Abstract
Background
Cockroaches are one of the most important and frequent insects responsible for harboring, transmission and dissemination of human pathogens in the hospital environment. The present research was done to study the phenotypic and genotypic characterization of antibiotic resistance in the Methicillin-resistant Staphylococcus aureus strains isolated from hospital cockroaches.
Methods
Five-hundred and thirty Periplanets americana and Blattella germanica cockroaches were collected and their gut content and external washing samples were subjected to bacterial isolation. MRSA strains were subjected to disk diffusion and PCR amplification of antibiotic resistance genes.
Results
Prevalence of MRSA strains in P. americana and B. germanica cockroaches were 52.77 and 43.33%, respectively. External washing samples of P. americana cockroaches had the highest prevalence of MRSA strains (59.57%). MRSA isolates of external washing samples harbored the highest prevalence of resistance against penicillin (100%), ceftaroline (100%), tetracycline (100%), gentamicin (83.33%) and trimethoprim-sulfamethoxazole (80.55%). MRSA strains isolated from gut content samples harbored the highest prevalence of resistance against penicillin (100%), ceftaroline (100%), tetracycline (100%), trimethoprim-sulfamethoxazole (80%) and gentamicin (73.33%). BlaZ, aacA-D, tetK, msrA, dfrA, ermA, gyrA, grlA and rpoB were the most commonly detected antibiotic resistance genes amongst the MRSA strains.
Conclusions
The present investigation is the first report of the phenotypic and genotypic evaluation of antibiotic resistance in the MRSA strains isolated from P. americana and B. germanica hospital cockroaches. Hospital cockroaches are considered as a potential mechanical vector for MRSA strains.
Keywords: Methicillin-resistant Staphylococcus aureus, Antibiotic resistance, Antibiotic resistance genes, Hospital cockroaches
Introduction
There are several ways to increase the survival of microorganisms in the environment and their transmission to human population. Cockroaches are among the most common insects found in industrial and residential environments such as hospitals. Indeed, the species has been highly successful in exploiting niches within human habitation. In addition to being a persistent pest and owing to their omnivorous habit of feeding and indiscriminate deposition of fecal materials, cockroaches are well-known agents for harboring, transmission and dissemination of human pathogens, thereby representing a public health risk [1, 2].
There are two cockroach species commonly found infesting in domestic, industrial and residential environments, these are the German cockroach (Blattella germanica (B. germanica)) and the American cockroach (Periplanets americana (P. americana)) (Dictyoptera; Blattidae). P. americana is the largest shiny reddish brown peridomestic cockroaches measuring on average 4–5 cm in length, while B. germanica is light yellowish brown with length ranging from 1.0–2.5 cm [1, 2]. B. germanica are the most abundantly distributed cockroach species. B. germanica are more prevalent inside the home, while the P. americana are common around the home and associated with water drainage systems and water pipes. Both of them can disseminate through the sewage system especially in public places such as hospitals [1, 2].
Over 100 species of virulent and resistant bacteria have been isolated from or passed through cockroaches. Microbiological and epidemiological investigations reported that Staphylococcus aureus (S. aureus) has commonly been isolated from P. americana and B. germanica found in the public places and especially hospital environment [3, 4]. S. aureus is commonly found in nose and respiratory system and on the skin [5]. It is responsible for the occurrence of nosocomial and community-acquired infections, food-borne diseases and food poisoning [5].
S. aureus strains are usually resist against several types of antibiotics [6–9]. Nowadays, methicillin-resistant S. aureus (MRSA) has become a serious problem in hospitals [6–9]. Documented data revealed that about 50–70% of the S. aureus strains isolated from the hospital environment were MRSA [6–9]. MRSA strains are responsible for about 100,000 cases of infections with around 20% mortality rate each year in the United States [10]. High pathogenicity of MRSA strains, its high resistance to several types of antibiotics and its nosocomial aspects have increased the importance of isolation of MRSA from the hospital cockroaches. MRSA strains are believed to serve as important reservoirs of antimicrobial resistance genes which can transfer and integrate into the MRSA genome leading to the emergence of new and potentially more resistant strains [6–9]. Documented data revealed that presence of certain antibiotic resistance genes is responsible for occurrence of severe antibiotic resistance [6–9]. Reports showed the high presence of rpoB, blaZ, mecA, aacA-D, tetK and tetM, ermA and msrA, linA, vatA and vatB, dfrA, gyrA and grlA and cat1 antibiotic resistance genes in the S. aureus strains caused severe occurrence of resistance against ansamycins, penicillins, methicillin, aminoglycosides, tetracyclines, macrolides–lincosamide-streptogramin B, lincosamides, streptogramins, folate inhibitors, fluoroquinolones and phenicols groups of antibiotics, respectively [6–9]. Reports of methicillin-resistant strains are challenging due to the large proportion of methicillin-resistant strains and increasing numbers of isolates reinforcing the need to revise their importance to hospital [6–9]. Therefore, screening of these elements is important for public health and despite the importance of such a screen, limited data are available for MRSA at the species level among the hospital cockroach samples.
MRSA strains have been tested in hospital cockroach samples to assess microbiological safety, sanitation conditions and finally epidemiological roles of these insects in survival and transmission of bacteria. High pathogenicity of MRSA strains and general weakness of hospitalized patients make it necessary to assess the presence of MRSA strains in hospital cockroach samples. Thus, the current research was done to study the phenotypic and genotypic properties of antibiotic resistance in the MRSA strains isolated from P. americana and B. germanica cockroaches in Iranian hospitals.
Materials and methods
Samples
The present descriptive study was conducted during 2016 and 2017 at the tertiary hospitals of the Tehran province, Iran. Five-hundred and thirty cockroaches were collected using sticky traps, vacuum cleaners and hand catch methods from human dwellings. Traps were manufactured according to the design of Reierson and Rust (1997) [11]. The traps were placed on the floor under beds, cupboards, wooden racks, and/or benches, for two consecutive days. Each trapped cockroach was placed in a sterile test tube before sending to the laboratory. After immobilization by freezing at 0 °C for 5 min, the species of cockroaches were identified under a dissecting microscope according to Harwood and James (1979) [12]. P. americana and B. germanica cockroaches were subjected to further parts of the study.
Sample preparation and isolation of S. aureus
Body surface of cockroaches were washed with physiological saline after vortexing for 2 min and taken as a homogenate sample. Before gastrointestinal tract (GIT) dissection, each cockroach was decontaminated with 95% ethanol, and the residue of ethanol was removed with saline solution. The gut was dissected aseptically using sterile needles and washed with 5-mL normal saline solution. Caution was taken to reduce the number of cut off or break in the gut. One milliliter of each homogenate was inoculated separately into 9-mL of buffered peptone water (Merck, Germany) for primary enrichment, and incubated at 37 °C for 18–24 h. Samples were then inoculated in screw cap test tube containing nutrient broth (NB, Merck, Germany) for the isolation of S. aureus, and enriched overnight at 37 °C. A loopful of inoculum from the enriched culture was streaked on to Blood Agar (BA, Merck, Germany) and Mannitol Salt Agar (MSA, Merck, Germany) incubated at 37 °C for 24 h for the observation of hemolysis and mannitol fermentation, respectively. Colonies with beta-hemolysis reaction and mannitol fermentation were further identified on the basis of Gram staining, catalase activity, coagulated test (rabbit plasma), oxidase test, glucose O/F test, resistance to bacitracin (0.04 U), urease activity, nitrate reduction, phosphatase, deoxyribonuclease (DNase, Merck, Germany) test, voges-proskaver (Merck, Germany) test and carbohydrate (xylose, sucrose, trehalose and maltose, fructose, lactose, mannose) fermentation tests [13].
Identification of methicillin-resistant Staphylococcus aureus strains
Cefoxitin (30 μg) and oxacillin (1 μg) susceptibility tests were used to distinguish the MRSA strains from S. aureus isolates of hospital cockroaches. All tests were performed using the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [14].
MRSA isolates were identified another time using the PCR-based amplification of mecA gene. MRSA strains were sub-cultured on TSB media (Merck, Germany) and further incubated for 48 h at 37 °C. Genomic DNA was extracted from bacterial colonies using the DNA extraction kit (Thermo Fisher Scientific, St. Leon-Rot, Germany) according to manufacturer’s instruction. Purity (A260/A280) and concentration of extracted DNA were then checked (NanoDrop, Thermo Scientific, Waltham, MA, USA). The truth of the DNA was assessed on a 2% agarose gel stained with ethidium bromide (0.5 μg/mL) (Thermo Fisher Scientific, St. Leon-Rot, Germany).
The PCR reactions were performed in a total volume of 25 μL, including 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 200 μM dNTPs each (Thermo Fisher Scientific, St. Leon-Rot, Germany), 2.5 μL PCR buffer (10X), 2.5 mM of each primer mecA1 (5′-ACGAGTAGATGCTCAATATAA-3′) and mecA2 (5′-CTTAGTTCTTTAGCGATTGC-3′) (Gen Bank Accession Number NC_003923M, 293 bp), 1.5 U of Taq DNA polymerase (Thermo Fisher Scientific, St. Leon-Rot, Germany) and 5 μL of the extracted DNA template of the MRSA isolates. The PCR cycling conditions were as follows: initial denaturation at 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s, followed by an extra cycle of annealing at 60 °C for 30 s, and a final extension at 72 °C for 5 min.
Phenotypic evaluation of antibiotic resistance
Patterns of antimicrobial resistance of the MRSA strains isolated from hospital cockroaches were studied using the simple disk diffusion according to the Kirby-Baur disc diffusion technique. The Mueller–Hinton agar (Merck, Germany) medium was used for this purpose. Susceptibility of MRSA isolates were tested against several types of antibiotic groups including Penicillins (penicillin (10 μg/disk)), Cephems (ceftaroline (30 μg/disk),), Aminoglycosides (gentamicin (10 μg/disk), amikacin (30 μg/disk), Macrolides (azithromycin (15 μg/disk) and erythromycin (15 μg/disk)), Tetracyclines (tetracycline (30 μg/disk), doxycycline (30 μg/disk)), Fluoroquinolones (ciprofloxacin (5 μg/disk) and levofloxacin (5 μg/disk)), Lincosamides (clindamycin (2 μg/disk)), Folate pathway inhibitors (trimethoprim-sulfamethoxazole (25 μg/disk)), Phenicols (chloramphenicol (30 μg/disk)) and Ansamycins (rifampin (5 μg/disk)) antibiotic agents (Oxoid, UK) using the instruction of Clinical and Laboratory Standards Institute [15]. The plates containing the discs were allowed to stand for at least 30 min before incubated at 37 °C for 24 h. The diameter of the zone of inhibition produced by each antibiotic disc was measured and interpreted using the CLSI zone diameter interpretative standards [15]. Staphylococcus aureus ATCC 25923 was used as quality control organism in antimicrobial susceptibility determination.
Genotypic evaluation of antibiotic resistance
Table 1 represents the list of primers and PCR conditions used for amplification of antibiotic resistance genes in the MRSA strains isolated from hospital cockroaches [16–22]. A programmable DNA thermo-cycler (Eppendorf Mastercycler 5330, Eppendorf-Nethel-Hinz GmbH, Hamburg, Germany) was used in all PCR reactions. Amplified samples were analyzed by electrophoresis (120 V/208 mA) in 2.5% agarose gel. The gel was stained with 0.1% ethidium bromide (0.4 μg/ml). The UVI doc gel documentation systems (Grade GB004, Jencons PLC, London, UK) was applied for analysis of images.
Table 1.
Target genes, oligonucleotide primers and PCR conditions used for detection of antibiotic resistance genes in the MRSA strains isolated from hospital cockroaches (16–22)
| Target gene | Primer sequence (5′-3′) | PCR product (bp) | PCR programs | PCR volume (50 μL) |
|---|---|---|---|---|
| AacA-D | F: TAATCCAAGAGCAATAAGGGC R: GCCACACTATCATAACCACTA |
227 | 1 cycle: 94 0C ------------ 5 min. 25 cycle: 94 0C ------------ 60 s 55 0C ------------ 70 s 72 0C ------------ 60 s 1 cycle: 72 0C ------------ 10 min |
5 μL PCR buffer 10X 1.5 mM Mgcl2 200 μM dNTP (Fermentas) 0.5 μM of each primers F & R 1.25 U Taq DNA polymerase (Fermentas) 2.5 μL DNA template |
| ermA | F: AAGCGGTAAACCCCTCTGA R: TTCGCAAATCCCTTCTCAAC |
190 | ||
| tetK | F: GTAGCGACAATAGGTAATAGT R: GTAGTGACAATAAACCTCCTA |
360 | ||
| tetM | F: AGTGGAGCGATTACAGAA R: CATATGTCCTGGCGTGTCTA |
158 | 1 cycle: 94 0C ------------ 6 min. 34 cycle: 95 0C ------------ 50 s 55 0C ------------ 70 s 72 0C ------------ 60 s 1 cycle: 72 0C ------------ 8 min |
5 μL PCR buffer 10X 2 mM Mgcl2 200 μM dNTP (Fermentas) 0.5 μM of each primers F & R 1.5 U Taq DNA polymerase (Fermentas) 5 μL DNA template |
| vatA | F: TGGTCCCGGAACAACATTTAT R: TCCACCGACAATAGAATAGGG |
268 | ||
| msrA | F: GGCACAATAAGAGTGTTTAAAGG R: AAGTTATATCATGAATAGATTGTCCTGTT |
940 | 1 cycle: 94 0C ------------ 6 min. 34 cycle: 95 0C ------------ 60 s 50 0C ------------ 70 s 72 0C ------------ 70 s 1 cycle: 72 0C ------------ 8 min |
5 μL PCR buffer 10X 2 mM Mgcl2 150 μM dNTP (Fermentas) 0.75 μM of each primers F & R 1.5 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
| vatB | F: GCTGCGAATTCAGTTGTTACA R: CTGACCAATCCCACCATTTTA |
136 | 1 cycle: 94 0C ------------ 6 min. 35 cycle: 95 0C ------------ 50 s 55 0C ------------ 70 s 72 0C ------------ 80 s 1 cycle: 72 0C ------------ 10 min |
5 μL PCR buffer 10X 2 mM Mgcl2 150 μM dNTP (Fermentas) 0.75 μM of each primers F & R 1.5 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
| linA | F: GGTGGCTGGGGGGTAGATGTATTAACTGG R: GCTTCTTTTGAAATACATGGTATTTTTCGA |
323 | 1 cycle: 94 0C ------------ 6 min. 30 cycle: 95 0C ------------ 60 s 57 0C ------------ 60 s 72 0C ------------ 60 s 1 cycle: 72 0C ------------ 10 min |
5 μL PCR buffer 10X 2 mM Mgcl2 150 μM dNTP (Fermentas) 0.75 μM of each primers F & R 1.5 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
| blaZ | F: ACTTCAACACCTGCTGCTTTC R: TGACCACTTTTATCA CAACC |
490 | 1 cycle: 94 0C ------------ 5 min. 30 cycle: 94 0C ------------ 20 s 60 0C ------------ 30 s 72 0C ------------ 90 s 1 cycle: 72 0C ------------ 5 min |
5 μL PCR buffer 10X 2 mM Mgcl2 150 μM dNTP (Fermentas) 0.75 μM of each primers F & R 1.5 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
| cat1 | F: AGTTGCTCAATGTACCTATAACC R: TTGTAATTCATTAAGCATTCTGCC |
547 | 1 cycle: 94 0C ------------ 8 min. 32 cycle: 95 0C ------------ 60 s 55 0C ------------ 70 s 72 0C ------------ 2 min 1 cycle: 72 0C ------------ 8 min |
5 μL PCR buffer 10X 2 mM Mgcl2 150 μM dNTP (Fermentas) 0.75 μM of each primers F & R 1.5 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
| gyrA | F: AATGAACAAGGTATGACACC R: TACGCGCTTCAGTATAACGC |
223 | 1 cycle: 94 0C ------------ 10 min. 25 cycle: 94 0C ------------ 20 s 52 0C ------------ 20 s 72 0C ------------ 50 s 1 cycle: 72 0C ------------ 5 min |
5 μL PCR buffer 10X 2 mM Mgcl2 150 μM dNTP (Fermentas) 0.75 μM of each primers F & R 1.5 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
| grlA | F: ACTTGAAGATGTTTTAGGTGAT R: TTAGG AAATCTTGATGGCAA |
459 | ||
| dfrA |
F: CTCACGATAAACAAAGAGTCA
R: CAATCATTGCTTCGTATAACG |
201 | 1 cycle: 94 0C ------------ 2 min. 30 cycle: 94 0C ------------ 60 s 50 0C ------------ 60 s 72 0C ------------ 60 s 1 cycle: 72 0C ------------ 5 min |
5 μL PCR buffer 10X 2 mM Mgcl2 150 μM dNTP (Fermentas) 0.75 μM of each primers F & R 1.5 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
| rpoB | F: ACCGTCGTTTACGTTCTGTA R: TCAGTGATAGCATGTGTATC |
460 | 1 cycle: 94 0C ------------ 5 min. 32 cycle: 94 0C ------------ 60 s 56 0C ------------ 45 s 72 0C ------------ 60 s 1 cycle: 72 0C ------------ 10 min |
5 μL PCR buffer 10X 2 mM Mgcl2 150 μM dNTP (Fermentas) 0.75 μM of each primers F & R 1.5 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
Statistical analysis
Statistical analysis was done using the SPSS 21.0 statistical software (SPSS Inc., Chicago, IL, USA). Chi-square test and Fisher’s exact two-tailed test were used to assess any significant relationship between prevalence of S. aureus and MRSA strains and their phenotypic and genotypic properties of antibiotic resistance. P value < 0.05 was considered as statistical significant level.
Results
Table 2 represents the prevalence of S. aureus and MRSA strains in different types of hospital cockroaches. Sixty-five out of 530 (12.26%) external washing samples of hospital cockroaches and thirty-seven out of 530 (6.98%) gut content samples of hospital cockroaches were positive for S. aureus strains. Prevalence of S. aureus strains in P. americana and B. germanica hospital cockroaches were 11.61% (72/620) and 6.81% (30/440), respectively. External washing samples of P. americana hospital cockroaches had the highest prevalence of S. aureus strains (15.16%), while gut content samples of P. americana hospital cockroaches had the lowest (8.06%). Prevalence of S. aureus strains in external washing and gut content samples of B. germanica hospital cockroaches were 8.18 and 5.45%, respectively. Statistically significant differences were seen between types of hospital cockroaches and prevalence of S. aureus (P < 0.05).
Table 2.
Prevalence of S. aureus and MRSA strains in different types of hospital cockroaches
| Types of cockroaches | No samples collected | N (%) of S. aureus positive samples | N (%) of MRSA positive samples | |
|---|---|---|---|---|
| P. americana | External washing | 310 | 47 (15.16) | 28 (59.57) |
| Gut contents | 310 | 25 (8.06) | 10 (40) | |
| B. germanica | External washing | 220 | 18 (8.18) | 8 (44.44) |
| Gut contents | 220 | 12 (5.45) | 5 (41.66) | |
| Total | External washing | 530 | 65 (12.26) | 36 (55.38) |
| Gut contents | 530 | 37 (6.98) | 15 (40.54) | |
MRSA strains were further identified using the mecA gene PCR amplification. Figure 1 shows the gel electrophoresis of the mecA gene of the MRSA strains in PCR reaction. Thirty-six out of 65 (55.38%) S. aureus strains isolated from external washing samples of hospital cockroaches and fifteen out of 37 (40.54%) S. aureus strains isolated from gut content samples of hospital cockroaches were confirmed to be MRSA strains. Prevalence of MRSA strains in P. americana and B. germanica hospital cockroaches were 52.77% (38/72) and 43.33% (13/30), respectively. External washing samples of P. americana hospital cockroaches had the highest prevalence of MRSA strains (59.57%), while gut content samples of P. americana hospital cockroaches had the lowest (40%). Prevalence of MRSA strains in external washing and gut content samples of B. germanica hospital cockroaches were 44.44 and 41.66%, respectively. Statistically significant differences were seen between types of hospital cockroaches and prevalence of MRSA (P < 0.05).
Fig. 1.
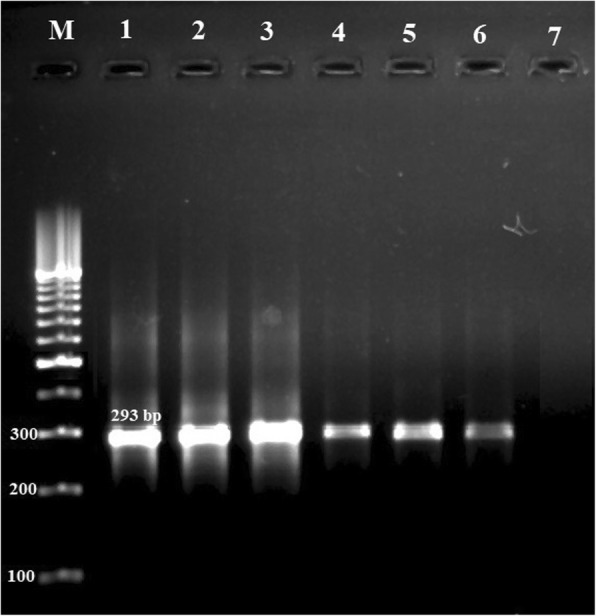
Gel electrophoresis of the mecA gene of the MRSA strains in PCR reaction. M: 100 bp ladder (Thermo Fisher Scientific, St. Leon-Rot, Germany), Lane 1: Positive control (MRSA ATCC 43300) Lanes 2–6: Positive samples for the mecA gene (293 bp) and Lane 7: Negative control (PCR grade water (Thermo Fisher Scientific, St. Leon-Rot, Germany))
MRSA isolates were subjected to disk diffusion to assess the antibiotic resistance pattern. Table 3 represents the phenotypic pattern of antibiotic resistance of MRSA strains isolated from hospital cockroaches. MRSA strains isolated from external washing samples of hospital cockroaches harbored the highest prevalence of resistance against penicillin (100%), ceftaroline (100%), tetracycline (100%), gentamicin (83.33%) and trimethoprim-sulfamethoxazole (80.55%) antibiotic agents. MRSA strains isolated from gut content samples of hospital cockroaches harbored the highest prevalence of resistance against penicillin (100%), ceftaroline (100%), tetracycline (100%), trimethoprim-sulfamethoxazole (80%) and gentamicin (73.33%) antibiotic agents. MRSA strains isolated from external washing samples of hospital cockroaches harbored the lowest prevalence of resistance against chloramphenicol (25%), rifampin (33.33%), levofloxacin (38.88%) and ciprofloxacin (44.44%) antibiotic agents. MRSA strains isolated from gut content samples of hospital cockroaches harbored the lowest prevalence of resistance against chloramphenicol (20%), rifampin (33.33%), levofloxacin (40%), ciprofloxacin (46.66%) and clindamycin (46.66%) antibiotic agents. MRSA strains isolated from P. americana harbored the higher prevalence of resistance against almost all antibiotic agents (P < 0.05). Additionally, those of external washing samples had the higher prevalence of resistance against almost all tested antibiotic agents (P < 0.05). Statistically significant differences were seen between types of samples and prevalence of antibiotic resistance (P < 0.05).
Table 3.
Phenotypic pattern of antibiotic resistance of MRSA strains isolated from different types of hospital cockroaches
| Origins (N of MRSA strains) | N (%) isolates resistant to each antibiotic | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Penicillins | Cephems | Aminoglycosides | Macrolides | Tetracyclines | Fluoroquinolones | Lincosamides | Folate inhibitors | Phenicols | Ansamycins | ||||||
| P10a | Cft | Gen | Amk | Azi | Ert | Tet | Dox | Cip | Lev | Clin | Tr-Sul | C30 | Rif | ||
| P. americana | External washing (28) | 28 (100) | 28 (100) | 24 (85.71) | 21 (75) | 15 (53.57) | 18 (64.28) | 28 (100) | 21 (75) | 12 (42.85) | 11 (39.28) | 14 (50) | 23 (82.14) | 7 (25) | 9 (36) |
| Gut contents (10) | 10 (100) | 10 (100) | 8 (80) | 7 (70) | 5 (50) | 6 (60) | 10 (100) | 6 (60) | 5 (50) | 4 (40) | 5 (50) | 8 (80) | 2 (20) | 3 (30) | |
| B. germanica | External washing (8) | 8 (100) | 8 (100) | 6 (75) | 5 (62.50) | 4 (50) | 5 (62.50) | 8 (100) | 5 (62.50) | 4 (50) | 3 (37.50) | 4 (50) | 6 (75) | 2 (25) | 3 (37.50) |
| Gut contents (5) | 5 (100) | 5 (100) | 3 (60) | 2 (40) | 3 (60) | 3 (60) | 5 (100) | 2 (40) | 2 (40) | 2 (40) | 2 (40) | 4 (80) | 1 (20) | 2 (40) | |
| Total | External washing (36) | 36 (100) | 36 (100) | 30 (83.33) | 26 (72.22) | 19 (52.77) | 23 (63.88) | 36 (100) | 26 (72.22) | 16 (44.44) | 14 (38.88) | 18 (50) | 29 (80.55) | 9 (25) | 12 (33.33) |
| Gut contents (15) | 15 (100) | 15 (100) | 11 (73.33) | 9 (60) | 8 (53.33) | 9 (60) | 15 (100) | 8 (53.33) | 7 (46.66) | 6 (40) | 7 (46.66) | 12 (80) | 3 (20) | 5 (33.33) | |
aP10: penicillin (10 μg/disk), Cft: ceftaroline (30 μg/disk), Gen: gentamicin (10 μg/disk), Amk: amikacin (30 μg/disk), Azi: azithromycin (15 μg/disk), Ert: erythromycin (15 μg/disk), Tet: tetracycline (30 μg/disk), Do: doxycycline (30 μg/disk), Cip: ciprofloxacin (5 μg/disk), Lev: levofloxacin (5 μg/disk), Clin: clindamycin (2 μg/disk), Tr-Sul: trimethoprim-sulfamethoxazole (25 μg/disk), C30: chloramphenicol (30 μg/disk), Rif: rifampin (5 μg/disk)
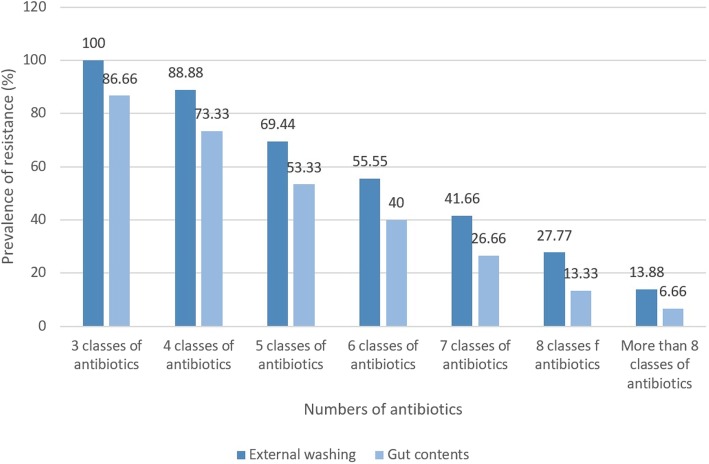
Figure 2 represents the prevalence of resistant MRSA strains in different types of hospital cockroaches. Multi-drug resistant strains were determined as those who had at least simultaneous resistance against 3 or more than 3 classes of antibiotics. All of the MRSA strains isolated from external washing samples of hospital cockroaches at least had resistance against 3 different classes of antibiotics, while prevalence of resistance against more than 8 classes of antibiotics was 13.88%. Additionally, 86.66% of the MRSA strains isolated from gut content samples of hospital cockroaches at least had resistance against 3 different classes of antibiotics, while prevalence of resistance against more than 8 classes of antibiotics was 6.66%.
Fig. 2.
Prevalence of resistant MRSA strains in external washing and gut content samples of hospital cockroaches. Results were analyzed based on the total of 36 MRSA strains isolated from external washing and 15 MRSA strains isolated from gut content samples of hospital cockroaches
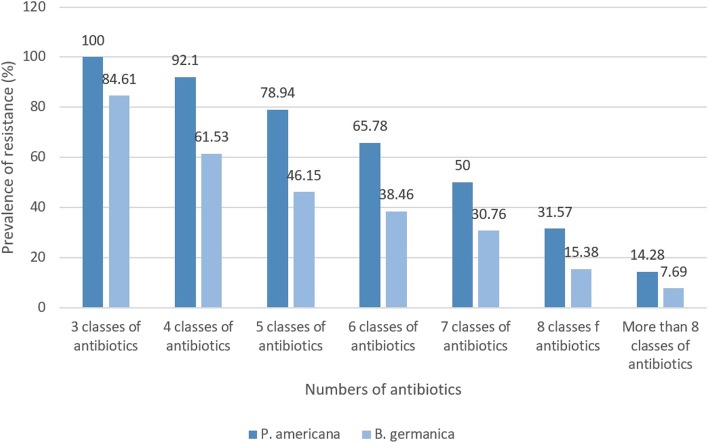
Figure 3 represents the prevalence of resistant MRSA strains in P. americana and B. germanica hospital cockroaches. All of the MRSA strains isolated from P. americana hospital cockroaches at least had resistance against 3 different classes of antibiotics, while prevalence of resistance against more than 8 classes of antibiotics was 14.28%. Additionally, 84.61% of the MRSA strains isolated from B. germanica hospital cockroaches at least had resistance against 3 different classes of antibiotics, while prevalence of resistance against more than 8 classes of antibiotics was 7.69%.
Fig. 3.
Prevalence of resistant MRSA strains in P. americana and B. germanica hospital cockroaches. Results were analyzed based on the total of 38 MRSA strains isolated from P. americana and 13 MRSA strains isolated from B. germanica hospital cockroaches
Table 4 represents the genotypic pattern of antibiotic resistance amongst the MRSA strain isolated from different types of hospital cockroaches. We found that blaZ (100%), aacA-D (88.88%), tetK (72.22%), msrA (63.88%), and dfrA (63.88%) were the most commonly detected antibiotic resistance genes amongst the MRSA strains isolated from external washing of hospital cockroaches. Additionally, blaZ (100%), aacA-D (73.33%), msrA (66.66%), tetK (60%) and dfrA (60%) were the most commonly detected antibiotic resistance genes amongst the MRSA strains isolated from gut contents of hospital cockroaches. We found that vatB (11.11%), cat1 (19.44%), tetM (27.77%), grlA (27.77%), rpoB (27.77%) and vatA (30.55%) had the lowest distribution amongst the antibiotic resistance genes in MRSA strains isolated from external washing of hospital cockroaches. Additionally, tetM (20%), linA (20%), ermA (20%), vatB (20%), grlA (20%), rpoB (20%) and cat1 (20%) had the lowest distribution amongst the antibiotic resistance genes in MRSA strains isolated from gut contents of hospital cockroaches. MRSA strains isolated from P. americana harbored the higher prevalence of antibiotic resistance genes (P < 0.05). Additionally, those of external washing had the higher prevalence of antibiotic resistance genes (P < 0.05). Statistically significant differences were seen between types of samples and prevalence of antibiotic resistance genes (P < 0.05).
Table 4.
Genotypic pattern of antibiotic resistance amongst the MRSA strain isolated from different types of hospital cockroaches
| Origins (N of MRSA strains) | N (%) isolates resistant to each antibiotic | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aminoglycosides | Tetracyclines | Macrolides | Lincosamides | Streptogramins | Penicillins | Folate inhibitors | Fluoroquinolones | Ansamycins | Phenicols | ||||||
| aacA-D | tetK | tetM | msrA | ermA | linA | vatA | vatB | blaZ | dfrA | gyrA | grlA | rpoB | cat1 | ||
| P. americana | External washing (28) | 26 (92.85) | 20 (71.42) | 7 (25) | 18 (64.28) | 15 (53.57) | 11 (39.28) | 7 (25) | 2 (7.14) | 28 (100) | 18 (64.28) | 10 (35.71) | 8 (28.57) | 7 (25) | 5 (17.85) |
| Gut contents (10) | 8 (80) | 6 (60) | 3 (30) | 7 (70) | 6 (60) | 3 (30) | 4 (40) | 1 (10) | 10 (100) | 6 (60) | 4 (40) | 2 (20) | 2 (20) | 2 (20) | |
| B. germanica | External washing (8) | 6 (75) | 6 (75) | 3 (37.50) | 5 (62.50) | 3 (37.50) | 2 (25) | 4 (50) | 2 (25) | 8 (100) | 5 (62.50) | 4 (50) | 2 (25) | 3 (37.50) | 2 (25) |
| Gut contents (5) | 3 (60) | 3 (60) | 1 (20) | 3 (60) | 1 (20) | 1 (20) | 3 (60) | 1 (20) | 5 (100) | 3 (60) | 3 (60) | 1 (20) | 1 (20) | 1 (20) | |
| Total | External washing (36) | 32 (88.88) | 26 (72.22) | 10 (27.77) | 23 (63.88) | 18 (50) | 13 (36.11) | 11 (30.55) | 4 (11.11) | 36 (100) | 23 (63.88) | 14 (38.88) | 10 (27.77) | 10 (27.77) | 7 (19.44) |
| Gut contents (15) | 11 (73.33) | 9 (60) | 4 (26.66) | 10 (66.66) | 7 (46.66) | 4 (26.66) | 7 (46.66) | 2 (13.33) | 15 (100) | 9 (60) | 7 (46.66) | 3 (20) | 3 (20) | 3 (20) | |
Discussion
Cockroaches are among the most notorious pests, having nocturnal and filthy habits, which do not only contaminate food by indiscriminate deposition of fecal materials and cause food poisoning but also transmit bacteria and other pathogenic microorganisms in infested areas. Cockroaches are common in many of human habitations, particularly in place where food is stored, processed, prepared, or served. Apart from that, they are also frequently detected in hospital environments, such as wards, operational rooms, area of intensive care units, and laboratory rooms. Indeed, cockroaches are found everywhere and possess nocturnal and omnivorous features; these characteristics make them the ideal carriers of pathogenic, virulent and resistant microorganisms and especially S. aureus [23].
The present investigation showed that 12.26% of external washing and 6.98% of gut content samples of hospital cockroaches were positive for S. aureus strains. Additionally, the prevalence of MRSA strains in external washing and gut content samples of hospital cockroaches were 6.79% (36/530) and 2.83% (15/530), respectively. Furthermore, P. americana cockroaches had the higher prevalence of S. aureus and also MRSA strains. Findings represents an important public health issue regarding the presence of B. germanica and P. americana cockroaches in hospital environment and also their high importance as a dangerous vector for survival and dissemination of S. aureus and also MRSA strains in the hospital environment. Findings also represent the higher prevalence of S. aureus and also MRSA strains in external washing samples of studied hospital cockroaches which may signify the higher role of external parts of the hospital cockroaches for dissemination of S. aureus and also MRSA strains. Moreover, S. aureus is not intestinal bacterium with high distribution in gut contents of the hospital cockroaches. However, presence of S. aureus in gut content samples of hospital cockroaches may be due to the permanent presence of S. aureus in hospital food samples [6, 7, 24] which may be consumed by cockroaches. Higher prevalence of S. aureus and MRSA in P. americana is may be due to their higher presence in hospital sewage than B. germanica cockroaches. Although P. americana is three to four folds larger than B. germanica in length, the capability of harboring microorganisms in cockroaches is not only related to their sizes but may also depend on the sanitation conditions of the environment.
S. aureus has been introduced as one of the most prevalent pathogenic bacteria isolated from the external washing and gut content samples of both B. germanica and P. americana cockroaches all-around the world [3, 4, 25–27]. Prevalence of S. aureus strains amongst the cockroach samples collected from Bangladesh [25], Iran [28], Nigeria [29], Ethiopia [1], Algeria [4], Brazil [30] and China [31] were 38, 62.86, 7.70, 69.20, 13.80, 49 and 5%, respectively.
Research on S. aureus carried by cockroaches in hospital environments is very important in relation to the control of nosocomial infections, which are becoming a major challenge. These infections cause high morbidity and mortality and increased hospitalization time having a consequent increase in costs. Cockroaches are able to explore various spaces in a hospital environment, making them a potential health risk, due to their ability to disperse pathogenic strains. Their growth is facilitated by fluids and food, as well as structural flaws in the hospital environment. Hospital infection is a major challenge for health professionals working in this area. The need to control and limit cockroaches has been stressed by most researchers.
MRSA strains isolated from hospital cockroaches harbored relatively high prevalence of resistance against tested antibiotic agents. MRSA strains harbored the highest prevalence of resistance against penicillin, ceftaroline, tetracycline, gentamicin, trimethoprim-sulfamethoxazole antibiotic agents. Additionally, prevalence of blaZ, aacA-D, tetK, msrA and dfrA was relatively higher than other tested antibiotic resistance genes. We found that phenotypic properties of antibiotic resistance were approved using the genotypic characterization of antibiotic resistance. MRSA strains isolated from P. americana cockroaches and also external washing samples had the higher prevalence of antibiotic resistance and antibiotic resistance genes. We also found the higher prevalence of resistance against human-based antibiotic agents in the MRSA strains isolated from hospital cockroaches which may indirectly show transmission of resistant MRSA strains from infected patients hospitalized in hospitals and health care units to resident cockroaches of hospitals. Antibiotics are not normally applied on cockroaches, but it is known that high resistance rates were reported among pathogens associated foods. In fact, a great association between cockroaches and foods could be the probable reason for isolation of resistant strains from cockroaches. Our findings revealed that more than 50% of S. aureus isolates were MRSA. High prevalence of antibiotic resistance reported in the present study is may be due to the unauthorized and indiscriminate prescription of antibiotic agents in Iranian hospitals and health care units. The circumstances in developing countries like Iran may be inflated by easy convenience of antimicrobials at a cheaper price and their extensive use in medicine.
According to the reports, burst of MRSA is increasing in Europe. In Austria, 21.60%; Belgium, 25.10%; Spain, 30.30%; and France, 33.60% of isolated S. aureus strains are methicillin resistant [32]. Higher pathogenicity and resistance of MRSA strains have also been reported in different types of human clinical infections [33, 34]. Similar antibiotic resistance patterns of the MRSA strains have also been reported against aminoglycosides [6–9, 35–38], cephems [6–9, 35–38], penicillins [6–9, 35–38], macrolides [35–38], tetracyclines [6–9, 36, 37], fluoroquinolones [6–9, 35–38], lincosamides [6–9, 35–38], folate inhibitors [6–9, 35–38], phenicols [6–9, 36, 37] and ansamycins [6–9, 36, 37] groups of antibiotics. Islam et al. (2016) [25] reported that S. aureus strains isolated from cockroach samples harbored the highest prevalence of resistance against erythromycin (58%), kanamycin (23%), penicillin (71%), oxacillin (45.50%), cephalothin (11%) and clindamycin (38%) antibiotic agents. Prado et al. (2006) [39] reported that S. aureus strains isolated from cockroach samples harbored the highest prevalence of resistance against ampicillin (30.80%), cephalexin (30.80%), cefepime (23%), and oxacillin (38.50%) antibiotic agents. Fowoyo and Ogunbanwo (2017) [40] reported that the S. aureus strains exhibited the high prevalence of resistance against ampicillin (86.70%), trimethoprim–sulfamethoxazole (74.90%), amoxicillin–clavulanic acid (52.50%), cefotaxime (3.50%), oxacillin (35.70%), ciprofloxacin (23.90%), erythromycin (15.70%), gentamicin (11.40%) and ofloxacin (7.10%). Rong et al. (2017) [41] reported that the prevalence of antibiotic resistance in the S. aureus strains against ampicillin, penicillin, amoxicillin–clavulanic acid, cefoxitin, ceftazidime, cefepime, kanamycin, streptomycin, amikacin, gentamicin, norfloxacin, ciprofloxacin, erythromycin, tetracycline, clindamycin, chloramphenicol, trimethoprim-sulfamethoxazole, vancomycin and rifampicin were 88.20, 88.20, 73.90, 8.40, 10.90, 8.40, 22.70, 14.30, 1.70, 4.20, 6.70, 5.00, 53.80, 26.90, 12.60, 7.50, 7.50, 0 and 2.50%, respectively.
We found that all of the MRSA strains were positive for the mecA gene. Most of the tetracycline-resistant MRSA isolates harbored tetK and tetM genes. Prevalence of aacA-D gene was high among gentamicin and amikacin-resistant MRSA strains. Prevalence of msrA, ermA and linA antibiotic resistance genes were also significant among the macrolide, erythromycin and clindamycin-resistant MRSA strains. Additionally, high prevalence of dfrA, rpoB and cat1 antibiotic resistance genes were also found amongst the trimethoprim-sulfamethoxazole, rifampin and chloramphenicol-resistant MRSA strains. Finally, high distribution of gyrA and grlA antibiotic resistance genes were found amongst the ciprofloxacin and levofloxacin-resistant MRSA strains. Therefore, the pattern of the antibiotic resistance of the MRSA strains of hospital cockroach samples was confirmed by the PCR amplification of the specific antibiotic resistance genes. MRSA strains of our study had considerable prevalence of resistance against clindamycin (44.44 to 46.66%). The most imperative mechanism involving resistance against clindamycin is modulated by methylase enzyme which often encoded by ermA gene [42]. Prevalence of ermA antibiotic resistance genes among the MRSA strains of our research were 46.66 to 50%. Majority of our isolates carried two tetracyclines, two erythromycins, one macrolide and several streptogramin resistance determinants reveals a great diffusion of these types of resistance. TetK, ermA, msrA and aacA-D which encode resistance against tetracycline, erythromycin, macrolides and aminoglycosides were the most commonly detected antibiotic resistance genes in the MRSA strains isolated of hospital cockroach samples. Furthermore, prevalence of blaZ, dfrA and gyrA antibiotic resistance genes which encode resistance against penicillins, folate inhibitors and fluoroquinolones, respectively. The literature survey did not indicate any report on the prevalence of blaZ, rpoB, gyrA, grlA, vatA, vatB, vatC, dfrA, cat1, msrA, ermA, linA, aacA-D, tetK and tetM genes among the MRSA strains of hospital cockroach samples. Kumar et al. (2010) [43] reported that the most commonly identified antibiotic resistance genes among the S. aureus isolates were linA (51.60%), msrB (46.10%), tetK (34.40%), tetM (34.40%), msrA (26.60%) and aacA-D (26.60%). Karataş et al. (2017) [44] revealed the higher prevalence of ermA than ermc antibiotic resistance genes among the clindamycin, erythromycin and telithromycin-resistant and also higher prevalence of tetM than tetK antibiotic resistance genes among the tetracycline-resistant MRSA strains which were similar to our findings. Our results were also similar with those of the previous research which was conducted by Simeoni et al. (2008) [45]. They reported that the prevalence of tetM, tetO, tetK, ermA, ermB, ermC, aac, blaZ and mecA antibiotic resistance genes amongst the S. aureus strains isolated from meat samples were 100, 0, 91.66, 16.66, 33.33, 58.33, 0, 100 and 58.33%, respectively. High prevalence of tetK and tetM antibiotic resistance genes in the MRSA isolates can be clarified by their usual genetic locations. Presence of tetK gene on small multicopy plasmids and tetM on conjugative transposons contributes to the spread of these determinants [46]. Some of the MRSA strains harbored ermA gene. This gene is often located on small multicopy plasmids which are present in many different staphylococcal species [46]. The ermA gene is usually carried by transposons which could explain its high prevalence amongst the MRSA strains. Resistance to aminoglycosides (60 to 83.33%) which encode by the aacA-D gene (73.33 to 88.88%) is also prevalent. Johler et al. (2011) [46] reported that prevalence of ermA, tetK and tetM antibiotic resistance genes among the S. aureus strains isolated from cases of food poisoning, milk and pork were 25, 4.87 and 0%, 50, 0 and 12.82% and 0, 12.19, and 53.84%, respectively. Podkowik et al. (2012) [47] reported that the prevalence of tetracycline resistance genes (tetO, tetK and tetM) and erythromycin resistance methylase gene (ermA, ermB and ermC) among the S. aureus strains were 44 and 60%, respectively. Prevalence of blaZ, rpoB, dfrA, gyrA, grlA and cat1 antibiotic resistance genes amongst the MRSA strains isolated from hospital cockroaches were 100%, 20–27.77%, 60–63.88%, 38.88–46.66%, 20–27.77% and 19.44–20%, respectively. High prevalence of blaZ, rpoB, dfrA, gyrA, grlA and cat1 antibiotic resistance genes was also reported in the S. aureus strains isolated from human clinical infection samples [48–50]. Resistance to benzylpenicillin is mainly caused by the blaZ gene encoding production of beta-lactamases, which hydrolytically destroy beta-lactams. Our results suggest that blaZ may play a major role in occurrence of resistance against penicillins but cannot be used alone as an indicator for penicillin resistance. Rifampin acts by interacting specifically with the β subunit of the bacterial RNA polymerase encoded by the rpoB gene. RpoB expression will increase the prevalence of resistance against rifampin in the S. aureus strains. Low prevalence of cat1 antibiotic resistance gene is may be due to the low levels of prescription of chloramphenicol in veterinary and medical sciences. Additionally, chloramphenicol is forbidden antibiotic agent in Iran. We also found the high prevalence of MRSA strains in the hospital cockroach samples. The multi-drug resistant S. aureus in cockroach was reported earlier from Nigeria, Ethiopia, Iran, and Brazil [3, 4, 51–53]. Though this study tested multi-drug resistant against single bacterium, in the absence of prior work, the findings of this study would signify the emergence of MRSA in the environment and the prospective likelihood of dissemination of such strains through mechanical vector B. germanica and P. americana cockroaches in Iran.
MRSA strains are considered as important foodborne pathogens all-around the world. Thus, their high prevalence in hospital cockroaches may influence on their presence in food samples of hospitals. It is because of the fact that cockroaches can easily penetrate into the hospital food store units. High prevalence of S. aureus and other foodborne pathogens have also been reported previously [54-64].
Conclusions
The present investigation is the first report of the phenotypic and genotypic evaluation of antibiotic resistance in the MRSA strains isolated from external washing and gut content samples of B. germanica and P. americana hospital cockroaches. High prevalence of S. aureus and MRSA strains, higher prevalence of bacteria in P. americana cockroaches, higher prevalence of bacteria in external washing samples of cockroaches, high prevalence of resistance against penicillin, ceftaroline tetracycline, gentamicin and trimethoprim-sulfamethoxazole, high distribution of blaZ, aacA-D, tetK, msrA, dfrA, ermA, gyrA and grlA antibiotic resistance genes and higher prevalence of MRSA strains in external washing samples of P. americana cockroaches were the most important findings of the present study. The present study shows the high importance of hospital cockroaches as dangerous reservoirs for harboring of virulent and resistant MRSA strains in the hospital environment and their transmission to human population. Otherwise, external surface of the P. americana cockroaches can act as a mechanical vector for transmission of MRSA strains in the hospital environment in Iran. However, further studies are required to find additional knowledge about the microbiological and epidemiological roles of the hospital cockroaches in survival and transmission of antibiotic-resistant bacteria.
Acknowledgements
The authors would like to thank Prof. Jamshidi from the Department of Microbiology, Science and Research Branch, Islamic Azad University, Tehran, Iran for his important roles in the research project. Authors would also thank from the Research Deputy of the Islkamic Azad University, Science and Research Branch, Tehran, Iran for financial support of the currenbt research.
Funding
This work was supported by the Faculty of Veterinary Medicine, Islamic Azad University, Karaj Branch, Karaj, Iran (grant no. 1159706100005).
Availability of data and materials
All data generated or analyzed throughout this research are included in this published article.
Abbreviations
- Blattella germanica
B. germanica
- MRSA
Methicillin-resistant Staphylococcus aureus
- PCR
Polymerase Chain Reaction
- Periplanets americana
P. americana
- S. aureus
Staphylococcus aureus
- SPSS
Statistical Package for the Social Sciences
Authors’ contributions
FSD designed the study and carried out the culture-based identification and PCR genetic alignment. ZA and ZM supported the study and carried out the samples collection, disk diffusion and statistical analysis. ZA carried out the writing and drafting of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethical Council of Research of the Faculty of Veterinary Medicine, Karaj Branch, Islamic Azad University, Karaj, Iran (Consent Ref Number 1159706100005). Verification of this research project and the licenses related to sampling process were approved by the Dr. Zohreh Abdolmaleki and Dr. Zohreh Mashak (Approval Ref Number 25273).
Consent for publication
There was no consent for publication.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zohreh Abdolmaleki, Phone: +982634182540, Email: zohreh.abdolmaleki@kiau.ac.ir.
Zohreh Mashak, Email: Mashak@kiau.ac.ir.
Farhad Safarpoor Dehkordi, Email: F.safarpoor@ut.ac.ir.
References
- 1.Moges F, Eshetie S, Endris M, Huruy K, Muluye D, Feleke T, et al. Cockroaches as a source of high bacterial pathogens with multidrug resistant strains in Gondar town, Ethiopia. Biomed Res Int. 2016;2016:1–7. doi: 10.1155/2016/2825056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fakoorziba M, Eghbal F, Hassanzadeh J, Moemenbellah-Fard M. Cockroaches (Periplaneta americana and Blattella germanica) as potential vectors of the pathogenic bacteria found in nosocomial infections. Ann Tropic Med Parasit. 2010;104:521–528. doi: 10.1179/136485910X12786389891326. [DOI] [PubMed] [Google Scholar]
- 3.Akinjogunla OJ, Odeyemi A, Udoinyang E. Cockroaches (Periplaneta americana and Blattella germanica): reservoirs of multi drug resistant (MDR) bacteria in Uyo, Akwa Ibom state. Sci J Bio Sci. 2012;1:19–30. [Google Scholar]
- 4.Menasria T, Moussa F, El-Hamza S, Tine S, Megri R, Chenchouni H. Bacterial load of German cockroach (Blattella germanica) found in hospital environment. Pathog Glob Health. 2014;108:141–147. doi: 10.1179/2047773214Y.0000000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Ruan S. Modeling nosocomial infections of methicillin-resistant Staphylococcus aureus with environment contamination. Sci Rep. 2017;7:580. doi: 10.1038/s41598-017-00261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Safarpoor Dehkordi F, Gandomi H, Akhondzadeh Basti A, Misaghi A, Rahimi E. Phenotypic and genotypic characterization of antibiotic resistance of methicillin-resistant Staphylococcus aureus isolated from hospital food. Antimicrob Resist Infect Control. 2017;6:104. doi: 10.1186/s13756-017-0257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Safarpoor Dehkordi F, Akhondzadeh Basti A, Gandomi H, Misaghi A, Rahimi E. Pathogenic Staphylococcus aureus in hospital food samples; prevalence and antimicrobial resistance properties. J Food Saf. 2018;38:e12501. doi: 10.1111/jfs.12501. [DOI] [Google Scholar]
- 8.Momtaz H, Dehkordi FS, Rahimi E, Asgarifar A, Momeni M. Virulence genes and antimicrobial resistance profiles of Staphylococcus aureus isolated from chicken meat in Isfahan province, Iran. J Appl Poult Res. 2013;22:913–921. doi: 10.3382/japr.2012-00673. [DOI] [Google Scholar]
- 9.Hasanpour Dehkordi A, Khaji L, Sakhaei Shahreza M, Mashak Z, Safarpoor Dehkordi F, Safaee Y, et al. One-year prevalence of antimicrobial susceptibility pattern of methicillin-resistant Staphylococcus aureus recovered from raw meat. Tropic Biomed. 2017;34:396–404. [PubMed] [Google Scholar]
- 10.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 11.Reierson DA, Rust MK. Trapping, flushing, counting German roaches. Pest Control. 1997;40:42–44. [Google Scholar]
- 12.Harwood RF, James MT. Entomology in human and animal health. seventh ed. New York, NY: Macmillan Publishing; 1979. [Google Scholar]
- 13.Fijałkowski K, Peitler D, Karakulska J. Staphylococci isolated from ready-to-eat meat–identification, antibiotic resistance and toxin gene profile. Int J Food Microb. 2016;238:113–120. doi: 10.1016/j.ijfoodmicro.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 14.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; 17th Informational Supplement. In: CLSI document M100-S17. Wayne: Clinical and laboratory standards institute; 2007.
- 15.CLSI . CLSI document M100-S25. Wayne: Clinical and Laboratory Standards Institute; 2015. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. [Google Scholar]
- 16.Lina G, Quaglia A, Reverdy M-E, Leclercq R, Vandenesch F, Etienne J. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother. 1999;43:1062–1066. doi: 10.1128/AAC.43.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strommenger B, Kettlitz C, Werner G, Witte W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J Clin Microb. 2003;41:4089–4094. doi: 10.1128/JCM.41.9.4089-4094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baddour M, AbuElKheir M, Fatani A. Comparison of mecA polymerase chain reaction with phenotypic methods for the detection of methicillin-resistant Staphylococcus aureus. Current Microb. 2007;55:473. doi: 10.1007/s00284-007-9015-6. [DOI] [PubMed] [Google Scholar]
- 19.Van TTH, Chin J, Chapman T, Tran LT, Coloe PJ. Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int J Food Microb. 2008;124:217–223. doi: 10.1016/j.ijfoodmicro.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 20.Schmitz FJ, Köhrer K, Schering S, Verhoef J, Fluit A, Heinz HP, et al. The stability of grlA, grlB, gyrA, gyrB and norA mutations and MIC values of five fluoroquinolones in three different clonal populations of methicillin-resistant Staphylococcus aureus. Clin Microb Infect. 1999;5:287–290. doi: 10.1111/j.1469-0691.1999.tb00143.x. [DOI] [PubMed] [Google Scholar]
- 21.Shittu AO, Okon K, Adesida S, Oyedara O, Witte W, Strommenger B, et al. Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol. 2011;11:92. doi: 10.1186/1471-2180-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aboshkiwa M, Rowland G, Coleman G. Nucleotide sequence of the Staphylococcus aureus RNA polymerase rpoB gene and comparison of its predicted amino acid sequence with those of other bacteria. Biochim Biophys Acta. 1995;1262:73–78. doi: 10.1016/0167-4781(95)00054-K. [DOI] [PubMed] [Google Scholar]
- 23.Pai H-H, Chen W-C, Peng C-F. Isolation of bacteria with antibiotic resistance from household cockroaches (Periplaneta americana and Blattella germanica) Acta Trop. 2005;93:259–265. doi: 10.1016/j.actatropica.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Ranjbar R, Masoudimanesh M, Dehkordi FS, Jonaidi-Jafari N, Rahimi E. Shiga (Vero)-toxin producing Escherichia coli isolated from the hospital foods; virulence factors, o-serogroups and antimicrobial resistance properties. Antimicrob Resist Infect Control. 2017;6:4. doi: 10.1186/s13756-016-0163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Islam A, Nath AD, Islam K, Islam S, Chakma S, Hossain MB, et al. Isolation, identification and antimicrobial resistance profile of Staphylococcus aureus in cockroaches (Periplaneta americana) J Adv Vet Animal Res. 2016;3:221–228. doi: 10.5455/javar.2016.c153. [DOI] [Google Scholar]
- 26.Feizhaddad M-H, Kassiri H, Sepand M-R, Ghasemi F. Bacteriological survey of American cockroaches in hospitals. Middle-East J Sci Res. 2012;12:985–989. [Google Scholar]
- 27.Hamid K, Shahnaz K. Cockroaches [periplaneta Americana (L.), dictyoptera; blattidae] as carriers of bacterial pathogens, Khorramshahr County, Iran. Jundishapur J Microb. 2012;5:320–322. [Google Scholar]
- 28.Heidari Soureshjani E, Doosti A. Examination methicillin-resistant Staphylococcus aureus (MRSA) prevalence in cockroaches from hospital in Chaharmahal-va-Bakhtiari province, Iran by polymerase chain reaction (PCR) Int J Med Med Sci. 2013;5:324–329. [Google Scholar]
- 29.Isaac C, Orue PO, Iyamu MI, Ehiaghe JI, Isaac O. Comparative analysis of pathogenic organisms in cockroaches from different community settings in Edo state, Nigeria. Korean J Parasitol. 2014;52:177. doi: 10.3347/kjp.2014.52.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliveira PS, Souza SG, Campos GB, da Silva DC, Sousa DS, Araújo SP, et al. Isolation, pathogenicity and disinfection of Staphylococcus aureus carried by insects in two public hospitals of Vitória da Conquista, Bahia, Brazil. Brazilian J Infect Dis. 2014;18:129–136. doi: 10.1016/j.bjid.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pai H-H, Chen W-C, Peng C-F. Cockroaches as potential vectors of nosocomial infections. Nfect. Control Hosp Epidemiol. 2004;25:979–984. doi: 10.1086/502330. [DOI] [PubMed] [Google Scholar]
- 32.Ekrami A, Samarbafzadeh A, Alavi M, Kalantar E, Hamzeloi F. Prevalence of methicillin resistant Staphylococcus species isolated from burn patients in a burn center, Ahvaz, Iran. Jundishapur J Microbiol. 2010;3:84–91. [Google Scholar]
- 33.Jani M, Sengupta S, Hu K, Azad RK. Deciphering pathogenicity and antibiotic resistance islands in methicillin-resistant Staphylococcus aureus genomes. Open Bio. 2017;7:170094. doi: 10.1098/rsob.170094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Espadinha D, Faria NA, Miragaia M, Lito LM, Melo-Cristino J, de Lencastre H, et al. Extensive dissemination of methicillin-resistant Staphylococcus aureus (MRSA) between the hospital and the community in a country with a high prevalence of nosocomial MRSA. PLoS One. 2013;8:e59960. doi: 10.1371/journal.pone.0059960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paludi D, Vergara A, Festino AR, Di Ciccio P, Costanzo C, Conter M, et al. Antimicrobial resistance pattern of methicillin-resistant Staphylococcus aureus in the food industry. J Biol Regul Homeost Agents. 2011;25:671. [PubMed] [Google Scholar]
- 36.Sallam KI, Abd-Elghany SM, Elhadidy M, Tamura T. Molecular characterization and antimicrobial resistance profile of methicillin-resistant Staphylococcus aureus in retail chicken. J Food Protect. 2015;78:1879–1884. doi: 10.4315/0362-028X.JFP-15-150. [DOI] [PubMed] [Google Scholar]
- 37.Jackson CR, Davis JA, Barrett JB. Prevalence and characterization of methicillin-resistant Staphylococcus aureus isolates from retail meat and humans in Georgia. J Clin Microbiol. 2013;51:1199–1207. doi: 10.1128/JCM.03166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daka D, Yihdego D. Antibiotic-resistance Staphylococcus aureus isolated from cow’s milk in the Hawassa area, South Ethiopia. Ann Clin Microbiol Antimicrob. 2012;11:26. doi: 10.1186/1476-0711-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prado MA, Gir E, Pereira MS, Reis C, Pimenta FC. Profile of antimicrobial resistance of bacteria isolated from cockroaches (Periplaneta americana) in a Brazilian health care institution. Braz J Infect Dis. 2006;10:26–32. doi: 10.1590/S1413-86702006000100006. [DOI] [PubMed] [Google Scholar]
- 40.Fowoyo P, Ogunbanwo S. Antimicrobial resistance in coagulase-negative staphylococci from Nigerian traditional fermented foods. Ann Clin Microb Antimicrob. 2017;16:4. doi: 10.1186/s12941-017-0181-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rong D, Wu Q, Xu M, Zhang J, Yu S. Prevalence, virulence genes, antimicrobial susceptibility, and genetic diversity of Staphylococcus aureus from retail aquatic products in China. Front Microbiol. 2017;8:714. doi: 10.3389/fmicb.2017.00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zelazny AM, Ferraro MJ, Glennen A, Hindler JF, Mann LM, Munro S, et al. Selection of strains for quality assessment of the disk induction method for detection of inducible clindamycin resistance in staphylococci: a CLSI collaborative study. J Clin Microbiol. 2005;43:2613–2615. doi: 10.1128/JCM.43.6.2613-2615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar R, Yadav B, Singh R. Genetic determinants of antibiotic resistance in Staphylococcus aureus isolates from milk of mastitic crossbred cattle. Current Microbiol. 2010;60:379–386. doi: 10.1007/s00284-009-9553-1. [DOI] [PubMed] [Google Scholar]
- 44.Karataş E, Fahriye EI, Yasemin Z. Investigation of antibiotic resistance genes and panton-valentine leucocidin in Staphylococcus aureus strains isolated from various clinical samples. Acta Med Mediterranea. 2017;33:341–349. [Google Scholar]
- 45.Simeoni D, Rizzotti L, Cocconcelli P, Gazzola S, Dellaglio F, Torriani S. Antibiotic resistance genes and identification of staphylococci collected from the production chain of swine meat commodities. Food Microbiol. 2008;25:196–201. doi: 10.1016/j.fm.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 46.Johler S, Layer F, Stephan R. Comparison of virulence and antibiotic resistance genes of food poisoning outbreak isolates of Staphylococcus aureus with isolates obtained from bovine mastitis milk and pig carcasses. J Food Protect. 2011;74:1852–1859. doi: 10.4315/0362-028X.JFP-11-192. [DOI] [PubMed] [Google Scholar]
- 47.Podkowik M, Bystroń J, Bania J. Prevalence of antibiotic resistance genes in staphylococci isolated from ready-to-eat meat products. Polish J Vet Sci. 2012;15:233–237. doi: 10.2478/v10181-011-0139-z. [DOI] [PubMed] [Google Scholar]
- 48.Akanbi OE, Njom HA, Fri J, Otigbu AC, Clarke AM. Antimicrobial susceptibility of Staphylococcus aureus isolated from recreational waters and beach sand in eastern Cape Province of South Africa. Int J Environ Res Public Health. 2017;14:1001. doi: 10.3390/ijerph14091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duran N, Ozer B, Duran GG, Onlen Y, Demir C. Antibiotic resistance genes & susceptibility patterns in staphylococci. Indian J Med Res. 2012;135:389. [PMC free article] [PubMed] [Google Scholar]
- 50.Gordon N, Price J, Cole K, Everitt R, Morgan M, Finney J, et al. Prediction of Staphylococcus aureus antimicrobial resistance from whole genome sequencing. J Clin Microbiol. 2014;52:1182–1191. doi: 10.1128/JCM.03117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tachbele E, Erku W, Gebre-Michael T, Ashenafi M. Cockroach-associated food-borne bacterial pathogens from some hospitals and restaurants in Addis Ababa, Ethiopia: distribution and antibiograms. J Rural Trop Public Health. 2006;5:34–41. [Google Scholar]
- 52.Salehzadeh A, Tavacol P, Mahjub H. Bacterial, fungal and parasitic contamination of cockroaches in public hospitals of Hamadan, Iran. J Vector Borne Dis. 2007;44:105–110. [PubMed] [Google Scholar]
- 53.Pai H-H. Multidrug resistant bacteria isolated from cockroaches in long-term care facilities and nursing homes. Acta Tropic. 2013;125:18–22. doi: 10.1016/j.actatropica.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 54.Dehkordi FS, Haghighi N, Momtaz H, Rafsanjani MS, Momeni M. Conventional vs real-time PCR for detection of bovine herpes virus type 1 in aborted bovine, buffalo and camel foetuses. Bulgar J Vet Med. 2013;16:102–11. [Google Scholar]
- 55.Dehkordi FS, Khamesipour F, Momeni M. Brucella abortus and Brucella melitensis in Iranian bovine and buffalo semen samples: The first clinical trial on seasonal, Senile and geographical distribution using culture, conventional and real-time polymerase chain reaction assays. Kafkas Univ Vet Fak Dergisi. 2014;20:821-8.
- 56.Dehkordi FS, Parsaei P, Saberian S, Moshkelani S, Hajshafiei P, Hosseini SR, et al Prevalence study of Theileria annulata by comparison of four diagnostic techniques in southwest Iran. Bulgar J Vet Med. 2012;15:123-30.
- 57.Dehkordi FS, Valizadeh Y, Birgani TA, Dehkordi KG. Prevalence Study of Brucella melitensis and Brucella abortus in cow's milk using dot enzyme linked immuno sorbent assay and duplex polymerase chain reaction. J Pure Appl Microbiol. 2014;8:1065-9.
- 58.Ghorbani F, Gheisari E, Dehkordi FS. Genotyping of vacA alleles of Helicobacter pylori strains recovered from some Iranian food items. Trop J Pharm Res. 2016;15;1631-6.
- 59.Hemmatinezhad B, Khamesipour F, Mohammadi M, Safarpoor Dehkordi F, Mashak Z. Microbiological investigation of O‐ serogroups, virulence factors and antimicrobial resistance properties of shiga toxin‐ producing Escherichia coli isolated from ostrich, turkey and quail meats. J Food Safety. 2015;35:491-500.
- 60.Madahi H, Rostami F, Rahimi E, Dehkordi FS. Prevalence of enterotoxigenic Staphylococcus aureus isolated from chicken nugget in Iran. Jundishapur J Microbiol. 2013;7:e10237. [DOI] [PMC free article] [PubMed]
- 61.Momtaz H, Davood Rahimian M, Safarpoor Dehkordi F. Identification and characterization of Yersinia enterocolitica isolated from raw chicken meat based on molecular and biological techniques. J Appl Poultry Res. 2013;22:137-45.
- 62.Rahimi E, Sepehri S, Dehkordi FS, Shaygan S, Momtaz H. Prevalence of Yersinia species in traditional and commercial dairy products in Isfahan Province. Iran. Jundishapur J Microbiol. 2014;7:e9249. doi: 10.5812/jjm.9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rahimi E, Yazdanpour S, Dehkordi FS. Detection of Toxoplasma gondii antibodies in various poultry meat samples using enzyme linked immuno sorbent assay and its confirmation by polymerase chain reaction. J Pure Appl Microbiol. 2014;8:421-7.
- 64.Safarpoor Dehkordi F, Barati S, Momtaz H, Hosseini Ahari SN, Nejat Dehkordi S. Comparison of shedding, and antibiotic resistance properties of Listeria monocytogenes isolated from milk, feces, urine, and vaginal secretion of bovine, ovine, caprine, buffalo, and camel species in Iran. Jundishapur J Microbiol. 2013;6:284-94.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed throughout this research are included in this published article.