 Growth and biofilm inhibitory action of biomimetic green ZnONPs and dissolved zinc ions on a bacterial interface.
Growth and biofilm inhibitory action of biomimetic green ZnONPs and dissolved zinc ions on a bacterial interface.
Abstract
This study was aimed to fill the critical gap of knowledge regarding the interaction between green zinc oxide nanoparticles (ZnONPs) and bacterial interface. Wurtzite phase ZnONPs with a band gap energy of 3.28 eV were produced by exploiting a simple and green biosynthesis method using an inexpensive precursor of A. indica leaf extract and zinc nitrate. ZnONPs were characterized using UV-Vis spectroscopy, XRD, FTIR, SEM, EDX, DLS, TEM, and zeta-potential analysis. The primary size obtained was 26.3 nm (XRD) and 33.5 ± 6.5 nm (TEM), whereas, the secondary size was found to be 287 ± 5.2 nm with –32.8 ± 1.8 mV ζ-potential denoting the physical colloid chemistry of ZnONPs. Crystallinity and the spherical morphology of ZnONPs were also evident with some sort of particle agglomeration. ZnONPs retained plant functional groups endorsing their hydrophilic character. The antibacterial and antibiofilm activity of ZnONPs was significant (p ≤ 0.05) and the MIC/MBC against most frequent clinical isolates of Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Staphylococcus aureus ranged from 0.5 to 1.0 (MIC)/1.0 to 1.5 mg ml–1 (MBC). The dissolution of ZnONPs to Zn2+ ions in a nutrient medium increased as a result of interaction with the bacterial surface and metabolites. Substantial surface binding of ZnONPs followed by intracellular uptake disrupted the cell morphology and caused obvious injury to the cell membrane. Interrupted bacterial growth kinetics, loss of cell respiration, enhanced production of intracellular ROS, and the leakage of the cytoplasmic content unequivocally suggested a strong interaction of ZnONPs with the exterior cell surface and intracellular components, eventually leading to cell death and destruction of biofilms. Overall, the results elucidated eco-friendly production of ZnONPs expressing a prominent interfacial correlation with bacteria and hence, prospecting the use of green ZnONPs as effective nanoantibiotics.
1. Introduction
The developments in manufacturing and increasing applications of nanomaterials, in recent times, have affected almost every domain of human life.1 Metal and metal oxide nanoparticles (NPs) such as silver, gold, zinc oxide, iron oxide, copper oxide etc. are extensively used in a range of consumer goods. Recently, they have also been widely used in agriculture, medical, and pharmaceutical products.2–4 When desired, NPs have been synthesized using physical and chemical methods, for instance, microwave irradiation, thermal evaporation, metal–organic chemical vapour deposition, sputter deposition, hydrothermal growth, laser ablation, ion beam assisted deposition, chemical precipitation, electro-chemical deposition, sol–gel growth, pH mediated growth, sol–gel process, and flame transport synthesis.5–7 Apart from these, NPs have also been generated in recent times using biological or biomimetic approaches which have shown greater advantages over other methods. Such advantages include- (i) swiftness of NPs (ii) economical/eco-friendly nature and (iii) no involvement of toxic chemicals, high energy, and pressure.8 Moreover, the metal oxide (MO) NPs, sometimes called nano-antibiotics have been realized as a supplement of conventional antibacterial drugs because of their significant antibacterial potential. This has prompted immense research interest due to the growing emergence of multiple drug resistance (MDR) among clinical bacteria.9 The emergence and consequent spread of drug resistant bacterial strains pose greater risks to human health as accepted globally by clinicians, healthcare agencies, and drug manufacturers.10 Moreover, the frequent use of antibiotics in hospitals followed by their discharge into effluents and municipal sewage through direct deposition or patient excreta enforce a selection pressure upon the existing bacterial pathogens to become more resilient against different classes and generations of antibiotics.11 Very recently, the World Health Organization (WHO) has reported high resistance rates in Klebsiella pneumoniae and Escherichia coli to IIIrd generation cephalosporins which are used to control both hospital and community infections (WHO, 2014).12 To overcome these issues and to find ways, green NP synthesis using plants extracts without further involvement of capping agents and surfactants is being advocated around the world.13 This green chemistry approach principally involves the use of biomolecules present in plant extracts for efficiently reducing and stabilizing metal ions.14 The reduction of ions followed by capping provides post reduction surface modification in addition to controlling the NP aggregation to relatively bigger sizes.15 Among variously reported NPs, zinc oxide (ZnO) with a wide band gap (∼3.37 eV) has been preferred due to its multiple flexible properties such as electro-catalysis, high chemical stability, and electron transfer capability.16 Furthermore, the US FDA listed ZnO as “Generally Recognized as Safe” (GRAS) due to low toxicity, even though ZnO has been biomedically utilized since ancient times. For instance, fly ash of Zn, in particular, was used to treat many severe illnesses and was found quite efficient.16 Additionally, the strong protein adsorption activity of ZnONPs makes it more useful in the modulation of metabolism, cellular responses, and cytotoxicity.16 In other studies, specially designed tetrapods of ZnO with oxygen vacancies were used as nano-immunotherapeutic agents and when introduced intra-vaginally, suppressed the HSV-2 genital infection in female BALB/c mice.17 These tetrapods also neutralized the HSV-2 virions and thus HeLa and human epithelial cells exhibited much reduced infection with such virions incubated with ZnO tetrapods.19 Other products of ZnO like quantum dots having properties such as chemical stability, high durability, ease in the synthesis process, heat resistance, low cytotoxicity, more selectivity, etc. have been found superior compared to other conventional organic nanomaterials.18 Still, ZnONPs prepared from various plant extracts have been shown to possess antimicrobial and anti-diabetic activities.20,21 These vast and varied advantages of greener synthesis over other methods and the extensive use of ZnO have encouraged us to use Azadirachta indica (neem) leaf extract for fabricating neem leaf extract capped ZnONPs by reduction of Zn(NO3)2·6H2O without incorporating any physical method and toxic chemical agent.
A. indica is a member of Meliaceae and is found commonly in tropical and subtropical regions like India, Pakistan, Nepal and Bangladesh. A. indica contains some important chemicals like, nimbidin, nimbin, limnoids, and nimbolide which play important roles in disease management through the modulation of various genetic pathways.21 The extract from A. indica has been used for the synthesis of silver (Ag) NPs,22 gold (Au) NPs,23 and ZnONPs.24,25 Earlier studies with A. indica plant extract have reported the synthesis of ZnONPs, but did not study their agglomeration in aqueous media and biological features. Moreover, the quantity of leaf extract used in this study (5% w/v) for the production of ZnONPs is much less than that used by others which is as high as 20% w/v and 25% w/v.24,25 Beside these, the major interest of our study was to understand in detail the plausible antibacterial mechanism of A. indica extract synthesized ZnONPs and their behavior at various contact sites on the bacterial interface and cell interior, which has not previously been explored adequately.
Considering the multiple exceptional advantages and wide ranging utility of ZnONPs, a comprehensive and detailed systematic work was undertaken to achieve the following objectives: (i) green synthesis of ZnO NPs using A. indica leaf extract and its confirmation/validation, (ii) determination of the surface morphology, particle and crystal size of ZnONPs, (iii) assessment of biomolecules involved in the reduction and stabilization of biosynthesized ZnONPs, (iv) determination of Zn2+ ion dissolution from ZnONPs alone and in the presence of bacterial cells, (v) determination of the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of ZnONPs against Gram negative (Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae) and Gram positive (Staphylococcus aureus) clinical isolates, (vi) evaluation of nucleic acid release, β-galactosidase activity and cell membrane permeability damage, (vii) stimulation of reactive oxygen species (ROS) in bacterial cells and its impact on cellular respiration, (viii) ZnONP concentration and time dependent growth of clinical isolates, (ix) bacterial cell morphology, surface adsorption and intracellular uptake determination of ZnONPs by SEM/EDX equipped with atomic absorption spectrophotometry (AAS) and (x) assessment of the in situ biofilm inhibition ability of ZnONPs employing light microscopy, laser confocal microscopy, and SEM.
2. Materials and methods
2.1. Green synthesis of ZnONPs: preparation of leaf extract, synthesis and harvesting of ZnONPs
The method used for green ZnONP synthesis is depicted in ESI Fig. S1.† Fresh foliage of A. indica rinsed with running tap water followed by double distilled water (DDW) was allowed to dry at 37 °C and 20 g of sample was used for ZnONP extraction. The aqueous leaf extract was boiled for 25 min at 60 °C and was then allowed to stabilize at room temperature (RT). The extract was further processed as described elsewhere.26 Zn (NO3)2·6H2O was dissolved in 500 mL DDW to attain the final molarity of 50 mM. After complete dissolution using a magnetic stirrer, 100 mL of aqueous leaf extract was added to it and the pH of the mixture was adjusted to 8. Then the sample was vigorously stirred (≈450 rpm) at 80 °C for 1 h. The resulting pale yellow colored slurry was washed with DDW and allowed to dry at 400 °C for 2 h in a muffle furnace. Finally, a white color powder was obtained which was maintained and used for characterization and determination of biological activities.
2.2. UV-visible, FTIR, XRD, SEM-EDX, TEM, and DLS analysis of particle morphology, composition, shape, and the primary and secondary size
Surface plasmon resonance (SPR), crystal size, functional groups, surface morphology, shape, and the primary and secondary size of ZnONPs were analyzed by the methods described earlier3,26,27 (please see the ESI†).
2.3. Antibacterial activity of ZnONPs
2.3.1. Bacterial growth inhibition and MIC/MBC determination of ZnONPs
Agar well diffusion assay as described earlier27 was used to test the antibacterial activity of ZnONPs. For this, 100 μL of ZnONPs (from 20 mg mL–1 stock) was added to each well. Gentamicin (5 μg per disc) was included as the positive control. The MIC and MBC of ZnONPs against P. aeruginosa, E. coli, S. aureus, and K. pneumoniae sub sp. pneumoniae were determined by growing bacterial cells in Luria broth supplemented with various doses of ZnONPs ranging from 0.125 to 1.5 mg mL–1 following our previous method.27 The MIC is defined as the lowest concentration of antimicrobial agent (ZnONPs) that prevents microbial growth whereas the MBC is the minimum concentration required to kill micro-organisms.26
2.3.2. Concentration and time dependent growth of test strains under ZnONP stress
The concentration dependent effect of ZnONPs on P. aeruginosa, E. coli, S. aureus, and K. pneumoniae sub sp. pneumoniae growth was assessed by employing an optical method.27,28 Polystyrene wells filled with 200 μL of LB were supplemented with 0.125, 0.25, 0.5, 1.0, and 1.5 mg mL–1 of ZnONPs and then inoculated with 20 μL of exponentially grown young bacterial culture. The data were recorded and analyzed as described elsewhere.26
2.4. Integrity of cell membranes
2.4.1. Determination of nucleic acid release from ZnONP treated cells
As the permeability of the bacterial plasma membrane is compromised, release of the cytoplasmic content can be monitored. By recording the absorbance at 260 nm, the amount of nucleic acids (DNA and RNA) leaked from the inner cellular material could be estimated. The overnight grown cultures (20 ml) of P. aeruginosa, E. coli, S. aureus, and K. pneumoniae in 100 ml capacity conical flasks were supplemented with varying concentrations of ZnONPs (0.125–1.5 mg ml–1) and then incubated at 37 °C for another 4 h. Aliquots of 1.5 ml were taken from each treatment and control and then centrifuged at 5000 rpm for 5 min to remove bacterial cells. 100 μl of the resulting supernatant was diluted with 1.5 ml DDW and the spectra were recorded in the UV range.
2.4.2. Determination of the β-galactosidase activity
The activity of the β-galactosidase enzyme was determined using o-nitrophenyl-β-d-galactopyranoside (ONPG; HiMedia, India) as the substrate to check the inner membrane permeability of the bacterial cells. Bacterial cultures were grown in LB broth with added 2% lactose and the log phase cells were separated by centrifugation followed by their re-suspension in 0.02 M sodium phosphate buffer (pH 7.5) containing NaCl (0.1 M) to maintain 108–9 CFU ml–1 bacterial cell density. A 500 μl cell suspension from each treatment group was mixed with ZnONPs appropriately diluted from the stock to attain the final concentration ranging from 0.125 to 1.5 mg ml–1. The mixture was incubated at 37 °C overnight followed by centrifugation. In the resulting supernatant, the conversion of ONPG into galactose and o-nitrophenol was monitored by recording the absorbance of o-nitrophenol at 420 nm.
2.4.3. Fluorescence detection of cell permeabilization by CLSM
For this, PI was used as a fluorescent probe for the detection of cells with compromised cell membranes.28 Untreated and bacterial cells treated with the MIC of ZnONPs were washed at least three times with 1× PBS and stained with PI (100 μM) for 15 min at RT in a dark chamber. Cells of K. pneumoniae and S. aureus were visualized under a CLS microscope.26
2.5. Detection of cellular respiration and intracellular ROS under ZnONP stress
Inhibition of bacterial cellular respiration was determined by dehydrogenase assay, as described by Wahab et al. with modifications. Briefly, the cells of P. aeruginosa, E. coli, S. aureus, and K. pneumoniae in the early log phase were harvested by centrifugation at 5000 rpm for 10 min.29 The cells were re-suspended in 1× sterile phosphate buffer (pH 7.0) to achieve an absorbance (Abs600) of 0.4, and 200 μl of this cell suspension was then transferred to wells in a 96-well microtitre plate. To each well, the ZnONPs were added in increasing concentrations of 0.125, 0.25, 0.5, 1.0, and 1.5 mg ml–1. The cell suspension without ZnONPs was run in parallel as a control. Subsequently, 40 μl of the reagent 2,3,5-triphenyl-tetrazolium chloride (TTC) (0.5%, w/v) was added to each well followed by incubation at room temperature. The change in color from a colorless solution to red was measured at 450 nm using a microplate reader (Thermo Scientific Multiskan EX, REF 51118170, China). The intracellular ROS produced by the Gram positive (S. aureus) and Gram negative (K. pneumoniae) bacteria grown in the absence and presence of the MIC of ZnONPs was detected by the method used earlier.26
2.6. Assessment of the cellular morphology and uptake of ZnONPs by SEM-EDX and AAS
Non-stressed and S. aureus and K. pneumoniae cells grown under ZnONP stress were processed for SEM, EDX, and elemental mapping of the region of interest (ROI) following methods described previously.26,30 Samples were visualized under a JSM 6510LV SEM (JEOL, Tokyo, Japan) at an accelerating voltage of 10 kV. The attachment of ZnONPs onto root surfaces was detected by using an Oxford Instruments INCAx-sight EDX equipped SEM followed by the mapping of elements. In order to determine Zn2+ ion dissolution, ZnONPs were incubated with a nutrient medium for 24 h at 37 °C with constant stirring at 150 rpm and the supernatant collected after centrifugation (10 000 rpm, 20 min) was filtered through a 0.22 μm membrane filter. The concentration of soluble zinc was measured by AAS, (GBC 932B plus, Australia). Similarly, Zn2+ ions in the bacterial supernatant and internalized ZnONPs in the bacterial cell pellet after treating the cells of clinical isolates with 1000 μg ZnONPs per ml at 37 °C for 24 h were determined. The bacterial cell pellet was digested with 5 ml aqua regia and appropriately diluted with DDW up to 100 ml volume in volumetric flasks prior to AAS analysis.
2.7. Assay for biofilm inhibition under ZnONP stress
The impact of the sub MIC concentrations of ZnONPs on the biofilm forming ability of K. pneumoniae and S. aureus was determined by the crystal violet (CV) method.26
2.8. In situ visualization of biofilms
2.8.1. Light microscopy analysis
The biofilm inhibition by ZnONPs was also evaluated on a glass surface under a light microscope.27 For this, wells (6 well polystyrene plates) were filled with LB broth and the sub MIC concentrations of ZnONPs followed by the addition of young bacterial cells (1 × 107 cfu mL–1) of K. pneumoniae and S. aureus at 1% culture rate. Glass cover slips were set as described for ROS production followed by incubation at 37 °C for 24 h. Biofilms were then rinsed with PBS and stained with 0.1% CV and visualized.
2.8.2. Confocal laser scanning microscopy (CLSM) analysis of biofilms
The anti-biofilm potential of ZnONPs on clinical isolates belonging to the genera Klebsiella and Staphylococcus was assessed using the fluorescent probe acridine orange (AO): a dye which is used to stain DNA and intercalates at the major groove of dsDNA. After interaction, it produces green fluorescence upon excitation. For this, control and biofilms treated with the sub MICs of ZnONPs were washed three times with sterile PBS and stained with 0.01% AO (prepared in PBS) for 20 min at RT in the dark. Biofilms of K. pneumoniae and S. aureus after incubation with AO were gently rinsed three times with PBS and then mounted on a microscope slide and visualized using an LSM-780 confocal microscope (Zeiss, Germany).
2.8.3. Scanning electron microscopy (SEM) analysis of biofilms
The inhibition of biofilm formation was further assessed using SEM. The biofilms of K. pneumoniae and S. aureus grown in the presence and absence of ZnONPs were rinsed with sterile PBS (1×) and subsequently fixed in a mixture of 2.5% glutaraldehyde and 2% paraformaldehyde in a sterile polystyrene plate for 12 h at 4 °C. The fixed biofilms after three successive washes with PBS (1×) were dehydrated by an ethanol gradient (30, 50, 70, 90 and 100%) for 10 min each. The cover slips containing biofilms were air dried and sputter coated with a 2 nm thin layer of gold. The coated samples were observed under a JEOL JSM-6510 (JEOL, Tokyo, Japan).
2.9. Statistical analysis
The data presented in this study represent mean (N = 3) ± S.D. (standard deviation) of experiments done at least in duplicate. Sigma Plot 10.0, USA was used for statistical analysis and for preparing curves and graphs. The difference among the treatment means was compared using Student's t-test at 95% and 99% probability levels.
3. Results and discussion
3.1. Green synthesis of ZnONPs
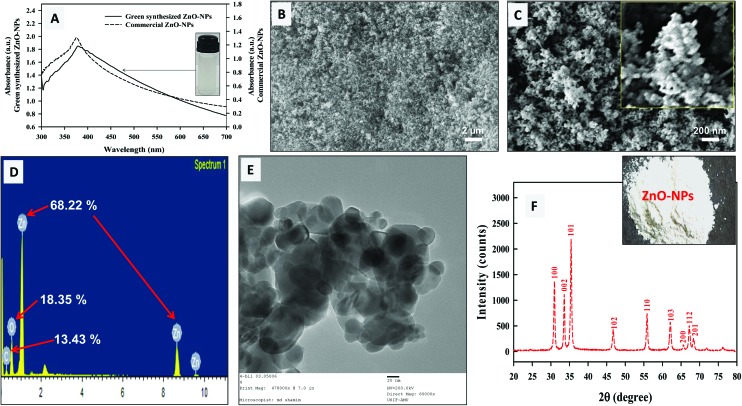
The reducing and capping efficiency of A. indica leaf extract was exploited for the green biosynthesis of ZnONPs. The qualitative observation of ZnONP formation was based on the development of a white colored precipitate as the end product of the reaction (ESI Fig. S1†). The major biomolecules present in A. indica leaves are terpenoids, flavanones, and sugars that act as reducing as well as stabilizing agents.31 The aldehyde groups in A. indica leaf broth have been suggested responsible for the production of ZnONPs along with other chemical groups.25 In a study, benzoquinones in plant extracts have also been reported as reducing agents which stimulate NP synthesis.32 Moreover, the proteomes of plant extracts are also known to alter the physicochemical properties of NPs by forming weaker associations with the nascent crystals of ZnONPs that ultimately lead to the isotropic growth of ZnONPs.33 Here, Fig. 1A shows the UV visible spectra of 0.1% (w/v) green synthesized ZnONPs and commercially available ZnONPs (Sigma Aldrich, USA) in an aqueous dispersion. A characteristic absorption of green synthesized ZnONPs was detected at 378 nm which was comparable to the Sigma Aldrich ZnONPs (374.5 nm). This strong excitonic absorption peak could be assigned to the intrinsic band gap of ZnONPs owing to the electronic transition from the valence to conduction band.24 No other absorption peaks were observed in the UV-vis spectra of green ZnONPs suggesting the maximum purity of ZnONPs (Fig. 1A). The band gap energy (E) of the green ZnONPs calculated using the equation E = hc/λ was 3.28 eV, where, h is Planck's constant (6.626 × 10–34 J s), c is the velocity of light (3 × 108 m s–1), and λ is the wavelength of ZnONPs. Indeed, ZnONPs are well known to absorb in the range of 300–400 nm, and a characteristic signal has been observed in various studies at 372 nm and 370 nm.34,35 Sangeetha et al. have also reported the absorption by ZnONPs in the range of 358–375 nm with increasing SPR under different synthesis conditions.36
Fig. 1. Characterization of ZnONPs; UV-visible spectrum (A) of green synthesized ZnO-NPs (0.1% w/v; solid line) and its comparison with commercially available ZnO-NPs (dashed line) which confirms the purity of ZnONPs. SEM analysis of ZnO-NPs at a direct magnification of ×10 000 (B) and ×50 000 (C). EDX spectrum of ZnONPs (D) and the TEM micrograph of at ×60 000 (E) with an average diameter of 33.5 ± 6.5. The inset in panel B depicts the morphology of ZnO-NPs. Panel F shows the X-ray diffractogram of ZnO nanopowder. The average crystal size by XRD was found to be 26.3 nm.
3.2. X-ray diffraction, FTIR, SEM, TEM, EDX, and DLS analysis of ZnONPs
The SEM images taken at 10 000× (Fig. 1B) and 50 000× (Fig. 1C) magnification exhibited the morphological features of ZnONPs. The micrographs clearly revealed the aggregates of variable sizes of ZnONPs in which the individual NPs were largely spherical in shape (Fig. 1B and C). The elemental analysis of the ZnONPs was revealed through EDX, and signals for Zn (68.22 wt%) along with C (13.43 wt%) and O (18.35 wt%) were found in the spectrum (Fig. 1D). The emission peaks of Zn with small signals for O confirmed the formation and purity of green synthesized ZnONPs. The even lower intensity signal for C was due to the carbon tape used in the scanning process. These results correlate with the report in which similar peaks have been observed in ZnONP synthesis using Acalypha indica leaf extract.8 The ZnO nanostructures also show different morphologies which vary from sheets to flowers with variable pH values (7–12).18 Moreover, the results of TEM analysis revealed predominantly spherical ZnONPs with an average particle diameter of 33.5 ± 6.5 nm (Fig. 1E). Fig. 1F represents the characteristics XRD pattern of ZnONPs with sharpened reflection peaks which indicate that the growth of crystalline ZnONPs has occurred. The Bragg reflections appearing at the 2θ scale of 31.76°, 34.46°, 36.26°, 47.57°, 56.59°, 62.92°, 66.37°, 66.97°, and 69.09° could be well indexed to the [100], [002], [101], [102], [110], [103], [200], [112], and [201] Miller indices (hkl) of ZnO crystal planes, respectively. This is in agreement with the wurtzite structure of ZnO (JCPDS file no.: 36-1451). The full-width-at-half-maximum (FWHM) value for the major crystalline planes of [100], [002], and [101] reflection was used to obtain the mean crystalline size of ZnONPs. The average particle size determined using Debye–Scherrer's equation was found to be 26.3 nm under our experimental conditions. The results of XRD are in good agreement with those reported for O. europa leaf extracts in which 2θ values were obtained at 31.84°, 34.50°, 36.32°, 47.59°, 56.63°, 66.89°, 67.98°, 69.09°, and 76.98°.37 The obtained XRD data clearly indicated the formation of ZnONPs and showed no other diffraction peaks, thus confirming the purity of the ZnO phase. The size obtained by TEM and XRD of ZnONPs corresponds to each other. The FT-IR spectra of the A. indica leaf extract alone and pure ZnONPs are shown in ESI Fig. S2A and B† and a tentative assignment of transmittance is presented in ESI Table 1.† The spectra showed a broad absorption band at 3440–3200 cm–1 assigned to the –OH phenolic group in the extract. The signal at 2970 cm–1 is attributed to the C–H stretching of aliphatic hydrocarbons. The peak shift at 3445 cm–1 is due to the change in the functional group present in the A. indica leaf extract, as a result of interaction with ZnONPs (ESI Fig. S2B†). Similar FT-IR bands have been suggested by Sangeetha et al. for Aloe barbadensis ZnONPs.36 The signals at 3445, 1760, and 1400 cm–1 in the ZnONP FT-IR spectrum indicate the participation of functional groups from polyols, terpenoids, and proteins such as amines, alcohols, ketones, alkanes, and carboxylic acids in the dual process of bioreduction and capping. In the bioreduction process, terpenoids are considered secondary due to their poor water soluble nature. However, proteins are considered less important in the biosynthesis of NPs.41 In contrast, the prime moieties in this process are water soluble flavonones and phenolics which are thought to act as the major reducing agents.38 The stability of ZnONPs may be due to the free amino and carboxylic groups that have interacted with the zinc surface. The bonds of functional groups such as –CO–C–, –C–O– and –C–C– are derived from heterocyclic compounds and the amide bands derived from the proteins are present in the leaf extract and they are the capping ligands of the NPs. Additionally, the amino (–NH) and carboxylic (–COO–) groups interact with the zinc surface and contribute to the formation and stability of ZnONPs.36 This adsorption of phyto-constituents on the ZnONP surface provides stability through supramolecular interactions such as H-bonding or reversible bond formation (Diels–Alder reaction), ionic, and π–π interactions.1,8,14,24 The signal at 495 cm–1 confirms the stretching vibrations of the Zn–O bond of ZnONPs. Similarly, the prominent IR signal at ∼440 cm–1 has been attributed to the stretching vibration of the Zn–O bond.39,40 The secondary size (hydrodynamic size) distribution measurement of ZnONPs in nutrient media showed particle aggregates of 287 ± 5.2 nm. The results for zeta potential of ZnONPs are shown in ESI Fig. S3.† The ζ-potential or electrokinetic potential of ZnONPs was found to be –32.8 ± 1.8 mV which denotes enough stability in solution to exert biological reactions. The average hydrodynamic diameter of ZnONPs in media suspension indicates particle aggregation. The hydrodynamic size and ζ-potential characterization data are in good agreement with a recent study on the agglomeration of ZnONPs in aqueous media.42 In an aqueous dispersion, the number of collisions due to inter-particular interactions increases whereas, the path average length travelled by particles between successive collisions falls.43 The increase of ZnONP size (287 ± 5.2 nm) could be suggested as a result of soft corona formation composed of nutrient broth constituents of different masses and charges.43 Hence, dispersed ZnONPs exhibited a hydrated surface which was wrapped within a cover of molecules not being the ingredients of ZnONPs itself.
3.3. Antibacterial activity of ZnONPs
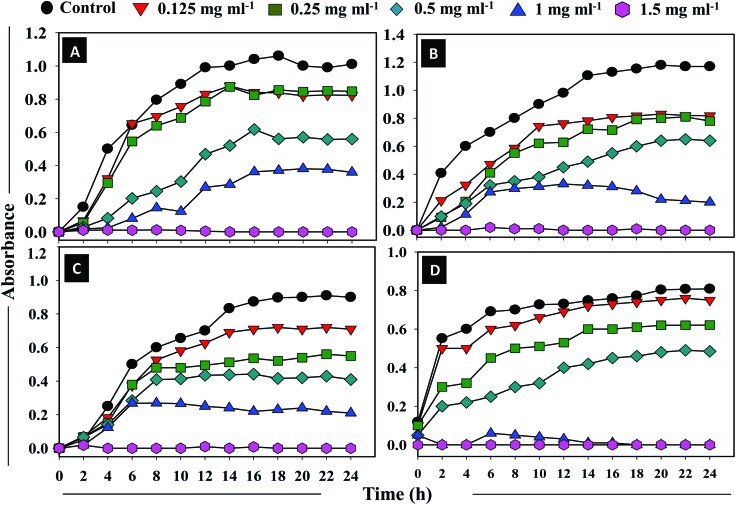
The ZnONPs were assessed for their antibacterial activity against Gram +ve S. aureus and Gram –ve E. coli, P. aeruginosa, and K. pneumoniae. The results of the well diffusion assay shown in ESI Fig. S4† are comparable to the gentamicin antibiotic (5 μg per disc) and are suggestive of the similar efficacy of ZnONPs towards the test bacteria. The MIC of ZnONPs for S. aureus, E. coli, and P. aeruginosa was found to be 1.0 mg ml–1, whereas, it was 0.5 mg ml–1 for K. pneumoniae. The MBC for S. aureus, E. coli, and P. aeruginosa was 1.5 mg ml–1, whereas, it was 1.0 mg ml–1 for K. pneumoniae under identical experimental conditions. Fig. 2A–D demonstrate the time (0–24 h) and ZnONP concentration dependent growth kinetics of S. aureus (Fig. 2A), E. coli (Fig. 2B), P. aeruginosa (Fig. 2C), and K. pneumoniae (Fig. 2D). With the increase in the concentration of ZnONPs (from 0.125 to 1.5 mg ml–1), a gradual increase in bacterial inhibition was recorded. The ZnONPs induced an obvious decline in bacterial viability which however, also increased with the exposure time. Time-resolved bacterial responses provided important information on toxic dynamics, i.e., ZnONPs rapidly killed bacterial strains within 6 h of incubation. The Gram –ve K. pneumoniae cells were observed most susceptible to ZnONPs compared to the other strains. In another study, the ZnO nanoparticles have shown attractive antibacterial properties due to the increased specific surface area.44 Additionally, the antimicrobial activities of ZnONPs towards bacteria depend on the particle size, concentration, crystal morphology etc. Mechanistically, the susceptibility of the bacterial cells to ZnONPs could possibly be due to the enhanced release of H2O2 from the surface of ZnONPs. The H2O2 so generated can penetrate the cell membrane and cause bacterial cell death.45 Since ZnONPs have been found biocompatible with human cells, they therefore have been used as antibacterial agents.18 Categorically, the results indicated that the ZnONPs synthesized from the A. indica leaf extract were effective enough and demonstrated considerable antibacterial activity against all the test bacterial species.
Fig. 2. Antibacterial activities of ZnONPs against the clinical isolates of bacteria. Comparison of the absorption spectra at 620 nm, as an index of the growth kinetics of clinical isolates, at an increasing ZnONP dose (0.125, 0.25, 0.5, 1, and 1.5 mg ml–1) and as a function of incubation time at 37 °C is shown in panels A–D. Panel A: Gram +ve S. aureus; panel B: Gram –ve E. coli; panel C: Gram –ve P. aeruginosa; and panel D: Gram –ve K. pneumoniae.
3.4. Determination of cell membrane integrity and porosity at the ZnONPs–bacteria interface
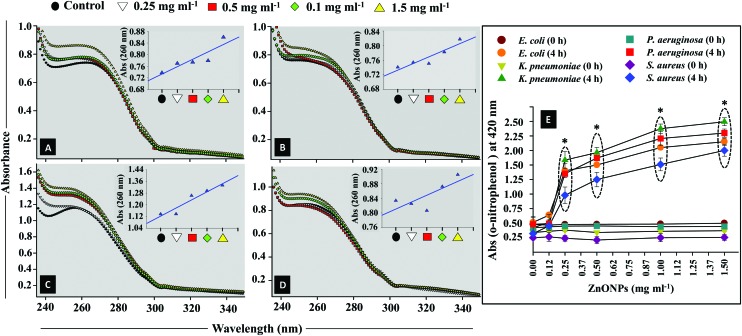
The plasma membrane of the bacterial cells allows them to communicate with the surrounding environment. It may become functionally ineffective under the influence of antibacterial agents.46 The plasma membrane is selectively permeable which facilitates the interfacial transport of molecules.47 In order to investigate the condition of the membrane, the bacterial isolates were exposed to different doses (0.125–1.5 mg ml–1) of ZnONPs and membrane disorganization caused by ZnONPs was observed through CLSM. Following the damage in the cellular membrane, small ions like potassium (K+) and phosphate (PO43–) tend to release first followed by the forceful discharge of larger ones such as nucleic acids (DNA and RNA) along with other molecules.26,48 Therefore, to check membrane disintegration, the extracellular release of nucleic acids (DNA and RNA) from ZnONP treated E. coli, K. pneumoniae, P. aeruginosa, and S. aureus was quantified (Fig. 3A–D). The data exhibited dose dependent enhancement in nucleic acid release and consequently the membrane permeability. The percent increase in nucleic acid release induced by ZnONPs was found to be 5–17%, 1–10%, 2–20%, and 1–8% from E. coli (Fig. 3A), K. pneumoniae (Fig. 3B), P. aeruginosa (Fig. 3C), and S. aureus (Fig. 3D) cells. To further confirm the alteration of cell membrane permeability more specifically the inner membrane (IM) permeability, the activity of the enzyme β-galactosidase was assayed. The enzyme β-galactosidase (an endoenzyme) is a frequently used stress marker to observe the injuries triggered by stressor molecules.49 The ability of ZnONPs to permeate the cellular membranes of clinical isolates in a lactose supplemented medium was evaluated in terms of the production of o-nitrophenol from the substrate ONPG (Fig. 3E). A maximal increase in the absorbance of o-nitrophenol was noticed which followed concentration dependent enhancement of ZnONPs: up to 1.9, 2.2, 2.05, 1.75 for E. coli, K. pneumoniae, P. aeruginosa, and S. aureus, respectively, at 1.5 mg ZnONPs per ml after 4 h of incubation. The enhanced release of β-galactosidase from the cell interior is in good agreement with those reported by others.50,51 However, the untreated bacterial cells showed negligible release of β-galactosidase (0.25–0.4) after 4 h growth.
Fig. 3. Mean absorbance spectra of nucleic acid released from the membrane compromised cells of the clinical strains of E. coli (panel A), K. pneumoniae (panel B), P. aeruginosa (panel C), and S. aureus (panel D) after exposure with various concentrations of ZnONPs (0–1.5 mg ml–1). The inset linear plots show enhancement in the optical density at 260 nm as a function of ZnO-NP concentration. Panel E depicts the extracellular β-glycosidase activity of bacterial cells as a result of inner plasma membrane damage. The absorbance change of o-nitrophenol at 420 nm has been plotted against various ZnONP concentrations (0.125–1.5 mg ml–1) at 0 and 4 h of incubation at 37 °C. Values from three replicates are expressed as mean ± SD (*p ≤ 0.05 vs. control at 4 h).
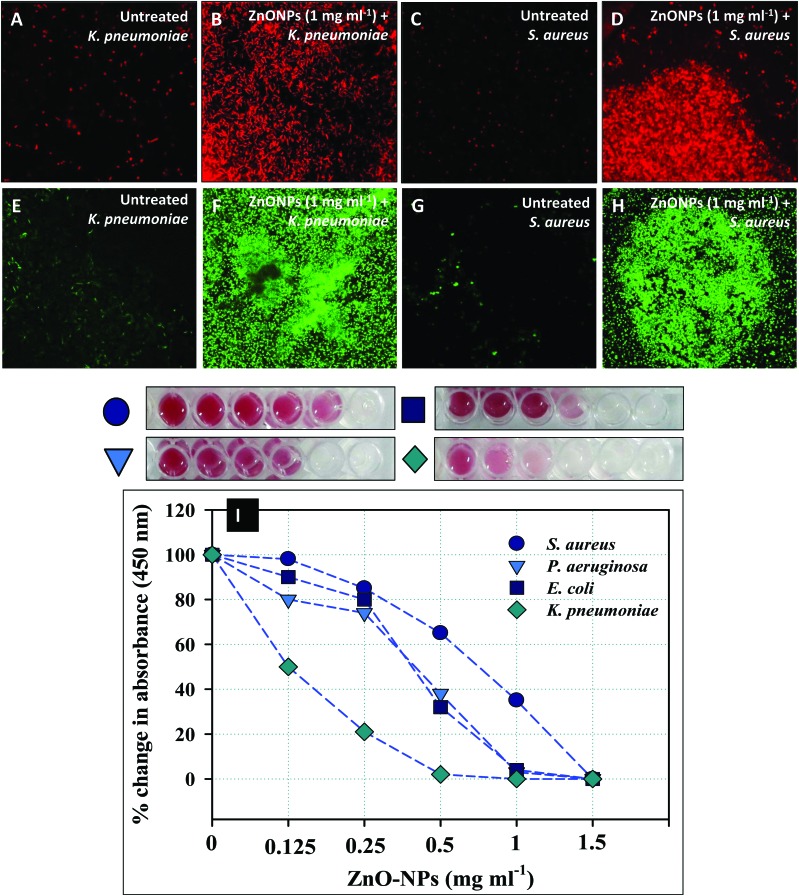
Qualitative measurement of the impact of ZnONPs on the permeability of K. pneumoniae and S. aureus cells was done using the fluorescent probe PI. PI is commonly used as a cell death marker due to its repulsion by an intact cellular membrane of untreated viable cells. Hence, the fluorescence emitted by PI can be effectively correlated to dead bacterial cells. Metabolically inactive cells of K. pneumoniae (Fig. 4B) and S. aureus (Fig. 4D) appeared red against the black background followed by the excitation at 532 nm (λexc) due to PI stained bacterial DNA. Control cells showed only a residual increase in the red fluorescence of PI (Fig. 4A and 5C). The damage caused by ZnONPs to cell membranes exhibited a concentration dependent effect and was coherent with the findings of the bactericidal activity. The attachment of ZnONPs to bacterial membranes at various locations has been reported to augment the level of ROS and superoxides (O2˙–), which creates new pores in cell membranes leading to an enhanced uptake of PI.52 The bacterial cell death due to ZnONPs could be attributed to the damage of the cell membrane and the extrusion of the cytoplasmic contents. The excessive leakage of solutes from inside to the exterior environment thereby results in cell death.53 The present findings therefore describe a clear toxicity of ZnONPs and further confirm the fact that the most critical target for any stressor molecule is the cell membrane.54,55 Similar to these findings, PI stained dead bacterial cells of E. coli, P. aeruginosa, and S. aureus observed under a CLSM have recently been reported.26,56,57
Fig. 4. ZnONP induced cell death, reactive oxygen species, and inhibition of cellular respiration in treated clinical isolates. The ZnONP (1.0 mg ml–1) mediated increase in the number of dead cells as a result of increased membrane permeability. Panels A and C show the residual fluorescence of PI in K. pneumoniae and S. aureus cells. Panel C and D present an increase in PI fluorescence due to the damaged cell membrane. Panels E and G show the residual fluorescence of DCF in K. pneumoniae and S. aureus cells. Panel F and H show a significant increase in intracellular ROS when exposed to 1 mg ml–1 of ZnONPs. Panel I shows the inhibition of cellular respiration of E. coli, S. aureus, K. pneumoniae, and P. aeruginosa by green synthesized ZnONPs at 0.125–1.5 mg ml–1. The decrease in red color intensity in microtitre wells represents the loss of metabolic activity of bacterial cells.
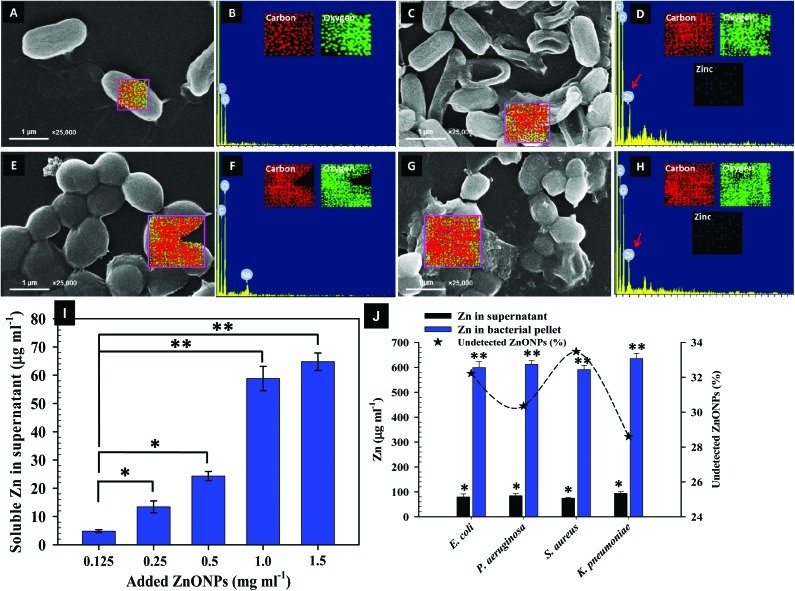
Fig. 5. SEM images and EDX spectra indicating ZnONP induced morphological damage in bacteria. Representative SEM micrographs depict cellular damage and structural distortion in ZnONP (1 mg ml–1) exposed cells: Panels C: Damage to K. pneumoniae cells while Panel G has S. aureus cells. Panels A and E show the untreated cells of K. pneumoniae and S. aureus cells, respectively. EDX spectra and elemental mapping of treated K. pneumoniae and S. aureus cells in panels D and H show the presence of Zn as peak signals and dots in mapping along with C and O; Panel B and F represent untreated cells. Panel I shows the release of soluble Zn ions from NPs. Panel J represents the concentration of Zn in the cell supernatant and bacterial pellets of E. coli, P. aeruginosa, S. aureus, and K. pneumoniae grown in 1000 μg ml–1 of ZnONPs (for 24 h at 37 °C. The loss of ZnONPs is shown as the undetected percentage of Zn. Values from three replicates are expressed as mean ± SD (*p ≤ 0.05, **p ≤ 0.005 vs. control).
3.5. Loss of cellular respiration and enhanced intracellular ROS production under ZnONP stress
The chemically reactive species containing oxygen such as H2O2, O2–, and OH are collectively known as reactive oxygen species (ROS) whose level in bacterial cells is affected by xenobiotics.58 Antibacterial agents modify the composition, structure and functions of bacterial cells leading to oxidative burst. Accordingly, the production of ROS due to ZnONP pressure was investigated in this study. ROS generation under ZnONP stress has been monitored using a fluorescent probe, DCFH-DA, which is a cell permeable stain. When cleaved by intracellular esterases, it gives the anion (H2DCF–) and after reaction with ROS, this low fluorescent reduced anion is converted to a highly fluorescent oxidized form (DCF) and serves as a ROS indicator.59Fig. 4E and G show the residual ROS formation by the cells of Gram –ve K. pneumoniae and Gram +ve S. aureus when grown in the absence of ZnONPs, since most of the stains are in the form of a reduced anion (H2DCF–) while some reacted with ROS present at the threshold levels for essential intracellular signaling.
Moreover, the ROS produced for intracellular signaling in untreated bacterial cultures is regularly scavenged by the ROS reacting enzyme system.60 However, in the presence of 1 mg ZnONPs per ml, significant ROS production was observed in K. pneumoniae (Fig. 4F) and S. aureus (Fig. 4H) cells. This ROS production in turn caused the loss of cellular metabolic activity (Fig. 4I) and substantially declined the population density (Fig. 3). The ROS also oxidize cellular components and disrupt the integrity of the cell membrane. Hence, the increased bacterial cell membrane porosity and the leakage of the cytoplasmic content (Fig. 3) can be correlated with the increased formation of ROS at the sub-lethal concentration of ZnONPs. Additionally, quantitative estimation of cell viability (Fig. 4I) and porosity (Fig. 3E) showed higher damage to E. coli, P. aeruginosa, and K. pneumoniae as compared to S. aureus cells. This behavioral difference of ZnONPs among the Gram –ve and Gram +ve bacteria can be rationalized on the basis of the varying magnitude of change in the interface potential. The neutralization of this interfacial potential by ZnONPs to exert toxicity has also been reported to induce ROS generation which is also a major cause of DNA, lipid, and protein damage.61,62
The ZnONP concentration dependent inhibition of respiration in E. coli, K. pneumoniae, P. aeruginosa, and S. aureus is shown in Fig. 4I. The inhibition of the dehydrogenase activity recorded in this study was obvious and visible by red color formation (red formazan) in untreated cells. In contrast, the cell metabolic activity was significantly reduced with increasing concentrations of ZnONPs (Fig. 4I). The percent change in absorbance (450 nm) was calculated and was found decreasing in a dose dependent manner. The percent change as compared to the untreated control (100%) was: 98%, 85%, 65% 35%, and 0% for S. aureus, 80%, 74%, 38%, 3%, and 0% for P. aeruginosa, 90%, 80%, 32%, 4%, and 0% for E. coli, and 50%, 21%, 2%, 0%, and 0% for K. pneumoniae at 0.12, 0.25, 0.5, 1.0, and 1.5 mg ml–1 ZnONPs, respectively. This kind of inhibition in bacterial cell respiration by some other NPs has also been reported by other workers in E. coli, P. aeruginosa, and nitrifying bacteria.29 Furthermore, it has been observed that the interaction of NPs with bacterial cell membrane components arrests respiration, causes cellular disintegration and produces oxidative damage.63,64 The main advantage associated with the use of ZnONPs as an antibacterial agent is its biocompatible nature and easy synthesis process. Due to these, ZnO nanostructures have also been found to function as prospective prophylactic agents against clinical infections.6 In similar experiments, many workers have concluded that ZnONPs could be used as anti-infective agents for chronic bacterial infections.61,62
3.6. Assessment of the bacterial morphology and uptake of ZnONPs
SEM-EDX and AAS were used to further assess the quantum of ZnONPs located on the bacterial surface. For instance, Fig. 5A–J show the SEM-EDX and AAS data related to Zn2+ ions released from ZnONPs in the presence and absence of bacterial cells under identical experimental conditions. SEM-EDX and data of elemental mapping reveal the interaction of ZnONPs at a sub lethal dose of 1000 μg ml–1 with bacterial surfaces and their adsorption followed by internalization in the representative strains of K. pneumoniae and S. aureus cells. Panels A and E in Fig. 5 show the control cultures of K. pneumoniae and S. aureus with their corresponding EDX spectra and element maps (Fig. 5B and F). The untreated cells appeared undamaged with a smooth and intact surface exhibiting the normal bacterial cell morphology. In contrast, cells with ZnONPs (Fig. 5C and G) had extensive cellular damage with a disorganized cell envelope and morphology. Moreover, the adsorption of Zn was also a noticeable sign of membrane injury (Fig. 5D and H). The SEM micrographs in this study, therefore, validated the ZnONP nuisance at the bacterial interface leading to a clear inhibition of bacterial growth. The ZnONP induced cellular damage in test isolates detected through SEM-EDX corroborated well with those obtained through CLSM (Fig. 4). In similar studies, Manzoor et al. and Dwivedi et al. have also noticed cellular destruction of bacterial cells due to the interaction with ZnONPs.66,67 Besides assessing the adsorption of ZnONPs on the bacterial surface, the release of Zn2+ ions as a result of interaction between the bacterial metabolites with ZnONPs was also assayed. The ZnONPs subjected to incubation (at 37 °C, 4 h) with the nutrient medium alone in the absence of bacterial cells showed a progressive release of soluble Zn2+ ions (Fig. 5I). After 24 h interaction with bacterial cells, the release of soluble zinc from ZnONPs (1000 μg ml–1) in the bacterial supernatant was found to be 79.28 ± 12.15 μg ml–1 for E. coli; 84.14 ± 8.4 μg ml–1 for P. aeruginosa; 74.56 ± 3.2 μg ml–1 for S. aureus; and 94.15 ± 6.2 μg ml–1 for K. pneumoniae, whereas, 598.4 ± 24.5, 612.3 ± 14.6, 590.6 ± 17.5, and 635.2 ± 21.2 μg ml–1, respectively, was recorded in the acid digested bacterial cell pellet (Fig. 5J).
The internalization propensity of NPs is a principal factor arbitrating the toxicity apart from the surface conditions of NPs.68 The bacterial products of metabolism are expected to be released in the growth medium. These metabolites in media interact with ZnONPs and biotransformation of ZnONPs occurs which ultimately liberates soluble Zn2+ ions from NPs. The soluble zinc is also suggested to form complexes with the inorganic and organic constituents of metabolites and subsequently be transported to spaces in bacterial cell walls, membranes, and taken up in the cell interior causing growth arrest. Similar interfacial behavior between zinc and the cell envelope has been advocated by Zhang et al.69 which is in good agreement with our findings. In spite of the additive role of a soluble metal on bacterial growth, ZnONPs–bacteria interaction accounts for their major contribution to the bacteriostatic impact after getting internalized and subsequently inducing intracellular ROS which inflicts cell death.70
3.7. Antibiofilm activity of ZnO-NPs
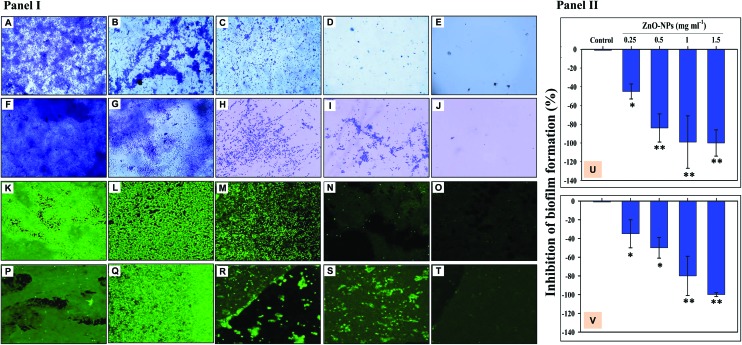
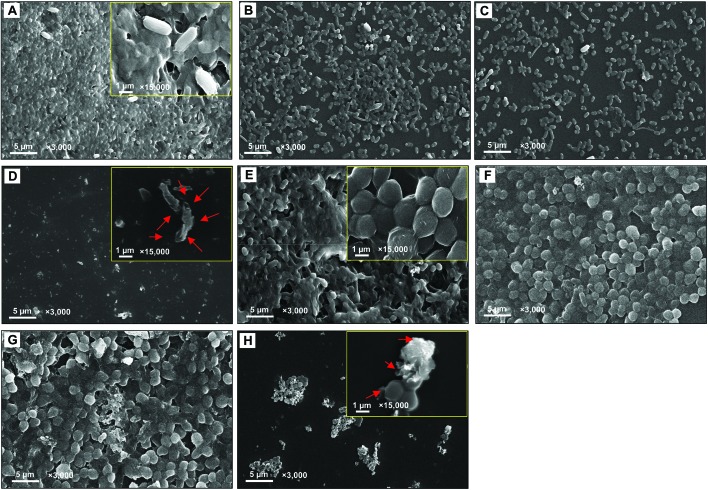
A dose dependent inhibition of the biofilm forming ability of K. pneumoniae and S. aureus was noticed while growing under ZnONPs and viewed under a light microscope (Fig. 6A–J), CLSM (Fig. 6K–T), and SEM (Fig. 7A–H). A maximum inhibition of 100 ± 2 and 100 ± 1.9% in biofilm formation was recorded for K. pneumoniae (Fig. 6U) and S. aureus (Fig. 6V), when these cultures were grown with ZnONPs. Moreover, the reduction in biofilm formation under ZnONP stress was also visible under a CLSM. Here also, the CLSM image of K. pneumoniae (Fig. 6K–O) and S. aureus (Fig. 6P–T) displayed enhanced biofilm inhibition which increased gradually with increasing concentrations of ZnONPs. The ZnONP induced reduction in biofilm formation was also clearly visible under an SEM (Fig. 6A–H). The alteration in bacterial biofilms due to ZnONP application caused morphological changes in bacterial cells. For example, both untreated (Fig. 7A) and biofilms treated with 0.25 mg ml–1 (Fig. 7B), 0.5 mg ml–1 (Fig. 7C) and 1 mg ml–1 ZnONPs (Fig. 7D) of K. pneumoniae revealed that the sessile cells with ZnONPs exhibited a different morphological appearance compared with the shape and intactness of the untreated sessile cells (Fig. 7A). The red arrows in Fig. 7D indicate the damaged bacterial cells due to the ZnONP antibiofilm activity. Likewise, the impact of the varying concentration of ZnONPs on S. aureus biofilms followed a trend similar to those observed for K. pneumoniae (Fig. 7F–H) which, however, significantly differed from the control (Fig. 7E). The antibiofilm activity of ZnONPs is suggested due to the uptake of ZnONPs by bacterial cells followed by cell membrane damage through enhanced ROS production and subsequent leakage of cellular materials.65 The clinical isolates used in this study are known biofilm formers, and hence their clinical management is difficult. Furthermore, successful eradication of infection becomes more puzzling with the increasing emergence of multi-drug resistance among microbial populations.71 Particularly, in chronic infections, the antibiotic inefficacy has been reported. Therefore, green synthesized ZnONP induced quorum quenching can be suggested as an alternative yet promising approach for treating biofilm established bacterial infections. Even so, it could be debated that the phytocompounds of A. indica involved in the entire process of ZnONP synthesis might also act as siderophores, chelating iron (Fe) from the growth media and causing the degradation of biofilms.72
Fig. 6. Inhibition of biofilm formation by ZnONP treated bacterial isolates (Panel I). Micrographs A–E represent the biofilm formed on the glass surface; untreated K. pneumoniae biofilm (A), K. pneumoniae + 0.25 mg ml–1 ZnONPs (B), K. pneumoniae + 0.5 mg ml–1 ZnONPs (C), K. pneumoniae + 1 mg ml–1 ZnONPs (D), and K. pneumoniae + 1.5 mg ml–1 ZnONPs (E). Images F–J represent the biofilm of S. aureus formed on the glass surface; untreated S. aureus biofilm (A), S. aureus + 0.25 mg ml–1 ZnONPs (B), S. aureus + 0.5 mg ml–1 ZnONPs (C), S. aureus + 1 mg ml–1 ZnONPs (D), and S. aureus + 1.5 mg ml–1 ZnONPs (E). CLSM analysis of the bacterial biofilm: images K–O represent the biofilm formed by K. pneumoniae on the glass surface; untreated K. pneumoniae biofilm (A), K. pneumoniae + 0.25 mg ml–1 ZnONPs (B), K. pneumoniae + 0.5 mg ml–1 ZnONPs (C), K. pneumoniae + 1 mg ml–1 ZnONPs (D), and K. pneumoniae + 1.5 mg ml–1 ZnONPs (E). Images P–T represent the biofilm formed by S. aureus on the glass surface; untreated S. aureus biofilm (A), S. aureus + 0.25 mg ml–1 ZnONPs (B), S. aureus + 0.5 mg ml–1 ZnONPs (C), S. aureus + 1 mg ml–1 ZnONPs (D), and S. aureus + 1.5 mg ml–1 ZnONPs (E). Bar diagrams in panel II show the percent inhibition in the biofilm formation of K. pneumoniae (U) and S. aureus (V) under ZnONP stress (0.25–1.5 mg ml–1). The bars represent the mean values of three independent replicates ± SD (*p ≤ 0.05, **p ≤ 0.005 vs. control).
Fig. 7. Scanning electron microimage of the bacterial biofilm. Images A–D represent the biofilm formed on a solid surface; untreated K. pneumoniae biofilm (A), K. pneumoniae + 0.25 mg ml–1 ZnONPs (B), K. pneumoniae + 0.5 mg ml–1 ZnONPs (C), and K. pneumoniae + 1 mg ml–1 ZnONPs (D). A magnified view at ×15 000 is shown (Panel A and D insets). Micrographs E–H show the biofilm of untreated S. aureus (E), S. aureus + 0.25 mg ml–1 ZnONPs (F), S. aureus + 0.5 mg ml–1 ZnONPs (G), and S. aureus + 1 mg ml–1 ZnONPs (H). A magnified view at ×15 000 is shown in Panel E and H insets. Red arrows indicate the distortion of the normal bacterial cell.
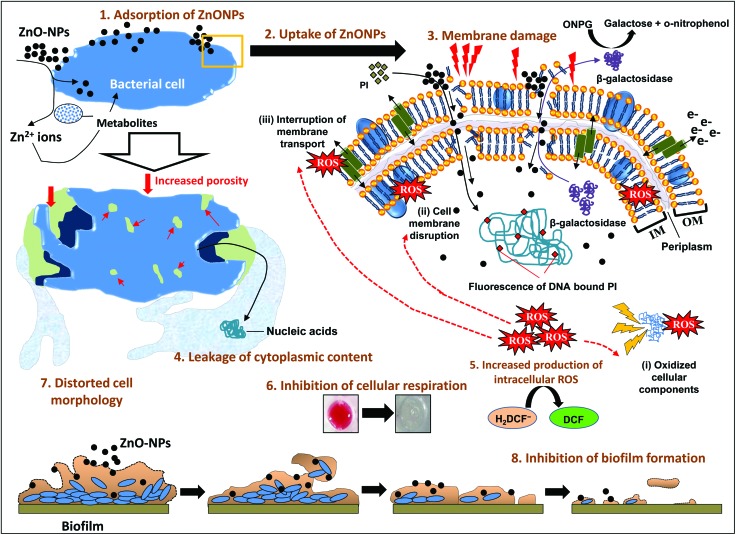
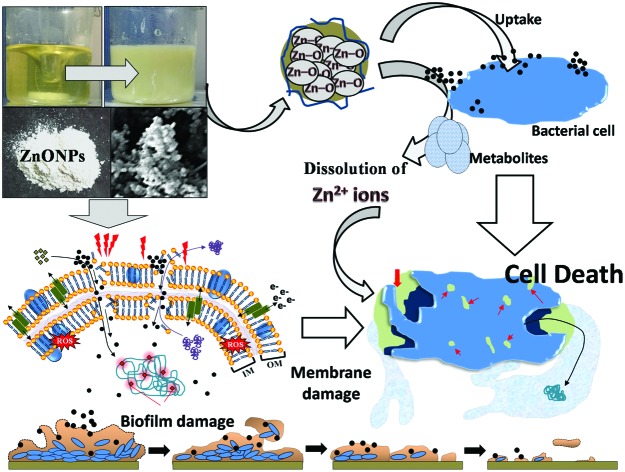
According to the results obtained in this study, a stepwise and systematic event of ZnONP action on- (i) the bacterial interface, (ii) cellular toxicity and (iii) biofilm inhibition is presented in Fig. 8. Broadly, the mechanism of bacterial cell suppression/inhibition due to the ZnONP action may involve the following steps- (i) adsorption of ZnONPs on the bacterial surface (wall and membranes) due to the surface potential, (ii) uptake of ZnONPs and their release into the periplasm and cytoplasm along with Zn2+ ions due to bacterium-assisted transformation of ZnONPs, (iii) membrane damage due to increased porosity, and structural and functional interruption, (iv) leakage of cytoplasmic and nuclear materials, (v) increased production of intracellular oxidative stress which further augments the magnitude of damage to cellular constituents and membranes, (vi) inhibition of cellular respiration, (vii) distortion of cell morphology, and (viii) eradication of biofilm formation due to one or the simultaneous impacts of the above stated actions. In general, the toxicity of NPs toward bacterial cells depends on- (a) bacterial and metal oxide species73 and (b) particle concentration, aggregation in growth media, and biotransformation by bacterial metabolites. The antibacterial performance of NPs lies in the composition of the outer membrane of bacteria which comprises nanoscale pores.73 The ZnONPs attach to the negatively charged cell membranes of the bacteria via electrostatic interactions.67,73 The surface adsorption and internalization triggers enhanced ROS formation. Besides NP penetration, the dissolution of metal ions is largely discussed to enhance the ROS response.74 Thus, the oxidative stress generated by the physical interaction of ZnONPs, their uptake, and Zn2+ ion dissolution lead to deformed cells and increased porosity of the cell membrane eventually leading to cell death. Moreover, the factors for metal oxide NP mediated oxidative stress include radical formation due to the activation of electron hole pairs by UV or visible light and reduction–oxidation cycles on the surface of transition metal based nanoparticles.74,75
Fig. 8. A proposed mechanistic illustration of the various steps of ZnONP action on the bacterial interface and initiation of the signaling cascade of bacterial cell death.
4. Conclusion
The results endorsed the development of rapid green bio-production of ZnONPs using the bio-reducing potential of a low cost precursor, A. indica leaf extract without using any extrinsic capping agent and surfactant. This study further unravels the interfacial mechanisms between ZnO-NPs and bacteria. The clear and distinct diffraction of X-rays confirmed the crystalline nature and purity of the wurtzite structure of hexagonal ZnONPs. The FTIR, SEM-EDX, and TEM validated the formation of ZnONPs and exhibited a spherical morphology. Agglomeration of ZnONPs in nutrient media detected by DLS and zeta-potential showed the hydrodynamic size in the nanoscale range with good stability. The role of functional groups present in A. indica leaf phytocompounds in ZnONP capping and stabilization was clearly evident. The data obtained for cell membrane activity and surface interaction through spectrophotometry, CLSM, SEM-EDX, elemental mapping and zinc uptake using AAS suggested the swift attachment and internalization of ZnONPs in both Gram –ve and Gram +ve genera in a dose dependent manner. The ZnONPs experienced enhanced bio-dissolution to Zn2+ ions in culture media when interacted with bacterial isolates. The dispersed ZnONPs along with soluble zinc in the nutrient medium caused severe bacterial damage. Differential growth kinetics of clinical isolates was observed under the influence of ZnONPs. The toxic impact of ZnONPs on K. pneumoniae, E. coli, and P. aeruginosa (G –ve bacteria) was largest compared to S. aureus (G +ve bacterium) which could possibly be due to the variation in the interfacial potential of two divergent groups of bacteria. This study also revealed a mechanism of strong antibiofilm and antibacterial activity of green ZnONPs, which makes them interesting and useful in treating specific bacterial infections and hence, can serve as a future nano-antibiotics. Overall, all experiments conducted in this study certainly add an important dimension to better understand the NPs–bacteria interaction and pave the way to explore ZnONPs in infection treatment.
Author contributions
J.M., M.S.K., and B.A. conceived and designed the research plan. B.S. and B.A. performed the experiments. B.A., M.S.K., and AZ prepared this manuscript and formatted it according to the style of the journal. B.S. and M.S.K. also contributed to data analysis. M.S.K., AZ and J.M. critically edited this manuscript. All authors have read and approved the final version of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Material
Acknowledgments
The authors appreciate the Council of Science and Technology, U. P., India for funding this research work through UPCST Research Project #372. One of the authors J. M. extends thanks to the International Scientific Partnership Program (ISPP) at King Saud University through ISPP# 0031. The instrumentation provided by the University Sophisticated Instruments Facility (USIF), Aligarh Muslim University, India for respective analysis is greatly acknowledged.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8tx00267c
References
- Tachikawa S., Noguchi A., Tsuge T., Hara M., Odawara O., Wada H. Materials. 2011;4:1132–1143. doi: 10.3390/ma4061132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu A. N. Int. J. Res. Ayurveda Pharm. 2013;4:472–474. [Google Scholar]
- Ahmed B., Shahid M., Khan M. S., Musarrat J. Metallomics. 2018;10:1315–1327. doi: 10.1039/c8mt00093j. [DOI] [PubMed] [Google Scholar]
- Ahmed B., Khan M. S., Saquib Q., Al-Shaeri M. and Musarrat J., Interplay between Engineered Nanomaterials (ENMs) and Edible Plants: A Current Perspective, in Phytotoxicity of Nanoparticles, ed. M. Faisal, et al., Springer Nature, 2018, pp. 63–102. [Google Scholar]
- Logeswari P., Silambarasan S., Abraham J. Sci. Iran. 2013;20:1049–1054. [Google Scholar]
- Papavlassopoulos H., Mishra Y. K., Kaps S., Paulowicz I., Abdelaziz R., Elbahri M., Maser E., Adelung R., Röhl C. PLoS One. doi: 10.1371/journal.pone.0084983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahab R., Khan F., Mishra Y. K., Musarrat J., Al-Khedhairy A. A. RSC Adv. 2016;6:32328–32339. [Google Scholar]
- Gnanasangeetha D., Saralathambavani D. J. Chem., Biol. Phys. Sci. 2014;4:238–246. [Google Scholar]
- Narayanan P. M., Wilson W. S., Abraham A. T., Sevanan M. Bionanoscience. 2012;4:329–335. [Google Scholar]
- Brolund A., Sandegren L. Infect. Dis. 2016;48:18–25. doi: 10.3109/23744235.2015.1062536. [DOI] [PubMed] [Google Scholar]
- Maheshwari M., Yaser N. H., Naz S., Fatima M., Ahmad I. J. Glob. Antimicrob. Resist. 2016;5:22–25. doi: 10.1016/j.jgar.2016.01.008. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO), Antimicrobial Resistance: Global report on surveillance, Geneva, 2014.
- Zhang Y., Yang D., Kong Y., Wang X., Pandoli O., Gao G. Nano Biomed. Eng. 2010;2:252–257. [Google Scholar]
- Ali K., Ahmed B., Khan M. S., Musarrat J. J. Photochem. Photobiol., B. 2018;188:146–158. doi: 10.1016/j.jphotobiol.2018.09.017. [DOI] [PubMed] [Google Scholar]
- Mallika Pathak G. J., Sharma M., Ojha H., Kumari R., Sharma N., Roy B. Green Chem. Technol. Lett. 2016;2:103–109. [Google Scholar]
- Mishra Y. K., Adelung R. Mater. Today. 2018;21:631–651. [Google Scholar]
- Antoine T. E., Hadigal S. R., Yakoub A. M., Mishra Y. K., Bhattacharya P., Haddad C., Valyi-Nagy T., Adelung R., Prabhakar B. S., Shukla D. J. Immunol. 2016;196:4566–4575. doi: 10.4049/jimmunol.1502373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahab R., Khan F., Yang Y. B., Hwang I. H., Shin H. S., Ahmad J., Dwivedi S., Khan S. T., Siddiqui M. A., Saquib Q., Musarrat J., Al-Khedhairy A. A., Mishra Y. K., Ali B. A. RSC Adv. 2016;6:26111–26120. [Google Scholar]
- Antoine T. E., Mishra Y. K., Trigilio J., Tiwari V., Adelung R., Shukla D. Antiviral Res. 2012;96:363–375. doi: 10.1016/j.antiviral.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bala N., Saha S., Chakraborty M., Maiti M., Das S., Basu R., Nandy P. RSC Adv. 2015;5:4993–5003. [Google Scholar]
- Alzohairy M. A. Altern. Med. 2016:7382506. doi: 10.1155/2016/7382506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A., Mehata M. S. J. Radiat. Res. Appl. Sci. 2016;9:109–115. [Google Scholar]
- Bindhani B. K., Panigrahi A. K. Int. J. Adv. Biotechnol. Res. 2014;5:457–464. [Google Scholar]
- Elumalai K., Velmurugan S. Appl. Surf. Sci. 2015;345:329–336. [Google Scholar]
- Bhuyan T., Mishra K., Khanuja M., Prasad R., Varma A. Mater. Sci. Semicond. Process. 2015;32:55–61. [Google Scholar]
- Saleem S., Ahmed B., Khan M. S., Al-Shaeri M., Musarrat J. Microb. Pathog. 2017;111:375–387. doi: 10.1016/j.micpath.2017.09.019. [DOI] [PubMed] [Google Scholar]
- Ahmed B., Hashmi A., Khan M. S., Musarrat J. Adv. Powder Technol. 2018;29:1601–1616. [Google Scholar]
- Shahid M., Ahmed B., Khan M. S. Biocatal. Agric. Biotechnol. 2018;14:96–108. [Google Scholar]
- Wahab R., Khan S. T., Dwivedi S., Ahamed M., Musarrat J., Al-Khedhairy A. A. Colloids Surf., B. 2013;111:211–217. doi: 10.1016/j.colsurfb.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Ahmed B., Khan M. S., Musarrat J. Environ. Pollut. 2018;240:802–816. doi: 10.1016/j.envpol.2018.05.015. [DOI] [PubMed] [Google Scholar]
- Nath D., Banerjee P. Environ. Toxicol. Pharmacol. 2013;36:997–1014. doi: 10.1016/j.etap.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Namratha N., Monica P. V. Asian J. Pharm. Tech. 2013;3:170–174. [Google Scholar]
- Lalitha A., Subbaiya R., Ponmurugan P. Int. J. Curr. Microbiol. Appl. Sci. 2013;2:228–235. [Google Scholar]
- Nagarajan S., Arumugam Kuppusamy K. J. Nanobiotechnol. 2013;11:39. doi: 10.1186/1477-3155-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed B., Dwivedi S., Abdin M. Z., Azam A., Al-Shaeri M., Khan M. S., Saquib Q., Al-Khedhairy A. A., Musarrat J. Sci. Rep. 2017;7:40685. doi: 10.1038/srep40685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangeetha G., Rajeshwari S., Venckatesh R. Mater. Res. Bull. 2011;46:2560–2566. [Google Scholar]
- Awwad A. M., Albiss B., Ahmad A. L. Adv. Mater. Lett. 2014;5:520–524. [Google Scholar]
- Raphael E. Global Adv. Res. J. Environ. Sci. Toxicol. 2012;1:14–17. [Google Scholar]
- Silva R. F., Zaniquelli M. E. D. Colloids Surf., A. 2002:551–558. [Google Scholar]
- Kwon Y. J., Kim K. H., Lim C. S., Shim K. B. J. Ceram. Process. Res. 2002;3:146–149. [Google Scholar]
- Li H., Wang J., Liu H., Yang C., Xu H., Li X., Cui H. Vacuum. 2004;77:57–62. [Google Scholar]
- Sharma V., Anderson D., Dhawan A. Apoptosis. 2012;17:852–870. doi: 10.1007/s10495-012-0705-6. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S. J. Controlled Release. 2016;235:337–351. doi: 10.1016/j.jconrel.2016.06.017. [DOI] [PubMed] [Google Scholar]
- Sirelkhatim A., Mahmud S., Seeni A., Kaus N. H. M., Ann L. C., Bakhori S. K. M., Hasan H., Mohamad D. Nano-Micro Lett. 2015;7:219–242. doi: 10.1007/s40820-015-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgile C., Hauk P., Wu H. C., Shang W., Tsao C. Y., Payne G. F., Bentley W. E. PLoS One. 2018;13 doi: 10.1371/journal.pone.0196999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand R. M., Walker C., Epand R. F., Magarvey N. A. Biochim. Biophys. Acta, Biomembr. 2016;1858:980–987. doi: 10.1016/j.bbamem.2015.10.018. [DOI] [PubMed] [Google Scholar]
- Kleanthous C., Armitage J. P. Philos. Trans. R. Soc., B. 2015;370:1769. doi: 10.1098/rstb.2015.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E., Yousef A. E. Appl. Environ. Microbiol. 2014;80:2700–2704. doi: 10.1128/AEM.03775-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padalia U. Int. J. Eng. Sci. Comput. 2016;6:2674–2676. [Google Scholar]
- Singh M. IET Nanobiotechnol. 2016;10:276–280. doi: 10.1049/iet-nbt.2015.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almaaytah A., Tarazi S., Alsheyab F., Al-Balas Q., Mukattash T. Int. J. Pept. Res. Ther. 2014;20:397–408. [Google Scholar]
- Saravanan M., Gopinath V., Chaurasia M. K., Syed A., Ameen F., Purushothaman N. Microb. Pathog. 2018;115:57–63. doi: 10.1016/j.micpath.2017.12.039. [DOI] [PubMed] [Google Scholar]
- Divyapriya S., Sowmia C., Sasikala S. World J. Pharm. Pharm. Sci. 2014;3:1635–1645. [Google Scholar]
- Jain S., Pillai J. Int. J. Nanomed. 2017;12:6329–6341. doi: 10.2147/IJN.S137368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi S., Saquib Q., Ahmad B., Ansari S. M., Azam A., Musarrat J. Adv. Exp. Med. Biol. 2018;1048:143–161. doi: 10.1007/978-3-319-72041-8_9. [DOI] [PubMed] [Google Scholar]
- Bankier C., Cheong Y., Mahalingam S., Edirisinghe M., Ren G., Cloutman-Green E., Ciric L. PLoS One. 2018;13 doi: 10.1371/journal.pone.0192093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyasi S., Majhi R. K., Kumar S., Mishra M., Ghosh A., Suar M., Satyam P. V., Mohapatra H., Goswami C., Goswami L. Sci. Rep. 2016;6 doi: 10.1038/srep24929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P., Huang B., Tsuji Y. Cell. Signalling. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Roper M. G. Anal. Methods. 2014;6:3019–3024. doi: 10.1039/C4AY00288A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Drlica K. Curr. Opin. Microbiol. 2014;21:1–6. doi: 10.1016/j.mib.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H. L., Chou C. C., Hung D. J., Lin S. H., Pao I. C., Lin J. H., Huang F. L., Dong R. X., Lin J. J. Biomaterials. 2009;30:5979–5987. doi: 10.1016/j.biomaterials.2009.07.030. [DOI] [PubMed] [Google Scholar]
- Wang D., Zhao L. X., Ma H. Y., Zhang H., Guo L.-H. H. Environ. Sci. Technol. 2017;51:10137–10145. doi: 10.1021/acs.est.7b00473. [DOI] [PubMed] [Google Scholar]
- Choi O., Deng K. K., Kim N.-J., Ross L., Surampalli R. Y., Hu Z., Ross Louis J., Surampalli R. Y., Hu Z. Water Res. 2008;42:3066–3074. doi: 10.1016/j.watres.2008.02.021. [DOI] [PubMed] [Google Scholar]
- Pati R., Mehta R. K., Mohanty S., Padhi A., Sengupta M., Vaseeharan B., Goswami C., Sonawane A. Nanomedicine. 2014;10:1195–1208. doi: 10.1016/j.nano.2014.02.012. [DOI] [PubMed] [Google Scholar]
- Khan S. T., Ahmad J., Ahamed M., Musarrat J., Al-Khedhairy A. A. J. Biol. Inorg. Chem. 2016;21:295–303. doi: 10.1007/s00775-016-1339-x. [DOI] [PubMed] [Google Scholar]
- Manzoor U., Siddique S., Ahmed R., Noreen Z., Bokhari H., Ahmad I. PLoS One. 2016;11 doi: 10.1371/journal.pone.0154704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi S., Wahab R., Khan F., Mishra Y. K., Musarrat J., Al-Khedhairy A. A. PLoS One. 2014;9 doi: 10.1371/journal.pone.0111289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawari A. H., Mulligan C. N. Process Biochem. 2007;42:1546–1552. [Google Scholar]
- Zhang L., Jiang Y., Ding Y., Povey M., York D. J. Nanopart. Res. 2007;9:479–489. [Google Scholar]
- Ren G., Hu D., Cheng E. W. C., Reus M. A. V., Reap P., Allaker R. P. Int. J. Antimicrob. Agents. 2009;33:587–590. doi: 10.1016/j.ijantimicag.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Mathur P., Singh S. J. Lab. Physicians. 2013;5:5. doi: 10.4103/0974-2727.115898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H. J. Antimicrob. Chemother. 2001;48:487–491. doi: 10.1093/jac/48.4.487. [DOI] [PubMed] [Google Scholar]
- Oves M., Khan M. S., Zaidi A. Saudi J. Biol. Sci. 2013;20:121–129. doi: 10.1016/j.sjbs.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvakkumar R., Suresh J., Nathanael A. J., Sundrarajan M., Hong S. I. Mater. Lett. 2014;128:170–174. doi: 10.1016/j.msec.2014.04.025. [DOI] [PubMed] [Google Scholar]
- Slavin Y. N., Asnis J., Häfeli U. O., Bach H. J. Nanobiotechnol. 2017;15:65. doi: 10.1186/s12951-017-0308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.