Lag is a temporary period of nonreplication seen in bacteria that are introduced to new media. Despite latency being described by Müller in 1895, only recently have we gained insights into the cellular processes characterizing lag phase.
KEYWORDS: antibiotic tolerance, bet-hedging, cell division, food safety, gene expression, host-pathogen interactions, oxidative stress, persister cells, phenotype switching, primary metabolism
ABSTRACT
Lag is a temporary period of nonreplication seen in bacteria that are introduced to new media. Despite latency being described by Müller in 1895, only recently have we gained insights into the cellular processes characterizing lag phase. This review covers literature to date on the transcriptomic, proteomic, metabolomic, physiological, biochemical, and evolutionary features of prokaryotic lag. Though lag is commonly described as a preparative phase that allows bacteria to harvest nutrients and adapt to new environments, the implications of recent studies indicate that a refinement of this view is well deserved. As shown, lag is a dynamic, organized, adaptive, and evolvable process that protects bacteria from threats, promotes reproductive fitness, and is broadly relevant to the study of bacterial evolution, host-pathogen interactions, antibiotic tolerance, environmental biology, molecular microbiology, and food safety.
HISTORY, DEFINITIONS, AND DETERMINANTS OF LAG-PHASE DURATION
Starving bacteria that encounter new nutrients do not immediately proliferate but first undergo a temporary period of nonreplication (1). This phenomenon was first observed by Müller in 1895 (2) and was identified as an explanation for inconsistent measurements on the growth rate of bacterial cultures. Termed “lag” or “latency” by bacteriologists, this phase was understood to be a temporary period of nonreplication when bacteria are introduced to new media (2–7). Despite lag phase being documented more than a century ago, little is known of the molecular and cellular events characterizing the latent period, nor are its implications to cell survival and proliferation clearly understood. This is because of the technical challenges associated with studying a small number of cells (8–10). A common description of lag phase is therefore relegated to an observational definition, that this is the time when bacteria have not yet started dividing. Nonetheless, recent experiments have begun to offer some insights into what is happening during this nonreplicative period, allowing bacteriologists to begin formulating an answer to a question first posed by Müller more than a century ago: what exactly is lag phase?
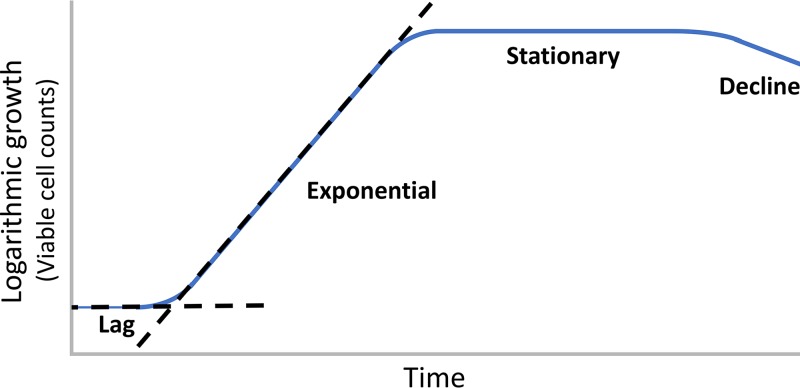
Two common methods for quantifying the growth of a bacterial culture over time are viable cell counts and optical density measurements (11). When data are plotted semilogarithmically, four growth phases are distinguishable, as follows: (i) lag phase, a nonreplicative period; (ii) exponential phase, a replicative period; (iii) stationary phase, the cessation of replication due to the exhaustion of nutrients; and (iv) decline/death phase, a gradual decline in viable cell counts due to starvation (Fig. 1). A long-term stationary phase consisting of a small number of surviving bacteria is known to follow the decline phase (12) but is not illustrated in Fig. 1. Numerous definitions have been proposed to define the endpoint of lag phase (11, 13). The most commonly adopted definition is the moment in the bacterial culture when the extrapolated slope of the logarithmic line on a growth plot intersects the starting inoculum concentration (Fig. 1). As shown below, biochemical, morphological, physiological, and transcriptional markers of lag phase are now available. Nonetheless, this common definition of lag is useful as a starting point for discussion and is adopted in this review for simplicity. When discussing an individual bacterium, the lag period refers to the time required for that bacterium to reach first cell division.
FIG 1.
A representative growth plot of a bacterial culture. Where the dotted lines cross is the commonly defined endpoint of the lag period for a bacterial culture. For individual cells, lag is defined as the time required to reach first cell division.
Due to the difficulty of studying small numbers of cells experimentally, bacteriologists have developed mathematical models to study lag phase (14–24). In addition to its importance to the environmental sciences and basic cellular research, this work is also important to the food industry. The duration of lag is the primary determinant in the spoilage of bacterially contaminated food. The purpose of refrigeration and other food preservation techniques is to prolong the lag period of any contaminating organisms that are present. Although these mathematical models are not reviewed here for the sake of brevity, it is emphasized that developing accurate growth models enables the food industry to minimize waste, develop risk management procedures, save money, and promote global food security (25).
To understand how bacteria respond to various conditions, bacteriologists have monitored the duration of lag in response to stress or injury. Thanks to this work, we know that the duration of the lag period is greatly influenced by the history of the inoculated culture. For example, bacteria that have been preadapted to extremes of pH, temperature, or osmolarity have shorter lag periods when encountering the identical stress than do bacteria that were not preadapted (26–37). In general, the magnitude of the change between old and new environments positively correlates with the duration of the lag period. When cells are placed under conditions approaching the threshold of survivability, a decrease in the number of viable cells may also occur before exponential phase begins. Bacteria that have been injured by freezing, starvation, heating, or desiccation or that have been subjected to offending chemicals require more time to exit the lag phase (38–46).
The number of bacteria present in a culture also influences the duration of lag phase. As the number of cells increases, lag duration decreases (41, 47–49). Studies of single cells have revealed that individual bacteria vary in the time required to reach first cell division. In addition, these studies indicate that when cells are injured or stressed, the average lag period becomes longer and the time points at which individual cells begin dividing show increased scattering (43–45, 50–57). Individual cells with exceptionally short lag times will begin dividing quickly and will therefore have a disproportionate effect on the culture growth plot. If short lag is considered an exceptional phenotype, increasing the total population of bacteria would have the expected effect of increasing the number of bacteria with exceptionally short lag phases. This mathematical reasoning could explain why larger populations of bacteria have shorter lag times.
Phenotypic differences among genetically identical cells, such as individual differences in stress tolerance and lag times, are known as phenotypic variance. In stressful and rapidly changing environments, such variance is advantageous to the survival of a kin population because it ensures that a subset of a bacterial population will survive an insult and later repopulate once conditions improve. This is a principle known as “bet-hedging” (58–60). Differences in the lag times and stress tolerance among individual cells may therefore help a kin population to proliferate within an unpredictable environment, for example, by having a contingent of cells that can rapidly divide in times of plenty and by having a contingent of cells that can survive when conditions worsen. As is shown in Lag Is An Evolvable Phenotype That Can Also Emerge De Novo, below, variations in the duration of lag phase among single cells are also relevant to the survival of a kin population in response to antibiotics. Cumulatively, these studies indicate that lag is an adaptive response to stress and injury, that bacteria can be preadapted to have shorter lag periods, and that individual cells vary in their lag times and stress tolerance.
BROAD EXPRESSION PROGRAMS PRODUCE CELLULAR MACHINERY ESSENTIAL FOR PROLIFERATION
What are the cellular conditions that bacteria inherit when initiating lag phase? The bacterial lifestyle is one of feasting and fasting. Bacteria spend most of their time in the stationary phase due to the general lack of abundant food in natural environments (61, 62). Adaptations during this period include thickening of the peptidoglycan layer, condensing of DNA, deactivation (but not destruction) of ribosomes, and reduction of cytoplasmic volume (63, 64). Oxidative damage to biomolecules accumulates during stationary phase (65–69). To begin dividing once more, a bacterium must alter its DNA superstructure, restructure its cellular morphology, reorganize its global metabolism, and repair oxidatively damaged biomolecules. In well-studied bacteria such as Escherichia coli and Salmonella enterica, onset of cell division can occur in as little as one or two hours. How is all of this done, and so quickly?
Bacteria entering the lag phase dramatically transform their transcriptome and proteome to produce the cellular components that are needed to accumulate biomass and divide. Isotope labeling and two-dimensional (2D) electrophoresis experiments in Lactobacillus delbrueckii provided some of the earliest evidence of a lag program, in this case revealing 47 proteins that were produced during this period (70). DNA microarray analyses of Bacillus licheniformis identified 75 genes during lag phase that were differentially expressed (induction or downregulation at least 2-fold) compared to their expression in the preceding stationary phase (71). A combined protein and gene expression analysis in Lactococcus lactis subsequently revealed 28 proteins that were highly and differentially expressed during lag phase (72). In these studies, the functions of the genes and proteins relate to diverse metabolic processes such as glycolysis, amino acid metabolism, nucleotide biosynthesis, gene transcription, protein translation, coenzyme biosynthesis, cell wall biosynthesis, phosphate transporters, stress response, respiration, and cell division (70–72). These experiments indicate that bacteria are producing new enzymes to digest food, build biomass, and prepare for cell division.
A landmark transcriptional profiling study of Salmonella enterica by Hinton and Rolfe (73) revealed that changes in gene expression can occur in as little as 4 min following inoculation into liquid medium. The expression of a total of 1,119 genes was altered within 4 min after inoculation. By the end of a 2-h lag period, more than half of all genes within the S. enterica genome were participating in a vast transcriptional program. Mutational studies in Salmonella spp. had previously identified 356 genes that are responsible for essential biological processes (74). The expression of most essential genes (60%) was altered within the first hour of S. enterica lag phase (73). The transcription initiation factor σ70 is the primary sigma factor in bacteria and is responsible for the induction of essential metabolic genes (75, 76). Many of the lag-associated genes identified in S. enterica possessed an upstream DNA motif consistent with the binding site of σ70, suggesting that this initiation factor could be responsible for regulating the expression of many of the genes seen within the lag transcriptome (73). The factor for inversion stimulation gene (fis) is a DNA recombinase and transcriptional regulator gene with growth phase-specific expression effects (77). The role of fis in lag phase was also investigated as a possible global stimulator of lag transcriptional programs. The deletion of fis in S. enterica produced longer lag phases when bacteria were inoculated into rich medium but had no apparent effect on lag phase duration when introduced to minimal medium (73, 78). The role of fis in lag phase remains unclear. In the stationary phase of Saccharomyces cerevisiae, RNA polymerase was found to be prepositioned upstream of genes in preparation for lag phase (79). A chromatin immunoprecipitation assay revealed that no such prepositioning occurred in S. enterica (73). This suggests that RNA polymerase in bacteria is recruited to genes during the lag phase itself. How RNA polymerase can transcribe hundreds of genes in 4 min and without an apparent prepositioning mechanism is a remarkable mystery.
Genes that display altered patterns of expression only during lag phase are designated lag-phase signature genes. A total of 39 lag-phase signature genes were identified in S. enterica (73). Among the 20 signature genes that were upregulated, 15 are involved in the uptake of iron and manganese as well as [Fe-S] cluster biosynthesis. The 19 downregulated genes included carbon-processing genes subject to catabolite control as well as various genes of unknown or speculative function. The fact that so many signature genes are involved in metal metabolism suggests that acquiring metals is an important feature of lag phase (73). Speculative reasons for iron absorption by bacteria, and its consequences, are discussed in Iron Influx during Lag Produces Oxidative Stress and Could Be Related to Immune Evasion, below.
To translate genes into proteins, ribosomes are necessary. Stationary-phase cells conserve energy by dimerizing 70S ribosomes into inactive 100S complexes (80). This is a process known as ribosome hibernation and serves to conserve energy while preserving ribosomes for reactivation once nutrients become available (81). In E. coli, two proteins are necessary to dimerize ribosomes, the hibernation promoting factor (HPF; encoded by hpf) and the ribosome modulation factor (RMF; encoded by rmf). In Bacillus subtilis, Staphylococcus aureus, and L. lactis, a larger version of HPF appears to be self-sufficient to dimerize ribosomes (81). The process of disassembling and reactivating hibernating ribosomes is less understood, though it is known to proceed quickly; inoculating stationary-phase E. coli cells into fresh medium results in the degradation of rmf mRNA, the disappearance of dimerized ribosomes within 2 min, and protein translation within 6 min (82, 83). This process of conserving translational machinery therefore allows bacteria entering lag phase to quickly produce the proteins they need.
One feature of lag phase appears to be the repair and replacement of damaged subcellular components. A repair program is indicated by the induction of genes associated with DNA repair, degradation of carbonylated proteins, reduction of disulfide bonds, and repair of oxidatively damaged aspartate residues (73). In polyploid cyanobacteria, rapid DNA replication and an increase in chromosome copy number are also observed (84). Biomass accumulates during lag phase, as immediately before first cell division occurs, these cells will be larger than those seen during their subsequent exponential period (73, 85). The speed and scale of cellular reorganization suggest that bacteria can quickly sense changes within their environments and initiate vast transcriptional programs for essential metabolism to occur. Additional experiments will be required to determine exactly how bacteria are able to sense extracellular conditions and trigger this global program in response.
BACTERIAL HISTORY REFLECTS A DYNAMIC TRANSCRIPTIONAL PROFILE
The nature of the lag transcriptome is dependent upon the history of the bacterial culture. Which genes are expressed and the intensity of the expression are influenced by preceding conditions. For example, variations in the production of rRNA during lag phase can be observed in response to excess heat or a prolonged stationary phase (86–88). Such variation is observable throughout the transcriptome. Pin and coworkers (89) performed a network analysis on the transcriptional profiles of E. coli cells that were transferred to fresh medium after being starved in stationary phase for 1 (“young”) or 16 (“old”) days. In both young and old E. coli cells, genes responsible for osmotolerance, acid resistance, and oxidative stress were downregulated, signifying the cessation of the stringent response (89). However, the differences between young and old profiles are numerous and unexpected. One would hypothesize that old cells would induce more genes to recover from prolonged starvation. One would also hypothesize that young and old cells would induce a similar array of genes to prepare for cell division. Neither is true. Compared to stationary phase, the number of genes that were differentially expressed in E. coli was much larger in young cells (467 genes) than in old cells (186 genes). Furthermore, only 62 of these genes between young and old E. coli were common, suggesting that young and old cells engage distinct transcriptional programs to prepare for cell division (Fig. 2). The 62 genes that were commonly up- or downregulated are related to glycine-betaine metabolism, glutamate metabolism, the acid resistance system, detoxification, response to oxidative stress, anaerobic respiration, fermentation, enterobactin biosynthesis, transport and binding proteins for cations and iron-carrying compounds, transport and binding proteins for carbohydrates, organic alcohols and acids, and transcription factors (89). The expression of genes related to DNA repair was only observed in young cells (89). How metabolism is reorganized also appears to be distinct between young and old cells, in that young cells upregulated genes involved in the citric acid cycle and aerobic respiration, whereas old cells upregulated the Entner-Doudoroff and gluconate pathways and downregulated the pentose phosphate pathway (89). These data suggest that the lag transcriptome is dynamic with respect to the cellular milieu. Providing that these data are correct, it is remarkable that young and old bacteria can achieve first cell division using what appears to be two distinct programs. In Potential Avenues of Research, below, some speculations are provided on the nature and rationale of these two distinct programs.
FIG 2.
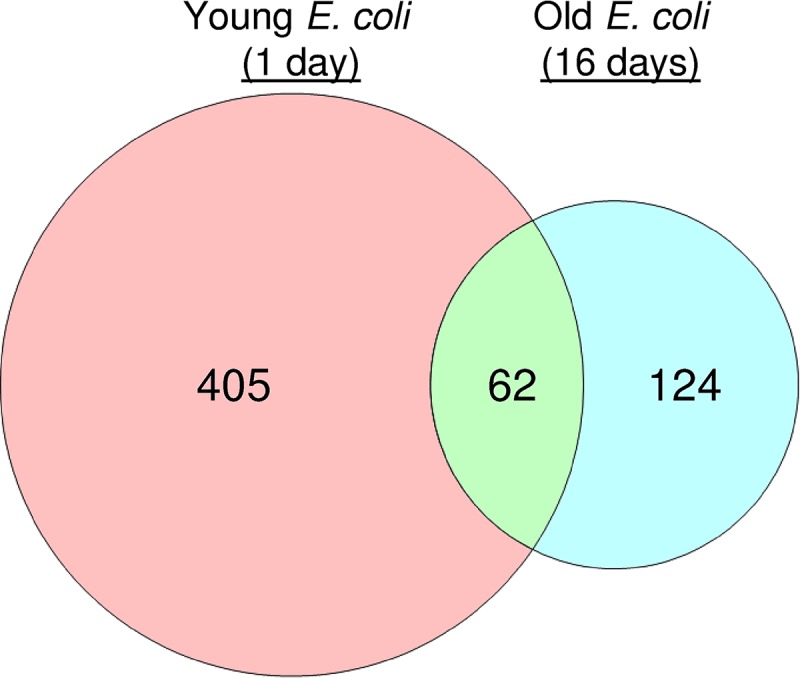
Total number and commonality genes in the lag phase of “young” (1-day-old) and “old” (16-day-old) E. coli cells that were differentially expressed compared to the preceding stationary phase. Diagram produced from data reported by Pin and coworkers (89).
OPTIMIZATION OF PRIMARY METABOLISM IS CHARACTERISTIC OF LAG PHASE
Carbohydrate metabolism is reorganized during the lag period to maximize carbon flow. This is made evident by the fact that intermediates of metabolic pathways rapidly accumulate during the lag period (84). It is now also apparent that bacteria can alter its enzymatic profile during lag phase in response to various nutrients. For example, if glucose is replaced with a less-preferred carbohydrate, such as arabinose, two metabolic stages are observable. Madar and coworkers (85) used fluorescent reporter cells and flow cytometry to monitor a library of E. coli strains in response to arabinose. Arabinose produced two phases which the authors termed “Lag1” and “Lag2” (85). Gene expression in Lag1 appears to be exclusively devoted to the biosynthesis of bottleneck carbon-processing proteins, in this case, enzymes necessary for the digestion of arabinose. Once a steady carbon flow is achieved, cells transition to Lag2, involving the biosynthesis of a broader array of metabolic enzymes required for biomass accumulation and cell division. If “preferred” carbohydrates such as glucose are used, cells skip immediately to Lag2 (85). What remains unclear is whether catabolite repression signaling plays a role in the transition from Lag1 to Lag2. A related question is how bacteria signal a transition between Lag1 and Lag2 as well as how bacteria discriminate between monophasic and biphasic programs. The second question is explored further in Potential Avenues of Research, below.
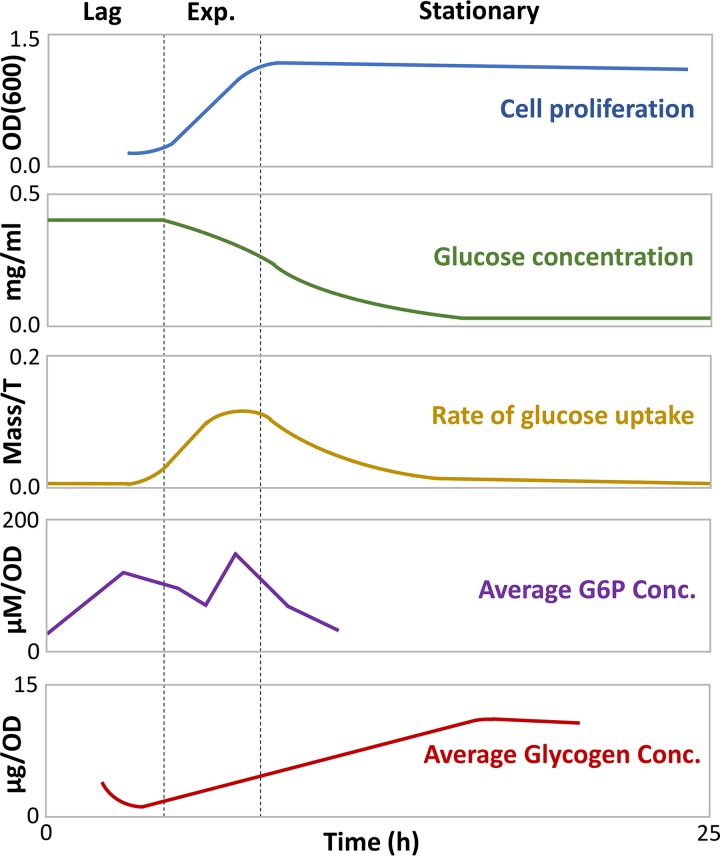
Lag phase is initiated when bacteria encounter new nutrients, and it involves the expression of primary metabolic pathway genes. One would logically expect that the predominant energy source priming cellular metabolism would come from extracellular carbon that is imported into the cell. A combined modeling and experimental study by Yamamotoya and coworkers (90) instead demonstrated that glycogen, a polysaccharide used by organisms as energy storage, is the primary source of energy during lag phase (Fig. 3). Although lag-phase cells have abundant intracellular glucose, most or all of it comes from the digestion of glycogen, as indicated by a decline in glycogen reserves (90). At this stage, glucose is not yet being rapidly imported into the cell, presumably because the proteins needed to do this are still being made. A rapid increase in the intracellular concentration of glucose-6-phosphate (G6P), the first intermediate of glycolysis, suggests that glycogen is being degraded to produce energy via the glycolysis pathway (90). In exponential phase, sugar flow is now established (Fig. 3). Imported glucose is shunted toward both glycolysis and glycogen assembly, and G6P concentration varies throughout exponential phase due to fluctuating energy demands (Fig. 3). As extracellular glucose depletes, cells transition to stationary phase, and glucose is predominantly devoted to glycogen assembly (90). An “energy bank” provides a suitable analogy for this relationship; once bacteria sense new nutrients are available, energy is first “loaned” from glycogen stores, creating an energy “debt” that is used to sponsor the absorption of glucose. Once a steady flow of carbon energy is established, this debt is “repaid” by shunting glucose back to the glycogen stores.
FIG 3.
Metabolic and growth trends of bacterial cultures from lag to early stationary phase. Adapted from the work of Yamamotoya and coworkers (90). Exp., exponential phase; OD, optical density; OD(600), OD at 600 nm; Conc., concentration.
CELLULAR EVENTS PROVIDE PHYSIOLOGICAL MARKERS OF LAG
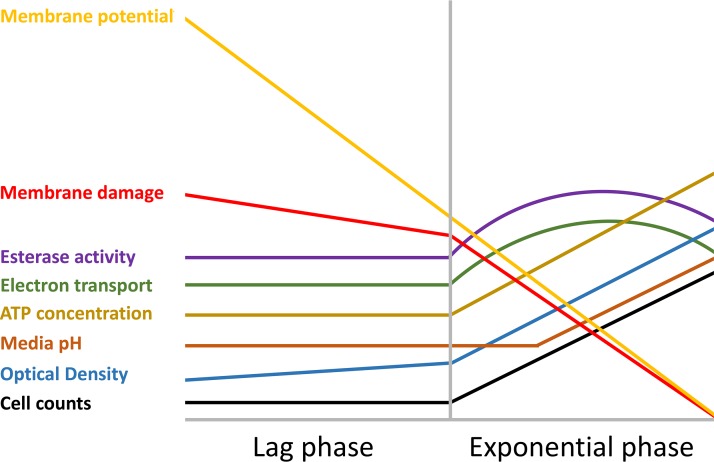
Physiological patterns of behavior can be identified by studying the metabolic, proteomic, and biochemical trends that emerge during latency. Though some of these trends are dependent upon and reflect changes in incubation conditions, others appear to occur independently of extracellular conditions and could therefore be characterized as physiological markers of lag. The physiology of Bacillus cereus in various levels of acid stress was studied by Biesta-Peters and coworkers (91). Decreasing the medium pH from 7.0 to 4.9 increased the duration of the lag phase from 1 h to 5 h. Several physiological trends remained consistent throughout these trials despite the variance in pH. An abstraction of these physiological trends is provided in Fig. 4. For example, under all pH conditions examined, ATP concentration remained stable throughout the lag phase at around 1 × 1018 molecules per cell. In exponential phase, ATP concentration increases about 50-fold (91). Cell size increased throughout lag phase, as suggested by a slight increase in optical density measurements. A maximum cell size of 2.5 μm was observed immediately before first cell division (91). Bacteria that were introduced to medium set to a pH between 4.9 and 7.0 did not change the pH of the medium during lag phase. Only when cell division was well under way were changes in medium pH observed (91). Esterase activity, monitored via fluorescent signal, was detectable at around 100 units throughout the lag periods under all pH conditions examined, thereafter increasing to approximately 30,000 units during exponential phase (Fig. 4). The membrane potential (difference in the interior versus exterior charge of a cell) decreased continuously under all pH conditions tested. By the time cells transitioned into exponential phase, approximately half of all cells had no measurable membrane potential (91). Electron transport chain activity, monitored using a fluorescent signal, remained constant during lag phase and increased in intensity upon initiation of exponential growth (Fig. 4). At the beginning of lag phase, propidium iodide staining indicated that an average of 20% of cells possessed compromised membranes. This percentage decreases during lag phase, suggesting that B. cereus can repair its membrane during lag phase. By mid-exponential phase, membrane defects are seen in only 3% of cells (91). Although the study was limited only to changes in pH, the universality of trends suggest that these could be physiological markers of lag phase and of lag- to exponential-phase transition (Fig. 4).
FIG 4.
An abstraction of the physiological trends in the lag and exponential phases of B. cereus. Adapted from the work of Biesta-Peters and coworkers (91).
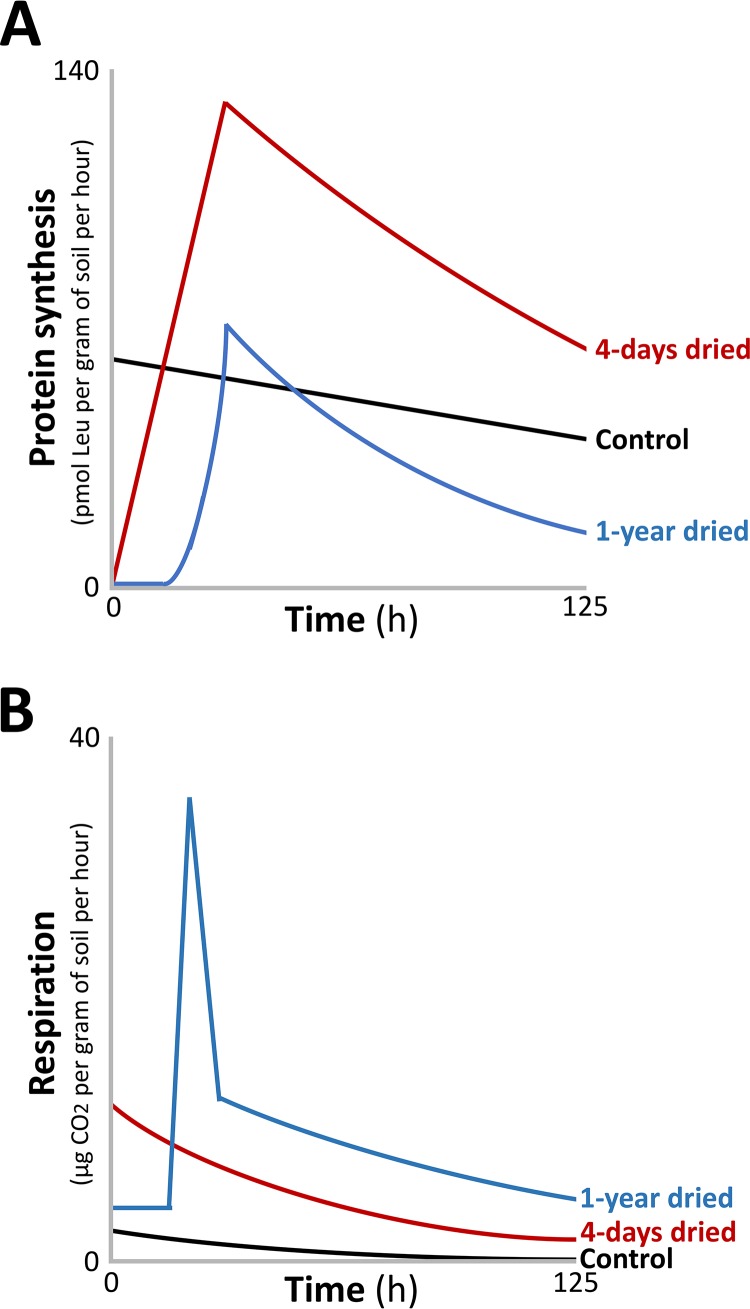
Distinct physiological trends can also be observed in response to various degrees of stress. In these cases, these trends comprise stimulus-dependent responses. In the example of soil biology, it is known that rewetting dried soil produces a burst of respiration from soil-dwelling bacteria (92). Meisner and coworkers (93) monitored soil-dwelling bacteria and quantified respiration (measured as CO2 production) and protein biosynthesis ([3H]leucine incorporation), following rewetting of soil after 4 days or 1 year of dryness. The purpose of monitoring protein biosynthesis was to estimate bacterial growth over time (93). A control sample, placed under continuous moisture, presented with linear and slightly decreasing rates of respiration and protein synthesis over time (Fig. 5). Bacteria rewetted after 4 days displayed no detectable lag phase, a steady increase in protein biosynthesis over time, and a respiratory burst that was maximal after 1 h. This was collectively termed a “type I pattern.” In contrast, cells dried for 1 year displayed a prolonged lag phase (∼16 h) followed by a respiratory burst that greatly exceeded the 4-day sample. This was collectively termed a “type II pattern” (Fig. 5). Although total protein biosynthesis was lower in the 1-year sample, the rate of protein biosynthesis was higher. Whether a type I or type II pattern is presented is dependent upon the degree and duration of desiccation. For example, longer periods of drought produce longer lag phases and greater cumulative respiration, aligning closely with a type II pattern (94). Soil that is not completely dry produces shorter lag periods and lower cumulative respiration, aligning with a type I pattern (95). Several factors remain unaccounted for that ought to be considered when interpreting these results. For example, it remains unknown how many species of bacteria are present in these soil samples as well as the relative contributions of each species toward producing these outcomes. It also remains unclear whether intra- and interspecies quorum sensing/communication is playing a role in changing the rates of respiration and proteinogenesis, nor whether the results are being conflated by germinating spores within the soil, a process that is distinct from lag phase. Nonetheless, these data are valuable because studying the physiology of lag phase under conditions approximating the natural habitats of bacteria is more likely to produce data that accurately reflect the physiology of lag phase as it is experienced by bacteria in natural settings. In summary, these soil studies reveal that changing conditions can stimulate lag phases in bacteria that are correlated with distinct respiratory and proteinogenic physiological outcomes.
FIG 5.
Physiological trends in protein synthesis (A) and respiration (B) in soil-dwelling bacteria. Soil was dried for 4 days or 1 year. The soil was then rewetted and monitored for 125 h. The control culture was incubated in moist soil. Adapted from the work of Meisner and coworkers (93).
IRON INFLUX DURING LAG PRODUCES OXIDATIVE STRESS AND COULD BE RELATED TO IMMUNE EVASION
Reactive oxygen species (ROS) are chemically reactive oxygen-containing molecules that damage biomolecules. Examples include superoxide (O2·−), hydrogen peroxide (H2O2), and hydroxyl radical (OH·). The majority of ROS are produced as by-products of cellular respiration (96–100). Enzymes that detoxify free radicals include catalase, superoxide dismutase, glutathione reductase, thioredoxins, and glutaredoxins (101, 102). Oxidative stress appears to be a feature of lag phase; placing E. coli cells onto solid agar results in the induction of heat shock regulons (RpoH, RpoE, and CpxAR) and oxidative stress regulons (SoxRS, OxyR, and Fur) (103). All three isozymes of superoxide dismutase, the enzyme that scavenges superoxide radicals, are induced (103). Similar patterns of oxidative stress response can be seen when S. enterica is placed in liquid medium (77). These data suggest that oxidative stress occurs during lag phase.
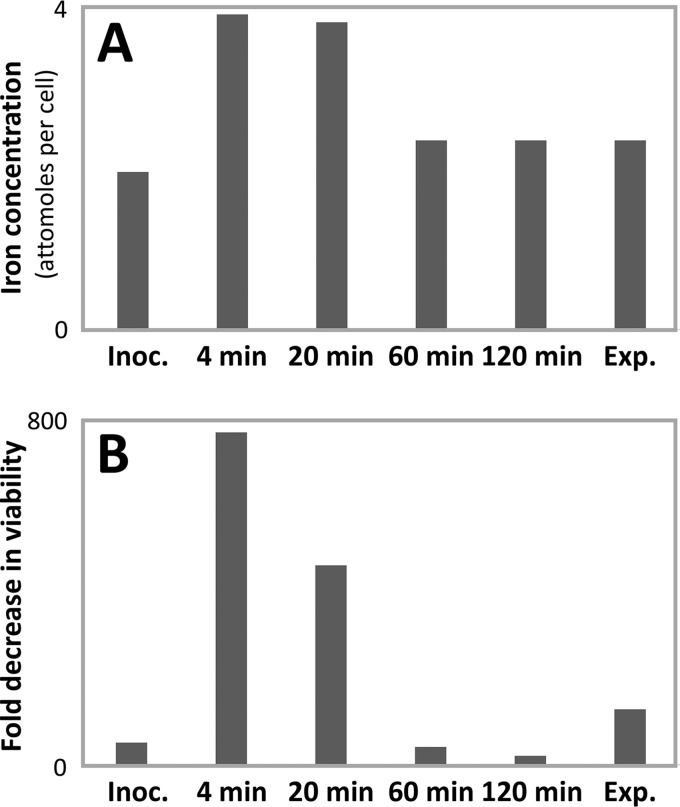
Metals are absorbed or expelled from S. enterica upon entry into lag phase. Increases in the intracellular concentrations of iron, manganese, and calcium were observed during the lag phase, whereas cobalt, nickel, sodium, and molybdenum concentrations decreased (73). This occurs quickly, as the total iron content in S. enterica was observed to double after only 4 min (73). The induction of genes responsible for iron recruitment and metabolism is observable in the lag phases of S. enterica and E. coli (73, 89). Iron is toxic in excess because it catalyzes the formation of deleterious hydroxyl radicals from hydrogen peroxide through the Fenton reaction (104). Prokaryotes have therefore evolved sophisticated regulatory mechanisms to provide iron for essential metabolism while mitigating iron toxicity (105). Rapid iron accumulation in S. enterica produced hypersensitivity to hydrogen peroxide, presumably a result of Fenton chemistry (Fig. 6). Challenging S. enterica with hydrogen peroxide during this iron influx decreased the viability of S. enterica by 800-fold (73). This suggests that iron accumulation in S. enterica produces hydrogen peroxide hypersensitivity and potentiates lethal oxidative damage.
FIG 6.
Quantification of the intracellular iron concentration (A) and sensitivity to H2O2 treatment (represented as fold decrease in viability) (B) during and after a 2-h lag phase in S. enterica. Adapted from the work of Rolfe and coworkers (73). Inoc., inoculation.
From a perspective of reproductive fitness and evolution, rapidly absorbing iron to the point of lethal hypersensitivity to hydrogen peroxide is a perplexing behavior. Why would bacteria suffer so much for the sake of iron? I advanced a hypothesis in relation to host-pathogen interactions (106), that vertebrates stymie infections by sequestering bioavailable iron, a process known as “nutritional immunity” (107–109). It would therefore be advantageous for pathogenic bacteria to preemptively acquire iron before the host forms an effective immune response (106). This hypothesis remains to be experimentally tested.
How iron accumulation occurs is also perplexing because iron load is strictly regulated. In Gram-negative bacteria such as S. enterica and E. coli, this is done through the ferric uptake regulator (Fur) via a negative-feedback mechanism (110). Normal Fur activity should therefore prevent rapid changes in iron loads. A possible explanation of how iron influx occurs in S. enterica could be offered by examining the expression of Fur-regulated proteins during lag phase. An enzyme regulated by Fur is superoxide dismutase (SOD). The two predominant isozymes in E. coli are a manganese-bound variant (MnSOD) and an iron-bound variant (FeSOD). By distinct mechanisms, Fur (when activated by ferrous iron) suppresses MnSOD and activates FeSOD (111). Adding ferrous iron to a culture therefore results in decreased MnSOD expression and increased FeSOD expression (112). In a series of experiments, our group used nondenaturing polyacrylamide electrophoresis and a quantitative enzyme activity assay to monitor changes in SOD isozyme expression in response to iron treatments during the lag, exponential, and stationary phases of E. coli (113–115). Whereas SOD isozyme expression responded predictably to iron treatment in exponential and stationary phases, no change in the enzymatic profile of SOD was observed during the lag phase. In the lag phase of E. coli, MnSOD expression was high, regardless of the absence or presence of iron, and FeSOD expression was low, regardless of the absence or presence of iron (113, 114). Transcriptional profiling performed elsewhere also observed high sodA (MnSOD) and low sodB (FeSOD) during lag phase of E. coli, providing congruence between transcriptional and translational data (89). We hypothesized that low or absent Fur protein could explain these observations, in that low Fur protein would result in the derepression of MnSOD and an increase in FeSOD (113, 114). If this is indeed the case, low Fur protein in the lag phase could also explain how S. enterica can quickly absorb iron without triggering a negative-feedback switch (106). It is notable that deleting fur in E. coli results in rapid iron accumulation and oxidative stress, features that are also seen in the lag phase of S. enterica (116). It would be useful to characterize Fur activity during the lag phases of S. enterica and E. coli to explore these hypotheses.
It ought to be cautioned that transcriptional or metabolic changes related to metals during lag have thus far only been demonstrated in S. enterica and E. coli. In both organisms, iron metabolism is regulated by Fur (110). In many other bacteria, regulators such as the diphtheria toxin repressor (DtxR) are used instead of Fur (117). Due to this variability, it should not be assumed that iron accumulation in the lag phase of S. enterica also occurs in other prokaryotes. Further research will be required to ascertain whether rapid changes in metal content during lag phase represent a generalized phenomenon among prokaryotes.
NUTRIENT SENSING IS COUPLED TO DIVISOME MACHINERY TO TRIGGER FIRST CELL DIVISION
Exponentially replicating bacteria in nutrient-rich media are typically used to study prokaryotic cell division (118–121). However, bacteria in natural environments spend most of their time in a nutrient-poor and nonreplicative state. How bacteria initiate first cell division at the end of lag phase following a prolonged period of starvation therefore remains poorly understood. One protein relevant to cell division is FtsZ (encoded by ftsZ), a prokaryotic cytoskeletal protein that is similar to tubulin in eukaryotes. FtsZ polymerizes to form a ring, known as a “Z ring,” in the middle of cells where future cell division is to take place (122). This Z ring is responsible for recruiting the division machinery that is responsible for synthesizing peptidoglycan and constricting the membrane, resulting in a septum that allows the scission of daughter cells (123).
Sekar and coworkers (124) recently demonstrated that the nutrient-dependent accumulation of FtsZ during the lag phase in E. coli is the trigger of first cell division following a period of starvation. Real-time metabolomics and microfluidic single-cell microscopy experiments revealed a pronounced rise in metabolic pathway intermediates as well as amino acid and nucleotide monomers in response to glucose pulse feeding (124). Pulse feeding of E. coli with isotope-labeled glucose demonstrated that the fed glucose was being used to build new proteins and DNA. A severalfold increase in the concentration of FtsZ was also observed in response to pulse feeding, suggesting that FtsZ is one of the proteins that was synthesized from glucose. As the rate of pulse feeding increased, the lag phase shortened (124).
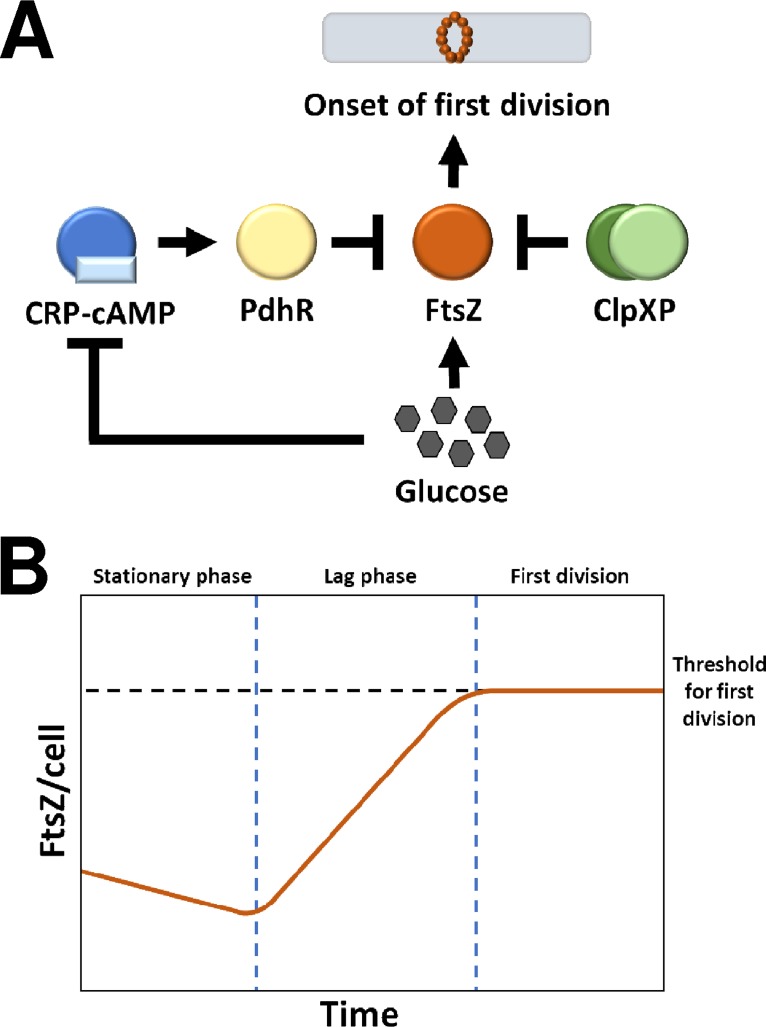
Transcription of ftsZ is repressed by the pyruvate dehydrogenase complex repressor (PdhR, encoded by pdhR) (125). The expression of PdhR is activated by the cAMP receptor protein (CRP-cAMP, encoded by crp) (126), a global transcription regulator that is highly active during carbon starvation in E. coli (127). As CRP-cAMP is downregulated in response to glucose (128), the biosynthesis of FtsZ is mediated by the availability of glucose. FtsZ concentration is therefore expected to decline when cells are starved and to rise when nutrients are found. Remarkably, the deletion of crp or pdhR in E. coli resulted in cells that immediately began dividing without a detectable lag period, whereas supplying PdhR via an expression plasmid restored the lag period. This suggests that the bioavailability of FtsZ mediates the timing of first cell division in E. coli (124). To substantiate this finding, the authors investigated the role of ClpXP (encoded by clpX and clpP), a complex that functions as both a protease of FtsZ and an inhibitor of FtsZ polymerization (129, 130). The deletion of clpX and clpP resulted in increased FtsZ concentration and a shorter lag period, whereas overexpression of ClpX decreased FtsZ abundance and produced a prolonged lag period (124). When a protease inhibitor cocktail was applied, the duration of lag was reduced by 30%, further exemplifying the role of ClpXP. The authors therefore proposed that the concentration of FtsZ, mediated by nutrient availability and proteolysis, serves as a general timing mechanism for triggering the first cell division and the conclusion of lag phase (124). This model is summarized in Fig. 7.
FIG 7.
(A) Activating and repressing elements determining the concentration of FtsZ and the triggering of first cell division at the end of lag phase. (B) Abstraction of the concentration of FtsZ during the stationary phase, lag phase, and first cell division. Adapted from the work of Sekar and coworkers (124).
Several intriguing questions emerge from this work. First, there are numerous factors that influence FtsZ biosynthesis and polymerization beyond CRP-cAMP and ClpXP. For example, in E. coli, the glucosyltransferase OpgH (UgtP in B. subtilis) is known to localize to the future division site to antagonize the assembly of FtsZ, thereby preventing premature division until the cell has accumulated enough biomass to divide (131, 132). In B. subtilis, pyruvate dehydrogenase appears to couple glucose metabolism to Z-ring formation, thereby coordinating the pace of division with nutrient availability (133). How elements such as these impact the timing of first cell division remains the subject of future research. Second, and more broadly, this study suggests that first cell division and the divisions seen in mid-exponential-phase cultures appear to be paced by distinct mechanisms. Whereas Sekar and coworkers (124) outlined a concentration-dependent role of FtsZ in dictating the onset of first division in E. coli, it is also known that exponentially replicating E. coli and B. subtilis cells present with a constant concentration of FtsZ throughout the cell cycle, and that inducing changes in FtsZ levels has little effect on the frequency of Z-ring formation (134, 135). Does this imply that the timing of first division is dictated by the concentration of FtsZ, whereas in the exponential phase, the timing is dictated by other factors related to FtsZ, for example, its cellular localization and ability to polymerize? Third, and unrelated to first cell division per se, the observation by Sekar and coworkers (124) that pulsing E. coli with 13C-labeled glucose results in isotope-labeled metabolites during the lag phase appears to contradict the energy bank model (see Cellular Events Provide Physiological Markers of Lag, above). As previously discussed, Yamamotoya and coworkers (90) argued that glycogen is the primary source of glucose during the lag phase of E. coli. Significant incorporation of isotope labels from [13C]glucose is an observation that appears to be inconsistent with this model. Additional investigative work will be required to reconcile these seemingly divergent observations. These considerations notwithstanding, the model provided by Sekar and coworkers (124) provides an elegant demonstration of how nutrient sensing can be coupled to the divisome machinery to trigger first cell division at the end of lag phase.
LAG IS AN EVOLVABLE PHENOTYPE THAT CAN ALSO EMERGE DE NOVO
A prolonged lag phase appears to be a defense mechanism that allows bacteria to tolerate stress. For example, incubation experiments of Streptococcus pneumoniae revealed that serotypes with a greater propensity for invasive disease were found to have longer lag phases. This suggests that longer lag phases may contribute to the ability of S. pneumoniae to evade host immune responses (136). A similar case is now being made for antibiotic tolerance. The first instance of a relationship between longer lag periods and antibiotic tolerance was observed by Dean and Hinshelwood in 1957 (137). Bacteria with longer lag phases are more tolerant to antibiotics (138–145). For example, a sublethal exposure of Enterococcus faecium to a variety of antibiotics produced a lag period of up to 30 h. Following a prolonged lag phase, the antibiotic-treated culture divided faster and had a greater final culture density than did an antibiotic-free control (143). This suggests that adaptive processes occurred within the lag period of E. faecium that not only conferred antibiotic tolerance but also enhanced subsequent proliferation. An alarming finding of this study was that some lag periods were longer than the standardized cultivation period of 16 h to 24 h recommended (at the time of publication) by the Clinical and Laboratory Standards Institute. Prolonged lag phases may therefore result in an overestimation of the efficacy of antibiotics in routine antibacterial susceptibility tests (143). In the highlighted case, it would be interesting to periodically reexpose E. faecium to the same antibiotics to conclusively rule out the possibility of an antibiotic resistance mechanism emerging in situ and to lend further empirical support to a protective function of a prolonged lag period. How a “prolonged-lag phenotype” can confer increased antibiotic tolerance is discussed in Potential Avenues of Research, below.
Whether prolonged lag is an evolvable phenotype was investigated by Fridman and coworkers (144). The authors hypothesized that bacteria can evolve to extend the lag period in response to a sustained program of antibiotic exposure. For example, by adjusting the exposure time of E. coli to ampicillin, the authors were able to establish an approximately correlative relationship between the duration of lag time and the duration of antibiotic exposure (144). When ampicillin-tolerant E. coli cells were exposed to norfloxacin (an antibiotic with a mode of action distinct from that of ampicillin), tolerance to norfloxacin was also observed (144). This suggests that a prolonged lag is itself the adaptive trait that confers tolerance to antibiotics, a phenomenon the authors termed “tolerance by lag.” The cross-protection of ampicillin-adapted E. coli strains to norfloxacin also indicates that the mechanism of tolerance does not involve developing resistance to specific antibiotics but instead appears to involve a generalized adaptive response to antibiotic stress. Genome sequencing and restoration of wild-type alleles identified mutations in three genes that caused the prolonged lag period. Of these three genes, the function of two are known and are associated with pathways involving toxin-antitoxin modules and aminoacyl-tRNA synthetases (144). Collectively, these observations suggest that prolonged lag is an evolvable phenotype that confers broad tolerance to antibiotics.
Prolonged lag also appears to be an adoptable phenotype that may emerge de novo in response to antibiotic therapies. The phenomenon of “persister cells” has been at the forefront of our concern over antibiotic resistance ever since Bigger first discovered its involvement in the resistance of Staphylococcus spp. to penicillin in 1944 (146). Persisters are cells that survive killing by antibiotics but do not have genetic changes conferring antibiotic resistance (147). Vulin and coworkers (148) investigated the relationship between prolonged lag and the persister phenomenon in S. aureus. When S. aureus cells are sampled from host body sites and plated on solid medium, colonies show variation in sizes. Small colonies, known as small colony variants, possess the antibiotic persistence phenotype. Compared to control cells preexposed to neutral pH, S. aureus preexposed to acidic pH or sampled from mouse abscesses presented with a greater number of small colony variants when plated on solid agar. Automated imaging and time-lapse microscopy revealed that these small colony variants are a result of cells needing more time to exit lag phase (148). Antibiotic exposure increased the number of small colony variants produced. Subculturing of small colonies resulted in both large and small colonies appearing on agar. This demonstrates that prolonged lag in S. aureus is not a fixed phenotype but emerges de novo in response to antibiotic exposure (148). Prolonged lag in S. aureus is therefore an adoptable phenotype that confers antibiotic tolerance in vitro and in vivo (148). Broadly speaking, the phenotypic variance of S. aureus follows the same bet-hedging principle that is known to play a role in the survival of a bacterial population in response to environmental stressors (see History, Definitions, and Determinants of Lag-Phase Duration, above). In the case of antibiotics, the prolonged-lag phenotype is an example of how bacteria can generate a diversity of phenotypes within a genetically identical population to ensure that a subset of a kin community will survive and repopulate in response to an antibiotic regimen (149–151).
In other circumstances, a short-lag phenotype could also be advantageous. Bacteria that can rapidly divide and take advantage of new-found nutrients may outcompete bacteria that cannot. The expression and catalytic proficiency of enzymes are fine-tuned by evolution to maximize fitness (152). In this case, the duration of lag appears to be a phenotype that can be altered by evolutionary changes to the catalytic proficiency of essential enzymes involved during lag. Adenylate kinase (AdK), a phosphotransferase that reversibly converts ADP to ATP and AMP, provides an example. A mutational study of AdK by Adkar and coworkers (153) revealed that the total catalytic capacity of this enzyme (defined as the combination of catalytic rate and enzyme abundance) is inversely correlated with the duration of lag phase in E. coli. This was demonstrated by producing a series of mutations in AdK that resulted in longer lag phases (153). As rapid metabolism and cell proliferation are considered markers of fitness among bacteria, this research suggests that a short lag phase is a desirable phenotype that can be produced through mutations that increase the catalytic capacity of essential enzymes (153).
POTENTIAL AVENUES OF RESEARCH
(i) What strategies do bacteria employ to rejuvenate their population?
Pin and coworkers (89) observed distinct transcriptional programs depending on the age of the inoculated cultures. Old cells expressed fewer genes, did not upregulate aerobic metabolic genes, and had longer lag periods. The authors hypothesized that old cultures employ a strategy of rejuvenation through replicative dilution. Cell division is asymmetrical because one daughter cell retains old and damaged subcellular components (“old pole”), whereas the other daughter cell receives newly built materials (“new pole”). As lineages inherit old poles over several generations, the rate of cell division decreases and eventually stops, which is the bacterial equivalent of aging and natural death in higher organisms (154, 155). As cells with new poles divide exponentially, old and damaged cells become serially diluted. The authors therefore hypothesized that old bacterial populations rejuvenate by diluting cellular damage to the point of insignificance (90). Case in point, only in young E. coli cultures were genes related to DNA damage observed to be upregulated (90). The transcriptional profiling study of S. enterica (these cells were not aged) also observed an upregulation of genes that are responsible for the repair of oxidative damage to DNA and proteins (see Broad Expression Programs Produce Cellular Machinery Essential for Proliferation, above). The physiological study of B. cereus (these cells were not aged) also indicated that these cells can repair membrane damage during the lag phase (see Cellular Events Provide Physiological Markers of Lag, above). This difference in approach could be characterized as a “dilution” versus “repair” strategy and may offer a rationale as to why young and old E. coli cells have seemingly distinct transcriptional programs (89). Under what conditions do bacteria rejuvenate populations by repairing damage, and under what conditions is a dilutive strategy the preferred route? Do both strategies operate in tandem, or are molecular “switches” in place that dictate one strategy over another given particular circumstances? Exploring such questions may provide fundamental insights into aging and longevity.
(ii) Does iron accumulation help or hinder survival from antibiotic therapies?
Multiple classes of antibiotics stimulate oxidative stress. This oxidative stress, when combined with the effects of the targeted mode of action, increases the efficacy of antibiotics (156–163). Iron, through the Fenton reaction, also contributes to antibiotic efficacy (164–166). Transcriptional and metabolic profiling revealed an upregulation of iron recruitment genes in E. coli and S. enterica and a doubling of iron content during the lag phase of S. enterica (73, 89). One could therefore hypothesize that iron accumulation is a maladaptive trait of pathogens that are exposed to antibiotics. Yet, there appears to be a purpose to iron accumulation, however presently unclear the reasons (73, 106). A guiding paradigm within the medical community is that restricting iron bioavailability retards pathogenic growth (167–169). Research in iron nanoparticles and iron transporters has also suggested that overloading antibiotic-resistant bacteria with redox-active iron is a viable adjunct strategy for bacterial clearance and restoring susceptibility to antibiotics (170–173). What is the appropriate role of iron in modern antibiotic regimens? A clear understanding of the interplay of iron and antibiotics during the lag phase may help us answer these questions and to determine how the toxicity of iron can be exploited without also contributing to the metabolic needs of rapidly dividing pathogens.
(iii) How do cells “choose” the optimal metabolic strategy?
Madar and coworkers (85) observed that bacteria fed with less-preferred carbon sources such as arabinose displayed a biphasic lag phase; Lag1 is devoted exclusively to the production of bottleneck carbon-processing enzymes, whereas Lag2 involves a broader array of biosynthetic activities in anticipation of cell division (85). The authors observed that this biphasic program is consistent with “bang-bang” optimal control theory and minimum-time problems. For example, to have an elevator move from one floor to another, the fastest way (though not necessarily the safest!) is to produce maximum acceleration followed by maximum deceleration. Similarly, biphasic cells produce the shortest possible lag period by first devoting resources to carbon flow before transforming the broader metabolome. Schultz and Kishony (174) opined that such optimization could be dysfunctional in some circumstances. For example, cells exposed to DNA synthesis inhibitors do not appropriately adjust ribosome biosynthesis, resulting in an imbalance between the availability of ribosomes and nucleic acid template (175). Are there circumstances in which the biphasic program is disadvantageous? For example, biphasic cells could be vulnerable to perturbations of heat, osmolarity, free radical stress, or antibiotics, because resources are devoted to producing bottleneck carbon-processing enzymes. In these circumstances, a better strategy would involve a longer lag period wherein more resources are devoted to producing proteins with protective functions. What are the molecular switches governing monophasic and biphasic programs? Do bacteria respond to environmental stressors by avoiding biphasic programs even if only secondary foods are available? Questions such as these are central to our understanding of how bacteria regulate global metabolism and promote their reproductive fitness within adverse environments.
(iv) How does the prolonged-lag phenotype confer antibiotic tolerance?
A key concept in antibiotic tolerance is that bactericidal antibiotics do not merely inhibit the function of its target molecule but corrupt its function (176). For example, aminoglycosides cause ribosomes to suffer proofreading errors and premature termination, resulting in energy waste and the accumulation of toxic peptides (177). The β-lactams not only inhibit peptidoglycan synthesis but trigger autolysin and cell wall destabilization (178). Quinolones inhibit the ligase function of gyrase and topoisomerase but do not inhibit its DNA strand-breaking function, thereby transforming these housekeeping enzymes into DNA denaturants (179). If a bacterium were to arrest the function of the target molecule by some means (e.g., do not produce the target molecule, convert it into an inactive form, etc.), this bacterium would avoid the worst consequences of the antibiotic, albeit at the cost of reduced metabolic activity. As cells in a prolonged state of lag have lower metabolic activity than do actively dividing cells, it is possible that the increased antibiotic tolerance that is associated with the prolonged-lag phenotype is a consequence of its lower metabolic state. This may at least partially explain the tolerance by lag phenomenon discovered by Fridman and coworkers (144) (see Lag Is An Evolvable Phenotype That Can Also Emerge De Novo, above), as ampicillin (cell wall synthesis) and norfloxacin (DNA replication) target processes which are not expected to be significantly occurring within a prolonged period of lag. Specific mechanisms of arresting the function of antibiotic targets are also possible. For example, the ability of cells to dimerize ribosomes into an inactive state is important for tolerating aminoglycosides (180). The hibernation promoting factor, the protein responsible for dimerizing ribosomes in Listeria monocytogenes, is induced in response to carbon starvation, heat shock, excess salt, and excess ethanol, suggesting that ribosome dimerization is used to confer tolerance to other sources of stress (181). It is plausible that ribosome dimerization could be playing a role in conferring antibiotic tolerance within the prolonged-lag phenotype. Fridman and coworkers (144) also observed that toxin-antitoxin modules were implicated in this phenotype (see Lag Is An Evolvable Phenotype That Can Also Emerge De Novo, above). This is interesting because toxin-antitoxin modules have been implicated in other models of antibiotic tolerance (182). Considering that antibiotic tolerance is a leading cause of poor therapeutic outcomes involving pathogenic infections (183), determining how prolonged lag confers antibiotic tolerance may help us develop tools to combat persistent bacterial infections.
TOWARD A MORE COMPLETE UNDERSTANDING OF LAG
What exactly is lag phase? A common description is that lag is a transient period of nonreplication wherein cells prepare to divide by adapting to stress and by rebuilding cellular components. This characterization of lag as a preparative period is a description that is well supported by the available evidence. However, it is incomplete. Consider the following.
The lag phase is dynamic.
The duration of latency is influenced by the history of the inoculum (see History, Definitions, and Determinants of Lag-Phase Duration, above). Young and old bacteria present with distinct transcriptional programs and strategies of rejuvenating microbial populations (see Bacterial History Reflects a Dynamic Transcriptional Profile, above). Bacteria respond to primary or secondary carbon sources with monophasic or biphasic programs to minimize the lag time (see Optimization of Primary Metabolism Is Characteristic of Lag Phase, above).
The lag phase is organized.
Bacteria rapidly initiate a transcriptional program involving more than half of all genes in the genome, starting mere minutes after inoculation (see Broad Expression Programs Produce Cellular Machinery Essential for Proliferation, above). These changes are energetically sponsored by the digestion of glycogen reserves in a manner analogous to an energy bank (see Optimization of Primary Metabolism Is Characteristic of Lag Phase, above). Bacteria sense when nutrients become available and trigger first cell division in response (see Nutrient Sensing Is Coupled to Divisome Machinery To Trigger First Cell Division, above).
The lag phase is adaptive.
The heterogeneity of lag times among individual cells follows bet-hedging principles allowing kin populations to survive environmental (see History, Definitions, and Determinants of Lag-Phase Duration, above) and antibiotic (see Lag Is an Evolvable Phenotype That Can Also Emerge De Novo, above) threats. Stress produces varied lag periods that are associated with distinct physiological patterns, and in some cases, enhanced markers of fitness (see Cellular Events Provide Physiological Markers of Lag, above). Metal accumulation is speculated to be a competitive behavior enabling bacteria to evade host nutritional immunity (see Iron Influx during Lag Produces Oxidative Stress and Could Be Related to Immune Evasion, above). Prolonged lag is a phenotype that may emerge de novo in response to antibiotic exposure (see Lag Is An Evolvable Phenotype That Can Also Emerge De Novo, above).
The lag phase is evolvable.
Prolonged lag is an evolvable phenotype that confers cross-tolerance to diverse antibiotics. The total metabolic capacity of primary enzymes is optimized by evolution to minimize the duration of lag (see Lag Is An Evolvable Phenotype That Can Also Emerge De Novo, above).
These insights are not yet well reflected in educational material. Consider the following passages, taken from textbooks ranging from 1937 to 2019. Despite advances in our understanding of lag phase, how this period is described to students has remained virtually unchanged across eight decades.
1937: “During the first or lag phase there is no appreciable growth of the bacteria…. The true explanation of this phase is somewhat controversial, but it doubtless represents an adjustment of the bacterium to the change of environment” (184).
1951: “Some cells do show metabolic and growth lag, especially those cells with more fastidious nutritive requirements. This period may coincide with the time necessary for the concentration of essential intermediates to accumulate” (185).
1959: “There is first a period of ‘lag’ (e.g., two hours) during which there is no multiplication though there is increase in cell size accompanied by intense metabolic activity” (186).
1974: “Lag phase represents a period during which the dormant organisms used as inoculum are probably imbibing water, restoring RNA…possibly producing inducible enzymes to cope with new nutrient substances, swelling, and otherwise becoming adjusted to the new environment” (187).
2008: “Although cell division does not take place right away and there is no net increase in mass, the cell is synthesizing new components…. [T]he cells retool, replicate their DNA, begin to increase in mass, and finally divide” (188).
2019: “Cells begin synthesizing enzymes required for growth…. [I]f cells are transferred into a medium that contains fewer nutrients, the lag phase will be longer” (189).
Offering a preparative explanation of lag is certainly understandable given what we still do not know about latency, the difficulty of studying small number of cells, and the relatively recent reporting of many of its most impressionable facts. Nonetheless, lag is evidently a more complex phenomenon, however little we understand it. Lag is not merely preparative but is also dynamic, organized, adaptive, and evolvable. As new insights unfold, a more complete understanding of this phase may yet emerge, one that reflects the remarkable complexity of bacterial life and its relevance to diverse scientific disciplines.
ACKNOWLEDGMENTS
This work is supported by the Natural Sciences and Engineering Research Council of Canada.
I thank John Sorensen, Lucile Jeusset, and three anonymous reviewers for their helpful suggestions. I offer my sincere apologies to authors of relevant works who were not included in this review; the omission was unintentional.
I declare no conflicts of interest.
REFERENCES
- 1.Monod J. 1949. The growth of bacterial cultures. Annu Rev Microbiol 3:371–394. doi: 10.1146/annurev.mi.03.100149.002103. [DOI] [Google Scholar]
- 2.Müller M. 1895. Ueber die Einfluss von Fieber Temperaturen auf die Wachstumsgeschwindigkeit und die Virulenz des Typhus-Bacillus. Z Hyg Infektionskr 20:245–280. doi: 10.1007/BF02216656. [DOI] [Google Scholar]
- 3.Coplans M. 1910. Influences affecting the growth of microorganisms–latency: inhibition: mass action. J Pathol 14:1–27. doi: 10.1002/path.1700140102. [DOI] [Google Scholar]
- 4.Lane-Claypon JE. 1909. Multiplication of bacteria and the influence of temperature and some other conditions thereon. J Hyg (Lond) 9:239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Penfold WJ. 1914. On the nature of bacterial lag. J Hyg (Lond) 14:215–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ledingham JCG, Penfold WJ. 1914. Mathematical analysis of the lag-phase in bacterial growth. J Hyg (Lond) 14:242–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchanan RE. 1918. Life phases in a bacterial culture. J Infect Dis 23:109–125. doi: 10.1086/infdis/23.2.109. [DOI] [Google Scholar]
- 8.Levin-Reisman I, Gefen O, Fridman O, Ronin I, Shwa D, Sheftel H, Balaban NQ. 2010. Automated imaging with ScanLag reveals previously undetectable bacterial growth phenotypes. Nat Methods 7:737–739. doi: 10.1038/nmeth.1485. [DOI] [PubMed] [Google Scholar]
- 9.Stylianidou S, Brennan C, Nissen SB, Kuwada NJ, Wiggins PA. 2016. SuperSegger: robust image segmentation, analysis and lineage tracking of bacterial cells. Mol Microbiol 102:690–700. doi: 10.1111/mmi.13486. [DOI] [PubMed] [Google Scholar]
- 10.Butler D, Goel N, Goodnight L, Tadigadapa S, Ebrahimi A. Detection of bacterial metabolism in lag-phase using impedance spectroscopy of agar-integrated 3D microelectrodes. Biosens Bioelectron, in press. doi: 10.1016/j.bios.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 11.Swinnen IAM, Bernaerts K, Dens EJJ, Geeraerd AH, Van Impe JF. 2004. Predictive modelling of the bacterial lag phase: a review. Int J Food Microbiol 94:137–159. doi: 10.1016/j.ijfoodmicro.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Finkel SE. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol 4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- 13.Baty F, Delignette-Muller ML. 2004. Estimating the bacterial lag time: which model, which precision? Int J Food Microbiol 91:261–277. doi: 10.1016/j.ijfoodmicro.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Zwietering MH, Jongenburger I, Rombouts FM, van ‘t Riet K. 1990. Modeling of the bacterial growth curve. Appl Environ Microbiol 56:1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baranyi J, Roberts TA. 1994. A dynamic approach to predicting bacterial growth in food. Int J Food Microbiol 23:277–294. [DOI] [PubMed] [Google Scholar]
- 16.McKellar RC. 1997. A heterogeneous population model for the analysis of bacterial growth kinetics. Int J Food Microbiol 36:179–186. [DOI] [PubMed] [Google Scholar]
- 17.Baranyi J. 1998. Comparison of stochastic and deterministic concepts of bacterial lag. J Theor Biol 192:403–408. doi: 10.1006/jtbi.1998.0673. [DOI] [PubMed] [Google Scholar]
- 18.McKellar RC, Knight K. 2000. A combined discrete-continuous model describing the lag phase of Listeria monocytogenes. Int J Food Microbiol 54:171–180. [DOI] [PubMed] [Google Scholar]
- 19.Baranyi J. 2002. Stochastic modelling of bacterial lag phase. Int J Food Microbiol 73:203–206. [DOI] [PubMed] [Google Scholar]
- 20.Dens EJ, Bernaerts K, Standaert AR, Van Impe JF. 2005. Cell division theory and individual-based modeling of microbial lag. Part I. The theory of cell division. Int J Food Microbiol 101:303–318. doi: 10.1016/j.ijfoodmicro.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 21.Dens EJ, Bernaerts K, Standaert AR, Kreft JU, Van Impe JF. 2005. Cell division theory and individual-based modeling of microbial lag. Part II. Modeling lag phenomena induced by temperature shifts. Int J Food Microbiol 101:319–332. doi: 10.1016/j.ijfoodmicro.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 22.Métris A, Le Marc Y, Elfwing A, Ballagi A, Baranyi J. 2005. Modelling the variability of lag times and the first generation times of single cells of E. coli. Int J Food Microbiol 100:13–19. doi: 10.1016/j.ijfoodmicro.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Yates GT, Smotzer T. 2007. On the lag phase and initial decline of microbial growth curves. J Theor Biol 244:511–517. doi: 10.1016/j.jtbi.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 24.Huang L. 2011. A new mechanistic growth model for simultaneous determination of lag phase duration and exponential growth rate and a new Bĕlehdrádek-type model for evaluating the effect of temperature on growth rate. Food Microbiol 28:770–776. doi: 10.1016/j.fm.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 25.Pérez-Rodríguez F. 2014. Development and application of predictive microbiology models in foods, p 321–361. In Granato D, Ares G (ed), Mathematical and statistical methods in food science and technology. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 26.Mackey BM, Kerridge AL. 1988. The effect of incubation temperature and inoculum size on growth of Salmonellae in minced beef. Int J Food Microbiol 6:57–65. [DOI] [PubMed] [Google Scholar]
- 27.Notermans S, Soentoro PSS, Bolder NM, Mulder RWAW. 1991. Adaptation of Listeria monocytogenes in liquid egg containing sucrose resulting in survival and outgrowth. Int J Food Microbiol 13:55–62. doi: 10.1016/0168-1605(91)90136-D. [DOI] [PubMed] [Google Scholar]
- 28.Zaika LL, Kim AH, Ford L. 1991. Effect of sodium nitrite on growth of Shigella flexneri. J Food Prot 54:424–428. doi: 10.4315/0362-028X-54.6.424. [DOI] [PubMed] [Google Scholar]
- 29.Buchanan RL, Klawitter LA. 1992. The effect of incubation temperature, initial pH, and sodium chloride on the growth kinetics of Escherichia coli O157:H7. Food Microbiol 9:185–196. doi: 10.1016/0740-0020(92)80046-7. [DOI] [Google Scholar]
- 30.Hudson JA. 1993. Effect of pre-incubation temperature on the lag time of Aeromonas hydrophila. Lett Appl Microbiol 16:274–276. doi: 10.1111/j.1472-765X.1993.tb01417.x. [DOI] [Google Scholar]
- 31.Dufrenne J, Bijwaard M, Te Giffel M, Beumer R, Notermans S. 1995. Characteristics of some psychrotrophic Bacillus cereus isolates. Int J Food Microbiol 27:175–183. [DOI] [PubMed] [Google Scholar]
- 32.Gay M, Cerf O, Davey KR. 1996. Significance of pre-incubation temperature and inoculum concentration on subsequent growth of Listeria monocytogenes at 14°C. J Appl Bacteriol 81:433–438. doi: 10.1111/j.1365-2672.1996.tb03530.x. [DOI] [PubMed] [Google Scholar]
- 33.Dufrenne J, Delfgou E, Ritmeester W, Notermans S. 1997. The effect of previous growth conditions on the lag phase time of some foodborne pathogenic micro-organisms. Int J Food Microbiol 34:89–94. [DOI] [PubMed] [Google Scholar]
- 34.Membré JM, Ross T, McMeekin T. 1999. Behaviour of Listeria monocytogenes under combined chilling processes. Lett Appl Microbiol 28:216–220. [DOI] [PubMed] [Google Scholar]
- 35.Mellefont LA, McMeekin TA, Ross T. 2003. The effect of abrupt osmotic shifts on the lag phase duration of foodborne bacteria. Int J Food Microbiol 83:281–293. doi: 10.1016/S0168-1605(02)00377-X. [DOI] [PubMed] [Google Scholar]
- 36.Mellefont LA, Ross T. 2003. The effects of abrupt shifts in temperature on the lag phase duration of Escherichia coli and Klebsiella oxytoca. Int J Food Microbiol 83:295–305. [DOI] [PubMed] [Google Scholar]
- 37.Zhou K, George SM, Métris A, Li PL, Baranyi J. 2011. Lag phase of Salmonella enterica under osmotic stress conditions. Appl Environ Microbiol 77:1758–1762. doi: 10.1128/AEM.02629-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackey BM, Derrick CM. 1982. The effect of sublethal injury by heating, freezing, drying and gamma-radiation on the duration of the lag phase of Salmonella Typhimurium. J Appl Bacteriol 53:243–251. [DOI] [PubMed] [Google Scholar]
- 39.Mackey BM, Derrick CM. 1984. Conductance measurements of the lag phase of injured Salmonella Typhimurium. J Appl Bacteriol 57:299–308. [DOI] [PubMed] [Google Scholar]
- 40.Stephens PJ, Joynson JA, Davies KW, Holbrook R, Lappin-Scott HM, Humphrey TJ. 1997. The use of an automated growth analyzer to measure recovery times of single heat-injured Salmonella cells. J Appl Microbiol 83:445–455. doi: 10.1046/j.1365-2672.1997.00255.x. [DOI] [PubMed] [Google Scholar]
- 41.Augustin JC, Brouillaud-Delattre A, Rosso L, Carlier V. 2000. Significance of inoculum size in the lag time of Listeria monocytogenes. Appl Environ Microbiol 66:1706–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whiting RC, Bagi LK. 2002. Modeling the lag phase of Listeria monocytogenes. Int J Food Microbiol 73:291–295. [DOI] [PubMed] [Google Scholar]
- 43.Métris A, George SM, Baranyi J. 2006. Use of optical density detection times to assess the effect of acetic acid on single-cell kinetics. Appl Environ Microbiol 72:6674–6679. doi: 10.1128/AEM.00914-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasch M, Métris A, Baranyi J, Bjørn Budde B. 2007. The effect of reuterin on the lag time of single cells of Listeria innocua grown on a solid agar surface at different pH and NaCl concentrations. Int J Food Microbiol 113:35–40. doi: 10.1016/j.ijfoodmicro.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 45.Pin C, Baranyi J. 2008. Single-cell and population lag times as a function of cell age. Appl Environ Microbiol 74:2534–2536. doi: 10.1128/AEM.02402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dupont C, Augustin JC. 2009. Influence of stress on single-cell lag time and growth probability for Listeria monocytogenes in half Fraser broth. Appl Environ Microbiol 75:3069–3076. doi: 10.1128/AEM.02864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pascual C, Robinson TP, Ocio MJ, Aboaba OO, Mackey BM. 2001. The effect of inoculum size and sublethal injury on the ability of Listeria monocytogenes to initiate growth under suboptimal conditions. Lett Appl Microbiol 33:357–361. [DOI] [PubMed] [Google Scholar]
- 48.Robinson TP, Aboaba OO, Kaloti A, Ocio MJ, Baranyi J, Mackey BM. 2001. The effect of inoculum size on the lag phase of Listeria monocytogenes. Int J Food Microbiol 70:163–173. [DOI] [PubMed] [Google Scholar]
- 49.Webb MD, Stringer SC, Le Marc Y, Baranyi J, Peck MW. 2012. Does proximity to neighbours affect germination of spores of non-proteolytic Clostridium botulinum? Food Microbiol 32:104–109. doi: 10.1016/j.fm.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 50.Smelt JP, Otten GD, Bos AP. 2002. Modelling the effect of sublethal injury on the distribution of the lag times of individual cells of Lactobacillus plantarum. Int J Food Microbiol 73:207–212. [DOI] [PubMed] [Google Scholar]
- 51.Elfwing A, LeMarc Y, Baranyi J, Ballagi A. 2004. Observing growth and division of large numbers of individual bacteria by image analysis. Appl Environ Microbiol 70:675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Francois K, Devlieghere F, Smet K, Standaert AR, Geeraerd AH, Van Impe JF, Debevere J. 2005. Modelling the individual cell lag phase: effect of temperature and pH on the individual cell lag distribution of Listeria monocytogenes. Int J Food Microbiol 100:41–53. doi: 10.1016/j.ijfoodmicro.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 53.Guillier L, Pardon P, Augustin JC. 2005. Influence of stress on individual lag time distributions of Listeria monocytogenes. Appl Environ Microbiol 71:2940–2948. doi: 10.1128/AEM.71.6.2940-2948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D’Arrigo M, Garcia de Fernando GD, Velasco de Diego R, Ordóñez JA, George SM, Pin C. 2006. Indirect measurement of the lag time distribution of single cells of Listeria innocua in food. Appl Environ Microbiol 72:2533–2538. doi: 10.1128/AEM.72.4.2533-2538.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Odumeru JA, Griffiths M, McKellar MC. 2006. Effect of environmental stresses on the mean and distribution of individual lag times of Escherichia coli O157:H7. Int J Food Microbiol 110:278–285. doi: 10.1016/j.ijfoodmicro.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 56.Webb MD, Pin C, Peck MW, Stringer SC. 2007. Historical and contemporary NaCl concentrations affect the duration and distribution of lag times from individual spores of nonproteolytic Clostridium botulinum. Appl Environ Microbiol 73:2118–2127. doi: 10.1128/AEM.01744-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Métris A, George SM, Mackey BM, Baranyi J. 2008. Modeling the variability of single-cell lag times for Listeria innocua populations after sublethal and lethal heat treatments. Appl Environ Microbiol 74:6949–6955. doi: 10.1128/AEM.01237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 59.Kussell E, Leibler S. 2005. Phenotypic diversity, population growth, and information on fluctuating environments. Science 309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 60.Geisel N, Vilar JMG, Rubi JM. 2011. Optimal resting-growth strategies of microbial populations in fluctuating environments. PLoS One 6:e18622. doi: 10.1371/journal.pone.0018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morita RY. 1988. Bioavailability of energy and its relationship to growth and starvation survival in nature. Can J Microbiol 34:436–441. doi: 10.1139/m88-076. [DOI] [Google Scholar]
- 62.Hobbie JE, Hobbie EA. 2013. Microbes in nature are limited by carbon and energy: the starving-survival lifestyle in soil and consequences for estimating microbial rates. Front Microbiol 4:324. doi: 10.3389/fmicb.2013.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pletnev P, Osterman I, Sergiev P, Bogdanov A, Dontsova O. 2015. Survival guide: Escherichia coli in the stationary phase. Acta Naturae 7:22–33. [PMC free article] [PubMed] [Google Scholar]
- 64.Jaishankar J, Srivastava P. 2017. Molecular basis of stationary phase survival and applications. Front Microbiol 8:2000. doi: 10.3389/fmicb.2017.02000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berlett BS, Stadtman ER. 1997. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem 272:20313–20316. [DOI] [PubMed] [Google Scholar]
- 66.Dukan S, Nyström T. 1998. Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev 12:3431–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dukan S, Nyström T. 1999. Oxidative stress defense and deterioration of growth-arrested Escherichia coli. J Biol Chem 274:26027–26032. [DOI] [PubMed] [Google Scholar]
- 68.Nyström T. 2003. The free-radical hypothesis of aging goes prokaryotic. Cell Mol Life Sci 60:1333–1341. doi: 10.1007/s00018-003-2310-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saint-Ruf C, Pesut J, Sopta M, Matic I. 2007. Causes and consequences of DNA repair activity modulation during stationary phase in Escherichia coli. Crit Rev Biochem Mol Biol 42:259–270. doi: 10.1080/10409230701495599. [DOI] [PubMed] [Google Scholar]
- 70.Rechinger KB, Siegumfeldt H, Svendsen I, Jakobsen M. 2000. Early” protein synthesis of Lactobacillus delbrueckii ssp. bulgaricus in milk revealed by [35S]-methionine labeling and two-dimensional gel electrophoresis. Electrophoresis 21:2660–2669. doi:. [DOI] [PubMed] [Google Scholar]
- 71.Hornbaek T, Jakobsen M, Dynesen J, Nielsen AK. 2004. Global transcription profiles and intracellular pH regulation measured in Bacillus licheniformis upon external pH upshifts. Arch Microbiol 182:467–474. doi: 10.1007/s00203-004-0729-6. [DOI] [PubMed] [Google Scholar]
- 72.Larsen N, Boye M, Siegumfeldt H, Jakobsen M. 2006. Differential expression of proteins and genes in the lag phase of Lactococcus lactis subsp. lactis grown in synthetic medium and reconstituted skim milk. Appl Environ Microbiol 72:1173–1179. doi: 10.1128/AEM.72.2.1173-1179.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rolfe MD, Rice CJ, Lucchini S, Pin C, Thompson A, Cameron ADS, Alston M, Stringer MF, Betts RP, Baranyi J, Peck MW, Hinton JCD. 2012. Lag phase is a distinct growth phase that prepares bacteria for exponential growth and involves transient metal accumulation. J Bacteriol 194:686–701. doi: 10.1128/JB.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feklístov A, Sharon BD, Darst SA, Gross CA. 2014. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 68:357–376. doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- 76.Paget MS. 2015. Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules 5:1245–1265. doi: 10.3390/biom5031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bradley MD, Beach MB, de Koning AP, Pratt TS, Osuna R. 2007. Effects of fis on Escherichia coli gene expression during different growth stages. Microbiology 153:2922–2940. doi: 10.1099/mic.0.2007/008565-0. [DOI] [PubMed] [Google Scholar]
- 78.Osuna R, Lienau D, Hughes KT, Johnson RC. 1995. Sequence, regulation, and function of fis in Salmonella typhimurium. J Bacteriol 177:2021–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. 2005. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell 18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 80.Beckert B, Turk M, Czech A, Berninghausen O, Beckmann R, Ignatova Z, Plitzko JM, Wilson DN. 2018. Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1. Nat Microbiol 3:1115–1121. doi: 10.1038/s41564-018-0237-0. [DOI] [PubMed] [Google Scholar]
- 81.Gohara DW, Yap MNF. 2018. Survival of the drowsiest: the hibernating 100S ribosome in bacterial stress management. Curr Genet 64:753–760. doi: 10.1007/s00294-017-0796-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aiso T, Yoshida H, Wada A, Ohki R. 2005. Modulation of mRNA stability participates in stationary-phase-specific expression of ribosome modulation factor. J Bacteriol 187:1951–1958. doi: 10.1128/JB.187.6.1951-1958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wada A. 1998. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells 3:203–208. [DOI] [PubMed] [Google Scholar]
- 84.Watanabe S, Ohbayashi R, Kanesaki Y, Saito N, Chibazakura T, Soga T, Yoshikawa H. 2015. Intensive DNA replication and metabolism during the lag phase of cyanobacteria. PLoS One 10:e0136800. doi: 10.1371/journal.pone.0136800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Madar D, Dekel E, Bren A, Zimmer A, Porat Z, Alon U. 2013. Promoter activity dynamics in the lag phase of Escherichia coli. BMC Syst Biol 7:136. doi: 10.1186/1752-0509-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McKellar RC. 2007. Effect of starvation on expression of the ribosomal RNA rrnB P2 promoter during the lag phase of Pseudomonas fluorescens. Int J Food Microbiol 114:307–315. doi: 10.1016/j.ijfoodmicro.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 87.McKellar RC. 2007. Effect of sub-lethal heating and growth temperature on expression of the ribosomal RNA rrnB P2 promoter during the lag phase of Pseudomonas fluorescens. Int J Food Microbiol 116:248–259. doi: 10.1016/j.ijfoodmicro.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 88.McKellar RC. 2008. Correlation between the change in the kinetics of the ribosomal RNA rrnB P2 promoter and the transition from lag to exponential phase with Pseudomonas fluorescens. Int J Food Microbiol 121:11–17. doi: 10.1016/j.ijfoodmicro.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 89.Pin C, Rolfe MD, Muñoz-Cuevas M, Hinton JCD, Peck MW, Walton NJ, Baranyi J. 2009. Network analysis of the transcriptional pattern of young and old cells of Escherichia coli during lag phase. BMC Syst Biol 3:108. doi: 10.1186/1752-0509-3-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yamamotoya T, Dose H, Tian Z, Fauré A, Toya Y, Honma M, Igarashi K, Nakahigashi K, Soga T, Mori H, Matsuno H. 2012. Glycogen is the primary source of glucose during the lag phase of E. coli proliferation. Biochim Biophys Acta 1824:1442–1448. doi: 10.1016/j.bbapap.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 91.Biesta-Peters EG, Mols M, Reij MW, Abee T. 2011. Physiological parameters of Bacllius cereus marking the end of acid-induced lag phases. Int J Food Microbiol 148:42–47. doi: 10.1016/j.ijfoodmicro.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 92.Iovieno P, Bååth E. 2008. Effect of drying and rewetting on bacterial growth rates in soil. FEMS Microbiol Ecol 65:400–407. doi: 10.1111/j.1574-6941.2008.00524.x. [DOI] [PubMed] [Google Scholar]
- 93.Meisner A, Bååth E, Rousk J. 2013. Microbial growth responses upon rewetting soil dried for four days or one year. Soil Biol Biochem 66:188–192. doi: 10.1016/j.soilbio.2013.07.014. [DOI] [Google Scholar]
- 94.Meisner A, Rousk J, Bååth E. 2015. Prolonged drought changes the bacterial growth response to rewetting. Soil Biol Biochem 88:314–322. doi: 10.1016/j.soilbio.2015.06.002. [DOI] [Google Scholar]
- 95.Meisner A, Leizeaga A, Rousk J, Bååth E. 2017. Partial drying accelerates bacterial growth recovery to rewetting. Soil Biol Biochem 112:269–276. doi: 10.1016/j.soilbio.2017.05.016. [DOI] [Google Scholar]
- 96.Korshunov S, Imlay JA. 2006. Detection and quantification of superoxide formed within the periplasm of Escherichia coli. J Bacteriol 188:6326–6334. doi: 10.1128/JB.00554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mailloux RJ. 2015. Teaching the fundamentals of electron transfer reactions in mitochondria and the production and detection of reactive oxygen species. Redox Biol 4:381–398. doi: 10.1016/j.redox.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brand MD. 2016. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic Biol Med 100:14–31. doi: 10.1016/j.freeradbiomed.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 101.Pisoschi AM, Pop A. 2015. The role of antioxidants in the chemistry of oxidative stress: a review. Eur J Med Chem 97:55–74. doi: 10.1016/j.ejmech.2015.04.040. [DOI] [PubMed] [Google Scholar]
- 102.Sies H, Berndt C, Jones DP. 2017. Oxidative stress. Annu Rev Biochem 86:715–748. doi: 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- 103.Cuny C, Lesbats M, Dukan S. 2007. Induction of a global stress response during the first step of Escherichia coli plate growth. Appl Environ Microbiol 73:885–889. doi: 10.1128/AEM.01874-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Henle ES, Linn S. 1997. Formation, prevention and repair of DNA damage by iron/hydrogen peroxide. J Biol Chem 272:19095–19098. [DOI] [PubMed] [Google Scholar]
- 105.Frawley ER, Fang FC. 2014. The ins and outs of bacterial iron metabolism. Mol Microbiol 93:609–616. doi: 10.1111/mmi.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bertrand RL. 2014. Lag phase-associated iron accumulation is likely a microbial counter-strategy to host iron sequestration: role of the ferric uptake regulator. J Theor Biol 359:72–79. doi: 10.1016/j.jtbi.2014.05.039. [DOI] [PubMed] [Google Scholar]
- 107.Skaar EP. 2010. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog 6:e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cassat JE, Skaar EP. 2013. Iron in infection and immunity. Cell Host Microbe 13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martins AC, Almeida JI, Lima IS, Kapitão AS, Gozzelino R. 2017. Iron metabolism and the inflammatory response. IUBMB Life 69:442–450. doi: 10.1002/iub.1635. [DOI] [PubMed] [Google Scholar]
- 110.Troxell B, Hassan HM. 2013. Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol 3:59. doi: 10.3389/fcimb.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Niederhoffer EC, Naranjo CM, Bradley KL, Fee JA. 1990. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J Bacteriol 172:1930–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moody CS, Hassan HM. 1984. Anaerobic biosynthesis of the manganese-containing superoxide dismutase in Escherichia coli. J Biol Chem 259:12821–12825. [PubMed] [Google Scholar]
- 113.Bertrand R, Danielson D, Gong V, Olynik B, Eze MO. 2012. Sodium nitroprusside may modulate Escherichia coli antioxidant enzyme expression by interacting with the ferric uptake regulator. Med Hypotheses 78:130–133. doi: 10.1016/j.mehy.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 114.Bertrand RL, Eze MO. 2013. Escherichia coli superoxide dismutase expression does not change in response to iron challenge during lag phase: Is the ferric uptake regulator to blame? Adv Enzyme Res 4:132–141. doi: 10.4236/aer.2013.14014. [DOI] [Google Scholar]
- 115.Bertrand RL, Eze MO. 2014. Modifying polyacrylamide background color for the nitroblue tetrazolium-based superoxide dismutase staining assay. Adv Enzyme Res 2:77–81. doi: 10.4236/aer.2014.22008. [DOI] [Google Scholar]
- 116.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. 1995. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: Protective role of superoxide dismutase. J Bacteriol 177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sheldon JR, Heinrichs DE. 2015. Recent developments in understanding the iron acquisition strategies of Gram-positive pathogens. FEMS Microbiol Rev 39:592–630. doi: 10.1093/femsre/fuv009. [DOI] [PubMed] [Google Scholar]
- 118.Willis L, Huang KC. 2017. Sizing up the bacterial cell cycle. Nat Rev Microbiol 15:606–620. doi: 10.1038/nrmicro.2017.79. [DOI] [PubMed] [Google Scholar]
- 119.Eswara PJ, Ramamurthi KS. 2017. Bacterial cell division: nonmodels poised to take the spotlight. Annu Rev Microbiol 71:393–411. doi: 10.1146/annurev-micro-102215-095657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Robert L. 2015. Size sensors in bacteria, cell cycle control, and size control. Front Microbiol 6:515. doi: 10.3389/fmicb.2015.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Adams DW, Wu LJ, Errington J. 2014. Cell cycle regulation by the bacterial nucleoid. Curr Opin Microbiol 22:94–101. doi: 10.1016/j.mib.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bi E, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 123.Haeusser DP, Margolin W. 2016. Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat Rev Microbiol 14:305–319. doi: 10.1038/nrmicro.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sekar K, Rusconi R, Sauls JT, Fuhrer T, Noor E, Nguyen J, Fernandez VI, Buffing MF, Berney M, Jun S, Stocker R, Sauer U. 2018. Synthesis and degradation of FtsZ quantitatively predict the first cell division in starved bacteria. Mol Syst Biol 14:e8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Göhler AK, Kökpinar Ö, Schmidt-Heck W, Geffers R, Guthke R, Rinas U, Schuster S, Jahreis K, Kaleta C. 2011. More than just a metabolic regulator–elucidation and validation of new targets of PdhR in Escherichia coli. BMC Syst Biol 5:197. doi: 10.1186/1752-0509-5-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Quail MA, Haydon DJ, Guest JR. 1994. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol Microbiol 12:95–104. [DOI] [PubMed] [Google Scholar]
- 127.Franchini AG, Ihssen J, Egli T. 2015. Effect of global regulators RpoS and cyclic-AMP/CRP on the catabolome and transcriptome of Escherichia coli K12 during carbon- and energy-limited growth. PLoS One 10:e0133793. doi: 10.1371/journal.pone.0133793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ishizuka H, Hanamura A, Inada T, Aiba H. 1994. Mechanism of the down-regulation of cAMP receptor protein by glucose in Escherichia coli: role of autoregulation of the crp gene. EMBO J 13:3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Camberg JL, Hoskins JR, Wickner S. 2009. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc Natl Acad Sci U S A 106:10614–10619. doi: 10.1073/pnas.0904886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sugimoto S, Yamanaka K, Nishikori S, Miyagi A, Ando T, Ogura T. 2010. AAA+ chaperone CplX regulates dynamics of prokaryotic cytoskeletal protein FtsZ. J Biol Chem 285:6648–6657. doi: 10.1074/jbc.M109.080739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hill NS, Buske PJ, Shi Y, Levin PA. 2013. A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLoS Genet 9:e1003663. doi: 10.1371/journal.pgen.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Weart RB, Lee AH, Chien AC, Haeusser DP, Hill NS, Levin PA. 2007. A metabolic sensor governing cell size in bacteria. Cell 130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Monahan LG, Hajduk IV, Blaber SP, Charles IG, Harry EJ. 2014. Coordinating bacterial cell division with nutrient availability: a role for glycolysis. mBio 5:e00935-14. doi: 10.1128/mBio.00935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weart RB, Levin PA. 2003. Growth rate-dependent regulation of medial FtsZ ring formation. J Bacteriol 185:2826–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rueda S, Vicente M, Mingorance J. 2003. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J Bacteriol 185:3344–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bättig P, Hathaway LJ, Hofer S, Mühlemann K. 2006. Serotype-specific invasiveness and colonization prevalence in Streptococcus pneumoniae correlate with the lag phase during in vitro growth. Microbes Infect 8:2612–2617. doi: 10.1016/j.micinf.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 137.Dean ACR, Hinshelwood CN. 1957. The adaptation of bacterial cultures during the lag phase in media containing new substrates or antibacterial agents. Proc R Soc Lond B Biol Sci 147:247–257. [DOI] [PubMed] [Google Scholar]
- 138.Frimodt-Møller N, Sebbesen O, Thomsen VF. 1983. The Pneumococcus and the mouse protection test: importance of the lag phase in vivo. Chemotherapy 29:128–134. doi: 10.1159/000238186. [DOI] [PubMed] [Google Scholar]
- 139.Drummond LJ, Smith DG, Poxton IR. 2003. Effects of sub-MIC concentrations of antibiotics on growth of and toxin production by Clostridium difficile. J Med Microbiol 52:1033–1038. doi: 10.1099/jmm.0.05387-0. [DOI] [PubMed] [Google Scholar]
- 140.Oikonomou O, Panopoulou M, Ikonomidis A. 2011. Investigation of carbapenem heteroresistance among different sequence types of Pseudomonas aeruginosa clinical isolates reveals further diversity. J Med Microbiol 60:1556–1558. doi: 10.1099/jmm.0.032276-0. [DOI] [PubMed] [Google Scholar]
- 141.Izutani N, Imazato S, Nakajo K, Takahashi N, Takahashi Y, Ebisu S, Russell RR. 2011. Effects of the antibacterial monomer 12-methacryloyloxydodecylpyridinium bromide (MDPB) on bacterial viability and metabolism. Eur J Oral Sci 119:175–181. doi: 10.1111/j.1600-0722.2011.00817.x. [DOI] [PubMed] [Google Scholar]
- 142.Ottosson JR, Jarnheimer P, Stenström TA, Olsen B. 2012. A longitudinal study of antimicrobial resistant faecal bacteria in sediments collected from a hospital wastewater system. Infect Ecol Epidemiol 2:7438. doi: 10.3402/iee.v2i0.7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Theophel K, Schacht VJ, Schlüter M, Schnell S, Stingu CS, Schaumann R, Bunge M. 2014. The importance of growth kinetic analysis in determining bacterial susceptibility against antibiotics and silver nanoparticles. Front Microbiol 5:544. doi: 10.3389/fmicb.2014.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. 2014. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- 145.Li B, Qiu Y, Shi H, Yin H. 2016. The importance of lag time extension in determining bacterial resistance to antibiotics. Analyst 141:3059–3067. doi: 10.1039/c5an02649k. [DOI] [PubMed] [Google Scholar]
- 146.Bigger JW. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilization. Lancet 244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 147.Fisher RA, Gollan B, Helaine S. 2017. Persistent bacterial infections and persister cells. Nat Rev Microbiol 15:453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 148.Vulin C, Leimer N, Huemer M, Ackermann M, Zinkernagel AS. 2018. Prolonged bacterial lag time results in small colony variants that represent a sub-population of persisters. Nat Commun 9:4074. doi: 10.1038/s41467-018-06527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dhar N, McKinney JD. 2007. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr Opin Microbiol 10:30–38. doi: 10.1016/j.mib.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 150.Gefen O, Balaban NQ. 2009. The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol Rev 33:704–717. doi: 10.1111/j.1574-6976.2008.00156.x. [DOI] [PubMed] [Google Scholar]
- 151.Sánchez-Romero MA, Casadesús J. 2014. Contribution of phenotypic heterogeneity to adaptive antibiotic resistance. Proc Natl Acad Sci U S A 111:355–360. doi: 10.1073/pnas.1316084111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dekel E, Alon U. 2005. Optimality and evolutionary tuning of the expression level of a protein. Nat Lett 436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- 153.Adkar BV, Manhart M, Bhattacharyya S, Tian J, Musharbash M, Shakhnovich EI. 2017. Optimization of lag phase shapes the evolution of a bacterial enzyme. Nat Ecol Evol 1:149. doi: 10.1038/s41559-017-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ackermann M, Stearns SC, Jenal U. 2003. Senescence in a bacterium with asymmetric division. Science 300:1920. doi: 10.1126/science.1083532. [DOI] [PubMed] [Google Scholar]
- 155.Stewart EJ, Madden R, Paul G, Taddei F. 2005. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol 3:e45. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Albesa I, Becerra MC, Battán PC, Páez PL. 2004. Oxidative stress in the antibacterial action of different antibiotics. Biochem Biophys Res Commun 317:605–609. doi: 10.1016/j.bbrc.2004.03.085. [DOI] [PubMed] [Google Scholar]
- 157.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. 2007. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol 3:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 159.Dwyer DJ, Kohanski MA, Collins JJ. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol 12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang X, Zhao X. 2009. Contribution of oxidative damage to antimicrobial lethality. Antimicrob Agents Chemother 53:1395–1402. doi: 10.1128/AAC.01087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dwyer DJ, Belenky P, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dwyer DJ, Collins JJ, Walker GC. 2015. Unraveling the physiological complexities of antibiotic lethality. Annu Rev Pharmacol Toxicol 55:313–332. doi: 10.1146/annurev-pharmtox-010814-124712. [DOI] [PubMed] [Google Scholar]
- 163.Van Acker H, Coenye T. 2017. The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol 25:456–466. doi: 10.1016/j.tim.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 164.Ajiboye TO, Skiebe E, Wilharm G. 2018. Contributions of ferric uptake regulator Fur to the sensitivity and oxidative response of Acinetobacter baumannii to antibiotics. Microb Pathog 119:35–41. doi: 10.1016/j.micpath.2018.03.060. [DOI] [PubMed] [Google Scholar]
- 165.Yeom J, Imlay JA, Park W. 2010. Iron homeostasis affects antibiotic-mediated cell death in Pseudomonas species. J Biol Chem 285:22689–22695. doi: 10.1074/jbc.M110.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ezraty B, Barras F. 2016. The ‘liaisons dangereuses’ between iron and antibiotics. FEMS Microbiol Rev 40:418–435. doi: 10.1093/femsre/fuw004. [DOI] [PubMed] [Google Scholar]
- 167.Foley TL, Simeonov A. 2012. Targeting iron assimilation to develop new antibacterials. Expert Opin Drug Discov 7:831–847. doi: 10.1517/17460441.2012.708335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cross JH, Bradbury RS, Fulford AJ, Jallow AT, Wegmüller R, Prentice AM, Cerami C. 2015. Oral iron acutely elevates bacterial growth in human serum. Sci Rep 5:16670. doi: 10.1038/srep16670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ganz T, Nemeth E. 2015. Iron homeostasis in host defence and inflammation. Nat Rev Immunol 15:500–510. doi: 10.1038/nri3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hussein-Al-Ali SH, Zowalaty Me E, Hussein MZ, Geilich BM, Webster TJ. 2014. Synthesis, characterization, and antimicrobial activity of an ampicillin-conjugated magnetic nanoantibiotic for medical applications. Int J Nanomedicine 9:3801–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ma L, Gao Y, Maresso AW. 2015. Escherichia coli free-radical-based killing mechanism driven by a unique combination of iron restriction and certain antibiotics. J Bacteriol 197:3708–3719. doi: 10.1128/JB.00758-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Aparicio-Caamaño M, Carrillo-Morales M, Olivares-Trejo JDJ. 2016. Iron oxide nanoparticle improves the antibacterial activity of erythromycin. J Bacteriol Parasitol 7:267. [Google Scholar]
- 173.Shi S, Jia J, Guo X, Zhao Y, Chen D, Guo Y, Zhang X. 2016. Reduced Staphylococcus aureus biofilm formation in the presence of chitosan-coated iron oxide nanoparticles. Int J Nanomedicine 11:6499–6506. doi: 10.2147/IJN.S41371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Schultz D, Kishony R. 2013. Optimization and control in bacterial lag phase. BMC Biol 11:120. doi: 10.1186/1741-7007-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bollenbach T, Quan S, Chait R, Kishony R. 2009. Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell 139:707–718. doi: 10.1016/j.cell.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 177.Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. 1999. Aminoglycosides: activity and resistance. Antimicrob Agents Chemother 43:727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kitano K, Tomasz A. 1979. Triggering of autolytic cell wall degradation in Escherichia coli by beta-lactam antibiotics. Antimicrob Agents Chemother 16:838–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Aldred KJ, Kerns RJ, Osheroff N. 2014. Mechanism of quinolone action and resistance. Biochemistry 53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.McKay SL, Portnoy DA. 2015. Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob Agents Chemother 59:6992–6999. doi: 10.1128/AAC.01532-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kline BC, McKay SL, Tang WW, Portnoy DA. 2015. The Listeria monocytogenes hibernation-promoting factor is required for the formation of 100S ribosomes, optimal fitness, and pathogenesis. J Bacteriol 197:581–591. doi: 10.1128/JB.02223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Harms A, Maisonneuve E, Gerdes K. 2016. Mechanism of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 183.Meylan S, Andrews IW, Collins JJ. 2018. Targeting antibiotic tolerance, pathogen by pathogen. Cell 172:1228–1238. doi: 10.1016/j.cell.2018.01.037. [DOI] [PubMed] [Google Scholar]
- 184.Fairbrother RW. 1937. A textbook of medical bacteriology. William Heinemann, London, United Kingdom, p 50. [Google Scholar]
- 185.Werkman CH, Wilson PW. 1951. Bacterial physiology. Academic Press, New York, NY, p 118. [Google Scholar]
- 186.Mackie TJ, McCartney JE. 1959. Handbook of practical bacteriology, 9th ed E&S Livingstone, London, United Kingdom, p 26. [Google Scholar]
- 187.Frobisher M, Hinsdill R, Crabtree KT, Goodheart CR. 1974. Fundamentals of microbiology, 9th ed WB Saunders, Philadelphia, PA, p 139. [Google Scholar]
- 188.Willey JM, Sherwood LM, Woolverton CJ. 2008. Prescott, Harley, and Klein’s microbiology, 7th ed McGraw Hill, New York, NY, p 123. [Google Scholar]
- 189.Anderson D, Salm S, Allen D. 2019. Nester’s microbiology: a human perspective, 9th ed McGraw Hill, New York, NY, p 97. [Google Scholar]