Abstract
Background and Aims
Vandenboschia speciosa is a highly vulnerable fern species, with a large genome (10.5 Gb). Haploid gametophytes and diploid sporophytes are perennial, can reproduce vegetatively, and certain populations are composed only of independent gametophytes. These features make this fern a good model: (1) for high-throughput analysis of satellite DNA (satDNA) to investigate possible evolutionary trends in satDNA sequence features; (2) to determine the relative contribution of satDNA and other repetitive DNAs to its large genome; and (3) to analyse whether the reproduction mode or phase alternation between long-lasting haploid and diploid stages influences satDNA abundance or divergence.
Methods
We analysed the repetitive fraction of the genome of this species in three different populations (one comprised only of independent gametophytes) using Illumina sequencing and bioinformatic analysis with RepeatExplorer and satMiner.
Key Results
The satellitome of V. speciosa is composed of 11 satDNA families, most of them showing a short repeat length and being A + T rich. Some satDNAs had complex repeats composed of sub-repeats, showing high similarity to shorter satDNAs. Three families had particular structural features and highly conserved motifs. SatDNA only amounts to approx. 0.4 % of its genome. Likewise, microsatellites do not represent more than 2 %, but transposable elements (TEs) represent approx. 50 % of the sporophytic genomes. We found high resemblance in satDNA abundance and divergence between both gametophyte and sporophyte samples from the same population and between populations.
Conclusions
(1) Longer (and older) satellites in V. speciosa have a higher A + T content and evolve from shorter ones and, in some cases, microsatellites were a source of new satDNAs; (2) the satellitome does not explain the huge genome size in this species while TEs are the major repetitive component of the V. speciosa genome and mostly contribute to its large genome; and (3) reproduction mode or phase alternation between gametophytes and sporophytes does not entail accumulation or divergence of satellites.
Keywords: Vandenboschia speciosa, ferns, satellitome, satDNA, satellite DNA evolution, phase alternation, reproduction mode, sporophyte, gametophyte, transposable elements
INTRODUCTION
Eukaryotic genomes contain large amounts of different classes of repetitive DNA sequences either arranged in tandem or dispersed (López-Flores and Garrido-Ramos, 2012; Biscotti et al., 2015). Among tandem repetitive DNA, moderately repetitive DNA includes rRNA and protein-coding gene families or short tandem telomeric repeats, while highly repetitive DNA includes non-coding microsatellite and satellite DNA (satDNA), including centromeric DNA. Among dispersed repeats, transposable elements (TEs) such as DNA transposons and retrotransposons [mainly long terminal repeat (LTR) retrotransposons and non-LTR retrotransposon or long interspersed elements (LINEs)] constitute a main fraction of highly repetitive DNA, also including short interspersed elements (SINEs; moderately to highly repetitive DNA), retrogenes and retropseudogenes, as well as several gene families composed of dispersed members (moderately repetitive DNA).
Repetitive DNA greatly contributes to pronounced differences in genome size between species (López-Flores and Garrido-Ramos, 2012; Biscotti et al., 2015). Among repetitive sequences, TEs are mostly responsible for these differences and retrotransposons are the most abundant, with a predominant presence of LTR retrotransposons in plants (López-Flores and Garrido-Ramos, 2012; Biscotti et al., 2015). Notwithstanding, satDNA also contributes greatly to genome size variation in some organisms (Plohl et al., 2012; Garrido-Ramos, 2015, 2017). While the repeatome (Kim et al., 2014) is the whole collection of repetitive DNA, the satellitome is the whole collection of different satDNA families in a genome (Ruiz-Ruano et al., 2016). For decades, access to satDNA families of the satellitome was based on isolation from restriction endonuclease treatment of genomic DNA (gDNA), a method that simplified and popularized the study of satDNA, but that has several drawbacks (reviewed in Garrido-Ramos, 2017). Today, next-generation sequencing (NGS) and high-throughput in silico analysis of the information contained in NGS reads have revolutionized the study of this and other repetitive fractions of eukaryotic genomes (Novák et al., 2010, 2013; Weiss-Schneeweiss et al., 2015).
For this purpose, an efficient pipeline called RepeatExplorer was developed by Novák et al. (2010, 2013) which allows for the de novo identification of repetitive DNA families in species lacking a reference genome, thus facilitating the analysis of both the repeatome (Kim et al., 2014), in general, and the satellitome (Ruiz-Ruano et al., 2016), in particular. Later, Ruiz-Ruano et al. (2016) implemented a bioinformatic toolkit (satMiner), based on consecutive rounds of clustering of Illumina reads by RepeatExplorer, which allows identification of satDNA families, alternating with filtering out of the already known families and thus increasing the likelihood of finding new rare satDNA families. This toolkit has proven useful in species with a high C-value and/or a small amount of satDNA (Ruiz-Ruano et al, 2016). In addition, Novák et al. (2017) developed Tandem Repeat Analyzer (TAREAN), a further improvement of RepeatExplorer which allows the automatic identification of satDNA repeats and reconstruction of representative monomer sequences for each satDNA family (Novák et al., 2017). All these genomic approaches have been used in the last few years for the analysis of TEs and satDNA content in many species, and provide an opportunity to uncover satDNA families whose isolation was elusive by other methods. Thus, the combination of NGS and computer analysis favours an in-depth global genomic analysis of the satellitome by revealing the different satDNA families making up a given genome, their relative abundance and variability, the core details of their evolution and their roles in different genetic and genomic processes (Weiss-Schneeweiss et al., 2015). In addition, this new perspective greatly contributes to the development of comparative genomics and phylogenomics (Weiss-Schneeweiss et al., 2015).
Vandenboschia speciosa (Willd.) G. Kunkel (= Trichomanes speciosum Willd.) is a fern species of the family Hymenophyllaceae, with a genome size of 10.496 Gb (Obermayer et al., 2002). Vandenboschia speciosa is an endangered rare European–Macaronesian endemism, the only representative of a genus which has a primarily tropical distribution, restricted to disjoined regions of the European Atlantic coast and the Macaronesian islands (Canaries, Madeira and Azores). The two phases of the V. speciosa life cycle are perennial and they can reproduce by vegetative propagation (Rumsey et al., 1999). The ‘floaty’ sporophyte (fronds made of a translucent single layer of cells) is rhizomatous and can spread by fragmentation of the rhizome. The gametophyte, unlike a heart-shaped prothallus, is epigeous and narrowly filamentous with specialized asexual propagules (gemmae), and can survive in some populations during long periods outside the range of sporophyte distribution (Rumsey et al, 1999). In the warmer climatic conditions in the South of the Iberian Peninsula and Macaronesian islands, this species usually undergoes a normal fern life cycle of two free-living generations but, as one goes further into north and east Europe, the sporophyte generation becomes increasingly rare (Rumsey et al, 1999). However, one out of eight populations of this species located in the south of the Iberian Peninsula shows only the gametophyte phase, which propagates vegetatively.
These biological, life history and genomic features, together with the phylogenetic position of ferns within vascular plants, make V. speciosa an attractive species for satellitome analysis. This part of the genome has been elusive in V. speciosa when employing conventional methods (M. A. Garrido-Ramos, pers. obs.). Here, we analyse four genomic libraries belonging to three different populations of this species, two of them showing alternation of haplo- and diploid generations and the other being composed only of haploid independent gametophytes. Our aim was to characterize the different satDNA families contributing to the satellitome of V. speciosa, to assess the relative contribution of satDNA to the large genome of V. speciosa in comparison with other repetitive DNAs and to ascertain whether the amount or divergence of satDNA changes between sporophytic and gametophytic stages or might be influenced by the mode of reproduction.
MATERIALS AND METHODS
Materials
Vandenboschia speciosa specimens were collected at three populations located in the Alcornocales Natural Park (Cádiz, Spain): Canuto de Ojén-Quesada (OJEN); Valdeinfierno (VALD); and La Almoraima (ALMO). Samples were OJENs (sporophyte phase from the OJEN population), VALDs (sporophyte phase from the VALD population), VALDg (gametophyte phase from the VALD population) and ALMOg (gametophyte phase from the ALMO population). While OJEN and VALD are two populations where this species alternates between the sporophyte and the gametophyte phases, ALMO is composed only of gametophytes. Sporophytes were frozen in liquid nitrogen in the field. Patches with gametophytes were taken in a Petri dish with soil to the laboratory where, under a binocular microscope, the filaments were separated one by one from the soil and from other visible plant and animal species, cleaned, and frozen in liquid nitrogen. All samples were stored at −80 °C, and gDNA was isolated from each population using the DNeasy plant Mini kit (Qiagen). Pools of DNAs were generated from sets of five specimen DNAs, and separated NGS of the samples was carried out based on the Illumina HiSeq 2000 PE 2 × 101 nucleotides (nt) for OJENs, VALDs and ALMOg, and on the Illumina HiSeq×Ten PE 2 × 151 nt for VALDg, yielding about 8 Gb (approx. 0.75× coverage) of data for VALDs, ALMOg and VALDg, and about 16 Gb (approx. 1.5× coverage) for OJENs. Illumina sequencing data can be accessed at the SRA-GenBank database in the BioProject PRJNA387541.
SatDNA mining
We applied the protocol satMiner (Ruiz-Ruano et al., 2016), which is based on consecutive rounds of clustering of Illumina reads by RepeatExplorer (Novák et al., 2013), using a sub-set of reads (100 000 per library), and subsequent filtering of the already assembled reads using DeconSeq (Schmieder and Edwards, 2011), in order to solve the computational problems of handling large data sets with RepeatExplorer.
We performed a quality trimming with Trimomatic (Bolger et al., 2014), and randomly selected 2 × 100 000 Illumina reads with SeqTK (https://github.com/lh3/seqtk), to run RepeatExplorer with default options. Clusters with spherical or ring-shaped structures and density values >0.1 are likely to be satDNA, and they were manually selected and inspected for tandem repeats using the dotplot tool in Geneious v4.8 (Drummond et al., 2009). The contigs with higher coverage were then split into monomers and aligned in order to generate a consensus monomer for each satDNA cluster.
We filtered out the reads showing homology with the already clustered contigs and the already identified satDNA using DeconSeq, and selected a new set of 2 × 100 000 reads from the filtered libraries, that were clustered with RepeatExplorer in a second round. This allows detection of satDNAs which are poorly represented in the raw reads. We repeated the filtering using the clusters in the second round, and selected 2 × 300 000 reads for a third round. We repeated the process another twice adding 2 × 600 000 reads each time, but no new satDNA was detected after the third round. This protocol was first performed in the OJEN population, where we obtained consensus sequences for 11 satDNAs. Later, we performed a similar analysis in the libraries from VALD and ALMO, adding the consensus of the satDNA sequences already known as a custom database in RepeatExplorer, in order to annotate them if the same satDNA was present in these two populations and also to detect whether new ring-shaped or spherical clusters (i.e. graph shape for satDNA) were specific to any of them.
SatDNA sequence analysis
To detect a representative number of sequences for each satDNA in each population, we selected the reads showing homology with the catalogue of satDNAs identified, using BLAT (Kent, 2002), as implemented in a custom script (https://github.com/fjruizruano/ngs-protocols/blob/master/mapping_blat_gs.py), and selected 2 × 10 000 reads from each population to run RepeatExplorer clustering using a custom database for annotating the sequences of all assembled satDNAs. The BLAT parameters we applied were -stepSize = 5 -repMatch = 2253 -minScore = 0 -minIdentity = 0.
None of the satDNAs contained telomere-like repeats. As TTTAGGG has been described as the telomere sequence for several species of pteridophytes (Suzuki, 2013), we performed a literal search of TTTAGGG in the raw reads using the grep tool (a command-line software to search for lines with a given pattern in plain-text data sets). Then, we selected those reads where this TTTAGGG was repeated. Alignments and comparison between populations were performed using ten repeats.
In addition, we estimated the abundance of short satDNAs with monomer length between 1 and 6 nt, also known as microsatellites, using RepeatMaksker with the ‘-int’ option.
Abundance and divergence for each satDNA in each population were calculated with RepeatMasker (Smit et al., 2015) with the cross-match search engine, mapping 2 × 5 000 000 reads from each population to the satDNA consensus sequences and (TTTAGGG)30 for the analysis of the telomeric repeats.
Repeat monomers were extracted from the reads and aligned, in order to perform sequence comparisons between populations. In the case of satDNAs with a repeat unit length <101 nt, monomers were directly extracted from Illumina read sequences. When repeat unit length surpassed read length, we used read pairs where the paired reads overlapped. MEGA v.6 (Tamura et al., 2013) was used to estimate intrapopulation genetic variation and interpopulation divergence, as well for phylogenetic analysis. Comparison between satDNA families was performed with the RepeatMasker method described below.
The EMBOSS suite of bioinformatics tools (Rice et al., 2000) was used for the detection of internal repeats (direct or inverted) as well as palindromes. The programs used from the package were MATCHER, ETANDEM, EINVERTED, POLYDOT and PALINDROME. Secondary structure estimation for repeat sequences with shorter inverted repeats and palindromes was made by using the RNAstructure Predict a Secondary Structure Web Server (Reuter and Mathews, 2010).
Our study also included an analysis of nucleotide diversity (π) per position for every satDNA using the sliding windows option of the DnaSP v.5.10 program (Librado and Rozas, 2009). Geneious v4.8 (Drummond et al., 2009) was used to generate sequence logos to convey the level of sequence divergence and display conserved motifs revealed by the DnaSP program.
The bendability/curvature propensity plots were made with the bend.it server (Vlahovicek et al., 2003), using the DNase I-based bendability parameters of Brukner et al. (1995) and the consensus bendability scale (Gabrielian and Pongor, 1996).
TE characterization from Illumina reads
The use of the RepeatExplorer pipeline (Novák et al., 2013) allowed us additionally to characterize the TE content of the V. speciosa genome. Repeat identification by a single similarity-based clustering of 250 000 read pairs from each of the two sporophyte libraries (1 000 000 reads in total) was performed using the RepeatExplorer pipeline. This software employs graph representation of read similarities to identify clusters of frequently overlapping reads representing various repetitive elements or their parts (Novák et al., 2010) and provides comparative information about repeat quantities estimated from the number of reads for each library in a cluster. In addition, it performed an annotation based on similarity searches to the RepeatMasker Viridiplantae database of repetitive elements.
Gametophyte contamination analysis
Since gametophytes are found in the ground in close contact with stream water, DNA extracted from them can conceivably be excessively contaminated by micro-organisms, even after careful cleaning. This contamination might bias satDNA abundance estimations in the gDNA libraries. For this reason, we checked the degree of contamination of every sample using the RNA-Seq Illumina sequencing data from a Valdeinfierno sporophyte and a gametophyte, previously used in Ruiz-Estévez et al. (2017a, b). We separately assembled the two transcriptomes using the Trinity v.2.5 de novo assembler (Haas et al., 2013) after trimming and in silico normalization under default options. Finally, we built SuperTranscripts to obtain a unique sequence per sequence graph using the Trinity’s tool.
We annotated the contigs for both assemblies utilizing the Trinotate v.5 software (https://trinotate.github.io/) using the SwissProt database (Bairoch and Apweiler, 2000) and aligning with BLASTX (Altschul et al., 1997). Then we compared the annotation terms between sporophytes and gametophytes to generate two pools of sequences: (1) a random selection of 500 sporophyte transcriptomic contigs with an annotation also found in the gametophyte transcriptome; and (2) a selection of the gametophyte transcriptomic contigs annotated with a term absent in the sporophyte transcriptome and, in addition, showing mappings in the two gametophytic genomic libraries, using SSAHA2 (http://www.sanger.ac.uk/science/tools/ssaha2-0). To assign the most similar species to the sequences in these two pools, we annotated them with Blast2GO (Conesa et al., 2005) against the NR database using BLASTX.
As we found a high level of contaminants in the gametophyte assembly, we designed an analysis to calculate the degree of contamination in the gametophyte libraries in order to estimate coefficients to correct satDNA abundance quantification in the gDNA libraries. For this purpose, we first selected, in the sporophyte transcriptome, those coding sequences (CDSs) which were longer than 3000 nt, in order to reduce bias due to the low coverage of gDNA libraries. Then we mapped the gDNA reads of the four V. speciosa libraries against these long CDS references, using SSAHA2 software. This allowed estimation of an approximate number of copies for each contig and library, using the protocol applied by Navarro-Dominguez et al. (2017), with 99 % minimum mapping identity for the SSAHA2 software to avoid mapping of the contaminant reads. To limit this analysis to genes showing a single copy per haploid genome, we restricted it to those contigs showing an average copy number in the sporophytes (OJENs and VALDs) lying between 0.7 and 1.3. We then calculated the average copy numbers per library and normalized them with respect to OJENs, i.e. the library showing the highest value. We finally used the resulting coefficients to correct satDNA abundance estimates in the gDNA libraries.
Statistical analyses
Shapiro–Wilk’s W test showed that the observed variation for unit length, A + T content, abundance and divergence in all populations fitted a normal distribution (Supplementary Data Table S1); for this reason we used parametric statistical tests, such as Pearson correlation analysis and Student t-test for dependent samples, using Statistica (Statsoft Inc.). Comparisons of satDNA abundance and divergence between genomic libraries were performed by the non-parametric Friedman analysis of variance (ANOVA).
RESULTS
High-throughput search for satDNA
After the first run of RepeatExplorer analysis and three additional runs of filtering + RepeatExplorer, performed on the OJENs library, we found 11 satDNAs, defined by monomer length and sequence (Table 1; Supplementary Data Fig. S1). Supplementary Data Fig. S2 shows the reconstruction of representative monomer sequences for each satDNA family. These sequences are deposited in the GenBank database under accession numbers MH048647–MH048657. The same 11 satDNAs were identified independently in the VALDs, VALDg and ALMOg libraries. SatDNA sequences showed no homology with any other DNA sequence deposited in DNA databases (BLAST search found no significant similarity with any other DNA sequence). Table 1 shows the main features of these satDNAs. Their repeat units were short in length (between 32 and 141 bp), with eight satDNAs being shorter than 100 bp. Likewise, most of them were A + T rich, with seven satDNAs showing this parameter between 56.6 and 70.0 % A + T, three between 51.1 and 52.7 % A + T, and only one actually being G+C rich by showing 45.4% A + T content. A correlation analysis showed that satDNAs with longer unit length showed a higher A + T content (r = 0.65, P = 0.031).
Table 1.
Characteristics of the 11 satDNAs found in V. speciosa
| Assembly/cluster | Satellite name | Abundance (%) | Divergence (%) | Monomer lenght (nt) | A + T (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OJENs | VALDs | VALDg | ALMOg | OJENs | VALDs | VALDg | ALMOg | OJENs | VALDs | VALDg | ALMOg | |||
| 3/CL11 | VspSat01-59 | 0.08 | 0.04 | 0.05 | 0.05 | 11.69 | 12.02 | 12.07 | 12.29 | 56–62 | 52–65 | 52–65 | 56–59 | 51.1 |
| 1/CL154 | VspSat02-101 | 0.08 | 0.08 | 0.07 | 0.08 | 10.43 | 10.43 | 11.03 | 10.61 | 100–104 | 99–103 | 99–103 | 100–102 | 56.6 |
| 3/CL11 | VspSat03-33 | 0.07 | 0.04 | 0.04 | 0.05 | 10.37 | 10.47 | 10.48 | 10.56 | 32–34 | 32–33 | 32–33 | 33–38 | 52.0 |
| 1/CL65 | VspSat04-107 | 0.06 | 0.07 | 0.06 | 0.06 | 14.76 | 15.08 | 14.87 | 14.81 | 107 | 96–107 | 96–107 | 106–107 | 63.2 |
| 1/CL167 | VspSat05-141 | 0.05 | 0.05 | 0.05 | 0.06 | 8.61 | 8.57 | 8.77 | 8.80 | 140–141 | 140–141 | 140–141 | 140–141 | 70.0 |
| 2/CL263 | VspSat06-67 | 0.01 | 0.01 | 0.01 | 0.01 | 6.66 | 7.46 | 7.81 | 7.38 | 66–69 | 64–69 | 64–69 | 66–69 | 52.7 |
| 2/CL65 | VspSat07-70 | 0.02 | 0.02 | 0.01 | 0.02 | 16.96 | 17.09 | 17.27 | 17.24 | 69–70 | 68–71 | 68–71 | 69–71 | 58.2 |
| 3/CL10 | VspSat08-82 | 0.02 | 0.02 | 0.01 | 0.02 | 13.43 | 12.67 | 14.03 | 12.63 | 79–82 | 81–84 | 81–84 | 81–82 | 61.7 |
| 2/CL286 | VspSat09-68 | 0.01 | 0.01 | 0.01 | 0.01 | 12.49 | 12.50 | 12.72 | 12.89 | 62–70 | 67–70 | 67–70 | 62–73 | 67.0 |
| 2/CL266 | VspSat10-62 | 0.01 | 0.01 | 0.01 | 0.01 | 10.16 | 9.87 | 10.32 | 10.36 | 56–67 | 61–64 | 61–64 | 59–63 | 69.2 |
| 3/CL3 | VspSat11-34 | 0.01 | 0.004 | 0.01 | 0.01 | 14.30 | 16.16 | 13.86 | 16.54 | 34–35 | 33–35 | 33–35 | 34 | 45.4 |
| Total | 0.43 | 0.36 | 0.33 | 0.37 | ||||||||||
Contamination coefficient calculation
The de novo assembly of the two RNA-Seq libraries yielded 85 246 contigs with N50 = 1953 nt for the sporophyte, and 347 685 contigs with N50 = 782 nt for the gametophyte, the latter showing four times more contigs than the former, thus demonstrating much higher diversity of transcripts in the gametophyte, which raised our suspicions of possible contamination.
Annotation with SwissProt revealed the existence of 17 224 contigs in the sporophyte sharing terms with those found in the gametophytic transcriptome and, to test the existence of putative contamination, we selected a random sample including 500 of these contigs. Blast2GO annotated 488 of them, 96 % of which showed the greatest similarity with Viriplantae, 1 % with Bacteria, 2 % with Fungi and 1 % with Metazoa. On the other hand, we obtained 77 772 gametophytic contigs with annotation terms absent in the sporophyte library, 672 of which mapped in both gametophytic gDNA libraries (VALDg and ALMOg). Blast2GO annotated 665 of them and, remarkably, only 16 % of them annotated with Viriplantae (in great contrast to the 96 % observed for the contigs shared with the sporophyte), whereas 7 % annotated with Bacteria, 16 % with Fungi, 45 % with Metazoa and 16 % with other Eukaryota. These results indicate that DNA extraction in the gametophytes resulted in severe contamination in spite of careful cleaning (see the Materials and Methods).
To obtain an estimate of the contamination level in the gametophyte gDNA libraries, we estimated the copy number for 623 sporophytic contigs longer than 3000 nt. The average copy number was 0.92, 0.91, 0.30 and 0.51 for OJENs, VALDs, VALDg and ALMOg, respectively (Supplementary Data Table S2), with remarkably lower values in the gametophyte libraries. Assuming that the lower copy number for these contigs in the gamethophytes is due to a decrease in coverage caused by the presence of contaminants, we normalized copy numbers with respect to the highest value (that in OJENs) and obtained coefficients (1 in OJENs, 0.98 in VALDs, 0.32 in VALDg and 0.55 in ALMOg) which were used for correcting the genomic abundance of satDNA (Table 1).
SatDNA abundance and divergence in the context of other repetitive DNAs
Collectively, all 11 satDNAs represent between 0.43 and 0.33 % of the genome of V. speciosa depending on the library (Table 1; Supplementary Data Fig. S1). There were some differences between libraries in the abundance of certain satDNAs, but a Friedman ANOVA comparing abundances for the 11 satDNAs between the four libraries did not reach significance (χ2 = 7.57, n =11, d.f. = 3, P = 0.056). This indicates that some specific satDNAs have recently been differentially amplified in a given population but not in others, but there is no general tendency for all satDNAs as a whole, except for OJENs showing higher abundance for five satDNAs (VspSat01-59, VspSat03-33, VspSat06-67, VspSat07-70 and VspSat11-34) but lower abundance for only one (VspSat04-107). Therefore, OJENs showed the highest figure for total satDNA abundance (0.43 % compared with 0.36, 0.33 and 0.37 % in VALDs, VALDg and ALMOg, respectively). However, we found significant differences between the four libraries with respect to satDNA divergence (Friedman ANOVA: χ2 = 13.73, n =11, d.f. = 3, P = 0.0033), with OJENs showing the lowest divergence (Supplementary Data Fig. S3). This result would be consistent with the fact that the higher abundance found in OJEN was due to recent amplifications decreasing the divergence for certain satDNAs.
No significant correlation was found between satDNA abundance and divergence within each library (P = 0.632 in OJENs, P = 0.697 in VALDs, P = 0.692 in VALDg and P = 0.563 in ALMOg). Likewise, abundance and divergence failed to show significant correlation with unit length or A + T content (see Supplementary Data Table S3).
Telomere sequences were not found through the SatMiner approach. Notwithstanding that, several thousands of telomeric repeats were found by searching TTTAGGG patterns among NGS reads. RepeatMasker analysis showed that the abundance of telomere sequences was 0.0256 and 0.0254 % in OJENs and VALDs, respectively.
In addition, by using the ‘simple repeats’ option of RepeatMasker, we found a 1.77 % microsatellite abundance in OJENs and 2.02 % in VALDs, but a t-test for dependent samples showed that this difference is not significant (t = 1.08, d.f. = 5, P = 0.33). Likewise, a comparison of microsatellite divergence between OJENs and VALDs failed to show a significant difference (t = 1.19, d.f. = 5, P = 0.29). In the case of telomeric and microsatellite repeats, we did not use the gametophyte gDNA libraries because contamination would yield misleading abundance estimates given that their extremely short motifs were not distinguishable from those of the contaminants.
On the other hand, the RepeatExplorer output showed 50.53 % TE genomic abundance in OJENs and 49.86 % in VALDs. A description of the different TE classes found within the genome of V. speciosa is listed in Table 2. Repeats classified as LTR retrotransposons represented the major fraction of the genome of V. speciosa, comprising up to approx. 28 % of their nuclear DNA. They were mostly represented by Ty3/gypsy elements (Table 2). Ty1/copia elements were generally less abundant (approx. 11.5 % of the genome). Other mobile elements detected included LINEs (non-LTR retrotransposons) which represent between 3.86 and 3.62 % of the genome in OJENs and VALDs, respectively. On the other hand, DNA transposons, mainly those of the CACTA superfamily, represent about 3.7 % of the genome. Unfortunately, there were another 14 % of repetitive sequences (TEs according to the graph-based clustering) that were not specifically annotated.
Table 2.
Contribution of TEs to the V. speciosa genome
| OJENs | VALDs | |
|---|---|---|
| DNA | 0.98 % | 1.13 % |
| DNA/CACTA | 2.44 % | 2.58 % |
| DNA/hAT | 0.16 % | 0.17 % |
| LINE | 3.86 % | 3.62 % |
| LTR | 0.76 % | 0.68 % |
| LTR/copia (Ty1) | 11.46 % | 11.47 % |
| LTR/gypsy (Ty3) | 16.53 % | 15.81 % |
| ND | 14.33 % | 14.41 % |
| Total | 50.53 % | 49.86 % |
Intrapopulation satDNA sequence variation
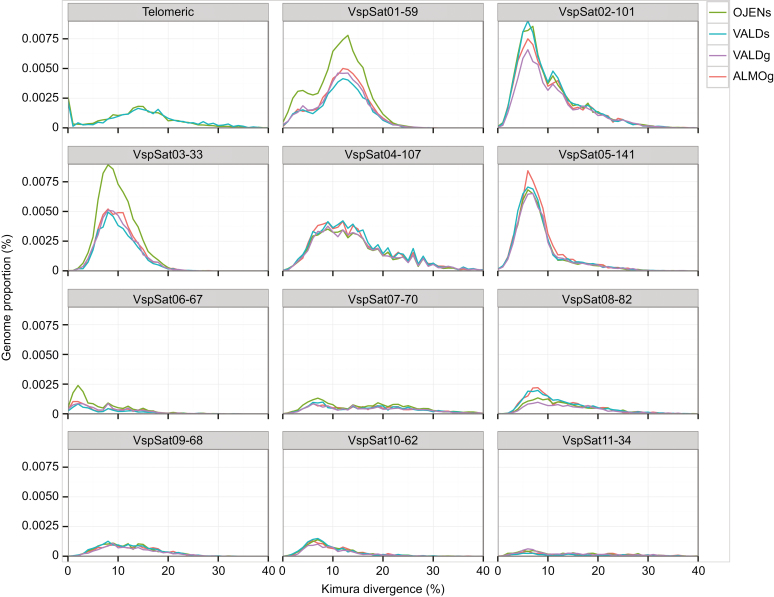
Figure 1 displays repeat landscape plots representing, for each satDNA, abundance (y-axis) and divergence (x-axis) with respect to a consensus sequence built for each satDNA repeat unit. Bearing in mind that satDNA evolution may be mainly marked by amplification and homogenization processes (both decreasing divergence) and point mutations (increasing divergence), the profiles of repeat landscapes are highly informative regarding the age of satDNA variants within the same family. It is thus reasonable to infer that peaks at lower divergence values are the product of recent amplification or homogenization, whereas those at higher divergence values are probably older variants degenerated by accumulation of mutations. Consistently, telomeric repeats showed two peaks, one corresponding to extremely low divergent sequences, as expected for sequences generated by the active role of telomerase, and the other, at about 15 % divergence, suggesting the existence of ectopic telomere tandem repeats which are not under telomerase action and thus manifest high divergence (Fig. 1). Likewise, VspSat01-59 showed two types of abundant repeats differing in divergence, suggesting that they might show different ages or else differential tendencies for homogenization. Of the remaining satDNAs, VspSat06-67 showed the lowest divergence (i.e. the highest homogenization), as indicated by a main peak below 5 % divergence. On the other hand, VspSat02-101, VspSat03-33, VspSat05-141 and VspSat08-82 showed a main peak between 5 and 10 % divergence, whereas VspSat04-107, VspSat07-70, VspSat09-68, VspSat10-62 and VspSat11-34 showed very flat distributions, suggesting the presence of a broad range of sequence variations with respect to the consensus (Fig. 1).
Fig. 1.
Repeat landscape plots representing, for each satDNA, abundance (y-axis) and divergence (x-axis) with respect to a consensus sequence built for each satDNA repeat unit.
Interestingly, microsatellite landscape plots also showed two peaks, one corresponding to extremely low divergent sequences in homogeneous loci, probably of recent origin, and the other at about 20 % divergence pointing to the existence of loci including old repeats degenerated by mutation (Supplementary Data Fig. S4).
SatDNA sequence divergence was low between populations
Due to the fact that most satDNA families in this species showed repeat unit lengths lower than the Illumina read length, we were able to extract a number of monomers directly from the reads, thus resembling a massive cloning experiment. This rendered a total of 1045 repeat units, even though the number of monomers obtained for the longest satDNAs (e.g. VspSat04-107 and VspSat05-141) was low because they depended on overlapping read pairs (Supplementary Data Table S4). With these data, differences between populations were similar, or even lower, than the intrapopulation variation observed for each satDNA (not shown). Likewise, phylogenies for repeat sequences did not display differentiation between populations (not shown). In fact, the sequences appeared intermingled in the phylogenetic trees independently of population of origin.
Conserved motifs and satDNA curvature
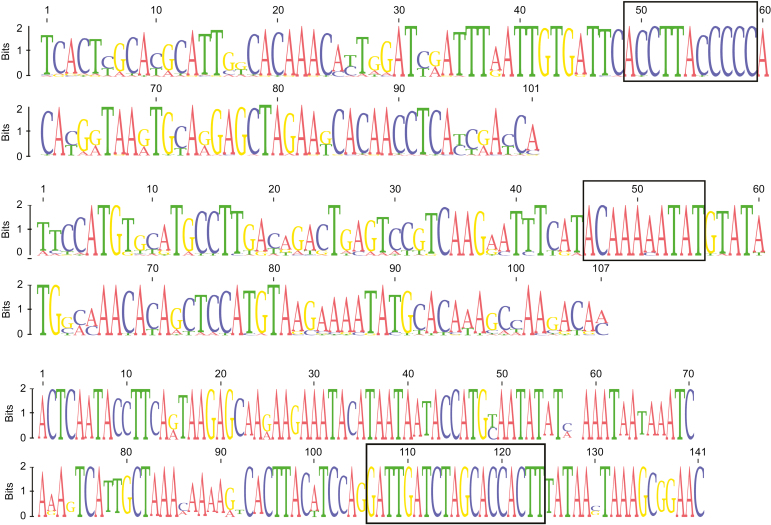
An analysis of nucleotide diversity (π) per position in each satDNA showed that π varied sharply between positions, with similarly alternating peaks and valleys. Interestingly, several satDNA graphs showed the existence of conserved parts, with little or no variation, within repeat units. For instance, positions 49–59 in VspSat02-101, 46–55 in VspSat04-107 and 106–124 in VspSat05-141 were extremely conserved compared with the remaining nucleotides in each of these satDNA units (Supplementary Data Table S5; Fig. S5). Figure 2 includes sequence logos for each satDNA which clearly convey the level of sequence divergence and reveal the conserved motifs. The position of the conserved motif coincided with a peak of DNA curvature in the case of the VspSat04-107 satDNA (Supplementary Data Fig. S6). The curvature propensity plot contains one peculiar maximum in this region whose magnitude (17.8°/10.5 bp helical turn) roughly corresponds to the value calculated for other highly curved motifs (Goodsell and Dickerson, 1994). Therefore, we believe that this region may adopt a curved conformation. The conserved regions in the VspSat02-101 and VspSat05-141 satDNAs were not within a peculiar maximum peak of curvature. However, the same plot drawn with the consensus bendability scale showed a conspicuous peak in this region in both cases, with a curvature propensity plot showing peaks in positions 20–40 (13.1°/10.5 bp helical turn) and 28–36 (10°/10.5 bp helical turn) within the VspSat02-101 and VspSat05-141 units, respectively (not shown). In addition, the VspSat08-82 satDNA showed two conspicuous peaks of curvature propensity >15°/10.5 bp helical turn. However, curvature propensity plots failed to show any value indicative of strong curvature for the remaining satDNA families. None of the conserved motifs was found to be significantly related to any other known DNA-binding motif.
Fig. 2.
Sequence logos showing the level of sequence divergence (from top to bottom: VspSat02-101, VspSat04-107 and VspSat05-141). Conserved motifs are enclosed in a square.
Similarity between satDNA families
Some satDNA families showed complex units including sub-repeats with high percentages of similarity to other shorter families. For instance, the VspSat06-67 unit includes two direct sub-repeats of 25 bp at positions 7–31 and 40–64 (76 % identity), each showing high similarity (64 and 68 %) to the core of the 34 bp repeat of the VspSat11-34 satDNA unit (Fig. 3 ). VspSat10-62 satDNA units are a rearranged version of the VspSat09-68 monomers with a 6 bp deletion (75 % identity; 83.3 % identity in the matched part of the sequence) (Fig. 4 ). The VspSat01-59 satDNA is also interesting in that it includes a complex combination of trinucleotides according to the formula: variable sequence–(GAT)2(GTG)(GAT)3(GTG)TC(GAT)4(GTG)TC–variable sequence, whereas the VspSat03-33 satDNA is composed of three repetitions of a simplified version of the former formula: (GAT)2(GTG)GC, showing 87 % identity with VspSat01-59 (Fig. 5 ). Finally, the VspSat08-82 satDNA showed units composed of two parts, each one being a palindrome (Supplementary Data Fig. S7). Prediction of the lowest free energy structure and the structure generated from highly probable base pairs for the VspSat08-82 sequence revealed a particular capability to acquire cruciform structures of VspSat08-82 monomers (Supplementary Data Fig. S7). Finally, the complex structure observed for most satDNAs analysed here, with the exception of VspSat02-101, VspSat04-107 and VspSat05-141, implied sequences showing a high probability to acquire particular secondary structures (not shown). These data support the consideration of the existence of some satDNA superfamilies (SFs), derived from a common ancestor satDNA, in V. speciosa by showing sequence homology, such as VspSat01-59 and VspSat03-33 (SF1), VspSat06-67 and VspSat11-34 (SF2) and VspSat09-69 and VspSat10-62 (SF3).
Fig. 3.
Sequence comparison between the two parts of VspSat06-67 monomers and the VspSat11-34 monomers. Shaded areas represent conserved nucleotides between the three sequences.
Fig. 4.
Sequence comparison between VspSat09-68 and VspSat10-62 monomers. Shaded areas represent conserved nucleotides between the two sequences.
Fig. 5.
Sequence comparison between VspSat01-59 and VspSat03-33 monomers. Shaded areas represent conserved nucleotides between the two sequences.
DISCUSSION
A catalogue of short but complex satDNAs in a large genome
Next-generation sequencing and high-throughput in silico analysis of the information contained in NGS reads (Novák et al., 2013, 2017; Ruiz-Ruano et al., 2016) have overtaken the limitation of conventional methods for the identification of the satDNA profile of V. speciosa which meant that the satellitome had been elusive for many years in this species (M. A. Garrido-Ramos, pers. obs.). This is probably because the percentage of each of these satDNAs in the genome is low enough to be inperceptible (Table 1; Supplementary Data Fig. S1). In fact, satDNA is not an abundant fraction of the repetitive part of the genome in V. speciosa (about 0.4 % of the genome), in spite of its large genome size (10.496 Gb; Obermayer et al., 2002) and in contrast to other species (Ambrožová et al., 2011; Melters et al., 2013; Garrido-Ramos, 2015, 2017). However, this value is even higher than those found in a set of six leptosporangiate ferns from across a range of three major clades (Polypodiales, Cyatheales and Gleicheniales), where Wolf et al. (2015) found that satDNA represents 0.1 ± 0.03 % (mean ± s.e.) of these fern genomes. These authors remarked that this satDNA abundance found in ferns is lower than that found in a group of six selected seed plants (0.8 ± 0.34 %), while the abundance of individual satDNA families in seed plants actually varies from 0.1 to 36 %, according to data gathered using non-genomic approaches (Garrido-Ramos, 2015, 2017). No satDNA family reaches 0.1 % of the genome in the case of V. speciosa (see Table 1). The presence of such a scarce set of satDNAs in a species with such a large genome is thus intriguing, and further research is needed on this aspect. A possible explanation is that satDNA families of V. speciosa are young and have not yet had time to increase in abundance. This is in contrast to the old age of this genus (Pryer et al., 2004), unless satDNA turnover is extremely fast in this species. This latter possibility appears to be unlikely given the resemblance of satDNA content and abundance between the three populations analysed here, and the homology found between several families constituting superfamilies, suggesting that satDNA families are long lived in this species. This is also supported by the existence of highly divergent variants for most satDNA families, evidenced by the repeat landscapes in Fig. 1, suggesting that the rate of satDNA degeneration through point mutation is high in this species. Another possibility might be related to the existence of genomic constraints impeding satDNA accumulation. Thus, the alternation of long-lasting haploid and diploid stages could run counter to the accumulation of high amounts of satDNA. For instance, satDNA amplification could be restrained during the haploid stage since unequal crossing-over would operate mainly during diploidy.
Alternatively, the active elimination of useless satDNA in V. speciosa might explain why this large genome contains such a low amount of satDNA. This might be due to a high rate of DNA removal in this species. Differential DNA loss is an interesting prospect which merits future investigation in V. speciosa, as it appears to be a programmed and regulated process in many eukaryote genomes involving satDNA diminution from germ to somatic lines (Wang and Davis, 2014). However, we have not found significant differences in satDNA amounts between the haploid and the diploid phases of V. speciosa.
An important contribution of TEs to the large genome size of V. speciosa
The large genome of the fern V. speciosa is mainly populated by TEs (approx. 50 % of the genome; Table 2), in contrast to the extremely low amounts of satDNA representing only approx. 0.4 % of the genome. TEs are highly ubiquitous elements found in all kingdoms of living organisms and are highly abundant in some genomes, reaching up to 85 % of some plant genomes, such as that of maize (Schnable et al., 2009). Furthermore, TE content can differ greatly even between related species, and this is the main factor responsible for genome size differences between them (Piegu et al., 2006; Hu et al., 2011). We have found a general landscape for repetitive DNA genomic composition in V. speciosa similar to that found in other plants (reviewed in López-Flores and Garrido-Ramos, 2012). In a recent analysis of six fern species from across a range of three major clades (Polypodiales, Cyatheales and Gleicheniales), Wolf et al. (2015) found that, compared with seed plants, ferns had a higher proportion of DNA transposons and LINEs in their genomes. Likewise, we have also found a higher proportion of these two types of elements in V. speciosa (approx. 3.7 % in both cases) compared with those in seed plants (mean percentages: approx. 0.83 and 0.89 %, respectively) and even higher than those found in other ferns (mean percentages: approx. 3.2 and 2.2 %, respectively). Wolf et al. (2015) also revealed that LTR/copia and LTR/gypsy retrotransposons represent 14 and 15 %, respectively, of fern genomes. Depending on the species, values ranged between 10 and 25 % of the genome for LTR/copia and between 8 and 25 % of the genome for LTR/gypsy. However, these authors did not find a correlation between genome sizes and amounts of LTR retrotransposons (Wolf et al., 2015). The fraction of the genome that is occupied by these elements in V. speciosa is within those ranges: approx. 11 5 % LTR/copia and 16 % LTR/gypsy.
A + T content is higher in longer repeat units of satDNA
SatDNA varies widely among species, not only in abundance but also in repeat length, repeat sequence and nucleotide composition (Plohl et al., 2012; Melters et al., 2013; Mehrotra and Goyal, 2014; Garrido-Ramos, 2015, 2017). Remarkably, all satDNA families found in V. speciosa are short and most of them are A + T rich (see Table 1). In fact, in general, it has been found that satellite repeats are generally A + T rich, especially in the case of centromeric satDNAs (Garrido-Ramos, 2015). According to theoretical models of satDNA evolution and on the basis of experimental evidence, any random sequence can lead to a family of tandem repeats (Smith, 1976; Melters et al., 2013). In this context, we should expect that the average A + T content of the monomer collection comprising the satellitome (ignoring differential abundances caused by processes subsequent to satDNA origin) would reflect A + T content in the genome as a whole. In V. speciosa, the average A + T content is 58.8 % in the satellitome and 61.5 % in the genome (inferred from the Illumina reads, not shown), i.e. 2.7 % lower in the former. In the grasshopper Locusta migratoria, these figures differed by 6 %, suggesting a tendency for satDNA to arise from G + C-rich regions (Ruiz-Ruano et al., 2016). In V. speciosa, A + T-rich satDNAs represent almost two-thirds of satDNA content (Table 1) and, likewise in L. migratoria (Ruiz-Ruano et al., 2016), there is a tendency for longer repeats to show a higher A + T content (see Table 1). Therefore, a tendency to increase length and A + T content with age would bring the satellitome A + T content close to the A + T genomic average even though satDNA showed a tendency to arise from G + C-rich regions.
On the other hand, satDNA is subject to epigenetic modification such as the methylation of cytosines, so that deamination of 5-methylcytosines might contribute to the relatively high A + T content of satDNAs in V. speciosa. Alternatively, the finding of a lower A + T content in the satellitome than in the genome of V. speciosa might be due to experimental procedures, as it appears that the standard protocol for preparing Illumina libraries (including PCR) might result in reduced representation of A + T-rich satDNA in the final sequences (Wei et al., 2018).
Repeat unit length tends to increase during satDNA evolution
Plant satDNA sequences commonly have unit lengths of 135–195 bp or 315–375 bp, but repeat length ranges between 58 bp for the pAm1 satDNA of Avena to 5.9 kb for the 2D8 repeats of Solanum bulbocastanum (Mehrotra and Goyal, 2014; Garrido-Ramos, 2015, 2017). A shorter monomer length has been found, for instance, for the 38 bp of the VicTR-B satDNA DNA in Vicia (Macas et al., 2006). The case of V. speciosa is exceptional in this respect because all satDNA families found show short repeat units (between 33 and 141 bp, with only three families surpassing 100 bp).
Among the collection of satDNAs in V. speciosa, we found several complex satDNA families that might guide us in the clarification of the apparently still ongoing evolutionary process for the formation of longer repeat units. The three superfamilies found illustrate this point. For instance, SF3 is composed of VspSat09-69 and VspSat10-62, two homologous satDNAs differing in a 7 bp indel. SF2 includes VspSat06-67 and VspSat11-34, the former being composed of two divergent VspSat11-34 units, thus being a higher order repeat (HOR) of the latter. Finally, SF1 is also composed of two satDNA families, so that VspSat01-59 includes two sub-repeats derived from divergent VspSat03-33 units, the former also being a HOR of the latter. This last case is enlightening as both satDNAs are composed of shorter sub-repeats of complex combinations of trinucleotides, an indication of the implication of two different mechanisms acting at different times: replication slippage (Garrido-Ramos, 2015, 2017) might be at the origin of the shorter units of VspSat03-33, whereas the longer satDNA (VspSat01-59) might have originated by unequal crossing-over (Garrido-Ramos, 2015, 2017). These two satDNAs, as well as VspSat06-67 and VspSat11-34, appear to be in an ongoing process of amplification in the OJEN population. The combination of short repeat units into longer units constituting HORs is a common trend in satDNA evolution (Plohl et al., 2008; Garrido-Ramos, 2017). We could thus postulate that longer satDNAs in V. speciosa evolve from shorter ones by means of alternate cycles of duplication and divergence, as previously proposed for other satDNA families (Navajas-Pérez et al., 2005; Macas et al., 2006; Emadzade et al., 2014).
We have checked the genomic content of microsatellites in V. speciosa to test whether it was possible that they were ‘seeds’ for longer satDNA repeat units. We found an important contribution of different types of microsatellites to the genome content of V. speciosa. Indeed, depending on the population, there was between four and five times more microsatellite DNA than satDNA content in this genome, suggesting that the former might be a source for new satDNAs. Recently, Wolf et al. (2015) found that microsatellites represent 15.5 ± 1.5 % of the genome in six fern species, a much higher figure than that reported for seed plants (1.19 ± 0.89 %). However, Portis et al. (2016) obtained even lower values for different seed plants (0.17–0.67 %). The case of V. speciosa (1.77–2.02 %) is thus closer to seed plants than to the six fern species analysed by Wolf et al. (2015).
Structural features in V. speciosa satDNAs
Structural features of satDNA might have implications for functional constraints. In this respect, VspSat02-101, VspSat04-107 and VspSat05-141 satDNAs share several interesting features. They are composed of longer satDNA repeat units, are A + T rich, are among the most abundant satDNAs and are among the most homogeneous satDNAs, at least as regards the VspSat02-101 and the VspSat05-141 satDNA families. In addition, they also share structural characteristics of bendability and curvature propensity, and, interestingly, conspicuous peaks of bending and curvature coincide in these repeats with the location of highly conserved motifs within VspSat02-101, VspSat04-107 and VspSat05-141 monomers.
Epigenetic control of heterochromatin assembly by transcriptional silencing complexes (RITS), recruitment of histone methyltransferases, histone H3 methylation (H3K9me2) and recruitment of heterochromatin protein 1 (HP1) are well documented (Pezer et al., 2012; Plohl et al., 2012; Johnson et al., 2017). The capability of DNA stretches to bend and curve into a superhelical tertiary structure is a sequence-dependent property that has been proposed many times as an additional element involved in specific recognition of DNA-binding protein components of heterochromatin, which might facilitate the tight packing of DNA into heterochromatin (reviewed in Pezer et al., 2012; Plohl et al., 2012).
On the other hand, the VspSat08-82 repeats are the result of two consecutive palindromes which, in combination, give the repeats the capability to acquire particular secondary structures such as cruciform structures. Cruciform structures are targets for many architectural and regulatory proteins, and are fundamentally important for a wide range of biological processes, including replication, regulation of gene expression, nucleosome structure and recombination (see, for example, the review by Brázda et al., 2011). The most remarkable detected inverted repeats are found in satDNAs from Tribolium species, characterized by complex monomers composed of inversely oriented sub-units capable of forming large dyad structures relevant in the formation of heterochromatin architecture in these species and in the amplification processes of these types of satDNAs (Plohl, 2010).
No apparent concerted evolution
Our data reveal no apparent genetic differentiation, i.e. concerted evolution (Garrido-Ramos, 2015, 2017), of satDNAs between the three populations analysed since the three populations share the same library with similar abundances and only decreased divergence in certain satDNAs in OJEN. Likewise, nucleotide diversity was roughly similar at intra- and interpopulation levels. The absence of concerted evolution might be due to recent common descent of the three populations and/or frequent gene flow between populations. Recently, Ben-Menni Schuler et al. (2017) have found support for migration–drift equilibrium in these populations, suggesting that gene flow had a key influence on population structure, although populations are currently predominantly influenced by genetic drift. In addition, as mentioned before, unequal crossing-over should operate mainly during diploidy, and the alternation of long-lasting haploid and diploid stages could run counter to sequence homogenization.
On the other hand, according to this model, it might be expected that, in the absence of sexual reproduction, the haploid population (ALMO) should have higher levels of sequence variation for satDNAs (Luchetti et al., 2003; Plohl et al., 2008). Data suggested that evolution of satDNA in ants follows a concerted evolution pattern but that this process is slow in relation to other organisms, probably due to the eusociality and haplodiploidy of these insects (Lorite et al., 2017). In the thelytokous partenogenetic species of the genus Bacillus, Luchetti et al. (2003) found that sexuality acts as a driving force in the fixation of sequence variants within a satDNA family, thus generating intrapopulation cohesiveness and interpopulation discontinuities, and that parthenogenesis has a slowing effect on molecular turnover processes. However, the analysis also proved the spreading of new variants in unisexual specimens by gene conversion events, arriving at the conclusion that given enough time, sequence homogenization can take place in a unisexual species. The study also confirmed the mitotic plasticity of tandem repeats since, to accomplish this, gene conversion events should preferentially take place during cell division. It appears that the haploid ALMO population seems to mirror this situation. A similar context is found for satDNAs of Y chromosomes of the plant Rumex acetosa. While the Y chromosomes of R. acetosa do not recombine, sister chromatid interchanges should explain gene conversion homogenizing events which should lead to concerted evolution of Y-linked satDNA DNA subfamilies (Navajas-Pérez et al., 2006).
A warning on possible contamination of DNA extractions
In this study, we have developed a reliable and successful protocol of evident utility for testing the possible existence of contamination during DNA extraction for high-throughput sequencing methods. As a preventive measure, we thought that the gametophyte samples, by existing in close association with the ground and water, could be contaminated even with very high standards of careful and exhaustive isolation and cleaning of the filaments. This allowed us to overcome any possible pitfall when quantifying satDNA abundance in these samples since, without the application of this corrective calculations, we would have estimated higher amounts of satDNA in sporophytes than in gametophytes, with the consequent impact on key concepts such as satDNA gain/loss between different life cycle phases. The application of our method to obtain correction coefficients, and their use in our calculations, allowed us to find more reliable estimates of satDNA abundance in every sample of V. speciosa. In addition, our estimates are independent of the percentage of contamination in each sample because the coefficients were derived in terms of the relative presence of a high number of long contigs (>3000 nt; thus increasing the likelihood of nucleotide mapping), which were used as reference, resembling the use of reference genes for quantitative PCR.
Conclusion
There is a trend for longer (and older) satellites in V. speciosa to show higher A + T content and to evolve from shorter ones. In this context, the role of microsatellites in the formation of some of these satDNAs is striking. The contributions of satDNA (approx. 0.4 %) and microsatellites (approx. 2 %) to the large genome of V. speciosa are almost negligible compared with that of TEs (approx. 50 %). The TE composition in this species, as in other ferns, was roughly similar to that in seed plant species, except for a higher proportion of DNA transposons and LINEs in the former. Our results also suggest that the reproductive mode or phase alternation between long-lasting haploid and diploid stages does not influence satDNA abundance or divergence. Finally, from a methodological point of view, our proposal of a reliable and successful protocol for testing, and disregarding in satDNA quantification, the existence of unavoidable contaminants might be useful for other NGS studies.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: proportion of each satellite DNA family within the genome of V. speciosa in each of the three populations. Figure S2: representative monomer sequences for each satDNA family. Figure S3: Friedman ANOVA for significant differences between the four libraries with respect to satDNA divergence. Figure S4: repeat landscape plots representing, for each microsatellite, abundance and divergence with respect to a consensus sequence built for each microsatellite repeat unit. Figure S5: sliding windows analysis showing that π varied greatly between positions. Figure S6: curvature propensity plot analysis of VspSat04-107 sequences. Figure S7: VspSat08-82 monomer composition and free energy structure. Table S1: normality analysis for the distributions observed for satDNA parameters in the four populations. Table S2: estimated copy number for 623 sporophytic contigs >3000 nt, to obtain an estimate of the contamination level in the gametophyte gDNA libraries. Table S3: correlation abundance and divergence of satDNA families in the four libraries analysed. Table S4: number of monomers analysed and mean variability for each satDNA fmily in three different populations of V. speciosa. Table S5: extremely conserved regions compared with the remaining nucleotides in each of three satDNA families.
ACKNOWLEDGMENTS
This research has been financed by the Spanish Ministerio de Economía y Competitividad and FEDER founds, grant: CGL2010-14856 (subprograma BOS). The Dirección General de Gestión del Medio Natural y Espacios Protegidos of the Consejería de Medio Ambiente y Ordenación del Territorio de la Junta de Andalucía authorized and facilitated the sampling of the material. We are highly indebted to Carmen Rodríguez Hiraldo and to Jaime Pereña Ortiz who, together with the team of Agentes de Medio Ambiente of the Consejería, helped us with the sampling procedure.
LITERATURE CITED
- Altschul SF, Madden TL, Schäffer AA, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrožová K, Mandáková T, Bureš P, et al. 2011. Diverse retrotransposon families and an AT-rich satellite DNA revealed in giant genomes of Fritillaria lilies. Annals of Botany 107: 255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R.. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Research 28: 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Menni Schuler SBM, García-López MC, López-Flores I, Nieto-Lugilde M, Suárez-Santiago VN.. 2017. Genetic diversity and population history of the Killarney fern, Vandenboschia speciosa (Hymenophyllaceae), at its southern distribution limit in continental Europe. Botanical Journal of the Linnean Society 183: 94–105. [Google Scholar]
- Biscotti MA, Olmo E, Heslop-Harrison JS.. 2015. Repetitive DNA in eukaryotic genomes. Chromosome Research 23: 415–420. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brázda V, Laister RC, Jagelská EB, Arrowsmith C.. 2011. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Molecular Biology 12: 33. doi: 10.1186/1471-2199-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukner I, Sánchez R, Suck D, Pongor S.. 1995. Sequence dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO Journal 14: 1812–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M.. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Cheung M, et al. 2009. Geneious v. 4.8. Auckland, New Zealand: Biomatters Ltd. [Google Scholar]
- Emadzade K, Jang TS, Macas J, et al. 2014. Differential amplification of satellite PaB6 in chromosomally hypervariable Prospero autumnale complex (Hyacinthaceae). Annals of Botany 114: 1597–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielian A, Pongor S.. 1996. Correlation of intrinsic DNA curvature with DNA property periodicity. FEBS Letters 393: 65–68. [DOI] [PubMed] [Google Scholar]
- Garrido-Ramos MA. 2015. Satellite DNA in plants: more than just rubbish. Cytogenetics and Genome Research 146: 153–170. [DOI] [PubMed] [Google Scholar]
- Garrido-Ramos MA. 2017. Satellite DNA: an evolving topic. Genes 8: 230. doi: 10.3390/genes8090230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell DS, Dickerson RE.. 1994. Bending and curvature calculations in B-DNA. Nucleic Acids Research 22: 5497–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols 8: 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, et al. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nature Genetics 43: 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WL, Straight AF.. 2017. RNA-mediated regulation of heterochromatin. Current Opinion in Cell Biology 46: 102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT – the BLAST-like alignment tool. Genome Research 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YB, Oh JH, McIver LJ, et al. 2014. Divergence of Drosophila melanogaster repeatomes in response to a sharp microclimate contrast in Evolution Canyon, Israel. Proceeding of the National Academy of Sciences, USA 111: 10630–10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J.. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- López-Flores I, Garrido-Ramos MA.. 2012. The repetitive DNA content of eukaryotic genomes. Genome Dynamics 7: 1–28. [DOI] [PubMed] [Google Scholar]
- Lorite P, Muñoz-López M, Carrillo JA, et al. 2017. Concerted evolution, a slow process for ant satellite DNA: study of the satellite DNA in the Aphaenogaster genus (Hymenoptera, Formicidae). Organisms Diversity and Evolution 17: 595–606. [Google Scholar]
- Luchetti A, Cesari M, Carrara G, et al. 2003. Unisexuality and molecular drive: Bag320 sequence diversity in Bacillus taxa (Insecta Phasmatodea). Journal of Molecular Evolution 56: 587–596. [DOI] [PubMed] [Google Scholar]
- Macas J, Navrátilová A, Koblížková A.. 2006. Sequence homogenization and chromosomal localization of VicTR-B satellites differ between closely related Vicia species. Chromosoma 115: 437–447. [DOI] [PubMed] [Google Scholar]
- Mehrotra S, Goyal V.. 2014. Repetitive sequences in plant nuclear DNA: types, distribution, evolution and function. Genomics, Proteomics and Bioinformatics 12: 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Bradnam KR, Young HA, et al. 2013. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biology 14: R10. doi: 10.1186/gb-2013-14-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navajas-Pérez R, de la Herrán R, Jamilena M, et al. 2005. Reduced rates of sequence evolution of Y-linked satellite DNA in Rumex (Polygonaceae). Journal of Molecular Evolution 60: 391–399. [DOI] [PubMed] [Google Scholar]
- Navajas-Pérez R, Schwarzacher T, de la Herrán R, Ruiz Rejón C, Ruiz Rejón M, Garrido-Ramos MA.. 2006. The origin and evolution of the variability in a Y-specific satellite-DNA of Rumex acetosa and its relatives. Gene 368: 61–71. [DOI] [PubMed] [Google Scholar]
- Navarro-Domínguez B, Ruiz-Ruano FJ, Cabrero J, et al. 2017. Protein-coding genes in B chromosomes of the grasshopper Eyprepocnemis plorans. Scientific Reports 7: 45200. doi: 10.1038/srep45200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Macas J.. 2010. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11: 378. doi: 10.1186/1471-2105-11-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Pech J, Steinhaisl J, Macas J.. 2013. RepeatExplorer: a galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29: 792–793. [DOI] [PubMed] [Google Scholar]
- Novák P, Ávila Robledillo P, Koblížková A, Vrbová I, Neumann P, Macas J.. 2017. TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Research 45: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermayer R, Leitch IJ, Hanson L, Bennett MD.. 2002. Nuclear DNA C-values in 30 species double the familial representation in pteridophytes. Annals of Botany 90: 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezer Z, Brajković J, Feliciello I, Ugarković Đ.. 2012. Satellite DNA-mediated effects on genome regulation. Genome Dynamics 7: 153–169. [DOI] [PubMed] [Google Scholar]
- Piegu B, Guyot R, Picault N, et al. 2006. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Research 16: 1262–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plohl M. 2010. Those mysterious sequences of satellite DNAs. Periodicum Biologorum 112: 403–410. [Google Scholar]
- Plohl M, Luchetti A, Meštrović N, Mantovani B.. 2008. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 409: 72–82. [DOI] [PubMed] [Google Scholar]
- Plohl M, Meštrović N, Mravinac B.. 2012. Satellite DNA evolution. Genome Dynamics 7: 126–152. [DOI] [PubMed] [Google Scholar]
- Portis E, Portis F, Valente L, et al. 2016. A genome-wide survey of the microsatellite content of the globe artichoke genome and the development of a web-based database. PLoS One 11: e0162841. doi: 10.1371/journal.pone.0162841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer KM, Schuettpelz E, Wolf PG, Schneider H, Smith AR, Cranfill R.. 2004. Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. American Journal of Botany 91: 1582–1598. [DOI] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH.. 2010. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A.. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends in Genetics 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Ruiz-Estévez M, Bakkali M, Martín-Blázquez R, Garrido-Ramos MA.. 2017. a Differential expression patterns of MIKCC-type MADS-box genes in the endangered fern Vandenboschia speciosa. Plant Gene 12: 50–56. [Google Scholar]
- Ruiz-Estévez M, Bakkali M, Martín-Blázquez R, Garrido-Ramos MA.. 2017. b Identification and characterization of TALE homeobox genes in the endangered fern Vandenboschia speciosa. Genes 8: 275. doi: 10.3390/genes8100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ruano FJ, López-León MD, Cabrero J, Camacho JPM.. 2016. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Scientific Reports 6: 28333. doi: 10.1038/srep28333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumsey FJ, Vogel JC, Russell SJ, Barrett JA, Gibby M.. 1999. Population genetics and conservation biology of the endangered fern Trichomanes speciosum (Hymenophyllaceae) in Scotland. Biological Journal of the Linnean Society 66: 333–344. [Google Scholar]
- Schmieder R, Edwards R.. 2011. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One 6: e17288. doi: 10.1371/journal.pone.0017288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P.. 2015. RepeatMasker Open-4.0. 2013–2015. http://repeatmasker.org. [Google Scholar]
- Smith GP. 1976. Evolution of repeated DNA sequences by unequal crossover. Science 191: 528–535. [DOI] [PubMed] [Google Scholar]
- Suzuki K. 2013. Characterization of telomere DNA among five species of pteridophytes and bryophytes. Journal of Bryology 26: 175–180. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahovicek K, Kaján , Pongor S.2003. DNA analysis servers: plot.it, bend.it, model.it and IS. Nucleic Acids Research 31: 3686–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Davis RE.. 2014. Programmed DNA elimination in multicellular organisms. Current Opinion in Genetics and Development 27: 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KH, Lower SE, Caldas IV, Sless TJ, Barbash DA, Clark AG.. 2018. Variable rates of simple satellite gains across the Drosophila phylogeny. Molecular Biology and Evolution 35: 925–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Leitch AR, McCann J, Jang TS, Macas J.. 2015. Employing next generation sequencing to explore the repeat landscape of the plant genome. In: Hörandl E, Appelhans M, eds. Next generation sequencing in plant systematics. Königstein, Germany: Koeltz Scientific Books, 155–179. [Google Scholar]
- Wolf PG, Sessa EB, Marchant DB, et al. 2015. An exploration into fern genome space. Genome Biology and Evolution 7: 2533–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.