Abstract
Background
Mutations in GLI2 have been associated with holoprosencephaly (HPE), a neuroanatomic anomaly resulting from incomplete cleavage of the developing forebrain, and an HPE-like phenotype involving pituitary anomalies and polydactyly.
Objective
To characterise the genotypic and phenotypic findings in individuals with GLI2 variants and clarify clinical findings in individuals with loss-of-function mutations.
Methods
Through the National Institutes of Health and collaborating centres, ~400 individuals with HPE spectrum disorders, endocrine disorders or craniofacial anomalies were screened for GLI2 mutations. Results were combined with all published cases. We compared the clinical and molecular features of individuals with truncating mutations to individuals with variants of unknown significance (defined as not resulting in protein truncation, reported in normal controls and/or deemed unlikely to be pathogenic by functional prediction software).
Results
112 individuals with variants in GLI2 were identified, with 43 having truncating mutations. Individuals with truncating mutations were more likely to have both pituitary anomalies and polydactyly versus those with variants of unknown significance (p<0.0001 by Fisher’s exact test); only 1 of 43 had frank HPE. These individuals were more likely to have recognised penetrance (polydactyly or pituitary anomalies or both) than those without truncating mutations (p=0.0036 by Fisher’s exact test). A common facial phenotype was seen in individuals (with midface hypoplasia, cleft lip/palate and hypotelorism) with truncating mutations.
Conclusions
Individuals with truncating mutations in GLI2 typically present with pituitary anomalies, polydactyly and subtle facial features rather than HPE. This will be helpful in screening populations for GLI2 mutations and for counselling affected patients.
Trial registration
98-HG-0249/04-HG-0093.
INTRODUCTION
Holoprosencephaly (HPE) is the most common fore-brain anomaly and results from incomplete separation of the cerebral hemispheres and midline brain structures early in gestation. Mutations in GLI2 have been described as associated with a range of phenotypes. These phenotypes include pituitary anomalies and postaxial polydactyly, as well as classic HPE.1–3 As a result of literature-based descriptions, GLI2 testing is often performed in patients with isolated, frank HPE. However, we show that pathogenic mutations in GLI2 result in a recognisable and clinically distinct phenotype that only rarely extends to HPE but rather includes more commonly, pituitary abnormalities and/or polydactyly.
There are three Gli proteins that are involved in the Sonic hedgehog signalling network. These interacting molecules encode zinc-finger transcription factors. Gli1 and Gli2 have activating affects, in contrast to Gli3, which has a repressive effect on Shh pathway activity.4 Alterations in this pathway resulting in activation or repression lead to several identified human diseases, including various cancer syndromes and birth defects.5 Some of these birth defects include brain abnormalities classically associated with Sonic Hedgehog signalling. Gli2−/− mouse embryos have been shown to exhibit severe ventral patterning defects in the hindbrain. GLI2 activator function is thought to be required for expression of a class of genes encoding specific progenitor cells that will become motoneurons and interneurons in the ventral neural tube.6
Gli2 has also been shown to play a specific role in pituitary development early in gestation, with variable loss of normal pituitary development in Gli2-deficient animal models.7 Gli2 mutant mice embryos have an essentially normally patterned but smaller pituitary, the latter finding secondary to a defect in cell proliferation. In this model, the Gli2 mutant cells were also shown to be less likely to contribute specifically to the anterior pituitary, as they had a decreased number of anterior pituitary secreting hormone cells: specifically corticotropes, somatotropes and lactotropes.8 In addition, the you-too (yot) zebrafish mutant has loss-of-function mutations in gli2, and these mutants have abnormal patterning of the ventral central nervous system as well as anterior pituitary hypoplasia.9
Mutations in GLI3 have been associated with various syndromes such as Greig cephalopolysyndactyly syndrome,10 Pallister–Hall syndrome11 and other polydactyly syndromes.12 These syndromes can involve polydactyly, characteristic facial anomalies and hypothalamic abnormalities, and individuals with HPE as part of the phenotypic spectrum have been reported.5 Interestingly, Gli1 mutant mice seem to be normal, and there is no current evidence that GLI1 mutations result in Mendelian forms of human congenital malformations.4
In this study, we aimed to analyse the phenotypic findings in individuals with GLI2 variants based on the variants’ potential pathogenicity, with the hypothesis that bona fide loss-of-function mutations result in a distinct and recognisable condition that does not typically include classic HPE.
METHODS
Samples from approximately 400 individuals with HPE spectrum disorders and their relatives were collected over 18 years in our laboratory at NIH,3,13 and these samples were screened or analysed for variants in the GLI2 gene (NP_005261) under the National Human Genome Research Institute/NIH Institutional Review Board approved brain research protocol, with appropriate consent obtained from all research participants. Patients identified through the NIH study had sequencing performed by previously published methods and were also tested for mutations in the major genes known to be associated with HPE (SHH, ZIC2, SIX3 and TGIF).14 These individuals’ features comprised the entire HPE spectrum, ranging from severely affected fetuses to very mildly affected individuals. Clinical details were supplied by the referring clinicians, which included items such as a patient summary, photographs and radiological imaging. Patients not seen at NIH were ascertained through their respective IRB-approved protocols (with appropriate consent obtained from research participants), but were not uniformly screened for mutations in other HPE associated genes.
Published cases were ascertained through a PubMed/Medline search using the search terms: GLI2, HPE, holoprosencephaly. These published cases were derived from a variety of patient cohorts. Some of the cohorts specifically tested patients with HPE;2,3,13,15–17 cleft lip/palate18; pituitary hormone deficiencies1,19; and craniofacial anomalies.20 Only cases with a proven variant involving GLI2 were included for analysis. Cases with involvement of other genes or chromosomes (eg, due to a large microdeletion including nearby genes) were excluded so as to not confound possible results.
We evaluated variants and sought to identify individuals with variants for which there was strong evidence for pathogenicity. Those variants predicted to result in truncation of the protein include nonsense, frameshift and splice-site variants, and those with deletions of all or nearly the entire gene were also included in this category. Variants were further evaluated through dbSNP and the Exome Variant Server21 and were binned as variants of unknown significance if they appeared in those databases. As the pattern of inheritance resulting from pathogenic GLI2 mutations has been shown to involve incomplete penetrance, identifying variants in such public databases does not necessarily exclude a functional role nor involvement in disease processes, especially due to the paucity of phenotypic information. Finally, the remaining variants were evaluated using Polyphen2,22 a software-based functional prediction algorithm, and were binned as variants of unknown significance unless predicted to be ‘probably damaging’.
In summary, for the genotype–phenotype analysis, in order to be maximally conservative, we only considered variants to have high evidence for pathogenicity if they resulted in truncation of the predicted protein, were not found in public databases and were predicted to be ‘probably damaging’ through software prediction. For the purposes of our analyses, we call these ‘mutations’. All others were considered to be variants of unknown significance.
Statistical comparisons were made using Fisher’s exact test.
RESULTS
We describe 112 individuals from 65 independent kindreds with variants affecting the GLI2 gene. Thirty of these individuals (27%) have not been previously reported in the literature. The patients ascertained through the medical literature were described in publications from 2003 to the present.
27/65 probands had more than one individual within the family identified with the variant, although familial testing was not uniformly available. Of those with known inheritance, maternal inheritance was found in 23/45 (51%), paternal inheritance was found in 18/45 (40%) and 4/45 (9%) had de novo mutations.
MUTATIONS
There were 53 distinct variants identified. 35/53 (66%) were (predicted) missense variants, 7/53 (13%) were nonsense variants, 8/53 (15%) were frameshift variants, 1/53 (2%) was a splice site variant and 2/53 (4%) were whole-gene or near whole-gene deletions.
One kindred had an additional variant in PTCH, and another kindred had an additional variant in ZIC2.2,16
Table 1 shows families with more than one variant found within GLI2.
Table 1.
Families with more than one GLI2 variant
| Family number |
Members affected | GLI2 variants | Other information | Reference |
|---|---|---|---|---|
| 25 | Proband and father | c.2081_2084del, p.Leu694fs*722; c.1760C>T, p.Pro608Leu |
19 | |
| 40 | Proband and mother | c.3555delC, p.Tyr1186Thrfs*34; c.3351C>A, p.Pro1184Gln |
Previously Unpublished |
|
| 30 | Proband, sister, mother, 3 maternal uncles, 2 female cousins, maternal grandmother |
c.2362_2368del, p.Leu788fsX794; c.4332G>C, p.Met144Ile; c.4333 C>T, p. Leu1445Phe |
Both missense variants were heterozygotes and occurred after the predicted stop codon on the same allele |
19 |
| 45 | Proband and father | c.3723G>A, p.Met1241Ile; c.4453C>G, p. Pro1485Ala |
1 |
There were also four apparently unrelated individuals with the same two GLI2 variants (c.4054A>G, p.Met1352Val; c.4558G>A, p.Asp1520Asn)1 and another four apparently unrelated individuals with the same GLI2 variants (c.4332G>A, p.Met1444Ile; c.4333C>T, p.Leu1445Phe).1 For the last two groups of individuals with more than one variant in GLI2, information is not available regarding the cis/trans orientation of the variants (eg, from familial or other testing), therefore rendering further hypotheses about multiple interacting variants moot.
CLINICAL FEATURES
3/112 individuals (3%) were described as having HPE, although only one of them had a pathogenic mutation (using the criteria described above, as opposed to a variant of unknown significance) in GLI2—see below for more clinical details. One individual had a nonsense mutation (c.1486C>T; p.Arg496*) with a clinical description of ‘HPE findings’ but no imaging or detailed findings were available such that, after review of the original clinical data and discussion with the authors of that publication, there was evidence that the individual may in fact be more accurately described as having findings consistent with microform HPE (subtle facial differences without the neuroanatomical anomalies seen in frank HPE).3 Another individual had a missense variant (c.677G>A; p.Arg226His) and had semilobar HPE; however, this individual was also found to have a pathogenic mutation in ZIC2, in which mutations are well established as a cause of HPE.16 Finally, one patient had a GLI2 frameshift mutation (c.864_866delCC; His289Profs*61) and had documented semilobar HPE. This individual’s phenotype included microcephaly, cleft lip/palate and bilateral postaxial polydactyly.20
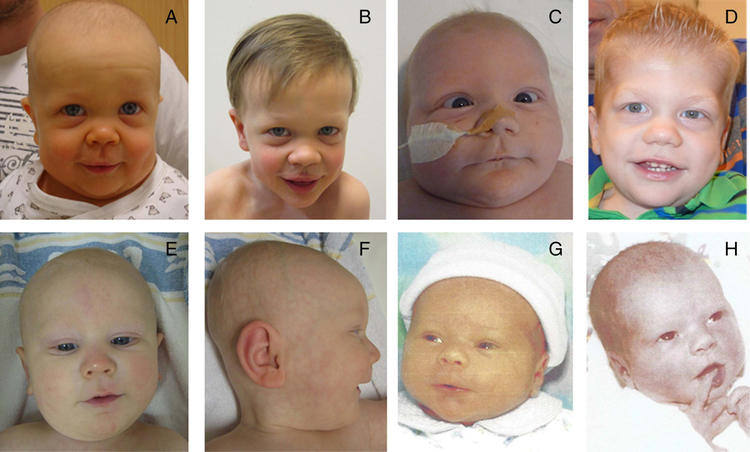
Patients with HPE with GLI2 variants were described as demonstrating typical facial features, with more severe facial features correlating with more severe brain abnormalities. These facial features include cyclopia, hypotelorism, proboscis, single nostril, flat nasal bridge, cleft lip and/or palate, iris coloboma and single central incisor.23 For our patients, 11/112 did not have facial features described in the medical literature. Of those with available clinical descriptions of facial features, 68/101 (67%) were described as being normal or non-dysmorphic. Of those with dysmorphic features reported, although not universal, the patients had a specific well-defined combination of facial features. Many of them had features of mild midface hypoplasia (13/101, 13%), cleft lip/palate (16/101, 16%) and/or hypotelorism (4/101, 4%). Specifically individuals with predicted loss of function mutations have these well-defined facial features, most often midface hypoplasia. Figure 1 shows four individuals with loss-of-function mutations. As shown for the individual for which there are both a neonatal and a later childhood photograph, the characteristic facial features become more obvious with age. It is important to note that midface hypoplasia (relative to older children and adults) can be seen in unaffected neonates as well. While one individual was described with a single nare, no other individuals had the more classic/severe HPE-related features such as cyclopia or proboscis.
Figure 1.
(A) Infant image of a patient with frameshift mutation in GLI2. (B) Same patient as (A) in early childhood. (C) Infant image of a patient with nonsense mutation in GLI2. (D) Same patient as (C) in early childhood. (E/F) Frontal and side views of infant with frameshift mutation in GLI2. (G) Infant with deletion of GLI2. (H) Mother of G (as an infant), with same deletion.
GENOTYPE–PHENOTYPE ANALYSIS
There is wide variability in the phenotype described in individuals with mutations in GLI2. After compiling all known cases of individuals with variants in the GLI2 gene, we compared the phenotypes of individuals with mutations predicted to lead to loss of function (such as nonsense or frameshift mutations, or large deletions) to those with variants of unknown significance. We specifically examined the presence of polydactyly and pituitary abnormalities both alone and in combination, as these features were frequently described in individuals with pathogenic mutations and were hypothesised to represent a core part of the phenotype. Online supplementary table S1 describes the phenotypes of all individuals with loss-of-function mutations, and online supplementary table S2 has more detailed information regarding all of the individuals included in this study.
98/112 individuals had information available regarding where polydactyly and pituitary abnormalities (eg, abnormal pituitary imaging or lab-based evidence of hormone deficiencies) were present. Of individuals with mutations predicted to result in protein truncation, 16/43 (37%) had both polydactyly and pituitary anomalies. Of those with non-truncating variants, 1/69 (1%) had both abnormalities. This difference was statistically significant (p<0.0001 by Fisher’s exact test).
102/112 individuals had information available regarding at least hand findings or pituitary abnormalities, though the available information varied, and it is possible that there are other, more subtle findings not uniformly reported. Despite this, we examined penetrance in these individuals and found that those with predicted truncating mutations had hand abnormalities, pituitary abnormalities or both in 36/43 (84%) individuals. Those with non-truncating variants had penetrance in 39/69 (57%) with 36/39 (92%) of them involving pituitary-only abnormalities. This difference was statistically significant (p=0.0036 by Fisher’s exact test).
Table 2 shows the number of individuals with pituitary abnormalities and hand abnormalities in those with truncating mutations and those with non-truncating variants.
Table 2.
Prevalence of pituitary anomalies and/or polydactyly associated with truncating and non-truncating variants
| Type of variant | Pituitary abnormalities only | Polydactyly only | Pituitary abnormalities and polydactyly |
No abnormalities | Either pituitary abnormalities, polydactyly, or both |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Truncating variants |
Non-truncating variants |
Truncating variants |
Non-truncating variants |
Truncating variants |
Non-truncating variants |
Truncating variants |
Non-truncating variants |
Truncating variants |
Non-truncating variants |
|
| Number with abnormality among individuals with available information |
7/29 (24%) | 36/62 (58%) | 13/39 (33%) | 2/64 (3%) | 16/26 (62%) | 1/58 (2%) | 4/26 (15%) | 23/58 (40%) | 36/42 (86%) | 39/68 (57%) |
| Number for which information was available |
29/43 (67%) | 62/69 (90%) | 39/43 (91 %) | 64/69 (93%) | 26/43 (60%) | 58/69 (84%) | 26/43 (60%) | 58/69 (84%) | 42/43 (98%) | 68/69 (99%) |
As information regarding polydactyly and pituitary abnormalities was not uniformly available for all individuals, the top row represents the prevalence of the abnormality among individuals with available information (ie, the denominator represents the number of individuals with these variants with available information). The second row demonstrates the availability of this clinical information among individuals in the cohort.
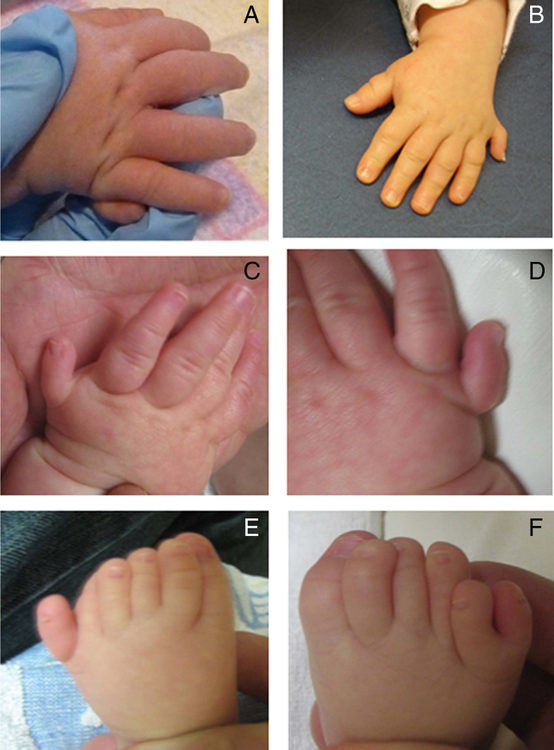
Of note, only two individuals with a non-truncating variant had polydactyly and one of them was the sole individual with preaxial polydactyly—all others with polydactyly had postaxial polydactyly. Figure 2 shows the postaxial polydactyly of both the hands and feet of three different individuals with truncating mutations in GLI2.
Figure 2.
(A) Image of right hand of an infant with a nonsense mutation in GLI2. (B) Image of left hand of a child with a frameshift mutation in GLI2. (C) Image of left hand of an infant with a frameshift mutation in GLI2. (D) Image of the right hand of the same infant (C). (E) Image of the left foot of the same infant (C). (F) Image of the right foot of the same infant (C).
DISCUSSION
GLI2 mutations have been described as associated with HPE for approximately the last decade. For this reason, GLI2 testing is often performed as part of a panel of tests for individuals with HPE. There are numerous ‘red herrings’ that may explain the supposition that mutations in GLI2 relatively frequently result in classic HPE. One reason may involve the identification of a GLI2 variant of unknown significance in a patient where an unidentified mutation (in a different gene) may account for the patient’s phenotype. For example, individual 5a, who had a GLI2 missense variant, had semilobar HPE; however, this patient was also (later) found to have a truncating mutation in the gene ZIC2, the latter of which has strong evidence as being the cause of HPE in this individual.24 Proband 10a was described as having semilobar HPE, and though there is not an alternate genetic explanation, it is suspected that another (as yet unknown) mutation may account for the neurological phenotype. The sister of proband 17 had classic, alobar HPE. However, HPE is overall not uncommon,25,26 and molecular testing on the deceased sister was never performed. Proband 20a was described as having HPE findings, but there was no further information given and the ‘HPE findings’ were based on facial features without any imaging available making the assignment of frank (neuroanatomical) HPE somewhat suspect.
This is not to say that GLI2 mutations do not result in a spectrum of severity. However, frank HPE does not appear to be a common part of this spectrum. Even severely affected patients (such as proband 12a in the online supplementary table) do not in fact appear to have HPE. In fact, the most severe neuroanatomical finding reliably reported appears to be agenesis of the genu of the corpus collosum, along with an abnormal cerebral periventricular venous system and abnormal gyri. Callosal anomalies are frequently described in HPE, but in HPE, they occur in conjunction with additional evidence of midline non-separation. Vaaralahti et al performed a study looking at individuals with Kallmann syndrome, which involves congenital hypogonadotropic hypogonadism and decreased or absent sense of smell, and reported one individual with a missense mutation in GLI2 (c.2509G>A; p.Glu837Lys). This demonstrates overlap of pituitary anomalies within two different genetic conditions.27 As in phenotypes related to GLI3 mutations, HPE may occur in individuals with GLI2 mutations in rare instances (in which HPE is somewhat more common than the occurrence of HPE in the general population) perhaps due to a multifactorial disease pattern involving multiple interacting genetic and environmental factors.
There are a few larger deletions including the GLI2 gene reported resulting in individuals with pituitary anomalies and/or polydactyly. Kevelam et al reported an individual with a 1.3 Mb submicroscopic heterozygous deletion in 2q14.2, which includes GLI2 along with four other genes. This individual had a bilateral cleft lip and palate and abnormal pituitary gland formation, along with panhypopituitarism and normal psychomotor development. He also had heterotaxy of the abdominal organs, thought to be related to the additional deletion of EPB4.1L5, which has been hypothesised as a candidate gene for heterotaxy.28 Gustavsson et al reported an individual with a balanced translocation with the karyotype 46,XY,t(2;20)(q21;p13) and a submicroscopic deletion on chromosome 2q14.2-q22.1, including 43 known genes, one of which was GLI2. This individual had hypospadias, postaxial polydactyly of the left hand, double-left-sided ureters and undescended testes, exotropia and amblyopia of the left eye, and growth hormone deficiency, as well as a history of deep vein thrombosis related to lupus anticoagulant factor.29 Both of these individuals have phenotypic agreement with our prediction of pituitary anomalies and/or postaxial polydactyly as being directly related to the ‘core GLI2 mutation phenotype’. It is possible, however, that given the complexity of the deletions in these individuals and the several other genes involved, their phenotypes could be secondary to other causes.
We found that truncating mutations may be identified along the length of the GLI2 gene and that there was no evidence for correlation between the location of these truncating mutations relative to known functional domains and the patient phenotype. As the activation domain is located at the distal portion of the molecule, all mutations leading to truncation would be expected to behave similarly on a functional level. That is, the encoded molecule would lose activation activity regardless of where the truncation occurs, as well as if a mutation fell within the zinc finger domain, as in this latter instance, it would then not be able to bind to targets. Our data support the presence of a well-defined phenotype in individuals with pathogenic GLI2 mutations. This phenotype includes anterior pituitary anomalies (as opposed to the posterior pituitary insufficiency frequent in typical HPE) and postaxial polydactyly. Although not all individuals with predicted pathogenic mutations have both findings, these findings are only described together once among all of the individuals with GLI2 variants of unknown significance. It is also possible that some individuals described here were not evaluated extensively enough to detect possible subtle hand findings or minor pituitary anomalies, which may further support our findings. We attempted to determine the degree of pathogenicity conservatively by our inclusion criteria for pathogenic mutations (see ‘Methods’), but we readily admit that sufficient bench-based functional data to better test our hypotheses are lacking.
The findings described here are relevant for several reasons. First, GLI2 is a large and polymorphic gene and rare familial variants are common. Sequencing any gene, especially one as polymorphic as GLI2, may result in the frequent identification of variants of unknown significance, which can make interpreting molecular findings difficult. Using the Exome Variant database, we identified 123 missense polymorphisms, one splice site change and one frameshift in GLI2. When comparing this with other major genes in which mutations are known to cause HPE, there are significantly more variants within GLI2, with SHH having five reported missense variants, ZIC2 having five, SIX3 having seven and TGIF having none (though findings related to TGIF may largely reflect sequencing issues related to this particular locus) (evs.gs.washington.edu/EVS/; 25 November 2013).
Another source of difficulty for our analysis is the admittedly important ascertainment bias. Individuals included in this study are derived from a variety of different clinical settings with a fairly large proportion coming from an endocrine clinic; this can be seen in the online supplementary table. This bias may at least partially explain why so many of the individuals with non-truncating variants had pituitary abnormalities, as that was the original reason why sequencing was performed. Future studies with whole exome or genome sequencing may expand the phenotype of patients with GLI2 mutations as well as under-cover variants in other genes that modulate the phenotype.
These findings also underscore the importance of the evaluation of documented genetic variants in terms of potential causation. This manuscript, like others, begs for further in-depth functional analyses of identified GLI2 variants in order to draw stronger conclusions about the consequences of variants.
Finally, in the age of high-throughput sequencing, in which sequencing a particular gene (or many genes simultaneously) becomes increasingly easy, it will be important to have a focused and phenotype-centred approach at least in the analysis phase so that more answers are given rather than questions raised. Specific to GLI2, our results suggest that an approach starting in the endocrinology clinic with close follow-up in the traditional dysmorphology/neurogenetic clinic may result in a higher yield of etiological explanations.
Supplementary Material
Acknowledgements
This research was supported by the Division of Intramural Research, National Human Genome Research Institute, National Institutes of Health, Department of Health and Human Services, USA. Pertaining to KAB, the views expressed in this publication are those of the author and do not reflect the official policy or position of the Department of the Army, Department of Defense, or the US Government. Pertaining to MW, the views expressed in this article are those of the author and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense or the United States Government. The authors would like to express their gratitude to the patients and families involved.
Footnotes
Competing interests None.
Patient consent Obtained.
Ethics approval Institutional Review Board.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.França MM, Jorge AAL, Carvalho LRS, Costalonga EF, Otto AP, Correa FA, Mendonca BB, Arnhold IJ. Relatively High Frequency of Non-synonymous GLI2 Variants in Patients with Congenital Hypopituitarism without Holoprosencephaly. Clin Endocrinol (Oxf) 2013;78:551–7. [DOI] [PubMed] [Google Scholar]
- 2.Rahimov F, Ribeiro LA, de Miranda E, Richieri-Costa A, Murray JC. GLI2 mutations in four Brazilian patients: how wide is the phenotypic spectrum? Am J Med Genet A 2006;140:2571–6. [DOI] [PubMed] [Google Scholar]
- 3.Roessler E, Du Y-Z, Mullor JL, Casas E, Allen WP, Gillessen-Kaesbach G, Roeder ER, Ming JE, Ruiz i Altaba A, Muenke M. Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features. Proc Natl Acad Sci USA 2003;100:13424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruiz i Altaba A, Palma V, Dahmane N. Hedgehog-Gli signalling and the growth of the brain. Nat Rev Neurosci 2002;3:24–33. [DOI] [PubMed] [Google Scholar]
- 5.Villavicencio EH, Walterhouse DO, Iannaccone PM. The sonic hedgehog-patched-gli pathway in human development and disease. Am J Hum Genet 2000;67:1047–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lebel M, Mo R, Shimamura K, Hui CC. Gli2 and Gli3 play distinct roles in the dorsoventral patterning of the mouse hindbrain. Dev Biol 2007;302:345–55. [DOI] [PubMed] [Google Scholar]
- 7.Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 2000;127:1593–605. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Martin JF, Bai CB. Direct and indirect requirements of Shh/Gli signaling in early pituitary development. Dev Biol 2010;348:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlstrom RO, Talbot WS, Schier AF. Comparative synteny cloning of zebrafish you-too: mutations in the Hedgehog target gli2 affect ventral forebrain patterning. Genes Dev 1999;13:388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vortkamp A, Gessler M, Grzeschik KH. GLI3 zinc-finger gene interrupted by translocations in Greig syndrome families. Nature 1991;352:539–40. [DOI] [PubMed] [Google Scholar]
- 11.Kang S, Graham JM, Olney AH, Biesecker LG. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat Genet 1997;15:266–8. [DOI] [PubMed] [Google Scholar]
- 12.Radhakrishna U, Bornholdt D, Scott HS, Patel UC, Rossier C, Engel H, Bottani A, Chandal D, Blouin JL, Solanki JV, Grzeschik KH, Antonarakis SE. The phenotypic spectrum of GLI3 morphopathies includes autosomal dominant preaxial polydactyly type-IV and postaxial polydactyly type-A/B; No phenotype prediction from the position of GLI3 mutations. Am J Hum Genet 1999;65:645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roessler E, Ermilov AN, Grange DK, Wang A, Grachtchouk M, Dlugosz AA, Muenke M. A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum Mol Genet 2005;14:2181–8. [DOI] [PubMed] [Google Scholar]
- 14.Solomon BD, Bear KA, Wyllie A, Keaton AA, Dubourg C, David V, Mercier S, Odent S, Hehr U, Paulussen A, Clegg NJ, Delgado MR, Bale SJ, Lacbawan F, Ardinger HH, Aylsworth AS, Bhengu NL, Braddock S, Brookhyser K, Burton B, Gaspar H, Grix A, Horovitz D, Kanetzke E, Kayserili H, Lev D, Nikkel SM, Norton M, Roberts R, Saal H, Schaefer GB, Schneider A, Smith EK, Sowry E, Spence MA, Shalev SA, Steiner CE, Thompson EM, Winder TL, Balog JZ, Hadley DW, Zhou N, Pineda-Alvarez DE, Roessler E, Muenke M. Genotypic and phenotypic analysis of 396 individuals with mutations in Sonic Hedgehog. J Med Genet 2012;49:473–9. [DOI] [PubMed] [Google Scholar]
- 15.Solomon BD, Pineda-Alvarez DE, Gropman AL, Willis MJ, Hadley DW, Muenke M. High Intellectual Function in Individuals with Mutation-Positive Microform Holoprosencephaly. Mol Syndromol 2012;3:140–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wannasilp N, Solomon BD, Warren-Mora N, Clegg NJ, Delgado MR, Lacbawan F, Hu P, Winder TL, Roessler E, Muenke M. Holoprosencephaly in a family segregating novel variants in ZIC2 and GLI2. Am J Med Genet A 2011;155A:860–4. [DOI] [PubMed] [Google Scholar]
- 17.Richieri-Costa A, Ribeiro LA. Holoprosencephaly-like phenotype: clinical and genetic perspectives. Am J Med Genet A 2006;140:2587–93. [DOI] [PubMed] [Google Scholar]
- 18.Vieira AR, Avila JR, Daack-Hirsch S, Dragan E, Félix TM, Rahimov F, Harrington J, Schultz RR, Watanabe Y, Johnson M, Fang J, O’Brien SE, Orioli IM, Castilla EE, Fitzpatrick DR, Jiang R, Marazita ML, Murray JC. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet 2005;1:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.França MM, Jorge AAL, Carvalho LRS, Costalonga EF, Vasques GA, Leite CC, Mendonca BB, Arnhold IJ. Novel heterozygous nonsense GLI2 mutations in patients with hypopituitarism and ectopic posterior pituitary lobe without holoprosencephaly. J Clin Endocrinol Metab 2010;95:E384–91. [DOI] [PubMed] [Google Scholar]
- 20.Bertolacini CDP, Ribeiro-Bicudo LA, Petrin A, Richieri-Costa A, Murray JC. Clinical findings in patients with GLI2 mutations—phenotypic variability. Clin Genet 2012;81:70–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Exome Variant Server NHLBI GO Exome Sequencing Project (ESP), Seattle, WA. http://evs.gs.washington.edu/EVS/ (accessed Nov 2013).
- 22.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen MM. Holoprosencephaly: clinical, anatomic, and molecular dimensions. Birth Defects Res Part A Clin Mol Teratol 2006;76:658–73. [DOI] [PubMed] [Google Scholar]
- 24.Solomon BD, Lacbawan F, Mercier S, Clegg NJ, Delgado MR, Rosenbaum K, Dubourg C, David V, Olney AH, Wehner LE, Hehr U, Bale S, Paulussen A, Smeets HJ, Hardisty E, Tylki-Szymanska A, Pronicka E, Clemens M, McPherson E, Hennekam RC, Hahn J, Stashinko E, Levey E, Wieczorek D, Roeder E, Schell-Apacik CC, Booth CW, Thomas RL, Kenwrick S, Cummings DA, Bous SM, Keaton A, Balog JZ, Hadley D, Zhou N, Long R, Vélez JI, Pineda-Alvarez DE, Odent S, Roessler E, Muenke M. Mutations in ZIC2 in human holoprosencephaly: description of a novel ZIC2 specific phenotype and comprehensive analysis of 157 individuals. J Med Genet 2010;47:513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsunaga E, Shiota K. Holoprosencephaly in human embryos: epidemiologic studies of 150 cases. Teratology 1977;16:261–72. [DOI] [PubMed] [Google Scholar]
- 26.Leoncini E, Baranello G, Orioli IM, Annerén G, Bakker M, Bianchi F, Bower C, Canfield MA, Castilla EE, Cocchi G, Correa A, De Vigan C, Doray B, Feldkamp ML, Gatt M, Irgens LM, Lowry RB, Maraschini A, Mc Donnell R, Morgan M, Mutchinick O, Poetzsch S, Riley M, Ritvanen A, Gnansia ER, Scarano G, Sipek A, Tenconi R, Mastroiacovo P. Frequency of holoprosencephaly in the International Clearinghouse Birth Defects Surveillance Systems: searching for population variations. Birth Defects Res Part A Clin Mol Teratol 2008;82:585–91. [DOI] [PubMed] [Google Scholar]
- 27.Vaaralahti K, Raivio T, Koivu R, Valanne L, Laitinen EM, Tommiska J. Genetic Overlap between Holoprosencephaly and Kallmann Syndrome. Mol Syndromol 2012;3:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kevelam SHG, van Harssel JJT, van der Zwaag B, Smeets HJ, Paulussen AD, Lichtenbelt KD. A patient with a mild holoprosencephaly spectrum phenotype and heterotaxy and a 1.3 Mb deletion encompassing GLI2. Am J Med Genet A 2012;158A:166–73. [DOI] [PubMed] [Google Scholar]
- 29.Gustavsson P, Schoumans J, Staaf J, Jönsson G, Carlsson F, Kristoffersson U, Borg A, Nordenskjöld M, Dahl N. Hemizygosity for chromosome 2q14.2-q22.1 spanning the GLI2 and PROC genes associated with growth hormone deficiency, polydactyly, deep vein thrombosis and urogenital abnormalities. Clin Genet 2006;69:441–3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.