Topological entanglement between daughter chromosomes has to be reduced to exactly zero every time an E. coli cell divides. The enzymatic agents that accomplish this task are the topoisomerases. E. coli possesses four topoisomerases. It has been thought that topoisomerase IV is primarily responsible for unlinking the daughter chromosomes during DNA replication. We show here that topoisomerase III also plays a role in this process and is specifically localized to the replisome, the multiprotein machine that duplicates the cell’s genome, in order to do so.
KEYWORDS: DNA replication, DNA topoisomerase, DNA topology, chromosome segregation
ABSTRACT
The role of DNA topoisomerase III (Topo III) in bacterial cells has proven elusive. Whereas eukaryotic Top IIIα homologs are clearly involved with homologs of the bacterial DNA helicase RecQ in unraveling double Holliday junctions, preventing crossover exchange of genetic information at unscheduled recombination intermediates, and Top IIIβ homologs have been shown to be involved in regulation of various mRNAs involved in neuronal function, there is little evidence for similar reactions in bacteria. Instead, most data point to Topo III playing a role supplemental to that of topoisomerase IV in unlinking daughter chromosomes during DNA replication. In support of this model, we show that Escherichia coli Topo III associates with the replication fork in vivo (likely via interactions with the single-stranded DNA-binding protein and the β clamp-loading DnaX complex of the DNA polymerase III holoenzyme), that the DnaX complex stimulates the ability of Topo III to unlink both catenated and precatenated DNA rings, and that ΔtopB cells show delayed and disorganized nucleoid segregation compared to that of wild-type cells. These data argue that Topo III normally assists topoisomerase IV in chromosome decatenation by removing excess positive topological linkages at or near the replication fork as they are converted into precatenanes.
IMPORTANCE Topological entanglement between daughter chromosomes has to be reduced to exactly zero every time an E. coli cell divides. The enzymatic agents that accomplish this task are the topoisomerases. E. coli possesses four topoisomerases. It has been thought that topoisomerase IV is primarily responsible for unlinking the daughter chromosomes during DNA replication. We show here that topoisomerase III also plays a role in this process and is specifically localized to the replisome, the multiprotein machine that duplicates the cell’s genome, in order to do so.
INTRODUCTION
DNA topoisomerases are critical components of essentially all macromolecular transactions on DNA, acting to untangle DNA strands either by passing either single-stranded DNA (ssDNA) (type IA topoisomerases) or double-stranded DNA (type II topoisomerases) through itself or by allowing free rotation of one DNA strand about the other (type IB topoisomerases) (1). Escherichia coli possesses four topoisomerases: two type IA (topoisomerase I [Topo I] and Topo III) and two type II (DNA gyrase and Topo IV). The roles of three of these topoisomerases are well established (2–4). DNA gyrase removes positive supercoils formed during DNA replication by converting them to negative supercoils and helps maintain an appropriate overall superhelical density of the chromosome. Topo IV also removes positive supercoils as well as precatenanes (windings of the two partially replicated sister duplexes about each other) during replication and decatenates the fully replicated sister chromosomes to ensure the complete removal of all topological linkages. Topo I, which interacts with RNA polymerase, removes excess negative supercoils formed during transcription and cooperates with DNA gyrase to maintain global supercoiling. The role of Topo III is less clear.
Topo III was identified initially in the Cozzarelli lab as a type I topoisomerase that had a DNA cleavage pattern different from that of Topo I (5). Srivenugopal et al. (6) also purified the enzyme as a type I topoisomerase that bound, oddly enough, to a novobiocin affinity column (the enzyme is not inhibited by this antibiotic). We subsequently purified Topo III as an activity detected in soluble extracts of E. coli that could resolve the late replication intermediates and catenated dimers formed in a reconstituted pBR322 DNA replication system in the presence of DNA gyrase (7) and cloned the gene, topB, encoding the enzyme (8). We demonstrated that Topo III had a strong preference for binding to ssDNA as its site of action, requiring single-strand gaps in the nascent strands to decatenate the linked sister chromosomes and being capable of efficiently relaxing negatively supercoiled DNA only at elevated temperature (8). A remarkable finding was that Topo III was able to form a covalent complex with RNA (9), and the enzyme was shown subsequently to also be an RNA topoisomerase, capable of knotting circular RNA (10). Interestingly, unlike the case for the metazoan homolog (see below), no role for the bacterial enzyme in RNA metabolism has emerged.
In both yeast and higher eukaryotes, the Topo III homolog Top3α was shown to physically and functionally associate with the RecQ homologs Sgs1 and BLM, respectively, to take apart double Holliday junctions that form between replicating sister chromosomes during unscheduled recombination likely stimulated by replication fork stalling, resulting in the suppression of genetic crossover events (11). More recently (12), it was shown that the Topo III metazoan homolog Top3β interacted with TDRD3 and that the Top3β-TDRD3 complex interacted with FMRP, an RNA-binding protein deficient in fragile X syndrome that regulates translation of mRNAs involved in neural function (13). Top3β can be found associated with polyribosomes, suggesting that Top3β might be involved in the regulation of gene expression, perhaps via its RNA Topo activity (14, 15).
Functional, but not physical, interaction between E. coli RecQ and Topo III has been demonstrated in several different assay systems. These enzymes can resolve opposing stalled replication forks (16) and catenate covalently closed DNA rings (17). Whereas these biochemical reactions support the concept that RecQ and Topo III could participate in resolving recombination intermediates in vivo as they do in eukaryotes, there is conflicting genetic evidence to this point.
Deletion of topB in a ΔtopA (encoding Topo I) mutant strain that also carried the DNA gyrase compensatory mutation gyrB225, which reduced the supercoiling activity of the enzyme, caused extensive filamentation and gave rise to abnormal nucleoid structures (18). This phenotype was suppressed by mutations in recA (encoding the strand exchange protein RecA), leading the authors to suggest unresolved recombination intermediates as the cause of the phenotype. Similarly, Lopez et al. (19) were unable to construct ΔtopB parE(Ts) (encoding the ParE subunit of Topo IV) strains by P1 transduction but could do so if recQ was mutated as well. However, we were able to construct the ΔtopB parE(Ts) strain, and none of its phenotypes were rescued by mutation in either recA or recQ (20). Interestingly, the growth and chromosome partition (par) phenotype of a ΔtopA gyrB(Ts) (encoding the GyrB subunit of DNA gyrase) mutant strain was relieved by mutation of recQ (21), suggesting a partnership between RecQ and Topo I, rather than Topo III.
Our biochemical studies have shown that Topo III can support both nascent chain elongation and decatenation of the sister chromosomes during DNA replication in vitro with purified proteins (22, 23). Furthermore, both Topo III and Topo IV, but not DNA gyrase, were capable of removing precatenanes (24), which are the result of the excess positive linkages that accumulate ahead of the replication fork distributing behind it, linking the two partially replicated sister chromosomes together (25, 26). We found that overexpression of Topo III in a parE(Ts) strain rescued both the temperature-sensitive growth and par phenotypes of the strain (24) and that ΔtopB was synthetically lethal at the permissive temperatures with both parE(Ts) and parC(Ts) (encoding the ParC subunit of Topo IV) mutations (20). Furthermore, ΔtopB strains exhibited increased sensitivity to novobiocin, an inhibitor of the type II topoisomerases, even in a gyrB(Novor) background (20). These observations led us to propose that Topo III was likely acting either at or close to the replication fork to remove precatenanes during DNA replication (20).
In support of our model for Topo III action, we show here that Topo III is found associated with the replication fork in vivo, that nucleoid separation is less orderly and takes somewhat longer in a ΔtopB strain than in the wild type, and that the DnaX complex of the DNA polymerase III holoenzyme (Pol III HE) stimulates the ability of Topo III to remove both precatenanes and catenanes from linked DNA rings.
RESULTS
Topo III colocalizes with the replication fork in vivo.
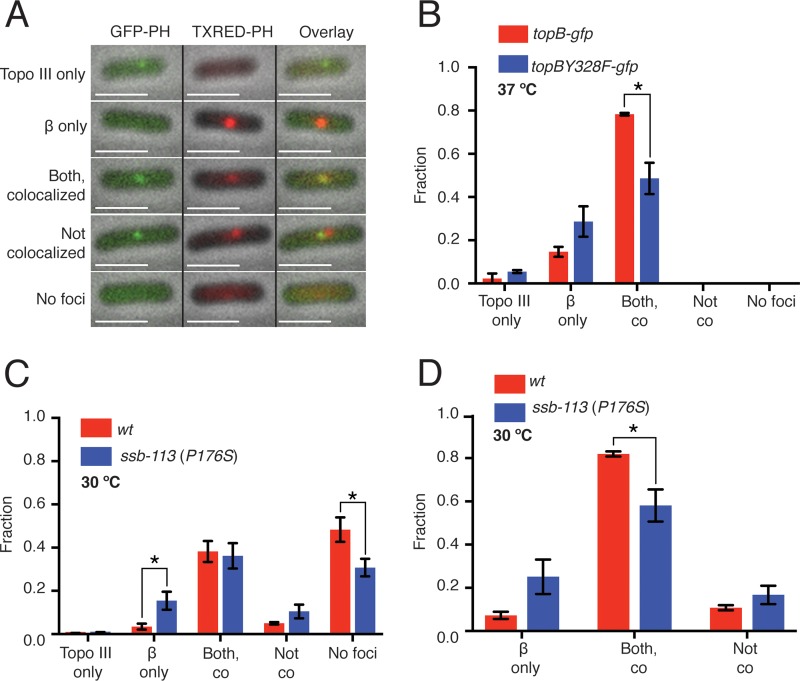
We suggested in a previous study that Topo III would act at or near the replication fork to remove precatenanes during DNA replication (20). To address this question directly by fluorescence microscopy, we constructed W3110 strains where Topo III was fused to an E. coli codon-optimized and straightened (27) version of green fluorescent protein (ecGFPS) (28) and the β processivity clamp of the DNA Pol III HE (encoded by dnaN [29]) was fused to the mCherry fluorescent protein, which marks the localization of the replisome (30). Accordingly, W3110 topB-ecgfpS mCherry-dnaN (CL109) was grown at 37°C in minimal medium (doubling time, 100 min) to early log phase and imaged directly on grooved agarose pads (Fig. 1A). In nearly 80% of the cells a single focus of Topo III-GFP colocalized with a single focus of mCherry-DnaN (Fig. 1B). We also examined the localization of a catalytically inactive version of Topo III where the catalytic Tyr had been substituted with Phe (Topo III Y328F). Localization of Topo III Y328F with β in W3110 topBY328F-ecgfpS mCherry-dnaN (CL113) was reduced somewhat but could still be observed in 50% of the cells (Fig. 1B), suggesting that the association of Topo III with the replisome was not dependent on its catalytic activity. Expression levels of both the wild-type and the catalytically inactive Topo III were equivalent (see Fig. S1 in the supplemental material). We do note that under the growth and imaging conditions used here, nearly all the cells contained a single focus of β, consistent with the observations of Mangiameli et al. (31) that the two replication forks remain associated during cell growth, as opposed to those of Reyes-Lamothe et al. (30) that argued the two replication forks separated.
FIG 1.
Topo III colocalizes with the β subunit of the DNA Pol III HE at the replication fork. CL109 (W3110 topB-gfp mCherry-dnaN), CL113 (W3110 topBY328F-gfp mCherry-dnaN), and CL281 [W3110 topB-gfp mCherry-dnaN ssb-113 (P176S)] were grown and imaged as described in Materials and Methods. (A) Classes of cell images. mCherry-β foci were consistently brighter than Topo III-GFP foci. Only the first three classes were observed with CL109 and CL113 growing at 37°C. “Both, colocalized” indicates that the GFP and mCherry foci colocalized in at least one of the Z planes. “Not colocalized” indicates that both GFP and mCherry foci were present but were not colocalized in any of the three Z planes. Left column, overlay of the GFP and phase channels. Middle column, overlay of the Texas Red and phase channels. Right column, overlay of the GFP, Texas Red, and phase channels. (B) Effect of the Topo III Y328F mutation on colocalization of Topo III and β. Greater than 200 cells were counted for each strain. Red, CL109; blue, CL113. (C) Effect of the SSB ProP176S mutation on colocalization of Topo III and β. (D) Replot of the data in panel C counting only cells that had Topo III and β foci. Greater than 200 cells were counted for each strain. Red, CL109; blue, CL281. wt, wild type. Error bars represent the standard deviation from the mean. *, P < 0.05.
Positioning of Topo III at the replication fork could occur by one of two mechanisms: Topo III could associate directly with a protein in the replisome, or Topo III could be binding to single-strand gaps on the nascent duplexes that are near the fork via its interaction with the single-stranded DNA-binding protein (SSB) (32). Topo III associates with the C-terminal eight amino acids of SSB (16). The interaction of many proteins with the SSB C-terminal tail is disrupted by mutation of the penultimate amino acid Pro176 to Ser (33). Strains carrying this mutation (ssb-113, originally lexC113) are temperature sensitive and quite compromised (34, 35). To investigate this question, we constructed W3110 topB-ecgfpS mCherry-dnaN ssb-113 (CL281) and compared Topo III and β localization with that in the wild-type strain (CL109) at 30°C in minimal medium (Fig. 1C). Overall, more cells had no visible foci of either Topo III or β than when CL109 was grown at 37°C (compare Fig. 1A and C), consistent with the reduction of growth rate at the lower temperature. When calculated using as a denominator all cells observed, the same fraction of cells of both the wild type (CL109) and the ssb-113 mutant strain (CL281) displayed colocalized foci of β and Topo III (Fig. 1C). There was an increase in the fraction of cells that displayed only a β focus that was concomitant with a decrease in the fraction of cells in the ssb-113 mutant that did not display any foci compared to the wild type (Fig. 1C). However, when only replicating cells were considered (i.e., eliminating cells that displayed no foci from the count), the fraction of ssb-113 mutant cells showing colocalization of β and Topo III declined by about 30% compared to that of the wild type (Fig. 1D). We interpret these data to mean that whereas SSB Pro176 plays some role in attracting Topo III to the replication fork, it is not the sole determinant nor is likely to be the major determinant.
Topo III interacts physically and functionally with the DnaX complex.
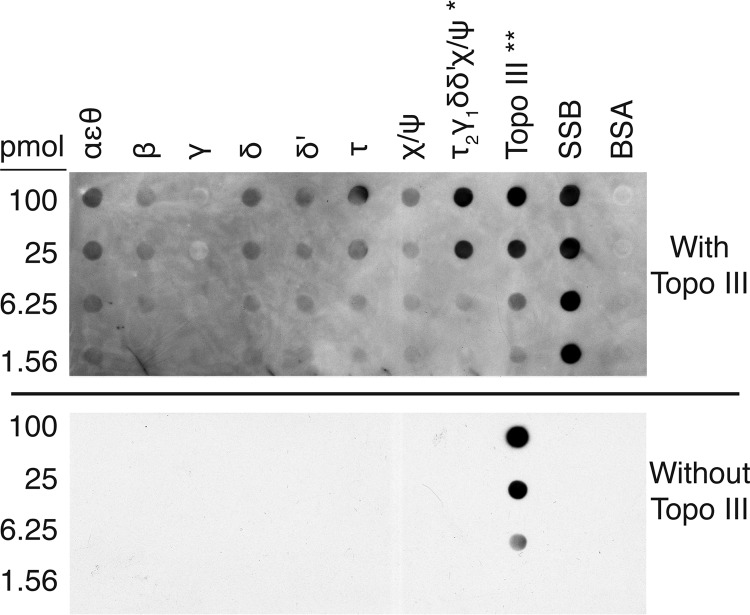
To determine whether there were other determinants of the association of Topo III with the replisome/replication fork, we performed immunoblotting with replisome components. Topo III showed a strong interaction with the DnaX complex of the Pol III HE (Fig. 2). The DnaX complex (τ2γδδ′χψ) forms the core of the cellular replicase, anchoring the two polymerase cores (αεθ) via their binding to the τ dimer and loading β to the 3′ end of the primer terminus as required to interact with the polymerase subunit (α) to engage processive DNA synthesis (36). The τ subunit also cements the replisome together by interacting with the DnaB replication fork DNA helicase (37). Topo III interacted with SSB as expected. Negligible interactions were detected between Topo III and the polymerase core, as well as with τ, δ, and δ′ individually. However, a fairly strong interaction was observed between the DnaX complex and Topo III, present in the same range of concentration as the interaction between Topo III and SSB. (Note that the amount of DnaX complex on the blots is 1/10 of that in all the other samples, except Topo III itself, which is present at 1/100 the level in the other samples.) We therefore investigated whether the DnaX complex functionally interacted with Topo III by stimulating its decatenating activity.
FIG 2.
Topo III interacts with the DnaX complex of the DNA Pol III HE. Immunoblotting was as described in Materials and Methods. Top panel, with Topo III; bottom panel, without Topo III. *, DnaX complex was applied to the membrane at 1/10 of the amounts indicated. **, Topo III was applied to the membrane at 1/100 of the amounts indicated. All quantities of proteins are given for the monomer.
We have examined this question in the past as a result of isolating dnaX as a high-copy-number suppressor of the temperature-sensitive growth and par phenotypes of the parE10 mutation (38). At that time, decatenation of Crithidia mitochondrial DNA was used as an assay, and we concluded that there was no effect of DnaX complex subunits on Topo III activity (39); however, that assay requires only one unlinking event to yield monomer product and as such is unlikely to detect stimulation of a processive reaction. We therefore turned to a more suitable assay where processive unlinking could be observed. We used two different substrates: (i) multiply linked dimers that represent fully catenated DNA sisters and (ii) precatenated DNA dimers. These substrates are purified from reconstituted DNA replication reactions. The multiply linked dimers (form II:form II dimers) are the product of replication reactions where DNA gyrase is the only topoisomerase present (40). DNA gyrase is a poor decatenating enzyme, so the products of the DNA replication reaction remain linked together. The precatenated DNA is the product of replication reactions where two Ter sites are present on the template DNA, separated by about 1 kbp, oriented to block the replication forks when bound by Tus. In this case, the excess positive linkages formed during replication distribute behind the forks, and the partially replicated DNA becomes precatenated (24, 26).
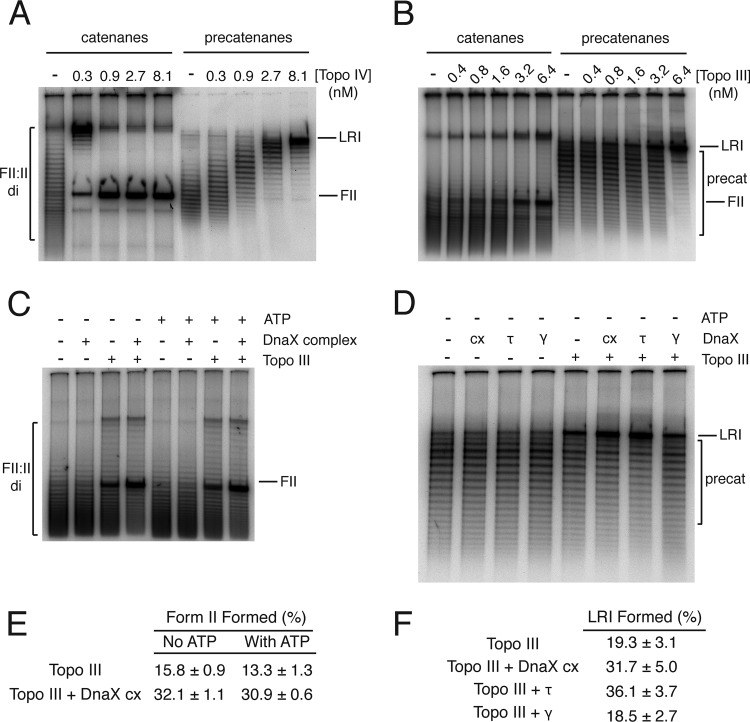
The gel shown in Fig. 3A shows the substrates and the expected products using reactions with Topo IV as a guide. The proteins required to join Okazaki fragments are omitted from the replication reactions used to generate these substrates; therefore, the sister molecules are not sealed and thus not supercoiled, allowing the linkages between the sister molecules to be visualized as steps in a ladder of bands when the substrates are electrophoresed through agarose gels. Each step in the ladder represents a difference of either one catenane or precatenane linkage between the two sister DNAs, with increasing electrophoretic mobility indicating species with increasing number of linkages between the sister molecules. Topo IV decatenates the catenated dimers processively (the final product appears while the starting material is still present), yielding the monomeric nicked (form II) DNA as the final product. On the other hand, unlinking of the precatenated DNA is distributive (all the DNA molecules are acted on as a group), and the final product is a late replicative intermediate (LRI) because the two sister molecules are still connected by 1 kbp of unreplicated DNA (Fig. 3A). Figure 3B shows that Topo III can unlink both substrates processively and is about 1/10 as potent a decatenase as Topo IV on the catenated substrate, but it is nearly as effective as Topo IV in unlinking precatenated DNA. The Okazaki fragments in the DNA substrates are separated by a mixture of nicks and small gaps; thus, Topo III can bind the DNAs to catalyze decatenation.
FIG 3.
The DnaX complex stimulates Topo III-catalyzed unlinking of catenanes and precatenanes. Assays were performed as described in Materials and Methods. Reaction products were analyzed by electrophoresis through 1% agarose gels at 1.5 V/cm for 16 h at room temperature using 50 mM Tris-HCl (pH 7.8 at 23°C), 40 mM sodium acetate (NaOAc), and 1 mM EDTA as the electrophoresis buffer. Gels were dried and autoradiographed. Reaction products were quantified using Fuji MacBas software. (A) Topo IV-catalyzed unlinking of catenated DNA dimers (left panel) and precatenated DNA (right panel). (B) Topo III-catalyzed unlinking of catenated DNA dimers (left panel) and precatenated DNA (right panel). (C) The DnaX complex stimulates Topo III-catalyzed unlinking of catenated DNA dimers. The indicated proteins were present at 40 nM. Topo III was present at 1.2 nM. (D) The DnaX complex stimulates Topo III-catalyzed unlinking of precatenated DNA. The indicated proteins were present at 40 nM. Topo III was present at 1.2 nM. (E) Quantification of the amount of final product (nicked, form II [FII] DNA) from panel C (mean and standard deviation from three experiments). (F) Quantification of the amount of final product (late replication intermediate [LRI]) from panel D (mean and standard deviation from three experiments).
The DnaX complex stimulated Topo III-catalyzed decatenation of the multiply linked DNA dimers 2-fold (Fig. 3C and E). The DnaX complex uses ATP to load β to the primer-template (36); however, ATP had no significant effect on Topo III either alone or in the presence of the DnaX complex. Similarly, the DnaX complex stimulated Topo III-catalyzed unlinking of precatenated DNA dimers (Fig. 3D and F), although the extent of stimulation was slightly less. This difference could relate to the fact that in the catenated DNA dimers the topological linkages can distribute over the surface of the torus formed by the linked DNA molecules and can freely slither (41), whereas in the precatenated DNA dimers the presence of the unreplicated region of parental DNA constrains slithering, possibly making any unlinking more difficult in general. Note that in Fig. 3A, Topo IV-catalyzed unlinking of the precatenated DNA becomes distributive. Interestingly, τ, not γ, appeared to be the major agent of stimulating Topo III activity (Fig. 3D and F), consistent with the results we observed by immunoblotting (Fig. 2), where some interaction between Topo III and τ alone was observed. This observation may relate to the region of DnaX (γ is a C-terminally truncated version of τ produced by ribosomal frameshifting [42]) that interacts with Topo III.
Abnormal nucleoid separation in the absence of Topo III.
Ensemble measurements of growth parameters show little difference between the ΔtopB strain and the wild type (20). Under the conditions of growth examined here, the C periods of the two strains as measured by flow cytometry was identical (see Fig. S2 in the supplemental material). To assess the effect of a topB deletion on nucleoid separation, we therefore examined this event directly by time-lapse imaging of wild-type (MG1655 hupA::mCherry:frtKan [CL269]) and topB deletion (MG1655 ΔtopB hupA::mCherry:frtKan [CL272]) strains where the nucleoid-associated protein HU had been tagged with the mCherry fluorescent protein to allow direct visualization of the nucleoids.
Cells growing logarithmically in a defined rich medium that eliminates autofluorescence were deposited on a grooved agarose pad and imaged at 2-min intervals (see Movies S1 [wild type] and S2 [ΔtopB mutant] in the supplemental material). Because these cells were growing rapidly, the newborn cells have two nucleoids present that then divide as the movies progress. A normal division process displays a regular progression of the center of the nucleoids away from each other, with the nucleoids roughly doubling in size just before division. Abnormal division can result in an increase in the time required for division and/or an elongation of the nucleoid mass during the course of division, as well as, in extreme cases, disruption of the nucleoid mass because of breakage, a complete failure of nucleoid division resulting in a par phenotype and cell filamentation, or cell division events resulting in cells with odd numbers of nucleoids.
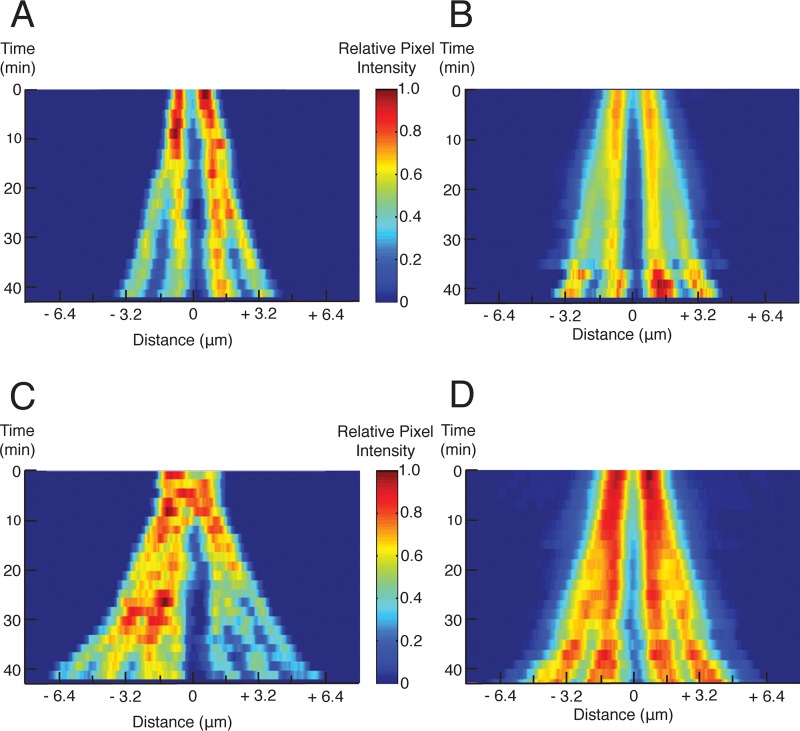
Examination of kymographs from single cells indicated that there were clear differences between the wild-type and ΔtopB cells (Fig. 4A and C, respectively). Complete nucleoid separation in the ΔtopB cells appeared to take longer and was more irregular than that in the wild type. To ensure that our observational impressions were correct, we wrote a MATLAB program to combine multiple kymographs and display the averaged information as a single image (Fig. 4B and D). The numbers of frames from movies of individual cells combined for each time point are given in Fig. S3A and B in the supplemental material. The averaged kymograph for the wild type (Fig. 4B) displayed a regular progression of one sister nucleoid away from the other, whereas that of the ΔtopB strain (Fig. 4D) displayed irregular and delayed separation, although the nucleoids did separate completely. This can be seen as increased intensity between the pairs of separating nucleoids in the ΔtopB strain compared to the wild type, particularly between the 16- and 26-min lines of the averaged kymographs (compare Fig. 4B and D). Pixel intensities from the same individual lines in the averaged kymographs are shown in Fig. S4 in the supplemental material, and here it can be clearly seen that with the wild type, two peaks that are separated emerge with time for each of the starting nucleoids. In contrast, separation is ill-defined and the nucleoid masses are more diffuse for the ΔtopB strain. We conclude that under wild-type conditions, Topo III is required to maximize the order and efficiency of the process of topological separation of the sister chromosomes.
FIG 4.
Topo III is required for orderly and efficient nucleoid separation. Time-lapse images of CL269 (MG1655 hupA::mCherry:frtKan) and CL272 (MG1655 ΔtopB hupA::mCherry:frtKan) growing in rich medium at 37°C were converted to kymographs as described in Materials and Methods. (A) Example of a single kymograph for CL269. (B) Averaged kymograph for CL269 cells. (C) Example of a single kymograph for CL272. (D) Averaged kymograph for CL272 cells. See Fig. S4A and B in the supplemental material for the number of individual frames of cells that were averaged for each line of the averaged kymographs shown. The color scale is set to 1 for the maximum intensity in each image presented.
DISCUSSION
The only two known phenotypes of topB mutations in an otherwise wild-type background are a slight defect in spontaneous deletion formation between short direct repeats, the mutR phenotype (43), and a hypersensitivity to the type II topoisomerase inhibitor novobiocin when ΔtopB strains are grown on rich medium (20). The latter phenotype, coupled with the observed synthetic lethality at permissive temperatures of ΔtopB with temperature-sensitive mutations in parE and parC, the genes encoding the subunits of Topo IV, formed the basis of our proposal that Topo III functioned under unstressed circumstances to assist Topo IV in unlinking the sister chromosomes during DNA replication, probably by the removal of precatenanes (20).
Consistent with our proposal, we found that Topo III-GFP was positioned at the replication fork, as marked by the location of mCherry-β. Localization of Topo III did not depend on its catalytic activity. Topo III is known to have a strong interaction with SSB that is partially mediated by the penultimate C-terminal Pro176 (16, 32). During DNA replication, there is almost always a gap in the nascent lagging strand as Okazaki fragment synthesis is ongoing. The ssDNA template in this gap will be coated with SSB. Similarly, recent observations indicate that gaps can form in the nascent leading strand as well because of polymerase pausing and continued unwinding by DnaB (44). Localization of Topo III to the replication fork could therefore be a result of its interaction with SSB. Whereas we did observe a statistically significant reduction in colocalization between Topo III-GFP and mCherry-β in replicating cells in the ssb-113 mutant background, significant colocalization persisted. Thus, although the Topo III-SSB interaction likely plays some role in localizing Topo III to the replisome, this interaction is clearly not the major determinant.
Not surprisingly, therefore, we also found an interaction between Topo III and subunits of the Pol III HE that could, of course, also account for the localization of Topo III with the replication fork. The DnaX complex interacted with Topo III in a concentration range similar to that of SSB. The individual protomers of the DnaX complex displayed weak to negligible interactions. The improved interaction of the DnaX complex compared to the individual subunits could be a result of how Topo III interacts with the seven-subunit clamp loader (45). Furthermore, we found that the DnaX complex stimulated the ability of Topo III to unlink catenated and precatenated DNA rings, the latter of which we presume to be the actual substrate in vivo. This 2-fold stimulation puts Topo III unlinking activity of precatenanes on par with that of Topo IV.
As we suggested some time ago, the only potential site of action for a type I topoisomerase to decatenate the replicating sisters was at a single-strand gap at the replication fork (46). Type IA topoisomerases, unlike type IB, are not able to unlink replicating sisters that are completely sealed. Thus, localization of Topo III to the replication fork is consonant with its activity. Topo IV, which can act anywhere on the DNA to unlink either catenanes or precatenanes, is seen, using fluorescently tagged derivatives, to be associated with the condensin MukB near the replication origin and otherwise uniformly distributed throughout the nucleoid (47). Topo I, which interacts with RNA polymerase (48) and is required to suppress hypernegative supercoiling and R loop formation (49–51), is found associated with the Bacillus subtilis SMC protein at regions of high negative supercoiling, whereas DNA gyrase was found associated with the replisome, presumably because of the high density of positive supercoils (52).
Our direct observations of nucleoid separation in wild-type and ΔtopB cells demonstrate that in the absence of Topo III, this process takes longer and can be significantly more irregular than in its presence. This observation indicates that Topo III plays a role in the process of untangling the sister chromosomes and is required for maximum speed and efficiency. However, even in the absence of Topo III, complete separation of most nucleoids occurred, as expected because Topo IV can act globally to remove linkages between the sister chromosomes.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Complete lists of bacterial strains and plasmids used in this study can be found in Tables 1 and 2, respectively. Oligonucleotides used for PCR amplification are listed in Table 3.
TABLE 1.
Strains
| Strain | Relevant characteristicsa | Construction, reference, and/or source |
|---|---|---|
| BP093 | W3110 ΔtopB::FRTkan Kmr | 20 |
| C600 | F− λ− tonA21 thi-1 thr-1 leuB6 lacY1 glnV44 rfbC1 fhuA1 | 60 |
| CL001 | W3110 parE10 parE(Ts) (parEC316Y) | 20 |
| CL002 | W3110 parE10ΔtopB::FRTkan Kmr | 20 |
| CL075 | C600 topB-ecgfpS:frt-kan-frt Kmr | This study |
| CL081 | W3110 topBY328F | This study |
| CL084 | W3110 topBY328F-ecgfpS:frt-kan-frt Kmr | This study |
| CL085 | W3110 topB-ecgfpS:frt-kan-frt Kmr | P1 (CL075) × W3110; this study |
| CL093 | W3110 topB-ecgfpS | Deletion of kan from CL085; this study |
| CL101 | W3110 topBY328F-ecgfpS | Deletion of kan from CL084; this study |
| CL103 | W3110 topB-ecgfpS mCherry:frt-kan-dnaN Kmr | P1 (RRL355) × CL093 this study |
| CL105 | W3110 topBY328F-ecgfpS mCherry:frt-kan-dnaN Kmr | P1 (RRL355) × CL101; this study |
| CL109 | W3110 topB-ecgfpS mCherry-dnaN | Deletion of kan from CL103; this study |
| CL113 | W3110 topBY328F-ecgfpS mCherry-dnaN | Deletion of kan from CL105; this study |
| CL143 | W3110 ΔtopB | Deletion of kan from BP093; this study |
| CL146 | MG1655 ΔtopB:FRTkan Kmr | P1 (TB28 ΔtopB::FRTkan) × MG1655; this study |
| CL151 | MG1655 ΔtopB | Deletion of kan from CL146; this study |
| CL257 | W3110 parE10 ΔtopB | Deletion of kan from CL002; this study |
| CL269 | MG1655 hupA::mCherry:frtKan Kmr | P1 (SS6282) × MG1655; this study |
| CL272 | MG1655 ΔtopB hupA::mCherry:frtKan Kmr | P1 (SS6282) × CL151; this study |
| CL281 | W3110 topB-ecgfpS mCherry-dnaN ssb-113 ssb-113(P176S) | This study |
| CL287 | W3110 parE10 hupA::mCherry:frtKan Kmr | P1 (SS6282) × CL001; this study |
| CL289 | W3110 parE10 ΔtopB hupA::mCherry:frtKan Kmr | P1 (SS6282) × CL257; this study |
| MG1655 | F− λ− rph-1 | 61 |
| PAM33 | F− sulA1 trpE65(Oc) [mal+]K-12(λS) ssb-113(P176S) | 56 |
| RRL355 | AB1157 mCherry::Frt-kan-dnaN | 30; gift from David Sherratt |
| RYH110 | W3110 hupA::mCherry:frtKan | P1 (SS628) × W3110; this study |
| SS6282 | JC13509 hupA::mCherry:frtKanΔ(attB)::sulAp-gfp Kmr | 62; gift From Steven Sandler |
| TB28ΔtopB::FRTkan | TB28 ΔtopB::FRTkan Kmr | 20 |
| W3110 | F− λ− IN(rrnD-rrnE)1 rph-1 | 61 |
Kmr, kanamycin resistance.
TABLE 2.
Plasmids
| Plasmid | Relevant characteristicsa | Reference(s) or source |
|---|---|---|
| pBP47 | pLEX5BA-topBY328F | 20 |
| pCP20 | Temperature-sensitive plasmid carrying heat-inducible FLP recombinase used to remove FRT-flanked antibiotic resistance genes | 54 |
| pKO3-topBY328F | pKO3 carrying the ORF for topBY328F; Cmr | This study |
| pKO3-ssb113 | pKO3 carrying nucleotides 4273659 to 4275647 from strain PAM33; Cmr | This study |
| pRC7-ssb | pRC7 carrying the ORF of ssb; Apr | This study |
| pRC7 | Low-copy-number, IPTG-inducible expression vector that expresses the target ORF as a LacZ fusion protein; Apr | 63 |
| pKO3 | Temperature-sensitive origin of replication; Cmr | 53 |
| pGBM2-ecgfps | pGBM2 carrying codon-optimized, straightened GFPuv; Kmr | 28, 64 |
Kmr, kanamycin resistance; Cmr, chloramphenicol resistance; Apr, ampicillin resistance; FRT, FLP recombinase target.
TABLE 3.
Oligonucleotides
| Name | Sequence (5′ → 3′) |
|---|---|
| BamHI-ssb 5′ | CATATTATATTGGATCCATGGCCAGCAGAGGCGTAAAC |
| HindIII-ssb 3′ | GATGGGGAGTAAGCTTTCAGAACGGAATGTCATCATC |
| BamHI-ssb113 5′ | GATTAGTATCGGATCCCGGCTTTTCCATCAGTGACAG |
| SalI-ssb113 3′ | CAATTAAATGTCGACCTGGCGAATGACCAGCTCGG |
| topB-BamHI 5′ | CTATGACCGGATCCATGCGGTTGTTTATTG |
| topB-SalI 3′ | CTATTCAAGGTCGACTTACGCTATCGCC |
C600 topB-ecgfpS:frt-kan-frt and W3110 topBY328F-ecgfpS:frt-kan-frt were constructed as described previously (28) for C-terminal fusion of Topo III and Topo III Y328F to E. coli codon-optimized and straightened GFPuv, resulting in strains CL075 and CL084, respectively. The sequences of the fusion protein alleles were confirmed.
For construction of W3110 topBY328F, gene replacement with pKO3-topBY328F, as described by Link et al. (53), was used to introduce the Y328F mutation into topB in W3110, resulting in strain CL081. The sequence of the mutated allele was confirmed.
For construction of W3110 topB-ecgfpS mCherry-dnaN ssb-113, the ssb-113 mutation was introduced into strain CL109 (W3110 topB-ecgfpS mCherry-dnaN ssb-113) as described for the construction of strain CL001 (W3110 parE10) (20). Plasmid pKO3-ssb113 served as the template for gene replacement in CL109, and plasmid pRC7-ssb expressed wild-type SSB during recombination at nonpermissive temperatures, resulting in strain CL281. The sequence of the mutated allele was confirmed.
As noted in Table 1, several strains were constructed by bacteriophage P1 transduction followed by selection for growth on medium containing the appropriate antibiotics. The sequences of all transferred alleles were confirmed. Antibiotic resistance genes were removed by utilizing the plasmid pCP20 and the FLP-FLP recombinase target (FLP-FRT) recombination system as described previously (54). Loss of the genes was confirmed by PCR and DNA sequencing.
For construction of pKO3-topBY328F, the topB open reading frame (ORF) was amplified from plasmid pBP47 (20) using primers that also introduced BamHI and SalI restriction enzyme sites at the 5′ and 3′ ends of the topBY328F gene, respectively (primers topB-BamHI 5′ and topB-SalI 3′ [Table 3]). The DNA product was cloned into the plasmid vector pKO3 (53) that had been digested with the same restriction enzymes.
For construction of pRC7-ssb, the ssb open reading frame was amplified from genomic DNA prepared from W3110 using primers that also introduced BamHI and HindIII restriction enzyme sites into the 5′ and 3′ ends, respectively (primers BamHI-ssb 5′ and HindIII-ssb 3′ [Table 3]). The DNA fragment was cloned into plasmid vector pRC7 (55) that had been digested with the same restriction enzymes.
For construction of pKO3-ssb113, nucleotides 4273659 to 4275647 were amplified from genomic DNA prepared from strain PAM33 (56) using primers that also introduced BamHI and SalI restriction enzyme sites to the 5′ and 3′ ends, respectively (primers BamHI-ssb113 5′ and SalI-ssb113 3′ [Table 3]), of chromosomal DNA that flanked the ssb-113 mutation by 1 kbp on each side. The DNA fragment was cloned into pKO3 as described for pKO3-topBY328F.
Microscopy and localization of Topo III and β.
Saturated cultures of strains CL109, CL113, and CL281 were diluted to an optical density at 600 nm (OD600) of 0.01 and grown at either 37°C (when CL109 and CL113 were compared) or 30°C (when CL109 and CL281 were compared) in M9 minimal medium supplemented with 20 μg/ml each of thiamine, thymine, and uracil and either 0.2% glucose (for the comparison of CL109 and CL113) or 0.2% glycerol (for the comparison of CL109 and CL281) to an OD600 of 0.1. Cells were viewed on grooved 4% agarose (NuSieve GTG; Lonza) pads prepared in 1× phosphate-buffered saline (PBS) as described previously (57). Cells were imaged in the phase, Texas Red, and GFP channels in three consecutive 0.2-μm Z-planes using a Nikon Eclipse Ti microscope with an Plan Apo 100×/1.40-numerical-aperture objective and an Andor Neo CMOS camera enclosed in an Okolab environmental chamber held at the indicated temperatures. Images were processed with Nikon Elements AR software (version 4.40.00) to remove background.
Microscopy and time-lapse imaging.
Strains CL269 and CL272 were grown in rich medium (MOPS [morpholinepropanesulfonic acid] EZ Media; Teknova) supplemented with 2% glucose. Saturated cultures were diluted to an OD600 of 0.005, grown at 37°C to an OD600 of 0.1, and viewed on grooved 4% agarose pads (prepared in the same medium) as described above. Cells were imaged in the phase and Texas Red channels every 2 min. The movies were cropped to the first ∼60 min of growth, and the phase and Texas Red signals were adjusted so that they were within the same dynamic range. The images were exported as stacked tif files and examined with Fiji software (ImageJ 2.0.0-rc-65/1.51w, Java 1.80 to 66) (58) to mark the frames where the cell cycle started and ended for the cells of interest. Kymographs of the stacked images were generated using a custom MATLAB R2014a (8.30.532)-based script called “analysis_kymographs” that allows the user to mark the cells’ boundaries on the x axis (cell length), find the midpoint of each cell (frames were aligned based on the midpoint of the cell boundaries), and measure the intensity of the fluorescent signal, as well as the location of any particular pixel relative to the boundaries of the cell. About 50 individual kymographs of individual cells for each strain were averaged together (see Fig. S3A and B in the supplemental material) using another custom MATLAB-based script called the “average_kymographs” script. The programs have the ability to select means or maximums of pixel intensity in individual kymographs and in the merged, averaged kymograph, as well as to rescale the information if desired for presentation. The images shown in Fig. 4 represent the mean pixel value in any particular line of an individual kymograph and the mean of the means in the averaged kymograph. The images were not rescaled (“kmean/mean/norescale”). The scale and color map were set in MATLAB. The custom MATLAB scripts used here are available on request.
Immunoblotting.
A nitrocellulose membrane (GE Healthcare) equilibrated in 20 mM Tris-HCl (pH 7.5)–500 mM NaCl was assembled on a Bio-Dot apparatus (Bio-Rad) and prepared for samples as per the manufacturer’s directions. Pol III core, β, Topo III, and SSB (lab stocks), τ, γ, δ, δ′, χ/ψ, and the DnaX complex (gifts of Charles McHenry, University of Colorado), and bovine serum albumin (BSA) (NEB) were serially diluted in 1× PBS such that 1.56 to 100 pmol of protein (as monomer) was applied to the membrane. The DnaX complex and Topo III were applied at 1/10 and 1/100, respectively, of these amounts for the other proteins. The proteins were allowed to flow through the membrane without a vacuum, and each well was washed twice with 1× PBS under gentle vacuum. The membrane was air dried for 10 min, blocked in 1× PBS–5% powdered milk for 1 h at 25°C, and then incubated in 40 mM HEPES-KOH (pH 7.5), 12 mM magnesium acetate [Mg(OAc)2], 5 mM dithiothreitol (DTT), 0.1 mg/ml BSA, and 4% sucrose with or without 2 μg/ml of Topo III at 25°C with slow rocking for 15 min. Glutaraldehyde (Sigma) was then added to a final concentration of 0.1%, and the membranes were incubated for an additional 45 min. The membranes were then washed in 25 mM Tris-HCl (pH 7.5)–137 mM NaCl–2.7 mM KCl for 15 min at 25°C, followed by four 15-min washes in 1× PBS–2% powdered milk. The membranes were then incubated with the primary anti-Topo III antiserum at a 1:5,000 dilution in 1× PBS for 24 h at 4°C, followed by four 15-min washes with 1× PBS–2% powdered milk. The membranes were then incubated with the secondary goat anti-rabbit antibody–horseradish peroxidase (HRP) conjugate (Bio-Rad) at a 1:10,000 dilution in 1× PBS–2% powdered milk for 1 h at 25°C and then washed with the primary antibody. Membranes were incubated with ECL reagents (GE Healthcare) prior to exposure to film.
Unlinking of catenated and precatenated DNA.
Preparation of precatenated DNA dimers was as described previously (24) except that pBROTB 535-I plasmid DNA (59) was used as the template and the dimers were isolated by sucrose gradient centrifugation. Catenated DNA dimers were prepared in the same manner except that Tus was omitted from the DNA replication reaction mixtures. Assay conditions were as described previously (24) except that incubation time was 10 min.
Supplementary Material
ACKNOWLEDGMENTS
These studies were supported by NIH grants 5R01GM34558 and 1R35GM126907 to K.J.M. and 1DP2GM105443-01 to A.P. and by Cancer Center Support Grant NCI P30CA008748 to the Memorial Sloan Kettering Cancer Center.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00563-18.
REFERENCES
- 1.Corbett KD, Berger JM. 2004. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct 33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 2.Vos SM, Tretter EM, Schmidt BH, Berger JM. 2011. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol 12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Espeli O, Marians KJ. 2004. Untangling intracellular DNA topology. Mol Microbiol 52:925–931. doi: 10.1111/j.1365-2958.2004.04047.x. [DOI] [PubMed] [Google Scholar]
- 4.Levine C, Hiasa H, Marians KJ. 1998. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim Biophys Acta 1400:29–43. [DOI] [PubMed] [Google Scholar]
- 5.Dean F, Krasnow MA, Otter R, Matzuk MM, Spengler SJ, Cozzarelli NR. 1983. Escherichia coli type-1 topoisomerases: identification, mechanism, and role in recombination. Cold Spring Harbor Symp Quant Biol 47:769–777. doi: 10.1101/SQB.1983.047.01.088. [DOI] [PubMed] [Google Scholar]
- 6.Srivenugopal KS, Lockshon D, Morris DR. 1984. Escherichia coli DNA topoisomerase III: purification and characterization of a new type I enzyme. Biochemistry 23:1899–1906. [DOI] [PubMed] [Google Scholar]
- 7.DiGate RJ, Marians KJ. 1988. Identification of a potent decatenating enzyme from Escherichia coli. J Biol Chem 263:13366–13373. [PubMed] [Google Scholar]
- 8.DiGate RJ, Marians KJ. 1989. Molecular cloning and DNA sequence analysis of Escherichia coli topB, the gene encoding topoisomerase III. J Biol Chem 264:17924–17930. [PubMed] [Google Scholar]
- 9.DiGate RJ, Marians KJ. 1992. Escherichia coli topoisomerase III-catalyzed cleavage of RNA. J Biol Chem 267:20532–20535. [PubMed] [Google Scholar]
- 10.Wang H, Di Gate RJ, Seeman NC. 1996. An RNA topoisomerase. Proc Natl Acad Sci U S A 93:9477–9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oakley TJ, Hickson ID. 2002. Defending genome integrity during S-phase: putative roles for RecQ helicases and topoisomerase III. DNA Repair (Amst) 1:175–207. [DOI] [PubMed] [Google Scholar]
- 12.Xu D, Shen W, Guo R, Xue Y, Peng W, Sima J, Yang J, Sharov A, Srikantan S, Yang J, Fox D, Qian Y, Martindale JL, Piao Y, Machamer J, Joshi SR, Mohanty S, Shaw AC, Lloyd TE, Brown GW, Ko MSH, Gorospe M, Zou S, Wang W. 2013. Top3β is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat Neurosci 16:1238–1247. doi: 10.1038/nn.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad M, Xue Y, Lee SK, Martindale JL, Shen W, Li W, Zou S, Ciaramella M, Debat H, Nadal M, Leng F, Zhang H, Wang Q, Siaw GE, Niu H, Pommier Y, Gorospe M, Hsieh TS, Tse-Dinh YC, Xu D, Wang W. 2016. RNA topoisomerase is prevalent in all domains of life and associates with polyribosomes in animals. Nucleic Acids Res 44:6335–6349. doi: 10.1093/nar/gkw508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad M, Shen W, Li W, Xue Y, Zou S, Xu D, Wang W. 2017. Topoisomerase 3beta is the major topoisomerase for mRNAs and linked to neurodevelopment and mental dysfunction. Nucleic Acids Res 45:2704–2713. doi: 10.1093/nar/gkw1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suski C, Marians KJ. 2008. Resolution of converging replication forks by RecQ and topoisomerase III. Mol Cell 30:779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harmon FG, DiGate RJ, Kowalczykowski SC. 1999. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol Cell 3:611–620. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Q, Pongpech P, DiGate RJ. 2001. Type I topoisomerase activity is required for proper chromosomal segregation in Escherichia coli. Proc Natl Acad Sci U S A 98:9766–9771. doi: 10.1073/pnas.171579898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez CR, Yang S, Deibler RW, Ray SA, Pennington JM, Digate RJ, Hastings PJ, Rosenberg SM, Zechiedrich EL. 2005. A role for topoisomerase III in a recombination pathway alternative to RuvABC. Mol Microbiol 58:80–101. doi: 10.1111/j.1365-2958.2005.04812.x. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Cheeks BA, Lee C, Hayama R, Marians KJ. 2012. A role for topoisomerase III in Escherichia coli chromosome segregation. Mol Microbiol 86:1007–1022. doi: 10.1111/mmi.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Usongo V, Drolet M. 2014. Roles of type 1A topoisomerases in genome maintenance in Escherichia coli. PLoS Genet 10:e1004543. doi: 10.1371/journal.pgen.1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiasa H, Marians KJ. 1994. Topoisomerase III, but not topoisomerase I, can support nascent chain elongation during theta-type DNA replication. J Biol Chem 269:32655–32659. [PubMed] [Google Scholar]
- 23.Hiasa H, DiGate RJ, Marians KJ. 1994. Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J Biol Chem 269:2093–2099. [PubMed] [Google Scholar]
- 24.Nurse P, Levine C, Hassing H, Marians KJ. 2003. Topoisomerase III can serve as the cellular decatenase in Escherichia coli. J Biol Chem 278:8653–8660. doi: 10.1074/jbc.M211211200. [DOI] [PubMed] [Google Scholar]
- 25.Champoux JJ, Been M. 1980. Topoisomerases and the swivel problem, p. 809–815. In Alberts B. (ed), Mechanistic studies of DNA replication and gentic recombination. Academic Press, New York, NY. [Google Scholar]
- 26.Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR. 1998. The structure of supercoiled intermediates in DNA replication. Cell 94:819–827. [DOI] [PubMed] [Google Scholar]
- 27.Corcoran CP, Cameron AD, Dorman CJ. 2010. H-NS silences gfp, the green fluorescent protein gene: gfpTCD is a genetically remastered gfp gene with reduced susceptibility to H-NS-mediated transcription silencing and with enhanced translation. J Bacteriol 192:4790–4793. doi: 10.1128/JB.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee C, Marians KJ. 2013. Characterization of the nucleoid-associated protein YejK. J Biol Chem 288:31503–31516. doi: 10.1074/jbc.M113.494237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgers PM, Kornberg A, Sakakibara Y. 1981. The dnaN gene codes for the beta subunit of DNA polymerase III holoenzyme of Escherichia coli. Proc Natl Acad Sci U S A 78:5391–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. 2008. Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangiameli SM, Veit BT, Merrikh H, Wiggins PA. 2017. The replisomes remain spatially proximal throughout the cell cycle in bacteria. PLoS Genet 13:e1006582. doi: 10.1371/journal.pgen.1006582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 33.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. 2008. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol 43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer RR, Glassberg J, Kornberg A. 1979. An Escherichia coli mutant defective in single-strand binding protein is defective in DNA replication. Proc Natl Acad Sci U S A 76:1702–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glassberg J, Meyer RR, Kornberg A. 1979. Mutant single-strand binding protein of Escherichia coli: genetic and physiological characterization. J Bacteriol 140:14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McHenry CS. 2011. DNA replicases from a bacterial perspective. Annu Rev Biochem 80:403–436. doi: 10.1146/annurev-biochem-061208-091655. [DOI] [PubMed] [Google Scholar]
- 37.Kim S, Dallmann HG, McHenry CS, Marians KJ. 1996. Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell 84:643–650. [DOI] [PubMed] [Google Scholar]
- 38.Levine C, Marians KJ. 1998. Identification of dnaX as a high-copy suppressor of the conditional lethal and partition phenotypes of the parE10 allele. J Bacteriol 180:1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levine C. 2000. The identification and analysis of high-copy suppressors of Topo IV mutations. Cornell University, Ithaca, NY. [Google Scholar]
- 40.Marians KJ. 1987. DNA gyrase-catalyzed decatenation of multiply linked DNA dimers. J Biol Chem 262:10362–10368. [PubMed] [Google Scholar]
- 41.Vologodskii AV, Cozzarelli NR. 1993. Monte Carlo analysis of the conformation of DNA catenanes. J Mol Biol 232:1130–1140. doi: 10.1006/jmbi.1993.1465. [DOI] [PubMed] [Google Scholar]
- 42.McHenry CS. 2011. Bacterial replicases and related polymerases. Curr Opin Chem Biol 15:587–594. doi: 10.1016/j.cbpa.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schofield MA, Agbunag R, Michaels ML, Miller JH. 1992. Cloning and sequencing of Escherichia coli mutR shows its identity to topB, encoding topoisomerase III. J Bacteriol 174:5168–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graham JE, Marians KJ, Kowalczykowski SC. 2017. Independent and stochastic action of DNA polymerases in the replisome. Cell 169:1201–1213. doi: 10.1016/j.cell.2017.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O'Donnell M, Kuriyan J. 2009. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell 137:659–671. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minden JS, Marians KJ. 1986. Escherichia coli topoisomerase I can segregate replicating pBR322 daughter DNA molecules in vitro. J Biol Chem 261:11906–11917. [PubMed] [Google Scholar]
- 47.Zawadzki P, Stracy M, Ginda K, Zawadzka K, Lesterlin C, Kapanidis AN, Sherratt DJ. 2015. The localization and action of topoisomerase IV in Escherichia coli chromosome segregation is coordinated by the SMC complex, MukBEF. Cell Rep 13:2587–2596. doi: 10.1016/j.celrep.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng B, Zhu CX, Ji C, Ahumada A, Tse-Dinh YC. 2003. Direct interaction between Escherichia coli RNA polymerase and the zinc ribbon domains of DNA topoisomerase I. J Biol Chem 278:30705–30710. doi: 10.1074/jbc.M303403200. [DOI] [PubMed] [Google Scholar]
- 49.Masse E, Drolet M. 1999. Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J Biol Chem 274:16659–16664. [DOI] [PubMed] [Google Scholar]
- 50.Drolet M, Bi X, Liu LF. 1994. Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J Biol Chem 269:2068–2074. [PubMed] [Google Scholar]
- 51.Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ. 1995. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci U S A 92:3526–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tadesse S, Graumann PL. 2006. Differential and dynamic localization of topoisomerases in Bacillus subtilis. J Bacteriol 188:3002–3011. doi: 10.1128/JB.188.8.3002-3011.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Link AJ, Phillips D, Church GM. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol 179:6228–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernhardt TG, de Boer PA. 2004. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol 52:1255–1269. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson BF. 1977. Genetic mapping of the lexC-113 mutation. Mol Gen Genet 157:91–97. [DOI] [PubMed] [Google Scholar]
- 57.Kumar R, Nurse P, Bahng S, Lee CM, Marians KJ. 2017. The MukB-topoisomerase IV interaction is required for proper chromosome compaction. J Biol Chem 292:16921–16932. doi: 10.1074/jbc.M117.803346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hiasa H, Marians KJ. 1994. Tus prevents overreplication of oriC plasmid DNA. J Biol Chem 269:26959–26968. [PubMed] [Google Scholar]
- 60.Appleyard RK. 1954. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics 39:440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jensen KF. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol 175:3401–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pelletier J, Halvorsen K, Ha BY, Paparcone R, Sandler SJ, Woldringh CL, Wong WP, Jun S. 2012. Physical manipulation of the Escherichia coli chromosome reveals its soft nature. Proc Natl Acad Sci U S A 109:E2649–E2656. doi: 10.1073/pnas.1208689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bernhardt TG, de Boer PA. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol 48:1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Espeli O, Lee C, Marians KJ. 2003. A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J Biol Chem 278:44639–44644. doi: 10.1074/jbc.M308926200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.