Abstract
MicroRNAs (miRNAs) control essential gene regulatory pathways in plants and animals. Serving as guides in silencing complexes, miRNAs direct Argonaute proteins to specific target messenger RNAs to repress protein expression. The mature, 22-nucleotide (nt) miRNA is the product of multiple processing steps, and recent studies have uncovered factors that directly control the stability of the functional RNA form. Although alteration of miRNA levels has been linked to numerous disease states, the mechanisms responsible for stabilized or reduced miRNA expression have been largely elusive. The discovery of specific cis-acting modifications and trans-acting proteins that affect miRNA half-life reveals new elements that contribute to the homeostasis of these vital regulatory molecules.
miRNAs eluded researchers for decades, stealthily participating in many of the most important biological pathways in eukaryotic cells. In recent years, our understanding of miRNAs has grown from the discovery of a single genetic oddity in worms to the recognition of an entirely new class of regulatory molecule with thousands of members1. The significance of miRNAs in normal development and cellular function is underscored by mounting evidence that misregulation of specific miRNA pathways is associated with complicated health afflictions, including cancer, heart disease and neurological disorders2–4. miRNAs are intertwined in complex regulatory pathways in plants as well5 and represent one of the most plentiful classes of gene regulators in multicellular organisms.
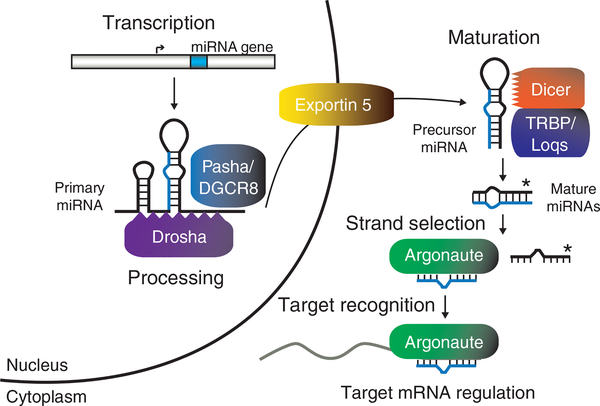
Production of the functional, ~22-nt mature miRNA involves multiple processing steps6–8 (Fig. 1). The general miRNA biogenesis pathway begins with synthesis of a primary transcript by RNA polymerase II (Pol II). Housed within the primary transcript is the hairpin precursor, which contains the sequence destined to be the mature miRNA in one arm of the stem. In animals, the Microprocessor complex, minimally composed of the Drosha RNase (RNase) and its RNA binding partner Pasha (also called DGCR8), releases the miRNA precursor from the primary transcript (Table 1). Exportin-5 delivers the precursor to the cytoplasm for final processing by the Dicer RNase and its double-stranded RNA binding cofactor TRBP (also called loquacious, Loqs) (Table 1). After loading onto Argonaute, one strand of the resulting partial duplex, designated the guide, is p referentially retained. This multistep pathway is shared in plants with a few exceptions: the Dicer-like (DCL) proteins catalyze both the primary and precursor processing steps in the nucleus, where the mature miRNA forms a complex with Argonaute and is transported to the cytoplasm (Table 1). miRNAs serve as guides to direct the Argonaute complex to target mRNAs through complementary base-pairing6,9. Typically, target recognition results in destabilization or translational repression, either of which ultimately silences gene expression.
Figure 1.
A general model of miRNA biogenesis and function6–9. After synthesis by RNA polymerase II, miRNA primary transcripts are recognized by Pasha/DGCR8 and Drosha, which excises the hairpin precursor. Exportin 5 delivers the miRNA precursor to Dicer and its RNA binding partner, TRBP/Loqs, for final processing to the mature 22-nt miRNAs. One strand is selected for stable association with Argonaute, where it serves as a guide to target and regulate specific mRNAs.
Table 1.
Enzymes that act on miRNAs
| Name | Type | Substrate | Activity |
|---|---|---|---|
| Drosha in animals DCL in plants |
RNase III endonuclease | Primary miRNAs | Generates hairpin precursor |
| Dicer in animals DCL in plants |
RNase III endonuclease | Precursor miRNAs | Removes loop from precursor to generate mature miRNA duplex |
| Argonaute | PIWI-RNase H endonuclease | Precursor miRNAs | Cleaves passenger strand of some miRNA precursors |
| HEN1 | Methyltransferase | Mature miRNAs and siRNAs in plants piRNAs and siRNAs in animals | Adds 2′-O-methyl group to the 3′ ends of small RNAs |
| GLD-2 in animals | Poly(A) polymerase | Mature single-stranded miRNAs | Adds adenosine to the 3′ end of miR-122 and possibly other miRNAs |
| TUT4/Zcchc11/ PUP-2 in animals | Uridyltransferase | Mature single-stranded and precursor miRNAs | Adds uridines to miRNA and precursor 3′ ends |
| SDN1 in plants | 3′-to-5′ exonuclease | Mature single-stranded miRNAs | Degrades mature miRNAs |
| XRN-2 in animals | 5′-to-3′exonuclease | Mature single-stranded miRNAs Primary miRNA cleavage products |
Degrades mature miRNAs Degrades 3′ sequence after Drosha cleavage |
| XRN2, XRN3 in plants | 5′-to-3′ exonuclease | Precursor miRNA loops | Degrades loop sequence released from miRNA precursors after Dicer cleavage |
Accumulation of a specific miRNA is dependent on the rates of transcription, processing and decay. Similar to the expression of many protein-coding genes, expression of miRNA primary transcripts is subject to regulation by specific transcription factors and chromatin marks6–8. Control of each processing step has also emerged as a key determinant of functional miRNA expression. The first global analysis of primary and mature miRNA levels revealed that extensive post-transcriptional regulation is involved in cellular miRNA homeostasis10. Several examples of proteins and mechanisms that govern processing of specific miRNAs are detailed in recent reviews7,8. Here, we focus on parameters that determine miRNA existence after maturation has been completed. The stability of mature miRNAs is controlled by cis-acting modifications, protein complex formation and exposure to nucleases. The recent discoveries of specific factors that mediate miRNA turnover offer new insights into mechanisms responsible for changes in the availability of these critical regulatory molecules.
Eluding the reapers: cis- and trans-acting stabilization elements
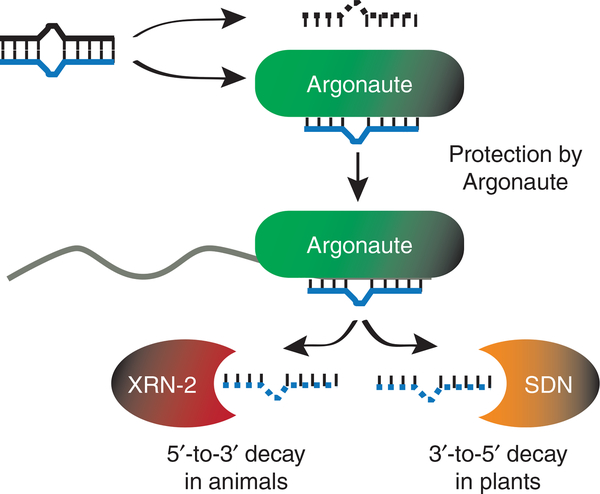
Unprotected 5′ or 3′ ends leave RNAs vulnerable to exonucleolytic decay pathways. To thwart degradation factors, 5′ cap structures and 3′ poly(A) tails are added to most protein-coding Pol II transcripts. Stable secondary structures help protect the ends of mature noncoding RNAs synthesized by Pol I (18S, 5.8S and 28S ribosomal RNA precursors) and Pol III (5S rRNA and tRNAs). miRNAs emerge as short, duplex RNAs with 5′ monophosphate and 3′ hydroxyl groups after processing by the sequential actions of Drosha and Dicer or DCL proteins6–8. Typically, one half of the hybrid, called the guide, is preferentially maintained, and the other strand, sometimes referred to as the star or passenger strand, disappears. This dichotomy has been attributed to biased Argonaute loading of the miRNA half that has weaker 5′ pairing interactions with its partner strand11,12. Presumably, the stably bound guide strand is protected by Argonaute while the passenger is vulnerable to degradation (Fig. 2). The profound imbalance in guide versus passenger for many miRNAs implies that an efficient decay pathway exists to clear unprotected miRNAs.
Figure 2.
Proteins that regulate miRNA stability. Incorporation into Argonaute stabilizes mature miRNAs and release from this complex leaves miRNAs vulnerable to decay by XRN-2 or SDN exonucleases13–16,19,31. In C. elegans, XRN-2 also facilitates release of miRNAs from Argonaute proteins that are not associated with targets31.
Mature miRNA abundance is sensitive to Argonaute protein levels, supporting a protective role for this core miRNA effector protein (Fig. 2). Downregulation or ectopic expression of Argonaute results in diminished or bolstered mature miRNA levels, respectively13–16. In some cases, depletion of Argonaute also results in impaired processing of precursor to mature miRNAs, indicating that Argonaute may function in biogenesis as well as stabilization of miRNAs14,15. In a screen for factors that are limiting for miRNA biogenesis in mammalian culture cells, ectopic expression of Argonaute proteins resulted in increased levels of mature miRNAs13. Other genes encoding proteins essential in the miRNA pathway, such as Drosha, Pasha/DGCR8 and Dicer, had no effect in this experimental system, indicating that the availability of Argonaute proteins largely influences cellular mature miRNA levels13.
In addition to taking refuge in protein complexes, mature miRNAs can undergo protective modifications (Table 2). In Arabidopsis thaliana, methyl groups are added to the 3′ ends of miRNAs by the HEN1 methyltransferase17. In hen1 mutants, levels of mature miRNAs are substantially reduced and the residual species often have 1–5 uracil residues appended to their 3′ ends18 (Table 2). Thus, 3′ methylation prevents uridylation and destabilization of miRNAs in Arabidopsis. In one model, unmodified miRNAs undergo uridylation, which serves as a tag to promote degradation. Alternatively, miRNAs that lack 3′ methyl groups could be exposed for direct exonucleolytic decay or U-tailing, which might instead serve as a protective modification. Interestingly, miRNAs with extra uracil residues were observed more frequently for the guide versus star strand of miR173 in hen1 mutants18. Thus, either uridylation of miRNAs with unmodified 3′ ends favors the Argonaute-bound form or the unselected passenger strands with U-tails are more efficiently degraded than nonuridylated species. Although the enzyme responsible for uridylation of plant miRNAs has yet to be identified, specific nucleases that degrade mature plant miRNAs were recently determined and will be discussed below19.
Table 2.
Modifications that affect miRNA stability
| Modification | Organism | Enzyme | Potential effects |
|---|---|---|---|
| 2′-O-methylation of miRNA 3′ ends | Plants | HEN1 | Stabilization Inhibition of 3′ uridylation |
| Uridylation of miRNA 3′ ends | Plants Animals |
Unknown TUT4/Zcchc11/PUP-2 |
Destabilization Inhibition of 3′-to-5′decay by SDN1 Destabilization of miR-122 Reduction of function, but not stability, of miR-26b |
| Adenylation of miRNA 3′ ends | Plants Animals |
Unknown GLD-2 |
Stabilization in P. trichocarpa extracts Stabilization of miR-122 Inhibition of 3′ uridylation |
In contrast to plants, a uniform modification of animal miRNAs has not been observed. The HEN1 methyltransferase is conserved in animals, but its substrates are piRNAs (PIWI-interacting RNAs) and, in some cases, siRNAs (small interfering RNAs), instead of miRNAs20–22. Diverse nucleotide substitutions, additions and deletions have been detected in animal miRNAs by massive sequencing approaches to probe deeply the miRNA composition of cells and organisms23–26. Despite the caveat that sequencing errors can also contribute to heterogeneity in apparent miRNA composition, the extent of these types of modifications appears substantial27. Although a change in mature miRNA sequence has clear implications for target recognition, possible effects on miRNA half-life are less predictable.
It was recently demonstrated that 3′ adenylation can have a stabilizing effect on animal miRNAs (Table 2). Although the addition of nontemplated adenines has been detected on many different animal miRNAs23,25,26,28, a functional consequence of this modification has so far only been established for miR-122 in liver cells29. The cytoplasmic poly(A) polymerase GLD-2 adds a single adenine residue to the 3′ end of mature miR-122; this modification appears to prevent shortening and to stabilize the miRNA29. Depletion of GLD-2 in liver cells resulted in disappearance of the 23-nt adenylated form of miR-122 with a concurrent increase in the 21-nt variant. Moreover, the total levels of mature, but not precursor, miR-122 miRNAs were substantially reduced in cells deficient in GLD-2 activity. Interestingly, the abundance of several other miRNAs expressed in liver cells was not affected by the loss of GLD-2, indicating that stabilization of miR-122 is specifically dependent on GLD-2 mediated adenylation. Because GLD-2 has other targets, including 7SL (the noncoding RNA component of the signal recognition particle) and select mRNAs, it is unclear what proportion of miRNAs is subject to adenylation by this factor. It is possible that GLD-2 nonspecifically adds adenine residues to miRNAs, but only a fraction of these miRNA species depend on the modification for stability.
Nontemplated addition of adenine residues has also been detected on plant miRNAs18,30. One to seven adenines were found attached to representatives of most miRNA families identified in Populus trichocarpa (black cottonwood)30 (Table 2). Adenylation was observed for both full-length as well as truncated miRNAs, suggesting that mature and partially degraded miRNAs are substrates for this modification. It remains to be determined whether adenylation has a functional consequence for miRNAs in vivo or whether this is a promiscuous activity on unprotected miRNAs. Supporting the first possibility, replacement of the 3′ nucleotide with an adenine residue resulted in slower miRNA degradation in an in vitro decay assay using extracts from P. trichocarpa30. The factors responsible for adenylation of plant miRNAs and the potential effect of this modification on plant miRNA homeostasis await elucidation.
The SDN slayers: an end attack
The first factors shown to degrade mature miRNAs are the appropriately named small RNA degrading nuclease (SDN) genes in Arabidopsis19 (Fig. 2). Members of this family of exonucleases catalyze 3′-to-5′ decay of single-stranded miRNAs, and depletion of SDN transcripts results in increased steady-state levels of mature miRNAs in vivo19 (Table 1). Consistent with a protective role for the 3′ methyl group on plant miRNAs, methylated miRNAs were less efficiently degraded than unmodified miRNAs by recombinant SDN1 in in vitro decay assays. Notably, miRNAs with two or five uracil residues added to the 3′ end were strikingly resistant to SDN1-mediated degradation (Table 2). Thus, unmethylated miRNAs in hen1 mutants may be subject to two opposing activities: 3′-to-5′ degradation by SDN proteins or uridylation by yet-to-be-identified factors. These findings prompt re-evaluation of the consequence of uridylation on plant miRNAs. If uridylation also impedes SDN-mediated degradation in vivo, then the addition of uracil residues to the 3′ ends of unmethylated miRNAs could function as a protective backup measure as opposed to a tag for destabilization.
SDN1 is related to four other predicted exonucleases in Arabidopsis that may have overlapping functions in regulating miRNA homeostasis19. Depletion of SDN1, SDN2 and SDN3 transcripts results in generally increased miRNA levels and pleiotropic developmental defects19. Overaccumulation of miRNAs presumably augments target downregulation, potentially reducing some targets below critical thresholds. Homologs of SDN genes are present in animals but roles in miRNA homeostasis or other pathways are yet to be discovered.
The XRN-2 executioners: beginning of the end
In animals, the 5′-to-3′ exonuclease XRN-2 (Rat1p in yeast) catalyzes degradation of mature miRNAs31 (Fig. 2 and Table 1). From a panel of eight candidate nucleases, which included homologs of SDN, Chatterjee and Grosshans identified XRN-2 as a factor involved in mature miRNA accumulation in Caenorhabditis elegans31. The failure to detect an effect of the SDN-related genes in the worm miRNA pathway could be due to the assay and/or to the potential redundancy of the worm homologs. Thus, a conserved role for these genes in plants and animals has not been ruled out. The finding that XRN-2 degrades single- but not double-stranded miRNAs in vitro indicates that after strand separation, miRNAs that fail to be incorporated into Argonaute or those that are released from the effector complex are the natural targets of XRN-2–mediated decay. Moreover, the vulnerability of a miRNA to degradation by XRN-2 in vitro is influenced by target availability. The evidence suggests that interaction of the miRNA–Argonaute complex with its target prevents release and subsequent destabilization of the miRNA31. At least in vitro, XRN-2 seems to both facilitate Argonaute unloading and catalyze degradation of miRNAs when target sequences are not available (Fig. 2). A relationship between miRNA homeostasis and functional utilization is an intriguing possibility and could contribute to changes in endogenous miRNA levels if this is also the case in vivo.
In addition to mature miRNAs, diverse RNA substrates are subject to 5′-to-3′ degradation by XRN-2. In plants, the XRN-2 related proteins XRN2 and XRN3 digest the loops resulting from miRNA precursor processing, an event that happens in the nucleus in Arabidopsis32. In mammalian cells, XRN-2 aids Pol II transcriptional termination of miRNA primary transcripts by catalyzing degradation of Drosha cleavage products downstream of the miRNA hairpin33,34. This role is similar to the function of XRN-2 in terminating Pol II transcription of mRNAs after cleavage by the polyadenylation machinery35. Additionally, XRN-2/Rat1 clears the nonfunctional products of numerous RNA processing events, including lariats from splicing, spacer regions from rRNA maturation and 5′ extensions of snoRNAs; this nuclease also targets aberrant RNAs that escape full maturation, such as hypomodified tRNAs and improperly processed mRNAs36. All of these functions require nuclear XRN-2 activity. Mature miRNAs, by contrast, reside primarily in cytoplasmic Argonaute complexes9. However, nuclear occupancy of Argonaute has been documented and is regulated by the import receptor protein Imp8 in mammalian culture cells37. The subcellular distribution of XRN-2 and its miRNA substrates has not yet been investigated. Nonetheless, it seems possible that cellular localization is another layer of regulation determining the turnover rate of mature miRNAs.
Outlook
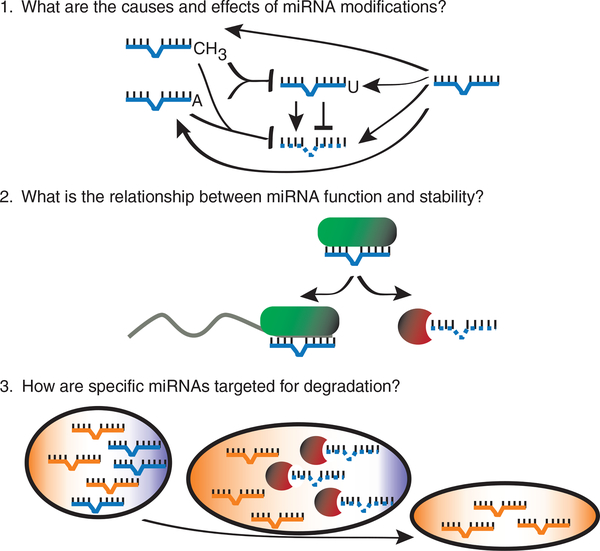
In the short history of their recognized existence, miRNAs have emerged as indispensable regulators of gene expression in plants and animals. Multilevel processing steps whittle miRNAs into precise mature forms that depend on base-pairing interactions to regulate specific target genes. Transcriptional and post-transcriptional regulatory mechanisms control where, when and how much of a particular miRNA accumulates. Although miRNAs have been considered to be generally stable molecules with half-lives that are often days long38–40, it is now clear that the absolute levels of mature miRNAs are also controlled by cis- and trans-acting factors that directly affect stability. Recent discoveries have established that specific modifications and exonucleases can profoundly influence miRNA existence. Stemming from these initial studies are three areas that warrant deeper investigation to further elucidate the causes and effects of altered miRNA homeostasis at the level of mature miRNA stability (Fig. 3).
Figure 3.
Outstanding questions regarding factors that regulate miRNA stability. First, some modifications appear general, such as methylation of plant miRNAs, whereas others may be targeted to specific miRNAs, such as adenylation of miR-122 (refs. 18,29). In several cases the modifying enzyme and effect of the modification on miRNA stability are not yet known. Second, evidence is mounting that Argonaute is a limiting factor for miRNA function and stability13–16. Target availability has been shown to influence the association of a miRNA with Argonaute and its protection from degradation in vitro31, but whether this parameter influences miRNA accumulation in vivo is not yet established. Third, the extent of targeted degradation of specific miRNAs as a means to transform the cellular miRNA population is unclear. Decay of select miRNAs could contribute to the dynamic changes in miRNA levels that often accompany differentiation.
Tagged for life or death
The 3′-methyl modification on plant miRNAs protects them against 3′-to-5′ degradation by SDN exonucleases18,19. This modification also appears to counter uridylation of plant miRNAs. However, it is less clear whether the addition of uracil residues marks the miRNA for destruction, or buffers against SDN activity, or instead is a spurious reaction on unmodified miRNAs that has no functional consequence. The first possibility is consistent with the observation that plants with mutations in the HEN1 methyltransferase gene have overall reduced miRNA levels and the residual species are heterogeneous in length due to the addition of 3′-uracil residues. Uridylation of other noncoding RNAs has been shown to stimulate their degradation. For example, the nucleotidyltransferase CDE-1 (cosuppression defective) was recently demonstrated to regulate the stability of endogenous siRNAs in C. elegans via 3′- terminal addition of uracil residues41. Additionally, targeted downregulation of the precursor form of let-7 miRNA has been associated with the appendage of uracil residues by the TUT4/Zcchc11/PUP-2 terminal uridyltransferase42–45. Another target of uridylation mediated by Zcchc11 is mature miR-26a (ref. 46). However, in this case the addition of uracil residues to miR-26a abrogates its function without obviously affecting its expression levels. Supporting the possibility that uridylation could also have a protective effect, miRNA substrates with uracil additions were degraded less efficiently by SDN1 in vitro19. Thus, uridylation is not a definitive tag for destruction of mature miRNAs.
Addition of a limited number of adenines seems to have a stabilizing effect on miRNAs in plants and animals29,30. Although adenylation of eukaryotic mRNAs has long been recognized for its importance in stabilization and translation, this same modification targets several noncoding RNAs, such as tRNAs, pre-rRNAs and snRNAs, for destruction by the TRAMP (Trf4/Air1–2/Mtr4 polyadenylation) complex47. The addition of a single adenine to the 3′ end of miR-122 in liver cells demonstrates a new role for the cytoplasmic poly(A) polymerase GLD-2 (ref. 29). The broadly conserved GLD-2 protein regulates the poly(A) tail length of specific mRNAs, which in turn influences their translational c ompetence47. GLD-2 participates in diverse biological pathways, including germline development and neuronal function47. The discovery of miRNAs as new substrates for this poly(A) polymerase raises the possibility that the adenylation and stabilization of specific miRNAs could be important for the biological outputs of GLD-2 activity29.
The consequence of modifications to mature miRNAs is likely to be dependent on context. Given the established examples in which adenylation can have opposite effects on RNA stability depending on the substrate and polymerase complex, a simple code that dictates miRNA half-life may not exist. The addition of an adenine residue to noncoding RNAs can prevent uridylation29,48. However, many mature miRNAs naturally end in adenine or uracil residues, so it is unclear whether the presence of these nucleotides per se or the act of modification itself elicits downstream effects on miRNA stability. Finally, an important consideration in studying the role of cis-acting modifications on miRNA homeostasis is the likelihood that some types of chemical changes are not apparent by current miRNA detection methods. Moreover, certain modifications would also interfere with standard miRNA cloning strategies. Thus, the extent and types of modifications as well as the possibility of yet-to-be-discovered miRNA species that have escaped detection are unknowns that await innovative chemical and molecular investigations.
Use it or lose it
The demonstration that target availability affects the release of miRNAs from Argonaute and their subsequent vulnerability to degradation by XRN-2 in vitro has important implications for endogenous miRNA function as well as for therapeutic use of small RNAs31. The generally poor correlation between expression of miRNA primary transcripts and mature miRNA forms has been attributed to processing regulation10. Given the study by Chatterjee and Grosshans, mature miRNA levels might also reflect their targeting activity within a cell31. Another clue that activity might influence miRNA accumulation is the finding that miRNA abundance correlates with the number of potential target sites bound by Argonaute in vivo49. If target association maintains miRNAs in the Argonaute-bound state, then the mechanism of target regulation could also influence miRNA stability: miRNAs that promote mRNA degradation would lose the stabilizing effect of target association more rapidly than miRNAs that remain bound to translationally repressed targets.
The influence of target recognition on Argonaute occupancy and stabilization of miRNAs in vivo is not yet established. However, the evidence that Argonaute is a limiting factor for endogenous miRNA accumulation implies that there is competition among small RNAs for Argonaute protection13–16. Notably, miRNA regulation of endogenous targets can be perturbed by transfection of siRNAs or miRNAs into culture cells50,51. The upregulation of predicted miRNA targets in cells introduced to exogenous siRNAs was attributed to titration of the silencing machinery50. Saturation of Argonaute-binding capacity is expected to limit the function and stability of endogenous miRN As13–16,50. Several miRNA targets related to oncogenic pathways were found to be commonly upregulated in response to unrelated siRNA transfections50. Thus, potential disruption of endogenous target regulation by Argonaute titration may have unexpected but profound biological consequences.
To be or not to be
The exonucleases SDN and XRN-2 degrade unprotected mature miRNAs. In Arabidopsis, miRNAs devoid of 3′ methyl modifications are subject to 3′-to-5′ degradation by SDN nucleases, and in C. elegans, release from Argonaute exposes miRNAs to 5′-to-3′ decay by XRN-2 (refs. 19,31). These exonucleases appear to generally act on miRNAs and could potentially be responsible for efficient clearance of the unselected passenger strand after separation from its guide-strand partner. A regulatory role for SDN or XRN-2 in the clearance of specific miRNAs has not been determined. Good candidates for miRNAs subject to regulated destabilization are the brain-enriched miRNAs miR-9 and miR-183, with short half-lives of about 1 h, and miR-124, whose mature but not precursor levels rapidly drop in response to serotonin treatment in neurons from Aplysia californica, a marine snail52,53. In another example, the extreme variations in mature miRNA levels for members of a common primary-transcript cluster during embryonic stem-cell differentiation could involve targeted degradation of individual miRNAs54. Presumably, cofactors that recognize specific miRNA sequences would be needed to recruit exonucleases to particular substrates. Some miRNAs show extensive sequence conservation beyond just the 5′ region important for target interaction. Maintenance of nucleotide identity may be important for recognition of certain miRNAs by sequence-specific RNA-binding proteins that regulate processing or stability. Curiously, the first cis-acting element shown to regulate mature miRNA fate is not broadly conserved. The 3′ terminal hexanucleotide sequence of human miR-29b promotes nuclear localization and subsequent destruction of this miRNA55. The trans-acting factors that recognize this sequence element and promote trafficking and degradation of miR-29b are yet undiscovered.
The connections between target availability, Argonaute capacity and miRNA accumulation underscore the exquisite regulation of mature miRNA expression. It is likely that some miRNAs have also evolved elements that influence Argonaute loading efficiency, recognition by modifying enzymes and vulnerability to nucleases, all of which may ultimately affect the lifespan of a miRNA. The birth and death of miRNAs have now come full circle; general features of miRNA biogenesis and degradation have been established. However, the regulatory mechanisms that govern transcription, processing and now destabilization of multitudes of different miRNAs are not yet fully defined. Determining how specific miRNAs are marked for death and identifying the assassins that do the job are vital challenges to understand the cause and consequence of dynamic changes in mature miRNA levels during development and disease.
ACKNOWLEDGMENTS
The authors thank D. Zisoulis for critical reading of the manuscript and our colleagues for helpful discussions. Z.S.K. is supported in part by a US National Institutes of Health Cellular and Molecular Graduate Student Training Grant, and research in the Pasquinelli laboratory is supported by grants from the US National Institutes of Health (GM071654-01) and the Keck, Searle, V, Emerald and Peter Gruber Foundations.
References
- 1.Griffiths-Jones S, Saini HK, van Dongen S & Enright AJ miRBase: tools for microRNA genomics. Nucleic Acids Res. 36, D154–D158 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hebert SS & De Strooper B Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 32, 199–206 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Latronico MV & Condorelli G MicroRNAs and cardiac pathology. Nat. Rev. Cardiol 6, 419–429 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Negrini M, Nicoloso MS & Calin GA MicroRNAs and cancer—new paradigms in molecular oncology. Curr. Opin. Cell Biol 21, 470–479 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Chuck G, Candela H & Hake S Big impacts by small RNAs in plant development. Curr. Opin. Plant Biol 12, 81–86 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Chen X Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol 25, 21–44 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis BN & Hata A Regulation of microRNA biogenesis: A miRiad of mechanisms. Cell Commun. Signal 7, 18 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winter J, Jung S, Keller S, Gregory RI & Diederichs S Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol 11, 228–234 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Chekulaeva M & Filipowicz W Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol 21, 452–460 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Thomson JM et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20, 2202–2207 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khvorova A, Reynolds A & Jayasena SD Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Schwarz DS et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Diederichs S & Haber DA Dual role for argonautes in microRNA processing and post-transcriptional regulation of microRNA expression. Cell 131, 1097–1108 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Grishok A et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34 (2001). [DOI] [PubMed] [Google Scholar]
- 15.O’Carroll D et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 21, 1999–2004 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaucheret H, Vazquez F, Crete P & Bartel DP The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu B et al. Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Yang Z, Yu B, Liu J & Chen X Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol 15, 1501–1507 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramachandran V & Chen X Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321, 1490–1492 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horwich MD et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol 17, 1265–1272 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Kirino Y & Mourelatos Z The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 13, 1397–1401 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito K et al. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev. 21, 1603–1608 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landgraf P et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morin RD et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 18, 610–621 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid JG et al. Mouse let-7 miRNA populations exhibit RNA editing that is constrained in the 5′-seed/cleavage/anchor regions and stabilize predicted mmu-let-7a–mRNA duplexes. Genome Res. 18, 1571–1581 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruby JG et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193–1207 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Ebhardt HA et al. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res. 37, 2461–2470 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azuma-Mukai A et al. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc. Natl. Acad. Sci. USA 105, 7964–7969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh T et al. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 23, 433–438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu S, Sun YH & Chiang VL Adenylation of plant miRNAs. Nucleic Acids Res. 37, 1878–1885 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee S & Grosshans H Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461, 546–549 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Gy I et al. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19, 3451–3461 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ballarino M et al. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol. Cell. Biol 29, 5632–5638 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morlando M et al. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol 15, 902–909 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buratowski S Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol 17, 257–261 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Doma MK & Parker R RNA quality control in eukaryotes. Cell 131, 660–668 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Weinmann L et al. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136, 496–507 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Gatfield D et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 23, 1313–1326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y et al. The role of PACT in the RNA silencing pathway. EMBO J. 25, 522–532 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Rooij E et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579 (2007). [DOI] [PubMed] [Google Scholar]
- 41.van Wolfswinkel JC et al. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139, 135–148 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Hagan JP, Piskounova E & Gregory RI Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol 16, 1021–1025 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heo I et al. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell 32, 276–284 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Heo I et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138, 696–708 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Lehrbach NJ et al. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol 16, 1016–1020 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones MR et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol 11, 1157–1163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin G & Keller W RNA-specific ribonucleotidyl transferases. RNA 13, 1834–1849 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y, Sinha K, Perumal K & Reddy R Effect of 3′ terminal adenylic acid residue on the uridylation of human small RNAs in vitro and in frog oocytes. RNA 6, 1277–1288 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chi SW, Zang JB, Mele A & Darnell RB Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan AA et al. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol 27, 549–555 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sood P, Krek A, Zavolan M, Macino G & Rajewsky N Cell-type–specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. USA 103, 2746–2751 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajasethupathy P et al. Characterization of small RNAs in aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 63, 803–817 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sethi P & Lukiw WJ Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci. Lett 459, 100–104 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Ciaudo C et al. Highly dynamic and sex-specific expression of microRNAs during early ES cell differentiation. PLoS Genet. 5, e1000620 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwang HW, Wentzel EA & Mendell JT A hexanucleotide element directs microRNA nuclear import. Science 315, 97–100 (2007). [DOI] [PubMed] [Google Scholar]