Abstract
Background:
A germline deletion in BIM (B cell lymphoma-2-like 11) gene has been shown to impair the apoptotic response to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in vitro but its impact on response to EGFR-TKIs in patients of nonsmall cell lung cancer (NSCLC) remains controversial.
Methods:
Eligible literature were searched and screened. Objective response rate (ORR) and disease control rate (DCR) were extracted and aggregated with odds ratio (OR). Hazard ratio (HR) and 95% confidence interval (CI) for progression-free survival (PFS) and overall survival (OS) were extracted and aggregated based on random-effect model.
Results:
Fourteen studies including 2694 NSCLC patients were eligible. Individuals harboring BIM deletion polymorphism had inferior ORR (OR = 0.49, 95% CI: 0.34–0.70, P < .001), inferior DCR (OR = 0.50, 95% CI: 0.30–0.84, P = .009). Patients with BIM deletion had shorter OS despite of the heterogeneity between countries (in subgroup of South Korea and Taiwan, HR = 1.34, 95% CI: 1.18–1.53, P < .001; in subgroup of other countries, HR = 2.43, 95% CI: 2.03–2.91, P < .001). The pooled analysis of PFS showed great heterogeneity (I2 = 79%). All the reported characteristics did not account for the heterogeneity. However, 2 subgroups could be obtained through sensitivity analysis. In one subgroup, patients with BIM deletion polymorphism had shorter PFS (HR = 2.03, 95% CI: 1.71–2.40, P < .001), while in the other subgroup, no significant difference was observed (HR = 0.92, 95% CI: 0.79–1.06, P = .25).
Conclusion:
NSCLC patients with BIM deletion polymorphism show poor ORR, DCR, and OS after EGFR-TKIs treatment. BIM deletion polymorphism indicates poor response to EGFR-TKIs, and it could be used as a predictor to identify those who would benefit from EGFR-TKIs in NSCLC patients.
Keywords: BIM deletion polymorphism, nonsmall cell lung cancer, response, tyrosine kinase inhibitor
1. Introduction
Nonsmall cell lung cancer (NSCLC) is a kinase-driven cancer, in which, epidermal growth factor receptor (EGFR), a common kind of tyrosine kinase, can bind to extracellular ligands and transfer a phosphate group from ATP to the tyrosine residues of target proteins to regulate survival of cancer cells.[1] EGFR-tyrosine kinase inhibitors (TKIs) can compete with ATP to bind to the intracellular catalytic domain of tyrosine kinase and consequently inhibit the process of cross-phosphorylation.[2] EGFR-TKIs, such as gefitinib, erlotinib, and afatinib, are widely used for treatment of EGFR-mutant NSCLC.[3] Despite high response rates with first-line EGFR-TKIs, a considerable portion of EGFR-mutant NSCLC patients develop acquired resistance after 9 to 18 months of treatment.[4,5] Approximately 30% of EGFR-mutant NSCLC patients displayed intrinsic resistance to EGFR-TKIs.[5,6]
Pro-apoptotic protein BIM (also known as B cell lymphoma-2-like 11) is a BH3-only protein of the BCL-2 family. The BH3 domain can bind and regulate the antiapoptotic bcl-2 proteins (bax, bak) to promote apoptosis.[7] Activation of BIM is essential for apoptosis triggered by EGFR-TKIs in EGFR-mutant NSCLC.[8–10] A 2903-bp germline deletion polymorphism in intron 2 of BIM gene was found in chronic myeloid leukemia (CML) and EGFR-mutant NSCLC in 2012.[11] BIM deletion polymorphism results in the generation of alternatively spliced isoforms of BIM that lack the crucial BH3 domain, thus impairs the apoptotic response to TKIs and confers NSCLC cells intrinsically resistant to TKIs in vitro.[11] Accumulating articles about the impact of BIM deletion polymorphism on response to EGFR-TKIs in NSCLC patients have been published. However, the results were contradictory. Some studies showed that NSCLC patients harboring BIM deletion polymorphism had inferior response to EGFR-TKIs than those with BIM wild after TKIs treatment,[11–20] while others argued that there was no difference in response to EGFR-TKIs in NSCLC patients with and without BIM deletion polymorphism.[21–24]
In order to obtain an objective and consistent conclusion, we conducted this comprehensive analysis to demonstrate the impact of BIM deletion on response to EGFR-TKIs in NSCLC patients.
2. Materials and methods
2.1. Ethic statement
This meta-analysis was performed based on previously published studies, ethical approval was not necessary.
2.2. Literature search
A comprehensive literature search was conducted in the database PubMed and Embase using the subject headings and text words of the following terms: BIM (“BCL2L11,” “B-cell lymphoma-like 11,” “BCL-2-like 11”), TKI (“Tyrosine kinase inhibitor,” “gefitinib,” “erlotinib,” “afatinib”), and NSCLC (“Non-Small Cell Lung Cancer,” “Nonsmall Cell Lung Cancer,” “non-small cell lung cancer,” “Lung Adenocarcinoma”) dating up to October 1, 2018. Manual retrieval was performed to obtain relevant studies by reviewing all the reference in the eligible studies. This study was approved by the Institution Ethics Commission of Weifang Medical University.
2.3. Eligibility criteria
Eligible literatures were identified in accordance with the following inclusion criteria: (1) Prospective or retrospective studies concerning the impact of BIM deletion polymorphism on response to EGFR-TKI therapy. (2) Studies in NSCLC patients with or without EGFR mutations. (3) Response data in studies were available. (4) Published full texts were available. However, review articles, systematic reviews, meta-analyses, and case studies were excluded.
2.4. Data extraction
Objective response rate (ORR), disease control rate (DCR), and overall survival (OS) were the primary outcomes and progression-free survival (PFS) was the second outcome for this meta-analysis. For ORR, the cases of events were extracted by response rate multiplied by number of patients with BIM deletion polymorphism or with BIM wild. For DCR, the cases of events were extracted by DCR multiplied by number of patients with BIM deletion polymorphism or with BIM wild. For OS and PFS, hazard ratios (HRs) and 95% confidence interval (CI) of patients with BIM polymorphism compared to those with BIM wild in EGFR-TKI-treated NSCLC were extracted. If HR and 95% CI for PFS or OS were not given in the study, the data were extracted from Kaplan–Meier curve by the method of Tierney.[25] Data from all eligible studies were extracted independently by 2 investigators. Any disagreement was discussed with the third investigator to reach a consensus.
2.5. Statistical analysis
Statistical analysis was performed using Revman 5.3 (The Nordic Cochrane Center, the Cochrane collaboration, Copenhagen, Denmark) and STATA 11.0 software (Stata Corporation, College Station, TX) and pooled odds ratios (ORs) for ORR and DCR were calculated by Mantel–Haenszel method with fixed-effect model, and pooled HRs for PFS and OS with 95% CIs were calculated by inverse variance method with random-effect model. Statistical significance was set at a 2-sided P < .05. A forest plot was applied for display of results.
For heterogeneity evaluation, chi-squared tests and I2 inconsistency statistics were used. A significant heterogeneity was considered when PH < .10. I2 values of 0% to 24.9%, 25% to 49.9%, 50% to 74%, and 75% to 100% were considered as none, low, moderate, and high heterogeneity, respectively.[26,27] Sensitivity analysis was performed to estimate stability of overall effect by omitting each eligible study or changing combination model. Publication bias was evaluated by Begg funnel plot and asymmetry of funnel plot was considered as an existence of publication bias.[28]
2.6. Meta-regression
Meta-regression was performed to assess the effect of the following given characteristics in the original articles on lnHR for PFS: gender (female vs. male), smoking history (without vs. with), Eastern Cooperative Oncology Group (ECOG) performance status (0–1 vs. 2–4), pathology (adenocarcinoma vs. nonadenocarcinoma), clinical stage (III B vs. IV, relapse), EGFR mutation (exon19 vs. L858R), TKI type (gefitinib vs. erlotinib, afatinib), and line of TKI treatment (first vs. second or more).
3. Results
3.1. Eligible studies
Three hundred forty-five records were identified using the search strategy, and 85 duplicated articles, 216 unrelated articles, 11 conference abstracts of the same original articles, 7 articles concerning BIM mRNA expression levels, 6 conference abstracts with data unavailable, 2 articles concerning the effect of other therapy, 1 article concerning lung cancer susceptibility, 1 in vitro study, 1 meta-analysis, and 1 report were excluded. Finally, 14 studies were eligible. ORR and DCR were available in 8 and 7 studies, respectively. PFS was available in 14 studies, and OS was available in 6 studies. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) study flow diagram was shown in Fig. 1.
Figure 1.
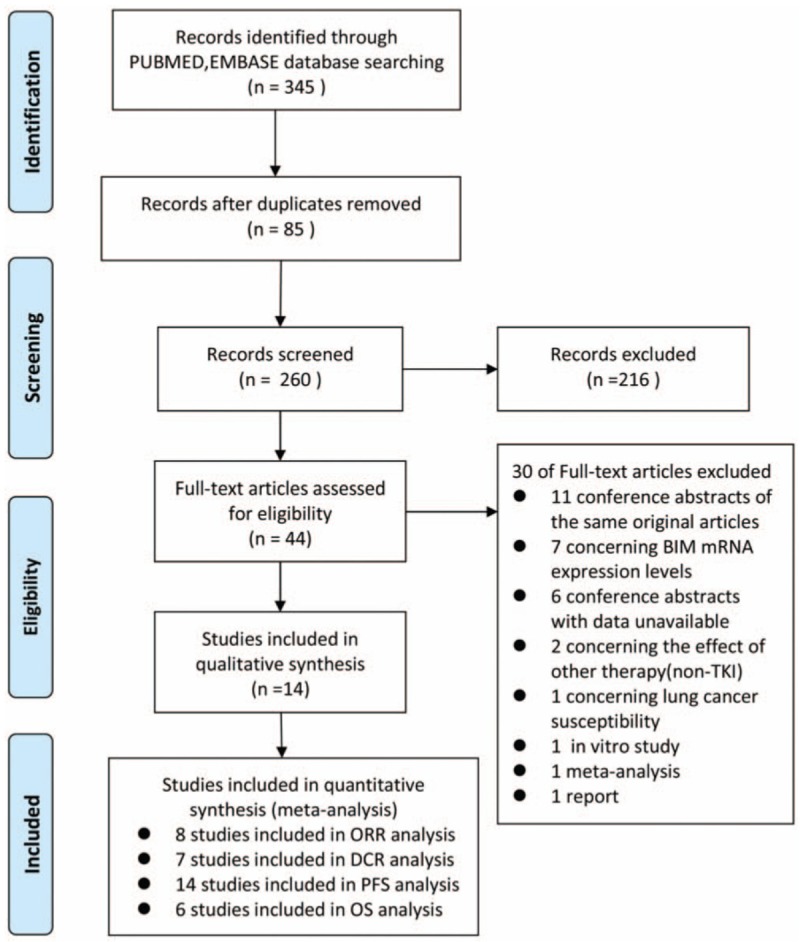
Flow diagram of eligible study selection.
3.2. Study characteristics
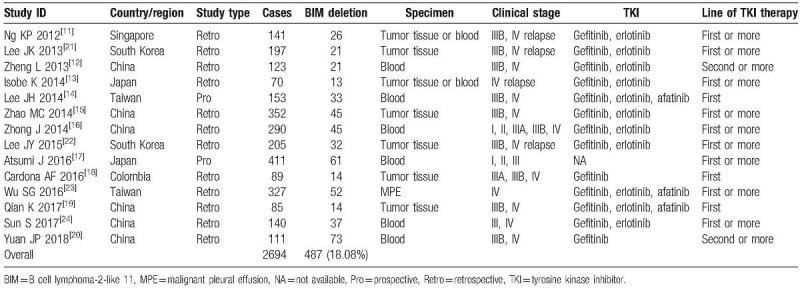
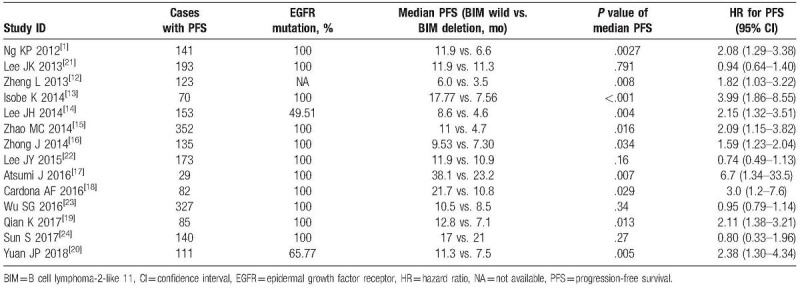
A total of 14 articles including 2 prospective studies and 12 retrospective studies with 2694 patients were included in this meta-analysis. Among them, 13 studies were in Asian populations and 1 study was in Latin American population. Gefitinib, erlotinib, and afatinib were investigated in 13, 11, and 2 studies, respectively. TKIs were used as first-line therapy in 3 studies, first-line or more-line therapy in 11 studies (Table 1).
Table 1.
Characteristics of studies included for meta-analysis.
3.3. ORR analysis
Eight studies including 1012 patients were pooled for ORR analysis. PH = .16, I2 = 33%, indicating low heterogeneity of these studies. The pooled estimate of the ORs in NSCLC patients harboring BIM deletion, compared with those harboring BIM wild was 0.49, 95% CI: 0.34 to 0.70, P < .001 (Fig. 2A). Sensitivity analysis ensured the consistent result and Begg test showed that there was no publication bias (Fig. 2C).
Figure 2.
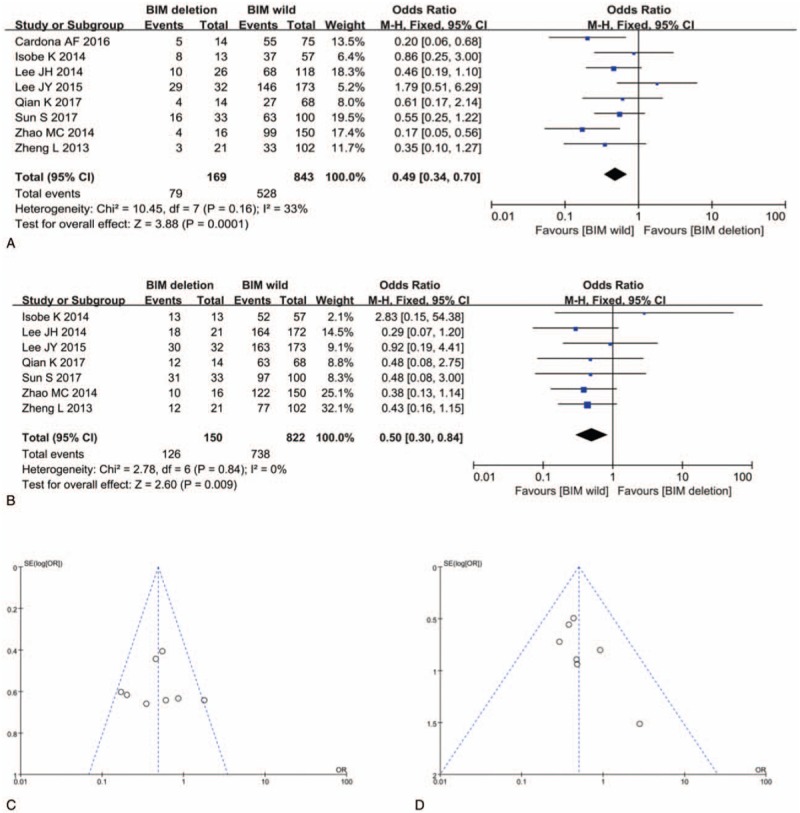
Impact of BIM deletion polymorphism on response to EGFR-TKIs. (A) Odds ratio (OR) for objective response rate (ORR) to EGFR-TKIs in NSCLC patients with BIM deletion polymorphism versus those with BIM wild. (B) OR for disease control rate (DCR) to EGFR-TKI in NSCLC patients with BIM deletion polymorphism versus those with BIM wild. (C) Funnel plot of ORR analysis. (D) Funnel plot of DCR analysis. BIM = B cell lymphoma-2-like 11, EGFR-TKIs = epidermal growth factor receptor-tyrosine kinase inhibitors, NSCLC = nonsmall cell lung cancer.
3.4. DCR analysis
Seven studies including 972 patients were pooled for DCR analysis. PH = 2.78, I2 = 0%, indicating low heterogeneity of these studies. NSCLC patients harboring BIM deletion polymorphism showed worse DCR (OR, 0.50; 95% CI: 0.30–0.84; P = .009) than those harboring BIM wild (Fig. 2B). Sensitivity analysis ensured the consistent result and Begg test showed that there was no publication bias (Fig. 2D).
3.5. OS analysis
Six studies including 655 patients were pooled for OS analysis. PH < .001, I2 = 82%, indicating high heterogeneity of these studies. Meta-regression revealed that the exp(b) for South Korea and Taiwan versus other countries was 1.80, 95% CI: 1.31 to 2.47, P = .007 (Fig. 3B); therefore, countries may be the source of heterogeneity. In the subgroup of South Korea and Taiwan, PH = .92, I2 = 0%, in the subgroup of other countries, PH = 0.74, I2 = 0%, suggesting the consistency of the studies in the 2 subgroups. In both subgroups, NSCLC patients with TKI therapy who harbored BIM deletion polymorphism had statistically significant shorter OS than those with BIM wild (in subgroup of South Korea and Taiwan, HR = 1.34, 95% CI: 1.18–1.53, P < .001; in subgroup of other countries, HR = 2.43, 95% CI: 2.03–2.91, P < .001) (Fig. 3A). Sensitivity analysis ensured the consistent result and Begg test showed that there was no publication bias (Fig. 3C).
Figure 3.
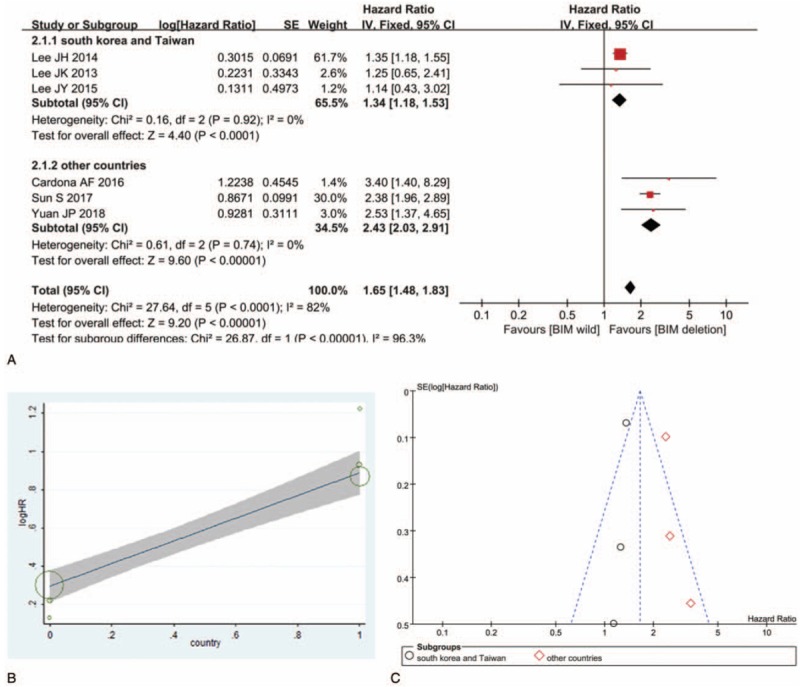
Impact of BIM deletion polymorphism on overall survival (OS) to EGFR-TKI. (A) Hazard ratio (HR) for overall survival (OS) to EGFR-TKI in NSCLC patients with BIM deletion polymorphism versus those with BIM wild. (B) Effect of country (South Korea and Taiwan vs. other countries) on heterogeneity across studies. (C) Funnel plot of OS analysis. BIM = B cell lymphoma-2-like 11, EGFR-TKIs = epidermal growth factor receptor-tyrosine kinase inhibitors, NSCLC = nonsmall cell lung cancer.
3.6. PFS analysis
Fourteen studies including 2114 patients were pooled for PFS analysis (Table 2). PH < .001, I2 = 79%, indicating high heterogeneity of these studies. To find out the sources of heterogeneity, meta-regression was performed using STATA 11.0 software. The exp(b) for male versus female was 0.88, 95% CI: 0.48 to 1.62, P = .65. The exp(b) for no smoking history versus smoking history was 0.79, 95% CI: 0.56 to 1.04, P = .09. The exp(b) for ECOG 0 to 1 versus ECOG 2 to 4 was 1.02, 95% CI: 0.99 to 1.04, P = .13. The exp(b) for adenocarcinoma versus nonadenocarcinoma was 0.11, 95% CI: 0.004 to 2.76, P = .16. The exp(b) for clinical stage III versus clinical stage IV and relapse was 1.76, 95% CI: 0.30 to 10.40, P = .50. The exp(b) for exon19 versus L858R was 1.36, 95% CI: 0.62 to 3.05, P = .41. The exp(b) for gefitinib versus erlotinib and afatinib was 1.01, 95% CI: 0.96 to 1.07, P = .49. The exp(b) for first-line therapy versus second-line or more line therapy was 0.93, 95% CI: 0.46 to 0.90, P = .82. Therefore, the reported clinical characteristics (i.e., sex, smoking history, ECOG, pathology, clinical stage, EGFR mutation, TKI type, and line of TKI treatment) in the original articles did not account for the heterogeneity.
Table 2.
Characteristics of studies included for PFS analysis.
Although the overall HR of the 14 studies is 1.66, 95% CI: 1.27 to 2.17, P < .001, this cannot be interpreted because the high heterogeneity. However, 2 subgroups could be obtained through sensitivity analysis. In subgroup A, PH = .36, I2 = 9%, NSCLC patients with TKI therapy who harbored BIM deletion polymorphism had statistically significant shorter PFS than those with BIM wild (HR = 2.03, 95% CI: 1.71–2.40, P < .001). In subgroup B, PH = .740, I2 = 0%, NSCLC patients with BIM deletion and with BIM wild had similar PFS (HR = 0.92, 95% CI: 0.79–1.07, P = .26) (Fig. 4). Begg test showed that there was publication bias (Fig. 5).
Figure 4.
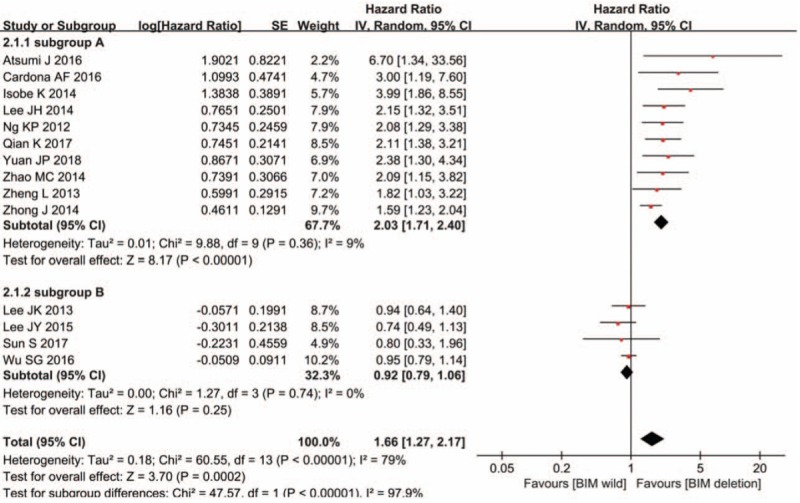
Impact of BIM deletion polymorphism on progression-free survival (PFS) to EGFR-TKIs. Hazard ratio (HR) for PFS to EGFR-TKIs in NSCLC patients with BIM deletion polymorphism versus those with BIM wild. BIM = B cell lymphoma-2-like 11, EGFR-TKIs = epidermal growth factor receptor-tyrosine kinase inhibitors, NSCLC = nonsmall cell lung cancer.
Figure 5.
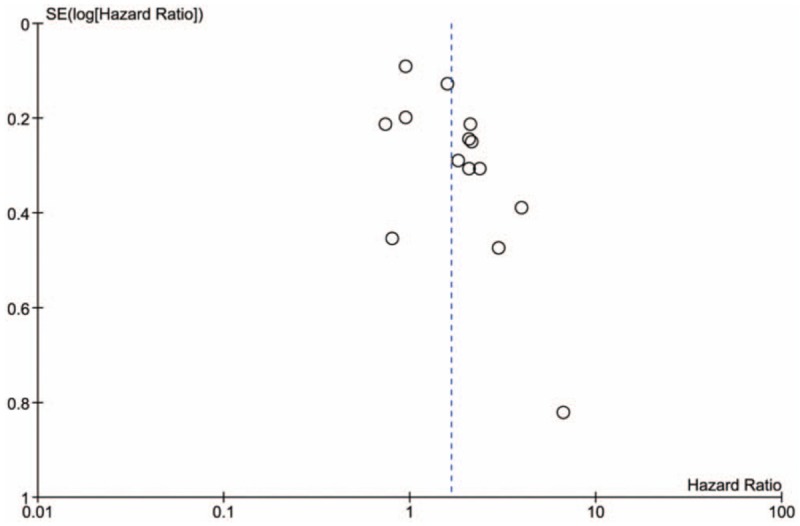
Funnel plot of progression-free survival analysis.
4. Discussion
Meta-analyses of the correlation of BIM deletion polymorphism and response to EGFR-TKIs in NSCLC patients have been conducted before the year 2016,[29–33] which were performed based on small number of studies and high heterogeneity. Therefore, the conclusions made by these meta-analyses should be interpreted cautiously. Since more original studies in this area have been published in recent 3 years,[17–20,23,24] we conducted this updated meta-analysis to obtain an objective and consistent conclusion. To the best of our knowledge, this updated meta-analysis collected the comprehensive literature and was more accurate as the heterogeneity in the analysis was low.
In 2012, using paired-end DNA sequencing, Ng et al[11] discovered a 2903-bp germline deletion polymorphism in intron 2 of BIM gene in East Asian populations. The polymorphism resulted in expression of BIM isoforms lacking the BH3 domain and lead to intrinsic TKI resistance in CML and EGFR-mutant NSCLC cell lines. In retrospective study in East Asian subjects from Singapore, Malaysia, and Japan, they found CML patients with BIM deletion polymorphism showed inferior DCR compared with controls after imatinib treatment and EGFR-mutant NSCLC patients with BIM deletion polymorphism showed shorter PFS compared with controls after gefitinib or erlotinib treatment. However, there was no influence of this polymorphism on response to imatinib in Chinese patients with CML.[34] Since BIM deletion polymorphism was found only in individuals of East Asian decent, the studies on the impact of BIM deletion polymorphism on the response of EGFR-TKIs in NSCLC were performed mainly in China, Japan, Korea, and South Korea. The results of these studies were contradictory.
By analysis of these studies, we found that NSCLC patients with BIM deletion polymorphism showed inferior ORR, DCR, and shorter OS than those without the polymorphism, which strongly suggested that BIM deletion polymorphism influenced the response to EGFR-TKIs and contributed to the resistance to EGFR-TKI in NSCLC patients. The EGFR-TKI-resistance due to BIM deletion can be circumvented by BH3 mimetics (ABT-737)[11] or histone deacetylase (HDAC) inhibitor (vorinostat).[35,36] Combined therapy of vorinostat and gefitinib to treat BIM deletion-associated resistance in EGFR-mutant NSCLC is under clinical trial in Japan.[37] If successful, EGFR-mutant NSCLC patients with BIM deletion polymorphism will benefit from the combined therapy.
Although this meta-analysis was performed with comprehensive literature and lower heterogeneity, the limitations cannot be neglected. First, PFS was taken as an important outcome in all the 14 original studies; it should be taken as the primary outcome in our analysis. However, the heterogeneity is high across studies, so it was taken as secondary outcome. Second, among the 14 studies, 10 studies were in favor of the correlation of BIM deletion polymorphism and poor response to EGFR-TKIs, while other 4 studies held that BIM deletion polymorphism had on influence on EGFR-TKIs response. Begg test showed that there was publication bias. Third, it was reported that smoking status and tumor histology are independent risk factors for the prediction of PFS to EGFR-TKIs therapy,[38,39] so we performed meta-regression with clinical characteristics to find out the source of heterogeneity across studies. However, the reported characteristics in original studies including sex, smoking history, ECOG performance status, pathology, clinical stage, EGFR mutation type, TKI type, line of TKI treatment, did not account for the heterogeneity. Song et al[40] reported that PFS of EGFR-TKI therapy in EGFR-mutant NSCLC could be stratified by the proposed 12-CT-phenotypic-feature-based signature, which may be the source of heterogeneity across studies.
5. Conclusions
In summary, this meta-analysis revealed that NSCLC patients with BIM deletion polymorphism showed poor response to EGFR-TKIs. BIM deletion polymorphism might be genetic cause of resistance to EGFR-TKIs therapy, and it could be used as a biomarker to predict the response to EGFR-TKIs in NSCLC patients, NSCLC patients without BIM deletion polymorphism will benefit from EGFR-TKIs therapy.
Author contributions
Conceptualization: Wenxia Su, Fengbin Wang.
Data curation: Xiaoyun Zhang, Xin Cai.
Formal analysis: Wenxia Su.
Funding acquisition: Wenxia Su, Meiyu Peng.
Investigation: Wenxia Su.
Methodology: Xiaoyun Zhang, Xin Cai, Fengbin Wang.
Software: Xiaoyun Zhang, Xin Cai, Meiyu Peng.
Validation: Yuliang Wang.
Writing – original draft: Wenxia Su.
Writing – review & editing: Fengbin Wang, Yuliang Wang.
Footnotes
Abbreviations: BIM = B cell lymphoma-2-like 11, CI = confidence interval, CML = chronic myeloid leukemia, ECOG = Eastern Cooperative Oncology Group, DCR = disease control rate, EGFR = epidermal growth factor receptor, HR = hazard ratio, NSCLC = nonsmall cell lung cancer, OR = odds ratio, ORR = objective response rate, OS = overall survival, PFS = progression-free survival, TKIs = tyrosine kinase inhibitors.
This work was supported by the Weifang Technology Development Program (grant number 2018YX030), the Medical Science Development Program of Shandong Province (grant number 2018WS058), and the National Natural Science Foundation of China (81502469).
The authors have no conflicts of interest to disclose.
References
- [1].Wang Z, Chu J. High efficacy of gefitinib in the treatment of EGFR mutation-positive advanced non-small cell lung adenocarcinoma: a case report. Oncol Lett 2014;8:1320–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Eckstein N, Roper L, Haas B, et al. Clinical pharmacology of tyrosine kinase inhibitors becoming generic drugs: the regulatory perspective. J Exp Clin Cancer Res 2014;33:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hanna N, Johnson D, Temin S, et al. Systemic therapy for stage IV non-small-cell lung cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol 2017;35:3484–515. [DOI] [PubMed] [Google Scholar]
- [4].Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735–42. [DOI] [PubMed] [Google Scholar]
- [5].Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380–8. [DOI] [PubMed] [Google Scholar]
- [6].Soria JC, Wu YL, Nakagawa K, et al. Gefitinib plus chemotherapy versus placebo plus chemotherapy in EGFR-mutation-positive non-small-cell lung cancer after progression on first-line gefitinib (IMPRESS): a phase 3 randomised trial. Lancet Oncol 2015;16:990–8. [DOI] [PubMed] [Google Scholar]
- [7].Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 2008;9:47–59. [DOI] [PubMed] [Google Scholar]
- [8].Costa DB, Halmos B, Kumar A, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med 2007;4:1669–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cragg MS, Kuroda J, Puthalakath H, et al. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med 2007;4:1681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gong Y, Somwar R, Politi K, et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med 2007;4:e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ng KP, Hillmer AM, Chuah CT, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med 2012;18:521–8. [DOI] [PubMed] [Google Scholar]
- [12].Zheng L, Lin B, Song Z, et al. Relationship between BIM gene polymorphism and therapeutic efficacy in the retreatment of advanced non-small cell lung cancer with tyrosine kinase inhibitor. Zhongguo Fei Ai Za Zhi 2013;16:632–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Isobe K, Hata Y, Tochigi N, et al. Clinical significance of BIM deletion polymorphism in non-small-cell lung cancer with epidermal growth factor receptor mutation. J Thorac Oncol 2014;9:483–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lee JH, Lin YL, Hsu WH, et al. Bcl-2-like protein 11 deletion polymorphism predicts survival in advanced non-small-cell lung cancer. J Thorac Oncol 2014;9:1385–92. [DOI] [PubMed] [Google Scholar]
- [15].Zhao M, Zhang Y, Cai W, et al. The Bim deletion polymorphism clinical profile and its relation with tyrosine kinase inhibitor resistance in Chinese patients with non-small cell lung cancer. Cancer 2014;120:2299–307. [DOI] [PubMed] [Google Scholar]
- [16].Zhong J, Li ZX, Zhao J, et al. Analysis of BIM (BCL-2 like 11 gene) deletion polymorphism in Chinese non-small cell lung cancer patients. Thorac Cancer 2014;5:509–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Atsumi J, Shimizu K, Ohtaki Y, et al. Impact of the Bim deletion polymorphism on survival among patients with completely resected non-small-cell lung carcinoma. J Glob Oncol 2016;2:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cardona AF, Rojas L, Wills B, et al. BIM deletion polymorphisms in Hispanic patients with non-small cell lung cancer carriers of EGFR mutations. Oncotarget 2016;7:68933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Qian K, Zhang Y, Zhi X. Retrospective study of efficacy in BIM gene polymorphism on first-line EGFR-TKIs treatment for advanced lung adenocarcinoma. Zhongguo Fei Ai Za Zhi 2017;20:543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yuan J, Li B, Zhang N, et al. Clinical implications of the BIM deletion polymorphism in advanced lung adenocarcinoma treated with gefitinib. Clin Lung Cancer 2018;19:e431–8. [DOI] [PubMed] [Google Scholar]
- [21].Lee JK, Shin JY, Kim S, et al. Primary resistance to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in patients with non-small-cell lung cancer harboring TKI-sensitive EGFR mutations: an exploratory study. Ann Oncol 2013;24:2080–7. [DOI] [PubMed] [Google Scholar]
- [22].Lee JY, Ku BM, Lim SH, et al. The BIM deletion polymorphism and its clinical implication in patients with EGFR- mutant non-small-cell lung cancer treated with EGFR tyrosine kinase inhibitors. J Thorac Oncol 2015;10:903–9. [DOI] [PubMed] [Google Scholar]
- [23].Wu SG, Liu YN, Yu CJ, et al. Association of BIM deletion polymorphism with intrinsic resistance to EGFR tyrosine kinase inhibitors in patients with lung adenocarcinoma. JAMA Oncol 2016;2:826–8. [DOI] [PubMed] [Google Scholar]
- [24].Sun S, Yu H, Wang H, et al. Exploratory cohort study and meta-analysis of BIM deletion polymorphism in patients with epidermal growth factor receptor-mutant non-small-cell lung cancer treated with epidermal growth factor receptor tyrosine kinase inhibitors. Onco Targets Ther 2017;10:1955–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. [DOI] [PubMed] [Google Scholar]
- [27].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zou Q, Zhan P, Lv T, et al. The relationship between BIM deletion polymorphism and clinical significance of epidermal growth factor receptor-mutated non-small cell lung cancer patients with epidermal growth factor receptor-tyrosine kinase inhibitor therapy: a meta-analysis. Transl Lung Cancer Res 2015;4:792–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ying HQ, Chen J, He BS, et al. The effect of BIM deletion polymorphism on intrinsic resistance and clinical outcome of cancer patient with kinase inhibitor therapy. Sci Rep 2015;5:11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nie W, Tao X, Wei H, et al. The BIM deletion polymorphism is a prognostic biomarker of EGFR-TKIs response in NSCLC: a systematic review and meta-analysis. Oncotarget 2015;6:25696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ma JY, Yan HJ, Gu W. Association between BIM deletion polymorphism and clinical outcome of EGFR-mutated NSCLC patient with EGFR-TKI therapy: a meta-analysis. J Cancer Res Ther 2015;11:397–402. [DOI] [PubMed] [Google Scholar]
- [33].Huang WF, Liu AH, Zhao HJ, et al. BIM gene polymorphism lowers the efficacy of EGFR-TKIs in advanced nonsmall cell lung cancer with sensitive EGFR mutations: a systematic review and meta-analysis. Medicine (Baltimore) 2015;94:e1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen X, Liu H, Xing H, et al. The BIM deletion polymorphism cannot account for intrinsic TKI resistance of Chinese individuals with chronic myeloid leukemia. Nat Med 2014;20:1090. [DOI] [PubMed] [Google Scholar]
- [35].Nakagawa T, Takeuchi S, Yamada T, et al. EGFR-TKI resistance due to BIM polymorphism can be circumvented in combination with HDAC inhibition. Cancer Res 2013;73:2428–34. [DOI] [PubMed] [Google Scholar]
- [36].Tanimoto A, Takeuchi S, Arai S, et al. Histone deacetylase 3 inhibition overcomes BIM deletion polymorphism-mediated osimertinib resistance in EGFR-mutant lung cancer. Clin Cancer Res 2017;23:3139–49. [DOI] [PubMed] [Google Scholar]
- [37].Takeuchi S, Yoshimura K, Fujiwara T, et al. Phase I study of combined therapy with vorinostat and gefitinib to treat BIM deletion polymorphism-associated resistance in EGFR-mutant lung cancer (VICTROY-J): a study protocol. J Med Invest 2017;64:321–5. [DOI] [PubMed] [Google Scholar]
- [38].Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005;353:123–32. [DOI] [PubMed] [Google Scholar]
- [39].Ho GYF, Zheng SL, Cushman M, et al. Associations of insulin and IGFBP-3 with lung cancer susceptibility in current smokers. J Natl Cancer Inst 2016;108: doi: 10.1093/jnci/djw012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Song J, Shi J, Dong D, et al. A new approach to predict progression-free survival in stage IV EGFR-mutant NSCLC patients with EGFR-TKI therapy. Clin Cancer Res 2018;24:3583–92. [DOI] [PubMed] [Google Scholar]


