Abstract
Neuromelanin sensitive magnetic resonance imaging (NMS-MRI) has been crucial in identifying abnormalities in the substantia nigra pars compacta (SNc) in Parkinson's disease (PD) as PD is characterized by loss of dopaminergic neurons in the SNc. Current techniques employ estimation of contrast ratios of the SNc, visualized on NMS-MRI, to discern PD patients from the healthy controls. However, the extraction of these features is time-consuming and laborious and moreover provides lower prediction accuracies. Furthermore, these do not account for patterns of subtle changes in PD in the SNc. To mitigate this, our work establishes a computer-based analysis technique that uses convolutional neural networks (CNNs) to create prognostic and diagnostic biomarkers of PD from NMS-MRI. Our technique not only performs with a superior testing accuracy (80%) as compared to contrast ratio-based classification (56.5% testing accuracy) and radiomics classifier (60.3% testing accuracy), but also supports discriminating PD from atypical parkinsonian syndromes (85.7% test accuracy). Moreover, it has the capability to locate the most discriminative regions on the neuromelanin contrast images. These discriminative activations demonstrate that the left SNc plays a key role in the classification in comparison to the right SNc, and are in agreement with the concept of asymmetry in PD. Overall, the proposed technique has the potential to support radiological diagnosis of PD while facilitating deeper understanding into the abnormalities in SNc.
Keywords: Parkinson's disease, Machine learning, Neuromelanin, Convolutional neural networks
Highlights
-
•
A novel convolutional neural net (CNN) based marker for Parkinson’s disease (PD) based on neuromelanin sensitive images
-
•
The classifier demonstrates high accuracy in delineating PD from healthy controls when compared to other techniques
-
•
The class activation maps demonstrated significant asymmetry reflecting the clinical asymmetry observed in PD
1. Introduction
Parkinson's disease (PD) is a progressive, neurodegenerative disorder characterized by loss of dopaminergic neurons in the substantia nigra pars compacta (SNc) (Mann and Yates, 1983). Neuromelanin which is a by-product of dopamine synthesis is a neuronal pigment which can be visualized in the SNc. Depigmentation of the SNc, secondary to loss of dopaminergic neurons is a conspicuous feature of PD, which although well visualized neuropathologically is poorly replicated by neuroimaging (Hutchinson and Raff, 2000). The role of conventional neuroimaging in the diagnosis of PD has been therefore limited, despite several methods to study the SNc as these techniques have been unable to directly visualize the SNc. The introduction of the “neuomelanin-sensitive MRI” (NMS-MRI), a 3T T1 weighted high-resolution fast spin-echo neuromelanin sensitive sequence by Sasaki et al., revolutionized the technique of visualizing the SNc (Sasaki et al., 2006). Following this, several studies have demonstrated the utility of this sequence to differentiate between patients with PD and controls using manually extracted features of the SNc that include the contrast ratio, area and volumes (Matsuura et al., 2013; Ogisu et al., 2013; Ohtsuka et al., 2013; Prasad et al., 2018b). Between these features the contrast ratios are well-accepted as they have demonstrated the highest discriminative power. However, these techniques have limited clinical utility and reproducibility owing to the time-consuming nature of computing these features, with a high scope for operator errors. Moreover, these features require SNc borders to be well defined which are difficult in patients with PD due to higher loss of neuromelanin containing neurons that may substantially decrease the contrast on the scan and may bias the analysis by excluding highly impacted areas of the SNc, leading to an over-estimate in contrast ratios (Sulzer et al., 2018).
At present the diagnosis of PD is highly dependent on clinical features and although dopamine transporter positron emitted topography is useful, it is not cost-effective and cannot be routinely employed. Hence, it is crucial to employ other neuro-imaging techniques to aid in the early or differential diagnosis of PD. Recent advances in the areas of machine learning and data-driven analysis have demonstrated the utility of different brain imaging modalities for automated diagnosis of PD. Table 1 provides a brief review of recent articles that have used machine learning and statistical learning techniques to predict PD from MRI modalities.
Table 1.
Brief review of methods employed by recent studies that have used machine learning and statistical learning techniques to predict PD from MRI modalities.
| Author, year | Number of subjects | Methods employed | Accuracy (%) |
|---|---|---|---|
| Salvatore et al., 2014 | PD (n = 28) | VBM | PD vs HC: 83.2 |
| PSP (n = 28) | Principal component analysis | PSP vs HC: 86.2 | |
| HC (n = 28) | SVM | PSP vs PD: 84.7 | |
| Cherubini et al., 2014a, Cherubini et al., 2014b | Tremor dominant PD (n = 15) | VBM, DTI | |
| ET with rest tremor (n = 15) | SVM | ||
| Cherubini et al., 2014a, Cherubini et al., 2014b | PD (n = 57) | VBM, DTI | 100 |
| PSP (n = 21) | SVM | ||
| Rana et al., 2015 | PD (n = 30) | Region of interest based | 86.67 |
| HC (n = 30) | SVM | ||
| Singh and Samavedham, 2015 | PPMI cohort | Self-organizing maps | 99.9 |
| PD (n = 518) | SVM | ||
| SWEDD (n = 68) | |||
| HC (n = 245) | |||
| Huppertz et al., 2016 | PD (n = 204) | Volumetry | 80 |
| PSP-RS (n = 106) | SVM | ||
| MSA-C (n = 21) | |||
| MSA-P (n = 60) | |||
| Adeli et al., 2016 | PPMI cohort | Joint feature-sample selection | 81.9 |
| PD (n = 374) | |||
| HC (n = 169) | |||
| Abos et al., 2017 | PD (n = 27) | Functional connectome | 80 |
| HC (n = 38) | SVM | ||
| Peran et al.,2018 | PD (n = 26) | VBM, T2* relaxometry, DTI | PD vs MSA: 96 |
| MSA-P (n = 16) | |||
| MSA-C (n = 13) | Self-organizing maps | ||
| HC (n = 26) | |||
| Amoroso et al., 2018 | PPMI cohort | Connectivity measures | 93 |
| PD (n = 374) | SVM | ||
| HC (n = 169) | |||
| Ariz et al., 2019 | PD (n = 40) | NM-MRI based atlas of | 79.9 |
| HC (n = 39) | Substantia nigra |
DTI: Diffusion tensor imaging; ET: Essential tremor; HC: Healthy controls: MSA-C: Multiple system atrophy with predominant cerebellar features; MSA-P: Multiple system atrophy with predominant parkinsonian features; NM-MRI: Neuromelanin sensitive magnetic resonance imaging; PD: Parkinson's disease; PPMI:Parkinson's Progression Markers Initiative; PSP: Progressive supranuclear palsy; PSP-RS: Progressive supranuclear palsy-Richardson syndrome; SVM: Support vector machine; SWEDD: Scans without evidence of dopaminergic deficit; VBM: Voxel based morphometry.
Considering the above-mentioned utility of the NMS-MRI in the differentiation of PD from healthy controls, and the relative ease of acquiring this sequence, we endeavor to utilize this sequence in an automated classification framework for PD prognosis and diagnosis. The main objective of the present study therefore is to create markers for PD using state-of-art deep convolution neural network (CNN) on NMS-MRI and compare it to classifiers based on (1) contrast ratios with machine learning (CR-ML) and (2) radiomics with machine learning (RA-ML) using regions of interest of the SNc. Our CNN based classifier is fully automated and is free of defining SNc borders as it uses a larger region of interest. Moreover, it employs the technique of discriminative localization to compute class activation maps (CAMs) that facilitate deeper understanding into the most important regions that participate in the classification.
2. Methodology
2.1. Subject recruitment and clinical evaluation
Forty-five patients with PD, 20 patients with atypical parkinsonian syndromes (APS) - multiple system atrophy (MSA), progressive supranuclear palsy (PSP), and 35 healthy controls (HCs) were recruited from the general outpatient clinic and movement disorder services at the Department of Neurology, National Institute of Mental Health and Neurosciences (NIMHANS), Bangalore, India. The diagnosis of idiopathic PD was based on the UK Parkinson's Disease Society Brain Bank criteria (Hughes et al., 1993), the diagnosis of MSA was based on the criteria by Gilman et al. (Gilman et al., 2008), and PSP was diagnosed based on the National Institute of Neurological Disorders and Stroke and Society for Progressive Supranuclear Palsy criteria (Litvan et al., 1996), and confirmed by a trained movement disorder specialist (author PKP). Patients included in this study have been part of other studies from this group (Prasad et al., 2018a; Prasad et al., 2018b; Shah et al., 2017; Shah et al., 2018) and all patients and controls provided informed consent prior to recruitment in the original projects.
Demographic and clinical details such as gender, age at presentation, age at onset of motor symptoms, disease duration, Unified Parkinson's Disease Rating Scale (UPDRS-III) OFF-state scores, and Hoehn and Yahr stage were recorded. Clinical laterality (CL) was established by measuring the difference between the right and left UPDRS-III sub scores, i.e., question 23–26 (Barrett et al., 2011). A difference of ≥1 was considered as CL. A subject with PD was said to have either right CL or left CL based on which side had a higher score. For patients with APS, i.e., MSA or PSP, gender, age at presentation, age at onset of motor symptoms, and disease duration were recorded. Age and gender matched HCs with no family history of parkinsonism or other movement disorder were recruited.
The demographic and clinical data of patients with PD, APS and HC were compared using a standard analysis of variance (ANOVA). The level of significance was set at p < .05.
2.2. Imaging
MR images were acquired on 3T Philips Achieva scanner with a 32-channel head coil at NIMHANS, Bangalore, India. High resolution 3D neuromelanin contrast sensitive sequence i.e., the spectral pre-saturation with inversion recovery (SPIR) sequence was acquired using TR/TE: 26/2.2 ms, flip angle: 20°, reconstructed matrix size: 512 × 512, field of view: 180 × 180 × 50 mm, voxel size: 0.9 × 0.9 × 1 mm, number of slices: 50 with an acquisition time 4 min 12.9 s. T1, T2, Fluid attenuation Inversion Recovery (FLAIR) and diffusion weighted images were acquired to eliminate the possibility of brain lesions. The MR images were retrieved from the archive and screened for gross cortical structural abnormalities by an experienced neuroradiologist (author JS), after which the images were set perpendicular to the fourth ventricle floor with coverage between the posterior commissure and inferior border of the pons.
2.3. Convolutional neural nets with discriminative localization (CNN-DL)
CNNs are a modern adaption of the traditional artificial neural network architecture where millions of 2D convolutional filter parameters are computed from multiple levels of granularity and transformed into the desired output by end-to-end optimization (Krizhevsky et al., 2012). The CNN can therefore be considered as an automated feature extraction tool with a classifier in the final stages.
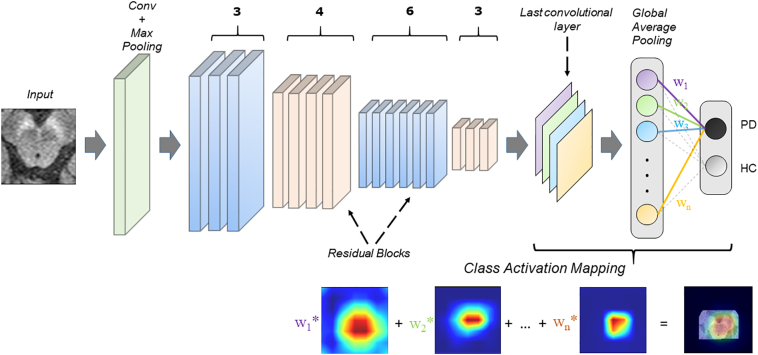
In our case, we employed a boxed region around the brain-stem on the axial slices of the NMS-MRI as input to the 2D CNN (Fig. 1). The CNN transforms these input images into an output vector with class probablities by passing them through a chain of convolutional layers. This is achieved by allowing the network to learn complex image features and by optmizing the weights of these features to facilitate better classification. The CNN architecture we employed was derived from ResNet50 design (He et al., 2016), which is a standard CNN architecture, and has been shown to work with superior performance on medical image classification tasks such as in tumor classification (Chang et al., 2018; Korfiatis et al., 2017). ResNets, unlike other CNN architectures which usually only stack multiple convolutional layers, consists of short-cut “residual” connections that facilitate effectively a deeper neural architecture without increasing the computational complexity. Our design (as shown in Fig. 1) consists of 16 residual blocks, each of which contains 3 convolutional layers and an identity connection. The convolutional layers contain variable sized kernels that capture the patterns and features of varying granularity depending upon the depth of the layer. A max-pooling layer that abstracts the maximum value and downsamples the image, is applied before the first block and after 3,4,6 and 3 residual blocks as shown in Fig. 1. The last convolutional layer acquires the most abstract markers that is given as input to the global average pooling (GAP) layer which averages across the output of the convolutional layer for both the classes resulting in 2 representations, one for each class using a softmax activation.
Fig. 1.
The figure displays a schematic diagram of the CNN architecture. ResNet50 architecture was employed with 16 blocks (50 layers in total). The class activation maps (CAMs) were computed using global average pooling as shown in the figure.
Throughout the CNN architecture, we employ rectified linear unit (ReLU) activations and with a learning rate of 0.0001 which decays based on the number of iterations. The weights of each of layers are updated to train the complete CNN model by minimizing the categorical cross entropy loss function as given in Eq. (1) where yn is target output probability, is the predicted output probability, N is number of testing samples and J(w) is categorical cross-entropy loss and is achieved by using the Adam optimizer.
| (1) |
Finally, to reduce the susceptibility to over-fitting, as a standard practice, data augmentation was performed which increased the dataset by several folds. The augmentation was achieved via random combinations of image translations, minor shifting as well as horizontal and vertical flipping to create 10 times the original data. The CNN-DL was implemented in Python 3.6 using the Keras library with the tensorflow library at the backend on a NVIDIA-quadroP6000 with 24GB GPU memory.
To obtain the discriminative activations, we forward propagated the input image and acquired the weights (w1, w2, w3… wn) at the output layer for the respective class, as given in Zhou et al. (Zhou et al., 2016). The feature maps (f1, f2, f3… fn) obtained from the last convolutional layer were upsampled to match the resolution of the original input image. To create the class activation map, the weights for the respective class were then multiplied with the corresponding feature maps and then added together, as shown in Fig. 1. The resulting map can therefore demonstrate the most discerning regions in the image for each subject.
Finally, to assess any contra-lateral deficits in PD, as have been shown in earlier studies, for each correctly classified subject the mean activations were computed separately on the left and right, by dividing the input image into two parts. Analysis of variance (ANOVA) was performed between the mean activations of left and right of all the subjects. The p-value threshold was retained at 0.05.
2.4. Contrast ratio classifier (CR-ML)
To compute the contrast ratio, technique described in Prasad et al. (Prasad et al., 2018b) was employed. A section of the midbrain at the midpoint of the mamillary bodies was selected for placement of ROIs. Signal intensity (SI) was measured by placing 10mm2 circular ROIs over the lateral part of bilateral SNc, and a normative SI (SIN) was obtained by placing a ROI anterior to the cerebral aqueduct. Contrast ratios (CRs) of the SNc were calculated based on the methodology described by Ohtsuka et al. (Ohtsuka et al., 2013). The following equation was used for the calculation of the CRs:
| (2) |
where SI is the value of the SI of either right or left SNc, and SIN is the SI of the region anterior to the cerebral aqueduct. CR for right and left SNc was computed separately. The left and right contrast ratios were employed in a random forest (Breiman, 2001) with XGBoost classification [https://xgboost.readthedocs.io/en/latest/], which is a gradient boosting framework, for comparison with our CNN model. The random forest classifier used 100 trees and the tree construction employed the ‘exact means exact greedy’ algorithm with a learning rate of 0.05 and a maximum depth of the trees = 6. It was implemented in Python 3.6 using scikit-learn and xgboost libraries. The complete schematic procedure is shown in the Supplementary Material Fig. 1.
2.5. Radiomics based classifier (RA-ML)
Radiomics involves extraction of a large number of quantitative measures such as textures, intensity features, grey-level co-occurrences, grey-level run lengths, statistical measures and energy features (Gillies et al., 2015). For our analysis, we employed the PyRadiomics (van Griethuysen et al., 2017) toolbox and computed 1470 number of features (details of which are provided in Supplementary Material Table 1) from the 3D SNc right and left region of interests. Bilateral SNc ROIs were created by manual segmentation of the NMS-MRI. An expert neurology PhD scholar (author -SP) who was blinded to the groups, delineated the right and left SNc on the axial slices and created a 3D binary mask. The radiomic measures were normalized using min-max normalization and were used as features in a random forest classifier (Breiman, 2001) with XGBoost for classification between PD and healthy controls. The classifier details are described in the earlier section. This was performed as a comparison with our proposed CNN model. Ten top-ranked (most discriminative) radiomic features were computed using the average information gain for each feature over all the decision trees (f-score), to facilitate deeper understanding of the patterns of abnormality from the NMS-MRI images in PD. Supplementary Material Fig. 1 shows the schematic diagram for the procedure employed.
2.6. Training and testing of the models
The dataset was randomly divided into training and testing while retaining the PD:HC ratio in both the classes. We used, 25 healthy controls and 30 PD subjects for training and cross-validating (5-fold validation) the models (our proposed CNN-DL and the comparative CR-ML and RA-ML) and the remaining 10 HCs and 15 PD patients (25 subjects) for testing the models. The receiver operation characteristics (ROC), accuracy, sensitivity and specificity (Powers, 2011) were computed on the cross-validation and test dataset and reported.
A separate CNN based classifier was constructed to discern PD from APS (MSA and PSP). The dataset was randomly divided into training and testing while retaining the PD:APS ratio. 30 PD and 14 APS were used for training and 5 fold cross-validation, while 15 PD and 6 APS were left out for testing. The receiver operation characteristics (ROC), accuracy, sensitivity and specificity were computed on the cross-validation and test dataset and reported. The heat-maps from the CNN were also computed and reported.
3. Results
Table 2 provides complete demographic and clinical information of the dataset under consideration. No significant difference was observed between the ages of PD in comparison to the controls. The mean age at onset of motor symptoms was 51.90 ± 8.64, with mean disease duration of 6.26 ± 4.06. The mean Hoehn and Yahr stages were 1.70 ± 0.54, and UPDRS-III (OFF) scores were 36.58 ± 13.66. Asymmetry in motor symptoms at evaluation was observed in all subjects with PD. Based on the UPDRS-III sub-scores, 60% (n = 27) patients with PD showed right CL and 40% (n = 18), showed left CL.
Table 2.
Demographic and clinical details of patients with Parkinson's disease, atypical parkinsonian syndromes and healthy controls.
| PD (n = 45) | APS (n = 20) | HC (n = 35) | PD vs HC | APS vs HC | PD vs APS | |
|---|---|---|---|---|---|---|
| Gender (M: F) | 32:13 | 13:07 | 22:13 | 0.47 | 1.00 | 0.77 |
| Age | 58.00 ± 8.70 | 53.65 ± 7.19 | 55.00 ± 5.40 | 0.07 | 0.43 | 0.06 |
| Age at onset | 51.90 ± 8.64 | 50.95 ± 7.59 | – | – | – | 0.67 |
| Duration of illness⁎ | 6.26 ± 4.06 | 2.80 ± 1.38 | – | – | – | 0.0005 |
| UPDRS III (OFF) | 36.58 ± 13.66 | NA | – | – | – | – |
| H & Y stage | 1.70 ± 0.54 | – | – | – | – | – |
APS: Atypical parkinsonian syndromes; F: Female; H & Y: Hoehn and Yahr; HC: Healthy controls; M: Male; NA: Not applicable; UPDRS: Unified Parkinson's disease rating scale.
PD vs Atypical parkinsonian syndromes p < .01.
The APS group was composed of 15 patients with MSA and 5 patients with PSP. The mean age of patients in this group was 53.65 ± 7.19, with mean disease duration of 2.80 ± 1.38. The duration of illness was significantly different between the PD and APS groups. With the exception of a single patient with PD, all other subjects (PD, HC and APS) were right-handed.
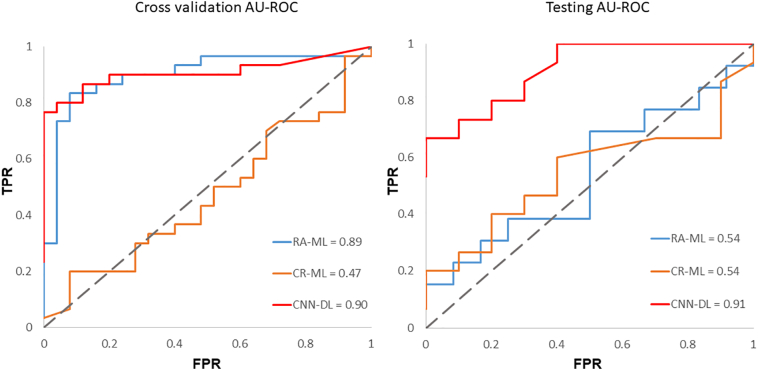
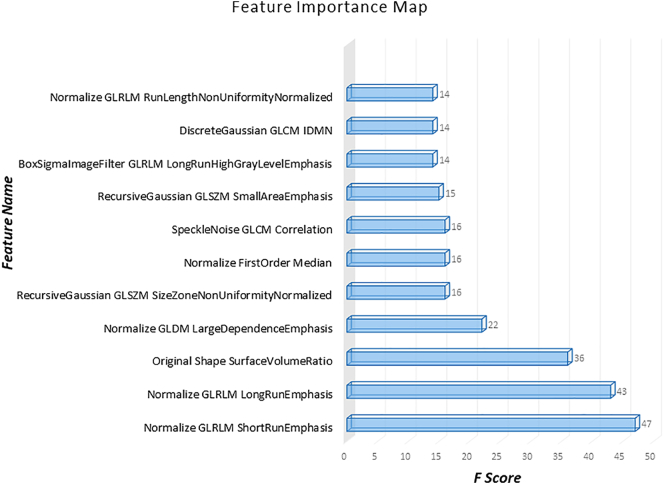
Our CNN-DL classifier performed with a cross-validation accuracy of 83.7% (AU-ROC = 0.90) and test accuracy of 80% (AU-ROC = 0.91), while the CR-ML performed with a cross-validation accuracy of 52.7% (AU-ROC = 0.47) and a test accuracy of 56.5% (0.54 AU-ROC), while RA-ML performed with a cross-validation accuracy of 81.1% (AU-ROC = 0.89) and 60.3% test accuracy (0.54 AU-ROC). Fig. 2 shows the ROC curves for the CNN-DL in comparison with the other two methods. The testing accuracy, sensitivity and specificity are reported in Table 3. The top-most radiomics based features included the run length features, non-uniformity, surface-volume ratio, grey level emphasis as shown in Fig. 3.
Fig. 2.
Receiver operating characteristics for all the three methods employed (a) cross-validation (b) testing.
Table 3.
Performance of CNNs compared to contrast ratios with machine learning (CR-ML) and radiomics with machine learning (RA-ML) and performance of CNNs on PD vs APS.
| CR-ML | RA-ML | CNN-DL | CNN-DL(PD-APS) | |
|---|---|---|---|---|
| Cross validation | ||||
| Accuracy | 52.7% | 81.8% | 83.6% | 81.8% |
| Sensitivity | 0.28 | 0.76 | 0.80 | 0.96 |
| Specificity | 0.73 | 0.86 | 0.88 | 0.50 |
| AU-ROC | 0.469 | 0.890 | 0.906 | 0.718 |
| Testing | ||||
| Accuracy | 56.5% | 60.3% | 80.0% | 85.7% |
| Sensitivity | 0.53 | 0.69 | 0.86 | 1.00 |
| Specificity | 0.60 | 0.50 | 0.70 | 0.50 |
| AU-ROC | 0.540 | 0.540 | 0.913 | 0.911 |
AU-ROC: Area under the receiver operating characteristic; APS: Atypical parkinsonian syndromes; PD: Parkinson's disease. First 3 columns report PD-HC classification results.
Fig. 3.
Radiomic features in order of their importance as plotted against the corresponding f-scores.
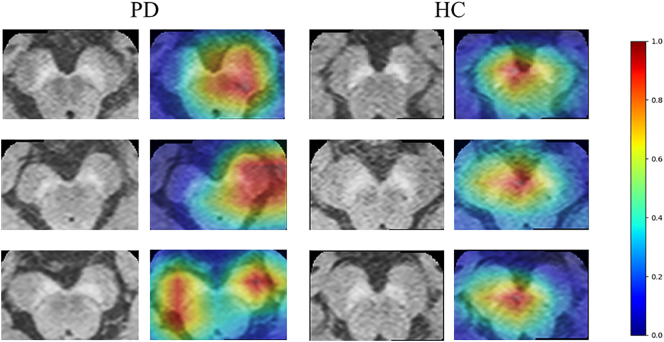
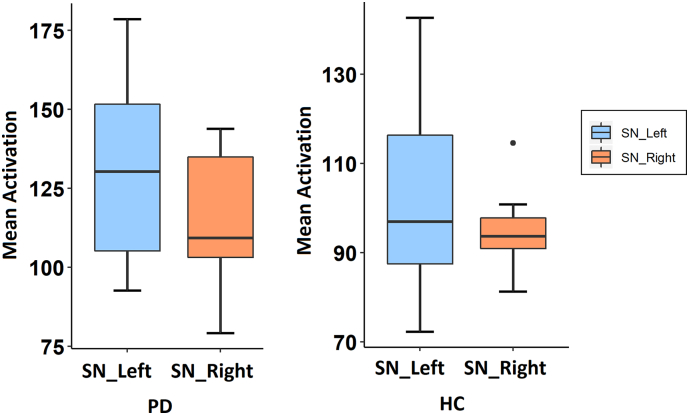
Fig. 4 demonstrates the class activation maps computed from CNNs for three patients. It was observed that in majority of the patients, the activations were more concentrated on the left SNc, which has been quantified and plotted in Fig. 5. The activations on the left demonstrated a significant trend as they were greater than the right (p-value = .09). Similarly, for HCs, the activations on the left were more concentrated than on the right, however were not significant (p-value = .35).
Fig. 4.
Examples of class Activation Maps of PD patients demonstrating that the SN area is highly activated while classifying PDs from Controls. In the first two subjects it can be observed that the left SNc is activated while in the third subject left and right SNc both are activated.
Fig. 5.
Boxplot demonstrating the asymmetry in activations computed from the CNNs.
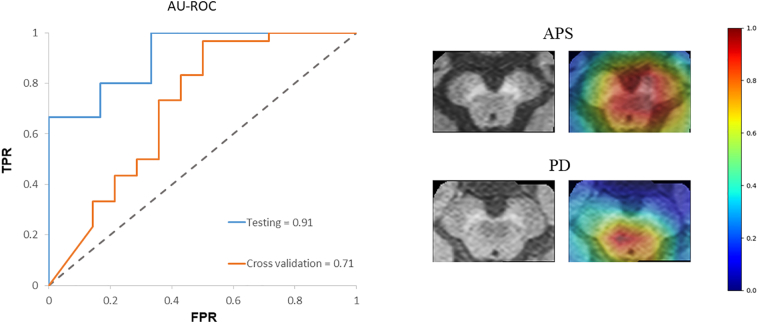
The classification model for PD vs. APS performed with a cross-validation accuracy of 81.8% and a test accuracy of 85.7%. (as shown in Table 3). Fig. 6 demonstrates the ROC for cross-validation and testing of the classifier as well as sample heatmaps for 2 subjects.
Fig. 6.
Figure showing the (a) ROC curves for PD-APS classifier (cross-validation and testing) (b) sample heatmaps for one APS subject and one PD subject.
4. Discussion
This work presented a framework for creating predictive markers of PD using state-of-art CNNs based on NMS-MRI. We obtained a superior accuracy in classifying PD from HCs as well as from APS and demonstrated that our technique was better than contrast ratio-based predictions as well as predictions based off radiomic features computed from the SNc region, and therefore, may be utilized to support a PD diagnosis. Moreover, our CNN based classifier did not require extracting SNc borders as it employed a larger region of interest while the class activation maps illustrated the most important regions and with a significant trend in asymmetry which is in concurrence with the usual clinical picture of PD.
NMS-MRI has been frequently utilized and has demonstrated potential to differentiate PD from healthy controls and other parkinsonian and tremor disorders. Studies in NMS-MRI have shown that this sequence can be used to measure the concentration of the neuromelanin in SNc and lower signal intensities have been observed in patients with PD in comparison to healthy subjects (Ohtsuka et al., 2013; Prasad et al., 2018b; Sasaki et al., 2006). However, these studies have been limited to contrast and volume features with group-wise uni-variate techniques used for analysis. Moreover, these features either require accurate boundaries of the SNc to be computed or placing manual ROIs to compute the contrast. We alleviated these setbacks in NMS-MRI analysis by employing an automated deep learning framework which once trained can be directly employed on every new patient facilitating a PD biomarker effortlessly, which may support in treatment planning and efficacy.
MR based machine learning for PD is not novel and multiple groups have made an effort in classifying PD (as shown in Table 1). These studies have utilized a host of techniques that include supervised predictive models such as support vector machines (SVMs) (Abos et al., 2017; Amoroso et al., 2018; Cherubini et al., 2014a; Cherubini et al., 2014b; Huppertz et al., 2016; Rana et al., 2015; Salvatore et al., 2014) as well as unsupervised models such as self-organizing maps (Peran et al., 2018; Singh and Samavedham, 2015) on data acquired from morphological T1 weighted MRI, functional MRI, diffusion tensor imaging, SPECT, etc. (Adeli et al., 2016; Ariz et al., 2019) and have reported high but variable accuracies. However, none of these studies employ NMS-MRI, a powerful imaging technique for PD diagnosis as described earlier. Furthermore, most of these studies employ millions of features from single or multiple modalities which include voxel level features, on a small sample size using SVMs that builds a hyperplane in the n-feature dimensional space. Even though such models achieve a good classification performance, it could be highly susceptible to over-fitting (Cherubini et al., 2014a). To account for this, some studies extract the most effective attributes from voxel-based morphometry (VBM) analysis and use the mean values from significant clusters as features (Cherubini et al., 2014b; Huppertz et al., 2016; Peran et al., 2018). Although this reduces the vulnerability to overfitting, extracting features off the population analysis may reduce the test accuracy as the pre-computed features from VBM may not be directly applicable to the test subjects. Unsupervised techniques such as self-organizing maps have also been employed on T1 MRIs (Singh and Samavedham, 2015). Our work is pioneering in terms of employing CNNs to classify PD, as CNNs can automatically extract relevant information from the images under consideration making the overall framework more applicable into a clinical setting. Such ability of CNNs in extracting discriminative features using local spatial coherence at various resolutions, is attractive, as it often accounts for better classification performance, especially on large datasets, when compared to a similar task on empirically drawn features such as CRs and/or Radiomic features in this case or VBM based clusters, diffusion measures etc. from earlier studies (Cherubini et al., 2014b; Huppertz et al., 2016; Peran et al., 2018). We demonstrate this in Fig. 2 where the AU-ROC of the CNN is superior to other techniques with manually extracted features. Such superior performance however may come with certain tradeoffs such as inadequate information about the internal operation and behavior of the CNNs. Gaining insights into the working of the CNN is highly crucial where the classes cannot be visually discerned and the classifier could be highly susceptible to overfitting. This is where our class activations computed using the GAP layer play a vital role as they not only illustrate the most discriminative regions but also facilitate deeper understanding of the CNN operation. Fig. 4, Fig. 5 together demonstrate a trend in activations that are more concentrated in the left SNc than the right as has been shown before using manually extracted CRs (Prasad et al., 2018a). In controls, the left mean activation was higher than the right however with no trend for significance. This observation of asymmetrical activation of the SNc is in concurrence with the observations of clinical asymmetry in PD. In the current cohort 60% of subjects demonstrated right CL suggestive of a higher extent of dopaminergic neuronal loss over the left SNc, and the observation of a higher activation, albeit with a trend for significance is in agreement with this observation. Furthermore, the observation of asymmetric activations even in controls is similar to previous observations (Prasad et al., 2018a). The mean activations in controls were lower than in patients suggesting that our model could effortlessly identify the regions which need to be highly weighted in patients with PD in comparison to healthy controls.
To compare our CNNs with other standard techniques we employed another novel method of extracting multiple radiomic features from the SNc region of interest. Radiomics, is a state-of-art technique more commonly employed in tumor classification problems. However, a recent study demonstrated the applicability of texture features in classifying PD using quantitative susceptibility maps (QSM) and R2* maps (Li et al., 2019). In our case, RA-ML displayed superior cross-validation accuracy, however could not perform at the same level on the test data (Table 3). Fig. 3 illustrates the most significant features that participated in the radiomics classification. These features involved grey level run length matrices that provide the size of homogenous runs for each grey level, non-uniformity measures, surface-volume ratios and grey level dependent matrix features that quantify the dependency of one voxel to another. These features overall captured the subtle changes in PD in the SNc that are revealed through NMS-MRI.
Although the NMS-MRI based CNN-DL classifier in the present study provided good accuracy in differentiating between PD and healthy controls, the clinical utility of this technique is dependent on testing the ability of the classifier to differentiate between PD and other parkinsonian disorders such as PSP and MSA. Earlier work by Matsuura et al. (Matsuura et al., 2013) and Ohtsuka et al. (Ohtsuka et al., 2014) have demonstrated the utility of the NMS-MRI to differentiate between PD, MSA-P and PSP. On similar lines, we performed a preliminary analysis on discerning PD from other parkinsonian disorders, using our proposed CNN-DL model. Our superior classification accuracy to discriminate PD from atypical parkinsonian disorders provides evidence that our technique is highly robust exhibiting great potential to be employed in a clinical setting. The heatmaps illustrated the areas that were highly discriminative in PD vs APS as shown in Fig. 6.
It is important to note that the regions of interest on the NMS-MRI that also included the brain stem were drawn manually, as input to the CNN, however, using deep learning-based techniques the boxed regions can be created automatically. Secondly, the APS group included multiple pathologies (MSA and PSP). However, owing to the small sample size we were unable to evaluate these as separate groups. Finally, the APS group had a shorter duration of illness in comparison to the PD group, this is secondary to the faster rate of progression of such patients, owing to which such patients are evaluated earlier in the course of illness. Nevertheless, a larger sample size is certainly required to establish the utility of the method.
In summary, we introduced a novel computer aided PD diagnostic framework using the neuromelanin signal. Our findings show a superior accuracy using CNNs as compared to radiomics, while the underlying activation maps (Fig. 4) confirm the involvement of SNc in the classification. This is commensurate with the idea of atypical neuromelanin contrast on the SNc in PDs, in comparison to HCs as well as to APS, thereby facilitating prediction of the underlying PD pathology.
Acknowledgments
Symbiosis International University has received partial support from DST SERB (ECR/2016/000808) for setting up the computing facility.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2019.101748.
Appendix A. Supplementary data
Supplementary Figure 1: Figure displaying the procedure for CR-ML and RA-ML; Supplementary Table 1: Radiomics Features.
References
- Abos A., Baggio H.C., Segura B., Garcia-Diaz A.I., Compta Y., Marti M.J., Valldeoriola F., Junque C. Discriminating cognitive status in Parkinson's disease through functional connectomics and machine learning. Sci. Rep. 2017;7 doi: 10.1038/srep45347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeli E., Shi F., An L., Wee C.Y., Wu G., Wang T., Shen D. Joint feature-sample selection and robust diagnosis of Parkinson's disease from MRI data. Neuroimage. 2016;141:206–219. doi: 10.1016/j.neuroimage.2016.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoroso N., La Rocca M., Monaco A., Bellotti R., Tangaro S. Complex networks reveal early MRI markers of Parkinson's disease. Med. Image Anal. 2018;48:12–24. doi: 10.1016/j.media.2018.05.004. [DOI] [PubMed] [Google Scholar]
- Ariz M., Abad R.C., Castellanos G., Martinez M., Munoz-Barrutia A., Fernandez-Seara M.A., Pastor P., Pastor M.A., Ortiz-de-Solorzano C. Dynamic atlas-based segmentation and quantification of Neuromelanin-rich brainstem structures in Parkinson disease. IEEE Trans. Med. Imaging. 2019;38:813–823. doi: 10.1109/TMI.2018.2872852. [DOI] [PubMed] [Google Scholar]
- Barrett M.J., Wylie S.A., Harrison M.B., Wooten G.F. Handedness and motor symptom asymmetry in Parkinson's disease. J. Neurol. Neurosurg. Psychiatry. 2011;82:1122–1124. doi: 10.1136/jnnp.2010.209783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Machine Learning. Vol. 45. 2001. Random forests; pp. 5–32. [Google Scholar]
- Chang K., Bai H.X., Zhou H., Su C., Bi W.L., Agbodza E., Kavouridis V.K., Senders J.T., Boaro A., Beers A. Residual convolutional neural network for the determination of IDH status in low-and high-grade gliomas from MR imaging. Clin. Cancer Res. 2018;24:1073–1081. doi: 10.1158/1078-0432.CCR-17-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherubini A., Morelli M., Nistico R., Salsone M., Arabia G., Vasta R., Augimeri A., Caligiuri M.E., Quattrone A. Magnetic resonance support vector machine discriminates between Parkinson disease and progressive supranuclear palsy. Mov. Disord. 2014;29:266–269. doi: 10.1002/mds.25737. [DOI] [PubMed] [Google Scholar]
- Cherubini A., Nistico R., Novellino F., Salsone M., Nigro S., Donzuso G., Quattrone A. Magnetic resonance support vector machine discriminates essential tremor with rest tremor from tremor-dominant Parkinson disease. Mov. Disord. 2014;29:1216–1219. doi: 10.1002/mds.25869. [DOI] [PubMed] [Google Scholar]
- Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2015;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman S., Wenning G.K., Low P.A., Brooks D.J., Mathias C.J., Trojanowski J.Q., Wood N.W., Colosimo C., Durr A., Fowler C.J., Kaufmann H., Klockgether T., Lees A., Poewe W., Quinn N., Revesz T., Robertson D., Sandroni P., Seppi K., Vidailhet M. Second consensus statement on the diagnosis of multiple system atrophy. Neurology. 2008;71:670–676. doi: 10.1212/01.wnl.0000324625.00404.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K., Zhang X., Ren S., Sun J. European Conference on Computer Vision. Springer; 2016. Identity mappings in deep residual networks; pp. 630–645. [Google Scholar]
- Hughes A.J., Daniel S.E., Blankson S., Lees A.J. A clinicopathologic study of 100 cases of Parkinson's disease. Arch. Neurol. 1993;50:140–148. doi: 10.1001/archneur.1993.00540020018011. [DOI] [PubMed] [Google Scholar]
- Huppertz H.J., Moller L., Sudmeyer M., Hilker R., Hattingen E., Egger K., Amtage F., Respondek G., Stamelou M., Schnitzler A., Pinkhardt E.H., Oertel W.H., Knake S., Kassubek J., Hoglinger G.U. Differentiation of neurodegenerative parkinsonian syndromes by volumetric magnetic resonance imaging analysis and support vector machine classification. Mov. Disord. 2016;31:1506–1517. doi: 10.1002/mds.26715. [DOI] [PubMed] [Google Scholar]
- Hutchinson M., Raff U. Structural changes of the substantia nigra in Parkinson's disease as revealed by MR imaging. AJNR Am. J. Neuroradiol. 2000;21:697–701. [PMC free article] [PubMed] [Google Scholar]
- Korfiatis P., Kline T.L., Lachance D.H., Parney I.F., Buckner J.C., Erickson B.J. Residual deep convolutional neural network predicts MGMT methylation status. J. Digit. Imaging. 2017;30:622–628. doi: 10.1007/s10278-017-0009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhevsky A., Sutskever I., Hinton G.E. Advances in Neural Information Processing Systems. 2012. Imagenet classification with deep convolutional neural networks; pp. 1097–1105. [Google Scholar]
- Li G., Zhai G., Zhao X., An H., Spincemaille P., Gillen K., Ku Y., Wang Y., Hunag D., Li J. 3D texture analyses within the substantia nigra of Parkinson's disease patients on quantitative susceptibility maps and R2* maps. Neuroimage. 2019;188:465–472. doi: 10.1016/j.neuroimage.2018.12.041. [DOI] [PubMed] [Google Scholar]
- Litvan I., Agid Y., Calne D., Campbell G., Dubois B., Duvoisin R.C., Goetz C.G., Golbe L.I., Grafman J., Growdon J.H., Hallett M., Jankovic J., Quinn N.P., Tolosa E., Zee D.S. Clinical research criteria for the diagnosis of progressive supranuclear palsy (Steele-Richardson-Olszewski syndrome): report of the NINDS-SPSP international workshop. Neurology. 1996;47:1–9. doi: 10.1212/wnl.47.1.1. [DOI] [PubMed] [Google Scholar]
- Mann D.M., Yates P.O. Pathological basis for neurotransmitter changes in Parkinson's disease. Neuropathol. Appl. Neurobiol. 1983;9:3–19. doi: 10.1111/j.1365-2990.1983.tb00320.x. [DOI] [PubMed] [Google Scholar]
- Matsuura K., Maeda M., Yata K., Ichiba Y., Yamaguchi T., Kanamaru K., Tomimoto H. Neuromelanin magnetic resonance imaging in Parkinson's disease and multiple system atrophy. Eur. Neurol. 2013;70:70–77. doi: 10.1159/000350291. [DOI] [PubMed] [Google Scholar]
- Ogisu K., Kudo K., Sasaki M., Sakushima K., Yabe I., Sasaki H., Terae S., Nakanishi M., Shirato H. 3D neuromelanin-sensitive magnetic resonance imaging with semi-automated volume measurement of the substantia nigra pars compacta for diagnosis of Parkinson's disease. Neuroradiology. 2013;55:719–724. doi: 10.1007/s00234-013-1171-8. [DOI] [PubMed] [Google Scholar]
- Ohtsuka C., Sasaki M., Konno K., Koide M., Kato K., Takahashi J., Takahashi S., Kudo K., Yamashita F., Terayama Y. Changes in substantia nigra and locus coeruleus in patients with early-stage Parkinson's disease using neuromelanin-sensitive MR imaging. Neurosci. Lett. 2013;541:93–98. doi: 10.1016/j.neulet.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Ohtsuka C., Sasaki M., Konno K., Kato K., Takahashi J., Yamashita F., Terayama Y. Differentiation of early-stage parkinsonisms using neuromelanin-sensitive magnetic resonance imaging. Parkinsonism Relat. Disord. 2014;20:755–760. doi: 10.1016/j.parkreldis.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Peran P., Barbagallo G., Nemmi F., Sierra M., Galitzky M., Traon A.P., Payoux P., Meissner W.G., Rascol O. MRI supervised and unsupervised classification of Parkinson's disease and multiple system atrophy. Mov. Disord. 2018;33:600–608. doi: 10.1002/mds.27307. [DOI] [PubMed] [Google Scholar]
- Powers, D. M. (2011). Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation.
- Prasad S., Saini J., Yadav R., Pal P.K. Motor asymmetry and neuromelanin imaging: concordance in Parkinson's disease. Parkinsonism Relat. Disord. 2018;53:28–32. doi: 10.1016/j.parkreldis.2018.04.022. [DOI] [PubMed] [Google Scholar]
- Prasad S., Stezin A., Lenka A., George L., Saini J., Yadav R., Pal P.K. 3D Neuromelanin-sensitive magnetic resonance imaging of the substantia nigra in Parkinson's disease. Eur. J. Neurol. 2018;25:680–686. doi: 10.1111/ene.13573. [DOI] [PubMed] [Google Scholar]
- Rana B., Juneja A., Saxena M., Gudwani S., Kumaran S.S., Agrawal R., Behari M. Regions-of-interest based automated diagnosis of Parkinson's disease using T1-weighted MRI. Expert Syst. Appl. 2015;42:4506–4516. [Google Scholar]
- Salvatore C., Cerasa A., Castiglioni I., Gallivanone F., Augimeri A., Lopez M., Arabia G., Morelli M., Gilardi M.C., Quattrone A. Machine learning on brain MRI data for differential diagnosis of Parkinson's disease and progressive Supranuclear palsy. J. Neurosci. Methods. 2014;222:230–237. doi: 10.1016/j.jneumeth.2013.11.016. [DOI] [PubMed] [Google Scholar]
- Sasaki M., Shibata E., Tohyama K., Takahashi J., Otsuka K., Tsuchiya K., Takahashi S., Ehara S., Terayama Y., Sakai A. Neuromelanin magnetic resonance imaging of locus ceruleus and substantia nigra in Parkinson's disease. Neuroreport. 2006;17:1215–1218. doi: 10.1097/01.wnr.0000227984.84927.a7. [DOI] [PubMed] [Google Scholar]
- Shah A., Lenka A., Saini J., Wagle S., Naduthota R.M., Yadav R., Pal P.K., Ingalhalikar M. Altered brain wiring in Parkinson's disease: a structural connectome-based analysis. Brain Connect. 2017;7:347–356. doi: 10.1089/brain.2017.0506. [DOI] [PubMed] [Google Scholar]
- Shah A., Prasad S., Rastogi B., Dash S., Saini J., Pal P.K., Ingalhalikar M. Altered structural connectivity of the motor subnetwork in multiple system atrophy with cerebellar features. Eur. Radiol. 2018 doi: 10.1007/s00330-018-5874-4. (ahead of print) [DOI] [PubMed] [Google Scholar]
- Singh G., Samavedham L. Unsupervised learning based feature extraction for differential diagnosis of neurodegenerative diseases: a case study on early-stage diagnosis of Parkinson disease. J. Neurosci. Methods. 2015;256:30–40. doi: 10.1016/j.jneumeth.2015.08.011. [DOI] [PubMed] [Google Scholar]
- Sulzer D., Cassidy C., Horga G., Kang U.J., Fahn S., Casella L., Pezzoli G., Langley J., Hu X.P., Zucca F.A., Isaias I.U., Zecca L. Neuromelanin detection by magnetic resonance imaging (MRI) and its promise as a biomarker for Parkinson's disease. NPJ Parkinsons Dis. 2018;4:11. doi: 10.1038/s41531-018-0047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Griethuysen J.J., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., Beets-Tan R.G., Fillion-Robin J.-C., Pieper S., Aerts H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Khosla A., Lapedriza A., Oliva A., Torralba A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Learning deep features for discriminative localization; pp. 2921–2929. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Figure displaying the procedure for CR-ML and RA-ML; Supplementary Table 1: Radiomics Features.