Abstract
Background and Aims
Identifying the processes that generate and maintain biodiversity requires understanding of how evolutionary processes interact with abiotic conditions to structure communities. Edaphic gradients are strongly associated with floristic patterns but, compared with climatic gradients, have received relatively little attention. We asked (1) How does the phylogenetic composition of palm communities vary along edaphic gradients within major habitat types? and (2) To what extent are phylogenetic patterns determined by (a) habitat specialists, (b) small versus large palms, and (c) hyperdiverse genera?
Methods
We paired data on palm community composition from 501 transects of 0.25 ha located in two main habitat types (non-inundated uplands and seasonally inundated floodplains) in western Amazonian rain forests with information on soil chemistry, climate, phylogeny and metrics of plant size. We focused on exchangeable base concentration (cmol+ kg−1) as a metric of soil fertility and a floristic index of inundation intensity. We used a null model approach to quantify the standard effect size of mean phylogenetic distance for each transect (a metric of phylogenetic community composition) and related this value to edaphic variables using generalized linear mixed models, including a term for spatial autocorrelation.
Key Results
Overall, we recorded 112 008 individuals belonging to 110 species. Palm communities in non-inundated upland transects (but not floodplain transects) were more phylogenetically clustered in areas of low soil fertility, measured as exchangeable base concentration. In contrast, floodplain transects with more severe flood regimes (as inferred from floristic structure) tended to be phylogenetically clustered. Nearly half of the species recorded (44 %) were upland specialists while 18 % were floodplain specialists. In both habitat types, phylogenetic clustering was largely due to the co-occurrence of small-sized habitat specialists belonging to two hyperdiverse genera (Bactris and Geonoma).
Conclusions
Edaphic conditions are associated with the phylogenetic community structure of palms across western Amazonia, and different factors (specifically, soil fertility and inundation intensity) appear to underlie diversity patterns in non-inundated upland versus floodplain habitats. By linking edaphic gradients with palm community phylogenetic structure, our study reinforces the need to integrate edaphic conditions in eco-evolutionary studies in order to better understand the processes that generate and maintain tropical forest diversity. Our results suggest a role for edaphic niche conservatism in the evolution and distribution of Amazonian palms, a finding with potential relevance for other clades.
Keywords: Amazon basin, Amazonian floodplains, Arecaceae, Bactris, edaphic gradients, Geonoma, habitat specialization, igapó, terra firme, tropical rain forest, várzea
INTRODUCTION
Environmental heterogeneity has long been considered a key driver of ecological and evolutionary processes promoting and maintaining biodiversity (Whittaker, 1960; Rosenzweig, 1995; Chesson, 2000; Antonelli and Sanmartín, 2011; Stein et al., 2014). In the tropics, various abiotic gradients have been shown to structure diversity patterns even within areas of similar climate (Clark et al., 1998; Higgins et al., 2011; Tuomisto et al., 2014; Lehtonen et al., 2015). Edaphic gradients, in particular, have major impacts on tropical plant distributions, community dynamics and diversity patterns (John et al., 2007; Andersen et al., 2010; Higgins et al., 2011; Quesada et al., 2012; Condit et al., 2013; Asner et al., 2015; Muscarella et al., 2016; Cámara-Leret et al., 2017). However, edaphic gradients remain poorly studied compared with climate gradients (Figueiredo et al., 2018), especially with respect to the ways they have interacted with evolutionary processes. As a result, we currently have a limited understanding of how edaphic heterogeneity within major habitat types is related to phylogenetic structure of tropical plant communities.
In lowland rain forests of Amazonia, for example, plant species distributions and community composition are associated with numerous edaphic conditions, including soil fertility, hydrology and physical properties (Dumont et al., 1990; Terborgh and Andresen, 1998; Tuomisto et al., 2003; Fine and Kembel, 2011; Kristiansen et al., 2012; Quesada et al., 2012; Fortunel et al., 2014; Schietti et al., 2014; Cámara-Leret et al., 2017; Myster, 2017). The tendency for a site to experience seasonal inundation is perhaps the most pronounced distinction between Amazonian habitat types, with profound effects on plant, animal and microbial communities (Dumont et al., 1990; Aleixo, 2002; Wittmann et al., 2006; Junk et al., 2010; Moulatlet et al., 2014; Schietti et al., 2014; Harvey et al., 2017; Ritter et al., 2018a). However, each of these coarse habitat categories (i.e. non-inundated ‘uplands’ and seasonally inundated floodplains) encompasses substantial heterogeneity that is likely to influence diversity patterns (Junk et al., 2010; Schietti et al., 2014; Cámara-Leret et al., 2017).
Among non-inundated ‘upland’ tropical forests (e.g. terra firme, terrace, white sands), soil fertility gradients strongly affect plant productivity and allocation strategies (Vitousek, 1984; Quesada et al., 2012), and have been linked to variation in community composition and dynamics (Tuomisto et al., 2003; Fine et al., 2006; John et al., 2007; Ruokolainen et al., 2007; Costa et al., 2009; Cámara-Leret et al., 2017; Ritter et al., 2018b). Across the Amazon basin, soils of the central and eastern regions are older and generally less fertile than those in western Amazonia, which are more recently derived from Andean sediments released during uplift and erosion and accumulated along the Andean foreland. Soils of western Amazonia are, however, highly variable, ranging from extremely nutrient-poor white sands to well-drained and fertile terra firme (Quesada et al., 2010; Higgins et al., 2011; Quesada et al., 2011).
Seasonally inundated floodplains (e.g. várzeas, igapós) also exhibit a wide range of soil fertility but many areas are more fertile than non-inundated uplands. This is in part because rivers deliver nutrients, especially during the flood season characteristic of several river systems, when the water is more enriched with nutrients compared with the low-flow season (Markewitz et al., 2001; Quesada et al., 2011; Asner et al., 2015). Inundation itself, however, represents a source of acute physiological stress (Myster, 2009; Junk et al., 2010). For instance, survival and reproduction under flooded conditions requires structural or physiological adaptations that have evolved in relatively few lineages (Kahn and de Granville, 1992; Pacheco, 2001; Parolin et al., 2004). Variation in the flood regime (e.g. duration, depth, water quality) represents a major source of environmental heterogeneity among floodplain forests, with strong implications for species distributions and community dynamics (Wittmann et al., 2006; Junk et al., 2010). In general, flood regimes have been shown to mediate floristic differences between inundated and nearby non-inundated sites (Balslev et al., 1987; Wittmann et al., 2006; Costa et al., 2009; de Freitas et al., 2014; Schietti et al., 2014).
Evolutionary processes can interact with edaphic factors to influence species distributions and patterns of diversity. On the one hand, phylogenetic conservatism of edaphic niches can lead to co-occurrence of closely related species in areas of similar conditions (Fine et al., 2006; Lehtonen et al., 2015; Bacon et al., 2018). For example, Eiserhardt et al. (2013a) suggested limited niche evolution as an explanation for patterns of geographical turnover of palms across South America. On the other hand, evolutionary divergence of lineages in terms of edaphic associations (‘edaphic niche partitioning’) can promote local phylogenetic diversity when distantly related species converge on similar edaphic niches (Cavender-Bares et al., 2004b; Russo et al., 2005; Silvertown et al., 2015; Zhang et al., 2017). These processes are generally considered to be scale-dependent rather than mutually exclusive. Specifically, niche conservatism is typically more pronounced at large spatial and phylogenetic scales, whereas niche partitioning tends to be more apparent at relatively fine scales (Kembel and Hubbell, 2006; Silvertown et al., 2006; Ackerly and Cornwell, 2007). Assessing evidence for these scenarios across edaphic gradients will improve our general understanding of the scale-dependent processes that generate and maintain tropical biodiversity.
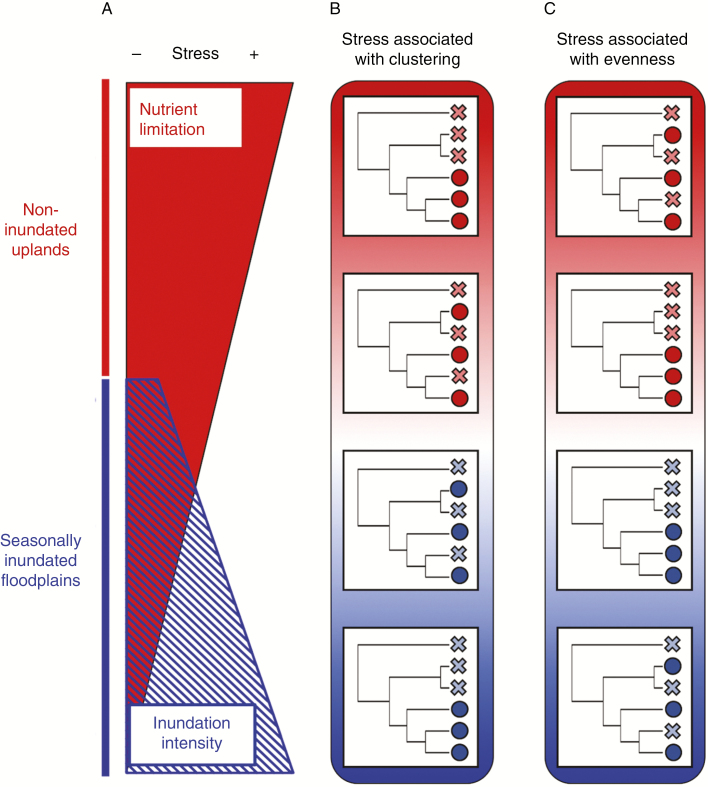
The patterns of environmental heterogeneity described above represent, in general, gradients of physiological stress within major habitat types (Fig. 1). One hypothesis that links these gradients with evolutionary processes is that high levels of abiotic stress (i.e. low soil fertility in non-inundated uplands and severe inundation intensity in seasonally inundated floodplains) exert selective pressure on multiple traits (including traits with phylogenetic signal), which can lead to phylogenetically clustered communities of closely related lineages (i.e. an extended version of the ‘stress dominance hypothesis’) (Swenson and Enquist, 2007; Fine and Kembel, 2011; Anacker and Harrison, 2012; Miller et al., 2013). In more favourable conditions (e.g. high fertility and more moderate levels of inundation), higher intensity of plant–plant competitive interactions could promote edaphic niche partitioning. As a result, communities may comprise relatively distantly related species in low-stress sites (i.e. phylogenetic over-dispersion, or ‘evenness’, via niche partitioning). In this scenario, we expect a trend from phylogenetic clustering to over-dispersion in communities that range from high to low levels of physiological stress (Fig. 1B). Alternatively, favourable conditions (i.e. fertile upland sites and less intensively inundated floodplains) could promote diversification (e.g. the ‘more individuals hypothesis’) (Srivastava and Lawton, 1998), which could lead to local assemblages comprising closely related lineages (Currie, 1991; Rosenzweig, 1995). At the same time, abiotic stress could promote co-occurrence of distantly related species (i.e. phylogenetic over-dispersion) if traits associated with stress tolerance are evolutionarily labile and evolve in a convergent way across the phylogenetic tree (Fine et al., 2005). In this case, we might expect a trend from phylogenetic over-dispersion to clustering in communities that range from high to low stress (Fig. 1C).
Fig. 1.
Physiological stress gradients can interact with evolutionary processes to mediate the phylogenetic composition of communities. (A) In lowland rain forests of western Amazonia, nutrient limitation and inundation intensity represent two sources of physiological stress in non-inundated ‘uplands’ and seasonally inundated floodplains, respectively. (B) If high levels of abiotic stress select for closely related lineages with traits enabling them to tolerate these conditions, we expect communities in relatively stressful sites to be phylogenetically clustered and communities in relatively favourable conditions to be more phylogenetically even (or ‘over-dispersed’). In this figure, species present in a given location are represented by circles and species absent from a location are represented by crosses. (C) In contrast, if low levels of abiotic stress are associated with diversification rates and evolutionary radiations, we expect communities in relatively favourable sites to be phylogenetically clustered and communities in more stressful conditions to be more phylogenetically even (or ‘over-dispersed’). See main text for additional details.
Characterizing community phylogenetic structure provides an entry point for understanding the evolutionary processes underlying diversity patterns, but additional information is required to draw robust inferences (Mason and Pavoine, 2013; Swenson, 2013; Gerhold et al., 2015). Attributes of species associated with variation of phylogenetic community composition along edaphic gradients can help clarify the processes that mediate diversity patterns. For example, a trade-off between stress tolerance and competition (Grime, 1977; Tilman and Pacala, 1993) could lead to variation in habitat specialization across species if stress-tolerant species are restricted to sites with unfavourable conditions because of their lower competitive ability across the broader range of conditions. In this case, we would expect habitat specialists to dominate communities structured by abiotic stress and generalist species to be more common in more favourable conditions. Additionally, we expect that abiotic stress gradients may select for species that attain different maximum sizes. In particular, taller species may be more common in fertile non-inundated sites, given the stronger levels of competition for light. It is also possible, however, that small understorey plants that are adapted to low-light conditions dominate fertile areas with generally taller canopies (Chazdon, 1986a, b). Floodplains, which are subject to relatively rapid rates of soil and turnover of woody stems (Hughes, 1997; Junk et al., 2010), may be dominated by small-sized species if they are able to reach reproductive maturity sooner (Lacey, 1986). Finally, phylogenetic clustering may be driven by co-occurring species of hyperdiverse lineages if, for example, these lineages have traits that have enabled them to persist and diversify under particular conditions (Anacker and Harrison, 2012; Miller et al., 2013).
Palms (Arecaceae) are an excellent group for studying the role of edaphic heterogeneity in determining floristic patterns because they are highly diverse and dominant components of Amazonian forests, and a wealth of previous research provides a foundation for interpreting diversity patterns (e.g. Svenning, 2001; Balslev et al., 2011; Eiserhardt et al., 2011; ter Steege et al., 2013; Bacon et al., 2018). In western Amazonia, for instance, Vormisto et al. (2004) and Kristiansen et al. (2012) reported associations between palm community variation and edaphic conditions. Cámara-Leret et al. (2017) reported strong associations between abundance of palm species and soil gradients in terra firme forests. Eiserhardt et al. (2013b) characterized phylogenetic structure of palm communities between major habitat types and suggested within-habitat abiotic filtering as a driver of phylogenetic community structure. While these studies have made progress in linking edaphic conditions, species distributions and community composition, we still know little about how continuous variation of edaphic conditions within major habitat types is associated with phylogenetic structure of entire palm communities. To help fill this knowledge gap, we asked two main questions.
(1) How does the phylogenetic composition of palm communities vary along edaphic gradients within major habitat types? If soil fertility and flood regime select for particular clades adapted for these conditions (e.g. via abiotic filtering), we expect phylogenetic clustering (i.e. relatively closely related co-occurring species) in non-inundated upland sites with low soil fertility and in floodplain sites with more severe flood regimes. In contrast, if physiological stress leads to stronger partitioning of edaphic niches, we expect greater phylogenetic clustering in more fertile uplands and floodplain sites with less severely flood regimes.
(2) To what extent are phylogenetic patterns determined by (a) habitat specialists, (b) small versus large palms and (c) hyperdiverse genera? (a) If a trade-off between stress tolerance and competition (Grime, 1977; Tilman and Pacala, 1993; Swenson and Enquist, 2007) affects habitat specialization by restricting stress-tolerant species to unfavourable sites, we expect phylogenetic clustering to be determined by the co-occurrence of habitat specialists in high-stress sites. Alternatively, favourable conditions (i.e. fertile upland sites and less intensively inundated floodplains) could promote diversification (e.g. the ‘more individuals hypothesis’), which could lead to local assemblages comprising closely related lineages. (b) Abiotic stress gradients may select for species that attain different maximum sizes. In relatively fertile upland sites, large palms may dominate, given the stronger levels of competition for light. In floodplains, we expect sites with more severe flood regimes to be dominated by small palms because of the rapid rates of soil and woody stem turnover. (c) Finally, patterns of phylogenetic clustering may be determined by co-occurring species of hyperdiverse lineages (e.g. Bactris and Geonoma) if these lineages have traits enabling them to succeed in stressful conditions.
MATERIALS AND METHODS
Study area and palm community transects
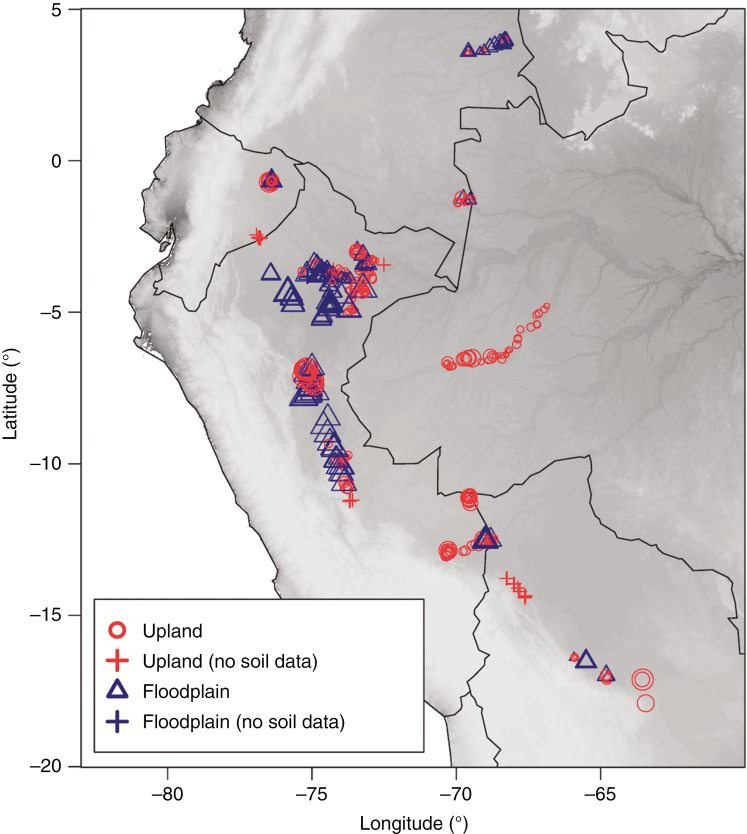
We assembled data from 501 transects of 5 × 500 m (0.25 ha) each, located throughout western Amazonia, representing a substantial expansion on previous work (Fig. 2; cf. Kristiansen et al., 2012; Eiserhardt et al., 2013b; Cámara-Leret et al., 2017). Transects were sampled between 1995 and 2012. Each transect was subdivided into 100 subunits of 5 × 5 m where all individual palms were identified, counted and assigned to a life-history stage (seedlings, juveniles, sub-adults, adults). In this study, we limited analyses to sub-adults and adults (defined as individuals of reproductive size for a species based on field knowledge) to decrease the chance of including individuals that dispersed into unfavourable locations (i.e. sink habitats). Species determinations were made by one of the authors (H.B.) with nomenclature following the World Checklist of Palms (Govaerts et al., 2011), although we do not differentiate varieties for this study. Vouchers were deposited primarily at the Aarhus University herbarium (AAU) and in the national herbaria (BOG, QCA, AMAZ, LPB, INPA). Table 1 summarizes environmental conditions across the transects.
Fig. 2.
Locations of 501 palm transects in western Amazonia in non-inundated upland (red circles, N = 325) and seasonally inundated floodplain (blue triangles, N = 176) habitats. The size of each symbol is proportional to the soil exchangeable base concentration (cmol+ kg−g) in the transect, while the grey-scale background shows elevation (darker shading indicates lower altitude). Transects without soil chemistry data are shown as + symbols.
Table 1.
Environmental variables for palm transects of the western Amazon. Values of mean annual precipitation are from Karger et al. (2017). Exchangeable base concentration (ExB conc.) is the sum of Ca, Mg and K. Georeferenced raw data are available upon request from the authors
| Habitat type | Variable | N | Unit | Mean ± s.d. | Range |
|---|---|---|---|---|---|
| Floodplain | Elevation | 325 | m a.s.l. | 172 ± 92 | 77–550 |
| Upland | Elevation | 176 | m a.s.l. | 144 ± 44 | 78–254 |
| Floodplain | Mean annual precipitation | 325 | mm yr−1 | 2,621 ± 1012 | 1088–7129 |
| Upland | Mean annual precipitation | 176 | mm yr−1 | 2,510 ± 707 | 1377 – 4338 |
| Floodplain | pH | 157 | Unitless | 4.47 ± 0.97 | 3.34–7.47 |
| Upland | pH | 245 | Unitless | 4.07 ± 0.77 | 3.29–7.33 |
| Floodplain | Al | 143 | cmol+ kg−k | 3.6 ± 4.29 | 0.00–18.57 |
| Upland | Al | 255 | cmol+ kg−k | 3.57 ± 3.05 | 0.00–14.52 |
| Floodplain | ExB conc. | 156 | cmol+ kg−k | 12.88 ± 12.63 | 0.16–101.54 |
| Upland | ExB conc. | 280 | cmol+ kg−k | 4.8 ± 9.45 | 0.04–54.22 |
| Floodplain | Ca | 156 | cmol+ kg−k | 10.04 ± 11.03 | 0.02–85.25 |
| Upland | Ca | 280 | cmol+ kg−k | 3.70 ± 8.13 | 0.01–47.62 |
| Floodplain | Mg | 156 | cmol+ kg−k | 2.45 ± 1.77 | 0.07–14.05 |
| Upland | Mg | 280 | cmol+ kg−k | 0.88 ± 1.36 | 0.00–7.42 |
| Floodplain | P | 144 | mg kg−k | 11.23 ± 7.53 | 1.15–40.20 |
| Upland | P | 280 | mg kg−k | 8.80 ± 8.78 | 0.43–89.27 |
| Floodplain | Na | 156 | cmol+ kg−k | 0.13 ± 0.10 | 0.00–0.51 |
| Upland | Na | 254 | cmol+ kg−k | 0.05 ± 0.04 | 0.00–0.36 |
| Floodplain | Clay | 120 | % | 15.73 ± 3.43 | 3.65–25.77 |
| Upland | Clay | 197 | % | 16.58 ± 12.00 | 2.79–57.58 |
| Floodplain | Silt | 120 | % | 71.49 ± 9.02 | 18.08–85.67 |
| Upland | Silt | 197 | % | 60.66 ± 15.17 | 9.21–80.86 |
| Floodplain | Sand | 120 | % | 12.77 ± 10.95 | 0.97–78.27 |
| Upland | Sand | 197 | % | 22.76 ± 17.43 | 0.00–88.00 |
Transects were assigned to two main habitat types based on field observations and interviews with local guides: non-inundated uplands (n = 325, including those classified more discretely as terra firme or terrace forest types) and seasonally inundated floodplains (n = 176, including black water, white water and intermediate floodplain types). We focus on continuous variation within (and major differences between) these two main habitat categories instead of distinguishing finer habitat types, which are often difficult to precisely define in the field. We excluded transects from high elevations (defined here as >600 m a.s.l.), white sand forests and permanently flooded swamps because we have few samples from these habitat types.
Soil chemistry and other environmental data
From a total of 406 transects (252 upland, 154 floodplain), surface soil samples (0–10 cm of mineral soil) were collected at several points (typically three samples, one from the beginning, one from the middle and one from the end of the transect). Each of the samples was a combination of five subsamples taken from the corners and the centre of the subunit. After roots and stones were removed, samples were air-dried and stored in cotton bags or plastic bottles. Exchangable nutrients were extracted by the standard ammonium acetate method and quantified using inductively coupled plasma-optical emission spectrometry (van Reeuwijk, 2002). The majority of these analyses (samlpes from 395 transects) were done at Aarhus University but samples from 11 transects were analysed at the Unversity of Turku. We focus on variation in exchangeable base concentration (i.e. sum of Mg, Ca and K; cmol+ kg−g) as a metric of soil fertility because previous work has shown this to be strongly associated with palm distributions and community patterns in western Amazonia, as well as floristic patterns of other groups in Amazonia more broadly (Ruokolainen et al., 2007; Quesada et al., 2012; Lehtonen et al., 2015; Cámara-Leret et al., 2017). Moreover, base cations are relatively mobile in the soil and do not tend to form complexes with soil organic material and clay particulates or have particular complicated nutrient cycles, as opposed to soil phosphorus (P). Nonetheless, soil P is often considered the limiting nutrient in tropical forests (Vitousek, 1984; Quesada et al., 2012; Turner et al., 2018), and so we repeated the analyses described below using soil P concentration instead of exchangeable base concentration. In our dataset, exchangeable base concentration and soil P are positively correlated (Pearson’s r = 0.40, across all samples). We provide results based on soil P in the Supplementary Data and discuss results from these analyses in the Discussion section. Although there is substantial variation in soil properties at fine spatial scales, the majority of variation in our database (94 and 88 % for exchangeable base concentration and soil P, respectively) is found among, rather than within, transects (Supplementary Data Table S1). Thus, we used transect median values for analyses because they capture broad-scale variation in soil fertility in our study area.
Because we lack direct measurements of flood regimes in our transects, we developed a floristic index of inundation intensity to test the hypothesis that phylogenetic composition is related to the severity of inundation. This index is based on previous work showing a decline in floristic similarity between inundated and non-inundated plant communities with increasing flood intensity of the inundated site (Wittmann et al., 2006; Drucker et al., 2008; Costa et al., 2009; Myster, 2009; Schietti et al., 2014). Specifically, we computed the incidence-based Sørensen dissimilarity (β diversity) of palm communities between each floodplain transect and its geographically nearest upland transect. Because floristic similarity also declines with geographical distance, we used the residuals from a regression between geographical distance between transects and the β diversity value for further analyses. High values of the index represent floodplain transects with palm communities more different from their nearest upland transect than expected by distance alone, and vice versa. Correlations between the inundation index and the measured soil variables are provided in Supplementary Data Table S2, and correlations among soil variables for upland sites are provided in Supplementary Data Table S3. Although we argue that this index captures major differences in the flood regimes of inundated sites, it is possible that the floristic differences underlying the index result from other (unmeasured) differences between sites (see Discussion section).
Phylogenetic community composition and statistical analyses
We quantified the mean phylogenetic pairwise distance (MPD) among species in each transect (Kembel et al., 2010) using the palm phylogeny of Faurby et al. (2016) (i.e. the Phylogeny_Con_Checklist.nex file provided in Appendix 3 of that paper). Although inter-generic phylogenetic resolution is modest, the metric of phylogenetic composition we used here (MPD) is not particularly sensitive to topology at the tips of the tree (Webb et al., 2002). Nonetheless, we propagated phylogenetic uncertainty through our analysis by making calculations on a set of 100 random trees from Faurby et al. (2016).
For each transect (and each of the 100 phylogenies), we calculated a standard effect size of MPD (SES.MPD) as: [robs − mean(rrand)]/s.d.(rrand), where r is the phylogenetic branch length among species in a transect. Note that we based our analysis on species presence/absence because we were most interested in the representation of particular clades across gradients as opposed to abundance patterns. robs is the observed value of MPD while rrand is a vector of MPD values based on a null model. For the null model, we generated 9999 random palm assemblages by randomizing species names across the phylogeny (separately for each of the 100 phylogenies) and calculating a value of rrand for each iteration. This procedure maintains the observed species occupancy rates and transect-level species richness values (we note that this null model gave results congruent to a set of other null models; Supplementary Data Fig. S1). The type of null model used here depends on how the regional species pool is defined (Eiserhardt et al., 2011), and we tested two different species pools (global and regional). The global species pool included the full set of species observed across all transects. To build regional pools, we used range maps from the Botanical Information and Ecology Network (BIEN) (Maitner et al., 2018) to make a list of species with ranges that overlap each transect. Analyses based on these two species pools gave very similar results (Pearson’s r = 0.96; Supplementary Data Fig. S2) and we present results based on the regional pools because this more accurately reflects species that could potentially occur in each transect. We used the median SES.MPD value across the 100 phylogenies for downstream analyses. Negative SES.MPD values indicate phylogenetic clustering (co-occurring species are more closely related than expected by chance) whereas positive values indicate phylogenetic over-dispersion (or ‘evenness’; co-occurring species are less closely related than expected by chance). To determine how the phylogenetic composition of palm communities varies along edaphic gradients within habitat types, we regressed SES.MPD values against log10 exchangeable base concentration and mean annual precipitation separately for transects in the two habitat types. We included mean annual precipitation to explore the extent to which soil fertility outperforms this climate variable in explaining phylogenetic patterns. For floodplain transects, we also included the index of inundation intensity as a model covariate.
We used three approaches to address our second question about attributes of palms responsible for driving patterns of phylogenetic clustering. First, we classified each species according to its degree of habitat specialization using the method of Chazdon et al. (2011). This analysis uses species abundances in two habitat types (i.e. uplands and floodplains) to statistically classify each species as an upland specialist, a floodplain specialist or a habitat generalist, or too rare to classify with confidence. Importantly, the method corrects for differences in sampling intensity across habitats (Chazdon et al., 2011). We used conservative settings (i.e. higher confidence required to assign species to habitat specialists) including a supermajority threshold (K = 2/3) and P = 0.0025 to correct for error rates (Chazdon et al., 2011). We then separately regressed the proportion of habitat specialists against values of SES.MPD, exchangeable base concentration and (for floodplain transects) the index of inundation of intensity. We tested for phylogenetic signal of habitat specialization using the D statistic of Fritz and Purvis (2010), a measure of phylogenetic signal for binary traits. The computation of D is based on simulating a continuous trait under a given model of evolution (e.g. Brownian motion) and then discretizing the trait based on a threshold value that gives the same prevalence of the trait as in the observed data (Fritz and Purvis, 2010). Values of D equal to zero correspond to trait evolution by Brownian motion; values equal to 1 represent random trait evolution. We also used Blomberg’s K statistic (Blomberg et al., 2003) to test for phylogenetic signal in the median value of exchangeable base concentration across transects where each species was recorded [note that measuring phylogenetic signal with Pagel’s λ (Pagel, 1992) gave congruent results and therefore here we only present results for Blomberg’s K]. Second, we used data on plant size to determine if phylogenetically clustered transects have predominantly large or small palms, on average, compared with more phylogenetically over-dispersed transects. For this, we used data on maximum stem diameter, maximum stem height and maximum leaf length from Henderson (2002), combined with data from the AAU herbarium and the Palmweb database (palmweb.org) (Göldel et al., 2015). We conducted a principal components analysis (PCA) based on these (log-transformed) size variables and extracted the first axis to use as a synthetic metric of plant size. This axis explained 74 % of the total variation and was positively associated with all three size variables (Pearson’s r ranged from 0.67 to 0.71). We then calculated the community mean value of this size metric as the mean across species present in each transect. We regressed these values against values of SES.MPD, exchangeable base concentration and (for floodplain transects) the index of inundation of intensity. Finally, we examined the relationship between the proportion of species in each transect belonging to two hyperdiverse genera (Bactris and Geonoma) and values of SES.MPD. Significant negative relationships indicate that phylogenetic clustering results from species belonging to a particular genus.
To account for spatial autocorrelation among transects, we included a Gaussian spatial correlation structure in all regression models using the gls and corGaus functions in the nlme R package (Pinheiro et al., 2013). In all cases, preliminary analyses suggested that including spatial terms improved model fit. We then used the Akaike information criterion (AIC) to compare each full model (with covariates) with an intercept-only null model that included only the spatial term. As another measure of model fit, we calculated a pseudo-R2 value based on a regression between the observed and fitted model values (Supplementary Data Table S4). All analyses were conducted in R 3.4.2 (R Development Core Team, 2018) and used functions from the packages ape (Paradis et al., 2004), caper (Orme et al., 2011), nlme (Pinheiro et al., 2013), phytools (Revell, 2012), picante (Kembel et al., 2010), raster (Hijmans and van Etten, 2013) and vegan (Oksanen et al., 2013). Data are available upon request from the authors.
RESULTS
Overall, we recorded 112 008 individuals belonging to 110 species (Supplementary Data Table S5). All except one species occurred in at least one non-inundated upland transect and 60 % of the species (n = 67) occurred in at least one floodplain transect. On average, upland transects had higher species richness than floodplain transects (mean number of species per 0.25 ha = 13.2 versus 9.1). The range of exchangeable base concentration recorded in the top 10 cm of the soil surface (0.04–101.54 cmol+ kg−k) corresponds to nearly the entire range documented in western Amazonia (Quesada et al., 2010, 2012). The median value of exchangeable base concentration was lower for upland compared with floodplain transects (0.49 versus 9.01 cmol+ kg−k), but the ranges were similar (Table 1). The median value of P concentration was also lower for upland compared with floodplain transects (8.8 versus 11.2 mg kg−k). The overall correlation between exchangeable base concentration and soil P was 0.40 and the correlation was much higher among upland transects (r = 0.44) than for floodplain transects (0.09).
Question 1: How does the phylogenetic structure of palm communities vary along edaphic gradients?
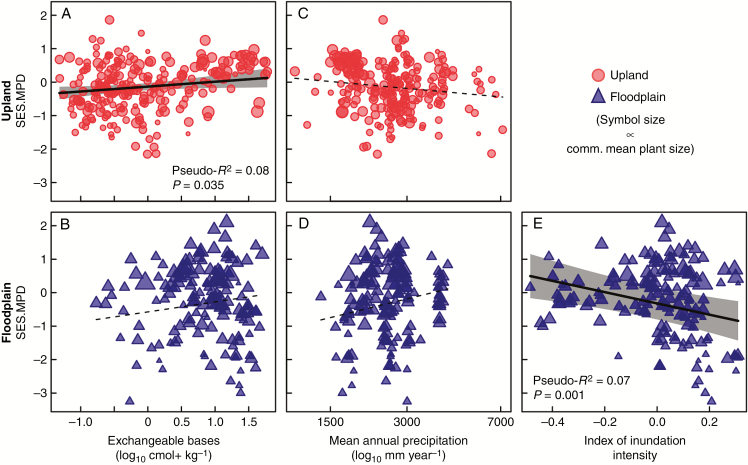
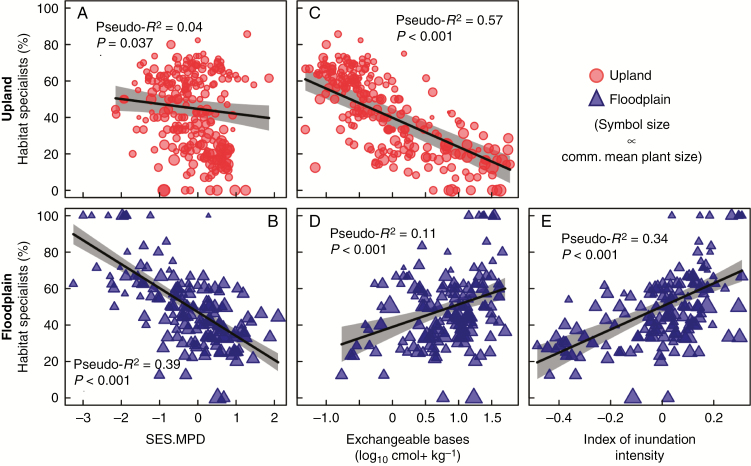
The phylogenetic composition of transects was differently associated with edaphic conditions in the two main habitat types. Specifically, among non-inundated upland transects, phylogenetic composition (SES.MPD) was positively associated with soil fertility (Fig. 3A). In other words, upland transects with poor soils tended to host phylogenetically clustered palm communities. In contrast, the phylogenetic composition of floodplain transects was not significantly related to soil fertility (Fig. 3B). Mean annual precipitation was not associated with phylogenetic composition in either habitat type (Fig. 3C, D). Among floodplain transects, there was a significant negative relationship between SES.MPD and the inundation index, suggesting that areas exposed to more severe inundation have relatively closely related palm species (Fig. 3E). Additional details are provided in Supplementary Data Table S5 and results based on soil P are presented in Supplementary Data Fig. S3.
Fig. 3.
Standard effect size of mean pairwise phylogenetic distance (SES.MPD) versus (A, B) exchangeable base concentration (log10 cmol+ kg−k), (C, D) mean annual precipitation (log10 mm year−1) and, for floodplain transects only, (E) the index of inundation intensity for western Amazonian palm communities. Top panels with (red) circles and bottom panels with (blue) triangles represent upland and floodplain transects, respectively. Symbol size is proportional to community mean plant size based on the size PCA axis 1 (see main text). Fitted model results with 95 % confidence envelopes (based on full models with spatial terms) are shown for significant (P < 0.05) relationships. Pseudo-R2 values shown for significant relationships represent the R2 value obtained from ordinary least squares regression analysis between the observed and fitted values. See Supplementary Data Table S4 for model fit statistics.
Question 2: To what extent are phylogenetic patterns determined by (a) habitat specialists, (b) small versus large palms and (c) hyperdiverse genera?
(a) Habitat specialists versus generalists
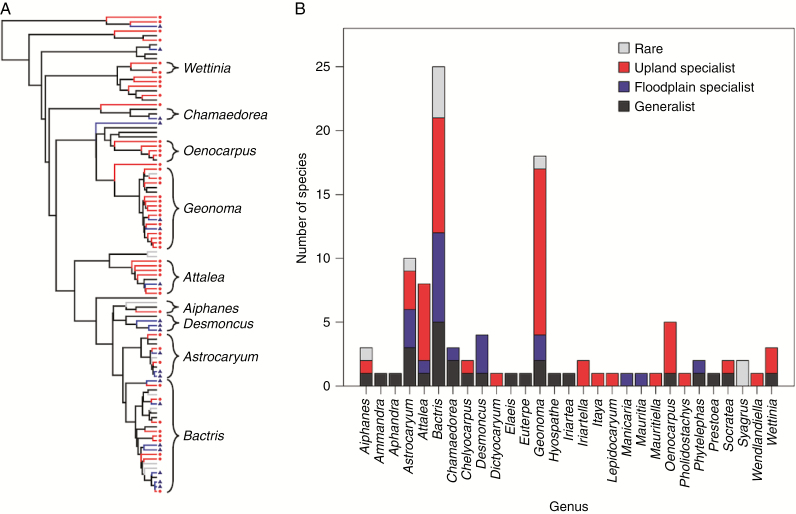
Overall, 48 species (44 %) were classified upland specialists, 20 (18 %) as floodplain specialists and 27 (25 %) as habitat generalists, and nine (8 %) were too rare to classify (Fig. 4). We briefly highlight patterns of habitat specialization for some key genera below; see Supplementary Data Table S5 for habitat classifications of all species. The diverse and widespread genus Geonoma, comprising ~68 species across Central and South America (Henderson, 2011), had a high proportion of upland specialists (13/18 species recorded in this study; 72 %) and only two floodplain specialists. Attalea and Oenocarpus were also both dominated by upland specialists (6/8 and 4/5 species, respectively). In Bactris (the genus with most species included in our study, n = 25), nine species (36 %) were classified as upland specialists and seven (28 %) as floodplain specialists, and the remaining nine (36 %) were either generalists (five species) or too rare to classify (four species). Three out of four species of Desmoncus (75 %) were classified as floodplain specialists. Based on the D statistic, specialization for both habitat types was randomly distributed across the phylogeny (D = 0.98 and 1.18 for upland and floodplain specialization, respectively; P < 0.001 for D > 1 and P > 0.1 for D = 1 for both groups). Based on Blomberg’s K statistic, the median value of exchangeable base concentration in transects where each species occurred did not exhibit significant phylogenetic signal (K = 0.09, P = 0.6; Supplementary Data Fig. S4).
Fig. 4.
(A) Phylogeny of palm species included in this study, with branches coloured for species classified as habitat specialists (upland specialists, red; floodplain specialists, blue; generalists, black; too rare to classify, grey). This panel represents a single random dichotomous tree of the phylogenetic tree sample from Faurby et al. (2016). Genera with three or more species are labelled (see Supplementary Data Table S5 for habitat classifications of all species). (B) Number of species in each genus recorded in this study coded by their habitat specialization category.
Despite the lack of tree-wide phylogenetic signal in habitat specialization and median base concentration in transects where each species occurred, phylogenetically clustered transects in both habitat types (especially floodplains) tended to have a high proportion of habitat specialists (Fig. 5A, B). Among upland transects, there was also a strong negative relationship between the proportion of upland specialists and exchangeable base concentration (Fig. 5C), indicating that upland sites with poor soils primarily have upland specialists, whereas more fertile upland sites tend to have both more generalist and more rare species. In contrast, high-fertility floodplain transects had a higher proportion of floodplain specialists (Fig. 5D). Additionally, the proportion of floodplain habitat specialists was positively associated with the index of inundation intensity (Fig. 5E). Results based on soil P are presented in Supplementary Data Fig. S5. Briefly, soil P and the percentage of habitat specialists were positively correlated in upland transects but not significantly correlated for floodplain transects, mirroring the relationship between habitat specialization and exchangeable base concentration.
Fig. 5.
Proportion of species identified as habitat specialists that occur in non-inundated upland or floodplain transects versus (A, B) phylogenetic community composition (standard effect size of mean pairwise phylogenetic distance, SES.MPD), (C, D) exchangeable base concentration, and, for floodplain transects only, (E) the index of inundation intensity. Top panels with (red) circles and bottom panels with (blue) triangles represent upland and floodplain transects, respectively. Symbol size is proportional to community mean plant size based on the size PCA axis 1 (see main text). Fitted model results with 95 % confidence envelopes (based on models with spatial terms) are shown for significant (P < 0.05) relationships. Pseudo-R2 values shown for significant relationships represent the R2 value obtained from ordinary least squares regression analysis between the observed and fitted values. See Supplementary Data Table S4 for model fit statistics.
(b) Small versus large palms
On average, upland specialists reached shorter maximum heights and had shorter maximum leaf lengths than floodplain specialists, but these groups had similar average maximum diameters (Supplementary Data Fig. S6). For transects in both habitat types, the community mean value of the size metric (i.e. the first axis of the PCA of size variables) increased with SES.MPD (Fig. 3, Supplementary Data Fig. S7). In other words, phylogenetically clustered transects in both habitat types contained, on average, small palm species. Among upland (but not floodplain) transects, the size metric was also positively associated with soil fertility. Contrary to our expectations, we did not find a significant negative relationship between the index of inundation intensity and the community mean value of palm size.
(c) Species belonging to hyperdiverse genera
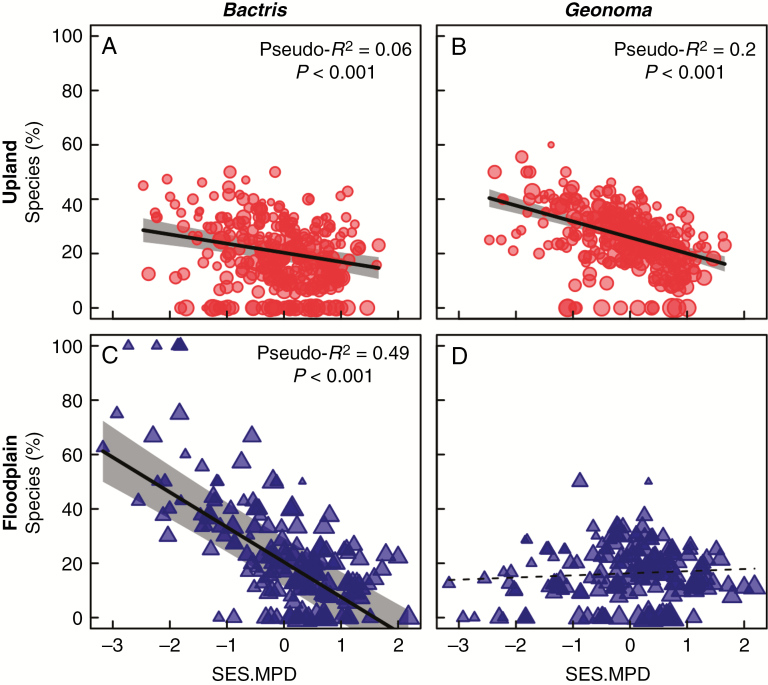
Co-occurrence of species in the two most diverse genera in our study [i.e. Bactris (25 species) and Geonoma (18 species)] largely determined the observed variation in phylogenetic community composition (Fig. 6). In particular, phylogenetically clustered upland transects had a high representation of both Geonoma and Bactris. Phylogenetically clustered floodplain transects had a high representation of Bactris, but not Geonoma. Excluding these species from the entire analysis generally led to more random values of SES.MPD and eliminated most of the significant trends presented here, which reinforces the idea that these genera dominate the observed patterns.
Fig. 6.
Percentage of species belonging to (A, C) Bactris and (B, D) Geonoma in western Amazonian palm transects versus standard effect size of mean pairwise phylogenetic distance (SES.MPD). Top panels with (red) circles and bottom panels with (blue) triangles represent upland and floodplain transects, respectively. Symbol size is proportional to community mean plant size based on the size PCA axis 1 (see main text). Fitted model results with 95 % confidence envelopes (based on models with spatial terms) are shown for significant (P < 0.05) relationships. Pseudo-R2 values shown for significant relationships represent the R2 value obtained from ordinary least squares regression analysis between the observed and fitted values. See Supplementary Data Table S4 for model fit statistics.
DISCUSSION
We found variation in the phylogenetic structure of palm communities along edaphic gradients within major habitat types across western Amazonia. Although tree-wide (or deep-time) metrics of phylogenetic signal did not support the scenario that edaphic niches are conserved across the entire phylogenetic tree, several other pieces of evidence do support more nuanced versions of edaphic niche conservatism. Specifically, (1) palm communities in low-fertility non-inundated uplands comprise phylogenetically clustered sets of small-sized upland specialists (especially Bactris and Geonoma), and (2) palm communities in severely inundated floodplains are dominated by closely related floodplain specialists (especially Bactris). Overall, strong associations between edaphic conditions and the phylogenetic composition of palm communities across western Amazonia suggest that, in general, physiological stress mediates evolutionary processes that have structured palm communities in this system. The most important sources of physiological stress, however, differ across habitat types. Our results increase our understanding of the factors that determine diversity patterns for palms and, more generally, advance the role of edaphic heterogeneity in promoting patterns of tropical plant diversity. Overall, our results support phylogenetic conservatism of edaphic niches and selection along abiotic gradients as key processes affecting community assembly for western Amazonian palms. Processes thought to promote edaphic niche partitioning (e.g. negative density dependence and microhabitat niche differences) may occur at finer spatial scales.
Community phylogenetic structure along edaphic gradients
Several studies have reported a lack of tree-wide (or deep-time) phylogenetic signal in soil and topographic associations for tropical tree species (Schreeg et al., 2010; Zhang et al., 2017). In fact, adaptive radiation across edaphic gradients has been demonstrated in a number of systems (e.g. Cavender-Bares et al., 2004a; Fine et al., 2005) and Swenson (2013) suggested that soil niches may be evolutionarily labile within tropical tree genera and families (i.e. often not similar among closely related species). On the one hand, some of our results are consistent with the conclusion that edaphic associations are not phylogenetically conserved. In particular, habitat specialization and the median values of soil fertility across transects where species occur did not show significant phylogenetic signal based on the D and K statistics, respectively. On the other hand, however, these metrics provide an all-or-nothing value of phylogenetic signal and do not describe more nuanced patterns of trait evolution among clades. It is certainly possible that edaphic niches are phylogenetically conserved in some clades and, indeed, several other patterns we found (detailed below) do imply a degree of phylogenetic conservatism of edaphic associations that appears to be important for the community structure of western Amazonian palms. A conclusion of phylogenetically conserved edaphic associations is generally consistent with Eiserhardt et al. (2013a), who found that the geographical turnover of palm clades across South America was driven by limited niche evolution (with respect to temperature and soil tolerances) in conjunction with limited dispersal.
In non-inundated upland sites, the association between phylogenetic clustering and soil fertility may reflect selection on phylogenetically conserved traits in low-fertility transects. In support of this scenario, we found that nutrient-poor upland sites had communities of closely related, small-sized upland specialists. Based on a portion of the dataset used here, Eiserhardt et al. (2013b) found that palm communities in both terra firme and floodplain transects tended to be phylogenetically clustered even with respect to the pool of species known to occur in each particular habitat type. They suggested an additional component of abiotic filtering within habitats, which we have identified in the current study as the within-habitat soil fertility gradient. Within-habitat edaphic gradients have also been shown to mediate palm phylogenetic composition in a central Amazonian terra firme forest (de Freitas et al., 2014). Specifically, palm communities in ‘bottomland’ sites that can be flooded for a few hours following heavy rains tended to be phylogenetically over-dispersed compared with sites higher above the nearest waterway (and with higher clay content). It is possible that the bottomland communities studied by de Freitas et al. (2014) comprise a mix of upland and floodplain-associated species, whereas less flood-prone sites are dominated by relatively closely related upland species. Alternatively, phylogenetic clustering in low-fertility upland sites may be more strongly related to particularly high rates of diversification of certain lineages (e.g. Geonoma) that are specialized to those conditions (Onstein et al., 2017). It is also possible that differences in soil age between low- and high-fertility sites mediate the phylogenetic patterns in ways that mirror patterns found more broadly across Amazonia (Honorio Coronado et al., 2015). Specifically, if there is a negative association between soil fertility and soil age within western Amazonia (as there is when comparing eastern and western Amazonia), low-fertility soils may have had a longer time to accumulate closely related lineages via in situ speciation. Western Amazonia has, however, experienced substantial geological dynamism since the Miocene and soils in the region are generally quite young (i.e. Pliocene or Pleistocene) compared with the eastern parts of the Amazon basin (Hoorn et al., 2010; Quesada et al., 2011; Roncal et al., 2013; Jaramillo et al., 2017). Future work examining relationships between geological time, uplift/soil formation and evolutionary patterns of specific taxonomic groups will help clarify these questions.
We focused on exchangeable base concentration as a metric of soil fertility, but we note that P is often considered the primary limiting nutrient in tropical soils (Vitousek, 1984; Quesada et al., 2012; Turner et al., 2018). In our dataset, as is more generally the case (Fyllas et al., 2009; Quesada et al., 2012), exchangeable base concentration and soil P are positively correlated. Nonetheless, models incorporating exchangeable base concentration performed better (based on the AIC) than models based on soil P. Results from models based on these two parameters differed mainly in that some relationships with exchangeable base concentration that were significant became non-significant when considering soil P (see Supplementary Data Table S4). This is consistent with previous work suggesting a stronger association between floristic patterns of Amazonian palms and exchangeable base concentration compared with soil P (e.g. Kristiansen et al., 2012; Cámara-Leret et al., 2017). Turner et al. (2018) recently showed a wide variation across tropical tree species in terms of P limitation and, while they did not include palms in their study, it is possible that palms may be more sensitive than other lineages to exchangeable base concentration compared with soil P. For example, Emilio et al. (2014) found a significant response by palms, but not trees, to a gradient of exchangeable base concentration in forests of central Amazonia. Since Ca is a key component of plant cell wall structure (Demarty et al., 1984), it is possible that this nutrient may be particularly important for maintaining the large, thick leaves characteristic of palms. Some other work suggests that palms are particularly rich in K (Vitousek and Sanford, 1986) and so it is possible that palms are sensitive to K availability. Literature related to these issues is, however, scarce and more work is needed to evaluate the mechanisms driving palm responses to different soil nutrients.
Regarding floodplain transects, we conclude that palm communities that differ more floristically from nearby upland palm communities (ranking high on our index of inundation) tend to be phylogenetically clustered. We interpret this result as providing support to the hypothesis that communities subjected to more severe flood regimes tend to comprise phylogenetically clustered communities of floodplain specialists. Our interpretation rests on the index of inundation intensity we developed from floristic patterns since we lack direct measurements of flood regimes in these transects. The justification for our approach is based on previous work showing strong links between Amazonian palm community composition and flood regimes (Balslev et al., 1987; Wittmann et al., 2006; Costa et al., 2009; de Freitas et al., 2014; Schietti et al., 2014). Nonetheless, it is possible that the patterns we attribute to inundation intensity are caused by other unmeasured factors associated with floristic differences between floodplain and upland sites. In any case, our results still support phylogenetic conservatism of floodplain associations. This supports the hypothesis that tolerance of inundation reflects an important axis of abiotic stress as well as phylogenetic niche conservatism of traits associated with flood tolerance (Junk et al., 2010). Unfortunately, we currently lack high-precision GPS coordinates as well as high-resolution remote sensing data (e.g. high-fidelity imaging spectroscopy and LiDAR data collected to account for seasonal variation) with which to accurately determine the flood regime or other related variables for our dataset (e.g. height above the nearest drainage; Rennó et al., 2008). Future work would benefit from additional data describing flood regimes in these forests, most likely obtained through remote sensing approaches (Rudorff et al., 2014; Hess et al., 2015). Recent development of various remote sensing technologies [e.g. synthetic aperture radar data (e.g. Sentinel) or polarimetric imagery (e.g. ALOS-2/PALSAR-2)] show promising results for mapping of flooded vegetation areas and accurately delineating areas of open water (Plank et al., 2017). Given the importance of hydrology in determining floristic patterns and dynamics in the region, future work will benefit from more detailed information on flood regimes and soil moisture.
Comparison with other plant groups
To what extent do the phylogenetic patterns we report for western Amazonian palms correspond to or contrast with results from other studies focusing on different plant groups? For example, work by Fine and colleagues suggests that habitat specialization in western Amazonian tree communities across soil fertility gradients is driven by phylogenetically constrained trade-offs in growth and defence (Fine et al., 2006; Fine and Kembel, 2011). Fine and Kembel (2011) found phylogenetic clustering of western Amazonian tree communities on nutrient-poor white sand soils, which is consistent with phylogenetic conservation of edaphic niches. They attributed their findings to a trade-off between herbivore defence and growth rate, which has been suggested to be a key driver of habitat specialization for trees on nutrient-poor white sand soils and more fertile upland soils (Fine et al., 2006). Similarly, for western Amazonian palm communities examined in our study, patterns of phylogenetic clustering were explained by a higher proportion of habitat specialists in low-fertility upland sites and areas with higher inundation intensity in flooded sites. Furthermore, low-fertility upland sites had smaller sized palms. Together, this suggests that phylogenetically constrained trade-offs in growth and defence may be a general driver of phylogenetic community composition across plant groups. Fine et al. (2005) reported evidence for repeated and independent evolution of soil specialization in western Amazonian trees (Burseraceae), proposing that multiple and rapid speciation events followed shifts in soil preference. To some extent, this finding is also consistent with our work showing that species belonging to two large genera (Geonoma and Bactris) dominate many patterns we report (also see below).
Our finding that upland palm communities are phylogenetically clustered in areas with low soil fertility contrasts with Lehtonen et al. (2015), who reported that Neotropical fern communities tend to be phylogenetically clustered in areas of high soil fertility. In contrast, older fern lineages were disproportionately represented in geologically older sites with poor soils. Lehtonen et al. (2015) attributed their results, in part, to the basal phylogenetic position of two key genera that specialized on infertile soils. In our study, the dominant phylogenetic patterns are mediated by co-occurrence of species in relatively shallow phylogenetic nodes (e.g. co-occurring species of Bactris and Geonoma). Even though we found the opposite pattern from Lehtonen et al. (2015) in terms of the association between phylogenetic clustering and soil fertility, both studies indicate a role for edaphic niche conservatism.
Phylogenetic patterns are dominated by small-sized habitat specialists belonging to hyperdiverse genera
Phylogenetic clustering in both habitat types resulted largely from the co-occurrence of habitat specialists. This further supports our conclusion that edaphic niches are phylogenetically conserved, at least across broad habitat types (e.g. non-inundated upland versus seasonally inundated floodplain conditions). Additionally, we found associations between phylogenetic patterns and community-mean values of palm size, suggesting that phylogenetic patterns emerge, in particular, from co-occurrence of small-sized (understorey) palms. Finally, patterns of phylogenetic clustering were strongly related to the proportion of co-occurring species belonging to the two most diverse genera included in our study: Bactris and Geonoma. These genera are both broadly distributed across the Neotropics and predominantly comprise small, understorey species. Together, these results provide insight not only into the factors underpinning community-wide patterns of diversity in western Amazonian palms, but also into the co-occurrence patterns of these two hyperdiverse genera.
Previous work on the distributions and evolutionary history of Bactris and Geonoma may clarify mechanisms underlying patterns of palm community composition in western Amazonia. For example, Cámara-Leret et al. (2017) modelled palm abundance distributions along edaphic gradients. They reported that congeners of Bactris and Geonoma reached their maximal abundance in transects with low values of exchangeable base concentrations, which is consistent with our results suggesting phylogenetic conservatism of edaphic associations. Roncal et al. (2012) found that the timing of Geonoma diversification was consistent with major geological events in Amazonia and that a high degree of relatedness among co-occurring species may reflect in situ diversification patterns. This may suggest that species of Geonoma are differentiated on resource axes other than edaphic conditions (e.g. light) (Chazdon, 1991; Roncal, 2014), which may also be consistent with our findings. In fact, Roncal et al. (2012) found phylogenetic signal in some morphological characters of Geonoma species but not for characters with clear relationships to edaphic conditions. It is also possible that fine-scale partitioning of edaphic niches could promote co-occurrence of closely related species. For example, associations with arbuscular mycorrhizal fungi across relatively fine-scale soil gradients appear to have contributed to sympatric speciation of Howea palms (Savolainen et al., 2006; Osborne et al., 2018).
In our dataset, upland specialists (which were over-represented in low-fertility sites) are smaller, on average, compared with floodplain specialists or generalists. This pattern is largely driven by high diversity of understorey palm species (e.g. species of Geonoma). We suspect that because more fertile soils promote taller and denser forests (and thus less understorey light availability), these sites select for larger-size palms both from the perspectives of competition for light and of nutrient supply. The tendency for small palms to occur in low-fertility sites, in addition to their shade-tolerant strategies, suggests that conservative resource strategies, in general, may be critical for this group (Chazdon, 1986b). In contrast, phylogenetically clustered floodplain transects tended to have particularly high proportions of small-sized Bactris species. This result is consistent with our original prediction based on the rapid rates of soil and woody stem turnover in sites that experience more severe flood regimes (see also Emilio et al., 2014).
Conclusions
Edaphic conditions have repeatedly been shown to influence the floristic composition and species distributions of many tropical plants, including palms (John et al., 2007; Andersen et al., 2010; Higgins et al., 2011; Quesada et al., 2012; Condit et al., 2013; Muscarella et al., 2016; Cámara-Leret et al., 2017). Our study links edaphic gradients with palm community phylogenetic structure and reinforces the need to integrate edaphic conditions in eco-evolutionary studies in order to better understand the processes that generate and maintain tropical forest diversity. Overall, our results do not support niche partitioning as the main process determining palm community composition across western Amazonia. Processes that promote niche partitioning are likely important for promoting co-occurrence of palm species at fine spatial scales. However, our results are more consistent with phylogenetic conservatism of edaphic niches and abiotic filtering as a key driver of community assembly for western Amazonian palms, because palm communities tended to be phylogenetically clustered in areas of high physiological stress.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: results of analysis comparing within- versus among-transect variation in exchangeable base concentration. Table S2: correlations among soil chemistry variables and index of inundation intensity for floodplain transects. Table S3: correlations between soil chemistry variables for upland transects. Table S4: model fit statistics. Table S5: details of species recorded in this study. Figure S1: correlation between SES.MPD values based on different null models. Figure S2: correlation between SES.MPD values based on the ‘global’ and ‘full’ species pools. Figure S3: standard effect size of mean pairwise phylogenetic distance versus soil P, mean annual precipitation and inundation index. Figure S4: median log10 exchangeable base and log10 phosphorus concentrations across transects where species occur, mapped onto a phylogeny. Figure S5: proportion of species identified as habitat specialists that occur in non-inundated upland or floodplain transects versus phylogenetic community composition (SES.MPD), soil P and the index of inundation intensity. Figure S6: maximum stem height for palm species assigned to four categories of habitat specialization. Figure S7: community mean value of palm size in relation to covariates.
ACKNOWLEDGEMENTS
We thank Birgitte Kretzschmar Tagesen for handling soil samples and local guides for making fieldwork possible. Three anonymous reviewers helped improve this manuscript. This work was supported by the Danish Council for Independent Research – Natural Sciences (grant 4181-00158 to H.B.) and the European Community (FP7 grant 212631 to H.B.). J.C.S. considers this work a contribution to his VILLUM Investigator project (VILLUM FONDEN, grant 16549). A.A. is funded by the Swedish Research Council (B0569601), a Wallenberg Academy Fellowship, the Wenner-Gren Foundation and the Swedish Foundation for Strategic Research. The authors declare no conflict of interest.
LITERATURE CITED
- Ackerly DD, Cornwell W. 2007. A trait-based approach to community assembly: partitioning of species trait values into within- and among-community components. Ecology Letters 10: 135–145. [DOI] [PubMed] [Google Scholar]
- Aleixo A. 2002. Molecular systematics and the role of the Várzea-Terra-Firme ecotone in the diversification of Xiphorhynchus woodcreepers (Aves: Dendrocolaptidae). Auk 119: 621–640. [Google Scholar]
- Anacker BL, Harrison SP. 2012. Historical and ecological controls on phylogenetic diversity in Californian plant communities. American Naturalist 180: 257–269. [DOI] [PubMed] [Google Scholar]
- Andersen KM, Turner BL, Dalling JW. 2010. Soil-based habitat partitioning in understorey palms in lower montane tropical forests. Journal of Biogeography 37: 278–292. [Google Scholar]
- Antonelli A, Sanmartín I. 2011. Why are there so many plant species in the Neotropics?Taxon 60: 403–414. [Google Scholar]
- Asner GP, Anderson CB, Martin RE, Tupayachi R, Knapp DE, Sinca F. 2015. Landscape biogeochemistry reflected in shifting distributions of chemical traits in the Amazon forest canopy. Nature Geoscience 8: 567–573. [Google Scholar]
- Bacon CD, Velásquez‐Puentes FJ, Hoorn C, Antonelli A. 2018. Iriarteeae palms tracked the uplift of Andean cordilleras. Journal of Biogeography 45: 1653–1663. [Google Scholar]
- Balslev H, Luteyn JL, Øllgaard B, Holm-Nielsen LB. 1987. Composition and structure of adjacent unflooded and floodplain forest in Amazonian Ecuador. Opera Botanica 92: 37–57. [Google Scholar]
- Balslev H, Kahn F, Millan B, et al. 2011. Species diversity and growth forms in tropical American palm communities. Botanical Review 77: 381–425. [Google Scholar]
- Blomberg SP, Garland T, Ives AR. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57: 717–745. [DOI] [PubMed] [Google Scholar]
- Cámara-Leret R, Tuomisto H, Ruokolainen K, Balslev H, Munch Kristiansen S. 2017. Modelling responses of western Amazonian palms to soil nutrients. Journal of Ecology 105: 367–381. [Google Scholar]
- Cavender-Bares J, Ackerly DD, Baum DA, Bazzaz FA. 2004a Phylogenetic overdispersion in Floridian oak communities. American Naturalist 163: 823–843. [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J, Kitajima K, Bazzaz FA. 2004b Multiple trait associations in relation to habitat differentiation among 17 Floridian oak species. Ecological Monographs 74: 635–662. [Google Scholar]
- Chazdon RL. 1986a Light variation and carbon gain in rain forest understorey palms. Journal of Ecology 74: 995–1012. [Google Scholar]
- Chazdon RL. 1986b Physiological and morphological basis of shade tolerance in rain forest understory palms. Principes 30: 92–99. [Google Scholar]
- Chazdon RL. 1991. Plant size and form in the understory palm genus Geonoma: are species variations on a theme?American Journal of Botany 78: 680–694. [Google Scholar]
- Chazdon RL, Chao A, Colwell RK, et al. 2011. A novel statistical method for classifying habitat generalists and specialists. Ecology 92: 1332–1343. [DOI] [PubMed] [Google Scholar]
- Chesson P. 2000. Mechanisms of maintenance of species diversity. Annual Review of Ecology and Systematics 31: 343–366. [Google Scholar]
- Clark DB, Clark DA, Read JM. 1998. Edaphic variation and the mesoscale distribution of tree species in a neotropical rain forest. Journal of Ecology 86: 101–112. [Google Scholar]
- Condit R, Engelbrecht BMJ, Pino D, Pérez R, Turner BL. 2013. Species distributions in response to individual soil nutrients and seasonal drought across a community of tropical trees. Proceedings of the National Academy of Sciences of the USA 110: 5064–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FRC, Guillaumet J-L, Lima AP, Pereira OS. 2009. Gradients within gradients: the mesoscale distribution patterns of palms in a central Amazonian forest. Journal of Vegetation Science 20: 69–78. [Google Scholar]
- Currie DJ. 1991. Energy and large-scale patterns of animal- and plant-species richness. American Naturalist 137: 27–49. [Google Scholar]
- Demarty M, Morvan C, Thellier M. 1984. Calcium and the cell wall. Plant, Cell and Environment 7: 441–448. [Google Scholar]
- Drucker DP, Costa FRC, Magnusson WE. 2008. How wide is the riparian zone of small streams in tropical forests? A test with terrestrial herbs. Journal of Tropical Ecology 24: 65–74. [Google Scholar]
- Dumont JF, Lamotte S, Kahn F. 1990. Wetland and upland forest ecosystems in Peruvian Amazonia: plant species diversity in the light of some geological and botanical evidence. Forest Ecology and Management 33: 125–139. [Google Scholar]
- Eiserhardt WL, Svenning J-C, Kissling WD, Balslev H. 2011. Geographical ecology of the palms (Arecaceae): determinants of diversity and distributions across spatial scales. Annals of Botany 108: 1391–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiserhardt WL, Svenning JC, Baker RJ, Couvreur TLP, Balslev H. 2013a Dispersal and niche evolution jointly shape the geographic turnover of phylogenetic clades across continents. Scientific Reports 3: doi: 10.1038/srep01164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiserhardt WL, Svenning JC, Borchsenius F, Kristiansen T, Balslev H. 2013b Separating environmental and geographical determinants of phylogenetic community structure in Amazonian palms (Arecaceae). Botanical Journal of the Linnean Society 171: 244–259. [Google Scholar]
- Emilio T, Quesada CA, Costa FRC, et al. 2014. Soil physical conditions limit palm and tree basal area in Amazonian forests. Plant Ecology & Diversity 7: 215–229. [Google Scholar]
- Faurby S, Eiserhardt WL, Baker WJ, Svenning J-C. 2016. An all-evidence species-level supertree for the palms (Arecaceae). Molecular Phylogenetics and Evolution 100: 57–69. [DOI] [PubMed] [Google Scholar]
- Figueiredo FOG, Zuquim G, Tuomisto H, Moulatlet GM, Balslev H, Costa FRC. 2018. Beyond climate control on species range: the importance of soil data to predict distribution of Amazonian plant species. Journal of Biogeography 45: 190–200. [Google Scholar]
- Fine PVA, Kembel SW. 2011. Phylogenetic community structure and phylogenetic turnover across space and edaphic gradients in western Amazonian tree communities. Ecography 34: 552–565. [Google Scholar]
- Fine PVA, Daly DC, Cameron KM. 2005. The contribution of edaphic heterogeneity to the evolution and diversity of Burseraceae trees in the western Amazon. Evolution 59: 1464–1478. [PubMed] [Google Scholar]
- Fine PVA, Miller ZJ, Mesones I, et al. 2006. The growth-defense trade-off and habitat specialization by plants in Amazonian forests. Ecology 87: S150–S162. [DOI] [PubMed] [Google Scholar]
- Fortunel C, Paine CET, Fine PVA, Kraft NJB, Baraloto C. 2014. Environmental factors predict community functional composition in Amazonian forests. Journal of Ecology 102: 145–155. [Google Scholar]
- de Freitas CG, de Sales Dambros C, Eiserhardt WL, Costa FRC, Svenning J-C, Balslev H. 2014. Phylogenetic structure of a palm community in the central Amazon: changes along a hydro-edaphic gradient. Plant Ecology 215: 1173–1185. [Google Scholar]
- Fritz SA, Purvis A. 2010. Selectivity in mammalian extinction risk and threat types: a new measure of phylogenetic signal strength in binary traits. Conservation Biology 24: 1042–1051. [DOI] [PubMed] [Google Scholar]
- Fyllas NM, Patiño S, Baker TR, et al. 2009. Basin-wide variations in foliar properties of Amazonian forest: phylogeny, soils and climate. Biogeosciences 6: 2677–2708. [Google Scholar]
- Gerhold P, Cahill JF, Winter M, Bartish IV, Prinzing A, Venail P. 2015. Phylogenetic patterns are not proxies of community assembly mechanisms (they are far better). Functional Ecology 29: 600–614. [Google Scholar]
- Göldel B, Kissling WD, Svenning J-C. 2015. Geographical variation and environmental correlates of functional trait distributions in palms (Arecaceae) across the New World. Botanical Journal of the Linnean Society 179: 602–617. [Google Scholar]
- Govaerts R, Dransfield J, Zona SF, Hodel DR, Henderson A. 2011. World checklist of Arecaceae. Facilitated by the Royal Botanic Gardens, Kew: http://apps.kew.org/wcsp. [Google Scholar]
- Grime JP. 1977. Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. American Naturalist 111: 1169–1194. [Google Scholar]
- Harvey MG, Aleixo A, Ribas CC, Brumfield RT. 2017. Habitat association predicts genetic diversity and population divergence in Amazonian birds. American Naturalist 190: 631–648. [DOI] [PubMed] [Google Scholar]
- Henderson A. 2002. Evolution and ecology of palms. New York: New York Botanical Garden Press. [Google Scholar]
- Henderson A. 2011. A revision of Geonoma (Arecaceae). Phytotaxa 17: 1–271. [Google Scholar]
- Hess LL, Melack JM, Affonso AG, Barbosa CCF, Gastil-Buhl M, Novo EMLM. 2015. LBA-ECO LC-07 wetland extent, vegetation, and inundation: lowland Amazon Basin. ORNL DAAC, Oak Ridge, TN, USA: 10.3334/ORNLDAAC/1284. [DOI] [Google Scholar]
- Higgins MA, Ruokolainen K, Tuomisto H, et al. 2011. Geological control of floristic composition in Amazonian forests. Journal of Biogeography 38: 2136–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, van Etten J. 2013. raster: Geographic data analysis and modeling. R package version 2.1–25. http://CRAN.R-project.org/package=raster. [Google Scholar]
- Honorio Coronado EN, Dexter KG, Pennington RT, et al. 2015. Phylogenetic diversity of Amazonian tree communities. Diversity and Distributions 21: 1295–1307. [Google Scholar]
- Hoorn C, Wesselingh FP, ter Steege H, et al. 2010. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science 330: 927–931. [DOI] [PubMed] [Google Scholar]
- Hughes FMR. 1997. Floodplain biogeomorphology. Progress in Physical Geography 21: 501–529. [Google Scholar]
- Jaramillo C, Romero I, D’Apolito C, et al. 2017. Miocene flooding events of western Amazonia. Science Advances 3: e1601693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John R, Dalling JW, Harms KE, et al. 2007. Soil nutrients influence spatial distributions of tropical tree species. Proceedings of the National Academy of Sciences of the USA 104: 864–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junk WJ, Piedade MTF, Wittmann F, Schöngart J, Parolin P. 2010. Amazonian floodplain forests: ecophysiology, biodiversity and sustainable management. New York: Springer. [Google Scholar]
- Kahn F, de Granville J-J. 1992. Palms in forest ecosystems of Amazonia. Berlin: Springer. [Google Scholar]
- Karger DN, Conrad O, Böhner J, et al. 2017. Climatologies at high resolution for the earth’s land surface areas. Scientific Data 4: 170122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel SW, Hubbell SP. 2006. The phylogenetic structure of a neotropical forest tree community. Ecology 87: 86–99. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, et al. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26: 1463–1464. [DOI] [PubMed] [Google Scholar]
- Kristiansen T, Svenning J-C, Eiserhardt WL, et al. 2012. Environment versus dispersal in the assembly of western Amazonian palm communities. Journal of Biogeography 39: 1318–1332. [Google Scholar]
- Lacey EP. 1986. Onset of reproduction in plants: size- versus age-dependency. Trends in Ecology & Evolution 1: 72–75. [DOI] [PubMed] [Google Scholar]
- Lehtonen S, Jones MM, Zuquim G, Prado J, Tuomisto H. 2015. Phylogenetic relatedness within Neotropical fern communities increases with soil fertility. Global Ecology and Biogeography 24: 695–705. [Google Scholar]
- Maitner BS, Boyle B, Casler N, et al. 2018. The BIEN R package: a tool to access the Botanical Information and Ecology Network (BIEN) database. Methods in Ecology and Evolution 9: 373–379. [Google Scholar]
- Markewitz D, Davidson EA, Figueiredo RDO, Victoria RL, Krusche AV. 2001. Control of cation concentrations in stream waters by surface soil processes in an Amazonian watershed. Nature 410: 802–805. [DOI] [PubMed] [Google Scholar]
- Mason NWH, Pavoine S. 2013. Does trait conservatism guarantee that indicators of phylogenetic community structure will reveal niche-based assembly processes along stress gradients?Journal of Vegetation Science 24: 820–833. [Google Scholar]
- Miller ET, Zanne AE, Ricklefs RE. 2013. Niche conservatism constrains Australian honeyeater assemblages in stressful environments. Ecology Letters 16: 1186–1194. [DOI] [PubMed] [Google Scholar]
- Moulatlet GM, Costa FRC, Rennó CD, Emilio T, Schietti J. 2014. Local hydrological conditions explain floristic composition in lowland Amazonian forests. Biotropica 46: 395–403. [Google Scholar]
- Muscarella R, Uriarte M, Erickson DL, Swenson NG, Kress WJ, Zimmerman JK. 2016. Variation of tropical forest assembly mechanisms across regional environmental gradients. Perspectives in Plant Ecology Evolution and Systematics 23: 52–62. [Google Scholar]
- Myster RW. 2009. Plant communities of western Amazonia. Botanical Review 75: 271–291. [Google Scholar]
- Myster RW. 2017. Forest structure, function and dynamics in Western Amazonia: West Sussex, UK: John Wiley & Sons. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, et al. 2013. vegan: community ecology package. R package version 2.0–7. http://CRAN.R-project.org/package=vegan. [Google Scholar]
- Onstein RE, Baker WJ, Couvreur TLP, Faurby S, Svenning J-C, Kissling WD. 2017. Frugivory-related traits promote speciation of tropical palms. Nature Ecology & Evolution 1: 1903–1911. [DOI] [PubMed] [Google Scholar]
- Orme D, Freckleton RP, Thomas G, Petzoldt T, Fritz S, Isaac N. 2011. caper: comparative analyses of phylogenetics and evolution in R. Version 0.4. London: Imperial College. [Google Scholar]
- Osborne OG, De-Kayne R, Bidartondo MI, et al. 2018. Arbuscular mycorrhizal fungi promote coexistence and niche divergence of sympatric palm species on a remote oceanic island. New Phytologist 217: 1254–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco MAW. 2001. Effects of flooding and herbivores on variation in recruitment of palms between habitats. Journal of Ecology 89: 358–366. [Google Scholar]
- Pagel MD. 1992. A method for the analysis of comparative data. Journal of Theoretical Biology 156: 431–442. [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Parolin P, De Simone O, Haase K, et al. 2004. Central Amazonian floodplain forests: tree adaptations in a pulsing system. Botanical Review 70: 357–380. [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D. 2013. nlme: linear and nonlinear mixed effects models. R package version 3.1–131. https://CRAN.R-project.org/package=nlme. [Google Scholar]
- Plank S, Jüssi M, Martinis S, Twele A. 2017. Mapping of flooded vegetation by means of polarimetric Sentinel-1 and ALOS-2/PALSAR-2 imagery. International Journal of Remote Sensing 38: 3831–3850. [Google Scholar]
- Quesada CA, Lloyd J, Schwarz M, et al. 2010. Variations in chemical and physical properties of Amazon forest soils in relation to their genesis. Biogeosciences 7: 1515–1541. [Google Scholar]
- Quesada CA, Lloyd J, Anderson LO, Fyllas NM, Schwarz M, Czimczik CI. 2011. Soils of Amazonia with particular reference to the RAINFOR sites. Biogeosciences 8: 1415–1440. [Google Scholar]
- Quesada CA, Phillips OL, Schwarz M, et al. 2012. Basin-wide variations in Amazon forest structure and function are mediated by both soils and climate. Biogeosciences 9: 2203–2246. [Google Scholar]
- R Development Core Team 2018. R: a language and environment for statistical computing. v 3.5.1 ed. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- van Reeuwijk LP. 2002. Procedures for soil analysis, 6th edition. ISRIC Technical Paper 9. Wageningen: International Soil Reference and Information Centre, Wageningen. [Google Scholar]
- Rennó CD, Nobre AD, Cuartas LA, et al. 2008. HAND, a new terrain descriptor using SRTM-DEM: mapping terra-firme rainforest environments in Amazonia. Remote Sensing of Environment 112: 3469–3481. [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. [Google Scholar]
- Ritter CD, Zizka A, Barnes C, Nilsson RH, Roger F, Antonelli A. 2018a High-throughput metabarcoding reveals the effect of physicochemical soil properties on soil and litter biodiversity and community turnover across Amazonia. Peer J 6:e5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter CD, Zizka A, Roger F, et al. 2018. b High-throughput metabarcoding reveals the effect of physicochemical soil properties on soil and litter biodiversity and community turnover across Amazonia. PeerJ Preprints 6: e27012v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncal J. 2014. Edaphic and light conditions of sympatric plant morphotypes in western Amazonia. Biodiversity Data Journal 2: e1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncal J, Henderson A, Borchsenius F, Cardoso SRS, Balslev H. 2012. Can phylogenetic signal, character displacement, or random phenotypic drift explain the morphological variation in the genus Geonoma (Arecaceae)?Biological Journal of the Linnean Society 106: 528–539. [Google Scholar]
- Roncal J, Kahn F, Millan B, Couvreur TLP, Pintaud J-C. 2013. Cenozoic colonization and diversification patterns of tropical American palms: evidence from Astrocaryum (Arecaceae). Botanical Journal of the Linnean Society 171: 120–139. [Google Scholar]
- Rosenzweig ML. 1995. Species diversity in space and time. Cambridge: Cambridge University Press. [Google Scholar]
- Rudorff CM, Melack JM, Bates PD. 2014. Flooding dynamics on the lower Amazon floodplain: 1. Hydraulic controls on water elevation, inundation extent, and river-floodplain discharge. Water Resources Research 50: 619–634. [Google Scholar]
- Ruokolainen K, Tuomisto H, Macía MJ, Higgins MA, Yli-Halla M. 2007. Are floristic and edaphic patterns in Amazonian rain forests congruent for trees, pteridophytes and Melastomataceae?Journal of Tropical Ecology 23: 13–25. [Google Scholar]
- Russo SE, Davies SJ, King DA, Tan S. 2005. Soil-related performance variation and distributions of tree species in a Bornean rain forest. Journal of Ecology 93: 879–889. [Google Scholar]
- Savolainen V, Anstett M-C, Lexer C, et al. 2006. Sympatric speciation in palms on an oceanic island. Nature 441: 210–213. [DOI] [PubMed] [Google Scholar]
- Schietti J, Emilio T, Rennó CD, et al. 2014. Vertical distance from drainage drives floristic composition changes in an Amazonian rainforest. Plant Ecology & Diversity 7: 241–253. [Google Scholar]
- Schreeg LA, Kress WJ, Erickson DL, Swenson NG. 2010. Phylogenetic analysis of local-scale tree soil associations in a lowland moist tropical forest. PLoS ONE 5: e13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvertown J, Dodd M, Gowing D, Lawson C, McConway K. 2006. Phylogeny and the hierarchical organization of plant diversity. Ecology 87: 39–49. [DOI] [PubMed] [Google Scholar]
- Silvertown J, Araya Y, Gowing D. 2015. Hydrological niches in terrestrial plant communities: a review. Journal of Ecology 103: 93–108. [Google Scholar]
- Srivastava DS, Lawton JH. 1998. Why more productive sites have more species: an experimental test of theory using tree hole communities. American Naturalist 152: 510–529. [DOI] [PubMed] [Google Scholar]
- Stein A, Gerstner K, Kreft H. 2014. Environmental heterogeneity as a universal driver of species richness across taxa, biomes and spatial scales. Ecology Letters 17: 866–880. [DOI] [PubMed] [Google Scholar]
- Svenning J-C. 2001. On the role of microenvironmental heterogeneity in the ecology and diversification of neotropical rain forest palms (Arecaceae). Botanical Review 67: 1–53. [Google Scholar]
- Swenson NG. 2013. The assembly of tropical tree communities – the advances and shortcomings of phylogenetic and functional trait analyses. Ecography 36: 264–276. [Google Scholar]
- Swenson NG, Enquist BJ. 2007. Ecological and evolutionary determinants of a key plant functional trait: wood density and its community-wide variation across latitude and elevation. American Journal of Botany 94: 451–459. [DOI] [PubMed] [Google Scholar]
- ter Steege H, Pitman NCA, Sabatier D, et al. 2013. Hyperdominance in the Amazonian tree flora. Science 342: 1243092. [DOI] [PubMed] [Google Scholar]
- Terborgh J, Andresen E. 1998. The composition of Amazonian forests: patterns at local and regional scales. Journal of Tropical Ecology 14: 645–664. [Google Scholar]
- Tilman D, Pacala S. 1993. The maintenance or species richness in plant communities. In: Ricklefs RE, Schluter D, eds. Species diversity in ecological communities. Chicago: University of Chicago Press. [Google Scholar]
- Tuomisto H, Ruokolainen K, Yli-Halla M. 2003. Dispersal, environment, and floristic variation of western Amazonian forests. Science 299: 241–244. [DOI] [PubMed] [Google Scholar]
- Tuomisto H, Zuquim G, Cárdenas G. 2014. Species richness and diversity along edaphic and climatic gradients in Amazonia. Ecography 37: 1034–1046. [Google Scholar]
- Turner BL, Brenes-Arguedas T, Condit R. 2018. Pervasive phosphorus limitation of tree species but not communities in tropical forests. Nature 555: 367–370. [DOI] [PubMed] [Google Scholar]
- Vitousek PM. 1984. Litterfall, nutrient cycling, and nutrient limitation in tropical forests. Ecology 65: 285–298. [Google Scholar]
- Vitousek PM, Sanford RL. 1986. Nutrient cycling in moist tropical forest. Annual Review of Ecology and Systematics 17: 137–167. [Google Scholar]
- Vormisto J, Svenning J-C, Hall P, Balslev H. 2004. Diversity and dominance in palm (Arecaceae) communities in terra firme forests in the western Amazon basin. Journal of Ecology 92: 577–588. [Google Scholar]
- Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. 2002. Phylogenies and community ecology. Annual Review of Ecology and Systematics 33: 475–505. [Google Scholar]
- Whittaker RH. 1960. Vegetation of the Siskiyou Mountains, Oregon and California. Ecological Monographs 30: 279–338. [Google Scholar]
- Wittmann F, Schöngart J, Montero JC, et al. 2006. Tree species composition and diversity gradients in white-water forests across the Amazon Basin. Journal of Biogeography 33: 1334–1347. [Google Scholar]
- Zhang C, Yang J, Sha L, et al. 2017. Lack of phylogenetic signals within environmental niches of tropical tree species across life stages. Scientific Reports 7: 42007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.