Abstract
Background
Dishevelled-associated activator of morphogenesis 1 (DAAM1) is a member of microfilament-related formins and mediates cell motility in breast cancer (BrCa). However, the genetic mutation status of DAAM1 mRNA and its correlation with pathological characteristics are still unclearly. Methods: A patient cohort and BrCa cells were recruited to demonstrate the role of functional SNP in microRNA-208a-5p binding site of DAAM1 3′-UTR and underlying mechanism in BrCa metastasis.
Methods
A patient cohort and BrCa cells were recruited to demonstrate the role of functional SNP in microRNA-208a-5p binding site of DAAM1 3′-UTR and underlying mechanism in BrCa metastasis.
Results
The expression and activation of DAAM1 increased markedly in lymphnode metastatic tissues. A genetic variant (rs79036859 A/G) was validated in the miR-208a-5p binding site of DAAM1 3′-UTR. The G genotype (AG/GG) was a risk genotype for the metastasis of BrCa by reducing binding affinity of miR-208a-5p for the DAAM1 3′-UTR. Furthermore, the miR-208a-5p expression level was significantly suppressed in lymphnode metastatic tissues compared with that in non-lymphnode metastatic tissues. Overexpression of miR-208a-5p inhibited DAAM1/RhoA signaling pathway, thereby leading to the decrease of the migratory ability.
Conclusion
Overall, the rs79036859 G variant of DAAM1 3′-UTR was identified as a relevant role in BrCa metastasis via the diversity of miR-208a-5p binding affinity.
Electronic supplementary material
The online version of this article (10.1186/s12935-019-0747-8) contains supplementary material, which is available to authorized users.
Keywords: DAAM1, 3′-UTR, rs79036859, miR-208a-5p, Metastasis
Background
Dishevelled-associated activator of morphogenesis 1 (DAAM1) is a member of microfilament-related formins and is involved in cell motility through mediating Wnt signaling pathway [1–3]. In the cytoplasm, DAAM1 exists in an autoinhibited state by intramolecular interaction between its N-terminal GBD and C-terminal DAD domains. When Dishevelled 2 binds to DAAM1, leading to disrupting the interaction between the GBD and DAD that mediates DAAM1 auto-inhibition, DAAM1 will exposure FH1 and FH2 domains to polymerize straight actin filaments [2, 4]. DAAM1 is essential for Wnt-11/Frizzled-induced activation of RhoA and Xenopus gastrulation [2]. DAAM1 directly collaborates to fascin in actin filaments and thus controls the formation of filopodia [5]. Our previous studies find that active DAAM1 is involved in Wnt5a/Dishevelled 2 and Collagen/Integrin αvβ3 signaling pathways, resulting in the elevation of the migratory and haptotatic ability of breast cancer (BrCa) cells [3, 6]. However, the genetic mutation status of DAAM1 mRNA and its correlation with pathological characteristics are still unknown in BrCa patients.
Single nucleotide polymorphisms (SNPs) located in untranslated region (UTR) have been reported to be associated with dysregulation of genes expression. A recent global transcriptional network study identifies mutations at somatic expression quantitative trait locus (eQTL) located 5′-UTR of DAAM1, directly regulating the expression of DAAM1 mRNA [7]. Nevertheless, noncoding mutations in the 3′-UTR of DAAM1 has not been reported. An increasing evidence revealed that functional 3′-UTR SNPs are participated in the progression of multiple cancers [8–11]. Most 3′-UTR of mRNAs function as the target sequences of microRNAs (miRNAs) by base pairing, thereby leading to the degradation of mRNAs and decrease of their translation [12, 13]. SNPs in the miRNA binding sites in the 3′-UTRs of target genes represent their differential binding affinities for corresponding miRNAs [14–16].
Here, we demonstrate that DAAM1 is highly expressed in lymphnode metastatic BrCa tissues. We also elucidate the functional role of SNP rs79036859 in the miR-208a-5p binding site of DAAM1 3′-UTR and its involvement in BrCa metastasis. Overall these data identify miR-208a-5p/DAAM1 axis as a potential therapeutic target in limiting BrCa metastasis and reducing death from this disease.
Methods
Clinical information
157 BrCa patients were recruited by the First Affiliated Hospital with Nanjing Medical University and Affiliated Cancer Hospital to Nanjing Medical University (NJMU) from 2015 to 2018. All cases had been diagnosed with BrCa by a pathologist on the basis of hematoxylin–eosin (HE) staining. Relevant clinicopathological characteristics record for each case were collected by review of patients’ medical files. Ethical approval for the study was obtained from the Clinical Research Ethics Committee, NJMU. Pathologic staging was determined by a pathologist based on AJCC Cancer Staging Manual 8th classification criteria. All the participants voluntarily joined this study with informed contents.
Immunohistochemistry (IHC)
A total of 46 BrCa sections were deparaffinised at 55 °C for 30 min. The sections were then washed with xylene for three 5-min. The sections were rehydrated by successive washes in 100, 90 and 70% graded ethanol. Hydrogen peroxidase (0.3%, ZSGB-Bio, Beijing, China) was used to block endogenous peroxidase activity for 20 min. The primary anti-DAAM1 (1:100 dilution, Cat. 14876-1-AP, ProteinTech, Wuhan, China) antibody and DAB and hematoxylin counterstain (ZSGB-Bio) were used to visualize its expression. The percentage of positively stained cells was scored as 0–4: 0 (< 5%), 1 (6–25%), 2 (26–50%), 3 (51–75%) and 4 (> 75%). The staining intensity was scored as 0–3: 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). The immunoreactivity score (IRS) equals to the percentages of positive cells multiplied with staining intensity. Immunostained sections were scanned by a microscope (Olympus Corporation, Tokyo, Japan).
Selection of SNPs and TaqMan genotyping
A next-generation sequencing of metastatic BrCa tissues (data not published) revealed some SNPs, including rs79036859 and rs45476291, locating in the 3′-UTR of DAAM1. After a review of dbSNP database (https://www.ncbi.nlm.nih.gov/snp) and PolymiRTS Database 3.0 (http://compbio.uthsc.edu/miRSNP), we selected the candidate SNP (rs79036859) suggested as a transcriptional regulation factor for the 3′-UTR of DAAM1. SNP genotyping was conducted by allelic discrimination using the TaqMan SNP Genotyping Assays according to the manufacturer’s instructions (Applied Biosystems). Specific primers (TATCTCCTGAAAGAGATAAGA, GTTTTTCCAACAACTCCAGT) and FAM/VIC-labeled TaqMan probes (FAM-labeled CAAACAAACAAAAAAAGCTTGCAAAATATTTT, VIC-labeled CAAACAAACAAAAAAGGCTTGCAAAATATTTT) were designed and supplied by Synbio Technologies (Soochow, China). The PCR conditions were as follows: initiation at 98 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 30 s and annealing/extension at 60 °C for 60 s. PCR application was undergoing in a StepOnePlus Real-Time PCR System (Applied Biosystems).
Cell culture and transfection
MCF-10A, MDA-MB-231, MCF-7, and COS-7 cell lines were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). MCF-7, MDA-MB-231, and COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, high glucose) (Hyclone, Thermo Scientific, Waltham, MA) supplemented with 10% (v/v) fetal bovine serum (FBS) (Hyclone) at 37 °C with 5% CO2. MCF-10A cells were cultured in DMEM/F12 media supplemented with 5% (v/v) horse serum, 20 ng/mL human EGF, 10 μg/mL insulin, 0.5 μg/mL hydrocortisone, penicillin, streptomycin and 100 ng/mL cholera toxin (Sigma-Aldrich, St. Louis, MO).
For subsequent assays, cells were transfected with miR-208a-5p mimic, miR-208a-5p inhibitor (an anti-sense of miR-208a-5p), or miRNA mimic control, which is synthesized in RiboBio Co., Ltd. (Guangzhou, China), using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
Western blotting analysis and pulldown assays
Cells were placed in 35-mm dishes (6 × 105 cells/dish) and transfected with synthesized miR-208a-5p mimic, miR-208a-5p inhibitor (anti-sense of miR-208a-5p), or miRNA mimic control. 72 h after transfection, all cells were harvested the proteins with lysis buffer. SDS–polyacrylamide gel electrophoresis and Western blotting analysis were performed as standard protocols. The primary antibodies for DAAM1 (1:1000 dilution, Cat. 14876-1-AP, ProteinTech), RhoA (1:1000 dilution, Cat. 10749-1-AP, ProteinTec), β-actin (1:5000 dilution, Cat. 60008-1-Ig, ProteinTec) were used. DAAM1 protein levels were normalized to β-actin for each sample.
To detect the active level of DAAM1, we employed GST-RhoA beads as bait. The activate level of DAAM1 was detected by the Pulldown assays and subsequently immunoblotted with anti-DAAM1 antibody (Cat. 14876-1-AP, ProteinTech) [6]. SDS-PAGE and Western blotting were performed using the above methods.
Dual-luciferase activity assay
The 3′-UTR of DAAM1 which contained the putative target site of miR-208a-5p was synthesized by Integrated Biotech Solutions Co., Ltd (Shanghai, China) and ligated into pGL3 construct (Promega, Madison, WI). The constructs pGL3-DAAM1-3′-UTR-WT or pGL3-DAAM1-3′-UTR-mutant (200 ng) and pRL-TK (80 ng, Promega) were co-transfected with 60 pmol miR-208a-5p mimic or miRNA mimic control by Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, Dual-Luciferase Reporter Assay System (Promega) was performed to measure luciferase activity.
Boyden chamber assays
Migratory ability of cancer cells was tested in a modified Boyden chamber (Cat. 3422, Costar, Corning, NY). The detail protocol was described as previously [6].
Immunofluorescence and actin cytoskeleton staining
Cells were placed on glass cover-slides and subjected to actin cytoskeleton staining. The detail protocol was described as previously [17]. The images were captured by a laser scanning confocal microscope (Zeiss LSM710, Oberkochen, Germany).
Quantitative real-time PCR
MiRNA of BrCa cells and tissues were extracted by using mirVana™ miRNA isolation kit (Ambion, Austin, TX). The primers for miRNA reverse transcription were synthesized in RiboBio Co., Ltd. (Guangzhou, China) called Bulge-Loop™ miRNA qRT-PCR Primer Set as previously described [17]. Primers used for DAAM1 amplification were GAPDH: 5′-TGAACGGGAAGCTCACTGG-3′ (sense) and 5′-TCCACCACCCTGTTGCTGTA-3′ (antisense); DAAM1: 5′-AAATTGAAACGGAATCGCAAAC-3′ (sense) and 5′-GCAAGGCAGTGTAATGAAACG-3′ (antisense). SYBR Green (SYBR® Premix Ex Taq™ II, TaKaRa, Dalian, China) was used to label the amplified genes. The 2−ΔΔCt method was used for miR-208a-5p or DAAM1 expression analysis.
RhoA GTPase activation assays (G-LISA small GTPase activation assays)
Total protein lysates extracted from BrCa cells were turned to measure RhoA activity by using RhoA GTPase activation assays (Cat. BK121, Cytoskeleton Inc., Denver, CO). RhoA activation was described as previously [6].
Kaplan–Meier plotter analysis
DAAM1 (Affy ID: 216060_s_at) mRNA expression data and survival information (progression free survival, overall survival, post progression survival and distant metastasis free survival) were collected from Kaplan–Meier plotter (http://kmplot.com). Then, the prognostic significance of DAAM1 mRNA was analyzed in BrCa. Kaplan–Meier survival plots were generated with survival curves compared by log-rank test.
Statistical analysis
All statistical analyses were calculated using SPSS 25.0 software (Chicago, IL). Most of the data were analyzed by Student’s t-test or one-way ANOVA followed by Dunnett’s multiple posthoc tests. All data are presented as means ± SDs of five independent experiments if not noted. The associations between DAAM1 mRNA expression levels, genotypes distribution of DAAM1 SNP, along with miR-208a-5p expression status and clinicopathological characteristics were performed using Pearson’s Chi squared test. The correlations between DAAM1 mRNA expression and miR-208a-5p expression or between DAAM1 mRNA level and protein level were assessed by Spearman’s correlation analysis.
Results
DAAM1 expression positively correlates with lymphnode metastasis and associates with prognosis in BrCa
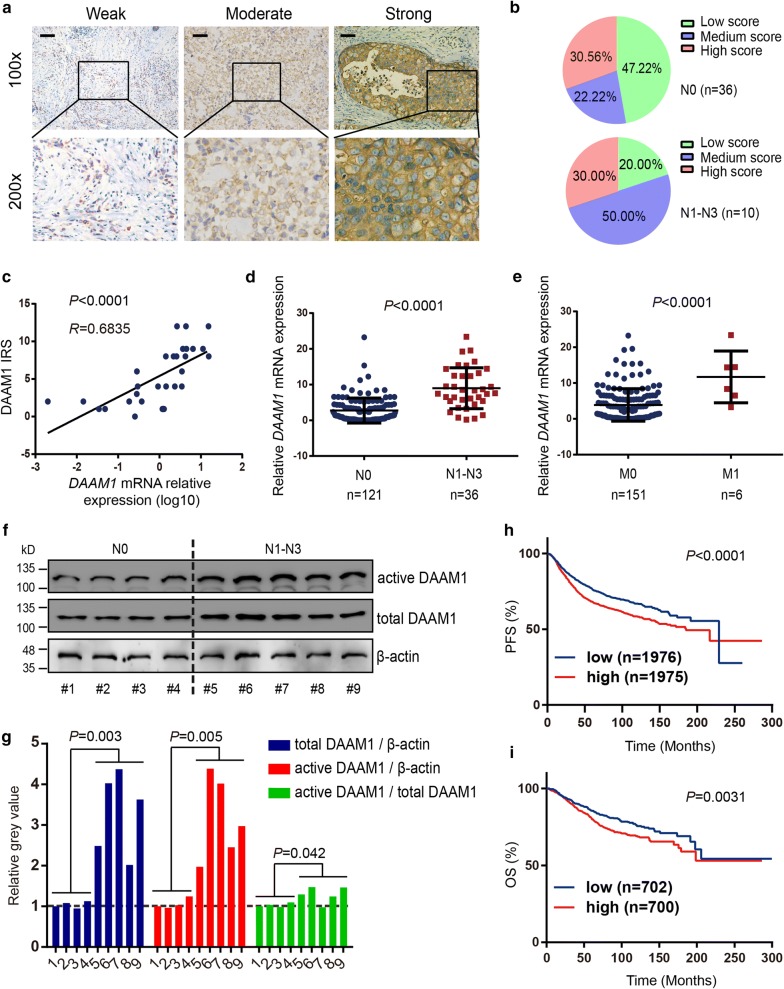
Our previous studies reported that DAAM1 regulates the re-organization of microfilaments for oriented the migration and haptotaxis of BrCa cells [3, 6]. However, it is still limited evidence demonstrating that DAAM1 is correlated with tumor metastasis in BrCa patients. To investigate whether DAAM1 acts as a metastatic promoter in BrCa, the immunohistochemistry (IHC) were performed to test the DAAM1 expression in 46 BrCa cases. The sections highly and moderately expressed DAAM1 accounted for a majority of lymphnode metastatic cases (Fig. 1a, b).
Fig. 1.
The expression of DAAM1 are relative with the high rate of lymphnode metastasis and associated with prognosis in BrCa. a Representative microphotographs revealing low, medium, and high DAAM1 expression using immunohistochemical (IHC) staining. Brown, DAAM1. Blue, haematoxylin. Bar = 100 μm. Objective lens, magnification, ×10; numerical aperture, 0.75. b DAAM1 protein expression intensity proportion based on lymphnode metastasis. N0, without lymphnode metastasis. N1–N3, with lymphnode metastasis. c Correlation between the expression levels of DAAM1 protein and DAAM1 mRNA in BrCa tissues. d, e The expression of DAAM1 mRNA according to lymphnode metastasis and distance metastasis. M0, without distance metastasis. M1, with distance metastasis. f The lysates of tumor tissues were assayed for the expression levels of total and active DAAM1 by Western blotting and pulldown assay using a GST-RhoA as a bait, respectively. β-actin was used as the loading control. g The expression ratios of total DAAM1/β-actin, active DAAM1/β-actin, and active DAAM1/total DAAM1. h, i Kaplan–Meier analysis showed survival curves of progression free survival (PFS) and overall survival (OS) and of patients with low DAAM1 mRNA expression vs high expression in BrCa
Because of limited amount of IHC sections, we further examined the transcriptional level of DAAM1 in 157 BrCa samples. We firstly determined that DAAM1 mRNA expression was positively correlated with DAAM1 protein expression (Fig. 1c). When we compared DAAM1 mRNA expression in lymphnode metastatic tumor tissues and non-lymphnode metastatic tumor tissues along with distance metastatic tumor tissues and non-distance metastatic tumor tissues, the expression of DAAM1 mRNA was significantly increased in metastatic tissues (Fig. 1d, e, Table 1). Furthermore, we tested DAAM1 expression and DAAM1 activity in fresh BrCa tissues (Fig. 1f). We found that both the expression and the activation of DAAM1 in lymphnode metastatic tumor tissues were significantly higher than these in non-lymphnode metastatic tumor tissues (Fig. 1f, g).
Table 1.
Associations between DAAM1 mRNA expression and clinicopathological characteristics in breast cancer
| Characteristics | n | DAAM1a | OR (95% Cl) | P valueb | |
|---|---|---|---|---|---|
| Low | High | ||||
| Tumor size (cm) | |||||
| ≤ 2 | 61 | 35 | 26 | 1.82 (0.94–3.53) | 0.075 |
| > 2 | 87 | 50 | 50 | ||
| Unknown | 9 | ||||
| Lymph node metastasis | |||||
| N0 | 121 | 73 | 48 | 9.43 (3.43–25.95) | < 0.001 |
| N1–N3 | 36 | 5 | 31 | ||
| Distant metastasis | |||||
| M0 | 151 | 78 | 73 | – | 0.039 |
| M1 | 6 | 0 | 6 | ||
| ER status | |||||
| Negative | 51 | 23 | 28 | 0.76 (0.39–1.49) | 0.426 |
| Positive | 106 | 55 | 51 | ||
| PR status | |||||
| Negative | 67 | 31 | 37 | 0.77 (0.41–1.45) | 0.419 |
| Positive | 89 | 47 | 42 | ||
| Unknown | 1 | ||||
| Her-2 status | |||||
| Negative | 94 | 45 | 49 | 0.89 (0.47–1.69) | 0.720 |
| Positive | 61 | 31 | 30 | ||
| Unknown | 2 | ||||
| P53 status | |||||
| Negative | 105 | 51 | 54 | 0.99 (0.48–2.04) | 0.982 |
| Positive | 41 | 20 | 21 | ||
| Unknown | 11 | ||||
| Ki67 status | |||||
| Negative | 33 | 19 | 14 | 1.53 (0.70–3.33) | 0.285 |
| Positive | 119 | 56 | 63 | ||
| Unknown | 5 | ||||
Italic values indicate significance of P value (P < 0.05)
aThe DAAM1 mRNA expression levels were divided at a cutoff point of 50%
bP value for χ2 test
Moreover, both progression free survival (PFS) and overall survival (OS) rate of BrCa patients with high expression of DAAM1 mRNA was significantly worse than that of patients with low DAAM1 expression according to survival information from Kaplan–Meier plotter (Fig. 1h, i). Overall, the clinical data reveals that DAAM1 functions as a metastatic promoter and associates with prognosis in BrCa.
G genotype of rs79036859 enhances the risk of metastasis in BrCa
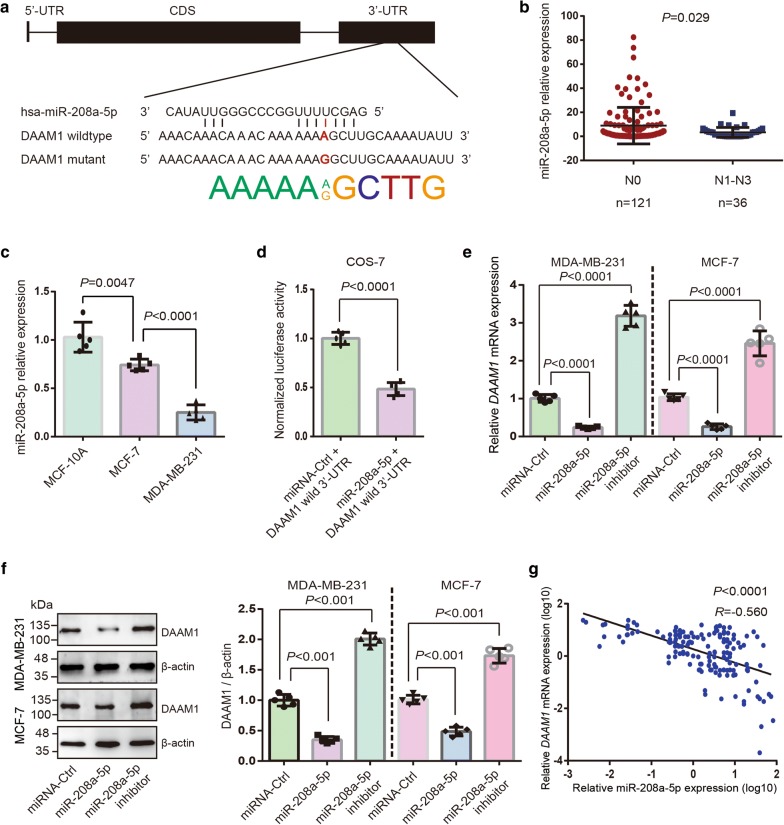
In view of a fact that DAAM1 showed the increased expression and activation level in lymphnode metastatic tumor tissues, we speculated that the regulation of genomic and/or transcriptional level may be heterogeneous in individual BrCa tissues. After a review of dbSNP database and PolymiRTS Database 3.0 [18], SNP rs79036859 was predicted as the regulatory element for the post-transcription of DAAM1 expression (Fig. 2a).
Fig. 2.
MiR-208a-5p is a metastatic suppressor and downregulates DAAM1. a DAAM1 gene structure and A/G polymorphism in the DAAM1 3′-UTR at the miR-208a-5p binding sites. b The miR-208a-5p expression in lymphnode metastatic BrCa tissues and non-metastatic BrCa tissues. N0, without lymphnode metastasis. N1–N3, with lymphnode metastasis. c MiR-208a-5p exhibited the lower expression level in MCF-7 and MDA-MB-231 BrCa cells than that in MCF-10A mammary epithelial cells. d The luciferase activity in COS-7 cells transfected with wild type DAAM1 3′-UTR (DAAM1 wild 3′-UTR) and miR-208a-5p or miRNA-Ctrl. e, f The downregulated DAAM1 both in mRNA and protein levels after miR-208a-5p overexpression, and the upregulated DAAM1 protein expression after transfection of miR-208a-5p inhibitor (anti-sense of miR-208a-5p) in MDA-MB-231 and MCF-7 BrCa cells. β-actin was used as the loading control. g Correlation between the expression levels of miR-208a-5p and DAAM1 mRNA in BrCa tissues
To further verify the potential effect of the DAAM1 3′-UTR SNP rs79036859 in BrCa, we performed a case-based method to assess the function of rs79036859 in tumor metastasis. A metastatic cases-control study including 157 BrCa cases (121 cases without lymphnode metastasis and 36 cases with lymphnode metastasis) was proceeded. BrCa patients with the G (AG/GG) allele genotypes had a prominently higher risk of metastasis compared with the AA genotype carriers (OR = 3.90, 95% CI 1.77–8.59; P < 0.001; Table 2). These findings demonstrate that G genotype in rs79036859 indicates a high risk of metastasis in BrCa.
Table 2.
Clinicopathological characteristics, allelic, and genotypes distribution in breast cancer patients
| Characteristics | n | rs79036859a | OR (95% Cl) | P valueb | |
|---|---|---|---|---|---|
| AA | AG/GG | ||||
| Tumor size (cm) | |||||
| ≤ 2 | 61 | 37 | 24 | 1.20 (0.61–2.33) | 0.599 |
| > 2 | 87 | 49 | 38 | ||
| Unknown | 9 | ||||
| Lymph node metastasis | |||||
| N0 | 121 | 70 | 41 | 3.90 (1.77–8.59) | < 0.001 |
| N1–N3 | 36 | 12 | 24 | ||
| Distant metastasis | |||||
| M0 | 151 | 90 | 61 | 2.95 (0.52–16.62) | 0.200 |
| M1 | 6 | 2 | 4 | ||
| ER status | |||||
| Negative | 51 | 29 | 22 | 0.90 (0.46–1.77) | 0.759 |
| Positive | 106 | 63 | 43 | ||
| PR status | |||||
| Negative | 67 | 38 | 29 | 0.85 (0.45–1.62) | 0.619 |
| Positive | 89 | 54 | 35 | ||
| Unknown | 1 | ||||
| Her-2 status | |||||
| Negative | 94 | 51 | 43 | 0.67 (0.35–1.30) | 0.233 |
| Positive | 61 | 39 | 22 | ||
| Unknown | 2 | ||||
| P53 status | |||||
| Negative | 105 | 63 | 42 | 1.30 (0.63–2.68) | 0.485 |
| Positive | 41 | 22 | 19 | ||
| Unknown | 11 | ||||
| Ki67 status | |||||
| Negative | 33 | 22 | 11 | 1.55 (0.69–3.49) | 0.285 |
| Positive | 119 | 67 | 52 | ||
| Unknown | 5 | ||||
Italic values indicate significance of P value (P < 0.05)
aAA genotype was used as a reference
bP value for χ2 test
SNP rs79036859 associated miR-208a-5p is downregulated in BrCa tissues and directly target DAAM1
Because of the SNP rs79036859 associated miR-208a-5p predicted as post-transcriptional regulatory factor of DAAM1 (Fig. 2a), we hypothesized that miR-208a-5p would be a metastatic suppressor by downregulating DAAM1. We examined miR-208a-5p expression in 36 metastatic BrCa tissues and 121 non-metastatic BrCa tissues using quantitative PCR. The miR-208a-5p expression level in lymphnode metastatic tumor tissues was significantly lower than that in non-lymphnode metastatic tumor tissues (Fig. 2b). The expression status of miR-208a-5p was not significantly associated with clinicopathological parameters of BrCa, including tumor size, lymph node metastasis, distant metastasis, ER, PR, Her2, P53, and Ki67 status by using χ2 test analysis (Additional file 1: Table S1). Furthermore, we examined miR-208a-5p expression in cell lines. MiR-208a-5p exhibited the lower expression level in MCF-7 and MDA-MB-231 BrCa cells than that in MCF-10A mammary epithelial cells (Fig. 2c).
To further verify the binding of miR-208a-5p and DAAM1 3′-UTR, we performed dual-luciferase activity assays. A notable decline of luciferase activity was shown in DAAM1 wild 3′-UTR and miR-208a-5p mimic overexpressed group, and disappeared in miRNA-control groups (Fig. 2d). Next, MDA-MB-231 and MCF-7 cells were transfected with FAM-labelled miR-208a-5p mimic, miR-208a-5p inhibitor (anti-sense of miR-208a-5p), or miRNA mimic control. The mRNA and protein level of DAAM1 was significantly decreased in miR-208a-5p-overexpressed in MCF-7 and MDA-MB-231 cells, but upregulated in miR-208a-5p inhibitor-overexpressed cancer cells (Fig. 2e, f). Besides, miR-208a-5p expression was inversely correlated with DAAM1 expression in 157 clinical samples (Fig. 2g). These results indicated that miR-208a-5p functions as metastatic suppressor and directly targets DAAM1 in BrCa.
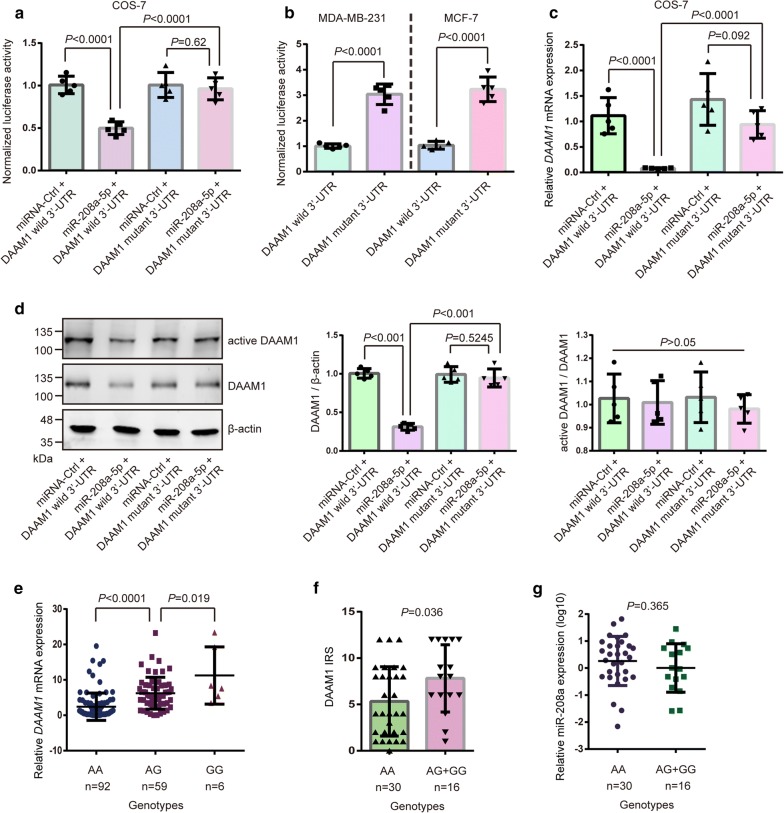
SNP rs79036859 A/G eliminates miR-208a-5p-mediated decrease of DAAM1 expression
Rs79036859 may regulate DAAM1 expression via altering the binding affinity of miR-208a-5p for two DAAM1 3′-UTR genotypes. A notable decline of luciferase activity was shown in DAAM1 wild 3′-UTR and miR-208a-5p mimic transfected group, but vanished in DAAM1 mutant 3′-UTR groups (Fig. 3a). Moreover, DAAM1 wild 3′-UTR or DAAM1 mutant 3′-UTR transfected into MDA-MB-231 and MCF-7 cells. We found that the relative luciferase activity in DAAM1 mutant 3′-UTR-expressed cells was higher than that in DAAM1 wild 3′-UTR-expressed cells (Fig. 3b). Compared with the DAAM1 mutant 3′-UTR group, DAAM1 expression was suppressed both in mRNA and protein level in cells transfected with DAAM1 wild 3′-UTR and miR-208a-5p mimic (Fig. 3c, d). Besides, the level of active DAAM1 was also significantly downregulated in cells transfected with DAAM1 wild 3′-UTR and miR-208a-5p mimic, but non-significant difference in the ratio of active DAAM1/total DAAM1 were found in above group (Fig. 3d).
Fig. 3.
SNP rs79036859 A/G eliminates miR-208a-5p-mediated decrease of DAAM1 expression. a The luciferase activity in COS-7 cells transfected with wild type DAAM1 3′-UTR (DAAM1 wild 3′-UTR) or the mutant of DAAM1 3′-UTR SNP rs79036859 A → G (DAAM1 mutant 3′-UTR) and miR-208a-5p. b The luciferase activity in both MDA-MB-231 and MCF-7 cells transfected with the A allele construct (wild type DAAM1) and the reporter bearing the G allele (mutant DAAM1). c, d The downregulated DAAM1 both in mRNA and protein levels COS-7 cells transfected with wild type DAAM1 3′-UTR (DAAM1 wild 3′-UTR) or the mutant of DAAM1 3′-UTR SNP rs79036859 A → G (DAAM1 mutant 3′-UTR) and miR-208a-5p. The level of active DAAM1 was measured and non-significant difference in ratio of active DAAM1/total DAAM1 were found in above group. β-actin was used as the loading control. e The expression levels of DAAM1 mRNA in BrCa samples with AA, AG, and GG genotypes. f According to the DAAM1 expression using IHC staining, G (AG + GG) genotypes had a higher IRS level of DAAM1 than the wild type AA genotype. g The non-significant difference in miR-208a-5p expression levels were found in wild type AA genotype and mutant G (AG + GG) genotypes in 46 samples
To assess whether rs79036859 A/G was related to DAAM1 expression in clinical samples, we detected DAAM1 mRNA levels in cancer tissues using quantitative PCR in 157 cases and examined DAAM1 protein expression by IHC in 46 patients. The results showed that DAAM1 mutant tumor tissues (AG/GG genotypes) expressed higher levels of DAAM1 mRNA and protein than DAAM1 wildtype tumor tissues (AA genotype) (Fig. 3e, f). MiR-208a-5p levels were measured in the above cancer tissues and shown non-significant differences between the wildtype and mutant genotype groups (Fig. 3g). Taken together, these results demonstrated that rs79036859 in DAAM1 3′-UTR decreases the binding affinity with miR-208a-5p and eliminates miR-208a-5p-mediated decrease of DAAM1 expression in mammalian cells.
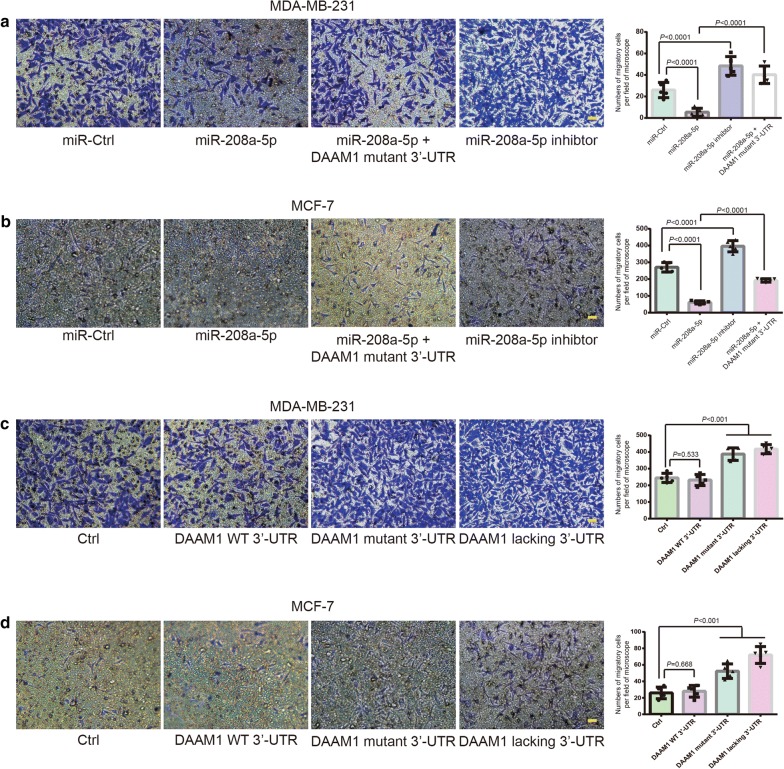
MiR-208a-5p suppresses the migration of BrCa cells
Next, we evaluated the role of miR-208a-5p on the migration of BrCa cells which carry wildtype DAAM1 3′-UTR. Boyden chamber assays revealed that miR-208a-5p overexpression suppressed the migratory ability of MDA-MB-231/MCF-7 cells (Fig. 4a, b). On the contrary, impediment of miR-208a-5p binding DAAM1 3′-UTR by the overexpression of miR-208a-5p inhibitor (anti-sense of miR-208a-5p) or DAAM1 linking with the G genotype of DAAM1 3′-UTR rs79036859 (DAAM1 mutant 3′-UTR) promoted the migration of BrCa cells (Fig. 4a, b). Besides, the overexpression of DAAM1 linking with the G genotype of DAAM1 3′-UTR rs79036859 and DAAM1 lacking 3′-UTR promoted the migration of MDA-MB-231/MCF-7 cells (Fig. 4c, d). DAAM1 linking with wildtype 3′-UTR failed to enhance the migratory rate of BrCa cells (Fig. 4c, d). Thus, these results suggested that miR-208a-5p suppresses cell migration via downregulating DAAM1 in BrCa cells.
Fig. 4.
MiR-208a-5p inhibits the migratory ability of BrCa cells. a, b miR-208a-5p suppressed the migration of MDA-MB-231 and MCF-7 cells, which could be rescued by the overexpression of miR-208a-5p inhibitor (anti-sense of miR-208a-5p) or DAAM1 linking with the G genotype of DAAM1 3′-UTR rs79036859 (DAAM1 mutant 3′-UTR). Migratory cells on the lower-side of Boyden chambers were counted per field of microscope. Bar = 20 μm. Objective lens, magnification, × 20; numerical aperture, 0.75. c, d The overexpression of DAAM1 linking with the G genotype of DAAM1 3′-UTR rs79036859 (DAAM1 mutant 3′-UTR) and DAAM1 lacking 3′-UTR elevated the migration of MDA-MB-231 and MCF-7 cells. DAAM1 linking with wildtype 3′-UTR (DAAM1 WT 3′-UTR) failed to promote the migratory rate of MDA-MB-231 and MCF-7 cells. MDA-MB-231 and MCF-7 cells transfected with DAAM1 WT 3′-UTR, DAAM1 mutant 3′-UTR, or DAAM1 lacking 3′-UTR were examined the migration for 8 h in 8.0-µm porous Boyden chamber membranes
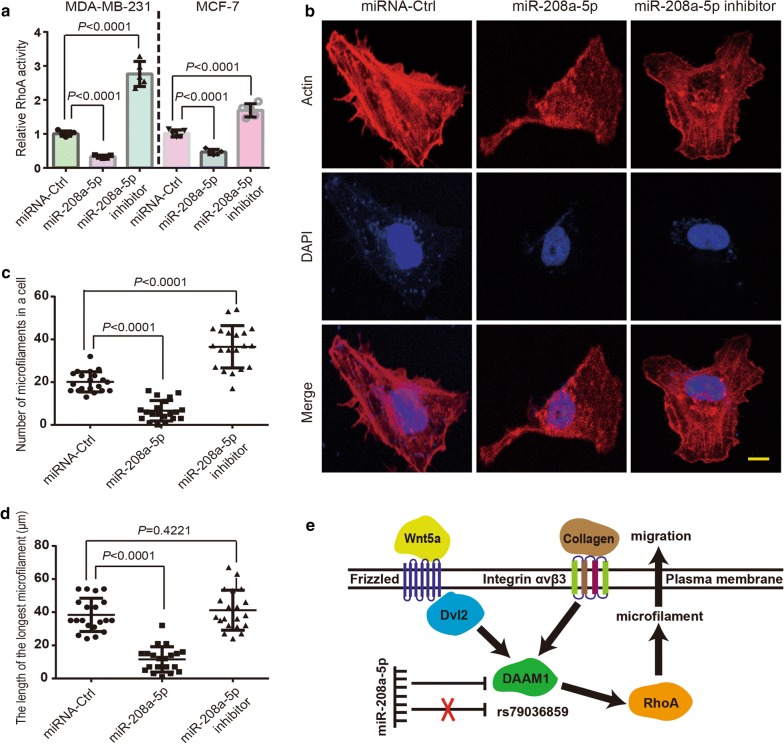
MiR-208a-5p inhibits the RhoA activity and disrupts the formation of microfilament in BrCa cells
DAAM1 is reported to be involved in Wnt5a-induced and collagen-induced signaling pathways [3, 6]. RhoA is a direct downstream target of DAAM1, which functions in actin assemblage [1, 3, 6]. We investigated the fact that whether miR-208a-5p inhibited the activation of RhoA and the inhibition was reversed by the transfection of miR-208a-5p inhibitor (anti-sense of miR-208a-5p). MiR-208a-5p overexpression notably suppressed RhoA activation, that could be reversed by miR-208a-5p inhibitor overexpression (Fig. 5a). Next, fluorescent phalloidin was stained to display the arrangement of microfilaments in BrCa cells. MiR-208a-5p overexpression obviously blocked the microfilament formation, and miR-208a-5p inhibitors largely enhanced the microfilament assemblage in MDA-MB-231 cells (Fig. 5b–d). Thus, the findings illuminated that active RhoA induced formation of microfilament is disrupted by miR-208a-5p through downregulating DAAM1. Collectively, the findings suggested that the formation of microfilament is disrupted by miR-208a-5p overexpression via targeting DAAM1/RhoA signaling in BrCa cells (Fig. 5e).
Fig. 5.
MiR-208a-5p inhibits the RhoA activity and disrupts the formation of microfilament in BrCa cells. a RhoA GTPase activation assays showing the downregulated RhoA activity in MDA-MB-231 and MCF-7 cells after miR-208a-5p overexpression. MiR-208a-5p inhibitor (anti-sense of miR-208a-5p) elevated the RhoA activity in MDA-MB-231 and MCF-7 cells. b miR-208a-5p overexpression disrupted the formation of microfilament in MDA-MB-231 cells. MiR-208a-5p inhibitor (anti-sense of miR-208a-5p) promoted the formation of microfilament. Bar = 10 μm. Objective lens, magnification, ×40; numerical aperture, 0.95. c, d The number of microfilaments in each MDA-MB-231 cell and the length of the longest microfilament were measured (n = 20). e A schematic diagram of DAAM1 (SNP rs79036859) associated signal transduction pathway and miR-208a-5p locating the upstream of DAAM1
Discussion
The first finding in this study is that the high level of DAAM1 expresses in lymphnode metastatic BrCa tissues. These clinicopathological and biochemical characteristics imply DAAM1 is likely to function as a metastatic promoter in BrCa, which is consistent with our previous findings verified on the cellular levels [3, 6, 17].
Protein is translated from RNA, which is strictly conformed to the genetic central dogma [19]. The mRNA modification exists extensively in eukaryotic cells and leads to the diversity of protein expression. Zhang et al. has analyzed the big data from the Cancer Genome Atlas (TCGA) and identified novel mutations at eQTL situated in DAAM1 promotor (− 91 bp), which are able to increase DAAM1 mRNA expression levels [7]. In this study, we focus on the polymorphism of 3′-UTR of DAAM1 gene. Patients with the G (GG/GA) allele genotypes have the prominently higher risk of metastasis in BrCa than AA genotype of SNP rs79036859. These results demonstrate that G genotype of rs79036859 in DAAM1 3′-UTR enhances the risk of metastasis in BrCa.
The most common post-transcriptional regulation is that miRNAs bind to 3′-UTR of target gene and induce the degradation of whole mRNAs. We find that miR-208a-5p targets to the 3′-UTR of DAAM1 gene. Wang et al. reported the downregulation of miR-208a/miR-208b significantly suppresses the harmful effect of extracellular vesicles on hypoxia/reoxygenation damage in cardiac myoblasts [20]. The regulatory feedback loop of miR-208a-SOX2/β-catenin-LIN28-let-7a-DICER1 mediates the renovation of BrCa stem cells [21]. MiR-208a is capable of promoting gastric cancer progression by suppressing SFRP1 and downregulating MEG3 [22]. Expression analysis demonstrate that miR-208a-5p is significantly suppressed in lymphnode metastatic tumor samples than that in non-lymphnode metastatic tumor samples, indicating the potential role of miR-208a-5p on anti-tumor metastasis via DAAM1 signaling.
DAAM1, an element of cellular actin cytoskeleton, transduces Wnt/Dishevelled signaling to RhoA and then assemble microfilaments in mammalian cells [2, 3, 23]. Actin polymerization is the key step of the assemblage of microfilament in the process of cancer metastasis [24]. The functional and dynamic microfilaments are generally provided a possible route for the degradation of extracellular matrix and available for cancer cells escaping from the primary lesion [24, 25]. Our previous studies find that active DAAM1 is elevated by the treatment of Wnt5a or type IV collagen and participates in the tumor cell migration and haptotaxis [3, 6]. Here, we demonstrate that miR-208a-5p overexpression decreases DAAM1 mRNA and protein expression levels, resulting in the decrease of the migratory ability of BrCa cells. We also find that miR-208a-5p downregulates the RhoA activity, disrupts the formation of microfilaments of BrCa cells. We summarize the potential DAAM1 signaling pathways in Fig. 5e. Thus, miR-208a-5p/DAAM1/RhoA axis is the potential therapeutic target in regulating microfilament assemblage and cell migration.
Conclusions
These results indicate that SNP rs79036859 G variant of DAAM1 3′-UTR contributes to the likelihood of BrCa metastasis via miR-208a-5p binding capacity. Cell migration is inhibited by miR-208a-5p overexpression via targeting DAAM1/RhoA signaling in BrCa cells. Overall these data identify miR-208a-5p/DAAM1/RhoA axis as the novel therapeutic target in limiting BrCa metastasis.
Additional file
Additional file 1. Associations between miR-208a-5p expression and clinicopathological characteristics in breast cancer.
Authors’ contributions
YZ and TY conceived the study and participated in the study design, performance, coordination and manuscript writing. JM, TY, YH, TX, LH, SS, YC, ZW, XJ, and BX carried out the assays and analysis. YZ, TY and JM revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank the Laboratory Center for Basic Medical Sciences (Nanjing Medical University) for providing technical support. We also thank Ms. Xiaoyu Dong (Nanjing Medical University) for giving the technical guidance to operate the microplate reader.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its additional files.
Consent for publication
All authors agreed on the manuscript.
Ethics approval and consent to participate
All experiments were approved by the Ethics Committee of Nanjing Medical University.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81472703) to Yichao Zhu, the Natural Science Foundation of Jiangsu Province (Grant No. BK20181367) to Yichao Zhu, and the Science and Technology Foundation of Nanjing Medical University (Grant No. 2017NJMU001) to Ting Yan.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- DAAM1
dishevelled-associated activator of morphogenesis 1
- BrCa
breast cancer
- SNP
single nucleotide polymorphism
- UTR
untranslated region
- eQTL
expression quantitative trait locus
- miRNA
microRNA
- IHC
immunohistochemistry
- IRS
immunoreactive score
- DFS
disease free survival
- OS
overall survival
- TCGA
The Cancer Genome Atlas
Contributor Information
Jie Mei, Email: meijie199621@163.com.
Ting Yan, Email: yanting@njmu.edu.cn.
Yifu Huang, Email: huangyifu07yfbj@sina.com.
Tiansong Xia, Email: xiatsswms@163.com.
Fei Chang, Email: changf1987@sina.com.
Shuning Shen, Email: shuningshen@163.com.
Leiyu Hao, Email: leiyuhao316@163.com.
Yin Chen, Email: arsenechen98@gmail.com.
Zhongyuan Wang, Email: wangzy@njmu.edu.cn.
Xiaozheng Jiang, Email: xiaoyubanana@163.com.
Bujie Xu, Email: bujiexu192@163.com.
Yichao Zhu, Phone: +86-25-86868318, Email: zhuyichao@njmu.edu.cn.
References
- 1.Aspenstrom P, Richnau N, Johansson AS. The diaphanous-related formin DAAM1 collaborates with the Rho GTPases RhoA and Cdc42, CIP4 and Src in regulating cell morphogenesis and actin dynamics. Exp Cell Res. 2006;312(12):2180–2194. doi: 10.1016/j.yexcr.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107(7):843–854. doi: 10.1016/S0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- 3.Zhu Y, Tian Y, Du J, Hu Z, Yang L, Liu J, et al. Dvl2-dependent activation of Daam1 and RhoA regulates Wnt5a-induced breast cancer cell migration. PLoS ONE. 2012;7(5):e37823. doi: 10.1371/journal.pone.0037823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu W, Sato A, Khadka D, Bharti R, Diaz H, Runnels LW, et al. Mechanism of activation of the Formin protein Daam1. Proc Natl Acad Sci USA. 2008;105(1):210–215. doi: 10.1073/pnas.0707277105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaiswal R, Breitsprecher D, Collins A, Correa IR, Jr, Xu MQ, Goode BL. The formin Daam1 and fascin directly collaborate to promote filopodia formation. Curr Biol. 2013;23(14):1373–1379. doi: 10.1016/j.cub.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu G, Yan T, Li X, Sun J, Zhang B, Wang H, et al. Daam1 activates RhoA to regulate Wnt5a induced glioblastoma cell invasion. Oncol Rep. 2018;39(2):465–472. doi: 10.3892/or.2017.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Bojorquez-Gomez A, Velez DO, Xu G, Sanchez KS, Shen JP, et al. A global transcriptional network connecting noncoding mutations to changes in tumor gene expression. Nat Genet. 2018;50(4):613–620. doi: 10.1038/s41588-018-0091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pirooz HJ, Jafari N, Rastegari M, Fathi-Roudsari M, Tasharrofi N, Shokri G, et al. Functional SNP in microRNA-491-5p binding site of MMP9 3′-UTR affects cancer susceptibility. J Cell Biochem. 2018;119(7):5126–5134. doi: 10.1002/jcb.26471. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Liu Y, Song F, Zheng H, Hu L, Lu H, et al. Functional SNP in the microRNA-367 binding site in the 3′UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc Natl Acad Sci USA. 2011;108(33):13653–13658. doi: 10.1073/pnas.1103360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y, Chen X, Xi B, Yu X, Ouyang J, Han C, et al. SNP rs3202538 in 3′UTR region of ErbB3 regulated by miR-204 and miR-211 promote gastric cancer development in Chinese population. Cancer Cell Int. 2017;17:81. doi: 10.1186/s12935-017-0449-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang H, Ge F, Hu B, Wu L, Yang H, Wang H. rs35301225 polymorphism in miR-34a promotes development of human colon cancer by deregulation of 3′UTR in E2F1 in Chinese population. Cancer Cell Int. 2017;17:39. doi: 10.1186/s12935-017-0402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431(7006):343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Lei C, He Q, Pan Z, Xiao D, Tao Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol Cancer. 2018;17(1):64. doi: 10.1186/s12943-018-0765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu S, Rong H, Zhang G, Kang L, Yang M, Guan H. Functional SNP in 3′-UTR microRNA-binding site of ZNF350 confers risk for age-related cataract. Hum Mutat. 2016;37(11):1223–1230. doi: 10.1002/humu.23073. [DOI] [PubMed] [Google Scholar]
- 15.Iannaccone M, Cosenza G, Pauciullo A, Martino G, Capparelli R. The SNP g.4667G>A at 3′-UTR of IFNG gene is associated with susceptibility to bovine tuberculosis in Mediterranean water buffalo (Bubalus bubalis) Anim Genet. 2018;49:496–497. doi: 10.1111/age.12698. [DOI] [PubMed] [Google Scholar]
- 16.Hua Y, Wang J, Yuan DL, Qi Y, Tang Z, Zhu X, et al. A tag SNP in syncytin-2 3-UTR significantly correlates with the risk of severe preeclampsia. Clin Chim Acta. 2018;483:265–270. doi: 10.1016/j.cca.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Xiong H, Yan T, Zhang W, Shi F, Jiang X, Wang X, et al. miR-613 inhibits cell migration and invasion by downregulating Daam1 in triple-negative breast cancer. Cell Signal. 2018;44:33–42. doi: 10.1016/j.cellsig.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucl Acids Res. 2014;42(Database issue):D86–D91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crick F. Central dogma of molecular biology. Nature. 1970;227(5258):561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 20.Wang F, Yuan Y, Yang P, Li X. Extracellular vesicles-mediated transfer of miR-208a/b exaggerate hypoxia/reoxygenation injury in cardiomyocytes by reducing QKI expression. Mol Cell Biochem. 2017;431(1–2):187–195. doi: 10.1007/s11010-017-2990-4. [DOI] [PubMed] [Google Scholar]
- 21.Sun X, Jiang S, Liu J, Wang H, Zhang Y, Tang SC, et al. MiR-208a stimulates the cocktail of SOX2 and beta-catenin to inhibit the let-7 induction of self-renewal repression of breast cancer stem cells and formed miR208a/let-7 feedback loop via LIN28 and DICER1. Oncotarget. 2015;6(32):32944–32954. doi: 10.18632/oncotarget.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui HB, Ge HE, Wang YS, Bai XY. MiR-208a enhances cell proliferation and invasion of gastric cancer by targeting SFRP1 and negatively regulating MEG3. Int J Biochem Cell Biol. 2018;102:31–39. doi: 10.1016/j.biocel.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Kerkhoff E. Actin dynamics at intracellular membranes: the Spir/formin nucleator complex. Eur J Cell Biol. 2011;90(11):922–925. doi: 10.1016/j.ejcb.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Schoumacher M, Goldman RD, Louvard D, Vignjevic DM. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J Cell Biol. 2010;189(3):541–556. doi: 10.1083/jcb.200909113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohmer LL, Clay MR, Naegeli KM, Chi Q, Ziel JW, Hagedorn EJ, et al. A sensitized screen for genes promoting invadopodia function in vivo: CDC-42 and Rab GDI-1 direct distinct aspects of invadopodia formation. PLoS Genet. 2016;12(1):e1005786. doi: 10.1371/journal.pgen.1005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Associations between miR-208a-5p expression and clinicopathological characteristics in breast cancer.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its additional files.