Abstract
Background
Neratinib is an irreversible pan-HER tyrosine kinase inhibitor that inhibits PI3K/Akt and MAPK signaling pathways after HER2 receptor activation. The ExteNET study showed that neratinib significantly improved 5-year invasive disease-free survival (iDFS) in women who completed trastuzumab-based adjuvant therapy for early breast cancer (EBC). We assessed the prognostic and predictive significance of PIK3CA alterations in patients in ExteNET.
Methods
Participants were women aged ≥ 18 years (≥ 20 years in Japan) with stage 1–3c (modified to stage 2–3c in February 2010) operable breast cancer, who had completed (neo)adjuvant chemotherapy plus trastuzumab ≤ 2 years before randomization, with no evidence of disease recurrence or metastatic disease at study entry. Patients were randomized to oral neratinib 240 mg/day or placebo for 1 year. Formalin-fixed, paraffin-embedded primary tumor specimens underwent polymerase chain reaction (PCR) PIK3CA testing for two hotspot mutations in exon 9, one hot-spot mutation in exon 20, and fluorescence in situ hybridization (FISH) analysis for PIK3CA amplification. The primary endpoint (iDFS) was tested with log-rank test and hazard ratios (HRs) estimated using Cox proportional-hazards models.
Results
Among the intent-to-treat population (n = 2840), tumor specimens were available for PCR testing (991 patients) and PIK3CA FISH (702 patients). Overall, 262 samples were PIK3CA altered: 201 were mutated (77%), 52 (20%) were amplified, and 9 (3%) were mutated and amplified. iDFS was non-significantly worse in placebo-treated patients with altered vs wild-type PIK3CA (HR 1.34; 95% CI 0.72–2.50; P = 0.357). Neratinib’s effect over placebo was significant in patients with PIK3CA-altered tumors (HR 0.41; 95% CI 0.17–0.90, P = 0.028) but not PIK3CA wild-type tumors (HR 0.72; 95% CI 0.36–1.41; P = 0.34). The interaction test was non-significant (P = 0.309).
Conclusions
Although there was a greater absolute risk reduction associated with neratinib treatment of patients with PIK3CA-altered tumors in ExteNET, current data do not support PIK3CA alteration as a predictive biomarker of response to neratinib in HER2-positive EBC.
Trial registration
ClinicalTrials.gov, NCT00878709. Trial registered April 9, 2009.
Electronic supplementary material
The online version of this article (10.1186/s13058-019-1115-2) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Drug targets, Neratinib, PIK3CA, Prognostic, Predictive
Introduction
Randomized phase III studies have demonstrated that 12 months’ treatment with the anti-HER2 antibody trastuzumab plus adjuvant chemotherapy significantly improves clinical outcomes in women with HER2-positive early-stage breast cancer (BC) [1–3]. Despite the initial dramatic benefits with adjuvant trastuzumab, relapses are observed on longer follow-up. At the first combined analysis of the NSABP B-31 and NCCTG N9831 trials, disease-free survival (DFS) was 87.1% in trastuzumab-treated patients after a median follow-up of 2 years [4]; however, DFS was 73.7% at the 10-year follow-up [2]. In the HERA study, 2-year DFS was 85.8% in trastuzumab-treated patients (median follow-up 1 year) [5], whereas 10-year DFS after 11 years’ follow-up was estimated at 69% [1]. Longer follow-up in the BCIRG 006 trial demonstrated a similar ongoing relapse rate to earlier data in the trastuzumab-treated arms (10-year DFS 73–74.6%), particularly in patients with hormone receptor-positive and HER2-positive BC [3, 6].
Multiple mechanisms of trastuzumab resistance have been explored in early- and advanced-stage BC [7]. The phosphatidylinositol 3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) pathway is crucial to many physiological processes, including cell-cycle progression, apoptosis, motility, autophagy, protein and lipid synthesis, and metabolism [8]. Dysregulation of this pathway is frequently implicated in tumorigenesis and plays a key role in breast carcinoma growth and regulation [9]. Activating PIK3CA gene mutations occur in approximately 30–40% of BCs [10] and 22% of HER2-positive BCs [11, 12]. PIK3CA mutational hotspots occurring in exon 9 (E542, E545) and exon 20 (H1047) comprise approximately 70% of reported PIK3CA mutations in BC [10]. The significance of PIK3CA mutations as a biomarker of resistance to HER2-targeted therapies is inconsistent, appearing to depend on the endpoints studied and disease stage [12–21]. PIK3CA amplification is less frequent, with gene copy number amplification occurring in approximately 3% of BCs [10].
Neratinib, an irreversible small-molecule tyrosine kinase inhibitor of HER1, HER2, and HER4 [22], has established single-agent efficacy in patients with trastuzumab-pretreated HER2-positive metastatic BC [23, 24]. In the neoadjuvant adaptive I-SPY 2 study, neratinib plus a taxane demonstrated similar pathological complete response (pCR) rates to trastuzumab plus a taxane in patients with HER2-positive BC [25]. The ExteNET study compared 1 year of neratinib vs placebo given after standard trastuzumab-based (neo) adjuvant therapy in patients with early-stage HER2-positive BC. The primary analysis, performed after 2 years’ follow-up, showed significantly improved invasive DFS (iDFS) for neratinib vs placebo (stratified hazard ratio [HR] 0.67; 95% confidence interval [CI] 0.50–0.91; P = 0.0091) [26]. A recent update with 5-year median follow-up reported that this benefit was maintained [27]. Pre-clinically, neratinib has demonstrated potent anti-proliferative activity in HER2-amplified, PIK3CA-mutant tumor cell lines [28]. Furthermore, neratinib significantly inhibited tumor growth in a HER2-positive, PIK3CA-mutated patient-derived xenograft BC model [29]. Based on this rationale, we assessed the prognostic and predictive significance of PIK3CA alterations in an exploratory analysis of ExteNET.
Patients and methods
Study design, randomization, and masking
Details of the multicenter, randomized, double-blind, placebo-controlled phase III ExteNET study have been described previously [26]. In brief, 2840 women with histologically confirmed stage 2–3c (1–3c in original protocol) HER2-positive BC were enrolled from academic- and community-based centers in 40 countries between July 2009 and October 2011. Patients were randomized (1:1) to neratinib or placebo, given for 1 year after standard locoregional treatment, chemotherapy, and (neo)adjuvant trastuzumab. Patients, investigators, and study sponsors were masked to treatment allocation. Clinical and radiologic assessments had to be negative for recurrence or metastatic disease at study entry. (Neo)adjuvant trastuzumab was completed up to 1 year (2 years in original protocol) before randomization. All patients (intention-to-treat [ITT] population) provided written informed consent; patients in the correlative cohort signed an optional consent form relating to primary tumor collection for exploratory biomarker analyses.
Three different sponsors were involved over the course of the study, resulting in three global amendments to the study design [26]; the correlative study was developed with amendment 3 and with the initial study sponsor. The Independent Data Monitoring Committee (IDMC) remained consistent throughout the study to preserve blinding integrity; the infrastructure for study conduct and monitoring remained in place to preserve operational consistency. The IDMC reviewed the data at least twice yearly. Additional study design details are described in Additional file 1.
The aim of the present analysis was to assess the prognostic and predictive significance of PIK3CA alterations in ExteNET.
Procedures
Patients were randomized to neratinib (Puma Biotechnology, Los Angeles, CA, USA) 240 mg orally once daily continuously or matching placebo for 1 year. Concurrent adjuvant endocrine therapy for women with locally determined hormone receptor-positive disease was permitted and recommended.
Tumor blocks or freshly cut, unstained sections mounted on positively charged slides were sent to a central certified laboratory. PIK3CA gene testing by reverse transcriptase polymerase chain reaction (RT-PCR) and/or fluorescence in situ hybridization (FISH) was performed as materials were received from August 2009 through June 2011. PIK3CA FISH was used to quantify PIK3CA gene copy number in relation to the number of copies of chromosome 3 using Vysis LSI PIK3CA Spectrum Green and Vysis CEP 3 Spectrum Orange Probes (Abbott Molecular, Abbott Park, IL, USA). Slides were deparaffinized and pretreated using Vysis Paraffin Pretreatment Reagent Kit II (Abbott Molecular). Probe mixes were hybridized at 37 °C for 17 h. Hybridized slides were counterstained with DAPI II. A minimum of 50 nuclei of invasive tumor cells were scored by a board-certified pathologist using a LEICA microscope at × 100 objective. Amplification status was determined from the ratio of PIK3CA to chromosome 3 centromere probe (CEP3) signals. A ratio ≥ 2.2 was considered FISH positive and < 2.2 was considered normal. The pathologist was blinded to the clinical data, randomized arm, and outcome throughout the pre-analytical and analytical process.
The proportion of tumor nuclei present was determined by a board-certified pathologist. Macro-dissection for tumor enrichment was performed if the tumor burden was < 20%. Genomic DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tumor slides (QiaAmp DNA FFPE tissue kit, Qiagen, Valencia, CA, USA). Genomic DNA (5 ng/μL) was tested for PIK3CA mutations (DxS PI3K Mutation Test, DxS, Manchester, UK) using RT-PCR to qualitatively detect E542K, E545D/E545K, and H1047R at a minimum detection level of 1% H1047R, E542K, and E545K or 2% E545D relative to the background. E545D and E545K mutations were detected in a single reaction; both were detectable but not distinguishable. PCR was performed using an ABI 7900HT machine and analyzed using SDS version 2.2.2 software (Applied Biosystems, Foster City, CA, USA).
Statistical analysis
Biomarker results were based on 5-year iDFS data. iDFS was the time from randomization to the first occurrence of invasive ipsilateral BC recurrence, invasive contralateral BC, local/regional invasive recurrence, distant recurrence, or death from any cause. Patients without DFS events were censored at the date of last physical examination. Exploratory PIK3CA biomarker analyses were performed in the correlative cohort, which included patients with samples tested for PIK3CA mutation and/or PIK3CA amplification.
Data from the placebo arm were used for the prognostic evaluation of PIK3CA alterations. The PIK3CA-altered group comprised patients with mutated and/or amplified PIK3CA. The PIK3CA wild-type group comprised of patients with no measurable PIK3CA mutation or amplification. Kaplan-Meier methods were used to estimate annual event-free survival rates. Log-rank tests were used to compare 5-year iDFS between the PIK3CA-altered and wild-type groups in the placebo arm. HRs were estimated using the Cox regression model.
For sensitivity analysis, a multivariate Cox model was fitted to estimate the treatment effect within the subgroups, adjusting for baseline prognostic factors including age, baseline Eastern Cooperative Oncology Group performance status, race, region, menopausal status, nodal status, hormone receptor status, histological grade, radiotherapy, prior trastuzumab, time from last trastuzumab, prior surgery, and prior neoadjuvant therapy. In addition, the interaction between PIK3CA status (altered vs wild-type) and treatment (neratinib vs placebo) was tested using a Cox regression model. All analyses were exploratory, with no adjustment for multiplicity, and were performed using SAS statistical software (version 9.3; SAS Institute Inc., Cary, NC, USA).
Results
Participants
The ITT population included 2840 women randomly assigned to the study treatment (neratinib, n = 1420; placebo, n = 1420) between July 9, 2009, and October 24, 2011; 1201 patients (42.3%) consented to the inclusion in the correlative cohort and had sufficient tumor material for biomarker testing using at least 1 of the testing methods (neratinib, n = 593; placebo, n = 608) (Additional file 1: Figure S1). Baseline demographics and disease characteristics were well balanced between correlative cohort treatment arms and were similar to the ITT population (Table 1). In the ITT population, 5-year iDFS was 90.2% (95% CI 88.3–91.8%) for neratinib and 87.7% (95% CI 85.7–89.4%) for placebo, giving an absolute benefit of 2.5% for neratinib vs placebo (HR 0.73; 95% CI 0.57–0.92; P = 0.008) [24]. In the correlative cohort, 5-year iDFS was 91.0% (95% CI 88.1–93.2%) for neratinib vs 87.8% (95% CI 84.8–90.3%) for placebo (HR 0.67; 95% CI 0.45–0.96; P = 0.032). Follow-up continues until the final analysis of the overall survival (OS) is reported.
Table 1.
Baseline demographics and disease characteristics for the ITT population and the correlative cohort
| Characteristic | ITT population (n = 2840) | Correlative cohort (n = 1201) | ||
|---|---|---|---|---|
| Neratinib (n = 1420) | Placebo (n = 1420) | Neratinib (n = 593) | Placebo (n = 608) | |
| Median age, years (range) | 52 (25–83) | 52 (23–82) | 53 (26–83) | 53 (26–81) |
| Menopausal status at diagnosis, n (%) | ||||
| Premenopausal | 663 (46.7) | 664 (46.8) | 277 (46.7) | 267 (43.9) |
| Postmenopausal | 757 (53.3) | 756 (53.2) | 316 (53.3) | 341 (56.1) |
| Prior trastuzumab, n (%) | ||||
| Concurrent to chemotherapy | 884 (62.3) | 886 (62.4) | 374 (63.1) | 400 (65.8) |
| Sequential after chemotherapy | 536 (37.7) | 534 (37.6) | 219 (36.9) | 208 (34.2) |
| Median time since trastuzumab, months (range) | 4.4 (0.2–30.9) | 4.6 (0.3–40.6) | 4.9 (0.3–24.0) | 6.5 (0.4–24.2) |
| Median time from diagnosis to randomization (range), months | 21.82 (7.69–73.69) | 22.29 (7.82–103.03) | 22.44 (7.79–73.69) | 23.64 (7.82–66.96) |
| Nodal status, n (%) | ||||
| Negative | 335 (23.6) | 336 (23.7) | 160 (27.0) | 177 (29.1) |
| 1–3 positive nodes | 664 (46.8) | 664 (46.8) | 265 (44.7) | 266 (43.8) |
| ≥ 4 positive nodes | 421 (29.6) | 420 (29.6) | 168 (28.3) | 165 (27.1) |
| Hormone receptor status, n (%) | ||||
| Positive | 816 (57.5) | 815 (57.4) | 339 (57.2) | 361 (59.4) |
| Negative | 604 (42.5) | 605 (42.6) | 254 (42.8) | 247 (40.6) |
Distribution of PIK3CA alterations
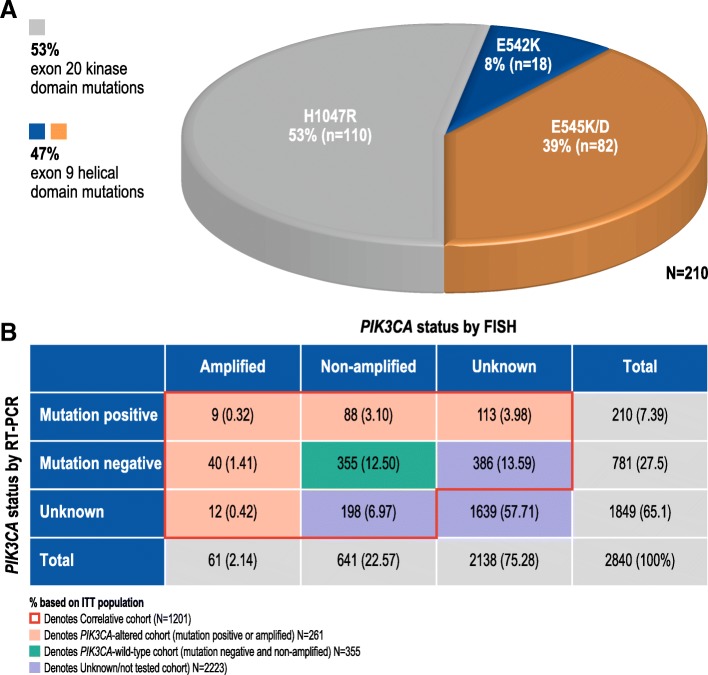
Tumor samples from 991 correlative cohort patients successfully underwent RT-PCR for the assessment of exon 9 E542K and E545K/D mutations and exon 20 H1047R mutation. A total of 210 tumors (21.2%) had 1 of these mutations, consistent with previous reports [10]: 110 mutations (53%) in exon 20 (H1047R), 18 (8%) in exon 9 E542K, and 82 (39%) in exon 9 E545K/D (Fig. 1a). PIK3CA mutations were detected in 129 of 576 (22.4%) hormone receptor-positive patients and in 81 of 415 (19.5%) hormone receptor-negative patients. Furthermore, 702 samples underwent FISH analyses, 61 (8.7%) of which were deemed PIK3CA amplified (PIK3CA:CEP3 ≥ 2.2). Nine samples were both mutation-positive and PIK3CA amplified; however, over half of the mutation-positive samples had unknown PIK3CA amplification status by FISH (Fig. 1b). PIK3CA gene amplification was detected in 28 of 408 patients (6.9%) with hormone receptor-positive disease and 33 of 294 (11.2%) with hormone receptor-negative disease.
Fig. 1.
PIK3CA mutations and PIK3CA amplifications in the correlative cohort. a Distribution of hotspot variants in PIK3CA mutation-positive subgroups. b Distribution of PIK3CA amplification or mutation status in the correlative cohort
Prognostic effects
The prognostic effect of PIK3CA alterations was assessed in 312 patients in the placebo arm of the correlative cohort who had sufficient tumor material for the testing of PIK3CA mutations and PIK3CA amplification. There was no significant association in iDFS observed in patients harboring PIK3CA alteration (HR 1.34; 95% CI 0.72–2.50) (P = 0.357). Estimated 5-year iDFS was 88.8% in the PIK3CA wild-type group vs 84.5% in the PIK3CA-altered group (Additional file 1: Figure S2).
Treatment effects
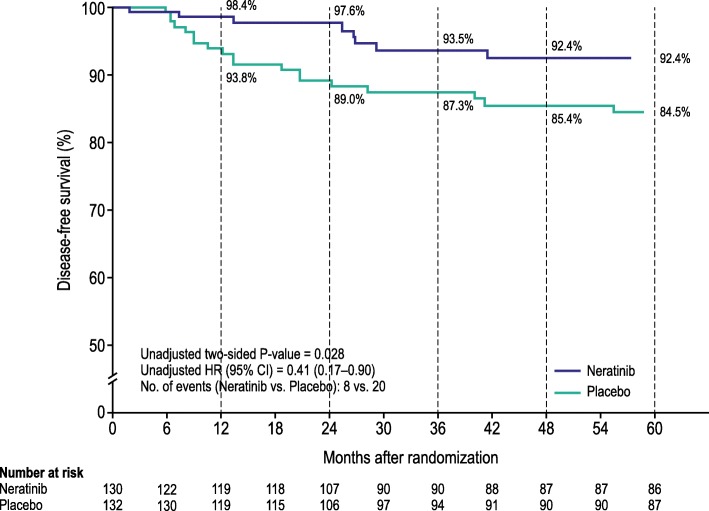
The predictive effect of PIK3CA alterations on response to neratinib was assessed by comparing 5-year iDFS rates for neratinib-treated and placebo-treated PIK3CA-altered patients. In PIK3CA-altered patients, 5-year iDFS was 92.4% for neratinib vs 84.5% for placebo (HR 0.41, 95% CI 0.17–0.90; P = 0.028) (Table 2; Fig. 2). In PIK3CA wild-type patients (mutation-negative and non-amplified), the treatment effect of neratinib was less pronounced (5-year iDFS 90.6% vs 88.8%; HR 0.72, 95% CI 0.36–1.41; P = 0.340) (Additional file 1: Figure S3). The interaction test for the PIK3CA subgroups was not statistically significant (P = 0.309).
Table 2.
Five-year iDFS events in patients treated with neratinib and placebo relative to PIK3CA status
| Population | Neratinib | Placebo | ||||
|---|---|---|---|---|---|---|
| n | iDFS events, n | n | iDFS events, n | HR (95% CI) | Two-sided P valuea | |
| ITT | 1420 | 116 | 1420 | 163 | 0.73 (0.57–0.92)b | 0.008b |
| Correlative cohort | 593 | 45 | 608 | 70 | 0.67 (0.45–0.96) | 0.032 |
| PIK3CA mutation positive | 104 | 7 | 106 | 17 | 0.43 (0.17–1.01) | 0.056 |
| PIK3CA mutation negative | 385 | 27 | 396 | 42 | 0.66 (0.40–1.06) | 0.089 |
| PIK3CA amplified | 33 | 1 | 28 | 4 | 0.20 (0.01–1.33) | 0.106 |
| PIK3CA non-amplified | 316 | 29 | 325 | 36 | 0.85 (0.52–1.39) | 0.521 |
| PIK3CA alteredc | 130 | 8 | 132 | 20 | 0.41 (0.17–0.90) | 0.028 |
A total of nine patients (seven patients in the neratinib arm and two patients in the placebo arm) had tumors harboring both a mutation and amplification
aLog-rank test
bStratified analysis
cTumors harboring either a PIK3CA mutation or PIK3CA amplification
Fig. 2.
Kaplan-Meier plot of 5-year invasive disease-free survival for patients with PIK3CA-altered tumors (correlative cohort)
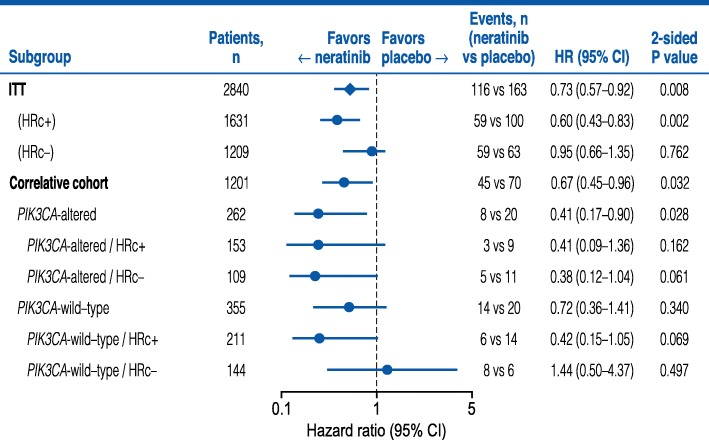
A multivariate analysis adjusting for clinical prognostic covariates revealed a statistically significant benefit for neratinib vs placebo in patients with PIK3CA-altered tumors (HR 0.40, 95% CI 0.15–0.92; P = 0.041) (Additional file 1: Table S1). The proportion of patients with hormone receptor-positive tumors was similar in PIK3CA-altered (58.4%) and wild-type subgroups (59.4%). The observed benefit for neratinib in patients with PIK3CA-altered tumors was independent of hormone receptor status (Fig. 3).
Fig. 3.
Forest plot of neratinib vs placebo in PIK3CA-altered and hormone receptor subgroups. HRc, hormone receptor
Discussion
Studies have demonstrated that PIK3CA mutations and amplifications contribute to the hyperactivation of the PI3K/Akt pathways in breast and other human cancers [30–36]. The contribution of PIK3CA gene amplifications in breast cancer tumorigenesis is not well studied, and gene amplifications are relatively infrequent in breast cancers compared to somatic mutations. In the present analysis of data from the ExteNET study, we evaluated PIK3CA amplifications and somatic mutations to obtain a comprehensive assessment of the prognostic and predictive significance of PIK3CA alterations in patients with early-stage HER2-positive breast cancer.
The ExteNET study demonstrated that 1 year of neratinib, given after standard trastuzumab-based adjuvant therapy, significantly improved iDFS in women with early-stage HER2-positive BC, with a 27% relative reduction in the risk of an iDFS event (stratified HR 0.73, 95% CI 0.57–0.92; P = 0.008), corresponding to an absolute improvement of 2.5% vs placebo [27]. In patients with hormone receptor-positive disease, most of whom received concurrent adjuvant hormonal therapy with neratinib/placebo, the magnitude of benefit was more pronounced (HR 0.60, 95% CI 0.43–0.83; P = 0.002) with an absolute improvement of 4.4%. The present correlative study attempted to further refine the HER2-positive patient population that may benefit based on PIK3CA alterations within their primary tumor. Patients with PIK3CA-altered tumors derived numerically greater benefit from neratinib than placebo: 5-year iDFS was numerically better with neratinib vs placebo, equating to an absolute improvement of 7.9% in favor of neratinib. Furthermore, neratinib’s benefit appeared similar in patients with hormone receptor-positive and hormone receptor-negative PIK3CA-altered tumors; however, as the interaction test between PIK3CA status and neratinib benefit was not statistically significant, we cannot definitively conclude that PIK3CA status is predictive in nature for selection of neratinib therapy.
PIK3CA alterations are among the most common genomic aberrations seen in patients with HER2-positive BC; data suggest that PIK3CA is altered in 36% of patients [10], with mutations in 30.7%, amplifications in 3.1%, and multiple alterations in 2.1% of cases [37]. Similar rates of PIK3CA alterations (35.9%) were reported in another cohort study, primarily comprising advanced-stage BC [38]. In their recent large pooled analysis of outcomes in patients with early-stage HER2-positive BC, Zavardas and colleagues reported an overall PIK3CA mutation rate of 22% [11], similar to the 21% mutation rate we report. We observed a higher amplification rate (8.7%) than previously reported [10], which may be related to discrepancies in gene amplification measurement methods (FISH vs copy number alterations by next-generation sequencing). Within The Cancer Genome Atlas dataset, 70% of PIK3CA mutations were at hotspots E542, E545, or H1047 [10]. Although available techniques at the time of measurement (2009–2011) only allowed us to profile two hotspot mutations in exon 9 (E542K, E545K/D) and one hotspot mutation in exon 20 (H1047R), these appear to account for over two thirds of the reported mutations across the PIK3CA gene [10].
The prognostic and predictive significance of PIK3CA alterations, particularly mutations, have been extensively studied in BC. In metastatic BC, PIK3CA mutation status showed the greatest prognostic effect of those analyzed in the CLEOPATRA study [19]. PIK3CA status was an independent poor prognostic factor, with 8.6 months’ median progression-free survival (PFS) for mutated PIK3CA vs 13.8 months for wild-type PIK3CA in the control arm, and 12.5 months for mutated PIK3CA vs 21.8 months for wild-type PIK3CA in the pertuzumab arm. In the EMILIA study, PIK3CA mutations were associated with shorter PFS and OS in capecitabine plus lapatinib-treated patients, but interestingly not in T-DM1-treated patients [20].
The prognostic effect of PIK3CA mutations is less clear in early-stage disease, with seeming variability between short- and long-term outcome and across different clinical trials [11–13, 17, 18]. In our correlative analysis of ExteNET, patients with PIK3CA-mutant tumors did not have a statistically significant difference in survival compared with patients with PIK3CA wild-type tumors.
The presence of PIK3CA alterations and a statistically significant differential benefit from HER2-directed therapies has not been demonstrated in prior studies. In the pooled analysis of five neoadjuvant studies, the interaction test was not significant for PIK3CA status and benefit from combination vs single-agent HER2-directed treatments [12]. Results from FinHER [17] and NSABP B-31 [18] also failed to demonstrate a significant interaction between PIK3CA mutation and the degree of benefit from adjuvant trastuzumab relative to control. Although we observed a greater absolute improvement in iDFS for neratinib in PIK3CA-altered vs wild-type tumors in the present study, the interaction test was not statistically significant (P = 0.1842), and thus, we cannot conclude that PIK3CA status is predictive of benefit for neratinib following 1 year of trastuzumab in patients with early-stage HER2-positive BC.
One limitation of this study is that only three PIK3CA mutation hotspots were assessed, although the PCR methodology used provided good sensitivity for detecting mutations even at low allele frequencies. Today, however, a more comprehensive, whole-exome, or next-generation sequencing might be performed to detect the full spectrum of genomic aberrations across the PIK3CA gene. It is unclear if other known PIK3CA variants would trend toward a similar benefit for neratinib as seen for the E542K, E545K/D, and H1047R variants. A second limitation is that validated guidelines for a definition for PI3K amplification to be associated with biological relevance have not yet been established. Since guidelines for PIK3CA amplification have not been established, we used the same cutoff that was defined for HER2 FISH amplification by ASCO/CAP at the time of sample testing. Another limitation is that FFPE tumor tissue was only collected from 42% of the ITT population, and only 41% of these samples were tested for both PIK3CA amplification and mutation. These tissues were archival and the primary resected breast specimen, and thus reflect the state of the tumor prior to receiving any systemic therapy. Although the study results were essentially unchanged after adjusting for important prognostic factors, the possibility of bias cannot be excluded. A larger sample size would have improved the likelihood of demonstrating a statistically significant interaction test, if one truly exists. A final limitation was the fact that patients with early relapse during adjuvant trastuzumab-based therapy were excluded from ExteNET as patients had to be clinically disease-free at the study entry, with a median time from diagnosis to randomization of 22–24 months. Thus, the results and hypotheses associated with our study may not be relevant in those tumors with primary resistance to adjuvant trastuzumab.
Conclusions
The use of biomarkers to identify patient populations likely to benefit from targeted therapies is a key challenge in the treatment of patients with breast cancer. The clinical significance of PIK3CA mutation as a biomarker of resistance to HER2-targeted therapies has not been clearly established. This exploratory correlative study reports a trend toward the absolute risk reduction for neratinib treatment being greater in patients with PIK3CA-altered vs wild-type HER2-positive BC following standard adjuvant chemotherapy and trastuzumab; however, this observation was not statistically significant. Therefore, these results do not support PIK3CA alterations as a predictive biomarker for benefit from neratinib treatment. Consideration of PIK3CA status as a definitive factor for the use of neratinib in the adjuvant setting would require a prospective clinical study with sufficient power to assess predictive significance for the biomarker.
Additional file
Supplementary methods. Table S1. Multivariate analyses adjusting for clinical prognostic covariates. Figure S1. ExteNET: CONSORT diagram of the correlative cohort. Figure S2. Kaplan-Meier plots of 5-year invasive disease-free survival for PIK3CA-altered vs wild-type tumors in the placebo arm of the correlative cohort, for assessment of prognostic effect. Figure S3. Kaplan-Meier plot of 5-year invasive disease-free survival for PIK3CA wild-type patients in the correlative cohort. (DOCX 404 kb)
Acknowledgements
We would like to acknowledge the contribution of the IDMC, study investigators, research staff, clinical research organizations, and other vendors, as well as the patients who participated in the ExteNET study.
Statistical analyses for the study were performed by, or with assistance from, Bin Yao, Feng Xu, and Ju RueyJiuan Lee, with programming support from Yan Yan and Janine Lu.
Funding
ExteNET was sponsored by Wyeth, Pfizer and Puma Biotechnology. Puma Biotechnology Inc. also funded the design of the current analysis, interpretation of the data and provision of editorial/medical writing support provided by Lee Miller and Deirdre Carman of Miller Medical Communications.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- BC
Breast cancer
- CEP3
Chromosome 3 centromere probe
- CI
Confidence interval
- DFS
Disease-free survival
- FFPE
Formalin-fixed, paraffin-embedded
- FISH
Fluorescence in situ hybridization
- HR
Hazards ratio
- IDMC
Independent Data Monitoring Committee
- ITT
Intention-to-treat
- mTOR
Mammalian target of rapamycin
- OS
Overall survival
- PI3K
Phosphatidylinositol 3-kinase
- RT-PCR
Reverse transcriptase polymerase chain reaction
Authors’ contributions
SKLC, MM, FAH, BE, SD, BM, HI, GvM, JM, CHB, MG, MB, and AC conceived and designed the study. BZ, LDE, YY, and ASL provided administrative support throughout the study. All authors acquired the data and provided the study materials or patients. SKLC, BZ, LDE, YY, and ASL analyzed the data. All authors were involved in the interpretation and critical review of the data, drafted or revised the manuscript for important intellectual content, approved the final version of the manuscript, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The study protocol was approved by the institutional ethics committee at participating sites; the study was conducted in accordance with the 2008 Declaration of Helsinki. All patients (intention-to-treat [ITT] population) provided written informed consent; patients in the correlative cohort signed an optional consent form relating to primary tumor collection for exploratory biomarker analyses.
Consent for publication
Not applicable
Competing interests
SKLC received honoraria from Novartis, Hoffmann-La Roche, Pfizer, and AstraZeneca. MM received grants and personal fees from Roche/Genentech, Novartis, Amgen, AstraZeneca, Pfizer, Pharmamar, and Lilly. BE received research funding from NanoString, Novartis, and Roche, and travel, accommodations, or expenses from AstraZeneca. SD received honoraria from Puma, Pfizer, AstraZeneca, Roche, and Lilly; consulting or advisory role from Puma, Pfizer, AstraZeneca, Roche, and Lilly; research funding from Puma, Pfizer, AstraZeneca, Roche, and Lilly; and travel and accommodations expenses from Pfizer, Roche, and AstraZeneca. BM acted in a consulting or advisory role (immediate family member) for Motus GI. HI received honoraria from Chugai, AstraZeneca, and Eisai, and consulting or advisory role for Chugai, Pfizer, Daiichi-Sankyo, AstraZeneca, and Lilly. GvM received research funding from Pfizer, Amgen, Roche, Teva, Novartis, AstraZeneca, Celgene, Myriad, AbbVie, and Vifor. JM received research funding from Roche, Macrogenics, and Puma Biotechnology Inc. CHB received grants and personal fees from GlaxoSmithKline, Roche, and Novartis outside the submitted work. MG received research funding from Sanofi-Aventis, Pfizer, Smith Medical, Novartis, Roche, GlaxoSmithKline, and personal fees from Novartis, Roche, GlaxoSmithKline, AstraZeneca, Nanostring Technologies, and Accelsiors. ND received research funding from Amgen and Genentech. SBK received research funding from Novartis, Sanofi-Aventis, Kyowa-Kirin Inc., and Dongkook Pharma Co. Ltd. NR acted in a consulting or advisory role for New Century Health and Bristol-Myers Squibb and received research funding from Side-Out Foundation. JS II acted in a consulting or advisory role for BMS and Novartis. BZ, LDE, and ASL are employed by Puma Biotechnology, Inc. MB is employed by the International Drug Development Institute (IDDI) and holds stock of IDDI. YY was employed by Puma Biotechnology at the time of the research but is now employed by QED Therapeutics Inc. FAH, ZT, RS, EHJ, VH, GH, and AC declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cameron D, Piccart-Gebhart MJ, Gelber RD, Procter M, Goldhirsch A, de Azambuja E, et al. 11 years’ follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive early breast cancer: final analysis of the HERceptin Adjuvant (HERA) trial. Lancet. 2017;389(10075):1195–1205. doi: 10.1016/S0140-6736(16)32616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez EA, Romond EH, Suman VJ, Jeong JH, Sledge G, Geyer CE, Jr, et al. Trastuzumab plus adjuvant chemotherapy for human epidermal growth factor receptor 2-positive breast cancer: planned joint analysis of overall survival from NSABP B-31 and NCCTG N9831. J Clin Oncol. 2014;32(33):3744–3752. doi: 10.1200/JCO.2014.55.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slamon DJ, Eiermann W, Robert NJ, Giermek J, Martin M, Jasiowka M, et al. Ten year follow-up of BCIRG-006 comparing doxorubicin plus cyclophosphamide followed by docetaxel (AC→T) with doxorubicin plus cyclophosphamide followed by docetaxel and trastuzumab (AC→TH) with docetaxel, carboplatin and trastuzumab (TCH) in HER2+ early breast cancer. Oral presentation at San Antonio breast Cancer Symposium; December 11, 2015; San Antonio, TX.
- 4.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353(16):1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 5.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353(16):1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 6.Slamon D, Eiermann W, Robert N, Pienkowski T, Martin M, Press M, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365(14):1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scaltriti M, Rojo F, Ocaña A, Anido J, Guzman M, Cortes J, et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J Natl Cancer Inst. 2007;99(8):628–638. doi: 10.1093/jnci/djk134. [DOI] [PubMed] [Google Scholar]
- 8.Saini KS, Loi S, de Azambuja E, Metzger-Filho O, Saini ML, et al. Targeting the PI3K/AKT/mTOR and Raf/MEK/ERK pathways in the treatment of breast cancer. Cancer Treat Rev. 2013;39(8):935–946. doi: 10.1016/j.ctrv.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Castaneda CA, Cortes-Funes H, Gomez HL, Ciruelos EM. The phosphatidyl inositol 3-kinase/AKT signaling pathway in breast cancer. Cancer Metastasis Rev. 2010;29(4):751–759. doi: 10.1007/s10555-010-9261-0. [DOI] [PubMed] [Google Scholar]
- 10.Breast Invasive Carcinoma TCGA Provisional Database, cBioPortal for Cancer Genomics. http://www.cbioportal.org/study?id=brca_tcga#summary. Accessed 21 Feb 2019.
- 11.Zardavas D, Te Marvelde L, Milne RL, Fumagalli D, Fountzilas G, Kotoula V, et al. Tumor PIK3CA genotype and prognosis in early-stage breast cancer: a pooled analysis of individual patient data. J Clin Oncol. 2018;36(10):981–990. doi: 10.1200/JCO.2017.74.8301. [DOI] [PubMed] [Google Scholar]
- 12.Loibl S, Majewski I, Guarneri V, Nekljudova V, Holmes E, Bria E, et al. PIK3CA mutations are associated with reduced pathological complete response rates in primary HER2-positive breast cancer: pooled analysis of 967 patients from five prospective trials investigating lapatinib and trastuzumab. Ann Oncol. 2016;27(8):1519–1525. doi: 10.1093/annonc/mdw197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bianchini G, Kiermaier A, Bianchi GV, Im YH, Pienkowski T, Liu MC, et al. Biomarker analysis of the NeoSphere study: pertuzumab, trastuzumab, and docetaxel versus trastuzumab plus docetaxel, pertuzumab plus trastuzumab, or pertuzumab plus docetaxel for the neoadjuvant treatment of HER2-positive breast cancer. Breast Cancer Res. 2017;19(1):16. doi: 10.1186/s13058-017-0806-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loibl S, von Minckwitz G, Schneeweiss A, Holmes E, Bria E, Denkert C, et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (HER2) therapy in primary HER2-overexpressing breast cancer. J Clin Oncol. 2014;32(29):3212–3220. doi: 10.1200/JCO.2014.55.7876. [DOI] [PubMed] [Google Scholar]
- 15.Majewski IJ, Nuciforo P, Mittempergher L, Bosma AJ, Eidtmann H, Holmes E, et al. PIK3CA mutations are associated with decreased benefit to neoadjuvant human epidermal growth factor receptor 2-targeted therapies in breast cancer. J Clin Oncol. 2015;33(12):1334–1339. doi: 10.1200/JCO.2014.55.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guarneri V, Dieci MV, Frassoldati A, Maiorana A, Ficarra G, Bettelli S, et al. Prospective biomarker analysis of the randomized CHER-LOB study evaluating the dual anti-HER2 treatment with trastuzumab and lapatinib plus chemotherapy as neoadjuvant therapy for HER2-positive breast cancer. Oncologist. 2015;20(9):1001–1010. doi: 10.1634/theoncologist.2015-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loi S, Michiels S, Lambrechts D, Fumagalli D, Claes B, Kellokumpu-Lehtinen PL, et al. Somatic mutation profiling and associations with prognosis and trastuzumab benefit in early breast cancer. J Natl Cancer Inst. 2013;105(13):960–967. doi: 10.1093/jnci/djt121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pogue-Geile KL, Song N, Jeong JH, Gavin PG, Kim SR, Blackmon NL, et al. Intrinsic subtypes, PIK3CA mutation, and the degree of benefit from adjuvant trastuzumab in the NSABP B-31 trial. J Clin Oncol. 2015;33(12):1340–1347. doi: 10.1200/JCO.2014.56.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baselga J, Cortés J, Im SA, Clark E, Ross G, Kiermaier A, et al. Biomarker analyses in CLEOPATRA: a phase III, placebo-controlled study of pertuzumab in human epidermal growth factor receptor 2-positive, first-line metastatic breast cancer. J Clin Oncol. 2014;32(33):3753–3761. doi: 10.1200/JCO.2013.54.5384. [DOI] [PubMed] [Google Scholar]
- 20.Baselga J, Lewis Phillips GD, Verma S, Ro J, Huober J, Guardino AE, et al. Relationship between tumor biomarkers and efficacy in EMILIA, a phase III study of trastuzumab emtansine in HER2-positive metastatic breast cancer. Clin Cancer Res. 2016;22(15):3755–3763. doi: 10.1158/1078-0432.CCR-15-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu B, Guan Z, Shen Z, Tong Z, Jiang Z, Yang J, et al. Association of phosphatase and tensin homolog low and phosphatidylinositol 3-kinase catalytic subunit alpha gene mutations on outcome in human epidermal growth factor receptor 2-positive metastatic breast cancer patients treated with first-line lapatinib plus paclitaxel or paclitaxel alone. Breast Cancer Res. 2014;16(4):405. doi: 10.1186/s13058-014-0405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabindran SK, Discafani CM, Rosfjord EC, Baxter M, Floyd MB, Golas J, et al. Antitumor activity of HKI-272, an orally active, irreversible inhibitor of the HER-2 tyrosine kinase. Cancer Res. 2004;64(11):3958–3965. doi: 10.1158/0008-5472.CAN-03-2868. [DOI] [PubMed] [Google Scholar]
- 23.Burstein HJ, Sun Y, Dirix LY, Jiang Z, Paridaens R, Tan AR, et al. Neratinib, an irreversible ErbB receptor tyrosine kinase inhibitor, in patients with advanced ErbB2-positive breast cancer. J Clin Oncol. 2010;28(8):1301–1307. doi: 10.1200/JCO.2009.25.8707. [DOI] [PubMed] [Google Scholar]
- 24.Martin M, Bonneterre J, Geyer CE, Jr, Ito Y, Ro J, Lang I, et al. A phase two randomised trial of neratinib monotherapy versus lapatinib plus capecitabine combination therapy in patients with HER2+ advanced breast cancer. Eur J Cancer. 2013;49(18):3763–3772. doi: 10.1016/j.ejca.2013.07.142. [DOI] [PubMed] [Google Scholar]
- 25.Park JW, Liu MC, Yee D, Yau C, van ‘t Veer LJ, Symmans WF, et al. Adaptive randomization of neratinib in early breast cancer. N Engl J Med. 2016;375(1):11–22. doi: 10.1056/NEJMoa1513750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan A, Delaloge S, Holmes FA, Moy B, Iwata H, Harvey VJ, et al. Neratinib after trastuzumab-based adjuvant therapy in patients with HER2-positive breast cancer (ExteNET): a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2016;17(3):367–377. doi: 10.1016/S1470-2045(15)00551-3. [DOI] [PubMed] [Google Scholar]
- 27.Martin M, Holmes FA, Ejlertsen B, Delaloge S, Moy B, Iwata H, et al. Neratinib after trastuzumab-based adjuvant therapy in HER2-positive breast cancer (ExteNET): 5-year analysis of a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18(12):1688–1700. doi: 10.1016/S1470-2045(17)30717-9. [DOI] [PubMed] [Google Scholar]
- 28.Canonici A, Gijsen M, Mullooly M, Bennett R, Bouguern N, Pedersen K, et al. Neratinib overcomes trastuzumab resistance in HER2 amplified breast cancer. Oncotarget. 2013;4:1592–1605. doi: 10.18632/oncotarget.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao M, Scott S, Evans K, Yuka E, Murthy R, Avogadri-Connors F, et al. Exploring optimal targeted combination therapies with neratinib for HER2+breast cancer. AACR Annual Meeting; Washington, DC; April 1–5, 2017; Abstract 4038.
- 30.Wu G, Xing M, Mambo E, Huang X, Liu J, Guo Z, et al. Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res. 2005;7(5):R609–R616. doi: 10.1186/bcr1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez-Angulo AM, Chen H, Karuturi MS, Chavez-MacGregor M, Tsavachidis S, Meric-Bernstam F, et al. Frequency of mesenchymal-epithelial transition factor gene (MET) and the catalytic subunit of phosphoinositide-3-kinase (PIK3CA) copy number elevation and correlation with outcome in patients with early stage breast cancer. Cancer. 2013;119(1):7–15. doi: 10.1002/cncr.27608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 33.Ma YY, Wei SJ, Lin YC, Lung JC, Chang TC, Whang-Peng J, et al. PIK3CA as an oncogene in cervical cancer. Oncogene. 2000;19:2739–2744. doi: 10.1038/sj.onc.1203597. [DOI] [PubMed] [Google Scholar]
- 34.Hou P, Liu D, Shan Y, Hu S, Studeman K, Condouris S, et al. Genetic alterations and their relationship in the phosphatidylinositol 3-kinase/Akt pathway in thyroid cancer. Clin Cancer Res. 2007;13:1161–1170. doi: 10.1158/1078-0432.CCR-06-1125. [DOI] [PubMed] [Google Scholar]
- 35.Ji M, Guan H, Gao C, Shi B, Hou P. Highly frequent promoter methylation and PIK3CA amplification in non-small cell lung cancer (NSCLC) BMC Cancer. 2011;11:147. doi: 10.1186/1471-2407-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi J, Yao D, Liu W, Wang N, Lv H, Zhang G, et al. Highly frequent PIK3CA amplification is associated with poor prognosis in gastric cancer. BMC Cancer. 2012;12:50. doi: 10.1186/1471-2407-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods. Table S1. Multivariate analyses adjusting for clinical prognostic covariates. Figure S1. ExteNET: CONSORT diagram of the correlative cohort. Figure S2. Kaplan-Meier plots of 5-year invasive disease-free survival for PIK3CA-altered vs wild-type tumors in the placebo arm of the correlative cohort, for assessment of prognostic effect. Figure S3. Kaplan-Meier plot of 5-year invasive disease-free survival for PIK3CA wild-type patients in the correlative cohort. (DOCX 404 kb)
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.