, et al. The effect of daily bathing with chlorhexidine on the acquisition of methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus, and healthcare-associated bloodstream infections: results of a quasi-experimental multicenter trial. Crit Care Med 2009;37:1858–1865. [DOI] [PubMed] [Google Scholar]
- 11.Smet AM de, Kluytmans JA, Cooper BS, et al. Decontamination of the digestive tract and oropharynx in ICU patients. N Engl J Med 2009;360:20–31. [DOI] [PubMed] [Google Scholar]
- 12.Reddy T, Chopra T, Marchaim D, et al. Trends in antimicrobial resistance ofAcinetobacter baumannii isolates from a metropolitan Detroit health system. Antimicrob Agents Chemother 2010;54: 2235–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huskins WC, Huckabee CM, O’Grady NP, et al. Intervention to reduce transmission of resistant bacteria in intensive care. N Engl J Med 2011;364:1407–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziakas PD, Thapa R, Rice LB, Mylonakis E. Trends and significance of VRE colonization in the ICU: a meta-analysis of published studies. PloS One 2013;8:e75658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swaminathan M, Sharma S, Blash SP, et al. Prevalence and risk factors for acquisition of carbapenem-resistant Enterobacteriaceae in the setting of endemicity. Infect Control Hosp Epidemiol 2013;34:809–817. [DOI] [PubMed] [Google Scholar]
- 16.Harris AD, Pineles L, Belton B, et al. Universal glove and gown use and acquisition of antibiotic-resistant bacteria in the ICU: a randomized trial. JAMA 2013;310:1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhar S, Marchaim D, Tansek R, et al. Contact precautions: more is not necessarily better. Infect Control Hosp Epidemiol 2014;35:213–221. [DOI] [PubMed] [Google Scholar]
- 18.Morgan DJ, Murthy R, Munoz-Price LS, et al. Reconsidering contact precautions for endemic methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus. Infect Control Hosp Epidemiol 2015;36:1163–1172. [DOI] [PubMed] [Google Scholar]
- 19.Render ML, Kim HM, Deddens J, et al. Variation in outcomes in Veterans Affairs intensive care units with a computerized severity measure. Crit Care Med 2005;33:930–939. [DOI] [PubMed] [Google Scholar]
- 20.Vasilevskis EE, Kuzniewicz MW, Cason BA, et al. Mortality probability model III and simplified acute physiology score II: assessing their value in predicting length of stay and comparison to APACHE IV. Chest 2009;136:89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan BA, Hui KY, Hui SL, et al. Time-motion analysis of health care workers’ contact with patients and workers’ hand hygiene: open vs closed units. Am J Crit Care Off Publ Am Assoc Crit-Care Nurses 2011;20:e75–e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgan DJ, Pineles L, Shardell M, et al. The effect of contact precautions on healthcare worker activity in acute care hospitals. Infect Control Hosp Epidemiol 2013;34:69–73. [DOI] [PubMed] [Google Scholar]
- 23.Hornbeck T, Naylor D, Segre AM, Thomas G, Herman T, Polgreen PM. Using sensor networks to study the effect of peripatetic healthcare workers on the spread of hospital-associated infections. J Infect Dis 2012;206:1549–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erasmus V, Daha TJ, Brug H, et al. Systematic review of studies on compliance with hand hygiene guidelines in hospital care. Infect Control Hosp Epidemiol 2010;31:283–294. [DOI] [PubMed] [Google Scholar]
- 25.Girou E, Loyeau S, Legrand P, Oppein F, Brun-Buisson C. Efficacy of handrubbing with alcohol based solution versus standard handwashing with antiseptic soap: randomised clinical trial. BMJ 2002;325:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence SJ, Puzniak LA, Shadel BN, Gillespie KN, Kollef MH, Mundy LM. Clostridium difficile in the intensive care unit: epidemiology, costs, and colonization pressure. Infect Control Hosp Epidemiol Off J Soc Hosp Epidemiol Am 2007;28:123–130. [DOI] [PubMed] [Google Scholar]
- 27.Vincent JL, Rello J, Marshall J, et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA 2009;302:2323–2329. [DOI] [PubMed] [Google Scholar]
- 28.Candeloro CL, Kelly LM, Bohdanowicz E, Martin CM, Bombassaro AM. Antimicrobial use in a critical care unit: a prospective observational study. Int J Pharm Pract 2012;20: 164–171. [DOI] [PubMed] [Google Scholar]
- 29.Lilly CM, Zuckerman IH, Badawi O, Riker RR. Benchmark data from more than 240,000 adults that reflect the current practice of critical care in the United States. Chest 2011;140:1232–1242. [DOI] [PubMed] [Google Scholar]
- 30.Get smart for healthcare. Core elements of hospital antibiotic stewardship programs. Centers for Disease Control and Prevention website. https://www.cdc.gov/getsmart/healthcare/implementation/core-elements.html. Accessed January 19, 2017.
- 31.File TM, Srinivasan A, Bartlett JG. Antimicrobial stewardship: importance for patient and public health. Clin Infect Dis Off Publ Infect Dis Soc Am 2014;59:S93–S96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilholm H, Holmstrand L, Ahl J, et al. An audit-based, infectious disease specialist-guided antimicrobial stewardship program profoundly reduced antibiotic use without negatively affecting patient outcomes. Open Forum Infect Dis 2015;2:ofv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malani AN, Richards PG, Kapila S, Otto MH, Czerwinski J, Singal B. Clinical and economic outcomes from a community hospital’s antimicrobial stewardship program. Am J Infect Control 2013; 41:145–148. [DOI] [PubMed] [Google Scholar]
- 34.Willmann M, Marschal M, Hölzl F, Schröppel K, Autenrieth IB, Peter S. Time series analysis as a tool to predict the impact of antimicrobial restriction in antibiotic stewardship programs using the example of multidrug-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 2013;57:1797–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelat C, Kardaś-Słoma L, Birgand G, et al. Hand hygiene, cohorting, or antibiotic restriction to control outbreaks of multidrug-resistant Enterobacteriaceae. Infect Control Hosp Epidemiol 2016;37:272–280. [DOI] [PubMed] [Google Scholar]
- 36.Dickstein Y, Geffen Y, Leibovici L, Paul M. Comparison of antibiotic susceptibility patterns of bacterial isolates based on time from hospitalization and culture source: implications for hospital antibiograms. Infect Control Hosp Epidemiol 2016; 37:212–214. [DOI] [PubMed] [Google Scholar]
- 37.McGregor JC, Furuno JP. Optimizing research methods used for the evaluation of antimicrobial stewardship programs. Clin Infect Dis Off Publ Infect Dis Soc Am 2014;59:S185–S192. [DOI] [PubMed] [Google Scholar]
- 38.Schechner V, Temkin E, Harbarth S, Carmeli Y, Schwaber MJ. Epidemiological interpretation of studies examining the effect of antibiotic usage on resistance. Clin Microbiol Rev 2013;26:289–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malhotra Kumar S, Lammens C, Coenen S, Van Herck K, Goossens H. Effect of azithromycin and clarithromycin therapy on pharyngeal carriage of macrolide-resistant streptococci in healthy volunteers: a randomised, double-blind, placebo-controlled study. Lancet 2007;369:482–490. [DOI] [PubMed] [Google Scholar]
- 40.Hay AD, Thomas M, Montgomery A, et al. The relationship between primary care antibiotic prescribing and bacterial resistance in adults in the community: a controlled observational study using individual patient data. J Antimicrob Chemother 2005;56:146–153. [DOI] [PubMed] [Google Scholar]
- 41.Cotter PD, Stanton C, Ross RP, Hill C. The impact of antibiotics on the gut microbiota as revealed by high-throughput DNA sequencing. Discov Med 2012;13:193–199. [PubMed] [Google Scholar]
- 42.Drusano GL, Louie A, Deziel M, Gumbo T. The crisis of resistance: identifying drug exposures to suppress amplification of resistant mutant subpopulations. Clin Infect Dis Off Publ Infect Dis Soc Am 2006;42:525–532. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.