Abstract
Human homologue of yeast UV excision repair protein Rad23b (HR23B) inclusions are found in a number of neurodegenerative diseases, including frontotemporal dementia (FTD), Huntington’s disease (HD), spinocerebellar ataxia type 3 and 7 (SCA3/7), fragile X associated tremor/ataxia syndrome (FXTAS) and Parkinson’s disease (PD). Here, we describe HR23B pathology in C9ORF72 linked FTD and amyotrophic lateral sclerosis (ALS) cases. HR23B presented in neuropils, intranuclear inclusions and cytoplasmic and perinuclear inclusions and was predominantly found in cortices (frontal, temporal and motor), spinal cord and hippocampal dentate gyrus. HR23B co-localized with poly-GA-, pTDP-43- and p62-positive inclusions in frontal cortex and in hippocampal dentate gyrus, the latter showing higher co-localization percentages. HR23B binding partners XPC, 20S and ataxin-3, which are involved in nucleotide excision repair (NER) and the ubiquitin-proteasome system (UPS), did not show an aberrant distribution. However, C9ORF72 fibroblasts were more sensitive for UV-C damage than healthy control fibroblasts, even though all factors involved in NER localized normally to DNA damage and the efficiency of DNA repair was not reduced. HR23Bs other binding partner NGly1/PNGase, involved in ER-associated degradation (ERAD) of misfolded proteins, was not expressed in the majority of neurons in C9FTD/ALS brain sections compared to non-demented controls. Our results suggest a difference in HR23B aggregation and co-localization pattern with DPRs, pTDP-43 and p62 between different brain areas from C9FTD/ALS cases. We hypothesize that HR23B may play a role in C9ORF72 pathogenesis, possibly by aberrant ERAD functioning.
Electronic supplementary material
The online version of this article (10.1186/s40478-019-0694-6) contains supplementary material, which is available to authorized users.
Keywords: C9ORF72, ALS, FTD, HR23B, ERAD, NGly1, DPRs, Poly-GA
Introduction
The hexanucleotide (G4C2) repeat expansion in the chromosome 9 open reading frame 72 (C9ORF72) gene is the most common genetic cause of FTD and ALS [12, 33]. FTD is characterized by the degeneration of the frontal and temporal parts of the brain, leading to abnormalities in behavior, language and personality [49]. ALS affects motor neurons in the brain and spinal cord, leading to loss of motor function, muscle weakness, breathing problems and eventually paralysis [52]. Clinical, pathological and genetic factors connect FTD and ALS, and in families often patients present with symptoms of both disorders [4]. The discovery of the C9ORF72 hexanucleotide repeat expansion confirmed the genetic overlap between FTD and ALS, collectively referred to as C9FTD/ALS. Pathologically, both diseases present with inclusions of autophagy protein p62/sequestosome 1 (p62) and inclusions of phosphorylated 43 kDa TAR DNA-binding protein (pTDP-43), of which the latter is predominantly found in areas that are known to display substantial neurodegeneration [24, 44].
The possible pathological mechanisms by which the C9ORF72 repeat expansion can lead to FTD and ALS are: 1) hypermethylation of the repeat expansion and the CpG promoter region of the C9ORF72 gene leading to haploinsufficiency [5, 43], 2) retention of repeat containing intron 1 in mRNAs causing RNA foci to appear in both nucleus and cytoplasm that sequester RNA-binding proteins [11, 27] or 3) the production of dipeptide repeats (DPR) by unconventional repeat-associated non-AUG (RAN) translation of the repeat [2, 18, 31]. These DPRs are produced from both sense and antisense transcripts resulting in 5 possibly toxic peptides (poly-GA, −GP, −GR, −PR and -PA) and are found as inclusions in post-mortem brain material of C9ORF72 carriers [28, 38].
How these three mechanisms – alone or in combination – cause neurodegeneration is currently under investigation. Several studies have indicated that especially the DPRs are toxic in both cell culture and in vivo models, with the arginine-containing poly-GR and -PR DPRs being the most detrimental [3, 30, 47]. And poly-GR has been associated with neurodegeneration in human post-mortem brain sections [35, 36]. DPRs seem to cause various types of stress to the cell, including ER-stress, mitochondrial stress and nucleolar stress [3]. They can disturb the formation of membrane-less organelles, including RNA granules, nucleoli, spliceosomes and the nuclear pore complex (NPC) and facilitate the formation of stress granules [26]. Furthermore, nucleocytoplasmic transport and autophagy defects have been reported [40, 53, 56]. In addition to the list of disturbed pathways found in model systems of C9FTD/ALS, there have also been a substantial number of proteins found to interact, bind or aggregate with repeat-containing RNA or DPRs [21, 48]. Currently, the primary affected pathways involved in the pathogeneses of C9FTD/ALS are under debate [3, 17, 19]. Constituents of inclusions in human C9FTD/ALS brain material might provide a tool to identify key players in neurodegeneration.
Here, we tested a set of proteins implicated in aberrant pathways in C9ORF72 disease models for their presence in pathology or abnormal localization in post-mortem human C9FTD/ALS brain sections. We selected Ran-GAP for its implication in nucleocytoplasmic transport defects [54], ADARB2 for its role in RNA binding and editing [13] and HR23B for its dual function in both DNA repair and the UPS [51]. FMRP and Pur-alpha were selected for their binding to C9ORF72 mRNA, their localization in stress granules and their rescue effect in multiple C9ORF72 models [34]. Surprisingly, we found only HR23B protein to be a constituent of inclusions observed in C9FTD/ALS cases. In this report, we describe HR23B distribution and its co-localization with known C9ORF72 pathological hallmarks (DPRs, p62 and pTDP-43). Furthermore, we analyze HR23B function in DNA damage repair, the ubiquitin-proteasome system and ER-associated degradation. Disturbances of these pathways may contribute to the disease onset and/or progression of C9FTD/ALS.
Materials and methods
Five C9ORF72 FTD, two GRN FTD, two MAPT FTD, three sporadic ALS and three non-demented control human brain sections were provided by the Dutch Brain Bank. C9ORF72 ALS brain material of two patients was collected post-mortem at the department of Neuropathology of Amsterdam UMC, University of Amsterdam, according to local legal and ethical regulations. Patients or relatives gave informed consent for autopsy and use of brain tissue for research purposes. Information about our patient cohort can be found in Table 1. Human fibroblasts lines were provided by the cell repository of the department of clinical genetics. All participants gave written informed consent for all obtained materials. The study was approved by the Medical and Ethical Review Committee of the Erasmus Medical Center. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Table 1.
Patient characteristics
| Patient | Clinical diagnosis | Family history | Genetic diagnosis | Age of onset | Disease duration | Male/ Female | Brain weight |
|---|---|---|---|---|---|---|---|
| 1 | bvFTD | FTD | C9ORF72 | 51,8 | 8,7 | Male | 960 g |
| 2 | bvFTD | FTD | C9ORF72 | 55,8 | 9,1 | Male | 1184 g |
| 3 | bvFTD | FTD and ALS | C9ORF72 | 66,4 | 8,1 | Female | 1060 g |
| 4 | bvFTD | ALS and dementia | C9ORF72 | 63,2 | 6,8 | Female | 958 g |
| 5 | bvFTD | FTD and ALS | C9ORF72 | 55,2 | 9,5 | Male | 1075 g |
| 6 | FTD | N/A | Progranulin (Gln200X) | 60,6 | 5,5 | Female | 894 g |
| 7 | FTD | FTD | Progranulin (Ser82ValfsX174) | 47,4 | 4,3 | Female | unknown |
| 8 | FTD | FTD | MAPT (G272 V) | 42,6 | 8,4 | Male | 962 g |
| 9 | FTD | FTD | MAPT (P301L) | 51,1 | 9,7 | Male | 887 g |
| 10 | ALS | N/A | unknown | 70 | 1 | Male | 1428 g |
| 11 | ALS | N/A | unknown | 65 | 2,2 | Female | 1125 g |
| 12 | ALS | N/A | unknown | 75 | 1,1 | Male | 1255 g |
| 13 | ALS | N/A | C9ORF72 | 60 | 4,4 | Female | 1390 g |
| 14 | ALS | N/A | C9ORF72 | 66 | 3,5 | Male | 1275 g |
| 15 | ALS | N/A | C9ORF72 | 71 | 2,4 | Female | 1080 g |
| 16 | Non-demented | N/A | unknown | N/A | N/A | Female | 1080 g |
| 17 | Non-demented | N/A | unknown | N/A | N/A | Male | 1215 g |
| 18 | Non-demented | N/A | unknown | N/A | N/A | Female | 1139 g |
Immunohistochemistry on human brain sections
Human brain sections (6 μm) were deparaffinized in xylene and rehydrated (100%–96%-90%–80%-70%–50% EtOH serie). Antigen retrieval was done in 0.01 M sodium citrate, pH 6.0 using pressure cooker treatment. Endogenous peroxidase activity was blocked with 0.6% H2O2 and 1,25% sodium azide in 0.1 M PBS. Immunostaining was performed overnight at 4 °C in PSB block buffer (0.1 M PBS/0.5%protifar/0.15%glycine). Antibodies used in this study are listed in Additional file 1: Table S3, including concentration and brand/catalogue number. Antigen-antibody complexes were visualized by incubation with DAB substrate (DAKO) after incubation with Brightvision poly-HRP-linker (Immunologic) or anti-mouse/rabbit HRP (DAKO). Slides were counterstained with Mayer’s haematoxylin and mounted with Entellan (Merck Millipore International). The slides were then left to dry in the fume hood for an hour and thereafter put in an 37 °C incubater overnight. Pictures were taken by using an Olympus BX40 microscope (Olympus).
Immunofluorescence staining on human brain sections
Human brain sections were treated as described above. After incubation with the primary antibody, sections were washed with PBS block buffer (1xPBS/0.5%protifar/0.15%glycine) and incubated with secondary anti-mouse/rabbit Cy2/3 linked antibodies (Jackson). To remove background staining, a 10 min incubation with Sudan Black (Sigma, 0.1 g in 100 ml 70% ethanol, filtered) was done. To visualize nuclei, slides were incubated for 10 min with Hoechst 33342(Invitrogen). Slides were mounted with ProLongGold (Invitrogen) and kept at 4 °C until imaging at a Zeiss LSM700 Confocal microscope.
Assessment of neurodegeneration and protein pathology
For the neuropathological assessment we used brain sections from 5 C9ORF72 FTD cases (see Table 1). Five different brain regions (frontal cortex, temporal cortex, motor cortex, hippocampus and cerebellum) per patient were semi-quantified on neurodegeneration and pathological score of p62, pTDP-43 and HR23B (Additional file 2: Table S1). Counting was not performed in a blinded fashion. Neurodegeneration was assessed on haematoxylin and eosin (HE) sections and graded as absent (0), mild (1), moderate (2) or severe (3) based on the presence of neuronal loss. The neurodegenerative score from the pathological report was also taken into account. Pathological scores were rated as (0) if completely absent, rare (1) if only a few could be found one brain section, occasional (2) if they not present in every microscopic field, moderate (3) if at least a few examples were present in most microscopic fields, and numerous (4) when many were present in every microscopic field [28]. We reported the overall number of immunoreactive inclusions (total score) as well as the number of neuronal cytoplasmic inclusions (NCI), neuronal intranuclear inclusions (NII) and dystrophic neurites (DN) (Additional file 2: Table S1). Total scores were assessed independent from NCI, NII and DN scores and represent an impression of the global load of pathology and the number of inclusions using the same grading system as described above. Quantification of co-localization of HR23B with DPRs, p62 and pTDP-43 was not performed in a blinded fashion.
Colony forming assays
Human fibroblast lines from 4 C9ORF72 carriers (13E634, 13E659, 17E0225, 17E0278) and 4 controls (81E253, 86E1375, 06E0717 and 99E0774) were obtained from the cell repository of the department of clinical genetics and the XP25RO fibroblast line was provided by the department of molecular genetics. Fibroblasts were cultured in DMEM medium (Gibco) with 10% fetal calf serum, 1% penicillin/streptavidin and 1% non-essential amino acids. To determine UV-sensitivity, human fibroblasts were seeded in triplicate in 10 cm plates (Greiner Bio-one) in a density of 2000 cells/plate. After 24 h, cells were treated with increasing doses of UV-C (254 nm UV-C lamp, Philips). After 5–7 days, colonies were fixed with 0.1% w/v Coomassie Blue (Bio-Rad) in a 50% Methanol, 10% Acetic Acid solution. Colonies were counted with the integrated colony counter GelCount (Oxford Optronix). Counting was performed automatically with the same settings for each fibroblast cell line, but not in a blinded fashion.
Immunofluorescence on human fibroblasts
To determine DNA damage recruitment of NER proteins, human fibroblasts were seeded on coverslips and after 1 week in culture irradiated with 60 J/m2 (254 nm UVC lamp, Philips) through an 8 μm microporous filter (Millipore) to induce sub-nuclear local DNA damage. Cells were fixed after 30 min with 2% paraformaldehyde and permeabilized with 0.1% Triton X-100 for 20 min. Next, cells were incubated with fresh 0.07 M NaOH in PBS for 5 min to denature DNA and enable CPD staining. Cells were then washed with PBS containing 0.15% glycine and 0.5% BSA and incubated with primary antibodies (see Additional file 1: Table S3) overnight at 4 °C. The next day, cells were washed with 0.1% Triton X-100 and incubated with Alexa Fluor conjugated secondary antibodies (488, 555 and 633; Invitrogen) for 1 h at RT. Coverslips were mounted using ProLongGold with DAPI (Invitrogen) and imaged using a Zeiss LSM700 microscope.
Unscheduled DNA synthesis (UDS)
To measure NER capacity, human fibroblasts were grown on coverslips for 1 week in culture and irradiated with 16 J/m2 UV-C. After irradiation, cells were incubated for 1 h in UDS medium (F10, 1% PS, 10% dialyzed serum) with 2% HEPES and 1% 5-ethynyl-2′-deoxyuridine (EdU, Invitrogen). After 1 h, medium was changed to normal DMEM medium (10% FCS, 1% NEAA, 1% PS) for 10 min. Next, cells were fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Blocking was done using 1.5% BSA in PBS for 30 min. EdU incorporation was visualized by incubating cells for 1 h at room temperature with Click-it reaction cocktail containing Atto 594 Azide (60 μM, Atto Tec.), Tris-HCl (50 mM, pH 7.6), CuSO4*5H2O (4 mM, Sigma) and ascorbic acid (10 mM, Sigma). After the Click-it reaction, cells were washed with 0.1% Triton X-100, incubated with Hoechst 33342 (Invitrogen) for 10 min and mounted with ProLongGold (Invitrogen). Images were acquired using a Zeiss LSM700 microscope. UDS levels were quantified by measuring the total nuclear fluorescence intensities (in at least 50 cells per experiment) with FIJI image analysis software. Intensity levels were averaged and normalized to the fluorescence levels in unirradiated cells. Quantification of intensity levels was done automatically with FIJI but was not performed in a blinded fashion.
Results
Characterizing Ran-GAP, ADARB2, HR23B, FMRP and Pur-alpha in a cohort of C9FTD patients and non-demented controls
We started with assessing the localization of ras-related nuclear protein GTPase activating protein (Ran-GAP) in C9FTD patient brain sections. We studied five FTLD-TDP cases with the C9ORF72 repeat expansion and three non-demented cases (for information about our cohort, see Table 1). We predominantly found a diffuse nuclear staining or nuclear envelop staining of Ran-GAP. Most nuclei were round-shaped but occasionally we observed misfolded nuclei or the invagination of the nuclear membrane (Additional file 3: Figure S1). The number of oddly-shaped nuclei did not clearly differ between C9FTD cases and non-demented controls. Next, we investigated Adenosine Deaminase, RNA Specific B2 (ADARB2) localization. ADARB2 immunostaining showed both nuclear and cytoplasmic localization in both C9FTD and non-demented controls. We could detect some intranuclear inclusions in the hippocampus dentate gyrus of C9FTD patients (Additional file 4: Figure S2A). However, IF double staining of ADARB2 with p62 showed similar results between hippocampal nuclei of C9FTD patients and non-demented cases (Additional file 4: Figure S2B).
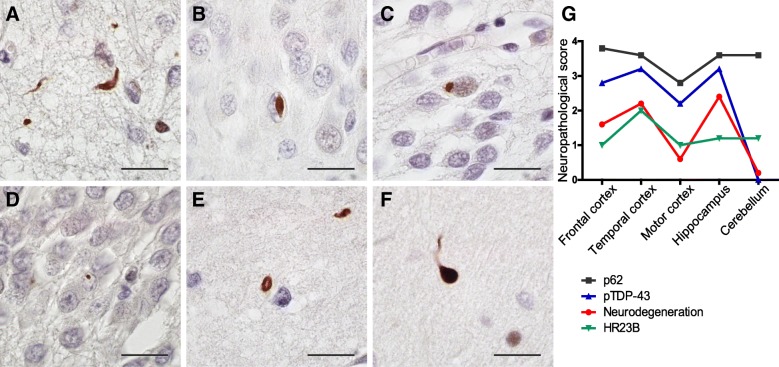
Next, we stained for HR23B and found different types of inclusions in cortical areas, hippocampus and cerebellum of C9FTD cases (Fig. 1 and Additional file 5: Figure S5). Most cytoplasmic inclusions showed a round and perinuclear appearance (Fig. 1a and c), while some inclusions had a negative central core surrounded by a positive halo labeling (Fig. 1e). Neuropils were mostly found in layer 2 of cortical areas (Fig. 1a). Intranuclear inclusions were also observed, including cat-eye inclusions (Fig. 1b). Overall, layer 2/3 and layer 5/6 of frontal and temporal cortices showed the highest HR23B pathological burden in C9FTD cases, followed by motor cortex, hippocampus and cerebellum (Fig. 1g and Additional file 2: Table S1). Hippocampus dentate gyrus (DG) harbored some perinuclear inclusions and hippocampus cornu ammonis (CA) showed some cells with strong nuclear staining (Additional file 5: Figure S5). Cerebellum showed low HR23B staining with some nuclear and perinuclear inclusions in the granular layer and nearly absent in the molecular layer (Additional file 5: Figure S5). Non-demented controls showed normal nuclear localization of HR23B (Additional file 5: Figure S5).
Fig. 1.
Type and spreading of HR23B pathology found in C9FTD cases. Different types of HR23B pathology in C9FTD cases: a) neuropils and puncta in frontal cortex layer 2. b) intranuclear (cat eye) inclusion in hippocampus dentate gyrus. c) perinuclear inclusion in hippocampus dentate gyrus. d) round intranuclear inclusion in hippocampus dentate gyrus. e) round or oval inclusion with a hole in frontal cortex f) dystrophic neuron in cerebellum molecular layer. g) Spreading of HR23B compared to known p62 and pTDP-43 pathology. Depicted are semi-quantitive measures of neurodegeneration and pathological score in C9FTD. Neuronal loss score was based on hematoxylin and eosin (HE) staining and pathological report and scored as absent (0), mild (1), moderate (2) or severe (3). Pathological scores were based on the degree of pathology as absent (0), rare (1), occasional (2), moderate (3), or numerous (4). See also Additional file 9: Table S2 for details of pathological quantifications. All scale bars are 20μm
Furthermore, we assessed fragile X mental retardation protein (FMRP) localization in C9FTD cases. Punctuated FMRP staining in the cytoplasm indicative of the formation of stress granules was absent in our C9FTD cases (Additional file 6: Figure S3). FMRP was evenly distributed in the cytoplasm in both C9FTD cases and controls. Very occasionally, we could detect inclusions in the hippocampus dentate gyrus in both C9FTD cases and controls (Additional file 6: Figure S3). Finally, we set out to test Pur-alpha staining, which showed to be located in stress granules in both C9FTD cases and controls (Additional file 7: Figure S4). We did not find Pur-alpha intranuclear inclusions in the cerebellum or other brain areas of C9FTD cases nor controls (Additional file 7: Figure S4).
HR23B pathology is also present in C9ALS and GRN FTD post-mortem brain tissue
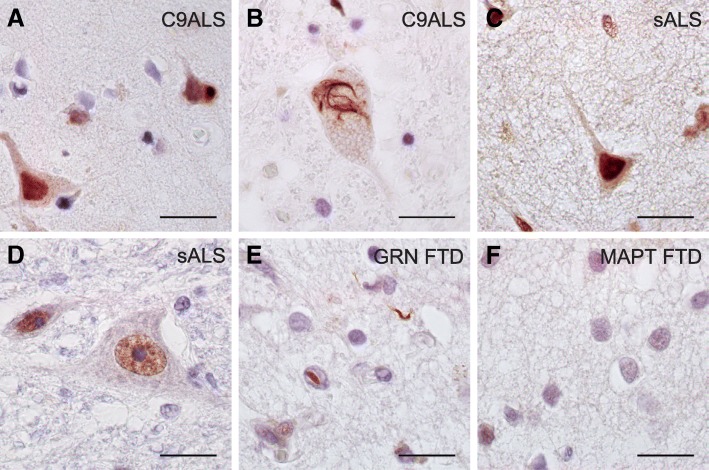
In order to validate the observed HR23B pathology, we used a second independent HR23B antibody (for details see methods) that revealed similar results (Additional file 8: Figure S6). Furthermore, we expanded our cohort with three ALS cases with the C9ORF72 repeat expansion, three sporadic ALS cases with unknown genetic cause, two FTD with GRN mutation and two FTD-MAPT cases. In non-demented controls, we observed immunoreactivity for HR23B in nuclei (Additional file 5: Figure S5 and Additional file 8: Figure S6). C9ALS cases showed very strong HR23B staining in nuclei and some cytoplasmic inclusions in motor cortex (Fig. 2a) and spinal cord sections (Fig. 2b). Sporadic ALS cases also showed very strong nuclear HR23B staining but no pathology in motor cortex (Fig. 2c) nor spinal cord (Fig. 2d). GRN FTD cases showed the same extend of HR23B pathology as C9FTD in frontal cortex, consisting of some intranuclear inclusions, cytoplasmic inclusions and neuropils (Fig. 2e). HR23B pathology was absent in MAPT FTD cases (Fig. 2f).
Fig. 2.
HR23B pathology is also present in C9ALS and GRN FTD cases. a) HR23B staining in C9ALS motor cortex shows strong staining in the nucleus and a cytoplasmic inclusion. b) C9ALS spinal cord section with cytoplasmic HR23B pathology c) Sporadic ALS with very strong nuclear HR23B staining in motor cortex d) and in spinal cord. e) GRN FTD case with an intranuclear inclusion and neurites positive for HR23B in frontal cortex. f) HR23B pathology is absent in frontal cortex of MAPT FTD. All scale bars are 20 μm
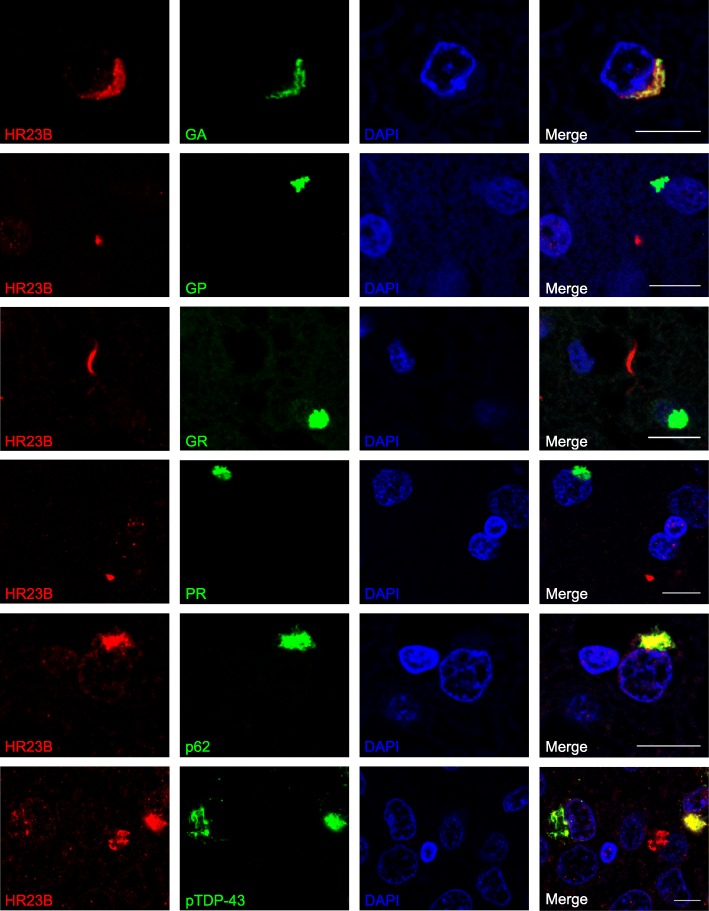
HR23B co-localizes with poly-GA, pTDP-43 and p62 in C9FTD cases
To evaluate HR23B’s aggregation process in C9FTD, we performed double labeling with known pathological hallmarks, including DPRs, pTDP-43 and p62 in five C9FTD frontal cortices (Fig. 3). HR23B was mostly found to be co-localized with p62 (Fig. 3), as 66% of all HR23B inclusions were also positive for p62 (Additional file 9: Table S2). Next is pTDP-43 with 22.6% of HR23B inclusions being positive for pTDP-43 (Fig. 3 and Additional file 9: Table S2). From all DPRs, HR23B showed partial co-localization with poly-GA (Fig. 3), for 6.6% (Additional file 9: Table S2). The other DPRs only co-stained with 0–3% of all HR23B inclusions. We did not evaluate poly-PA because we found too few inclusions. Interestingly, hippocampus dentate gyrus showed a much higher co-localization between DPRs and HR23B than frontal cortex (Fig. 4 and Additional file 9: Table S2). We found 60.6% of HR23B inclusions being positive for poly-GA in the dentate gyrus, which is nearly a 10-fold increase compared to the frontal cortex (Fig. 4 and Additional file 9: Table S2). Poly-GP increased from 0.8% in frontal cortex to 4.7% in hippocampus and poly-GR also showed a 10-fold increase from 1% in frontal cortex to 10.5% in hippocampus (Fig. 4 and Additional file 9: Table S2). Only poly-PR stayed fairly undetectable with a slight increase from 0.65% in frontal cortex to almost 1% in hippocampus. HR23B co-localization with p62 in dentate gyrus was also higher in hippocampus than in frontal cortex (87.4% vs 66%) (Fig. 4 and Additional file 9: Table S2). We performed a 2-way ANOVA to compare co-localization percentages of HR23B with pathological hallmarks between different brain areas (p < 0.0001). Post Bonferroni test indicated that percentages of poly-GA and p62 were significantly different between hippocampus DG and frontal cortex (Fig. 4 and Additional file 9: Table S2). For the other DPRs and pTDP-43, differences were not significant (Fig. 4 and Additional file 9: Table S2). These data suggest a difference in aggregation formation and co-localization patterns between different brain areas or cell types.
Fig. 3.
HR23B co-localizes with p62, TDP-43 and poly-GA in C9FTD cases. Immunofluorescent staining for HR23B (shown in red) in combination with DPRs (poly-GA, −GP, −GR and -PR) or p62 or pTDP-43 (shown in green). Poly-PA was not evaluated because too few inclusions were found. All pictures are from frontal cortex of C9FTD cases. All scale bars are 10 μm
Fig. 4.
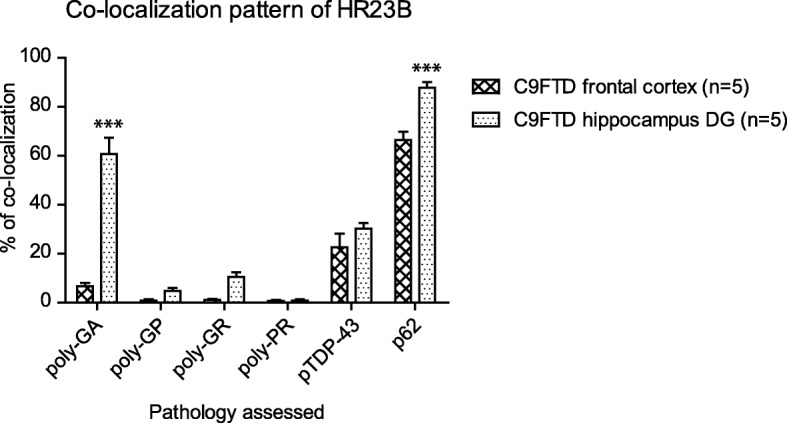
HR23B co-localization percentages with poly-GA and p62 differ between frontal cortex and hippocampus DG. Semi-quantification of co-localization of HR23B with pathological hallmarks such as DPRs, p62 and pTDP-43 based on raw data in Additional file 9: Table S2. Two-way ANOVA is significant (p < 0.0001) for pathology, brain area and interaction. Bonferroni test indicates that only poly-GA and p62 are significantly different between frontal cortex and hippocampus DG (both p < 0.001)
Nucleotide excision repair is not affected in C9ORF72 patient fibroblasts
To assess any changes in the normal cellular function of HR23B, we first focused on its role in DNA damage repair. HR23B interacts with and stabilizes Xeroderma pigmentosum, complementation group C (XPC) protein [32], which is involved in the recognition of bulky DNA adducts in global genome nucleotide excision repair (GG-NER). Staining with XPC antibody did not reveal any gross differences between C9FTD and non-demented control post-mortem brain sections (Fig. 5). To assess nucleotide excision repair capacity in living cells, we performed UV-sensitivity assays with four C9ORF72 patient fibroblast lines and compared these to four healthy control fibroblast lines. Intriguingly, C9ORF72 fibroblasts were more sensitive to UV-C damage than healthy control fibroblasts (Additional file 10: Figure S7A), but not as sensitive as a fibroblast line that is fully deficient in NER (XP25RO, homozygous for 619C > T causing an ARG207X change in exon 5 of the XPA gene). The GG-NER pathway is initiated by recognition of DNA damage by the HR23B/XPC/CETN2 complex, which is then followed by multiple downstream steps to verify and excise the damage [29]. We therefore assessed whether factors involved in each of the steps of the NER pathway were recruited normally to DNA damage in C9ORF72 fibroblasts. To do so, we evoked local DNA damage by UV-C irradiation through a microporous filter in our fibroblast lines and performed immunofluorescence to visualize DNA damage (by cyclobutane pyrimidine dimers (CPD) antibody) and DNA damage recruitment of NER factors XPC, XPB, XPA, XPF and XPG. All tested NER proteins clearly co-localized with DNA damage in C9ORF72 patient fibroblasts, indicating that recognition and processing of CPDs is still functional in these cells (Additional file 10: Figure S7B). To finally verify that NER is fully operational, we measured incorporation of the thymidine analogue 5-ethynyl-2-deoxiuridine (EdU) after UV-C irradiation to quantify the efficiency of DNA repair in our fibroblast lines. The NER-deficient XP25RO cell line, which we used as negative control, did not show any EdU incorporation. In contrast, we still observed efficient EdU incorporation in our four C9ORF72 patients cell lines to similar levels as in four healthy control lines (Additional file 10: Figure S7C), indicating that NER is not deficient in C9ORF72 patient fibroblasts.
Fig. 5.
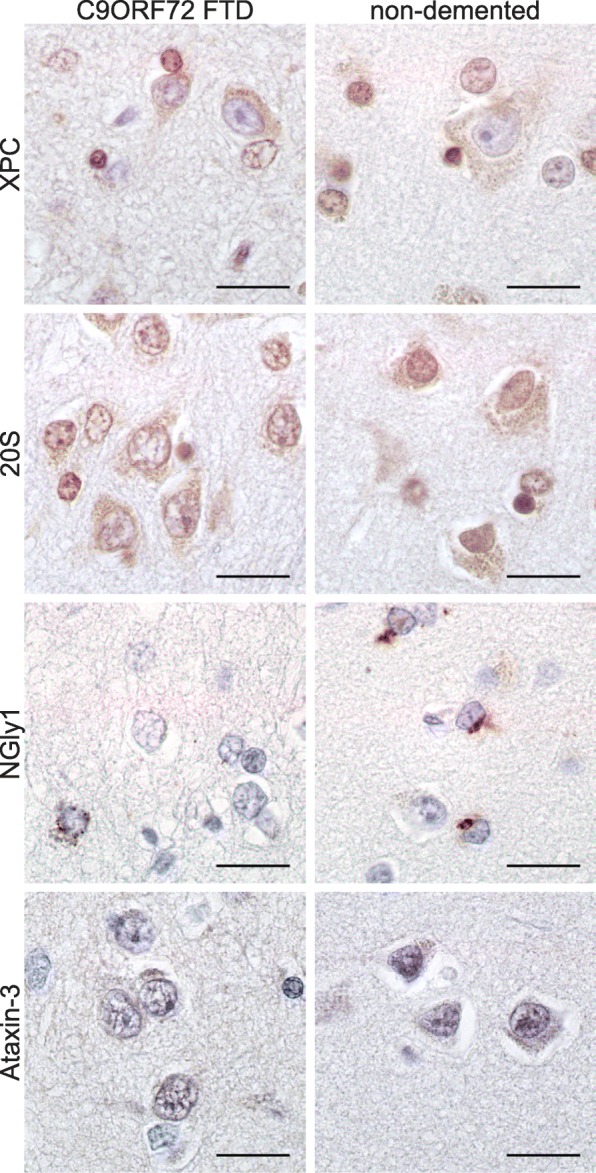
HR23B does not sequester its bindings partners into inclusions. XPC, 20S and ataxin-3 stainings do not reveal any differences between C9FTD patients and non-demented controls. For NGly1, we observed less nuclei with perinuclear staining in C9ORF72 FTD brains than in non-demented controls. All pictures are from frontal cortex. All scale bars are 20 μm
NGly1, ERAD factor and HR23B binding partner, is less abundant in C9FTD brain sections
Besides its role in DNA damage repair, HR23B is also known for its function in the ubiquitin-proteasome system. Various HR23B binding partners have been identified which are involved in the unfolded protein response (UPR), transcriptional regulation, cell cycle control and ER-associated degradation (ERAD) [51]. To assess if HR23B aggregation also evokes changes in the localization of the proteasome, we stained our sections for proteasome subunit 20S. However, we did not observe any obvious changes in 20S normal localization (Fig. 5). Another binding partner of HR23B is ataxin-3, a deubiquitinase enzyme in which a poly-glutamine expansion is linked to SCA3 [46]. HR23B-positive inclusions have been found in post-mortem brain material of SCA3 patients [6] however we could not detect ataxin-3 pathology in C9FTD cases (Fig. 5). Besides the proteasome, we wondered if we could find any changes in ERAD. HR23B did not sequester NGly1, one of its known binding partners involved in ERAD, into protein inclusions. Strikingly, however, we did observe clearance of NGly1 staining in C9FTD frontal cortex (Fig. 5). Although some neurons showed the same strong peri-nuclear staining as almost all non-demented control cells, in the majority of patient neurons no NGly1 staining was observed, indicating reduced expression of NGly1 in C9FTD brains. This may suggest a partial loss of function of ERAD in a subset of neurons in the brain of C9FTD patients.
Discussion
In the present study, we characterize HR23B pathology distribution and its co-localization pattern with pathological hallmarks. To our knowledge, we are the first to show that HR23B co-localizes with pTDP-43 pathology in brain tissue of C9FTD patients. We could also demonstrate HR23B pathology in C9ALS and sporadic ALS. HR23B pathology has been described before in HD, SCA3, SCA7, FXTAS and PD [6]. HR23B pathology was also present in both C9ORF72 and GRN FTD cases, but not in MAPT FTD cases, in contrast to a previous report of HR23B pathology described for FTDP-17 (FTD with parkinsonism with Pick bodies consisting of tau protein) [6]. Also Alzheimer’s disease (AD) brain material did not show HR23B pathology [6]. Why AD and some FTLD-tau patients are an exception and do not present with HR23B pathology is unclear and requires further investigation. It should be noted that our study cohort was rather small (see Table 1), which might explain why we could not confirm HR23B pathology in MAPT FTD cases. Next to HR23B, HR23A inclusions have been reported in FXTAS and in C9FTD post-mortem brain tissue [6, 55]. In general, HR23 pathology seems to be widespread among neurodegenerative diseases, which underscores its relevance in a common disease pathogenesis of these disorders.
In our semi-quantitative co-localization studies, we found HR23B to be co-localized predominantly with p62, followed by pTDP-43 and poly-GA. HR23B inclusions were nearly always negative for the other DPRs. Overexpression of poly-GA in mouse models is enough to sequester HR23B [37, 55] and in a co-immunoprecipitation experiment of lysates from cells expressing GFP-tagged poly-GP, −GR and -GA, only poly-GA was found to bind HR23B [55]. This could also be a stochastic event, given the fact that poly-GA inclusions are the most abundant DPR in C9FTD/ALS brains [28]. It could also be caused by the aggregation-prone nature of the poly-GA peptide itself [9], causing HR23B to bind easier or faster to poly-GA peptides than other DPRs. Binding of HR23B to DPRs also differs between brain areas; in the frontal cortex, only 6.6% of HR23B inclusions were poly-GA positive, while this increased to 60.6% in the hippocampus dentate gyrus. The same is true for poly-GP and -GR; co-localization with HR23B increased 5- to 10-fold between frontal cortex and hippocampus, although it remained low (1–10%). Subtypes of neurons may differ in their vulnerability for DPRs or there might be differences in the expression level and availability of HR23B and/or its binding partners between frontal cortex and hippocampus dentate gyrus. Levels of HR23B can influence aggregation of poly-GA and toxicity of mutant TDP-43 and mutant SOD1 [23, 55]. Overexpression of HR23B protected for the formation of poly-GA inclusions in mouse primary neuronal cultures [55]. In contrast, loss of HR23B seems to protect against motor neuron disease by enhancing mutant protein clearance [23]. Levels of HR23B seem to have opposite effects on aggregation, clearance and solubility of several proteins, which could result in differences in aggregation and co-localization patterns observed between different brain areas.
HR23B can directly bind the proteasome and ataxin-3, a deubiquitinase enzyme that binds ubiquitinated proteins and can also bind to the proteasome [14, 46]. Even though HR23B and proteasome subunits are sequestered into intranuclear inclusions of ataxin-3 in SCA3 patients brain tissue [6, 39], ataxin-3 and 20S do not show an aberrant localization in C9FTD patients post-mortem brain tissue. HR23B can also bind PNGase, a deglycosylation hydrolase involved in ERAD of misfolded glycoproteins. The affinity of PNGase for the proteasome is HR23B-dependent, which makes HR23B essential for the shuttling of misfolded proteins to the proteasome [25]. If HR23B is sequestrated into inclusions and becomes unavailable for PNGase, this might cause loss of initiation of ERAD. This is in line with our observation that a substantial number of neurons of C9FTD/ALS patients show less abundant NGly1 staining. Many FTD-causing mutations are associated with protein degradation pathways [20]. In addition, mutations in NGLY1, the gene encoding PNGase, are linked to motor impairment, intellectual disability, and neuropathy in humans [8].
HR23B is well known for its role in global genome nucleotide excision repair (GG-NER) and genetic polymorphisms in RAD23B are modifiers of laryngeal cancer risk in human [1]. The DNA damage response can be induced by the C9ORF72 repeat expansion [15] and elevated levels of R-loops (DNA-RNA hybrids), double strand breaks and ATM-mediated DNA repair signaling defects have been described before in rat neurons, human cells and C9ALS spinal cord tissue [15, 45]. Furthermore, ALS and C9ORF72 repeat carriers have an increased risk for melanoma [16, 42], suggesting they may have an reduced response to DNA damage. XPC, the binding partner of HR23B in NER, was found in inclusions in a poly-GA mouse model of C9FTD/ALS [55]. Nonetheless, we could not find XPC pathology in our human brain sections nor deficits in the NER pathway in C9ORF72 patient fibroblasts, even though C9ORF72 patient fibroblasts seem to be more sensitive for UV-C damage than healthy control fibroblasts. Why we do not find a clear impairment of NER in our study is unknown. Species-specific factors, overexpression of poly-GA in the mouse model or difference between fibroblasts and neurons might explain a part of the absence of an effect. In addition, it could be possible that HR23A takes over the DNA repair function of HR23B when the latter is sequestered or dysfunctional. This has been demonstrated in mHr23b knockout (KO) mice that show no impairment in NER [51]. Still, mHr23b KO mice display impaired embryonic development, retarded growth and facial dysmorphologies that are not observed in mouse models deficient in other NER genes [51], which suggests a second function of HR23B. Although HR23A and HR23B have similar functions in DNA repair, they form distinct interactions with various cellular factors, including proteasomes, multi-ubiquitinated proteins and stress-related factors [10].
Here, we set out to validate the aggregation of several proteins that have been described to mis-localize or bind RNA foci in C9FTD/ALS. Strikingly, we could not reproduce earlier published pathology for Ran-GAP, ADARB2, Pur-alpha and FMRP. The differences observed between our study and previous publications can be explained by multiple factors. First of all, we used post-mortem brain material that only presents the end-stage of the disease, so changes in localization of proteins in early stages of the disease can be missed. Also, the number of cells presenting with stress granules varies a lot between subjects and might be attributed to autolytic processes during human brain preservation, which can make it hard to detect subtle differences. Secondly, this study focused on FTD rather than ALS. This could especially be important for ADARB2, as one of the targets of ADAR proteins is the Q/R site of the GluR2 AMPA receptor [22]. Changes in ADARB2 localization could therefore mostly affect ALS cases and may be missed in our FTD cohort. Thirdly, pathology observed in cell culture and in vivo models could be due to overexpression of C9ORF72 RNA or DPRs in these models. Changes in patient neurons with endogenous expression can be more subtle but still act disturbing over time. Most model systems used so far do not include haploinsufficiency, which can be a modifying factor for cellular toxicity as well [41]. Finally, effects might be missed due to our small cohort. For example, differences in oddly-shaped nuclei in the Ran-GAP staining might only become evident when quantifying large number of cells. However, Saberi et al. also were unable to confirm Ran-GAP pathology [35], which strengthens our findings and illustrates the need for validation studies in biomedical research. Even though we did not observe any pathology of Ran-GAP, ADARB2, FMRP and Pur-alpha, their levels could still have a modifying effect on disease progression of FTD, as has been shown in iPSC-derived neurons and Drosophila models [7, 13, 50, 54].
Conclusion
In this study, we confirm HR23B aggregation and its implication in C9FTD/ALS. HR23B has an important function in both the DNA damage response and the degradation of proteins via the UPS, UPR and ERAD. Our results in human postmortem brain tissue suggests that especially the degradation of proteins via ERAD may be involved in the pathogenesis of ALS and FTD. The exact role and timing of HR23B in disease onset and progression needs further investigation, including its interaction with and possible degradation of proteins implicated in neurodegenerative disorders.
Additional files
Table S3. Antibody information. (DOCX 16 kb)
Table S1. Neuropathological scores of C9ORF72 FTD patients. Neuronal loss score was based on hematoxylin and eosin (HE) staining and pathological report and scored as absent (0), mild (1), moderate (2) or severe (3). Pathological scores were based on the degree of pathology as absent (0), rare (1), occasional (2), moderate (3), or numerous (4). Brain areas: F = frontal cortex, T = temporal cortex, M = motor cortex, H = hippocampus dentate gyrus, C = cerebellum. NCI = neuronal cytoplasmic inclusion, NII = neuronal intranuclear inclusion, DNs = dystrophic neurites. (DOCX 23 kb)
Figure S1. Ran-GAP staining in C9FTD cases and non-demented controls. Ran-GAP is predominantly localized to the nucleus and nuclear membrane. Unevenly shaped nuclear membranes occur in both C9ORF72 FTD cases (n = 5) and non-demented controls (n = 3). All scale bars are 20 μm. (PDF 2202 kb)
Figure S2. ADARB2 staining in C9FTD cases and non-demented controls. A) Staining of ADARB2 in C9ORF72 FTD cases (n = 5) and non-demented control (n = 3) post-mortem brain sections shows some intranuclear inclusions in hippocampus CA and DG. All scale bars are 20 μm B) Immunofluorescence staining of ADARB2 (red) and p62 (green) in hippocampal dentate gyrus reveals ADARB2 punctuated staining and some intranuclear inclusions in both C9ORF72 FTD cases (n = 5) and non-demented controls (n = 3). Scale bars in fluorescent pictures are 10 μm. (PDF 2721 kb)
Figure S5. HR23B pathology in different brain areas of C9FTD cases. Staining of HR23B in several brain areas of C9FTD cases and non-demented controls. Pathology burden was highest in cortices (frontal, temporal and motor) and was mostly cytoplasmic (inclusions and neuropils) and intranuclear (cateye). Hippocampus dentate gyrus (DG) harbors perinuclear inclusions, and hippocampus cornu ammonis (CA) had some cells with strong nuclear staining. Pathology was low in cerebellum granular layer with only some nuclear and perinuclear inclusions and almost absent in cerebellum molecular layer. Our C9FTD cases did not show HR23B pathology in spinal cord neurons. All scale bars are 20 μm (PDF 2260 kb)
Figure S3. FMRP staining in C9FTD cases and non-demented controls. FMRP staining does not reveal any differences between C9ORF72 FTD cases (n = 5) and non-demented control (n = 3) post-mortem brain sections. Occasional inclusions are found in the hippocampus dentate gyrus in both C9FTD cases and controls. All scale bars are 20 μm. (PDF 1883 kb)
Figure S4 Pur-alpha staining in C9FTD cases and non-demented controls. Pur-alpha staining reveals abundant stress granules in both C9ORF72 FTD cases (n = 5) and non-demented controls (n = 3) post-mortem brain sections. All scale bars are 20 μm. (PDF 1826 kb)
Figure S6 Validation of HR23B pathology by a second independent antibody. HR23B staining using Abcam antibody in C9ORF72 FTD cases (n = 5) and non-demented control (n = 3) post-mortem brain sections. Staining pattern is consistent with HR23B GeneTex antibody (see Figs. 1 and 2). All scale bars are 20 μm (PDF 1543 kb)
Table S2. Co-localization of HR23B with DPRs/pTDP-43/p62 differs between brain areas. The percentage is the amount of HR23B inclusions also positive for other pathological hallmarks (not the other way around). For example 11/153 for poly-GA in C9FTD patient 1 means that out of 153 HR23B inclusions, 11 were also positive forpoly- GA, which is 7%. F = frontal cortex. H = hippocampus dentate gyrus. *1 = All co-localizations are fibrils of poly-GP and HR23B in frontal cortex. Perinuclear inclusions of poly-GP did not stain positive for HR23B. *2 = In total 2 poly-PA inclusions have been found in frontal cortex of 5 C9FTD patients, too less to quantify. We therefore state N/A = non-applicable in this table. *3 = Non-demented cases had some p62 and some HR23B inclusions per person per section, which sometimes overlapped. No pTDP-43 inclusions were found so no co-localization of pTDP-43 with HR23B in non-demented cases. (DOCX 19 kb)
Figure S7. Nucleotide excision repair is not affected in C9ORF72 human fibroblasts. A) Dose-response curve for 4 healthy control human fibroblast lines (81E253, 86E1375, 06E0717 and 99E0774) and 4 C9ORF72 human fibroblast lines (13E634, 13E659, 17E0225, 17E0278) and the NER deficient XP25RO human fibroblast line treated with increasing dose of UV-C light (0–12 J/m2). B) Immunofluorescence staining showing the recruitment of NER factors XPC, XPB, XPA, XPF and XPG to local DNA damage (visualized by CPD antibody), induced by 60 J/m2 UV-C irradiation through a microporous filter. Fibroblast lines used for pictures: 13E634, 13E659 and 81E253 C) Human fibroblasts lines were treated with 16 J/m2 UV-C light and incubated with EdU for 1 h to measure unscheduled DNA synthesis (UDS) as measure of DNA repair. The NER-deficient XPC25RO cell line is shown as negative control. Ctrl 1–4 are lines 81E253, 86E1375, 06E0717 and 99E0774 in this order. C9 1–4 are lines 13E634, 13E659, 17E0225, 17E0278 in this order. (PDF 934 kb)
Acknowledgements
Authors would like to thank Arjan F. Theil for his practical advice on colony forming assay experiments and Michiel Kooreman from the Dutch Brain Bank for his assistance in selecting and sending C9ORF72 FTD patient and sporadic ALS patient brain sections. We also would like to thank Ronald Buijsen for providing the ataxin-3 antibody. We thank the team who helped in the collection of ALS tissue samples (Prof. dr. D. Troost, Prof. dr. M. de Visser, Dr. A.J. van der Kooi and Dr. J. Raaphorst).
Funding
This study was supported by the European Joint Programme - Neurodegenerative Disease Research and the Netherlands Organization for Health Research and Development (PreFrontALS: 733051042; RW and JS). European Research Council Advanced Grant to WV (grant 340988-ERC-ID). ALS Stichting (2014–28, TOTALS) to RJP and the Netherlands ALS foundation (“The Dutch ALS Tissue Bank”) to EA.
Availability of data and materials
The data supporting the conclusions of this article is included within the article (and its additional file). Raw datasets (including UV-C treatment of human fibroblasts) analyzed during the study are available from the corresponding author on reasonable request.
Abbreviations
- AD
Alzheimer’s disease
- ADARB2
Adenosine Deaminase, RNA Specific B2
- ALS
Amyotrophic lateral sclerosis
- C9ORF72
Chromosome 9 open reading frame 72
- CPD
Cyclobutane pyrimidine dimers
- DN
Dystrophic neurites
- DPRs
Dipeptide repeat proteins
- EdU
5-ethynyl-2-deoxiuridine
- ERAD
Endoplasmic reticulum associated degradation
- FMRP
Fragile X mental retardation protein
- FTD
Frontotemporal dementia
- FXTAS
Fragile X associated tremor/ataxia syndrome
- GG-NER
Global genome nucleotide excision repair
- HD
Huntington’s Disease
- HE
Haematoxylin and eosin
- HR23B
Human homologue of yeast UV excision repair protein Rad23b
- IF
Immunofluorescence
- IHC
Immunohistochemistry
- NCI
Neuronal cytoplasmic inclusions
- NCT
Nucleocytoplasmic transport
- NER
Nucleotide excision repair
- NII
Neuronal intranuclear inclusion
- NPC
Nuclear pore complex
- p62
Sequestosome 1
- PD
Parkinson’s disease
- poly-GA
Poly-glycine-alanine
- poly-GP
Poly-glycine-proline
- poly-GR
Poly-glycine-arginine
- poly-PA
Poly-proline-alanine
- poly-PR
Poly-proline-arginine
- pTDP-43
Phosphorylated 43 kDa TAR DNA-binding protein
- RAN
Repeat-associated-non-AUG
- Ran-GAP
ras-related nuclear protein GTPase activating protein
- SCA3/7
Spinocerebellar ataxia type 3 and 7
- UDS
Unscheduled DNA Synthesis
- UPR
Unfolded-protein response
- UPS
Ubiquitin-proteasome system
- XPC
Xeroderma pigmentosum complementation group C
Authors’ contributions
FWR has designed, performed and analyzed experiments and drafted the manuscript. HL has designed, performed and analyzed experiments. HS has designed, performed and analyzed experiments. L.S and S.M have helped performing experiments. WV, EA, JP and JCS provided human fibroblasts and post-mortem brain samples and helped interpreted data. JCS and RW guided study design, have interpreted data, and substantively revised the work. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All participants gave written informed consent for all obtained materials. The study was approved by the Medical and Ethical Review Committee of the Erasmus Medical Center.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Frederike W. Riemslagh, Phone: 0031107044542, Email: f.w.riemslagh@erasmusmc.nl
Hannes Lans, Email: w.lans@erasmusmc.nl.
Harro Seelaar, Email: h.seelaar@erasmusmc.nl.
Lies-Anne W. F. M. Severijnen, Email: e.severijnen@erasmusmc.nl
Shamiram Melhem, Email: s.melhem@erasmusmc.nl.
Wim Vermeulen, Email: w.vermeulen@erasmusmc.nl.
Eleonora Aronica, Email: e.aronica@amc.uva.nl.
R. Jeroen Pasterkamp, Email: r.j.pasterkamp@umcutrecht.nl.
John C. van Swieten, Email: j.c.vanswieten@erasmusmc.nl
Rob Willemsen, Email: r.willemsen@erasmusmc.nl.
References
- 1.Abbasi R, Ramroth H, Becher H, Dietz A, Schmezer P, Popanda O. Laryngeal cancer risk associated with smoking and alcohol consumption is modified by genetic polymorphisms in ERCC5, ERCC6 and RAD23B but not by polymorphisms in five other nucleotide excision repair genes. Int J Cancer. 2009;125:1431–1439. doi: 10.1002/ijc.24442. [DOI] [PubMed] [Google Scholar]
- 2.Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balendra R, Isaacs AM (2018) C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol 14:544–558. 10.1038/s41582-018-0047-2 [DOI] [PMC free article] [PubMed]
- 4.Beeldman E, Raaphorst J, Klein Twennaar M, Govaarts R, Pijnenburg YAL, de Haan RJ, de Visser M, Schmand BA. The cognitive profile of behavioural variant FTD and its similarities with ALS: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2018;89:995–1002. doi: 10.1136/jnnp-2017-317459. [DOI] [PubMed] [Google Scholar]
- 5.Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, Pregent L, Daughrity L, Baker MC, Rademakers R, Boylan K, Patel TC, Dickson DW, Petrucelli L. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126:895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergink S, Severijnen LA, Wijgers N, Sugasawa K, Yousaf H, Kros JM, van Swieten J, Oostra BA, Hoeijmakers JH, Vermeulen W, Willemsen R. The DNA repair-ubiquitin-associated HR23 proteins are constituents of neuronal inclusions in specific neurodegenerative disorders without hampering DNA repair. Neurobiol Dis. 2006;23:708–716. doi: 10.1016/j.nbd.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Burguete AS, Almeida S, Gao FB, Kalb R, Akins MR, Bonini NM. GGGGCC microsatellite RNA is neuritically localized, induces branching defects, and perturbs transport granule function. Elife. 2015;4:e08881. doi: 10.7554/eLife.08881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caglayan AO, Comu S, Baranoski JF, Parman Y, Kaymakcalan H, Akgumus GT, Caglar C, Dolen D, Erson-Omay EZ, Harmanci AS, Mishra-Gorur K, Freeze HH, Yasuno K, Bilguvar K, Gunel M. NGLY1 mutation causes neuromotor impairment, intellectual disability, and neuropathy. Eur J Med Genet. 2015;58:39–43. doi: 10.1016/j.ejmg.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang YJ, Jeng US, Chiang YL, Hwang IS, Chen YR. The Glycine-alanine dipeptide repeat from C9orf72 Hexanucleotide expansions forms toxic amyloids possessing cell-to-cell transmission properties. J Biol Chem. 2016;291:4903–4911. doi: 10.1074/jbc.M115.694273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Madura K. Evidence for distinct functions for human DNA repair factors hHR23A and hHR23B. FEBS Lett. 2006;580:3401–3408. doi: 10.1016/j.febslet.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Cooper-Knock J, Walsh MJ, Higginbottom A, Robin Highley J, Dickman MJ, Edbauer D, Ince PG, Wharton SB, Wilson SA, Kirby J, Hautbergue GM, Shaw PJ. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain. 2014;137:2040–2051. doi: 10.1093/brain/awu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, Maragakis N, Tienari PJ, Petrucelli L, Traynor BJ, Wang J, Rigo F, Bennett CF, Blackshaw S, Sattler R, Rothstein JD. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doss-Pepe EW, Stenroos ES, Johnson WG, Madura K. Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol Cell Biol. 2003;23:6469–6483. doi: 10.1128/MCB.23.18.6469-6483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farg MA, Konopka A, Soo KY, Ito D, Atkin JD. The DNA damage response (DDR) is induced by the C9orf72 repeat expansion in amyotrophic lateral sclerosis. Hum Mol Genet. 2017;26:2882–2896. doi: 10.1093/hmg/ddx170. [DOI] [PubMed] [Google Scholar]
- 16.Freedman DM, Curtis RE, Daugherty SE, Goedert JJ, Kuncl RW, Tucker MA. The association between cancer and amyotrophic lateral sclerosis. Cancer Causes Control. 2013;24:55–60. doi: 10.1007/s10552-012-0089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao FB, Almeida S, Lopez-Gonzalez R. Dysregulated molecular pathways in amyotrophic lateral sclerosis-frontotemporal dementia spectrum disorder. EMBO J. 2017;36:2931–2950. doi: 10.15252/embj.201797568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, Cosio DM, van Blitterswijk M, Lee WC, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gendron TF, Petrucelli L (2018) Disease mechanisms of C9ORF72 repeat expansions. Cold Spring Harb Perspect Med 8. 10.1101/cshperspect.a024224 [DOI] [PMC free article] [PubMed]
- 20.Gotzl JK, Lang CM, Haass C, Capell A. Impaired protein degradation in FTLD and related disorders. Ageing Res Rev. 2016;32:122–139. doi: 10.1016/j.arr.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Haeusler AR, Donnelly CJ, Rothstein JD. The expanding biology of the C9orf72 nucleotide repeat expansion in neurodegenerative disease. Nat Rev Neurosci. 2016;17:383–395. doi: 10.1038/nrn.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hideyama T, Yamashita T, Aizawa H, Tsuji S, Kakita A, Takahashi H, Kwak S. Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol Dis. 2012;45:1121–1128. doi: 10.1016/j.nbd.2011.12.033. [DOI] [PubMed] [Google Scholar]
- 23.Jablonski AM, Lamitina T, Liachko NF, Sabatella M, Lu J, Zhang L, Ostrow LW, Gupta P, Wu CY, Doshi S, Mojsilovic-Petrovic J, Lans H, Wang J, Kraemer B, Kalb RG. Loss of RAD-23 protects against models of motor neuron disease by enhancing mutant protein clearance. J Neurosci. 2015;35:14286–14306. doi: 10.1523/JNEUROSCI.0642-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lashley T, Rohrer JD, Mead S, Revesz T. Review: an update on clinical, genetic and pathological aspects of frontotemporal lobar degenerations. Neuropathol Appl Neurobiol. 2015;41:858–881. doi: 10.1111/nan.12250. [DOI] [PubMed] [Google Scholar]
- 25.Lee JH, Choi JM, Lee C, Yi KJ, Cho Y. Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins. Proc Natl Acad Sci U S A. 2005;102:9144–9149. doi: 10.1073/pnas.0502082102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, Maxwell BA, Kim NC, Temirov J, Moore J, Kolaitis RM, Shaw TI, Bai B, Peng J, Kriwacki RW, Taylor JP. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167:774–788 e717. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, Adachi Y, Sardone V, Miller JW, Smith BN, Gallo JM, Ule J, Hirth F, Rogelj B, Houart C, Shaw CE. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackenzie IR, Frick P, Grasser FA, Gendron TF, Petrucelli L, Cashman NR, Edbauer D, Kremmer E, Prudlo J, Troost D, Neumann M. Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol. 2015;130:845–861. doi: 10.1007/s00401-015-1476-2. [DOI] [PubMed] [Google Scholar]
- 29.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 30.Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J, Cleverley K, Nicoll AJ, Pickering-Brown S, Dols J, Cabecinha M, Hendrich O, Fratta P, Fisher EM, Partridge L, Isaacs AM. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C, Edbauer D. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 32.Ng JM, Vermeulen W, van der Horst GT, Bergink S, Sugasawa K, Vrieling H, Hoeijmakers JH. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003;17:1630–1645. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, Consortium I, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi S, Serrano A, Gerbino V, Giorgi A, Di Francesco L, Nencini M, Bozzo F, Schinina ME, Bagni C, Cestra G, Carri MT, Achsel T, Cozzolino M. Nuclear accumulation of mRNAs underlies G4C2-repeat-induced translational repression in a cellular model of C9orf72 ALS. J Cell Sci. 2015;128:1787–1799. doi: 10.1242/jcs.165332. [DOI] [PubMed] [Google Scholar]
- 35.Saberi S, Stauffer JE, Jiang J, Garcia SD, Taylor AE, Schulte D, Ohkubo T, Schloffman CL, Maldonado M, Baughn M, Rodriguez MJ, Pizzo D, Cleveland D, Ravits J. Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol. 2018;135:459–474. doi: 10.1007/s00401-017-1793-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakae N, Bieniek KF, Zhang YJ, Ross K, Gendron TF, Murray ME, Rademakers R, Petrucelli L, Dickson DW. Poly-GR dipeptide repeat polymers correlate with neurodegeneration and Clinicopathological subtypes in C9ORF72-related brain disease. Acta Neuropathol Commun. 2018;6:63. doi: 10.1186/s40478-018-0564-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schludi MH, Becker L, Garrett L, Gendron TF, Zhou Q, Schreiber F, Popper B, Dimou L, Strom TM, Winkelmann J, von Thaden A, Rentzsch K, May S, Michaelsen M, Schwenk BM, Tan J, Schoser B, Dieterich M, Petrucelli L, Holter SM, Wurst W, Fuchs H, Gailus-Durner V, de Angelis MH, Klopstock T, Arzberger T, Edbauer D. Spinal poly-GA inclusions in a C9orf72 mouse model trigger motor deficits and inflammation without neuron loss. Acta Neuropathol. 2017;134:241–254. doi: 10.1007/s00401-017-1711-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, German Consortium for Frontotemporal Lobar D, Bavarian Brain Banking A. Arzberger T, Edbauer D. Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. 2015;130:537–555. doi: 10.1007/s00401-015-1450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt T, Lindenberg KS, Krebs A, Schols L, Laccone F, Herms J, Rechsteiner M, Riess O, Landwehrmeyer GB. Protein surveillance machinery in brains with spinocerebellar ataxia type 3: redistribution and differential recruitment of 26S proteasome subunits and chaperones to neuronal intranuclear inclusions. Ann Neurol. 2002;51:302–310. doi: 10.1002/ana.10101. [DOI] [PubMed] [Google Scholar]
- 40.Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang S, Wu LC, Ding M, Yu Y, Gall JG, McKnight SL. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc Natl Acad Sci U S A. 2017;114:E1111–E1117. doi: 10.1073/pnas.1620293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Y, Lin S, Staats KA, Li Y, Chang WH, Hung ST, Hendricks E, Linares GR, Wang Y, Son EY, Wen X, Kisler K, Wilkinson B, Menendez L, Sugawara T, Woolwine P, Huang M, Cowan MJ, Ge B, Koutsodendris N, Sandor KP, Komberg J, Vangoor VR, Senthilkumar K, Hennes V, Seah C, Nelson AR, Cheng TY, Lee SJ, August PR, Chen JA, Wisniewski N, Hanson-Smith V, Belgard TG, Zhang A, Coba M, Grunseich C, Ward ME, van den Berg LH, Pasterkamp RJ, Trotti D, Zlokovic BV, Ichida JK. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med. 2018;24:313–325. doi: 10.1038/nm.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabuas-Pereira M, Almendra L, Almeida MR, Duraes J, Pinho A, Matos A, Negrao L, Geraldo A, Santana I (2018) Increased risk of melanoma in C9ORF72 repeat expansion carriers: a case-control study. Muscle Nerve. 10.1002/mus.26383 [DOI] [PubMed]
- 43.van Blitterswijk M, Gendron TF, Baker MC, DeJesus-Hernandez M, Finch NA, Brown PH, Daughrity LM, Murray ME, Heckman MG, Jiang J, Lagier-Tourenne C, Edbauer D, Cleveland DW, Josephs KA, Parisi JE, Knopman DS, Petersen RC, Petrucelli L, Boeve BF, Graff-Radford NR, Boylan KB, Dickson DW, Rademakers R. Novel clinical associations with specific C9ORF72 transcripts in patients with repeat expansions in C9ORF72. Acta Neuropathol. 2015;130:863–876. doi: 10.1007/s00401-015-1480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vatsavayai SC, Nana AL, Yokoyama JS, Seeley WW (2018) C9orf72-FTD/ALS pathogenesis: evidence from human neuropathological studies. Acta Neuropathol. 10.1007/s00401-018-1921-0 [DOI] [PMC free article] [PubMed]
- 45.Walker C, Herranz-Martin S, Karyka E, Liao C, Lewis K, Elsayed W, Lukashchuk V, Chiang SC, Ray S, Mulcahy PJ, Jurga M, Tsagakis I, Iannitti T, Chandran J, Coldicott I, De Vos KJ, Hassan MK, Higginbottom A, Shaw PJ, Hautbergue GM, Azzouz M, El-Khamisy SF. C9orf72 expansion disrupts ATM-mediated chromosomal break repair. Nat Neurosci. 2017;20:1225–1235. doi: 10.1038/nn.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang G, Sawai N, Kotliarova S, Kanazawa I, Nukina N. Ataxin-3, the MJD1 gene product, interacts with the two human homologs of yeast DNA repair protein RAD23, HHR23A and HHR23B. Hum Mol Genet. 2000;9:1795–1803. doi: 10.1093/hmg/9.12.1795. [DOI] [PubMed] [Google Scholar]
- 47.Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, Pasinelli P, Ichida JK, Trotti D. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen X, Westergard T, Pasinelli P, Trotti D. Pathogenic determinants and mechanisms of ALS/FTD linked to hexanucleotide repeat expansions in the C9orf72 gene. Neurosci Lett. 2017;636:16–26. doi: 10.1016/j.neulet.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woollacott IO, Rohrer JD. The clinical spectrum of sporadic and familial forms of frontotemporal dementia. J Neurochem. 2016;138(Suppl 1):6–31. doi: 10.1111/jnc.13654. [DOI] [PubMed] [Google Scholar]
- 50.Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, Jin P. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci U S A. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yokoi M, Hanaoka F. Two mammalian homologs of yeast Rad23, HR23A and HR23B, as multifunctional proteins. Gene. 2017;597:1–9. doi: 10.1016/j.gene.2016.10.027. [DOI] [PubMed] [Google Scholar]
- 52.Zarei S, Carr K, Reiley L, Diaz K, Guerra O, Altamirano PF, Pagani W, Lodin D, Orozco G, Chinea A. A comprehensive review of amyotrophic lateral sclerosis. Surg Neurol Int. 2015;6:171. doi: 10.4103/2152-7806.169561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, Bowen KE, Wadhwa H, Yang P, Rigo F, Taylor JP, Gitler AD, Rothstein JD, Lloyd TE. Stress granule assembly disrupts nucleocytoplasmic transport. Cell. 2018;173:958–971. doi: 10.1016/j.cell.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, Gupta S, Thomas MA, Hong I, Chiu SL, Huganir RL, Ostrow LW, Matunis MJ, Wang J, Sattler R, Lloyd TE, Rothstein JD. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M, Lin WL, Garrett A, Stankowski JN, Daughrity L, Tong J, Perkerson EA, Yue M, Chew J, Castanedes-Casey M, Kurti A, Wang ZS, Liesinger AM, Baker JD, Jiang J, Lagier-Tourenne C, Edbauer D, Cleveland DW, Rademakers R, Boylan KB, Bu G, Link CD, Dickey CA, Rothstein JD, Dickson DW, Fryer JD, Petrucelli L. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci. 2016;19:668–677. doi: 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, Sasaguri H, Caulfield T, Hubbard J, Daughrity L, Chew J, Belzil VV, Prudencio M, Stankowski JN, Castanedes-Casey M, Whitelaw E, Ash PE, DeTure M, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 2014;128:505–524. doi: 10.1007/s00401-014-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. Antibody information. (DOCX 16 kb)
Table S1. Neuropathological scores of C9ORF72 FTD patients. Neuronal loss score was based on hematoxylin and eosin (HE) staining and pathological report and scored as absent (0), mild (1), moderate (2) or severe (3). Pathological scores were based on the degree of pathology as absent (0), rare (1), occasional (2), moderate (3), or numerous (4). Brain areas: F = frontal cortex, T = temporal cortex, M = motor cortex, H = hippocampus dentate gyrus, C = cerebellum. NCI = neuronal cytoplasmic inclusion, NII = neuronal intranuclear inclusion, DNs = dystrophic neurites. (DOCX 23 kb)
Figure S1. Ran-GAP staining in C9FTD cases and non-demented controls. Ran-GAP is predominantly localized to the nucleus and nuclear membrane. Unevenly shaped nuclear membranes occur in both C9ORF72 FTD cases (n = 5) and non-demented controls (n = 3). All scale bars are 20 μm. (PDF 2202 kb)
Figure S2. ADARB2 staining in C9FTD cases and non-demented controls. A) Staining of ADARB2 in C9ORF72 FTD cases (n = 5) and non-demented control (n = 3) post-mortem brain sections shows some intranuclear inclusions in hippocampus CA and DG. All scale bars are 20 μm B) Immunofluorescence staining of ADARB2 (red) and p62 (green) in hippocampal dentate gyrus reveals ADARB2 punctuated staining and some intranuclear inclusions in both C9ORF72 FTD cases (n = 5) and non-demented controls (n = 3). Scale bars in fluorescent pictures are 10 μm. (PDF 2721 kb)
Figure S5. HR23B pathology in different brain areas of C9FTD cases. Staining of HR23B in several brain areas of C9FTD cases and non-demented controls. Pathology burden was highest in cortices (frontal, temporal and motor) and was mostly cytoplasmic (inclusions and neuropils) and intranuclear (cateye). Hippocampus dentate gyrus (DG) harbors perinuclear inclusions, and hippocampus cornu ammonis (CA) had some cells with strong nuclear staining. Pathology was low in cerebellum granular layer with only some nuclear and perinuclear inclusions and almost absent in cerebellum molecular layer. Our C9FTD cases did not show HR23B pathology in spinal cord neurons. All scale bars are 20 μm (PDF 2260 kb)
Figure S3. FMRP staining in C9FTD cases and non-demented controls. FMRP staining does not reveal any differences between C9ORF72 FTD cases (n = 5) and non-demented control (n = 3) post-mortem brain sections. Occasional inclusions are found in the hippocampus dentate gyrus in both C9FTD cases and controls. All scale bars are 20 μm. (PDF 1883 kb)
Figure S4 Pur-alpha staining in C9FTD cases and non-demented controls. Pur-alpha staining reveals abundant stress granules in both C9ORF72 FTD cases (n = 5) and non-demented controls (n = 3) post-mortem brain sections. All scale bars are 20 μm. (PDF 1826 kb)
Figure S6 Validation of HR23B pathology by a second independent antibody. HR23B staining using Abcam antibody in C9ORF72 FTD cases (n = 5) and non-demented control (n = 3) post-mortem brain sections. Staining pattern is consistent with HR23B GeneTex antibody (see Figs. 1 and 2). All scale bars are 20 μm (PDF 1543 kb)
Table S2. Co-localization of HR23B with DPRs/pTDP-43/p62 differs between brain areas. The percentage is the amount of HR23B inclusions also positive for other pathological hallmarks (not the other way around). For example 11/153 for poly-GA in C9FTD patient 1 means that out of 153 HR23B inclusions, 11 were also positive forpoly- GA, which is 7%. F = frontal cortex. H = hippocampus dentate gyrus. *1 = All co-localizations are fibrils of poly-GP and HR23B in frontal cortex. Perinuclear inclusions of poly-GP did not stain positive for HR23B. *2 = In total 2 poly-PA inclusions have been found in frontal cortex of 5 C9FTD patients, too less to quantify. We therefore state N/A = non-applicable in this table. *3 = Non-demented cases had some p62 and some HR23B inclusions per person per section, which sometimes overlapped. No pTDP-43 inclusions were found so no co-localization of pTDP-43 with HR23B in non-demented cases. (DOCX 19 kb)
Figure S7. Nucleotide excision repair is not affected in C9ORF72 human fibroblasts. A) Dose-response curve for 4 healthy control human fibroblast lines (81E253, 86E1375, 06E0717 and 99E0774) and 4 C9ORF72 human fibroblast lines (13E634, 13E659, 17E0225, 17E0278) and the NER deficient XP25RO human fibroblast line treated with increasing dose of UV-C light (0–12 J/m2). B) Immunofluorescence staining showing the recruitment of NER factors XPC, XPB, XPA, XPF and XPG to local DNA damage (visualized by CPD antibody), induced by 60 J/m2 UV-C irradiation through a microporous filter. Fibroblast lines used for pictures: 13E634, 13E659 and 81E253 C) Human fibroblasts lines were treated with 16 J/m2 UV-C light and incubated with EdU for 1 h to measure unscheduled DNA synthesis (UDS) as measure of DNA repair. The NER-deficient XPC25RO cell line is shown as negative control. Ctrl 1–4 are lines 81E253, 86E1375, 06E0717 and 99E0774 in this order. C9 1–4 are lines 13E634, 13E659, 17E0225, 17E0278 in this order. (PDF 934 kb)
Data Availability Statement
The data supporting the conclusions of this article is included within the article (and its additional file). Raw datasets (including UV-C treatment of human fibroblasts) analyzed during the study are available from the corresponding author on reasonable request.