Abstract
Coding variants in the triggering receptor expressed on myeloid cells 2 (TREM2) are associated with late onset Alzheimer’s disease (AD). We demonstrate that amyloid plaque seeding is increased in the absence of functional Trem2. Increased seeding is accompanied by decreased microglial clustering around newly seeded plaques and reduced plaque associated Apolipoprotein E (ApoE). Reduced ApoE deposition in plaques is also observed in brains of AD patients carrying TREM2 coding variants. Proteomic analyses and microglia depletion experiments revealed microglia as one origin of plaque associated ApoE. Longitudinal amyloid small animal positron emission tomography demonstrates accelerated amyloidogenesis in Trem2 loss of function mutants at early stages, which progressed at a lower rate with aging. These findings suggest that in the absence of functional Trem2 early amyloidogenesis is accelerated due to reduced phagocytic clearance of amyloid seeds despite reduced plaque associated ApoE.
Keywords: Alzheimer’s disease, amyloid plaque seeding, ApoE, microglia, neurodegeneration, TREM2
In many neurodegenerative disorders, misfolded amyloidogenic proteins propagate and spread throughout the brain in a disease characteristic manner reminiscent to prion-like disorders1. Seeding and spreading can be experimentally induced by intracerebral injection of homogenates derived from brains of humans with neurodegenerative disorders or respective mouse models2. Although little is known about the cellular mechanisms controlling and modifying prion-like propagation, it is conceivable that amyloid β-peptide (Aβ) clearance may be involved. Microgliosis is well known since a long time as a disease overarching pathological phenomenon characteristic for almost all neurodegenerative disorders3. The recent identification of coding variants in genes, which are selectively expressed in microglial cells and their genetic association with late onset Alzheimer’s disease (AD) and other neurodegenerative disorders, further pinpoint a pivotal disease modifying role of microglia4, 5. Among these microglial expressed disease associated genes, sequence variants in the gene encoding the triggering receptor expressed on myeloid cells 2 (TREM2) are associated with an extraordinarily increased risk for AD comparable to that conferred by the single Apolipoprotein E (APOE) ε4 allele6, 7. Trem2 is functionally required for a wide variety of important cellular functions including chemotaxis8, maintenance of energy metabolism9, 10, engulfment of dead cells8 and phagocytosis of Aβ fibrils11. Recent findings indicated that microglia switch from a homeostatic state to a neurodegenerative phenotype in several different disease models by suppressing their homeostatic mRNA profile and increasing a disease associated transcriptional profile12–14. Trem2 and ApoE are both robustly upregulated in microglia responding to brain lesions13, 14. Moreover, ApoE is a major constituent of amyloid plaques and ApoE promotes their aggregation and deposition15–17. Furthermore, anti-ApoE immunotherapy inhibits amyloid accumulation and deposition further supporting a role of ApoE in Aβ aggregation and clearance18. Conflicting data on the potential role of Trem2 in amyloid metabolism have been explained by the different model systems used, the stage of neuropathology and the artificially increased production and rapid deposition of Aβ in transgenic mouse models highly overexpressing the amyloid precursor protein (APP) along with autosomal dominant APP and presenilin mutations5, 19–22. To elucidate the role of Trem2 at the earliest accessible time point in the amyloid cascade, we used an in vivo seeding paradigm and performed intra-hippocampal injections of Aβ-containing brain extracts into APPPS1 transgenic mice that previously have been shown to robustly induce Aβ plaque formation in vivo 2. By additionally analyzing non-experimentally seeded cortical Aβ plaques, we could directly compare the effect of Trem2 on more mature plaques within the same mice. Furthermore, we used longitudinal small animal positron tomography (μPET) imaging to monitor microglial activation and the kinetics of amyloid deposition up to an age of twelve months. Finally, we investigated microglial clustering and ApoE deposition in amyloid plaques in loss of Trem2 function mice as well as in AD cases with TREM2 coding variants.
Results
Increased amyloid plaque seeding in the absence of functional Trem2
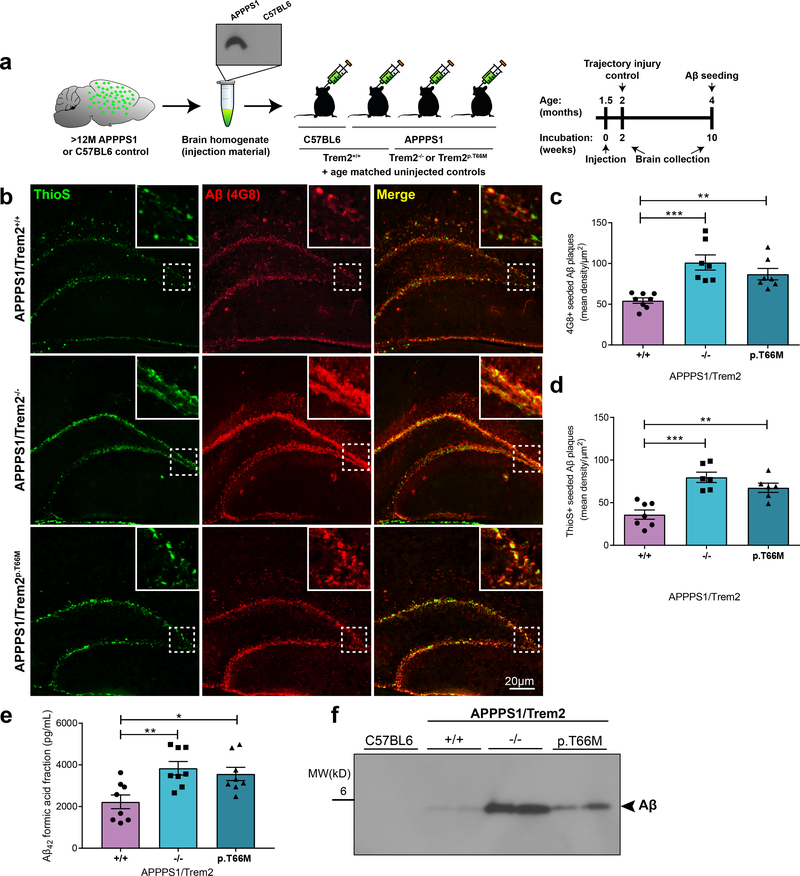
Amyloid plaques can be induced experimentally in vivo by a prion-like mechanism via inoculation of Aβ-containing homogenates in transgenic mice. Intracerebral injection of Aβ-rich brain extracts from Alzheimer’s disease patients or from aged APP transgenic mice promotes early formation and seeding of plaques 2, 23. Compared to non-experimentally seeded APP transgenic mice, the seeding model provides the unique opportunity to study plaque formation within a very defined and early time window with minimal variation from one mouse to another 2, 23. In contrast, uninjected non-seeded APP mice do not develop any seeded plaques and have an unpredictable onset of plaque formation at young age. Therefore to examine the very early effects of loss of Trem2 function on the premature formation of Aβ plaques as well as on the capacity of microglia to remove Aβ seeds, we stereotactically injected brain homogenates derived from APP transgenic mice expressing APP695KM670/671NL and PSEN1L166P (APPPS1 mice)24 into hippocampi of six weeks old APPPS1/Trem2+/+, APPPS1/Trem2−/− 25, or APPPS1/Trem2p.T66M 10 mice as well as non-transgenic C57BL6 littermates (Fig. 1a; Supplementary Fig. 1a). Ten weeks later, at a time point when normally little to no plaques are observed within the hippocampus (Supplementary Fig. 1a & b)2, mice were sacrificed and brains were prepared for immunohistochemistry and protein analysis. No seeded plaques were observed within the hippocampi of non-transgenic C57BL6 mice (Supplementary Fig. 1a & b), two weeks post injection controls (Supplementary Fig. 1c), or uninjected APPPS1/Trem2+/+, APPPS1/Trem2−/− or APPPS1/Trem2p.T66M mice (Supplementary Fig. 1d). In contrast, intense amyloid plaque seeding was observed in the subgranular layer of the hippocampi of APPPS1/Trem2+/+ mice upon injection of Aβ containing brain homogenates as previously published 2, 23 (Fig. 1b; Supplementary Fig. 1a & b). No seeding was observed upon injection of lysates from Aβ-free C57BL6 mice (Supplementary Fig. 1a & b). As previously described by Meyer-Luehmann et al.2, seeded amyloid plaques had a coarse and punctate appearance (Fig. 1b & Supplementary Fig. 1a) in the presence of Trem2. In contrast, in the absence of Trem2, the area occupied by seeded amyloid plaques not only increased (Fig. 1b), but also showed a more diffuse 4G8 staining as well as less separation of individual ThioS-positive fibrillar Aβ puncta (Fig. 1b). While all seeded amyloid plaques were detected immunopositively by 4G8 antibody, ThioS stained the dense cored plaques only. Although the T66M mutation has been strongly linked to Nasu-Hakola disease and FTD-like syndrome rather than AD26, we used the APPPS1/Trem2p.T66M model to investigate Trem2 dependent microglial effects on amyloid seeding in a second independent loss of function model10, 11. Similar findings were obtained when we injected Aβ containing brain homogenates in hippocampi of APPPS1 mice endogenously expressing the Trem2 p.T66M loss of function mutation (APPPS1/Trem2p.T66M)10 (Fig. 1b). Quantitative image analysis, Aβ42-ELISA as well as western blotting confirmed increased amyloidogenesis in the absence of functional Trem2 (Fig. 1c-f). These findings show that Trem2 dependent microglial functions limit early amyloidogenesis.
Figure 1. Increased amyloidogenesis upon loss of Trem2 function.
(a) Schematic illustration of the study design and timeline. (b) In contrast to APPPS1/Trem2+/+, Trem2 loss of function mice (APPPS1/Trem2−/− and APPPS1/Trem2p.T66M) show increased amyloid seeding in the dentate gyrus at four months when injected with APPPS1 brain homogenates. Insets show seeding pathology at higher magnification as indicated by the area in dotted white box. (c) Mice were analyzed for 4G8-positive immunoreactivity (n+/+=8 mice, n−/−=7 mice, np.T66M=7 mice; F2,19=13.13, p=0.0003) and (d) ThioS-positive Aβ deposits (n+/+=7 mice, n−/−=6 mice, np.T66M=6 mice; F2,16=16.73, p=0.0001). (e) Aβ42 levels quantified in formic acid fractions using Meso-Scale discovery electrochemiluminescence assay confirm increased amyloid seeding in APPPS1/Trem2−/− and APPPS1/Trem2p.T66M mice compared to APPPS1/Trem2+/+ mice (n=8 mice/genotype; F2,21=9.016, p=0.0015). (f) Immunoblotting with the anti-Aβ antibody 6E10 demonstrates increased Aβ in the formic acid fraction of APPPS1/Trem2−/− and APPPS1/Trem2p.T66M as compared to APPPS1/Trem2+/+ mice. Western blots were independently repeated five times to analyze n=10 mice/genotype. Full image of immunoblots are shown in Supplementary Fig. 7. Data represent mean ± SEM. One-way ANOVA, Dunnetts’s post hoc analysis; *p<0.05, **p<0.005, ***p<0.0005.
Reduced microglial clustering around seeded plaques in the absence of functional Trem2
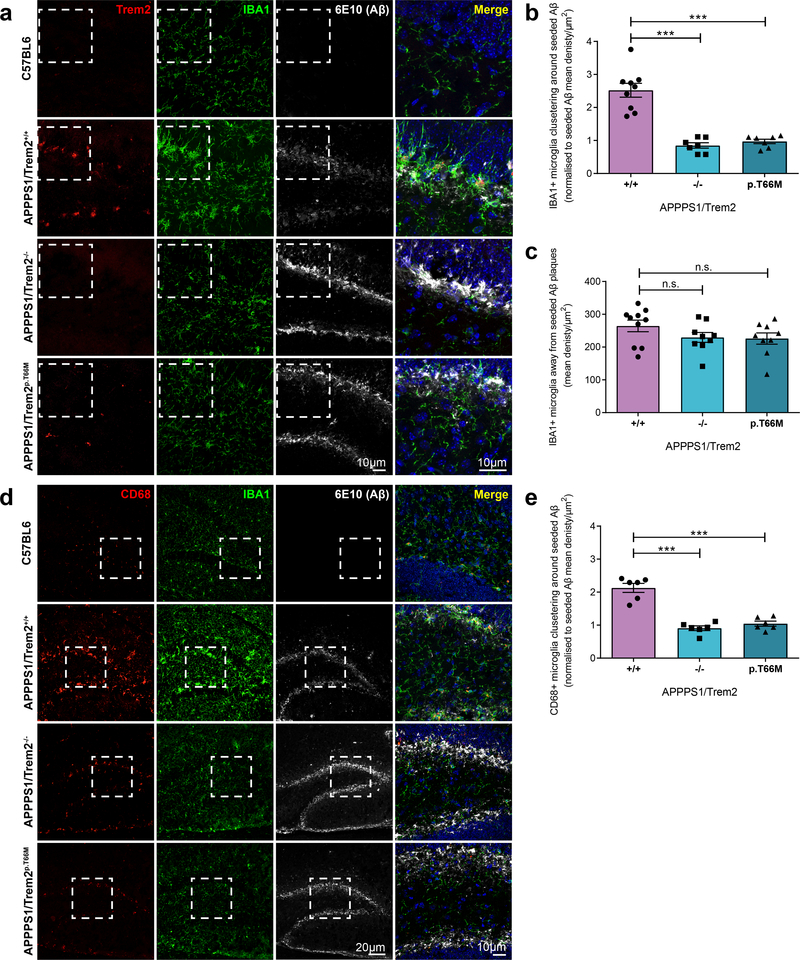
Since chemotaxis and plaque association of microglia is disturbed upon loss of Trem2 function8, 20, 27, we next investigated clustering of microglia around newly seeded plaques. In C57BL6 control mice injected with APPPS1 mouse brain homogenate, microglia were randomly distributed throughout the hippocampus and microglial Trem2 expression was below the detection limit (Fig. 2a). In contrast, microglia clustered around seeded plaques upon injection of Aβ containing brain homogenates into APPPS1/Trem2+/+ mice and increased Trem2 expression (Fig. 2a). Moreover, microglia clustering was severely reduced in APPPS1/Trem2−/− or APPPS1/Trem2p.T66M mice (Fig. 2a & b), although the density of IBA1-positive microglia outside the seeded plaque laden dentate gyrus was not significantly different (Fig. 2c). Clustering of microglia around seeded plaques was accompanied by increased CD68 staining, a marker for lysosomal activity, which was significantly reduced in APPPS1/Trem2−/− or APPPS1/Trem2p.T66M mice (Fig. 2d & e). Contrary to Trem2 loss of function mice, intracellular 6E10-positive Aβ staining was observed in plaque associated IBA1, Trem2 and CD68-positive microglia in APPPS1/Trem2+/+ mice, indicating microglial engulfment and phagocytosis of amyloid plaques (Fig. 2a & d).
Figure 2. Reduced microglial clustering, CD68 and Trem2 expression in microglia around newly seeded plaques in Trem2 loss of function mice.
(a) First panel: Random distribution of IBA1-positive microglia in C57BL6 mice. Second panel: Clustering of IBA1 and Trem2-positive microglia around seeded plaques in the hippocampus of APPPS1/Trem2+/+. Third and fourth panel: Loss of Trem2 function reduces microglia clustering around seeded plaques. (b) IBA1-positive microglia clustering around seeded amyloid plaques (n+/+=9 mice, n−/−=7 mice, np.T66M=7 mice, F2,20=38.87, p=1.5E-7) normalized to seeded amyloid pathology shown in Fig. 1c. (c) IBA1 immunoreactivity quantified in the hippocampus excluding seeded dentate gyrus (n+/+=10 mice, n−/−=9 mice, np.T66M=9 mice, F2,25=1.657, p=0.2109) (d) First panel: CD68 expression is at the detection limit in C57BL6 mice. Second panel: Increased CD68 staining in IBA1-positive microglia around seeded plaques in the presence of functional Trem2. Third and fourth panel: reduced CD68 expression in microglia around seeded plaques in the absence of functional Trem2. (e) Quantification of CD68-positive microglia in the seeded dentate gyrus (n=6 mice/genotype, F2,15=44.87, p=4.6E-7) normalized to seeded amyloid pathology shown in Fig. 1a. Dotted white boxes indicate the area in each staining that is merged and shown at higher magnification. Data represent mean ± SEM. One-way ANOVA, Dunnett’s post hoc analysis; n.s. p>0.05; ***p<0.0005.
In line with previous data9, 19, 20 microglia also clustered around non-experimentally seeded cortical Aβ plaques (Supplementary Fig. 2a & b), and exhibited a robust increase of Trem2 (Supplementary Fig. 2a) and CD68 expression (Supplementary Fig. 2c). Again, clustering of microglia as well as CD68 expression were both reduced in Trem2 loss of function mice (Supplementary Fig. 2a-d), whereas the density of IBA1-positive microglia in a non-plaque bearing area such as the thalamic nuclei was not significantly different (Supplementary Fig. 2e).
Loss of Trem2 function changes the kinetics of amyloidogenesis
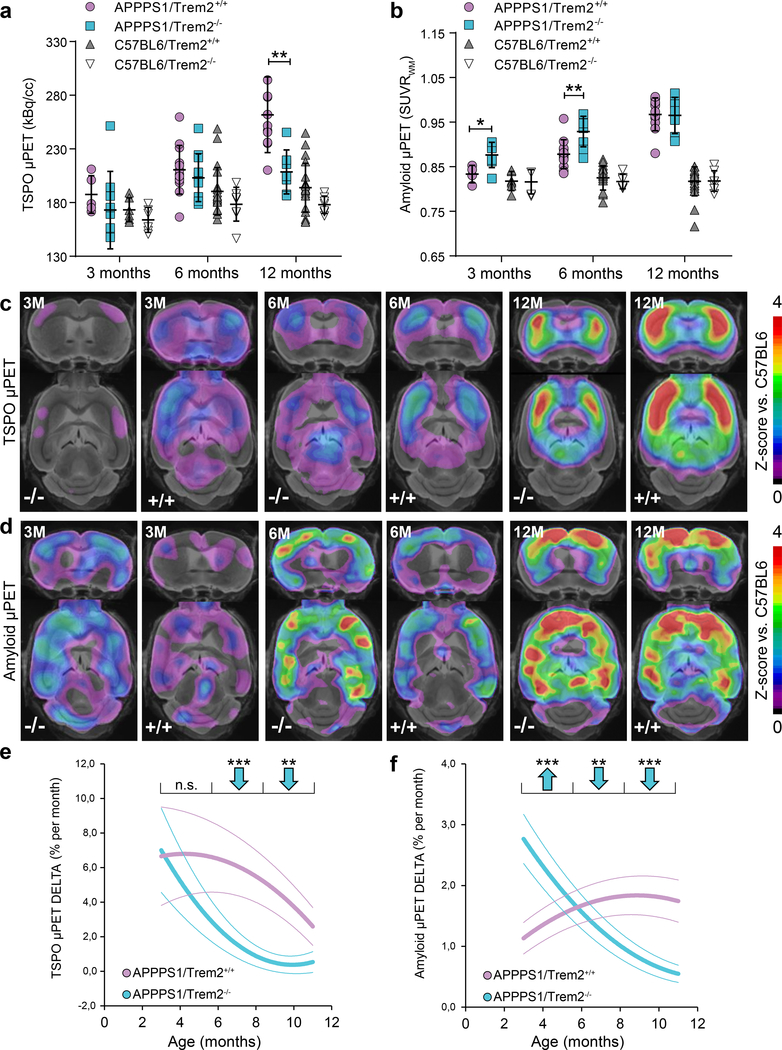
Conventional immunohistochemical analyses of brain region specific amyloid plaque burden in Trem2 deficient APP transgenic mice at different ages have yielded conflicting results5, 19–22. Therefore, to longitudinally investigate if amyloid plaque accumulation is affected by Trem2 loss of function during disease development in the entire brain, we performed dual-tracer longitudinal μPET imaging using tracers for amyloidogenesis (Florbetaben) and microglia activation (TSPO)28 (Fig. 3 and Supplementary Fig. 3). APPPS1 as well as C57BL6 mice with and without Trem2 expression were subjected to μPET imaging from three to twelve months of age. Both μPET tracers demonstrated their feasibility to track changes over time in individual mice (Supplementary Fig. 3a-d). However, due to limited resolution μPET was not appropriate for in vivo assessment of small hippocampal plaques in the seeding model. Additionally, individual μPET data of each imaging time point were validated by corresponding immunohistochemistry, which gave strong correlations between x34 staining and the amyloid μPET signal as well as between CD68 staining and the TSPO μPET signal (Supplementary Fig. 3e-l).
Figure 3. Increased amyloidogenesis in young Trem2 deficient mice is followed by lower amyloid accumulation rates during aging.
(a) Cortical microglial activity measured by in vivo TSPO μPET at ages of three (p=0.418), six (p=0.426) and twelve months (p=0.002) for Trem2 deficient APPPS1 mice compared to APPPS1/Trem2+/+. Age matched results for C57BL6 mice with and without Trem2 deficiency are implemented for comparison with mice lacking amyloid pathology. (b) Corresponding results for cortical fibrillar amyloidogensis assessed by in vivo Amyloid μPET at three (p=0.026), six (p=0.007) and twelve months (p=0.895). (APPPS1: three months Aβ PET n+/+=4 mice, TSPO PET n+/+=5 mice, Aβ PET n−/−=7 mice, TSPO PET n−/−=9 mice; six months Aβ PET n+/+=12 mice, TSPO PET n+/+=13 mice, Aβ PET n−/−=9 mice, TSPO PET n−/−=11 mice; twelve months Aβ PET n+/+=11 mice, TSPO PET n+/+=11 mice, Aβ PET n−/−=7 mice, TSPO PET n−/−=7 mice. C57BL6: three months Aβ PET n+/+=6 mice, TSPO PET n+/+=6 mice, Aβ PET n−/−=5 mice, TSPO PET n−/−=6 mice; six months Aβ PET n+/+=25 mice, TSPO PET n+/+=20 mice, Aβ PET n−/−=6 mice, TSPO PET n−/−=8 mice; twelve months Aβ PET n+/+=21 mice, TSPO PET n+/+=20 mice, Aβ PET n−/−=7 mice, TSPO PET n−/−=7 mice). Error bars represent mean ± SD. Two-sided t-test. (c) Coronal and axial slices show Z-scores of increased TSPO μPET against age matched C57BL6 (n=18 mice) to compare APPPS1/Trem2−/− and APPPS1/Trem2+/+ at three, six and twelve months of age using an MRI template. (d) Coronal and axial slices show Z-scores of increased Amyloid μPET against age matched C57BL6 (n=18 mice) to compare APPPS1/Trem2−/− and APPPS1/Trem2+/+ at three, six and twelve months. (e) Serial imaging shows distinctly lower monthly increases for TSPO μPET during aging (6–12 months) in APPPS1/Trem2−/− mice compared to APPPS1/Trem2+/+ mice (n+/+=11 mice, n−/−=11 mice, 3–5 months p=0.756, 6–8 months p=8.03E-7, 9–11 months p=0.003). (f) Serial Amyloid μPET indicates an increased accumulation rate of fibrillar amyloidogenesis in young APPPS1/Trem2−/− mice (3–5 months) compared to age matched APPPS1/Trem2+/+ mice. Importantly, the accumulation rate of fibrillar amyloid in APPPS1/Trem2−/− mice declines below those of APPPS1/Trem2+/+ during aging (6–12 months) n+/+=10 mice, n−/−=9 mice, 3–5 months p=1.52E-4, 6–8 months p=0.003, 9–11 months p=1.48E-4). Thick lines in e and f represent polynomic functions of longitudinal changes, whereas dotted lines represent functions of SEM. Two-sided t-test. *p<0.05; **p<0.01; ***p<0.001.
As expected, microglial activation was highly increased in APPPS1/Trem2+/+ mice at twelve months compared to age matched C57BL6 mice without amyloidogenesis (Fig. 3a & c). Trem2 loss of function APPPS1 mice mainly displayed a reduced ability of microglial activation at twelve months of age when compared to age matched APPPS1/Trem2+/+ (Fig. 3a & c). Importantly, we observed a significantly reduced longitudinal progression of microglial activity (Fig. 3e; Supplementary Fig. 3a & c) from three to twelve months of age in APPPS1/Trem2−/− compared to APPPS1/Trem2+/+ mice. Increased microglial activation in APPPS1/Trem2+/+ compared to APPPS1/Trem2−/− or C57BL6 mice reflected the previously observed high 18[F]-GE180 uptake in microglial cells and low retention in the non-inflamed and non-diseased brain29.
Interestingly, amyloid μPET imaging revealed an increased fibrillar amyloidogenesis early during pathogenesis in APPPS1/Trem2−/− mice at three and six months, which leveled out during late disease progression at twelve months of age (Fig. 3b & d). Noteworthy, fibrillar amyloidogenesis was already significantly elevated in three months old APPPS1/Trem2−/− compared to age matched APPPS1/Trem2+/+ mice. Furthermore, we observed a significantly lower accumulation rate of fibrillar amyloid from six to twelve months of age (Fig. 3f; Supplementary Fig. 3b & d). In contrast, monthly rates of fibrillar amyloid accumulation were elevated in APPPS1/Trem2−/− compared to APPPS1/Trem2+/+ mice between three and six months of age (Fig 3f). This suggests an initial phase of enhanced fibrillar amyloidogenesis is followed by a later phase of slower amyloid deposition or alternatively, an earlier saturation of amyloid deposition in Trem2 knockout mice as a result of an elevated baseline. Regardless of the mechanism involved, this may suggest that additional age dependent factors contribute to plaque growth and clearance during disease progression.
ApoE load of amyloid plaques depends on functional Trem2
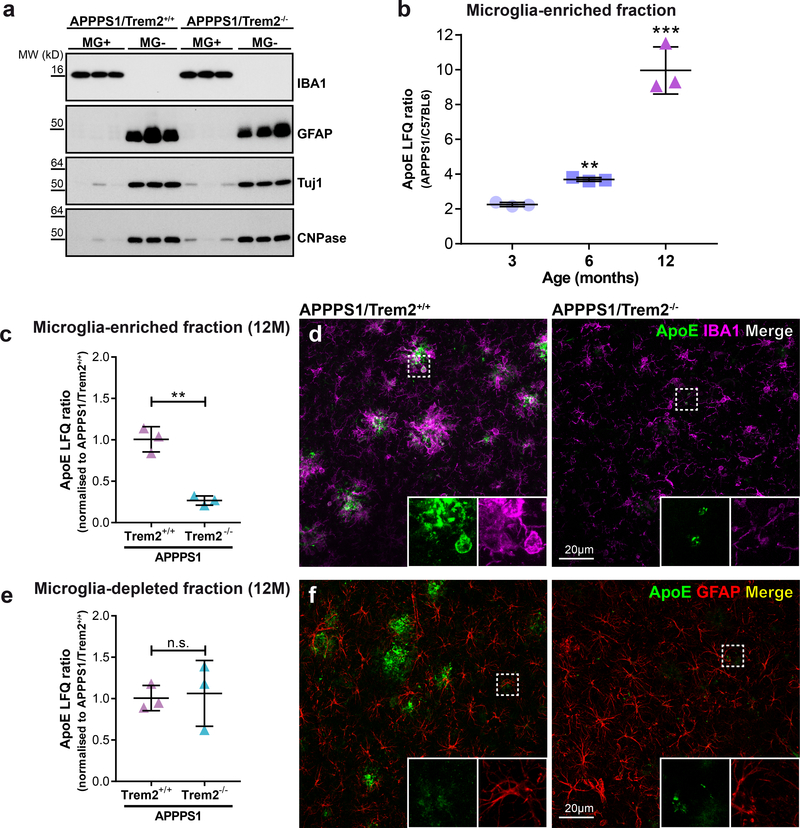
Since disease associated microglia migrate towards amyloid plaques and increase ApoE mRNA13, 14, 30, we wondered if ApoE expressing microglia might directly contribute to the ApoE load of amyloid deposits and therefore could contribute to the observed changes in the kinetics of amyloid plaque deposition. To find out if microglia or astrocytes and other neuronal cells selectively contribute to the entire pool of ApoE, we first compared ApoE protein abundance in lysates from microglia-enriched and microglia-depleted fractions obtained from twelve months old APPPS1 mice in the presence or absence of Trem2. Enrichment of microglia as well as depletion of microglia was confirmed in the corresponding fractions by western blotting with antibodies to microglial, astroglial, neuronal and oligodendrocytic marker proteins (Fig. 4a). Protein abundance of ApoE in microglia-enriched fractions increased from three to twelve months in APPPS1/Trem2+/+ mice (Fig. 4b). Moreover, proteomic analysis of APPPS1/Trem2+/+ mice revealed increased ApoE abundance selectively in the microglia-enriched fraction when compared to APPPS1/Trem2−/− mice (Fig. 4c). Increased production of ApoE in microglia was further confirmed by ApoE/IBA1 co-stainings (Fig. 4d). In contrast, the microglia-depleted fraction showed no Trem2 dependent change in ApoE production (Fig. 4e), which was also confirmed by immunohistochemistry (Fig. 4f). This suggests that microglia may be directly involved in the process of ApoE co-deposition in amyloid plaques, since the strong increase of microglial ApoE expression occurs only when microglia switch to the disease associated signature upon clustering around amyloid plaques13, 14. We therefore investigated deposition of ApoE in newly seeded amyloid plaques in the presence and absence of Trem2 dependent microglial clustering. Seeded plaques within the hippocampus were robustly co-stained with ApoE in APPPS1/Trem2+/+ mice (Fig. 5a). In contrast, upon loss of Trem2 function newly seeded hippocampal plaques showed a significant reduction of plaque associated ApoE (Fig. 5b-d) despite higher plaque load. Reduced plaque associated ApoE was confirmed by western blotting of plaque-enriched formic acid hippocampal extracts (Fig. 5e). Similar findings were obtained when we investigated non-experimentally seeded cortical Aβ plaques in the same mice (Fig. 6). Again, while APPPS1/Trem2+/+ mice showed robust ApoE association with amyloid plaques (Fig. 6a), the ApoE load in cortical plaques of APPPS1/Trem2−/− or APPPS1/Trem2p.T66M mice was significantly reduced (Fig. 6b-e). Similarly, co-immunostaining of ApoE and IBA1-positive microglia was significantly lower upon loss of Trem2 function compared to APPPS1/Trem2+/+ mice (Fig. 6f). This is in line with both the absence of ApoE mRNA induction in microglia upon loss of Trem2 function8, 14, and the age dependent increase in ApoE protein levels in microglia from APPPS1/Trem2+/+ mice (Fig. 4b). Indeed, in APPPS1/Trem2+/+ mice, microglia clustering around seeded plaques within the hippocampus (Fig. 5a) or around non-experimentally seeded cortical Aβ plaques (Fig. 6a) express enhanced levels of ApoE in contrast to microglia in Trem2 loss of function mice as shown by 3D reconstruction of high resolution confocal images (Fig. 5b-e; Fig. 6b-f). Thus, loss of Trem2 function leads to a rather dramatic reduction of ApoE co-deposition in amyloid plaques.
Figure 4. Relative protein quantification of ApoE in microglia-enriched and microglia-depleted lysates using mass spectrometry and label free quantification (LFQ).
(a) Immunoblotting of microglia-enriched (MG+) and microglia-depleted (MG-) lysates from twelve months old APPPS1/Trem2+/+ and APPPS1/Trem2−/− mice. Brain cell types in each fraction were identified by detection of IBA1 for microglia, GFAP for astrocytes, Tuj1 for neurons and CNPase for oligodendrocytes. Full image of each immunoblot is shown in Supplementary Fig. 7. (b) ApoE LFQ intensities of microglia-enriched lysates APPPS1/Trem2+/+ mice compared to age matched C57BL6 controls. Two-sided Student’s t-test comparing log2 transformed LFQ intensities of APPPS1 (n=3 mice) and C57BL6 (n=3 mice) separately for three (p=0.061), six (p=0.00372) and twelve months (p=1.78E-5) of age (**p<0.01; ***p<0.001). (c) ApoE LFQ intensities of microglia-enriched lysates from twelve months old APPPS1/Trem2−/− mice show significantly reduced ApoE compared to age matched APPPS1/Trem2−/− controls (n=3 mice/genotype). APPPS1 mice show significant reduction in ApoE after loss of Trem2 (p=0.00105). (d) ApoE and IBA1 costained in twelve months old APPPS1/Trem2+/+ and APPPS1/Trem2−/− mice. White boxes indicate the area in each staining that is magnified as inset. (e) Microglia-depleted lysates from twelve months old APPPS1 mice show no statistically significant changes regardless of Trem2 expression (n=3 mice/genotype; p=0.979). Data represent mean ± SD. (f) ApoE and GFAP costained in twelve months old APPPS1/Trem2+/+ and APPPS1/Trem2−/− mice. White boxes indicate the area in each staining that is magnified as inset.
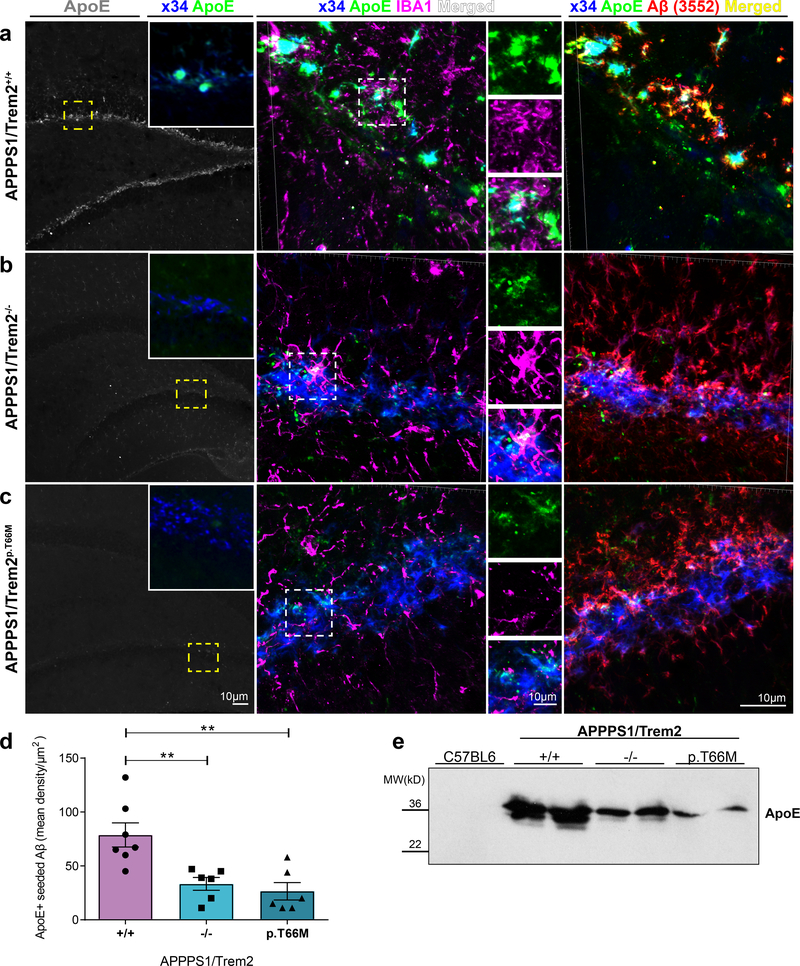
Figure 5. Decreased ApoE in newly seeded hippocampal plaques in the absence of functional Trem2.
(a) Left: ApoE (gray) staining in seeded hippocampus of APPPS1/Trem2+/+ mice. Inset shows a higher magnification of ApoE (green) colocalised with x34-positive seeded plaques (blue) in the same area shown in yellow dotted box. Middle: 3D reconstructed images of x34/ApoE/IBA1 stainings of seeded area in APPPS1/Trem2+/+ dentate gyrus. Enlarged areas of each staining (top - ApoE in green, middle – IBA1 in magenta, bottom - merged in white) are shown adjacently. Right: 3D reconstructed images of x34/ApoE/Aβ (3552) stainings of seeded area in APPPS1/Trem2+/+ dentate gyrus. (b) Left: Reduced ApoE staining (gray) in seeded APPPS1/Trem2−/− mice and (c, left) APPPS1/Trem2p.T66M mice compared to APPPS1/Trem2+/+ mice. Insets show a higher magnification of ApoE (green) with seeded amyloid pathology (x34; blue) in the same area indicated in yellow dotted box. Middle: 3D reconstructed images of x34/ApoE/IBA1 show reduced IBA1 and ApoE colocalisation in APPPS1/Trem2−/− mice and (c, middle) APPPS1/Trem2p.T66M mice. Right: 3D reconstructed images of x34/ApoE/Aβ (3552) stainings of seeded area in APPPS1/Trem2−/− and APPPS1/Trem2p.T66M dentate gyrus show increased amyloid pathology despite reduced ApoE levels. Note increased staining of non-fibrillar (x34 negative) Aβ upon Trem2 loss of function in (b) and (c) as compared to (a). (d) Quantification of mean ApoE density over seeded amyloid pathology area (n+/+=7 mice, n−/−=6 mice, np.T66M=6 mice; F2,16=10.35, p=0.0013). (e) Immunoblotting with the anti-ApoE antibody HJ6.3 demonstrates decreased hippocampal ApoE in the formic acid fraction of seeded APPPS1/Trem2−/− and APPPS1/Trem2p.T66M as compared to APPPS1/Trem2+/+ mice. Western blots were independently repeated three times to analyze n=6 mice/genotype. Full image of immunoblots are shown in Supplementary Fig. 7. Data represent mean ± SEM. One-way ANOVA, Dunnett’s post hoc analysis; **p<0.005.
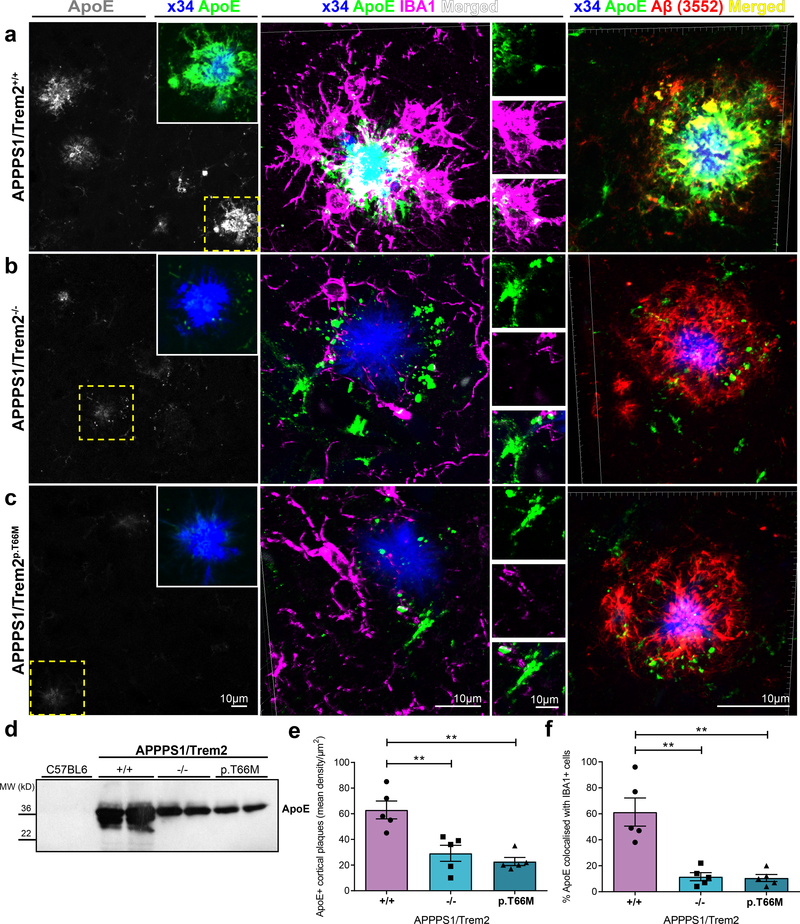
Figure 6. Decreased ApoE in non-experimentally seeded cortical Aβ plaques after loss of Trem2 function.
(a) Left: ApoE (gray) staining of non-experimentally seeded cortical Aβ plaques of APPPS1/Trem2+/+ mice. Inset demonstrates a higher magnification of ApoE (green) colocalised with x34-positive cortical plaque (blue) in the same area shown in yellow dotted box. Middle: High-resolution confocal images of x34/ApoE/IBA1 stained cortical amyloid plaque in APPPS1/Trem2+/+ were 3D reconstructed. Selected areas of each staining (top - ApoE in green, middle - IBA1 in magenta, bottom - merged in white) are shown adjacently. Right: 3D reconstructed high-resolution confocal images of x34/ApoE/Aβ (3552) stained amyloid plaque in APPPS1/Trem2+/+ cortex. Immunopositive Aβ (red) and ApoE (green) are strongly colocalised (yellow). (b) Left: Reduced ApoE staining (gray) in cortical amyloid plaques of APPPS1/Trem2−/− mice and (C, left) APPPS1/Trem2p.T66M mice. Dotted cyan box indicates the area that is magnified as inset (ApoE – green, x34 – blue). Middle: 3D reconstructed images of x34/ApoE/IBA1 show reduced IBA1 and ApoE colocalisation in APPPS1/Trem2−/− mice and (c, middle) APPPS1/Trem2p.T66M mice. Right: 3D reconstructed images of x34/ApoE/Aβ (3552) stained amyloid plaque in APPPS1/Trem2−/− and APPPS1/Trem2p.T66M cortex show very little to no colocalisation between ApoE (green) and immunopositive-Aβ (red). (d) Immunoblotting with the anti-ApoE antibody HJ6.3 shows decreased cortical ApoE in the formic acid fraction of seeded APPPS1/Trem2−/− and APPPS1/Trem2p.T66M as compared to APPPS1/Trem2+/+ mice. Western blots were independently repeated four times to analyze at least n=8 mice/genotype. Full image of immunoblots are shown in Supplementary Fig. 7. (e) Quantification of ApoE density in cortex (n=5 mice/genotype; F2,12=14.31, p=0.0007). (f) Quantification of ApoE colocalisation in IBA1-positive microglia in APPPS1/Trem2+/+, APPPS1/Trem2−/− and APPPS1/Trem2p.T66M mice (n=5 mice/genotype; F2,12=18.82, p=0.0002). Data represent mean ± SEM. One-way ANOVA, Dunnett’s post hoc analysis; **p<0.005.
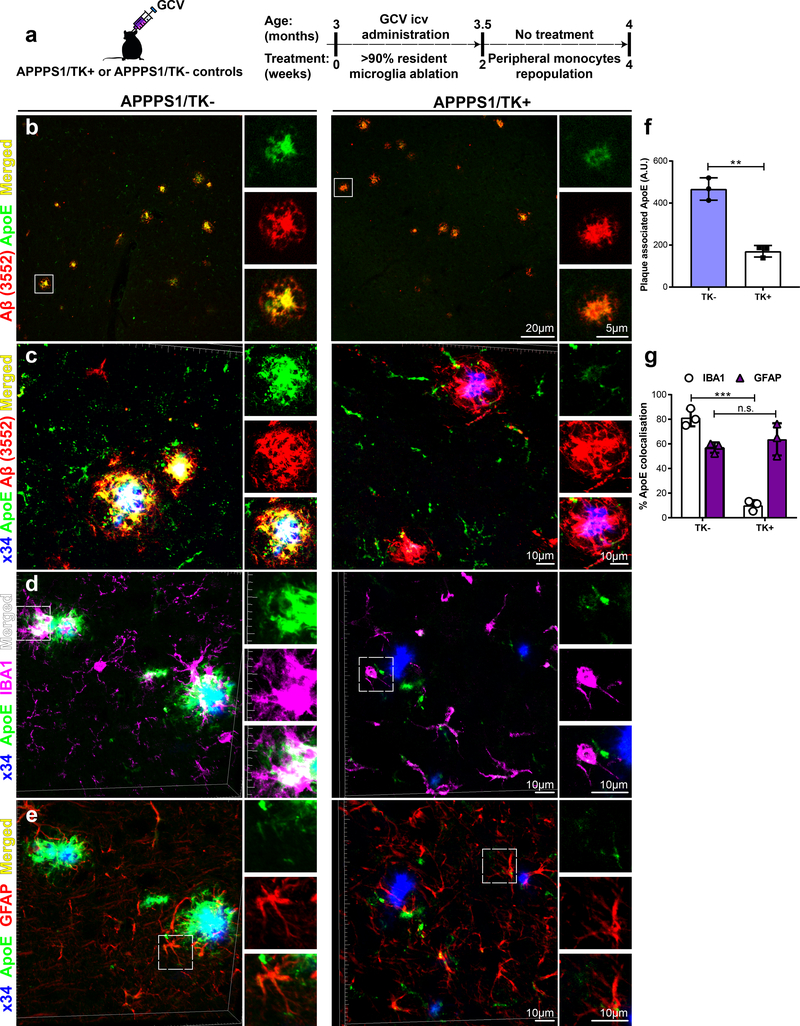
To independently verify microglia as one source of amyloid plaque associated ApoE, we investigated ApoE co-deposition in amyloid plaques in brains of microglia-depleted APPPS1 mice (Fig. 7). APPPS1 mice crossed to mice expressing thymidine kinase of herpes simplex virus (TK) under the CD11b promoter were used for this study31. TK as a suicide gene converts antiviral nucleotide analogue prodrugs such as ganciclovir (GCV) into a monophoshorylated form, which is subsequently transformed into a toxic triphosphate variant by cellular kinases. GCV treatment for longer than three weeks can lead to microhemorrhages in the brain as well as increased rate of lethality31. With these limitations in mind, we decided to include three months old APPPS1/TK+ mice treated with GCV for two weeks only to avoid side effects (Fig. 7a). Moreover, the subsequent two weeks during which no GCV was administered allowed repopulation of the brain parenchyma with peripheral monocytes, which failed to cluster around plaques32. Compared to APPPS1/TK- controls, immunohistochemical analyses demonstrated significantly reduced plaque associated ApoE upon microglial depletion (Fig. 7b-f; Supplementary Fig. 4a). Moreover, newly re-populated IBA1-positive cells in APPPS1/TK+ mice displayed decreased IBA1/ApoE costaining (Fig. 7d & g; Supplementary Fig. 4b). In contrast to microglia, GFAP-positive astrocytic staining in both APPPS1/TK mice showed no changes in GFAP/ApoE co-localization (Fig. 7e & g; Supplementary Fig. 4c). These findings provide further evidence for the pivotal role of microglia as one source of plaque associated ApoE.
Figure 7. Decreased ApoE in cortical Aβ plaques after microglia depletion.
(a) Schematic figure outlining study design and timeline of GCV application and subsequent repopulation phase. (b) An overview of fibrillar and immunopositive amyloid plaques costained with ApoE. Left: APPPS1/TK- mice show strong colocalisation of amyloid plaques and ApoE compared to APPPS1/TK+ mice (right). Right: Reduced plaque associated ApoE shown at larger magnification. (c) IMARIS 3D reconstructed high-resolution confocal images of costainings shown in b. White box indicates the area that is magnified for each immunostaining and placed adjacently. (d & e) 3D reconstructed images of x34/IBA1/ApoE/GFAP quadruple immunostaining in APPPS1/TK- and APPPS1/TK+ mice. Channels from the same confocal image were split to focus on costaining with IBA1 or GFAP individually. (d) Left: x34/ApoE/IBA1 costaining shows increased IBA1 and ApoE colocalisation with amyloid plaques in APPPS1/TK- mice compared to APPPS1/TK+ mice (right). Smaller images placed adjacently display each channel separately and merged channels (indicated by white box). (e) x34/ApoE/GFAP immunostaining demonstrates no qualitative differences in APPPS1/TK- mice (left) compared to APPPS1/TK+ mice (right). White box indicates the area that is magnified for each immunostaining and placed adjacently. (f) Plaque associated ApoE staining quantified from immunostainings shown in b and c (n=3 mice/genotype, p=0.0034). (g) Quantification of percentage IBA1/ApoE (nALL=3, p=0.0004) and GFAP/ApoE colocalisation (n=3 mice/genotype, p=0.4464) from stainings displayed in d and e, respectively. Two-tailed unpaired T-test with Welch’s correction; n.s. p>0.05; **p<0.005; ***p<0.001. Data represent mean ± SD.
Reduced ApoE in amyloid plaques of AD patients with TREM2 variants
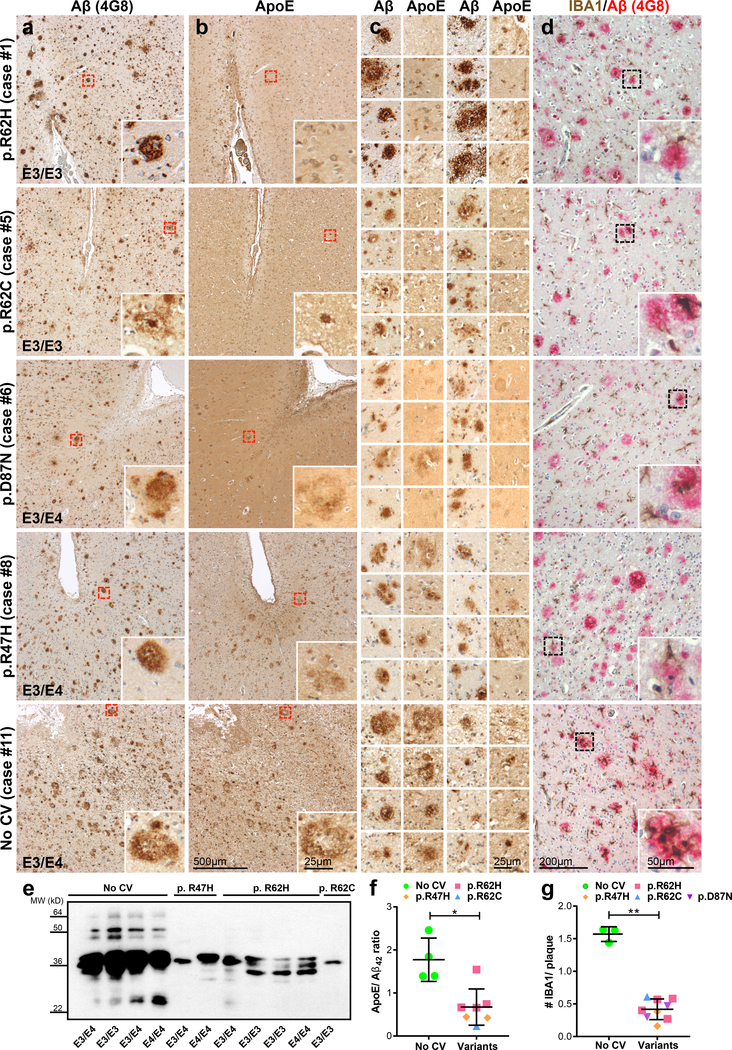
To test whether these preclinical findings have relevance for humans, we analyzed ApoE co-deposition in amyloid plaques in brains from AD patients with and without TREM2 variants (Fig. 8; Supplementary Fig. 5). Reduced ApoE colocalisation with amyloid plaques was also observed in AD patients with the TREM2 p.R47H, p.R62C, p.R62H and p.D87N loss of function variants when compared to AD cases without TREM2 variant (Fig. 8a-c; Supplementary Fig. 5 a-c). ApoE reduction was most pronounced in cases with TREM2 variants at amino acid 62, a position where an amino acid change was confirmed to result in an increased risk for developing late onset AD33. Reduced ApoE in amyloid plaques was further confirmed by western blotting of plaque-enriched formic acid extracts from brains with TREM2 variants compared to those without TREM2 coding variants, which was significant upon normalization to Aβ42 concentration (Fig. 8e & f; Supplementary Fig. 5f). Based on the findings in mouse brains (Fig. 2a & b; Supplementary Fig. 2), we therefore expected that microglia in brains of TREM2 mutation carriers should have a reduced capacity to cluster around amyloid plaques. Indeed large clusters of microglia around amyloid plaques were frequently found in AD cases without TREM2 variants but were far less frequently detected in AD patients with TREM2 loss of function variants (Fig. 8d, g; Supplementary Fig. 5d & g). While there was a trend in increased plaque associated Aβ42 with the number of E4 isoforms present, an ApoE genotype specific effect was not observed on plaque-associated microglia (Supplementary Fig. 5h & i). These data demonstrate that several independent TREM2 coding variants all cause a loss of function of TREM2 resulting in reduced microglial clustering and ApoE accumulation in amyloid plaques.
Figure 8. Reduced ApoE levels in Aβ plaques and impaired microglial clustering in TREM2 coding variants.
(a) Temporal neocortex of human AD patients with and without the indicated TREM2 variants stained for Aβ by 4G8 immunohistochemistry. (b) In sections consecutive to those of the left column, AD patients with TREM2 variants show reduced ApoE immunoreactivity within amyloid plaques compared to an AD case with no TREM2 coding variants (no CV). Note that the ApoE reduction is most pronounced in cases with an amino acid exchange at position 62. Red boxes indicate the area in each staining that is magnified as inset. (c) More examples of ApoE stainings comparing the same region in consecutive Aβ stained sections. (d) IBA1 (brown) and 4G8 (red) costaining in temporal neocortex. Microglia cells association with amyloid plaques as seen in a no CV case (last row) is severely impaired in AD cases with different TREM2 coding variants (rows 1–4). Furthermore, the overall density of microglial cells is reduced in TREM2 variant AD cases when compared to no CV. Dotted black boxes indicate the area magnified as inset. (e) ApoE immunoblotted in plaque-enriched formic acid fractions from cases shown in a-d and additional cases shown in Supplementary Fig. 5. Full image of the immunoblot is shown in Supplementary Fig. 7. (f) ApoE/Aβ42 ratio quantified from plaque-enriched formic acid fraction shows significantly decreased ApoE levels in AD cases with TREM2 coding variants (nR47H=2 cases, nR62H=4 cases; nR62C=1 case) compared to no CV (n=4 cases; p=0.0121). Of note, frozen material from p.D87N cases was not available and therefore not included. One of the no CV cases was excluded due to diagnosed Hepatitis. (g) Number of IBA1-positive microglia per plaque quantified from images shown in d and supplementary Fig. 5d (nR47H=2 cases, nR62H=4 cases; nR62C=1 case; nD87N=2 cases; nno CV=3 cases; p=3.9E-4). Noteworthy, sections from only three no CV cases were available in comparison to frozen material. No subjects were excluded in this analysis. Medial temporal cortex at the level of anterior hippocampus was used for all experiments. Data represent as mean ± SD. Unpaired Two-tailed T-test with Welch’s correction; *p<0.05; ***p<0.001.
Discussion
Our findings indicate that TREM2 dependent microglial functions limit amyloid plaque growth early but not late during the disease. For lowering amyloidogenesis, a TREM2 dependent activity is required that allows clustering of microglia around newly seeded amyloid plaques. In the presence of functional Trem2, phagocytic CD68-positive microglia accumulate around amyloid plaques and may initially promote Aβ clearance. Meanwhile, microglia that cluster around amyloid plaques increase ApoE expression12, 13, 30 and may generate a microenvironment, which could lead to an increased ApoE concentration in the immediate neighborhood of Aβ deposits (Supplementary Fig. 6). In line with the well described function of ApoE in amyloidogenesis15–17, this may further promote Aβ aggregation and deposition. Consequently, high levels of ApoE in amyloid plaques may support their fibrillization and compaction, which is reflected by the longitudinal amyloid μPET imaging data. This is further supported by the recent finding that ApoE deficient mice as well as mice expressing the AD associated p.R47H TREM2 variant exhibit a striking reduction of plaque compaction34, 35. In the absence of functional Trem2, microglia activation is dramatically reduced as shown by longitudinal TSPO μPET imaging. While using TSPO radiotracers to study neuroinflammation may present certain limitations such as the presence of non-binders and unspecific binding to multiple types of immune cells including astrocytes36, the high affinity for TSPO with better brain uptake, retention, clearance, and improved signal-to-noise ratio makes the enantiomerically stable tricyclic compound [18F]-GE180 a suitable agent for imaging neuroinflammation in vivo37–39. Furthermore, the specificity of microglial activation has also been confirmed by in vitro autoradiography and immunohistochemistry using different transgenic models with varying degrees of neuroinflammation10, 28, 40, 41. Nonetheless, future development of radioligands should facilitate in vivo detection of even more specific phenotypes of microglia activation.
Microglia clustering is abolished in Trem2 loss of function mice leading to reduced phagocytic amyloid plaque clearance but also to significantly less ApoE accumulation in amyloid plaques (both, in seeded and non-experimentally seeded cortical Aβ plaques; Supplementary Fig. 6). Reduced ApoE accumulation is in line with the finding that microglia lacking functional Trem2 suppress the disease associated mRNA signature and appear to be locked in a homeostatic state with repressed ApoE expression8, 12–14, 30. Thus, absence of ApoE expressing microglia in the immediate neighborhood of amyloid plaques may result in reduced ApoE co-aggregation in amyloid plaques (Supplementary Fig. 6), which is further supported by our finding that microglia depletion leads to a rather dramatic decline of plaque associated ApoE. This also demonstrates that Trem2 dependent microglial uptake of ApoE42 could not be the sole reason for increased ApoE in microglia, since upon microglia depletion amyloid plaques should then contain higher and not lower ApoE levels. Thus, microglia may be one source for plaque associated ApoE although an additional contribution of astrocytes cannot be excluded. For example, microglia may signal to astrocytes in a TREM2 dependent manner and may thus influence astroglial release of ApoE. Furthermore, the ApoE antibodies used may preferentially recognize a microglial ApoE variant. Future studies using ApoE antibodies that selectively recognize lipidated versus unlipidated ApoE could thus provide additional insights into potential mechanisms underlying our observations.
ApoE is predominantly synthesized in and secreted by astrocytes in non-diseased mouse43 and human brain44. However, our study demonstrates that ApoE protein levels are strongly induced in plaque associated microglia under disease conditions in a Trem2 dependent manner. This is in contrast to Trem2 loss of function microglia that are locked in a homeostatic state 8, 14. A limitation of our study is the use of murine Trem2 and ApoE for investigating the interaction between microglia and amyloid pathology. Future studies should include humanized TREM2 and APOE variants as these may influence Aβ metabolism and therefore amyloid pathogenesis45. With these limitations in mind, we studied the effect of TREM2 coding variants in the AD patients. The observed reduction in plaque associated microglia and ApoE in AD cases with TREM2 coding variants further supports our findings and raises the possibility that microglial ApoE is induced in a TREM2 dependent manner in humans as well. To finally address this question, it will be important to study human microglial cells by single cell sequencing.
Although we cannot fully exclude a saturation of amyloid plaque pathology late during disease progression in the Trem2 loss of function mutants, it is tempting to speculate that early reduced plaque clearance in the absence of functional Trem2 may be opposed by less ApoE driven amyloidogenesis later during disease progression. This could explain why late in the disease levels of amyloid deposition converge in the presence and absence of functional Trem2. The age dependent change in biphasic rate of amyloidogenesis observed in the absence of functional Trem2 is also consistent with the disease progression dependent effects of Trem2 deficiency on amyloid plaque deposition at late time points reported by Jay et al.19. However, at very early time points we observe differences in baseline levels of fibrillar amyloid pathology which could be explained by slightly different ages, the use of different technologies as well as regions of interest used to follow amyloidogenesis. In this regard, it needs to be considered that 18F-Florbetaben has a strong affinity to dense fibrillar amyloid plaques compared to diffuse amyloid deposition.
TREM2 dependent effects on ApoE load of amyloid plaques must be considered for any attempt to therapeutically modulate TREM2 and microglial function in general. Since we observe enhanced amyloidogenesis of TREM2 mutants only at early stages of the disease, and since these differences may be compensated later by the reduced microglial production of ApoE, our findings support the observation that the disease is manifested decades before clinical symptoms occur46. AD associated TREM2 loss of function variants may thus promote seeding selectively at an early phase of the disease. Seeded plaques have been reported to be neurotoxic by decreasing neurogenesis and increasing neuronal cell death in their immediate neighborhood47. Thus by eliminating newly seeded plaques TREM2 may have protective functions at least early during amyloidogenesis. A protective function of TREM2 is also supported by the finding that disease associated TREM2 variants cause a loss of function (e.g. ref.10, 11, 42, 48). This is consistent with the finding of increased soluble TREM2 in cerebrospinal fluids of patients of the DIAN (Dominantly inherited Alzheimer’s disease network) cohort already five years before onset of clinical symptoms, which may reflect a defensive microglial activation at an early state of the disease49. However, microglia not only modulate amyloid plaque clearance but also provide ApoE as an Aβ aggregation factor17, which could consequently facilitate amyloidogenesis. Finally, our finding that microglia influence amyloid plaque fibrillarity depending on their activation state is highly relevant for clinical studies as longitudinal amyloid μPET is a read-out for amyloid based therapies 50.
Online Methods
Transgenic mice
All animal experiments were approved by the Bavarian government and performed in accordance with local animal handling laws. Four months old male APPPS1 transgenic (Tg) mice (Thy1-APP695KM670/671NL; Thy-1PS1L166P)24 and age matched male C57BL6 littermate controls were used. Using male mice ensured reduced gender-dependent variability in plaque-deposition24.
APPPS1 mice were crossed to TREM2−/− mice25 and maintained on a C57BL6J background. Additionally, APPPS1 were crossed to achieve homozygous TREM2 p.T66M knock-in mice10, which were maintained on a mixed genetic background (N1 backcross to C57BL6N). The TREM2 p.T66M mice were compared to APPPS1/TREM2 wild-type mice from the same background for all experiments. No differences were observed between wild-type control mice from the TREM2−/− or TREM2 p.T66M mouse lines. For μPET experiments, three, six and twelve months old female APPPS1/TREM2+/+ and age matched APPPS1/TREM2−/− were used. As additional controls, non-transgenic age matched female C57BL7/TREM2+/+ as well as C57BL6/TREM2−/− were included.
Randomization and Blinding
For all animal experiments, mice were allocated randomly after genotyping. Mice of different genotype were allocated randomly to PET experiments with scan number and slot, as well as with blinded identity to the experimenter using chipping system. Investigator was not blinded during the intrahippocampal injection experiments. All the immunohistochemical analysis was initially performed blinded, however due the strong microglial clustering impairment phenotype of loss of Trem2 mice, complete blinding was not possible. Slides were imaged and saved with random numbers to identify them. Images were then quantified and unblinded to perform group statistics. No statistical methods were used to pre-determine sample sizes but our sample sizes were based on previous findings2, 10, 28, 31.
Preparation of brain homogenate for intracerebral injections
Brain homogenates for injection were prepared as previously described2. Mouse brain extracts were prepared by dissecting the neocortex of 15-month old female APPPS1 transgenic mice and age matched non-transgenic C57BL6 controls. Tissue samples were homogenized at 10% (w/v) in sterile phosphate-buffered saline (PBS), vortexed, sonicated 3 × 5 seconds (Bandelin Sonorex RK100H) and centrifuged at 3000× g for five mins at 4°C. The supernatant was aliquoted and frozen at −80°C until use.
Stereotaxic surgery
Six weeks old asymptomatic host mice were anaesthetized with a mixture of medetomidin (0.05 mg−1kg−1), midazolam (5 mg−1kg−1) and fentanyl (0.5 mg−1kg−1). As previously described2, a 0.5–1.0cm-long incision was made in the shaved and sterilized skin to expose the bregma. Bilateral stereotaxic injections of mouse brain homogenate (1.25μl min−1 over two min) were performed with a Hamilton syringe into the hippocampi and overlaying cortices at specific stereotaxic coordinates (AP −2.3mm, L +/−2.0mm, DV –0.8/−2.0mm). The needle was retained for additional two min before it was slowly withdrawn. The incision was cleaned with povidone-iodine antiseptic solution and sutured. Following this, mice were brought out of anaesthesia using a mixture of atipamezol (2.5 mg−1kg−1), flumazenil (0.5 mg−1kg−1) and naloxone (1.2 mg−1kg−1), returned to clean cages and monitored until recovery from anaesthesia. Mice were weighed and monitored daily for seven days following surgery. Food and water was provided ad libitum.
Immunofluorescence analyses of mouse brains
Brains were removed after transcardial perfusion with PBS and dissected into two hemispheres. The left hemibrain was immersion-fixed for 48h in 4% paraformaldehyde, followed by cryoprotection in 30% sucrose for 48h. After freezing, 25μm microtome sections were sequentially collected in phosphate buffer saline with 15% glycerol and kept at −80°C until further use. For staining, heat-induced sodium citrate antigen retrieval was performed using 25mM sodium citrate with 0.05% Tween-20 at 95°C for 30mins. Free-floating sections were incubated in x34 for one hour at room temperature in 60% PBS/40% EtOH mix; pH was adjusted with 1N NaOH 51. Sections were washed briefly with 60% PBS/40% EtOH before blocking. Primary antibodies (Supplementary table 1) were added and the sections were kept at 4°C overnight with slow agitation. For Trem2 staining, sections were incubated at 4°C for two nights. After applying the appropriate secondary antibodies, tissue sections were stained with Thioflavine S (ThioS, 0.01%) or 4′,6-Diamidin-2-phenylindol (DAPI, 5μg/mL) before mounting sections onto slides (Prolong™ Gold Antifade reagent, Thermo Fisher Scientific).
Confocal imaging and analyses
Images were acquired using a LSM 710 Confocal microscope (Zeiss) and the ZEN 2011 software package (black edition, Zeiss). Laser and detector settings were maintained constant for the acquisition of each immunostaining. For all analyses, at least three images were taken per brain region and slide using 20x objective and 63x oil differential interference contrast objective, respectively, at 2048 × 2048 pixel resolution, with z-step size of 2μm at 16μm thickness. High-resolution stacks imaged by confocal microscopy were 3D reconstructed using IMARIS 8 software.
For quantification of plaque densities acquired images were imported to Fiji software (ImageJ) and data channels were separated (Image\Color\Split Channels). Gaussian filtering was used to remove noise and intensity distribution for each image was equalized using rolling ball algorithm, which is implemented as background subtraction plugin in Fiji. For the feasibility of the quantification, all layers from a single image stack were projected on a single slice (Stack\Z projection). Next, the plaques were segmented using automatic thresholding methods in Fiji (with “Moments” thresholding setting for microglial stainings and “Otsu” for plaques and ApoE). Subsequently, the dentate gyrus was selected manually whereupon the area as well as the cumulative area of the segmented plaques were calculated in the region of interest. Finally, the coverage of segmented plaques over the area of dentate gyrus was calculated, indicating the distribution of segmented plaques in the dentate gyrus. Similar thresholding was used to quantify cortical plaques as well as IBA1, CD68 and ApoE stainings. The number of IBA1 and CD68-positive microglia around plaques were counted in the cortex over the length of layers 3–5 using ImageJ.
In vivo μPET imaging and quantification
All small animal positron emission tomography (μPET) procedures followed an established standardized protocol for radiochemistry, acquisition and post-processing 52, 53. In brief, 18F-GE180 TSPO μPET with an emission window of 60–90 mins post injection was used to measure cerebral microglial activity, and 18F-Florbetaben amyloid μPET with an emission window of 30–60 mins post injection was used for assessment of fibrillar cerebral amyloidogenesis. APPPS1/Trem2−/− and age matched APPPS1/TREM2+/+ mice were longitudinally studied from six to twelve months of age using dual μPET. All analyses were performed by PMOD (V3.4, PMOD technologies, Basel, Switzerland). Normalization of injected activity was performed by the previously validated myocardium correction method54 for TSPO μPET and by a white matter reference region for amyloid μPET.
TSPO and amyloid μPET estimates (absolute values and percentage changes over time) deriving from the same large neocortical target VOI (50 mm3) were extracted and compared between the two age and genotype groups by a student’s t-test. In house scans of C57BL6 mice served for generation of Z-score deviation images for both tracers (Z-score = (averageAPPPS1/TREM2−/− - averageC57BL6)/SDC57BL6) or (averageAPPPS1/TREM2+/+ - averageC57BL6)/SDC57BL6). Percentage changes with baseline as starting point was calculated such that the earliest time interval was set as 3–4 months and the latest at 11–12, depicted as 3 and 11 months.
Patient material
Tissue samples of patient autopsy cases were provided by the Neurobiobank Munich, Ludwig-Maximilians-University (LMU) Munich and Brain Bank, UCL. Detailed clinical characteristics were ascertained from an integrated autopsy database at respective Brain Banks. Written informed consent for autopsy and analysis of tissue sample data was obtained for all patients, either from the patients themselves or their next of kin. All samples were collected according to the guidelines of the local ethics committee following all ethical regulations. All requirements for written informed consent was waived by the Medical Ethics Committee that approved the study protocol. Information on cases regarding brain bank, clinical diagnosis age at death, post mortem delay, AD Braak & Braak stage, TREM2 coding variant and APOE status is given in Supplementary Table 2. Sample sizes were based on availability of patient material. For all experiments, temporal neocortex was used which included cortex of medial temporal gyrus at the level of anterior hippocampus.
Genotyping of AD cases
To identify TREM2 variant carriers within the Neurobiobank Munich TREM2 exon 2 was amplified from genomic DNA by polymerase chain reaction, purified using ExoSAP-IT (Thermo Fisher Scientific) and sequenced by standard Sanger sequencing (GATC Biotech). Sequences were aligned to the TREM2 reference sequence (UCSC genome browser, assembly GRCh38/hg38; chr6:41,158,506–41,163,186) and variants called manually by two investigators using the CLC Main Workbench Software (Qiagen). To identify pathogenic APP, PSEN1, PSEN2 mutations all cases were additionally sequenced using a TruSeq CustomAmplicon kit on a MiSeq (Illumina) according to the protocol from the manufacturer. Panel sequencing confirmed all TREM2 variants and was also used to determine the ApoE genotype.
Immunohistochemistry on human post-mortem brain tissue
Temporal neocortex sections of 5μm thickness were cut from paraffin embedded specimens. For immunohistochemistry, all slides were deparaffinized and rehydrated in a series of xylene and graded ethanol. The sections were subjected to citric acid antigen retrieval (1M sodium citrate in PBS, pH 6.0) and boiled in a microwave for 20 min. After cooling, endogenous peroxidase activity was quenched using 30% hydrogen peroxide for 20 min. Sections were blocked and incubated with primary antibody (Supplementary table 1) overnight at 4°C. Primary antibodies were detected with biotinylated anti-mouse and anti-rabbit IgG secondary antibodies and visualized with avidin-biotin-complex (ABC-Kit, Vector laboratories) followed by development with diaminobenzidine-HCl (DAB, Vector laboratories) for five min. Lastly, sections were counterstained with haematoxylin. Stainings were performed in serially cut sections to compare the same region of interest through all immunostainings. For IBA1/4G8 costainings, mouse AP conjugated and rabbit HRP conjugated secondary antibodies were incubated at room temperature for one hour. HRP staining was developed using DAB as mentioned above and AP staining was developed using Permanent AP Red Kit for 20 min at room temperature (Zytomed systems). We controlled for artifacts resulting from long formaldehyde fixation period by comparing different antigen retrieval methods as well as staining freshly fixed frozen tissue. Brightfield images were taken by CellD, Olympus BX50 Soft Imaging System (Olympus, Tokyo, Japan). Plaque associated ApoE in human temporal neocortex was quantified using Analyze Particles feature in ImageJ. All thresholded (“Otsu”) and masked plaques were selected from the regions of interest (ROI) manager, and used to measure ApoE in the ApoE stained serial section. Images from similar ROI were taken in all samples, including the superior, middle and inferior temporal gyri using 4x as well as 10x objectives for a better overview of the tissue. Amyloid plaques in the subcortical white matter were not analyzed. The number of IBA1-positive microglia per cortical plaque was scored and counted from 6–10 images per slide. A total of 2461 plaques were analyzed and only IBA1-positive microglia with visibly stained nuclei were counted.
Preparation of protein extracts from brain
After transcardial perfusion with PBS, the right hemibrain was further dissected on ice to isolate mouse hippocampi and cortices, which were frozen at −80°C until use. Frozen mouse or human tissue was weighed and homogenized with 500μl DEA buffer (50 mM NaCl, 0,2% Diethylamine, pH 10, supplemented with 2x protease inhibitor (P8340, Sigma-Aldrich)) using the Precellys 24 homogenizer (Bertin Instruments) with the CKMix Tissue Homogenizing Kit (2ml tubes, VWR) under vacuum for 30 seconds at 6500 rpm at 4°C. This homogenate was centrifuged for five min at 4°C at 5000 × g to pellet DEA insoluble material and the supernatant was ultracentrifuged for one hour at 100000 × g with a TLA55 rotor in an Optima MAX-XP ultracentrifuge (Beckman Coulter) to obtain the DEA extracts which were neutralized with 10% 1M Tris pH 6.8. From the remaining cellular pellet, proteins were extracted with RIPA buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate plus protease inhibitor) using the Precellys 24 under vacuum for ten seconds at 5000 rpm. After a five min clearing for RIPA insoluble material at 5000 × g at 4°C the supernatant was again ultracentrifuged as above to obtain the RIPA soluble protein fraction. The RIPA insoluble pellet was dissolved with ice cold 70% formic acid (FA) and sonicated for seven min at room temperature using a Sonorex RK100H (Bandelin) followed by a final ultracentrifugation as above. The FA soluble fractions were supplemented and neutralized using ten volumes of 1M Tris pH 9.5 plus protease inhibitor. Protein concentrations were measured for DEA and RIPA fractions using a BCA assay (Pierce). All samples were aliquoted and frozen at −80°C until use.
Aβ ELISA
Aβ was quantified by a sandwich immunoassay using the Meso Scale Discovery SECTOR Imager 2400 as described previously55. Samples were extracted in FA as described above and measured in duplicates.
Immunoblotting
For amyloid plaques and ApoE western blots, equal volumes of FA extracted sample were subjected to 10–20% Tris-tricine gels (1mm, Novex) and 10% Tris-glycine gels, respectively, under denaturing conditions. After separation, proteins were transferred onto nitrocellulose membranes (Protran BA85; GE Healthcare), boiled in PBS for five min and blocked in I-Block solution (0.2% Tropix I-Block (Applied Biosystems), 0.1% Tween-20 in PBS) for one hour at room temperature with agitation. To detect different brain cell types from microglia enriched and depleted lysates, 1μg of total protein was loaded on 12% Tris-glycine gels and run under denaturing conditions. Proteins were subsequently transferred onto polyvinylidene difluoride membranes (PVDF Immobilon-P; Merck Millipore) and blocked in I-Block solution for one hour at room temperature. Primary antibodies (HJ6.3, Murine ApoE; 6E10, Aβ1–16; IBA1; GFAP; Tuj1; CNPase; HJ15.7, Human ApoE) were diluted in I-Block solution or TBS-Tween and incubated overnight at 4°C with agitation. Blots were washed with TBS-Tween and incubated in corresponding HRP-conjugated secondary antibodies for one hour at room temperature and visualized using enhanced chemiluminescence technique (Pierce).
Relative protein quantification of ApoE in microglia-enriched and microglia-depleted fractions by mass spectrometry
Microglia isolation was performed using the Magnetic Activated Cell Sorting (MACS) technology was described previously56. Microglia-enriched and depleted samples were prepared from three APPPS1 as well as three wild type mice each at the age of three, six and twelve months. Additionally, microglia-enriched and depleted fractions were prepared for twelve months old APPPS1/Trem2+/+ and age matched APPPS1/Trem2−/− mice. Both fractions were lysed in lysis buffer (50 mM Tris, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, pH 7.5). A protein amount of 15 μg per sample was digested with LysC and trypsin using the filter-aided sample preparation protocol57. The resulting peptides were concentrated using self-made C18 Stage Tips 58. The eluted peptides were concentrated by vacuum centrifugation and analyzed by nLC-MS/MS using an Easy nLC 1000 coupled online via a Nanospray Flex ion source (both Thermo Fisher Scientific) equipped with a PRSO-V1 column oven (Sonation) to a Q Exactive high field mass spectrometer (Thermo Fisher Scientific). Briefly, 1.3 μg of peptides were separated on a self-packed 30 cm column (Reprosil-Pur 120 C18-AQ, 1.9 μm, Dr. Maisch) using a binary gradient of water (A) and acetonitrile (B) supplemented with 0.1% formic acid at 50°C and a flow rate of 250 nl/min (0 min., 2% B; 3:30 min., 5% B; 137:30 min., 25% B; 168:30 min, 35% B; 182:30 min, 60% B; 185 min, 95% B; 200 min, 95% B).
A data independent acquisition method using sequential window acquisition of all theoretical mass spectra from m/z 300–1400 was applied (25 windows). Full MS spectra were acquired at a resolution of 120,000, whereas fragment ion spectra were recorded at a resolution of 30,000. Data analysis and label free protein quantification was performed with the software Spectronaut (version 11.0.15038.4.29119, Biognosys) 59 using a microglia spectral library. The library was generated with the software Maxquant (version 1.5.5.1) 60 using a top 15 data dependent acquisition method on the basis of a reviewed canonical UniProt database of Mus musculus (download: 2017–01-11, 16844 entries). The false discovery rate for both, peptides and proteins, was adjusted to less than 1%. Two unique peptides were required for protein quantification using standard settings. Here, only ApoE label free quantification intensities were extracted and subjected to statistical analysis.
Statistical analysis
Data are presented as mean ± SEM unless otherwise stated. Data were checked for normality using the Shapiro-Wilk method, D’Agostino and Pearson normality test as well as KS normality test. Statistical significance was calculated by one-way ANOVA followed by Dunnett’s post hoc test for group-wise comparisons unless otherwise stated. The label free protein quantification intensities were log2 transformed and a Student’s t-test was applied to calculate statistical significance. Amyloid and microglia stainings were correlated with amyloid and TSPO μPET, respectively, using Pearson correlation coefficients and linear regression analyses. A P value of less than 0.05 was considered significant.
Life Sciences Reporting Summary.
Further information on experimental design is available in the Life Sciences Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) within the framework of the Munich Cluster for Systems Neurology (EXC 1010 SyNergy), a DFG funded Koselleck Project (HA1737/16-1 to C.H.), by the Helmholtz-Gemeinschaft Zukunftsthema “Immunology and Inflammation” (ZT-0027 to C.H.), the FOR2290 (to S.F. L. and C.H.), and by a dedicated PET imaging grant to M.B. & A.R. (BR4580/1-1 & RO5194/1-1). This project has received additional funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No. 115976. Startup funding for this project came from LMUexcellent. Additional funding came from the general legacy of Mrs. Ammer, the MetLife award, and the Cure Alzheimer’s fund. M.M.L. is supported by the Emmy Noether Program of the DFG (ME 3542/1-1). M.M.L. and S.F.L. are supported by the Hans and Ilse Breuer Foundation. S.T. got support from Ono Pharmaceuticals, Japan. D.M.H. is supported by NIH grants NS090934 and AG047644, the JPB Foundation, and the Cure Alzheimer’s Fund. S.F.L. is supported by the Centers of Excellence in neurodegeneration and the Helmholtz-Israel program. O.B. is supported by NIH grants NINDS (R01NS088137), NIH-NIA (R01AG051812), NIH-NIA (R01AG054672) and the Cure Alzheimer’s Fund. The APPPS1 colony was established from a breeding pair kindly provided by Mathias Jucker (Hertie-Institute for Clinical Brain Research, University of Tübingen and DZNE-Tübingen). The authors thank Marco Colonna for the Trem2−/− mice. Thanks to the Queen Square Brain Bank for access to tissue: this resource is funded in part by the Weston Foundation and the MRC. D.E. is supported by the European Community’s Health Seventh Framework Programme under grant agreement 617198 [DPR-MODELS].
Footnotes
Competing interests
C.H. collaborates with Denali and received a speaker honorarium of Roche and Novartis. D.M.H. co-founded and is on the scientific advisory board of C2N Diagnostics. D.M.H. consults for Denali, Eli Lilly, Glaxosmithkline, and AbbVie. A.R. received consultant and speaker honoria from Piramal Imaging and GE Healthcare. O.B. collaborates with Sanofi. S.T. collaborates with Ono Pharmaceuticals (Japan). S.A.G is an employee of Neurimmune AG. All other authors declare that they have no conflict of interest.
References
- 1.Jucker M & Walker LC Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501, 45–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer-Luehmann M, et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science 313, 1781–1784 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Butovsky O & Weiner HL Microglial signatures and their role in health and disease. Nat Rev Neurosci 19, 622–635 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song WM & Colonna M The identity and function of microglia in neurodegeneration. Nat Immunol 19, 1048–1058 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Ulrich JD, Ulland TK, Colonna M & Holtzman DM Elucidating the Role of TREM2 in Alzheimer’s Disease. Neuron 94, 237–248 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Guerreiro R, et al. TREM2 variants in Alzheimer’s disease. N Engl J Med 368, 117–127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonsson T, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med 368, 107–116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazaheri F, et al. TREM2 deficiency impairs chemotaxis and microglial responses to neuronal injury. EMBO Rep (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulland TK, et al. TREM2 Maintains Microglial Metabolic Fitness in Alzheimer’s Disease. Cell 170, 649–663 e613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinberger G, et al. The FTD-like syndrome causing TREM2 T66M mutation impairs microglia function, brain perfusion, and glucose metabolism. EMBO J 36, 1837–1853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleinberger G, et al. TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci Transl Med 6, 243ra286 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Butovsky O, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci 17, 131–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keren-Shaul H, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290 e1217 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Krasemann S, et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566–581 e569 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu CC, et al. ApoE4 Accelerates Early Seeding of Amyloid Pathology. Neuron 96, 1024–1032 e1023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huynh TV, et al. Age-Dependent Effects of apoE Reduction Using Antisense Oligonucleotides in a Model of beta-amyloidosis. Neuron 96, 1013–1023 e1014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma J, Yee A, Brewer HB Jr., Das S & Potter H Amyloid-associated proteins alpha 1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer beta-protein into filaments. Nature 372, 92–94 (1994). [DOI] [PubMed] [Google Scholar]
- 18.Liao F, et al. Targeting of nonlipidated, aggregated apoE with antibodies inhibits amyloid accumulation. J Clin Invest 128, 2144–2155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jay TR, et al. Disease Progression-Dependent Effects of TREM2 Deficiency in a Mouse Model of Alzheimer’s Disease. J Neurosci 37, 637–647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, et al. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J Exp Med 213, 667–675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jay TR, et al. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J Exp Med 212, 287–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160, 1061–1071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kane MD, et al. Evidence for seeding of beta -amyloid by intracerebral infusion of Alzheimer brain extracts in beta -amyloid precursor protein-transgenic mice. J Neurosci 20, 3606–3611 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radde R, et al. Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep 7, 940–946 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turnbull IR, et al. Cutting edge: TREM-2 attenuates macrophage activation. J Immunol 177, 3520–3524 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Guerreiro RJ, et al. Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol 70, 78–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulrich JD, et al. Altered microglial response to Abeta plaques in APPPS1–21 mice heterozygous for TREM2. Mol Neurodegener 9, 20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brendel M, et al. Increase of TREM2 during Aging of an Alzheimer’s Disease Mouse Model Is Paralleled by Microglial Activation and Amyloidosis. Front Aging Neurosci 9, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickens AM, et al. Detection of microglial activation in an acute model of neuroinflammation using PET and radiotracers 11C-(R)-PK11195 and 18F-GE-180. J Nucl Med 55, 466–472 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Holtman IR, et al. Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: a co-expression meta-analysis. Acta Neuropathol Commun 3, 31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grathwohl SA, et al. Formation and maintenance of Alzheimer’s disease beta-amyloid plaques in the absence of microglia. Nat Neurosci 12, 1361–1363 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heppner FL, et al. Experimental autoimmune encephalomyelitis repressed by microglial paralysis. Nat Med 11, 146–152 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Sims R, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet 49, 1373–1384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ulrich JD, et al. ApoE facilitates the microglial response to amyloid plaque pathology. J Exp Med 215, 1047–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng-Hathaway PJ, et al. The Trem2 R47H variant confers loss-of-function-like phenotypes in Alzheimer’s disease. Mol Neurodegener 13, 29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavisse S, et al. Reactive astrocytes overexpress TSPO and are detected by TSPO positron emission tomography imaging. J Neurosci 32, 10809–10818 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chau WF, et al. Exploration of the impact of stereochemistry on the identification of the novel translocator protein PET imaging agent [(18)F]GE-180. Nucl Med Biol 42, 711–719 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Feeney C, et al. Kinetic analysis of the translocator protein positron emission tomography ligand [(18)F]GE-180 in the human brain. Eur J Nucl Med Mol Imaging 43, 2201–2210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vomacka L, et al. TSPO imaging using the novel PET ligand [(18)F]GE-180: quantification approaches in patients with multiple sclerosis. EJNMMI Res 7, 89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boutin H, et al. 18F-GE-180: a novel TSPO radiotracer compared to 11C-R-PK11195 in a preclinical model of stroke. Eur J Nucl Med Mol Imaging 42, 503–511 (2015). [DOI] [PubMed] [Google Scholar]
- 41.James ML, et al. [(18)F]GE-180 PET Detects Reduced Microglia Activation After LM11A-31 Therapy in a Mouse Model of Alzheimer’s Disease. Theranostics 7, 1422–1436 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeh FL, Wang Y, Tom I, Gonzalez LC & Sheng M TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia. Neuron 91, 328–340 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89, 37–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fagan AM, et al. Human and murine ApoE markedly alters A beta metabolism before and after plaque formation in a mouse model of Alzheimer’s disease. Neurobiol Dis 9, 305–318 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Sperling R, Mormino E & Johnson K The evolution of preclinical Alzheimer’s disease: implications for prevention trials. Neuron 84, 608–622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ziegler-Waldkirch S, et al. Seed-induced Abeta deposition is modulated by microglia under environmental enrichment in a mouse model of Alzheimer’s disease. EMBO J 37, 167–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song WM, et al. Humanized TREM2 mice reveal microglia-intrinsic and -extrinsic effects of R47H polymorphism. J Exp Med 215, 745–760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suarez-Calvet M, et al. Early changes in CSF sTREM2 in dominantly inherited Alzheimer’s disease occur after amyloid deposition and neuronal injury. Sci Transl Med 8, 369ra178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sevigny J, et al. The antibody aducanumab reduces Abeta plaques in Alzheimer’s disease. Nature 537, 50–56 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Styren SD, Hamilton RL, Styren GC & Klunk WE X-34, a fluorescent derivative of Congo red: a novel histochemical stain for Alzheimer’s disease pathology. J Histochem Cytochem 48, 1223–1232 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Brendel M, et al. Glial Activation and Glucose Metabolism in a Transgenic Amyloid Mouse Model: A Triple-Tracer PET Study. J Nucl Med 57, 954–960 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Overhoff F, et al. Automated Spatial Brain Normalization and Hindbrain White Matter Reference Tissue Give Improved [(18)F]-Florbetaben PET Quantitation in Alzheimer’s Model Mice. Front Neurosci 10, 45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deussing M, et al. Coupling between physiological TSPO expression in brain and myocardium allows stabilization of late-phase cerebral [(18)F]GE180 PET quantification. NeuroImage 165, 83–91 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Page RM, et al. Generation of Abeta38 and Abeta42 is independently and differentially affected by familial Alzheimer disease-associated presenilin mutations and gamma-secretase modulation. J Biol Chem 283, 677–683 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Daria A, et al. Young microglia restore amyloid plaque clearance of aged microglia. EMBO J 36, 583–603 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wisniewski JR, Zougman A, Nagaraj N & Mann M Universal sample preparation method for proteome analysis. Nat Methods 6, 359–362 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Rappsilber J, Mann M & Ishihama Y Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2, 1896–1906 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Bruderer R, Bernhardt OM, Gandhi T & Reiter L High-precision iRT prediction in the targeted analysis of data-independent acquisition and its impact on identification and quantitation. Proteomics 16, 2246–2256 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13, 2513–2526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.