The experimental characterization of novel metabolic pathways is essential for realizing the value of genome sequences and improving our knowledge of the enzymatic capabilities of the bacterial world. However, 30% to 40% of genes of a typical genome remain unannotated or associated with a putative function. We used enzyme kinetics, liquid chromatography-mass spectroscopy (LC-MS)-based metabolomics, and mutant phenotyping for the characterization of the metabolism of l-carnitine in Sinorhizobium meliloti to provide an accurate annotation of the corresponding genes. The occurrence of conserved gene clusters for carnitine metabolism in soil, plant-associated, and marine bacteria underlines the environmental abundance of carnitine and suggests this molecule might make a significant contribution to ecosystem nitrogen and carbon cycling.
KEYWORDS: l-carnitine, LC-MS, orbitrap, bacterial metabolism, enzymology, functional genomics, metabolomics, trimethylammonium compounds
ABSTRACT
l-Carnitine is a trimethylammonium compound mostly known for its contribution to fatty acid transport into mitochondria. In bacteria, it is synthesized from γ-butyrobetaine (GBB) and can be used as a carbon source. l-Carnitine can be formed directly by GBB hydroxylation or synthesized via a biosynthetic route analogous to fatty acid degradation. However, this multistep pathway has not been experimentally characterized. In this work, we identified by gene context analysis a cluster of l-carnitine anabolic genes next to those involved in its catabolism and proceeded to the complete in vitro characterization of l-carnitine biosynthesis and degradation in Sinorhizobium meliloti. The five enzymes catalyzing the seven steps that convert GBB to glycine betaine are described. Metabolomic analysis confirmed the multistage synthesis of l-carnitine in GBB-grown cells but also revealed that GBB is synthesized by S. meliloti. To our knowledge, this is the first report of aerobic GBB synthesis in bacteria. The conservation of l-carnitine metabolism genes in different bacterial taxonomic classes underscores the role of l-carnitine as a ubiquitous nutrient.
IMPORTANCE The experimental characterization of novel metabolic pathways is essential for realizing the value of genome sequences and improving our knowledge of the enzymatic capabilities of the bacterial world. However, 30% to 40% of genes of a typical genome remain unannotated or associated with a putative function. We used enzyme kinetics, liquid chromatography-mass spectroscopy (LC-MS)-based metabolomics, and mutant phenotyping for the characterization of the metabolism of l-carnitine in Sinorhizobium meliloti to provide an accurate annotation of the corresponding genes. The occurrence of conserved gene clusters for carnitine metabolism in soil, plant-associated, and marine bacteria underlines the environmental abundance of carnitine and suggests this molecule might make a significant contribution to ecosystem nitrogen and carbon cycling.
INTRODUCTION
The quaternary amine l-carnitine is produced by all domains of life. It is synthesized from l-lysine and l-methionine (1). The genes of the carnitine biosynthesis pathway have been identified in numerous organisms such as mice and rats (2), yeasts (3, 4), and plants (5, 6). In eukaryotes, this compound is known to act as a carrier for the transport of esterified long-chain fatty acids from the cytosol into the mitochondrial matrix, where β-oxidation takes place (7). Another role for l-carnitine is helping bacteria to cope with the harsh environment in which they live. Bacteria face challenges such as heat, desiccation, freezing, and osmotic changes (8, 9). As a consequence, these cells can accumulate low-molecular-mass organic compounds known as compatible solutes. l-Carnitine is an archetypal osmolyte that can be imported or generated from direct precursors. Finally, this ubiquitous compound, released in the environment after the death of the producing organisms, can be degraded by bacteria. Two degradation pathways are reported. In Serratia marcescens (10) and Acinetobacter calcoaceticus (11), the carbon-nitrogen bond of carnitine is first cleaved to form trimethylamine and malate semialdehyde, which is oxidized to malate. In an alternative route, carnitine is metabolized into glycine betaine and acetoacetate (12). In this context, l-carnitine is first oxidized into 3-dehydrocarnitine (Fig. 1, step 6). This compound is next condensed with acetyl coenzyme A (acetyl-CoA) to yield acetoacetate and betainyl-CoA (step 7), which is eventually cleaved into glycine betaine and coenzyme A (step 8). These two aerobic carnitine catabolic pathways have only recently been fully described, from biochemical and genetic perspectives (12, 13). However, much less is known in prokaryotes about its synthesis. Synthesis of l-carnitine is not described as de novo but reported to occur from trimethylated precursors only (8). It can be formed from γ-butyrobetaine (GBB) via a single-stage route that involves a 2-oxoglutarate-dependent dioxygenase (Fig. 1, step 1) (14). An alternative way to convert GBB to l-carnitine was proposed by the Swiss company Lonza (15). Lonza developed an efficient method for the production of l-carnitine that involves a metabolic pathway analogous to fatty acid degradation. A synthetase forms γ-butyrobetainyl-CoA from GBB (step 2, Fig. 1) that is oxidized to crotonobetainyl-CoA (step 3) and hydrated to l-carnitinyl-CoA (step 4) before being cleaved to l-carnitine by a thioesterase (step 5). The sequences of the corresponding genes are not available but were reported to be located directly next to the gene coding for l-carnitine dehydrogenase (CDH) in HK4 (DSMZ-2903), a strain related to the Agrobacterium or Rhizobium genus (15). This gene was used to locate the bco operon, anticipated to contain the candidate genes for l-carnitine synthesis (16). The encoded enzymes, predicted as γ-butyrobetainyl-CoA/crotonobetainyl-CoA synthetase (BcoA/B), γ-butyrobetainyl-CoA dehydrogenase (BcoC), and crotonobetainyl-CoA hydratase (BcoD), were nonetheless not experimentally validated. Enzyme function is established only if two criteria are satisfied: the reaction catalyzed is described at the molecular level, and the biological dimension is considered fulfilled when the pathway in which the enzyme participates is characterized (17). To this end, we undertook the experimental characterization of l-carnitine metabolism in Sinorhizobium meliloti (strain 3D0a2; DSMZ-30135). We took advantage of the capacity of the bacterium to grow on GBB to investigate its metabolism via a metabolomic approach and detected the intermediates of the l-carnitine biosynthetic pathway. We selected the candidate genes by genome context analysis and validated the corresponding proteins by enzymatic analysis. Finally, we conducted a growth phenotype analysis of mutants deleted for genes involved in GBB utilization. The presence of these genes in more than 100 genomes of soil and marine organisms stresses the role of GBB and l-carnitine as ubiquitous nutrients.
FIG 1.
Metabolism of l-carnitine in bacteria. Enzymes involved are GBB dioxygenase (1), γ-butyrobetainyl-CoA synthetase (2), γ-butyrobetainyl-CoA dehydrogenase (3), crotonobetainyl-CoA hydratase (4), l-carnitinyl-CoA thiolase (5), l-carnitine dehydrogenase (6), dehydrocarnitine cleavage enzyme (BKACE) (7), and betainyl-CoA thiolase (8).
RESULTS
Metabolomic analysis of l-carnitine metabolism.
S. meliloti strain 3D0a2 (DSMZ-30135) readily grows with trimethylammonium compounds such as l-carnitine or GBB as carbon sources (see Fig. S1 in the supplemental material). Metabolomes were prepared from cells growing exponentially in a minimal medium containing GBB as the sole carbon source. Results are presented in Fig. 2A. We detected cations of m/z 895.2210, 893.2063, and 911.2149, matching the masses of the protonated forms ([M+H]+) of γ-butyrobetainyl-CoA, crotonobetainyl-CoA, and carnitinyl-CoA, respectively. The ion observed at m/z 162.1124 was identified as carnitine based on the comparison of its accurate mass, retention time, and mass spectrometry (MS2) spectrum with those of a reference compound (Fig. S2). However, these compounds were not detected in sucrose-grown cells (Fig. 2B). Together, these results are consistent with the multistage way of l-carnitine biosynthesis (Fig. 1).
FIG 2.

Detection of the biosynthetic intermediates of l-carnitine in S. meliloti strain 3D0a2 (DSMZ-30135). Metabolome analysis of cells grown on GBB (A) and sucrose (B). Samples were analyzed by LC-MS in the positive ionization mode. Extracted ion chromatograms correspond to the protonated forms ([M+H]+) at 5-ppm accuracy of γ-butyrobetainyl-CoA (1) at m/z 895.2222, crotonobetainyl-CoA (2) at m/z 893.2066, carnitinyl-CoA (3) at m/z 911.2171, and carnitine (4) at m/z 162.1125.
Identification of the genes involved in l-carnitine synthesis.
We recently elucidated the l-carnitine degradation pathway in Pseudomonas aeruginosa (12) and showed experimentally that it involves a member of the BKACE family (Fig. 1, step 7). Because the genome sequence of S. meliloti 3D0a2 was not available, we used that of S. meliloti Rm2011 to design oligonucleotides for gene cloning and also for genome context analyses (Fig. 3). In S. meliloti Rm2011, next to the gene orthologous to the one encoding BKACE (SM2011_c01637) are found genes annotated as “bifunctional 3-hydroxyacyl-CoA dehydrogenase/thioesterase” (SM2011_c01638), “putative acyl-CoA dehydrogenase” (SM2011_c01639), “putative acyl-CoA synthetase protein” (SM2011_c01640), and “carnitinyl-CoA dehydratase” (SM2011_c01641) genes. SM2011_c01640 shares 79% identity with the recently characterized γ-butyrobetainyl-CoA synthetase gene from Agrobacterium sp. strain 525a (18). PA5386 from P. aeruginosa, which oxidizes l-carnitine (12), shares 49% identity with the N-terminal part of the product of SM2011_c01638. PA5385, which hydrolyses betainyl-CoA (12), has 40% identity with the C-terminal part of the product of SM2011_c01638. The latter was thus anticipated to catalyze both l-carnitine oxidation and the formation of glycine betaine (Fig. 1, steps 6 and 8). Furthermore, since no other gene in the vicinity is annotated as a thiolase gene, we suspected that SM2011_c01638 encodes an enzyme to hydrolyze l-carnitinyl-CoA also (Fig. 1, step 5). The candidate genes were cloned from S. meliloti 3D0a2. The sequences were 100% identical to those from S. meliloti Rm2011 and were named Smc01637, Smc01638, Smc01639, Smc01640, and Smc01641. In summary, we considered that Smc01640 codes for BcoA/B, which forms γ-butyrobetainyl-CoA from GBB, ATP, and CoA, Smc01639 codes for BcoC, which oxidizes γ-butyrobetainyl-CoA to crotonobetainyl-CoA, Smc01641 codes for BcoD, which hydrates crotonobetainyl-CoA to l-carnitinyl-CoA, and Smc01638 codes for a fused CDH thioesterase, which finally hydrolyzes l-carnitinyl-CoA.
FIG 3.
l-Carnitine metabolism gene cluster in P. aeruginosa PAO1 and S. meliloti Rm2011. Physical colocalization of genetic loci was observed through the MicroScope platform (57). Transporter genes are colored gray and predicted transcriptional regulators are colored yellow. The uncolored gene symbol is used for a gene apparently unrelated to carnitine metabolism. PA, Pseudomonas aeruginosa PAO1; SM2011_c, Sinorhizobium meliloti Rm2011.
Purification of the candidate proteins.
The candidate genes were heterologously expressed in Escherichia coli and the corresponding proteins purified (see Fig. S3). Because the enzymes from HK4 were reported to be unstable (19), sorbitol and betaine were added to the culture medium to improve the yield of soluble and properly folded recombinant proteins (20). Under these conditions, pure and stable proteins were obtained. SDS-PAGE showed a major band with a molecular mass consistent with those of the candidate proteins.
Smc01640 is a γ-butyrobetainyl-CoA synthetase (BcoA/B).
According to gel filtration experiments, Smc01640 was purified as a homodimer, as previously reported (18). The enzyme was incubated in the presence of GBB, ATP, and CoA for 60 min and analyzed by liquid chromatography-mass spectroscopy (LC-MS). A cation of m/z 895.2205 was detected, with the same retention time and fragmentation pattern as the putative γ-butyrobetainyl-CoA observed in the metabolome of GBB-grown cells (Fig. 4). Contrary to what was previously proposed, we detected the formation of ADP and not AMP during the reaction (15, 16). This result is nevertheless in agreement with what was observed in Agrobacterium sp. 525a (18). Kinetic parameters of BcoA/B are presented in Table 1. While Km values of BcoA/B from S. meliloti for GBB, ATP, and CoA are in the same range as those reported for the only γ-butyrobetainyl-CoA synthetase characterized so far (18), the value for kcat is approximately 40 times higher in S. meliloti (25.3 s−1 versus 0.65 s−1). As stated by Zimmermann and Werlen (15), BcoA/B can use crotonobetaine as a substrate. Nevertheless, results showed that the enzyme is ∼1,400 times more efficient with GBB, as indicated by the ratios kcat/Km GBB and kcat/Km crotonobetaine (Table 1). This low efficiency probably prevents BcoA/B from participating in the growth of S. meliloti when crotonobetaine is the carbon source (Fig. S1). Crotonobetaine may instead be converted in vivo to crotonobetainyl-CoA by a CoA transferase, in a mechanism similar to what is reported for the metabolism of l-carnitine in E. coli (8), and further metabolized by Smc01641.
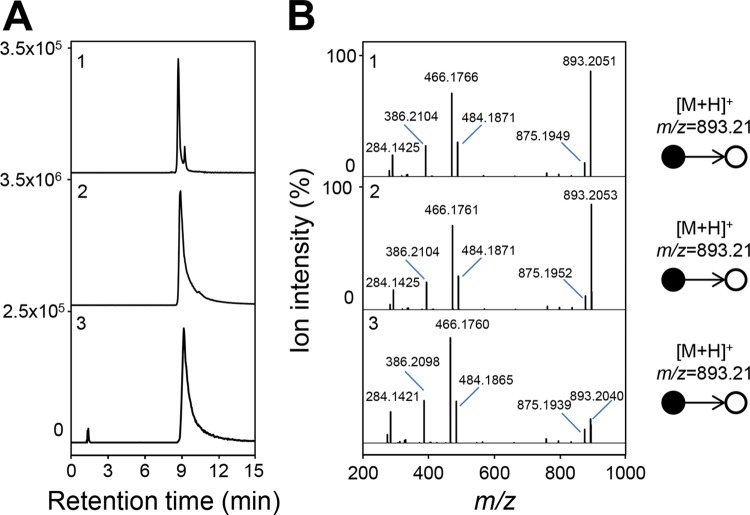
FIG 4.
Collision-induced dissociation γ-butyrobetainyl-CoA. (A) Extracted ion chromatograms correspond to the protonated form ([M+H]+) of γ-butyrobetainyl-CoA at m/z 895.2222 (5-ppm accuracy). (B) Collision-induced dissociation tandem mass spectra (25% normalized collision energy). (1) γ-Butyrobetainyl-CoA from the metabolome of GBB-grown cells. (2) Enzymatic formation of γ-butyrobetainyl-CoA analyzed after 60 min in 100 μl of Tris-HCl 100 mM (pH 8.0) containing 2.7 μg of BcoA/B, 5 mM GBB, 200 μM CoA, 2 mM ATP, and 10 mM MgCl2. LC-MS analyses were conducted in the positive ionization mode.
TABLE 1.
Kinetic parameters for the enzymes of the l-carnitine metabolic pathway
| Enzyme | Substrate | Km (μM)a | kcat (s−1)a | kcat/Km (s−1 · M−1)a |
|---|---|---|---|---|
| BcoA/B (Smc01640) | GBBb | 1,120 ± 303 | 25.3 ± 1.9 | 2.3 × 104 |
| ATPc | 96 ± 9 | 2.6 × 105 | ||
| CoAd | 45 ± 7 | 5.6 × 105 | ||
| Crotonobetaineb | 3,000 ± 693 | 0.05 ± 0.004 | 17 | |
| BcoC (Smc01639) | γ-Butyrobetainyl-CoAe | 4.1 ± 1.7 | 1.8 ± 0.12 | 4.4 × 105 |
| FADf | 0.8 ± 0.2 | 2.3 × 106 | ||
| BcoD (Smc01641) | Crotonobetainyl-CoA | 20.5 ± 2.1 | 8.5 ± 0.10 | 4.2 × 105 |
| CDH thioesterase (Smc01638) | l-Carnitineg | 940 ± 68 | 6.2 ± 0.10 | 6.6 × 103 |
| NAD+h | 154 ± 13 | 5.1 ± 0.10 | 3.3 × 104 | |
| l-Carnitinyl-CoA | 12 ± 3 | 0.9 ± 0.1 | 7.5 × 104 | |
| Betainyl-CoA | 12 ± 1 | 142.3 ± 3.0 | 1.2 × 107 | |
| BKACE (Smc01637) | Dehydrocarnitinei | 13 ± 2 | 0.24 ± 0.01 | 1.8 × 104 |
| Acetyl-CoAj | 118 ± 18 | 2.0 × 103 |
Values correspond to the averages from two replicates.
ATP and CoA concentrations were 2 mM and 200 μM, respectively.
GBB and CoA concentrations were 5 mM and 200 μM, respectively.
GBB and ATP concentrations were 5 mM and 2 mM, respectively.
FAD concentration was 50 μM.
γ-Butyrobetainyl-CoA concentration was 80 μM.
NAD+ concentration was 1.5 mM.
l-Carnitine concentration was 10 mM.
Acetyl-CoA concentration was 1.5 mM.
Dehydrocarnitine concentration was 100 μM.
Smc01639 is a γ-butyrobetainyl-CoA dehydrogenase (BcoC).
Consistently with previous studies on mammalian acyl-CoA dehydrogenases (21), the enzyme was purified as a homotetramer. After purification, it exhibited a 280/446 nm ratio of 23 (see Fig. S4), which is significantly higher than the value reported for mammalian acyl-CoA dehydrogenases (21). Using an extinction coefficient for bound FAD of 15.4 mM−1 · cm−1 at 446 nm (21) and of 39.8 mM−1 · cm−1 at 280 nm, we estimated that only 10% of purified BcoC was complexed to flavin. This suggests that either the enzyme lost FAD during purification or that the flavin binds weakly to the enzyme and acts as a substrate rather than a prosthetic group. Upon dilution for enzymatic characterization, the enzyme was inactive unless FAD was added. Its enzymatic behavior was thus studied in the presence of FAD plus ferrocenium hexafluorophosphate to ensure FAD reoxidation during catalysis (22). The reaction product was similar to the putative crotonobetainyl-CoA detected in GBB-grown cells and also to authentic crotonobetainyl-CoA (Fig. 5). Kinetic parameters are presented in Table 1. The Km value for γ-butyrobetainyl-CoA is in the micromolar range, as reported for medium acyl-CoA dehydrogenases (23–25). Using the approach described by Benziman and Galanter (26), we also determined the apparent Km for FAD by monitoring the activation of the apoenzyme with various concentrations of FAD and in the presence of saturating concentrations of γ-butyrobetainyl-CoA. The obtained value is in the same order of magnitude as the one for malic dehydrogenase from Acetobacter xylinum or Mycobacterium avium (26, 27) and d-amino acid oxidase from pig kidney (28).
FIG 5.
Collision-induced dissociation crotonobetainyl-CoA. (A) Extracted ion chromatograms correspond to the protonated form ([M+H]+) of crotonobetainyl-CoA at m/z 893.2066 (5-ppm accuracy). (B) Collision-induced dissociation tandem mass spectra (25% normalized collision energy). (1) Crotonobetainyl-CoA from the metabolome of GBB-grown cells. (2) Enzymatic formation of crotonobetainyl-CoA analyzed after 60 min in 100 μl of 50 mM Tris-HCl (pH 8.0) containing 3.9 μg of BcoC and 90 μM γ-butyrobetainyl-CoA 50, μM FAD, and 500 μM FC+PF−6. (3) Crotonobetainyl-CoA reference standard. LC-MS analyses were conducted in the positive ionization mode.
Lau et al. reported that the pig kidney medium-chain acyl-CoA dehydrogenase that oxidizes butyryl-CoA (a structural analog of γ-butyrobetainyl-CoA) into crotonyl-CoA exhibits an intrinsic hydratase activity toward crotonyl-CoA and forms l-3-hydroxybutyryl-CoA, the compound formed by the subsequent enzyme in β-oxidation pathway, the short-chain enoyl-CoA hydratase crotonase (29). We observed a related phenomenon. BcoC incubated in the presence of γ-butyrobetainyl-CoA, FAD, and ferrocenium hexafluorophosphate formed, in addition to crotonobetainyl-CoA, an ion with m/z 911.2159. This mass-to-charge ratio is consistent with l-carnitinyl-CoA, but the peak eluted with a higher retention time (9.96 versus 8.85 min). The same “l-carnitinyl-CoA”-like compound was observed in the metabolome of GBB-grown cells, but with a much lower intensity than l-carnitinyl-CoA (see Fig. S5). The fragmentation pattern of this minor compound is different from the one of authentic l-carnitinyl-CoA (Fig. 6). Thus, the hydration product of crotonobetainyl-CoA by BcoC is not l-carnitinyl-CoA but could be d-carnitinyl-CoA. The enantiomers l- and d-carnitine cannot be separated on an achiral stationary phase such as ZIC-pHILIC, but l- and d-carnitinyl-CoA, being diastereoisomers, are expected to have distinct retention times. According to this hypothesis, the structure of the active sites of BcoC and the crotonobetainyl-CoA hydratase should be different. The attack of the incoming hydroxide or water nucleophile seems to occur from the different faces of the carbon-carbon double bond of crotonobetainyl-CoA, depending on the presence of crotonobetainyl-CoA hydratase or BcoC.
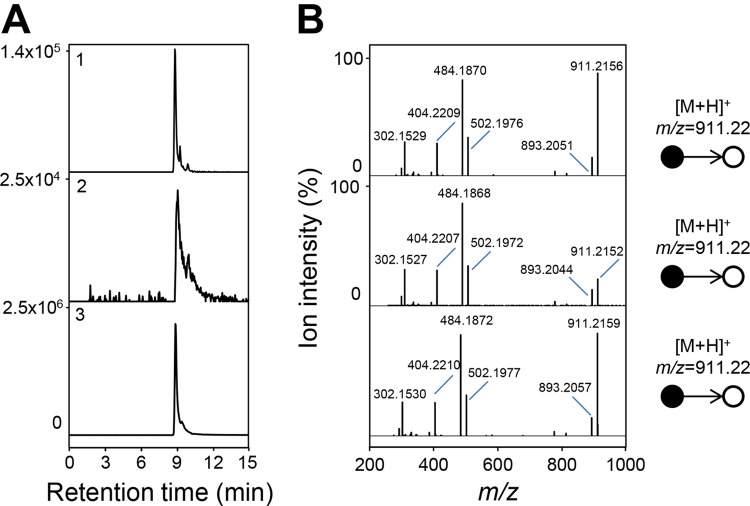
FIG 6.
Collision-induced dissociation l-carnitinyl-CoA. (A) Extracted ion chromatograms correspond to the protonated form ([M+H]+) of l-carnitinyl-CoA at m/z 911.2171 (5-ppm accuracy). (B) Collision-induced dissociation tandem mass spectra (25% normalized collision energy). (1) l-Carnitinyl-CoA from the metabolome of GBB-grown cells. (2) Enzymatic formation of l-carnitinyl-CoA analyzed after 60 min in 100 μl of 20 mM Tris-HCl (pH 8.0) containing 3.9 μg of BcoC and 1.7 μg of BcoD in the presence of 90 μM GBB, 50 μM FAD, and 500 μM FC+PF−6. (3) l-Carnitinyl-CoA synthesized by CaiC from E. coli by enzymatic conversion of l-carnitine; 2.6 μg of CaiC was incubated with 5 mM l-carnitine, 200 μM CoA, and 2 mM ATP in 100 μl of 100 mM Tris-HCl (pH 8.0) for 60 min. LC-MS analyses were conducted in the positive ionization mode.
Smc01641 is a crotonobetainyl-CoA hydratase (BcoD).
Smc01641 was purified as a homotetramer. The enzyme was incubated in the presence of enzymatically synthesized γ-butyrobetainyl-CoA plus BcoC and analyzed by LC-MS (Fig. 6). A cation of m/z 911.2152 was detected, with the same retention time and fragmentation pattern as the putative l-carnitinyl-CoA observed in the metabolome of GBB-grown cells, and also to reference l-carnitinyl-CoA synthesized according to Bernal et al. (30). BCoD activity was assayed following absorbance decrease at 260 nm due to the hydration of the Δ2,3-double bond of the substrate (31). Kinetic parameters are presented in Table 1. The Km value is in the same range as the one reported for the hydration of crotonobetainyl-CoA by CaiD, although the kcat value is 10-fold lower (32).
Smc01638 is a trifunctional enzyme (CDH thioesterase).
Smc01638 was purified as a homodimer. It was first assayed for l-carnitine dehydrogenase activity. Reactions were monitored following the formation of NADH at 340 nm (Table 1). Km values obtained for Smc01638 are close to those reported for the soil isolate Rhizobium sp. (940 versus 1,100 μM for l-carnitine and 154 versus 87 μM for NAD+) (33). The enzyme was next assayed for the hydrolysis of l-carnitinyl-CoA. l-Carnitinyl-CoA was prepared and assayed as described in Materials and Methods. The reaction was monitored following the increase of A412 upon l-carnitinyl-CoA cleavage and reaction with 5,5′-dithiobis-(2-nitrobenzoic) acid (DTNB) (Table 1). We also confirmed that CDH thioesterase is involved in the l-carnitine degradation pathway by hydrolyzing betainyl-CoA (Table 1). The latter compound was enzymatically synthesized from dehydrocarnitine, acetyl-CoA, and the BKACE Smc01637 (which was also kinetically characterized in this work). CDH thioesterase is approximately 100 times more efficient with betainyl-CoA, as estimated by the kcat/Km values obtained for the two CoA esters: 1.2 × 107 versus 7.5 × 104 s−1 · M−1 (Table 1). The difference lies in the higher kcat value for betainyl-CoA, as the Km values are not affected. The very high catalytic efficiency of CDH thioesterase toward betainyl-CoA probably precludes the participation of a CoA transferase in the metabolism of betainyl-CoA, such as the DhcAB enzyme in Pseudomonas aeruginosa, as previously suggested (8, 34). To support our hypothesis, no gene in the immediate vicinity of the cluster SM2011_c01637-SM2011_c01641 is annotated as “putative CoA transferase” gene.
Growth of S. meliloti on GBB is impaired in mutants deleted for Smc01638-Smc01640.
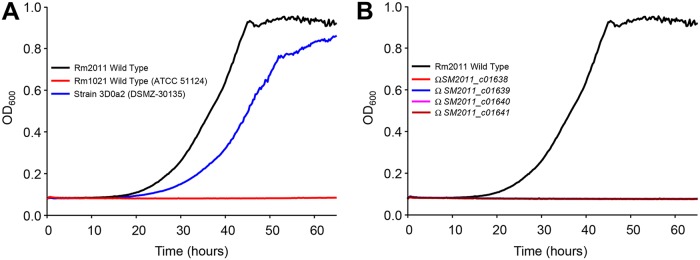
To demonstrate that each of these genes is required in vivo, we investigated the growth kinetics with GBB as the carbon source of mutants disrupted for SM2011_c01638, SM2011_c01639, SM2011_c01640, and SM2011_c01641 from the 3D0a2-related strain Rm2011. All these strains were successfully pregrown with succinate as the carbon source. While the wild-type strain grew as well as 3D0a2 (Fig. 7A), the growth of mutants was abolished (Fig. 7B). These results along with metabolomic and biochemical data definitively validate this novel metabolic pathway. However, another 3D0a2-related strain, Rm1021, did not grow on GBB (Fig. 7A). This result was unexpected, since DNA sequences between Rm1021 and Rm2011 are 100% identical in the 20-Kb genomic region encompassing the genes involved in GBB and l-carnitine metabolism (from nucleotides 2423202 to 2443202 of Rm2011; strand +1). It is noteworthy that Rm1021 strains from five different resources were tested for growth (including ATCC 51124). The comparison of the genome sequence of Rm1021 and 2011 nevertheless showed polymorphism (35). It is thus possible that a sequence variant(s) located outside the l-carnitine metabolism gene cluster affected the ability of Rm1021 to grow with GBB. Finally, it should be mentioned that in our laboratory, Rm2011 growth with GBB is significantly different from that observed by Goldmann et al. with the related compound carnitine (36). The poor growth obtained by Goldmann et al. (the optical density at 650 nm [OD650] value obtained after 50 h was <0.2) can be explained by the absence of biotin in the culture medium. In a paper by Watson et al., it is reported that biotin is essential for growth, since the bacterium does not have the known genes involved in biotin synthesis (37). It could be hypothesized that the partial growth obtained by Goldmann et al. is due to biotin carryover from complete culture medium used for precultures.
FIG 7.
Growth kinetics of S. meliloti strains on M9 mineral medium containing γ-butyrobetaine as the carbon source. (A) Growth behavior of wild-type S. meliloti strains. (B) Growth behavior of wild-type and mutant strains of S. meliloti Rm2011. All cultures were successfully pregrown in M9 with 10 mM succinate as the carbon source, washed twice, and finally inoculated at an OD600 of 0.08 in M9 medium containing 10 mM γ-butyrobetaine. OD600 was continuously recorded by an automated growth curve analysis system (Bioscreen-C; Thermo Fisher Scientific). Values correspond to the averages from three replicates. Cultures were supplemented with 5 μM biotin.
GBB is produced in sucrose-grown cells.
The metabolomic analysis of sucrose-grown cells revealed the unexpected presence of GBB (see Fig. S6A and B). GBB can thus be formed by S. meliloti as a metabolic intermediate. To our knowledge, this is the first report of aerobic GBB formation by a bacterial cell. In mammals, it is synthesized from N6-trimethyllysine (1). Since bacteria can produce N6-trimethyllysine (38, 39), it can be argued that they also can use it to synthesize GBB. Whether these enzymatic reactions exist in S. meliloti deserves attention. However, we did not detect N6-trimethyllysine in its metabolome. Eventually, since GBB is present in the cell, l-carnitine and glycine betaine were also detected (Fig. S6C to F). GBB formed by S. meliloti may then be metabolized in the same way as a carbon source. The CoA esters (γ-butyrobetainyl-CoA, crotonobetainyl-CoA, and l-carnitinyl-CoA), being probably too scarce, were not be observed.
DISCUSSION
Quaternary ammonium compounds such as GBB, carnitine, and acylated carnitine are ubiquitous metabolites. Carnitine was actually the most abundant quaternary ammonium compound (0.49 mM) found in the soil of a subalpine grassland and the third most abundant soluble nitrogen compound overall. The acetylcarnitine concentration was only slightly lower (0.33 mM) (40). In seawater, the concentrations of GBB and l-carnitine reach 10 and 128 pM, respectively (41). Logically, these abundant molecules are expected to be nutrients for bacteria (8, 42, 43). The data presented here indicate that S. meliloti degrades GBB to glycine betaine through seven enzymatic steps catalyzed by five distinct enzymes. This is consistent with previous work that indicated that glycine betaine is a carbon and nitrogen source for S. meliloti (44). Successive demethylations convert glycine betaine to glycine, which is eventually converted to pyruvate as the entry point in the central metabolism (44).
Among the enzymes characterized here, Smc01638 is unusual as it participates in three enzymatic steps. It is a fused protein composed of CDH and a thioesterase. Discovering that Smc01638 is trifunctional was not unexpected, because no gene coding for a CoA transferase or a thiolase was detected in its genomic neighborhood. As frequently observed in fusion proteins, Smc01638 catalyzes two consecutive steps of the same metabolic pathway: l-carnitinyl-CoA hydrolysis and l-carnitine oxidation. This organization can represent an advantage in vivo, for catalysis and its regulation. l-Carnitine formed from l-carnitinyl-CoA is probably channeled to the second active site that catalyzes its dehydrogenation. This should avoid free diffusion of the metabolite and increase the global reaction rate. One could thus, hypothesize that betainyl-CoA hydrolysis was acquired through substrate promiscuity. The acquisition of promiscuous activities generally serves as an evolutionary starting point, with promiscuous substrates that have little physiological relevance (45). Promiscuous enzymes usually exhibit kcat/Km values for their native substrate orders of magnitude higher than for promiscuous ones (45). But surprisingly, according to this postulate, betainyl-CoA hydrolysis (kcat/Km = 1.2 × 107 s−1 · M−1) should be the original activity and l-carnitinyl-CoA hydrolysis (kcat/Km = 7.5 × 104 s−1 · M−1) the activity acquired more recently. In any case, the multifunctionality of Smc01638 is representative of the optimization of metabolic efficiency.
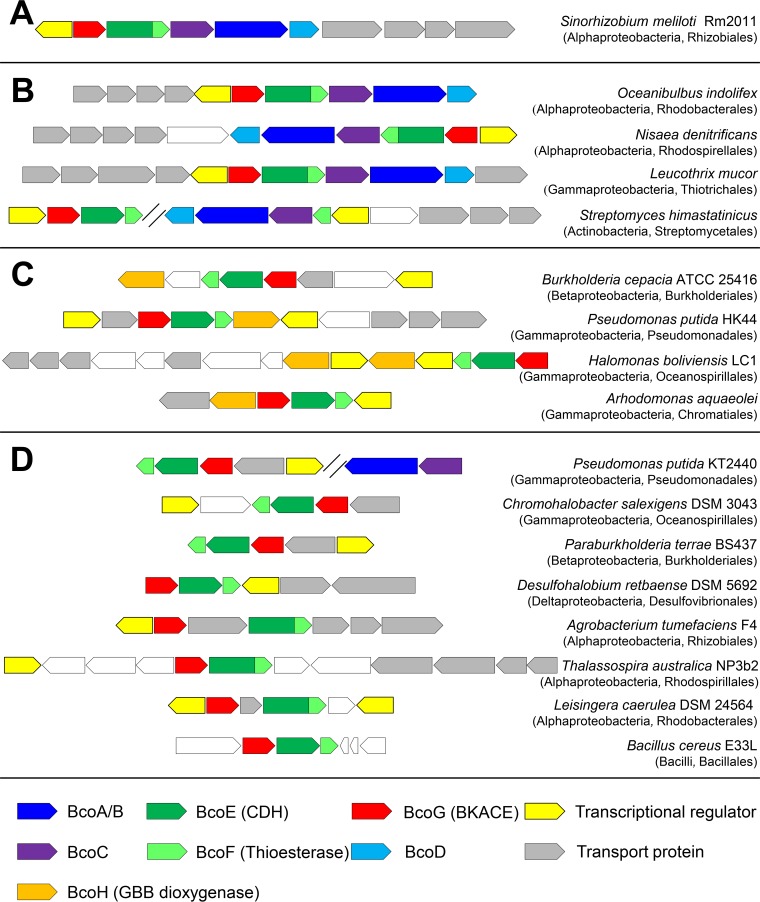
Comparative analyses were conducted on the MicroScope platform (https://www.genoscope.cns.fr/agc/microscope/home/index.php). To aid in future genome annotations, we propose to name genes coding for CDH, thioesterase, BKACE, and GBB dioxygenase bcoE, bcoF, bcoG, and bcoH, respectively (Fig. 8; see also Fig. S7 in the supplemental material). Analyses showed that clusters of genes orthologous to Smc01637-Smc01641 (Fig. 8A) are present in the chromosomes of more than 100 bacterial species (as of May 2018). Clusters representative of the taxonomic diversity are presented in Fig. 8B. These genes appear in three taxonomic classes (Alphaproteobacteria, Gammaproteobacteria, and Actinobacteria) but are essentially encountered in Proteobacteria. Their presence in the Gram-positive Actinobacteria, scarce and only observed in the order of Streptomycetales, suggests lateral gene transfer events. The cluster in Streptomycetales is split in two parts, with genes homologous to the l-carnitine catabolizing genes on one side and genes homologous to the biosynthetic genes on the other side. Concerning the direct l-carnitine biosynthesis pathway that involves a GBB hydroxylase, its coding gene is only detected in a few tens of Betaproteobacteria and Gammaproteobacteria (Fig. 8C). Finally, a last group of a few hundred bacterial species only possess genes homologous to those involved in the catabolism of l-carnitine. These organisms are mainly Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria (Fig. 8D). In this last group, Pseudomonas putida KT2440 contains elsewhere in its genome genes homologous to those coding for BcoA/B and BcoC, as observed for Streptomycetales (see above). However, we could not cultivate KT2440 with GBB as the sole carbon source. This suggests that these genes orthologous to bcoA/B and bcoC (as inferred from bidirectional best hits) do not participate in l-carnitine synthesis.
FIG 8.
Illustration of the taxonomic diversity of homologous predicted l-carnitine metabolism gene clusters. Homologous gene clusters in bacterial genomes were retrieved using MicroScope. (A) Gene cluster in S. meliloti Rm2011. (B) Gene clusters that contain homologous genes for both the multistep synthesis of l-carnitine and its degradation. (C) Gene clusters that contain the gene coding for the GBB hydroxylase and the genes for l-carnitine degradation. (D) Gene clusters that only contain genes for l-carnitine degradation. Taxonomic classes and orders are indicated in brackets. Candidate genes for transporters and transcriptional regulators were frequently found to be conserved within these clusters. Genes indicated in white are not predicted to be related to l-carnitine metabolism.
The genes involved in carnitine metabolism are anticipated in organisms that encounter this molecule in their habitat. Carnitine, found at high concentrations in animal tissues, is expected to be abundant in soil after animal decay. Logically, the gene cluster is present in soil organisms (Pseudomonas, Parabulkholderia, Chromohalobacter, etc.). Similarly, carnitine is detected in plants, and the gene cluster is observed in organisms that interact with them (Sinorhizobium, Burkholderia, Agrobacterium, and Azospirillum). Finally, the abundance of carnitine in sea can be linked to the presence of the gene cluster in genera such as Oceanibulbus, Nisaea, and Leucothrix. These observations, which indicate that l-carnitine is mainly metabolized by Alphaproteobacteria and Betaproteobacteria, are evocative of what was observed for the degradation of another environmental abundant quaternary ammonium osmolyte, trigonelline (46).
The physiological electron acceptor of BcoC remains to be identified. In E. coli, the use of carnitine as a terminal electron acceptor depends on the caiTABCDE operon. The adjacent operon, fixABCX, is required to provide electrons for carnitine reduction (47). More specifically, FixAB exhibits similarity to the electron transfer flavoprotein (ETF) and was hypothesized to bring reductant to the crotonobetainyl-CoA reductase CaiA (the reverse reaction of that catalyzed by BcoC). The fixABCX genes were originally identified in S. meliloti (48). Because mutations in any of these genes in Rhizobium meliloti completely abolished nitrogen fixation, they were proposed to participate in electron transport to nitrogenase (49). A similar role was proposed for fixABCX in Rhodospirillum rubrum (50). Nevertheless, it was observed that the fix genes represent a very heterogeneous class that may play a role in other processes not related to nitrogen fixation (51). Therefore, it seems unlikely that the bona fide fix operon in S. meliloti (Sma0816-Sma0819 and Sma0822) accepts electrons from BcoC. To support this hypothesis, bidirectional best hits of FixA (ECK0042) and FixB (ECK0043) from E. coli K-12 are the products of Smc00729 and Smc00728, respectively. Whether these genes accept electrons from BcoC deserves attention.
MATERIALS AND METHODS
Chemicals.
All chemicals and enzymes were purchased from Sigma-Aldrich. Reagents for molecular biology were from Invitrogen. Oligonucleotides were from Sigma Genosys. Proteinase inhibitor Pefabloc SC was purchased from Roche Diagnostics. Crotonobetaine and crotonobetainyl-CoA were synthesized by Synthenova (Saint Clair, France). γ-Butyrobetainyl-CoA was prepared by enzymatic conversion of GBB, using 1.4 μg of γ-butyrobetainyl-CoA synthetase in the presence of 25 mM GBB, 10 mM ATP, 1 mM CoA, and 10 mM MgCl2 in 1 ml 100 mM Tris-HCl (pH 8.0) for 90 min. The reaction was stopped by ultrafiltration (VWR centrifugal filter 3K). l-Carnitinyl-CoA was prepared according to Bernal et al. (30) by enzymatic conversion of l-carnitine by 115 μg of purified CaiC in the presence of 25 mM l-carnitine, 10 mM ATP, and 1 mM CoA in 1 ml 100 mM Tris-HCl (pH 8.0) for 90 min. The reaction was stopped by ultrafiltration. Betainyl-CoA was prepared by enzymatic conversion of dehydrocarnitine, using 6.9 μg Smc0137 (BKACE) in the presence of 1.5 mM acetyl-CoA in 2 ml 100 mM Tris-HCl (pH 8.0) for 90 min. The reaction was stopped by ultrafiltration. Dehydrocarnitine was prepared as previously described (12).
Strains and media.
S. meliloti strain 3D0a2 was from DSMZ (number 30135). S. meliloti Rm2011 wild-type and mutant strains were kindly provided by Anke Becker, LOEWE Center for Synthetic Microbiology, Marburg University, Germany. Briefly, Rm2011 mutants were constructed by Tn5 transposon insertion. Mutants were 2011mTn5STM.4.14.D01 (ΩSM2011_c01640), 2011mTn5STM.1.27.C11 (ΩSM2011_c01639), 2011mTn5STM.4.11.H01 (ΩSM2011_c01641), and 2011mTn5STM.2.02.D08 (ΩSM2011_c01638). Information about the S. meliloti mutant library is available at https://synmikro.com/research/chassis-and-genomes/anke-becker/resources/s.meliloti-mutant-libraries.html. Strain Rm1021 was from ATCC (number 51124) but was also kindly provided by Anke Becker and by Jacques Batut, Laboratory of Plant-Microbe Interactions, INRA, France. All strains were grown on M9 minimal medium (34 mM Na2HPO4, 22 mM KH2PO4, 8.6 mM NaCl, 18 mM NH4Cl, 41 μM nitrilotriacetic acid, 2 mM MgSO4, 0.45 mM CaCl2, 3 μM FeCl3, 1 μM MnCl2, 1 μM ZnCl2, and 0.3 μM each CrCl3, H3BO3, CoCl2, CuCl2, NiCl2, Na2MoO2, and Na2SeO3) supplemented with 5 μM biotin and 10 mM the desired carbon source. Pseudomonas putida KT2440 (ATCC 47054) was grown on a medium composed of 10.30 mM Na2HPO4, 4.76 mM KH2PO4, 1.67 mM MgSO4, 6.58 μM FeSO4, and 18 mM NH4Cl and supplemented with 20 mM desired carbon source (52).
Construction of the expression vectors.
The coding sequences of Smc01637, Smc01638, Smc01639, Smc01640, and Smc01641 from Sinorhizobium meliloti strain 3D0a2 (DSMZ-30135) and caiC from E. coli K-12 were amplified by PCR with primers shown in Table S1 in the supplemental material. The amplified sequences were inserted into the modified Novagen pET22b(+) as previously described (53). The sequence of the resulting plasmids was verified.
Expression and purification of the recombinant proteins.
Cell cultures and cell extracts were prepared as previously reported (54). Protein purification was performed using a preparative chromatography system (Äkta Pure, GE Healthcare Life Sciences). A fully automated two-step method was set up for each protein in which a His Trap FF-5ml (GE Healthcare Life Sciences) column was used in the first purification step. The eluted peak was redirected on a Hi load 16/600 Superdex 200-pg-size exclusion column (GE Healthcare Life Sciences) and collected in 50 mM Tris-HCl (pH 8.0), 0.15 M NaCl, and 10% glycerol. CaiC was purified using a nickel-nitrilotriacetic acid (Ni-NTA) spin column (Qiagen) from a 50-ml cell culture. Protein oligomerization state was determined by gel filtration experiments using a Superdex 200 Increase 10/300 GL column (GE Healthcare) calibrated with LMW and HMW Gel Filtration Calibration kits (GE Healthcare).
Analytical methods.
The concentrations of γ-butyrobetainyl-CoA and l-carnitinyl-CoA enzymatically synthesized were estimated by assaying the remaining CoA with 5,5′-dithiobis-(2-nitrobenzoic) acid (DTNB) (55). NADH was determined spectrophotometrically at 340 nm (Δε = 6,220 M−1 · cm−1). Crotonobetainyl-CoA was estimated using an extinction coefficient of 6,700 M−1 · cm−1 at 263 nm (31).
Enzyme assays.
Experiments were conducted in 100 μl of Tris-HCl 50 mM (pH 8.0). γ-Butyrobetainyl-CoA synthetase activity was determined in a coupled assay, following NADH oxidation. Reactions were conducted in the presence of 10 mM MgCl2, 300 μM NADH, with 13 ng Smc01640 (BcoA/B), 12 units lactate dehydrogenase, and 5 units pyruvate kinase, and initiated by the addition of GBB. γ-Butyrobetainyl-CoA dehydrogenase activity was monitored by following the reduction of ferrocenium hexafluorophosphate (FC+PF−6) at 300 nm (Δε = 4,300 M−1 · cm−1) according to Lehman et al. (22). Reactions were performed in the presence of 500 μM FC+PF−6 and 50 μM FAD and started by the addition of 0.4 μg Smc01639 (BcoC). For the oxidation of 1 mol γ-butyrobetainyl-CoA, 2 mol ferrocenium were required. Crotonobetainyl-CoA hydratase activity was characterized monitoring the disappearance at 260 nm of the Δ2,3-double bond of the substrate (31). Assays were initiated by the addition of 60 ng Smc01641 (BcoD). The thioesterase activities of Smc01638 (CDH thioesterase) were monitored following the release of CoA upon l-carnitinyl-CoA or betainyl-CoA cleavage at 412 nm (55). Reactions were performed in the presence of 450 μM DTNB and initiated by the addition of 30 to 75 ng Smc01638. l-Carnitine dehydrogenase activity of Smc01638 was monitored following NADH formation. Reactions were conducted in the presence of 1 mM NAD+ and initiated by the addition of 60 ng of enzyme. Dehydrocarnitine cleavage activity was assayed as previously reported (12) using 0.5 μg Smc01637 (BKACE). All enzymatic reactions were performed at 25°C in a Safas UV mc2 spectrophotometer.
Metabolome preparation.
Metabolomes from cells of S. meliloti 3D0a2 grown with GBB or sucrose as the sole carbon source were prepared according to Stuani et al. (56). Briefly, a saturated overnight minimal medium liquid culture was diluted in a fresh liquid medium containing the desired carbon source at an OD600 of 0.1, and further grown to an OD600 of 0.2. Five milliliters of this culture was filtered onto a 47-mm polytetrafluoroethylene (PTFE) filter (0.45 μm). The filter was then positioned with cells on top on an agarose plate containing the same medium. Cells were grown to log phase (OD600 of ∼0.8) until quenching of metabolism and extraction of metabolites.
Mass spectrometry analysis.
High-resolution measurements were obtained with an Orbitrap Elite hybrid mass spectrometer (Thermo Fisher Scientific) fitted with a heated electrospray ionization source (HESI) operating in the positive ionization mode. The ionization spray (IS) was set to +3.5 kV. For LC-MS and tandem mass spectrometry (MS/MS; collision induced dissociations [CID]) experiments, the resolving power was set at 60,000 m/Δm (full width at half maximum [FWHM] at m/z 400). Mass spectra were acquired over an m/z range from m/z 50 up to m/z 1,000.
High-pressure liquid chromatographic conditions.
Analyses were conducted using a Dionex Ultimate 3000 Rapid Separation LC (Thermo Fisher Scientific) using a ZIC-pHILIC column (150 by 2.1 mm; 5 μm; Merck). Elution was conducted at 40°C using a mobile phase gradient with a flow rate of 200 μl · min−1. Phase A consisted of 10 mM ammonium carbonate with pH adjusted to 9.9 with NH4OH and phase B was acetonitrile. The gradient started at 80% B for 2 min, followed by a linear gradient to 20% B for 15 min and remained 6 min at 20% B. The system returned to the initial solvent composition in 5 min and reequilibrated under these conditions for 16 min. Samples were diluted 4-fold in 80:20 acetonitrile-10 mM ammonium carbonate (pH 9.9) before analysis. Two microliters was injected. The autosampler was kept at 4°C.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Pellé and Peggy Sirvain for excellent technical assistance.
This work was supported by grants from the Commissariat à l’Energie Atomique et aux Energies Alternatives, CNRS, and Université Evry-Val-d’Essonne/Université Paris-Saclay.
We declare no conflicts of interest with the contents of this article.
M.S. and A.P. conceived the project. A.P. designed the study and wrote the paper. P.B. and N.P. performed gene cloning and biochemistry experiments. E.D., P.B., and C.L. conducted LC-MS experiments. All authors analyzed the results and approved the final version of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00772-18.
REFERENCES
- 1.Vaz FM, Wanders RJ. 2002. Carnitine biosynthesis in mammals. Biochem J 361:417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaz FM, Fouchier SW, Ofman R, Sommer M, Wanders RJ. 2000. Molecular and biochemical characterization of rat gamma-trimethylaminobutyraldehyde dehydrogenase and evidence for the involvement of human aldehyde dehydrogenase 9 in carnitine biosynthesis. J Biol Chem 275:7390–7394. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman RA, Broquist HP. 1977. Biosynthesis of carnitine in Neurospora crassa. J Biol Chem 252:7437–7439. [PubMed] [Google Scholar]
- 4.Strijbis K, van Roermund CW, Hardy GP, van den Burg J, Bloem K, de Haan J, van Vlies N, Wanders RJ, Vaz FM, Distel B. 2009. Identification and characterization of a complete carnitine biosynthesis pathway in Candida albicans. FASEB J 23:2349–2359. doi: 10.1096/fj.08-127985. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen P-J, Rippa S, Rossez Y, Perrin Y. 2016. Acylcarnitines participate in developmental processes associated to lipid metabolism in plants. Planta 243:1011–1022. doi: 10.1007/s00425-016-2465-y. [DOI] [PubMed] [Google Scholar]
- 6.Rippa S, Zhao Y, Merlier F, Charrier A, Perrin Y. 2012. The carnitine biosynthetic pathway in Arabidopsis thaliana shares similar features with the pathway of mammals and fungi. Plant Physiol Biochem 60:109–114. doi: 10.1016/j.plaphy.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Strijbis K, Vaz FM, Distel B. 2010. Enzymology of the carnitine biosynthesis pathway. IUBMB Life 62:357–362. doi: 10.1002/iub.323. [DOI] [PubMed] [Google Scholar]
- 8.Meadows JA, Wargo MJ. 2015. Carnitine in bacterial physiology and metabolism. Microbiology 161:1161–1174. doi: 10.1099/mic.0.000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peluso G, Petillo O, Barbarisi A, Melone MA, Reda E, Nicolai R, Calvani M. 2001. Carnitine protects the molecular chaperone activity of lens alpha-crystallin and decreases the post-translational protein modifications induced by oxidative stress. FASEB J 15:1604–1606. [DOI] [PubMed] [Google Scholar]
- 10.Unemoto T, Hayashi M, Miyaki K, Hayashi M. 1966. Formation of trimethylamine from dl-carnitine by Serratia marcescens. Biochim Biophys Acta 121:220–222. [DOI] [PubMed] [Google Scholar]
- 11.Miura-Fraboni J, Kleber H-P, Englard S. 1982. Assimilation of γ-butyrobetaine, and d-and l-carnitine by resting cell suspensions of Acinetobacter calcoaceticus and Pseudomonas putida. Arch Microbiol 133:217–221. doi: 10.1007/BF00415004. [DOI] [Google Scholar]
- 12.Bastard K, Smith AA, Vergne-Vaxelaire C, Perret A, Zaparucha A, De Melo-Minardi R, Mariage A, Boutard M, Debard A, Lechaplais C, Pelle C, Pellouin V, Perchat N, Petit JL, Kreimeyer A, Medigue C, Weissenbach J, Artiguenave F, De Berardinis V, Vallenet D, Salanoubat M. 2014. Revealing the hidden functional diversity of an enzyme family. Nat Chem Biol 10:42–49. doi: 10.1038/nchembio.1387. [DOI] [PubMed] [Google Scholar]
- 13.Zhu Y, Jameson E, Crosatti M, Schafer H, Rajakumar K, Bugg TD, Chen Y. 2014. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc Natl Acad Sci U S A 111:4268–4273. doi: 10.1073/pnas.1316569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruetschi U, Nordin I, Odelhog B, Jornvall H, Lindstedt S. 1993. Gamma-butyrobetaine hydroxylase. Structural characterization of the Pseudomonas enzyme. Eur J Biochem 213:1075–1080. [DOI] [PubMed] [Google Scholar]
- 15.Zimmermann T, Werlen J. July 1996. Genes for butyrobetaine/crotonobetaine-l-carnitine metabolism and their use for the microbiological production of l-carnitine. European patent EP0722500A1.
- 16.Uanschou C, Frieht R, Pittner F. 2005. What to learn from a comparative genomic sequence analysis of l-carnitine dehydrogenase. Monatsh Chem 136:1365–1381. doi: 10.1007/s00706-005-0331-x. [DOI] [Google Scholar]
- 17.Blaby-Haas CE, De Crecy-Lagard V. 2011. Mining high-throughput experimental data to link gene and function. Trends Biotechnol 29:174–182. doi: 10.1016/j.tibtech.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujimitsu H, Matsumoto A, Takubo S, Fukui A, Okada K, Mohamed Ahmed IA, Arima J, Mori N. 2016. Purification, gene cloning, and characterization of gamma-butyrobetainyl CoA synthetase from Agrobacterium sp. 525a. Biosci Biotechnol Biochem 80:1536–1545. doi: 10.1080/09168451.2016.1177447. [DOI] [PubMed] [Google Scholar]
- 19.Kulla HG. 1991. Enzymatic hydroxylations in industrial application. Chimia (Aarau) 45:81–85. [Google Scholar]
- 20.Blackwell JR, Horgan R. 1991. A novel strategy for production of a highly expressed recombinant protein in an active form. FEBS Lett 295:10–12. doi: 10.1016/0014-5793(91)81372-F. [DOI] [PubMed] [Google Scholar]
- 21.Thorpe C, Matthews RG, Williams CH Jr.. 1979. Acyl-coenzyme A dehydrogenase from pig kidney. Purification and properties. Biochemistry 18:331–337. [DOI] [PubMed] [Google Scholar]
- 22.Lehman TC, Hale DE, Bhala A, Thorpe C. 1990. An acyl-coenzyme A dehydrogenase assay utilizing the ferricenium ion. Anal Biochem 186:280–284. [DOI] [PubMed] [Google Scholar]
- 23.Dommes V, Kunau WH. 1984. Purification and properties of acyl coenzyme A dehydrogenases from bovine liver. Formation of 2-trans,4-cis-decadienoyl coenzyme A. J Biol Chem 259:1789–1797. [PubMed] [Google Scholar]
- 24.Hall CL. 1978. Acyl-CoA dehydrogenases and electron-transferring flavoprotein. Methods Enzymol 53:502–518. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda Y, Okamura-Ikeda K, Tanaka K. 1985. Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from rat liver mitochondria. Isolation of the holo- and apoenzymes and conversion of the apoenzyme to the holoenzyme. J Biol Chem 260:1311–1325. [PubMed] [Google Scholar]
- 26.Benziman M, Galanter Y. 1964. Flavine adenine dinucleotide-linked malic dehydrogenase from Acetobacter xylinum. J Bacteriol 88:1010–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura T, Tobari J. 1963. Participation of flavin-adenine dinucleotide in the activity of malate dehydrogenase from Mycobacterium avium. Biochim Biophys Acta 73:399–405. [DOI] [PubMed] [Google Scholar]
- 28.Massey V, Ganther H. 1965. On the interpretation of the absorption spectra of flavoproteins with special reference to d-amino acid oxidase. Biochemistry 4:1161–1173. [DOI] [PubMed] [Google Scholar]
- 29.Lau SM, Powell P, Buettner H, Ghisla S, Thorpe C. 1986. Medium-chain acyl coenzyme A dehydrogenase from pig kidney has intrinsic enoyl coenzyme A hydratase activity. Biochemistry 25:4184–4189. [DOI] [PubMed] [Google Scholar]
- 30.Bernal V, Arense P, Blatz V, Mandrand-Berthelot MA, Cánovas M, Iborra JL. 2008. Role of betaine:CoA ligase (CaiC) in the activation of betaines and the transfer of coenzyme A in Escherichia coli. J Appl Microbiol 105:42–50. doi: 10.1111/j.1365-2672.2008.03740.x. [DOI] [PubMed] [Google Scholar]
- 31.Fong JC, Schulz H. 1981. Short-chain and long-chain enoyl-CoA hydratases from pig heart muscle. Methods Enzymol 71(Pt C):390–398. [DOI] [PubMed] [Google Scholar]
- 32.Elssner T, Engemann C, Baumgart K, Kleber HP. 2001. Involvement of coenzyme A esters and two new enzymes, an enoyl-CoA hydratase and a CoA-transferase, in the hydration of crotonobetaine to l-carnitine by Escherichia coli. Biochemistry 40:11140–11148. [DOI] [PubMed] [Google Scholar]
- 33.Arima J, Uesumi A, Mitsuzumi H, Mori N. 2010. Biochemical characterization of l-carnitine dehydrogenases from Rhizobium sp. and Xanthomonas translucens. Biosci Biotechnol Biochem 74:1237–1242. doi: 10.1271/bbb.100072. [DOI] [PubMed] [Google Scholar]
- 34.Wargo MJ, Hogan DA. 2009. Identification of genes required for Pseudomonas aeruginosa carnitine catabolism. Microbiology 155:2411–2419. doi: 10.1099/mic.0.028787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sallet E, Roux B, Sauviac L, Jardinaud MF, Carrere S, Faraut T, de Carvalho-Niebel F, Gouzy J, Gamas P, Capela D, Bruand C, Schiex T. 2013. Next-generation annotation of prokaryotic genomes with EuGene-P: application to Sinorhizobium meliloti 2011. DNA Res 20:339–354. doi: 10.1093/dnares/dst014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldmann A, Boivin C, Fleury V, Message B, Lecoeur L, Maille M, Tepfer D. 1991. Betaine use by rhizosphere bacteria: genes essential for trigonelline, stachydrine, and carnitine catabolism in Rhizobium meliloti are located on pSym in the symbiotic region. Mol Plant Microbe Interact 4:571–578. [DOI] [PubMed] [Google Scholar]
- 37.Watson RJ, Heys R, Martin T, Savard M. 2001. Sinorhizobium meliloti cells require biotin and either cobalt or methionine for growth. Appl Environ Microbiol 67:3767–3770. doi: 10.1128/AEM.67.8.3767-3770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbier M, Owings JP, Martínez-Ramos I, Damron FH, Gomila R, Blázquez J, Goldberg JB, Albertí S. 2013. Lysine trimethylation of EF-Tu mimics platelet-activating factor to initiate Pseudomonas aeruginosa pneumonia. mBio 4:e00207-13. doi: 10.1128/mBio.00207-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klagsbrun M, Furano AV. 1975. Methylated amino acids in the proteins of bacterial and mammalian cells. Arch Biochem Biophys 169:529–539. [DOI] [PubMed] [Google Scholar]
- 40.Warren C. 2013. High diversity of small organic N observed in soil water. Soil Biol Biochem 57:444–450. doi: 10.1016/j.soilbio.2012.09.025. [DOI] [Google Scholar]
- 41.Muslin O. 2017. Quantification of osmolytes in the sargasso sea surface layer water column. M.S. thesis Oregon State University, Corvallis, OR: https://ir.library.oregonstate.edu/concern/graduate_thesis_or_dissertations/t435gh40k. [Google Scholar]
- 42.Kleber HP. 1997. Bacterial carnitine metabolism. FEMS Microbiol Lett 147:1–9. doi: 10.1111/j.1574-6968.1997.tb10212.x. [DOI] [PubMed] [Google Scholar]
- 43.Miura-Fraboni J, Englard S. 1983. Quantitative aspects of γ-butyrobetaine and d- and l-carnitine utilization by growing cell cultures of Acinetobacter calcoaceticus and Pseudomonas putida. FEMS Microbiol Lett 18:113–116. [Google Scholar]
- 44.Smith LT, Pocard JA, Bernard T, Le Rudulier D. 1988. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J Bacteriol 170:3142–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khersonsky O, Tawfik DS. 2010. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem 79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 46.Perchat N, Saaidi PL, Darii E, Pelle C, Petit JL, Besnard-Gonnet M, de Berardinis V, Dupont M, Gimbernat A, Salanoubat M, Fischer C, Perret A. 2018. Elucidation of the trigonelline degradation pathway reveals previously undescribed enzymes and metabolites. Proc Natl Acad Sci U S A 115:E4358–E4367. doi: 10.1073/pnas.1722368115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walt A, Kahn ML. 2002. The fixA and fixB genes are necessary for anaerobic carnitine reduction in Escherichia coli. J Bacteriol 184:4044–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corbin D, Barran L, Ditta G. 1983. Organization and expression of Rhizobium meliloti nitrogen fixation genes. Proc Natl Acad Sci U S A 80:3005–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Earl CD, Ronson CW, Ausubel FM. 1987. Genetic and structural analysis of the Rhizobium meliloti fixA, fixB, fixC, and fixX genes. J Bacteriol 169:1127–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgren T, Nordlund S. 2004. The fixABCX genes in Rhodospirillum rubrum encode a putative membrane complex participating in electron transfer to nitrogenase. J Bacteriol 186:2052–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischer HM. 1994. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev 58:352–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robert-Gero M, Poiret M, Cohen GN. 1970. The aspartate kinase of Pseudomonas putida. Regulation of synthesis and activity. Biochim Biophys Acta 206:17–30. [DOI] [PubMed] [Google Scholar]
- 53.Perret A, Lechaplais C, Tricot S, Perchat N, Vergne C, Pelle C, Bastard K, Kreimeyer A, Vallenet D, Zaparucha A, Weissenbach J, Salanoubat M. 2011. A novel acyl-CoA beta-transaminase characterized from a metagenome. PLoS One 6:e22918. doi: 10.1371/journal.pone.0022918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kreimeyer A, Perret A, Lechaplais C, Vallenet D, Medigue C, Salanoubat M, Weissenbach J. 2007. Identification of the last unknown genes in the fermentation pathway of lysine. J Biol Chem 282:7191–7197. doi: 10.1074/jbc.M609829200. [DOI] [PubMed] [Google Scholar]
- 55.Ellman GL. 1959. Tissue sulfhydryl groups. Arch Biochem Biophys 82:70–77. [DOI] [PubMed] [Google Scholar]
- 56.Stuani L, Lechaplais C, Salminen AV, Segurens B, Durot M, Castelli V, Pinet A, Labadie K, Cruveiller S, Weissenbach J, de Berardinis V, Salanoubat M, Perret A. 2014. Novel metabolic features in Acinetobacter baylyi ADP1 revealed by a multiomics approach. Metabolomics 10:1223–1238. doi: 10.1007/s11306-014-0662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vallenet D, Calteau A, Cruveiller S, Gachet M, Lajus A, Josso A, Mercier J, Renaux A, Rollin J, Rouy Z, Roche D, Scarpelli C, Medigue C. 2017. MicroScope in 2017: an expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res 45:D517–D528. doi: 10.1093/nar/gkw1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.