Abstract
Background
Cancer stem cells (CSCs) play an important role in the development of pancreatic cancer. We previously showed that the microRNA miR-137 is downregulated in clinical samples of pancreatic cancer, and its expression negatively regulates the proliferation and invasiveness of pancreatic cancer cells.
Methods
The stemness features of pancreatic cancer cells was detected by flow cytometry, immunofluorescence and sphere formation assay. Xenograft mouse models were used to assess the role of miR-137 in stemness features of pancreatic cancer cells in vivo. Dual-luciferase reporter assays were used to determine how miR-137 regulates KLF12. Bioinformatics and Chromatin immunoprecipitation analysis of KLF12 recruitment to the DVL2 promoters. Involvement of the Wnt/β-catenin pathways was investigated by western blot and Immunohistochemistry.
Results
miR-137 inhibits pancreatic cancer cell stemness in vitro and vivo. KLF12 as miR-137 target inhibits CSC phenotype in pancreatic cancer cells. Suppression of KLF12 by miR-137 inhibits Wnt/β-catenin signalling. KLF12 expression correlates with DVL2 and canonical Wnt pathway in clinical pancreatic cancer.
Conclusion
Our results suggest that miR-137 reduces stemness features of pancreatic cancer cells by Targeting KLF12-associated Wnt/β-catenin pathways and may identify new diagnostic and therapeutic targets in pancreatic cancer.
Electronic supplementary material
The online version of this article (10.1186/s13046-019-1105-3) contains supplementary material, which is available to authorized users.
Keywords: miR-137, Pancreatic cancer, KLF12, Stemness
Background
Pancreatic cancer is one of the most aggressive and lethal malignancies worldwide [1]. Despite effective surgical resection and systemic chemo-radiotherapy, the overall survival rate for pancreatic cancer remains very low [2]. Cancer stem cells (CSCs) represent a small subpopulation of cells within tumors; however, they have the ability to self-renew, play vital roles in tumor initiation and metastasis in multiple cancers, and are typically chemo-radiotherapy resistant [3]. In pancreatic tumors, CSCs account for 0.2–0.8% of the total cancer cell population and are regarded as vital for malignant behavior [4, 5]. Furthermore, a specific subpopulation of cells that contains CSC characteristics has been identified in human pancreatic cancers [6]. However, despite recent advances related to CSC isolation and characterization, the mechanisms that regulate CSC formation in pancreatic tumors remain largely unknown.
Various oncogenes and signaling pathways contribute to CSC maintenance and tumorigenicity. These include BMI1 (B-cell-specific Moloney murine leukemia virus insertion site 1), LGR5 (leucine-rich repeat-containing G-protein-coupled receptor 5), NANOG (Nanog homeobox), OCT4A (POU class 5 homeobox 1), and SOX2 (SRY-box 2), [7]. These pluripotency genes have important functions during embryonic development and participate in the maintenance of CSC phenotypes by directly targeting genes that control tumor stemness, survival, proliferation, invasion, and chemo-radiotherapy resistance [3, 8]. Transcripts of the above genes exert control over multiple morphogenetic signaling pathways, including Wnt/β-Catenin TGFβ, and Hedgehog, that are aberrantly active in many human tumors [9, 10]. β-catenin is the main effector of the Wnt pathway. It directly binds to TCF/LEF transcription factors to upregulate the expression of downstream genes, promoting cell proliferation, metastasis, and chemo-resistance [11]. β-catenin is phosphorylated by GSK3β within a ‘destruction complex’ formed with APC and AXIN1, resulting in β-catenin ubiquitination and degradation [12]. Therefore, suppressing β-catenin phosphorylation prevents formation of the destruction complex and β-catenin degradation. Non-phosphorylated β-catenin is translocated to the nucleus and interacts with TCF/LEF to regulate downstream gene expression [13]. Activation of β-catenin may increase the CSC population in pancreatic cancer, while inhibition of the Wnt/β-catenin pathway blocks the tumor-promoting activity of pancreatic CSCs [14]. When the Wnt/β-catenin pathway is activated, these receptors phosphorylate the cytoplasmic protein Dishevelled (Dvl), which protects β-catenin from a protein complex containing APC, GSK3, Axin. This promotes β-catenin nuclear translocation, as well as transcription of Wnt/signaling target genes [15–17]. Thus, exploring new ways to target Wnt/β-catenin might lead to novel and successful treatments for pancreatic cancer.
MicroRNAs (miRNAs) are short non-coding RNAs, 17–25 nucleotides long, that regulate target genes post-transcriptionally [18]. Multiple miRNAs have been shown to control CSC maintenance and tumorigenicity [19]. Among these miRNAs, It was reported that miR-137 exhibited both oncogenic and tumor-suppressive roles in the context of different cancer types. For example, Chang TH al. found that miR-137 is a Slug-induced miRNA that relays the pro-metastatic effects of Slug by targeting TFAP2C in in non-small cell lung cancer (NSCLC) [20]. miR-137 was also reported to play a tumor suppressor in various tumor. miR-137 can regulate XIAP via its 3′UTR sensitise ovarian cancer cells to cisplatin-induced apoptosis in ovarian cancer [21]. Our previous studies showed that the miR-137 inhibits pancreatic cancer cell proliferation, invasion and chemoresistance through targeting PTN in pancreatic cancer [22]. Recently, Neault M found that miR-137 inhibits the bypass of Ras-induced senescence and triggers the p53 and p16INK4A tumor suppressor pathways by targeting KDM4A in pancreatic cancer [23]. However, the mechanism of miR-137 in pancreatic cancer stem cells has not been fully elucidated. Therefore, it is of great significance to reveal the molecular mechanism of abnormal expression of miR-137 for understanding the pathogenesis of pancreatic cancer. In this study, We firstly demonstrated that KLF12 was a direct target of miR-137, and down-expression of miR-137 significantly promoted stemness by down-regulating KLF12 via activating Wnt/β-catenin pathway. Hence, our study identified a new molecular signaling pathway for understanding the pathogenesis and of pancreatic cancer.
Materials and methods
Chemicals and cell culture
All the antibodies were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA) unless otherwise noted. The human pancreatic cancer cell lines AsPC-1 and PANC-1 were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured at 37 °C in a humidified 5% CO2 incubator according to ATCC protocols. PANC-1 cells were grown in DMEM medium and AsPC-1 cells were grown in RPMI-1640 medium (Gibco, NY, USA). Culture media were supplemented with 10% fetal bovine serum (Gibco), 100 U/mL penicillin G, and 100 μg/ml streptomycin (Sigma, MO, United States).
RNA preparation and real-time PCR
Tissue harvesting and processing and PCR were conducted according to standard procedures. RNA was reverse-transcribed to cDNA using a qPCR RT Kit (Takara, Japan). Quantitative real-time PCR was performed using the SYBR Green Realtime PCR Premix (Takara, Japan), according to the manufacturer’s instructions. qRT-PCR primers as Supplementary Material 1:Table 1.
Table 1.
The characteristics of the primers used for real-time PCR
| GENE | Sequence (5′ - > 3′)Sequence (5′ - > 3′) | |
|---|---|---|
| BMI-1 | Forward Primer | CCACCTGATGTGTGTGCTTTG |
| Reverse Primer | TTCAGTAGTGGTCTGGTCTTGT | |
| ABCG2 | Forward Primer | CAGGTGGAGGCAAATCTTCGT |
| Reverse Primer | ACCCTGTTAATCCGTTCGTTTT | |
| NANOG | Forward Primer | TTTGTGGGCCTGAAGAAAACT |
| Reverse Primer | AGGGCTGTCCTGAATAAGCAG | |
| SOX2 | Forward Primer | GCCGAGTGGAAACTTTTGTCG |
| Reverse Primer | GGCAGCGTGTACTTATCCTTCT | |
| OCT4A | Forward Primer | CTGGGTTGATCCTCGGACCT |
| Reverse Primer | CCATCGGAGTTGCTCTCCA | |
| LGR5 | Forward Primer | CTCCCAGGTCTGGTGTGTTG |
| Reverse Primer | GAGGTCTAGGTAGGAGGTGAAG | |
| KLF12 | Forward Primer | CGGCAGTCAGAGTCAAAACAG |
| Reverse Primer | CGGCTTCCATATCGGGATAGT | |
| TCF4 | Forward Primer | CAAGCACTGCCGACTACAATA |
| Reverse Primer | CCAGGCTGATTCATCCCACTG | |
| CCND1 | Forward Primer | GCTGCGAAGTGGAAACCATC |
| Reverse Primer | CCTCCTTCTGCACACATTTGAA | |
| MYC | Forward Primer | GGCTCCTGGCAAAAGGTCA |
| Reverse Primer | CTGCGTAGTTGTGCTGATGT | |
| TWIST1 | Forward Primer | GTCCGCAGTCTTACGAGGAG |
| Reverse Primer | GCTTGAGGGTCTGAATCTTGCT | |
| CD44 | Forward Primer | CTGCCGCTTTGCAGGTGTA |
| Reverse Primer | CATTGTGGGCAAGGTGCTATT | |
| MMP7 | Forward Primer | GAGTGAGCTACAGTGGGAACA |
| Reverse Primer | CTATGACGCGGGAGTTTAACAT | |
| Control | UUAUUGCUUAAGAAUACGCGUAG | |
| UCACAACCUCCUAGAAAGAGUAGA | ||
| inhibitor | CTACGCGTATTCTTAAGCAATAA | |
| miR-137 mimic | UUAUUGCUUAAGAAUACGCGUAG | |
Generation of stable cell lines
Human miR-137 overexpressing (miR-137), knockdown (anti-miR-137), and negative-control (NC) lentiviruses were purchased from Genechem (Shanghai, China). All transfections were carried out according to the manufacturer’s instructions.
Plasmids, siRNAs, and transfection
Cells were seeded in 6-well plates at 50% confluence without antibiotics on the day before transfection with siRNAs (RiboBio, Guangzhou, China). For luciferase reporter assays, pairs of oligonucleotides containing the 3′-UTR binding site for miR-137 (RiboBio, Guangzhou, China) were used. Transfection of miRNAs, siRNAs, and plasmids was performed using Lipofectamine 3000 (Life Technologies Co., Carlsbad, CA, USA) according to the manufacturer’s instructions. The siRNA is used at a concentration of 50 nM and the transfection time is 48 h.The sequence of miR-137-control: 5′-UCACAACCUCCUAGAAAGAGUAGA-3′, sequence of miR-137-mimic: 5′-UUAUUGCUUAAGAAUACGCGUAG-3′, sequence of miR-137-5′-inhibitor: 5′-CTACGCGTATTCTTAAGCAATAA-3′. sequence of small-interfering RNA sequence of KLF12: 5′-GCAATCGAATGAATAATCA-3′.
Sphere formation assay
Cells (500/well) were seeded into 6-well ultra-low attachment cluster plates (Corning, NY, USA) and cultured in serum-free DMEM/F12 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 2% B27 (Invitrogen), 20 ng/ml EGF (PeproTech), 20 ng/ml bFGF (PeproTech), 0.4% BSA (Sigma-Aldrich), and 5 μg/ml insulin (Sigma-Aldrich). After two weeks, spheres were photographed and counted.
Flow cytometric analysis
Cells were dissociated with trypsin, resuspended at 1 × 106 cells per ml in 200 μl DMEM containing 2% FBS and subsequently incubated with PE-conjugated CD133 antibody (MiltenyiBiotec, Germany) for 90 min at 37 °C. Cells were then incubated on ice for 10 min and washed with ice-cold PBS before flow cytometric analysis. Isotype-matched immunoglobulins served as controls.
Tumor xenografts
All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Guizhou Medical University. Six-week-old BALB/c-nu mice were randomly divided into eight groups (n = 4 per group), and implanted with 1 × 106, 1 × 105, 1 × 104 or 1 × 103 cells previously transfected with negative control or miR-137-overexpressing lentiviruses. Cells were mixed with Matrigel (50% volume) and implanted subcutaneously into the inguinal folds of the nude mice. Tumor volume was determined using an external caliper and calculated using the eq. (L × W2)/2. Mice were sacrificed 35 days after inoculation. At this point, tumors were excised and subjected to downstream analyses.
Subcellular fractionation assay
Subcellular fraction was obtained from cultured cells and tissues using Subcellular Protein Fractionation Kit for Cultured Cells and Subcellular Protein Fractionation Kit for Tissues (Thermo Fisher Scientific).
Immunofluorescence
Cells grown on glass plates were fixed with 4% paraformaldehyde. Next, the cells were incubated with the primary antibody (1:100) at 4 °C overnight followed by Alexa Fluor 594-conjugated goat anti-rabbit IgG (1:250). Slides were counterstained with DAPI to visualize cell nuclei. Images were recorded using a confocal laser scanning microscope.
Histology and immunostaining
Immunohistochemistry assays were performed on the paraffin-embedded Subcutaneous implants tumor and pancreatic cancer tissue, using the primary antibodies as Supplementary materials. The degree of immunostaining of indicated proteins was evaluated and scored by two independent observers, who scored both the proportions of tumor cells that stained positively and the intensity of the staining.
Statistical analysis
All values are presented as means ± standard deviation (SD). Statistical analyses were performed using SPSS 22.0 software (SPSS, Chicago, USA). Student’s t-test was used to determine statistical differences. P < 0.05 was considered significant.
Results
miR-137 inhibits pancreatic cancer cell stemness in vitro
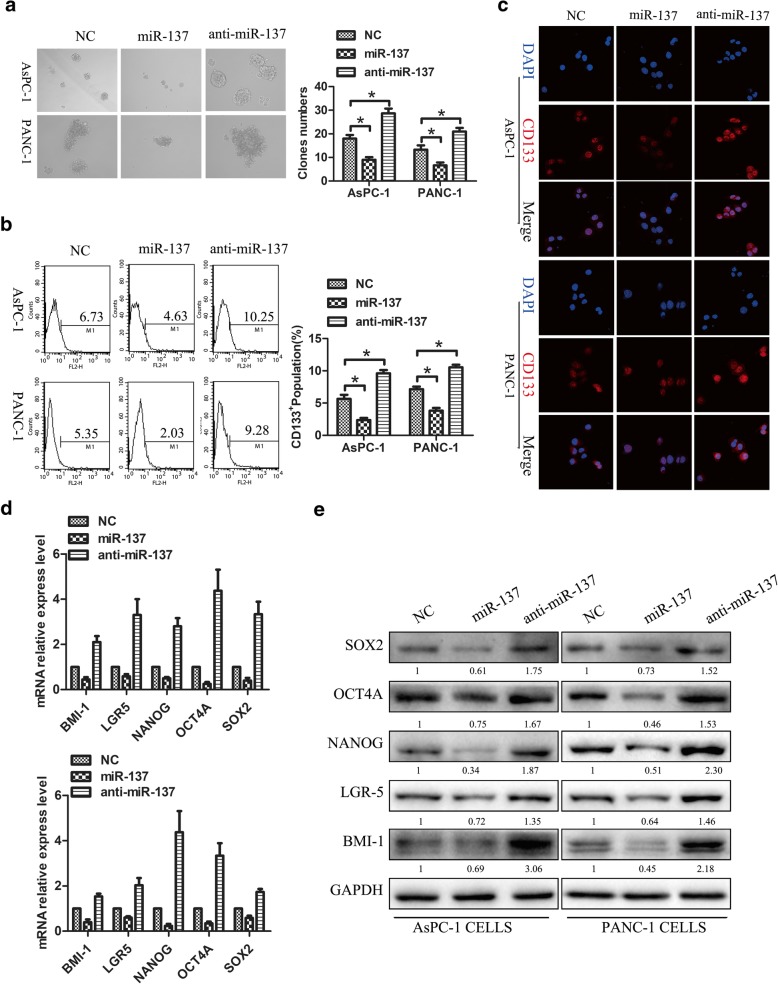
We reported that miR-137 is markedly downregulated in pancreatic cancer, contributing to tumor growth, invasion, and resistance to chemotherapy. To determine whether miR-137 levels affect the self-renewal of pancreatic CSCs, the human pancreatic cancer cell lines PANC-1 and AsPC-1 were transfected with miR-137 mimic (miR-137) or inhibitor (anti-miR-137) in vitro. As shown in Fig. 1a, upregulation of miR-137 dramatically decreased the size and number of spheres in both cell lines, while anti-miR-137 stimulated sphere formation. Since CD133 expression has been reported to characterize CSC-like populations in many tumors, including pancreatic cancer, the effect of miR-137 on the CD133+ populations of AsPC-1 and PANC-1 cells was examined by flow cytometry and immunofluorescence. Overexpression of miR-137 decreased CD133+ cell populations in both cell lines, while miR-137 downregulation had the opposite effect in Fig. 1b-c. These results indicate that miR-137 inhibits the CSC phenotype in human pancreatic cancer cells in vitro. Several transcription factors, including BMI1, LGR5, NANOG, OCT4, and SOX2, are upregulated in various cancer types and promote stemness maintenance, local tumor invasion, and distant metastasis. To assess whether alterations in miR-137 expression affect the expression of these markers, RT-PCR and Western-blot were conducted in AsPC-1 and PANC-1 cells treated with miR-137 mimics or inhibitors. As expected, overexpression of miR-137 significantly downregulated the expression of these genes both at the mRNA and protein levels, whereas miR-137 inhibition had the opposite effect in Fig. 1d-e. These results suggest that miR-137 inhibits CSC reprogramming by repressing the transcription of several pluripotency-related genes.
Fig. 1.
miR-137 inhibits pancreatic cancer cell stemness in vitro. a miR-137 upregulation dramatically decreased the size and number of spheres formed in AsPC-1 and PANC-1 cell cultures, while anti-miR-137 had the opposite effect (p < 0.05). b-c Overexpression of miR-137 decreased the CD133+ population in cultured AsPC-1 and PANC-1 cells. In contrast, anti-miR-137 increased the CD133+ population in both cell lines (p < 0.05).d-e Overexpression of miR-137 markedly downregulated the expression of pluripotency-related mRNAs and proteins (p < 0.05), whereas miR-137 knockdown significantly upregulated the expression of these markers (p < 0.05)
miR-137 inhibits pancreatic cancer cell tumorigenesis in vivo
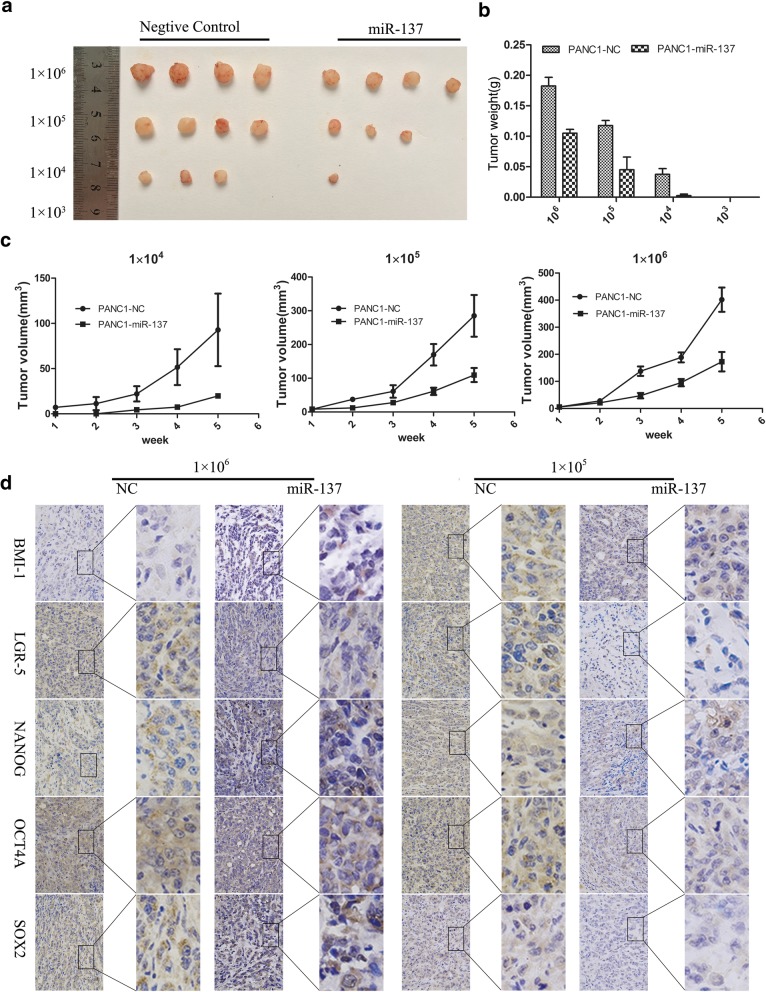
To test the involvement of miR-137 on human pancreatic cancer cells’ tumorigenesis in vivo, PANC-1 cells transfected with miR-137 overexpression or negative control lentivirus were implanted into nude mice at different seeding densities (1 × 106, 1 × 105, 1 × 104 or 1 × 103 cells). As shown in Fig. 2a-c, miR-137-overexpressing tumors grew more slowly than tumors in the negative control group. Moreover, whereas no tumors developed in either group after implantation of 1 × 103 cells, only negative control-transfected cells formed tumors when 1 × 104 cells were implanted. Ex-vivo tumor immunohistochemistry analyses revealed significantly higher levels of pluripotency-associated markers in negative control specimens compared with miR-137-overexpressing tumors in Fig. 2c. These results demonstrate that miR-137 reduces CSC marker expression and inhibits pancreatic cancer cell tumorigenesis in vivo.
Fig. 2.
miR-137 inhibits pancreatic cancer cell tumorigenesis in vivo. a Images of the subcutaneous xenografts from the miR-137-overexpressing and control groups. b Tumors weight were significantly lower in the miR-137-overexpressing group compared with the control group. c Tumors in the miR-137-overexpressing group grew more slowly than tumors in the control group. d Immunohistochemical detection of pluripotency-associated markers in excised tumors
KLF12 as miR-137 target inhibits CSC phenotype in pancreatic cancer cells
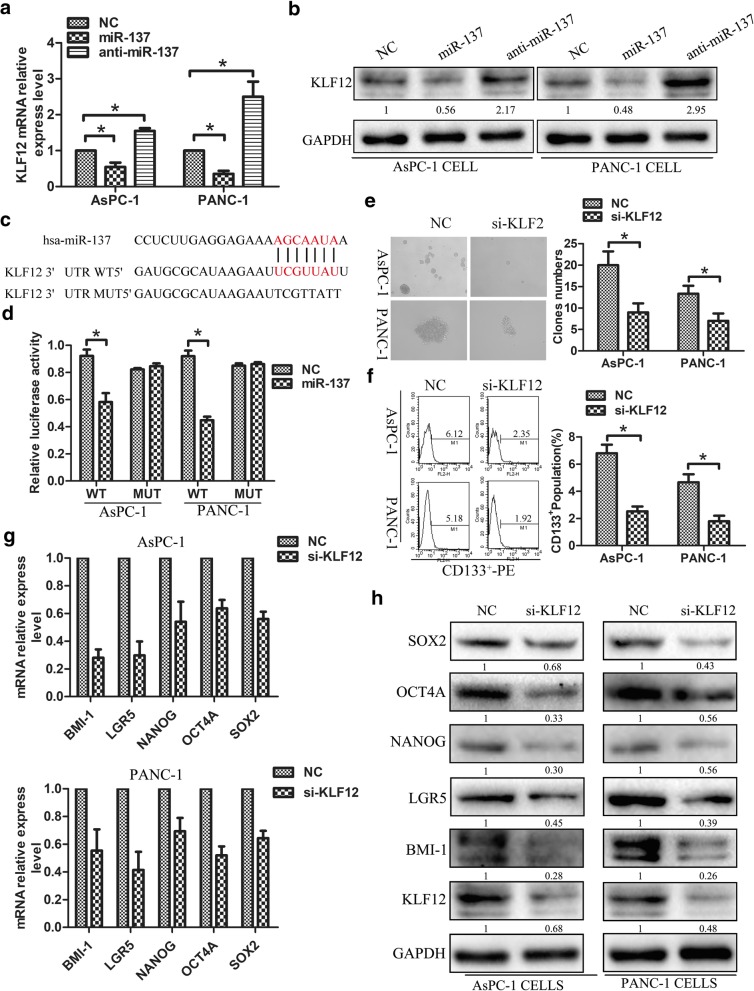
To explore potential targets through which miR-137 inhibits the CSC phenotype in pancreatic cancer cells, we searched publicly available algorithms (miRanda and TargetScan) and identified KLF12. Figure 3a-b revealed that KLF12 expression in AsPC-1 and PANC-1 cells was decreased after miR-137 upregulating,but was increased after miR-137 downregulating. To examine whether direct miR-137-mediated KLF12 downregulation occurs through a miR-137-binding site in KLF12 3′-UTR, the latter sequence was cloned into the pGL3 dual luciferase reporter vector. Results showed that miR-137 overexpression effectively reduced luciferase reporter activity of the KLF12 3′-UTR. In contrast, the activity of the KLF12 3′-UTR luciferase reporter containing point mutations (mut) in the miR-137-binding seed region was unaffected by miR-137 overexpression in Fig. 3c-d. These results indicate that miR-137 directly targets KLF12.To analyze the role of KLF12 in CSC formation and proliferation, PANC-1 and AsPC-1 cell lines were transfected with KLF12 siRNA. As expected, KLF12-knockdown significantly impaired the ability to form tumor spheres in Fig. 3e, and also decreased CD133+ populations in both cell lines in Fig. 3f. Furthermore, the expression of stemness markers, as assessed by RT-PCR and western blotting, was significantly downregulated by KLF12 knockdown in Fig. 3.g-h. These results suggest that miR-137 directly targets KLF12 and KLF12 activity contributes to the CSC phenotype in human pancreatic cancer cells.
Fig. 3.
KLF12 as miR-137 target inhibits CSC phenotype in pancreatic cancer cells. a-b PCR and western blot analysis the KLF12 expression in miR-137-mimic and miR-137-inhibitor transfected AsPC-1 and PANC-1 cells. c Predicted binding sites of miR-137 in the wild type 3-UTRs of KLF12,Mutations in these 3-UTRs are highlighted in red. d Luciferase activity of reporters with wild type or mutant 3-UTR of KLF12 in the indicated cells co-transfected with the indicated oligonucleotides e KLF12 knockdown significantly impaired tumor-sphere formation in both in AsPC-1 and PANC-1 cells. f KLF12 knockdown decreased the CD133+ population in AsPC-1 and PANC-1 cells g Real-time PCR and western blot analyses showed that downregulation of KLF12 inhibited the expression of pluripotency-associated markers in AsPC-1 and PANC-1 cells
KLF12 mediates CSC phenotype induction after miR-137 downregulation
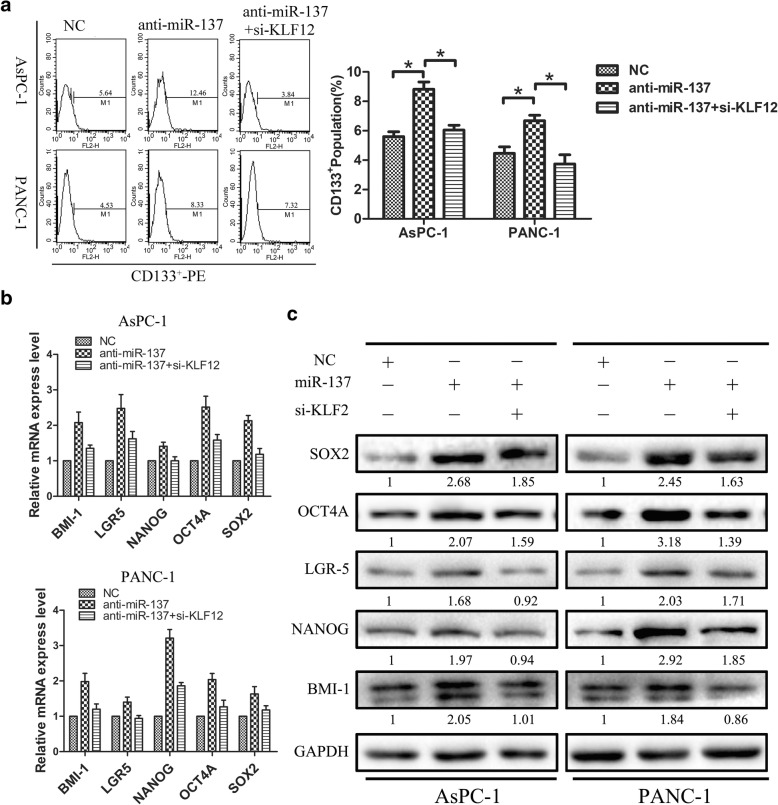
Next, we investigated whether KLF12 activity mediates miR-137-dependent CSC marker expression in in AsPC-1 and PANC-1 cells by silencing KLF12 gene expression in cells with downregulated expression of miR-137. As shown in Fig. 4a, KLF12 silencing significantly reversed CD133+ population increases induced by miR-137-downregulation. In addition, real-time PCR and western-blot analyses showed that the expression of the pluripotency-associated markers BMI1, NANOG, LGR5, OCT4A, and SOX2 was inhibited in these cells in Fig. 4b-c. Thus, our overall results suggest that miR-137 and KLF12 effectively interact to suppress CSC formation and proliferation in human pancreatic cancer cells.
Fig. 4.
KLF12 mediates CSC phenotype induction after miR-137 downregulation. a Silencing KLF12 reduced CD133+ populations after downregulation of miR-137. b Real-time PCR analyses showed that downregulation of KLF12 inhibited the expression of pluripotency-associated markers in AsPC-1 and PANC-1 cells. c Western blot analyses showed that downregulation of KLF12 inhibited the expression of pluripotency-associated markers in AsPC-1 and PANC-1 cells
Suppression of KLF12 by miR-137 inhibits Wnt/β-catenin signaling
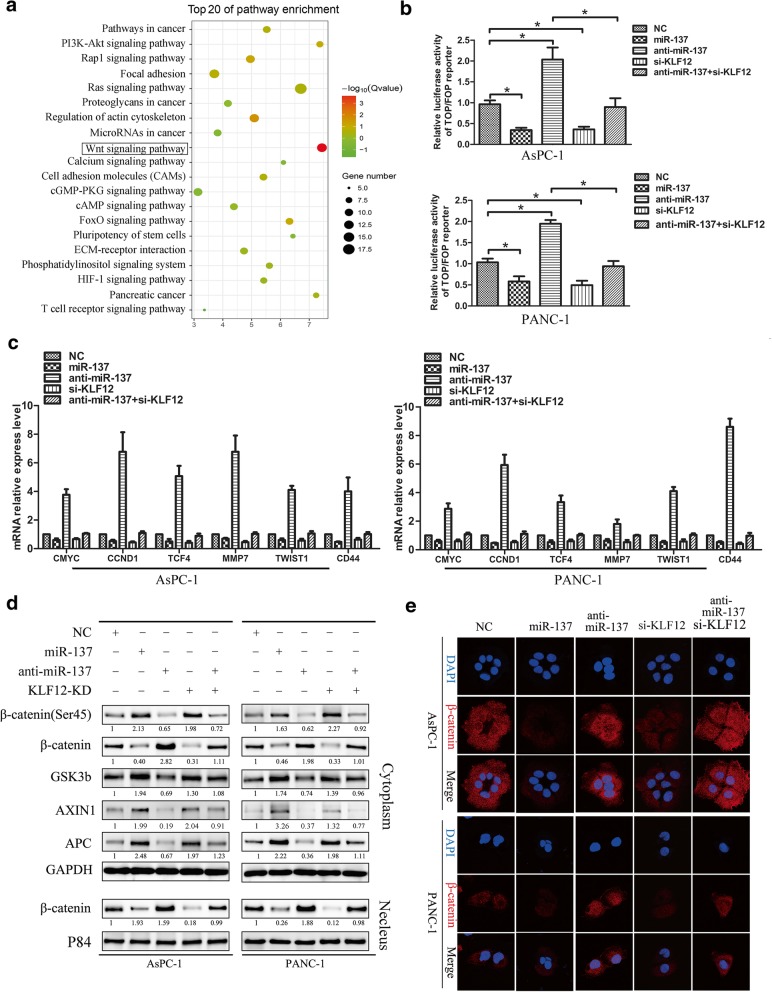
In order to study the molecular biological mechanism involving miR-137, and target gene KLF12 regulating pancreatic cancer cell stemness. By performing KEGG-pathway analysis in the TCGA Pancreatic adenocarcinoma data set, we found that the KLF12 level was positively correlated with Wnt-activated gene signatures (Fig. 5a), suggesting that KLF12 might be involved in Wnt/β-catenin signaling activation. Subsequently, we transfected PANC-1 and AsPC-1 cells with miR-137-control,miR-137-mimic, miR-137-inhibitor, si-KLF12, co-transfection with miR-137-inhibitor and si-KLF12 respectively, to further examine the effects of miR-137 and KLF12 on the Wnt/β-catenin pathway. As shown in Fig. 5b,c over-expression of miR-137 in PANC-1 and AsPC-1 cells significantly decreased the activity of the luciferase reporter driven by Wnt/β-catenin signals and the expression of several well-established downstream target genes of the Wnt/b-catenin pathway, whereas the transactivating activity of β-catenin was markedly increased in response to miR-137 inhibiting. Silencing KLF12 produced the consistent with miR-137-mimic,and down-regulation of KLF12 can partially inhibit the function of miR-137-inhibitor effect. Immunohistochemistry was used to detect the association between KLF12 and β-catenin expression in the subcutaneous implanted tumor. The results of immunohistochemistry indicated that the expression of KLF12 and β-catenin was higher in the control group, while the expression of KLF12 and β-catenin was lower in the miR-137 up-regulated group in the Additional file 1: Figure S1. As shown in Fig. 5d, miR-137-mimic significantly promoted AXIN1 and APC, GSK-3β, the phosphorylation of β-catenin on Ser45 expression, and reduced the β-catenin of cytoplasm and nuclear expression in pancreatic cancer cells. Meanwhile, the immunofluorescence staining assays showed that nuclear β-catenin expression decreased significantly in miR-137-mimic and si-KLF12 cell (Fig.5e). whereas decreased in miR-137-inhibitor cells, this effect of miR-137 inhibitor was significantly attenuated by KLF12 knockdown. These results demonstrate that miR-137 represses KLF12-mediated Wnt/β-catenin signaling activation in pancreatic cancer cell lines, and highlight a potential mechanism for miR-137-mediated suppression of stemness properties in pancreatic tumors.
Fig. 5.
Suppression of KLF12 activity by miR-137 prevents Wnt/β-catenin signaling in pancreatic cancer cells. a KEGG pathway analysis showing that KLF12 expression was positively correlated with Wnt-activated gene signatures in the TCGA pancreatic cancer data set. b The indicated cells were transfected with TOP or FOP reporter and Renilla pRL-TK plasmids and subjected to dual-luciferase assays 48 h after transfection. The detected reporter activity was normalized to the Renilla activity. c RT–qPCR analysis of the expression of the established downstream targets for the Wnt/β-catenin pathway, including TCF4, TWIST, CMYC, MMP7, CCND1 and CD44 in the indicated cells d Western blot analyses the expression levels of AXIN1, APC, GSK-3B, β-catenin, phosphorylation β-catenin on Ser45 and nuclear β-catenin in the indicated cells e Subcellular β-catenin localization in the indicated cells was assessed by immunofluorescence staining
KLF12 transcriptionally regulates DVL2–β-catenin signaling in pancreatic cancer cells
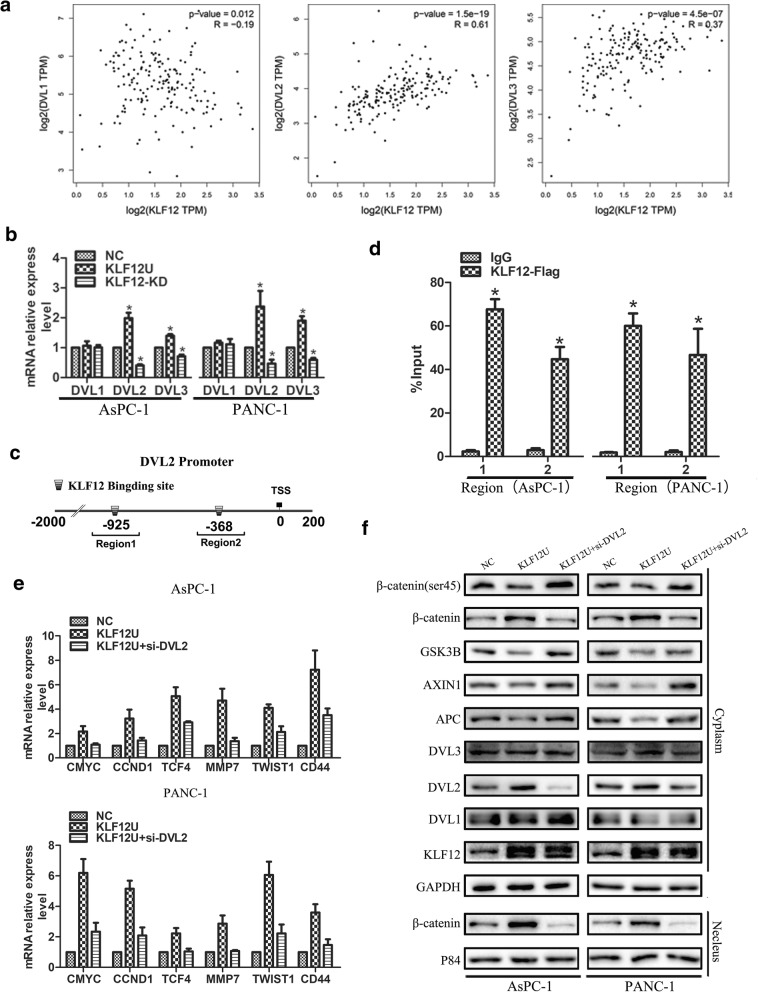
We next used bioinformatics analysis to identify genes that are regulated by KLF12 Additional file 2. The cytoplasmic protein Dishevelled (DVL1,DVL2,DVL3), which protects β-catenin from a protein complex containing APC, GSK3b, AXIN1. Therefore, we first examined which above candidates have positive correlation with KLF12 expression using analysis of TCGA, showing that level of DVL2,DVL3 associated significantly and positively with KLF12 expression (Fig. 6a). We verified the above results by q-PCR and the results were consistent with the above analysis, after overexpression of KLF12, there was no significant difference in the expression of DVL1, and the expression of DVL2 and DVL3 was significantly up-regulated (Fig. 6b). KLF12 has been reported to bind to the CACCC motif of target genes to regulate their expression [24]. Then, we examined which above candidates have CACCC motif in DVL2 and DVL3 promoter regions. Interestingly, the DVL2 promoter contains two putative KLF12 DNA-binding motifs (CACCC) located at -925 bp (Region 1) and -368 bp (Region 2) relative to the transcription start site (see Supplementary Material 2,3:DVL2 promoter, DVL3 promoter). The results indicated that KLF12 may bind to the promoter regions of DVL2. This was confirmed by chromatin immunoprecipitation assays (Fig. 6c,d). Our results demonstrate Additional file 3 that KLF12 transcriptionally regulates DVL2. Furthermore, small-interfering RNA (siRNA) for DVL2 (si-DVL2) inhibited the effect of increased KLF12 activity on downstream target genes levels in this pathway (Fig.6e), whereas the cytoplasm and nucleus β-catenin was markedly decreased (Fig. 6f).
Fig. 6.
KLF12 transcriptionally regulates DVL2–β-catenin signaling in pancreatic cancer cells. a TCGA analysis showing that the expression correlation of KLF12 and DVL1,DVL2,DVL3 in the TCGA pancreatic cancer data set. b RT–qPCR analysis of the expression of DVL1,DVL2,DVL3 in the indicated cells c Bioinformatics analysis showing that the DVL2 promoter contains two putative KLF12 DNA-binding motifs . d Chromatin Immunoprecipitation assays indicated that KLF12 may bind to the promoter regions of DVL2.e RT–qPCR analysis of the expression of the established downstream targets for the Wnt/β-catenin pathway in the indicated cells. e The expression of Wnt-activated /suppressed gene and nuclear β-catenin in the indicated cells, as determined by western blotting
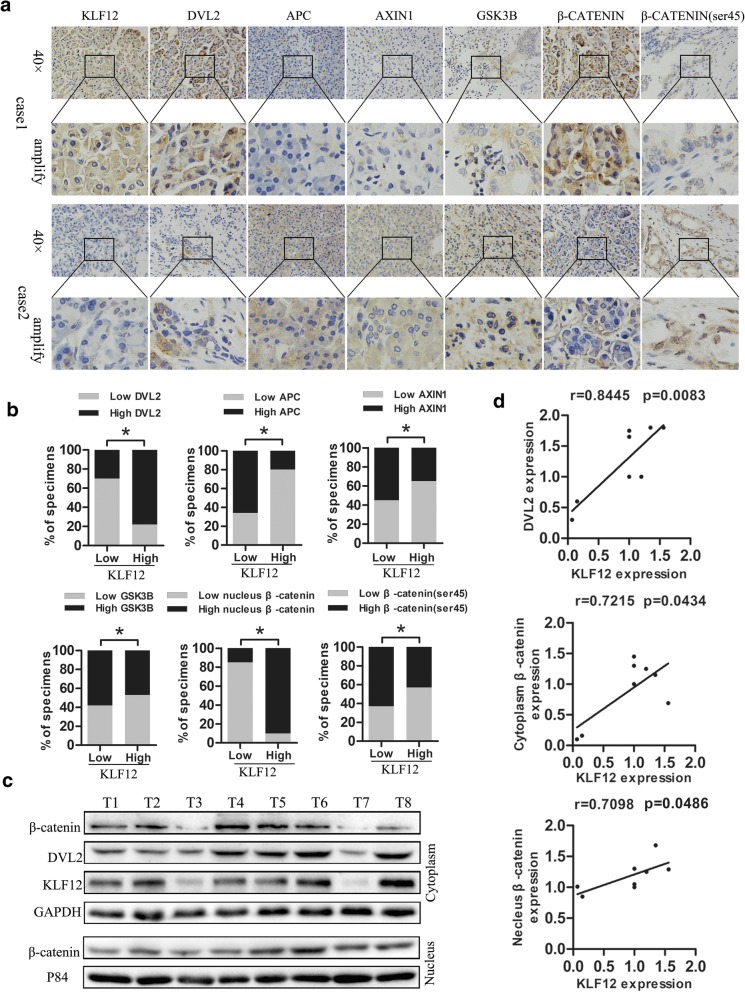
KLF12 expression correlates with DVL2 and canonical Wnt pathway in clinical pancreatic cancer
Next, we investigate the clinical relevance between KLF12 level and canonical Wnt pathway in human pancreatic cancer specimens. KLF12 level was correlated positively and significantly with DVL2 expression (p < 0.05), nuclear β-catenin expression (p < 0.05), demonstrated by IHC (n = 24; Fig. 7a, b). Eight freshly collected clinical pancreatic cancer specimens were examined using analysis of western blot, showing that level of KLF12 associated significantly and positively with DVL2 expression (r = 0.8445, p = 0.0083), cytoplasm β-catenin expression (r = 0.7098, p = 0.0434), as well as nuclear β-catenin expression (r = 0.7098, p = 0.0486) (Fig. 7c,d). Taken together, our findings further supported the notion that elevated KLF12 expression activates canonical Wnt pathway through transcriptional regulating Dvl2, which subsequently prevents β-catenin degradation and promotes β-catenin translocated to the nucleus in pancreatic cancer.
Fig. 7.
KLF12 expression correlates with DVL2 and canonical Wnt pathway in clinical pancreatic cancer. a. Immunohistochemical analysis of KLF12,DVL2, AXIN1, APC,GSK3B,β-catenin, the phosphorylation of β-catenin on Ser45 expression in serial sections of pancreatic cancer specimens.b.KLF12 level correlates with canonical Wnt pathway in clinical pancreatic cancer specimens. c-d. Eight freshly collected glioma specimens were subject to western blot, showing the correlation of KLF12 expression with DVL2, cytoplasm and nuclear β-catenin
Discussion
Pancreatic cancer continues to be a highly lethal malignancy, despite the use of multimodal treatment approaches [25]. The CSCs statement has been proposed to explain the high rate of relapse and subsequent resistance of cancer to current systemic treatments [26]. Although CSCs constitute a small subset of the total tumor cell population, they are thought to be responsible not only for tumorigenesis, metastasis, and recurrence, but also for tumor heterogeneity and drug resistance [27]. These features reveal that CSCs are involved in the development of tumors, indicating that molecular and pathological characterization of CSCs is important to improve the prognoses of cancer patients. CSCs have been identified in a variety of solid tumors, including pancreatic cancer [28]. However, the precise roles and association between CSCs and the prognoses of patients are accurately understood, especially in pancreatic cancer.
MicroRNAs (miRNAs) exert widespread gene regulation, affecting the expression of oncogenes and tumor suppressor genes that impact CSCs features and modulate tumor growth and resistance to therapy [29]. Therefore, elucidating the underlying mechanism of miRNAs in tumor development may provide valuable diagnostic and therapeutic strategies for malignancy. Several miRNAs, which are involved in stemness maintenance and differentiation, have been identified to play a critical role in regulating pancreatic cancer tumorigenesis signaling networks [30]. Therefore, targeting pancreatic CSCs and elucidating the underlying mechanisms of miRNA in pancreatic CSCs may improve diagnostic and therapeutic strategies for pancreatic cancer.
The miRNA-137 was previously identified to be epigenetically silenced in various types of cancer and to play important roles in tumor development and progression [21–23, 31, 32]. However, the clinical significance of the miRNA-137 family in pancreatic cancer, and the molecular mechanisms underlying the deactivation of its target genes in tumorigenesis and drug resistance derived by pancreatic CSCs still require elucidation.
As our previous work showed that miR-137 levels affect growth, invasion, and sensitivity to chemotherapy in pancreatic tumors [27], we carried out the present study to test the hypothesis that miR-137 is a stemness regulator of human pancreatic cancer cells. To uncover the underlying mechanism, three target gene prediction websites were used to forecast target genes of miRNA-137. KLF12, one of the target genes with a higher predictive score, has been shown to be regulated by miR-137 in gastric cancer [32]. And the potential candidate Krüppel-like factor (KLF) family member KLF12, which was regarded as a transcriptional factor, was selected and monitored in PANC-1 and AsPC-1 cells with knocked down and overexpressed miRNA-137. In this context, the mRNA and protein expression of KLF12 was markedly inhibited by miRNA-137. Owing to the typical seed sequence in the 3′-UTR of KLF12, a direct role of miRNA-137 was naturally surveyed next. As expected, KLF12 was a direct downstream target of miRNA-137, as demonstrated by luciferase assays. The members of the KLF family function as transcription factors and are widely expressed in a number of tissues and organs, they have been reported to control the expression of genes and possess many regulatory functions related to cellular proliferation, differentiation, anoikis, the cell cycle, and cell apoptosis [33–35]. In order to explore the function and downstream signal pathways of KLF12, combining with the TCGA pancreatic cancer data set analysis, our results revealed that KLF12 was significantly correlated with activation of the Wnt/β-catenin signaling pathway. Aberrant Wnt signaling activation has been identified in many solid tumors [36]. Notably, the expression or nuclear localization of β-catenin is often abnormal in cancer cell, indicating a constitutive activation of Wnt/β-catenin signaling [37]. However, how abnormal upstream signal molecule change in wnt/β-catenin signaling cascade, leading to constitutively activated Wnt/β-catenin remains unclear. Herein, we demonstrated that KLF12 as the target of miR-137 induced activation of wnt/β-catenin promotes the stemess of pancreatic cancer cells. To explore the impact of miR-137 and KLF12 on Wnt/β-catenin signaling pathway, we tested the activity of Wnt/β-catenin signaling pathway by performing TOP/FOP Luciferase assays. The activity of Wnt/β-catenin signaling and expression of downstream molecules was significantly decreased in miR-137-overexpress and KLF12-downregulated pancreatic cancer cells, but was increased in miR-137-inhibited cells. Therefore, our findings reveal a novel mechanism for activation of wnt/β-catenin pathway involving miR-137/KLF12 in pancreatic cancer.
When canonical Wnt activated, the Wnt receptors phosphorylate the cytoplasmic protein Dishevelled (Dvl), which protect β-catenin from ubiquitination degradation [38]. Upregulation of Dvl sustains Wnt/β-catenin signaling, whereas Dvl2 loss suppresses tumor development in the intestine of the APC-mutant mouse, suggesting Dvl2 may represent a potential target for colorectal cancer therapy [39]. DVL2 has been consider as an important intermediates of Wnt signaling pathways that inhibits GSK3B,APC,AXIN complex formation. DVL2 is highly expressed and plays an important role in the β-catenin mediated TCF-dependent transcriptional activity, promoting the proliferation of NSCLC cells [40]. Interestingly, We found that the expression of KLF12 was found to be significantly correlated with DVL2 through the TCGA database. Further, we found that KLF12 could transcriptional regulate the expression of DVL2 by q-PCR and Chip experiments. Therefore, we have reason to speculate that KLF12 may transcriptionally activate DVL2 and promote the sustained activation of WNT signaling pathway. Finally, we found a significant correlation between KLF12 and DVL2, β-catenin expression in clinical samples, further supporting the above hypothesis.
Accordingly, we found that miR-137 inhibits the expression and nuclear translocation of β-catenin, and this effect can be partially suppressed by KLF12. Therefore, we propose that miR-137 inhibits pancreatic cancer stemness properties and tumorigenicity by targeting KLF12 via preventing nuclear translocation of β-catenin and activation of Wnt signaling. Further exploration of the upstream molecular events modulating the miR-137/KLF12 interaction, and the resulting impact on Wnt/β-catenin signaling-mediated CSC induction, might shed new light on pancreatic cancer development and progression.
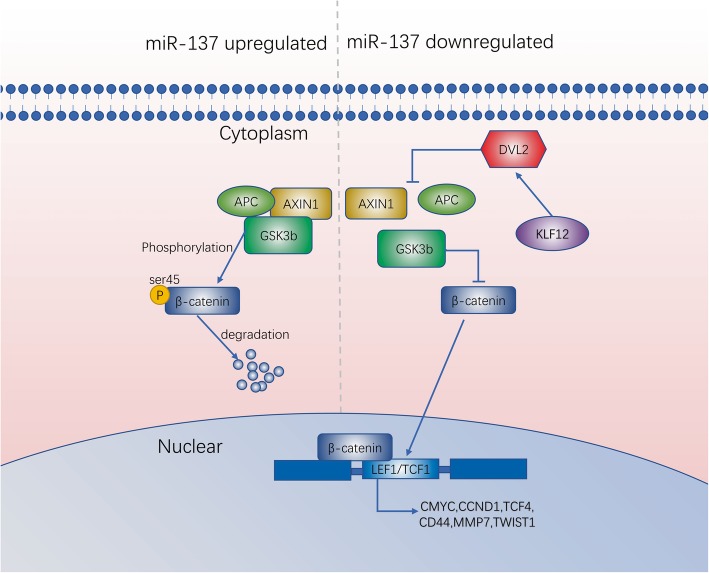
Conclusion
In summary, we have identified for the first time that the microRNA-137 reduces stemness features of pancreatic cancer cells by targeting KLF12 inhibit Wnt/β-catenin signaling pathway (Fig.8).
Fig. 8.
Illustration of suppression of KLF12 activity by miR-137 prevents Wnt/β-catenin signaling in pancreatic cancer cells
Additional file
Figure S1. Immunohistochemistry was used to detect the association between KLF12 and β-catenin expression in the subcutaneous implanted tumor. (TIF 4341 kb)
The sequence of DVL2 promoter. (DOCX 16 kb)
The sequence of DVL3 promoter. (DOCX 16 kb)
Acknowledgements
We thank Department of Hepatic-Biliary-Pancreatic Surgery, The Affiliated Hospital of Guizhou Medical University, Guiyang, China for providing the pancreatic cancer tissue samples and related anonymous clinical data.
Ethical approval
This study was approved by the Human Research Ethics Committee of Guizhou Medical University and was conducted in accordance with the principles of the Declaration of Helsinki. All patients gave written, informed consent to participate in the study. All animal experiments were approved by the Guizhou Medical University Committee on the Ethics of Animal Experiments (IRB). All treatments were in accordance with the US Public Health Service Policy on Humane Care and Use of Laboratory Animals. Surgery was performed using sodium pentobarbital anesthesia.
Funding
This study was funded by grants from The National Natural Science Foundation of China (81560477 and 81860505 to Chengyi Sun, 81572429 to Jianxin Jiang, 81602475 to Xingjun Guo, and 81660483 to Chao Yu), The United Fund of Science and Technology Department of Guizhou Province, The Affiliated Hospital Guizhou Medical University [Grant no. Qian Ke He LH Zi (2017) 5404 to Chengyi Sun, Qian Ke He LH Zi GZS (2016) 09 to Chengyi Sun, Qian Ke He LH Zi (2016) 5647 to Chengyi Sun].
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ChIP
Chromatin immunoprecipitation
- CSC
Cancer stem cell
- IHC
Immunohistochemical
- MiRNAs
MicroRNAs
- qRT-PCR
Quantitative real time polymerase chain reaction
- TCGA
The Cancer Genome Atlas
Authors’ contributions
ZH, XG carried out the experiments. CY analyzed the data. JJ and CS designed the experiments. All authors were involved in writing the paper and approved the submission and publication.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhiwei He, Email: 2268272794@qq.com.
Xingjun Guo, Email: xjguo@tjh.tjmu.edu.cn.
She Tian, Email: 87473942@qq.com.
Changhao Zhu, Email: 1789575543@qq.com.
Shiyu Chen, Email: 734517973@qq.com.
Chao Yu, Email: yuchao2002@gmc.edu.cn.
Jianxin Jiang, Email: rm002979@whu.edu.cn.
Chengyi Sun, Email: chengyisun2014@163.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Krampitz GW, George BM, Willingham SB, Volkmer JP, Weiskopf K, Jahchan N, Newman AM, Sahoo D, Zemek AJ, Yanovsky RL, et al. Identification of tumorigenic cells and therapeutic targets in pancreatic neuroendocrine tumors. Proc Natl Acad Sci U S A. 2016;113:4464–4469. doi: 10.1073/pnas.1600007113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang S, Huang X, Tian Y, Aimaiti S, Zhang J, Zhao J, Chen Y, Wang C. Clinicopathologic characteristics, laboratory parameters, treatment protocols, and outcomes of pancreatic cancer: a retrospective cohort study of 1433 patients in China. Peerj. 2018;6:e4893. doi: 10.7717/peerj.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lonardo E, Hermann PC, Heeschen C. Pancreatic cancer stem cells - update and future perspectives. Mol Oncol. 2010;4:431–442. doi: 10.1016/j.molonc.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sureban SM, Qu D, Houchen CW. Regulation of miRNAs by agents targeting the tumor stem cell markers DCLK1, MSI1, LGR5, and BMI1. Curr Pharmacol Rep. 2015;1:217–222. doi: 10.1007/s40495-014-0006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 10.Aval SF, Lotfi H, Sheervalilou R, Zarghami N. Tuning of major signaling networks (TGF-beta, Wnt, notch and hedgehog) by miRNAs in human stem cells commitment to different lineages: possible clinical application. Biomed Pharmacother. 2017;91:849–860. doi: 10.1016/j.biopha.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 11.Liu CC, Cai DL, Sun F, Wu ZH, Yue B, Zhao SL, Wu XS, Zhang M, Zhu XW, Peng ZH, Yan DW. FERMT1 mediates epithelial-mesenchymal transition to promote colon cancer metastasis via modulation of beta-catenin transcriptional activity. Oncogene. 2017;36:1779–1792. doi: 10.1038/onc.2016.339. [DOI] [PubMed] [Google Scholar]
- 12.Xia Y, Wu S. Tissue inhibitor of metalloproteinase 2 inhibits activation of the beta-catenin signaling in melanoma cells. Cell Cycle. 2015;14:1666–1674. doi: 10.1080/15384101.2015.1030557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tseng RC, Lin RK, Wen CK, Tseng C, Hsu HS, Hsu WH, Wang YC. Epigenetic silencing of AXIN2/betaTrCP and deregulation of p53-mediated control lead to wild-type beta-catenin nuclear accumulation in lung tumorigenesis. Oncogene. 2008;27:4488–4496. doi: 10.1038/onc.2008.83. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Q, Lou Y, Zhang J, Fu Q, Wei T, Sun X, Chen Q, Yang J, Bai X, Liang T. Hypoxia-inducible factor-2alpha promotes tumor progression and has crosstalk with Wnt/beta-catenin signaling in pancreatic cancer. Mol Cancer. 2017;16:119. doi: 10.1186/s12943-017-0689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang TW, Gao YH, Ma SY, Wu Q, Li ZF. Low-grade slightly elevated and polypoid colorectal adenomas display differential beta-catenin-TCF/LEF activity, c-Myc, and cyclin D1 expression. World J Gastroenterol. 2017;23:3066–3076. doi: 10.3748/wjg.v23.i17.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui J, Jiang W, Wang S, Wang L, Xie K. Role of Wnt/beta-catenin signaling in drug resistance of pancreatic cancer. Curr Pharm Des. 2012;18:2464–2471. doi: 10.2174/13816128112092464. [DOI] [PubMed] [Google Scholar]
- 17.Lee SH, Koo BS, Kim JM, Huang S, Rho YS, Bae WJ, Kang HJ, Kim YS, Moon JH, Lim YC. Wnt/beta-catenin signalling maintains self-renewal and tumourigenicity of head and neck squamous cell carcinoma stem-like cells by activating Oct4. J Pathol. 2014;234:99–107. doi: 10.1002/path.4383. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Bussing I, Slack FJ, Grosshans H. Let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Chang TH, Tsai MF, Gow CH, Wu SG, Liu YN, Chang YL, Yu SL, Tsai HC, Lin SW, Chen YW, et al. Upregulation of microRNA-137 expression by slug promotes tumor invasion and metastasis of non-small cell lung cancer cells through suppression of TFAP2C. Cancer Lett. 2017;402:190–202. doi: 10.1016/j.canlet.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Chen W, Zeng W, Wan C, Duan S, Jiang S. microRNA-137 promotes apoptosis in ovarian cancer cells via the regulation of XIAP. Br J Cancer. 2017;116:66–76. doi: 10.1038/bjc.2016.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao J, Peng F, Yu C, Wang M, Li X, Li Z, Jiang J, Sun C. microRNA-137 modulates pancreatic cancer cells tumor growth, invasion and sensitivity to chemotherapy. Int J Clin Exp Pathol. 2014;7:7442–7450. [PMC free article] [PubMed] [Google Scholar]
- 23.Neault M, Mallette FA, Richard S. miR-137 modulates a tumor suppressor network-inducing senescence in pancreatic Cancer cells. Cell Rep. 2016;14:1966–1978. doi: 10.1016/j.celrep.2016.01.068. [DOI] [PubMed] [Google Scholar]
- 24.Shen X, Hu Y, Jiang Y, Liu H, Zhu L, Jin X, Shan H, Zhen X, Sun L, Yan G, Sun H. Kruppel-like factor 12 negatively regulates human endometrial stromal cell decidualization. Biochem Biophys Res Commun. 2013;433:11–17. doi: 10.1016/j.bbrc.2013.02.078. [DOI] [PubMed] [Google Scholar]
- 25.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 26.Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 28.Raza U, Zhang JD, Sahin O. MicroRNAs: master regulators of drug resistance, stemness, and metastasis. J Mol Med (Berl). 2014;92:321–336. [DOI] [PubMed]
- 29.Aponte PM, Caicedo A: Stemness in Cancer: stem cells, Cancer stem cells, and their microenvironment. Stem Cells Int 2017, 2017:5619472. [DOI] [PMC free article] [PubMed]
- 30.Bhardwaj A, Arora S, Prajapati VK, Singh S, Singh AP. Cancer “stemness”- regulating microRNAs: role, mechanisms and therapeutic potential. Curr Drug Targets. 2013;14:1175–1184. doi: 10.2174/13894501113149990190. [DOI] [PubMed] [Google Scholar]
- 31.Li KK, Yang L, Pang JC, Chan AK, Zhou L, Mao Y, Wang Y, Lau KM, Poon WS, Shi Z, Ng HK. miR-137 suppresses growth and invasion, is downregulated in oligodendroglial tumors and targets CSE1L. Brain Pathol. 2013;23:426–439. doi: 10.1111/bpa.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du Y, Chen Y, Wang F, Gu L. miR-137 plays tumor suppressor roles in gastric cancer cell lines by targeting KLF12 and MYO1C. Tumour Biol. 2016;37:13557–13569. doi: 10.1007/s13277-016-5199-3. [DOI] [PubMed] [Google Scholar]
- 33.Mak CS, Yung MM, Hui LM, Leung LL, Liang R, Chen K, Liu SS, Qin Y, Leung TH, Lee KF, et al. MicroRNA-141 enhances anoikis resistance in metastatic progression of ovarian cancer through targeting KLF12/Sp1/survivin axis. Mol Cancer. 2017;16:11. doi: 10.1186/s12943-017-0582-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Godin-Heymann N, Brabetz S, Murillo MM, Saponaro M, Santos CR, Lobley A, East P, Chakravarty P, Matthews N, Kelly G, et al. Tumour-suppression function of KLF12 through regulation of anoikis. Oncogene. 2016;35:3324–3334. doi: 10.1038/onc.2015.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura Y, Migita T, Hosoda F, Okada N, Gotoh M, Arai Y, Fukushima M, Ohki M, Miyata S, Takeuchi K, et al. Kruppel-like factor 12 plays a significant role in poorly differentiated gastric cancer progression. Int J Cancer. 2009;125:1859–1867. doi: 10.1002/ijc.24538. [DOI] [PubMed] [Google Scholar]
- 36.Javadinia SA, Shahidsales S, Fanipakdel A, Joudi-Mashhad M, Mehramiz M, Talebian S, Maftouh M, Mardani R, Hassanian SM, Khazaei M, et al. Therapeutic potential of targeting the Wnt/beta-catenin pathway in the treatment of pancreatic cancer. J Cell Biochem. 2018. [DOI] [PubMed]
- 37.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao C, Chen YG. Dishevelled: the hub of Wnt signaling. Cell Signal. 2010;22:717–727. doi: 10.1016/j.cellsig.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 39.Metcalfe C, Ibrahim AE, Graeb M, de la Roche M, Schwarz-Romond T, Fiedler M, Winton DJ, Corfield A, Bienz M. Dvl2 promotes intestinal length and neoplasia in the ApcMin mouse model for colorectal cancer. Cancer Res. 2010;70:6629–6638. doi: 10.1158/0008-5472.CAN-10-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, Fu W, Zhang J, Wu W, Zhang X, Chen YG. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–790. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Immunohistochemistry was used to detect the association between KLF12 and β-catenin expression in the subcutaneous implanted tumor. (TIF 4341 kb)
The sequence of DVL2 promoter. (DOCX 16 kb)
The sequence of DVL3 promoter. (DOCX 16 kb)
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.