Abstract
Background:
The prognostic significance of PBK/TOPK overexpression in solid tumors remains controversial. Therefore, we carried out a meta-analysis to evaluate the impact of PBK/TOPK overexpression in solid tumors on patients’ overall survival (OS) and disease-free survival (DFS).
Methods:
Relevant articles were identified through searching the PubMed, Embase and Web of Science up to May 2017. The pooled hazard ratio (HR) with 95% confidence interval (CI) was used to estimate the effects.
Results:
In this meta-analysis, 12 studies involving 1571 participants were included, PBK/TOPK overexpression was significantly associated with poor OS (pooled HR = 1.91, 95%CI = 1.22-3.00, P = .005) and short DFS (pooled HR = 1.95, 95%CI = 1.46-2.58, P < .001).
Conclusions:
PBK/TOPK overexpression was associated with poor survival in human solid tumors which may be a valuable prognosis biomarker and a potential therapeutic target of solid tumors.
Keywords: meta-analysis, PBK/TOPK, prognosis, solid tumors, survival
1. Introduction
PDZ-binding kinase/T-LAK cell-originated protein kinase (PBK/TOPK) is a 322 amino-acid MAPKK-like serine/threonine kinase (mitogen-activated protein kinase kinase-like serine/threonine kinase) that is difficult to detect in normal tissues other than testicular and fetal samples.[1,2] PBK/TOPK is involved in many cellular functions, such as cell growth, DNA damage repair, apoptosis, immune responses and inflammation.[3–6] In recent years, it was confirmed that PBK/TOPK is overexpressed in proliferative cells. Expression of PBK/TOPK increased during mitosis.[7,8] During mitosis, the up-regulated PBK/TOPK directly binds cdk1/cyclin B1 complex and then threonine-9 of PBK/TOPK is phosphorylated by cdk1/cyclin B1 complex[9] which promotes cytokinesis through phosphorylation of a protein regulator of cytokinesis 1 (PRC1).[10,11]
A growing number of studies suggested that elevated PBK/TOPK expression in tumor tissue was correlated with poor survival of patients with various solid tumors such as lung adenocarcinoma,[12–14] colorectal cancer,[15,16] gastric cancer,[17] prostate cancer,[18] gastric carcinoma,[17] esophageal squamous cell carcinoma,[19] nasopharygneal carcinoma,[20] ovarian cancer;[21] however, other studies[22,23] could not confirm this.
Therefore, we conducted this comprehensive meta-analysis, which combined all the published evidence to clarify the prognostic value of PBK/TOPK in solid tumors. The results of this meta-analysis could potentially shed more light on the development of PBK/TOPK -targeted therapy and prognostic prediction in solid tumor.
2. Materials and methods
2.1. Literature search
We conducted a comprehensive literature search of Pubmed, Embase and Web of Science for studies measuring expression of PBK/TOPK and survival in patients with solid tumors from 1993 to May 2017 with the search terms: (PBK/TOPK OR PDZ Binding Kinase OR TOPK OR T-LAK Cell-Originated Protein Kinase) AND (cancer OR carcinoma OR neoplasm OR malignancy OR tumor). All potentially eligible studies were retrieved. The bibliographies in these studies were also carefully scanned to identify other eligible studies and extra studies. When multiple studies of the same patient population were identified, we included the published report with the largest sample size.
2.2. Inclusion criteria
To be eligible for inclusion in this meta-analysis, studies:
-
1.
should evaluate PBK/TOPK expression for prognostic value in cancer;
-
2.
should test PBK/TOPK expression by immunohistochemistry (IHC);
-
3.
should have hazard ratios (HRs) with 95% confidence intervals (CIs), or enable estimate these statistics from the data presented;
-
4.
should classify PBK/TOPK expression as “high” and “low” or “positive” and “negative”;
-
5.
should publish in English.
2.3. Exclusion criteria
Exclusion criteria were:
-
1.
literatures published as letters, editorials, abstracts, reviews, case reports and expert opinions;
-
2.
experiments performed in vitro or in vivo, but not associated with patients;
-
3.
articles without the HRs and 95% CI or K-M survival curves dealing with overall survival, disease-free survival;
-
4.
The follow-up duration was less than 3 years.
2.4. Data extraction
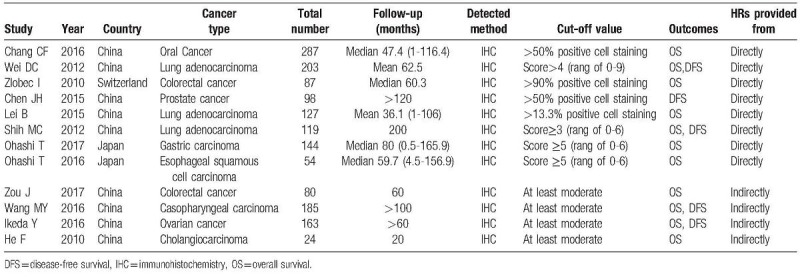
Two investigators independently extracted the data from eligible studies using a predefined form. The following data were extracted for each study: the first author's surname, publication year, country of origin, types of cancer, number of patients analyzed, follow-up time, detected method, cut-off values, outcome endpoint: OS and DFS. For studies that presented only Kaplan-Meier curves, Engauge Digitizer version 4.1 was used to extract the survival data. The estimated HRs and 95% CIs were calculated by Tierney method.[3] Multivariate HR and 95% CI of high PBK/TOPK expression group versus low were selected if both univariate and multivariate results were reported in an individual study. The main features of these selected studies were summarized in Table 1.
Table 1.
Characteristics of studies included in the meta-analysis.
2.5. Statistical analysis
Pooled HRs and 95% CIs for 2 outcome endpoints (OS and DFS) were calculated. Statistical heterogeneity was assessed through the Chi-square test and I-square test, which was checked through the Q test and a P value >.10 indicated a lack of heterogeneity. We also quantified the effect of heterogeneity via I2 = 100% × (Q−df)/Q. I2 values of <25% could be considered “low”, values of about 50% could be considered “moderate”, and values of over 75% could be considered “high”.[24] Without statistical heterogeneity, a fixed-effects model was employed to calculate the pooled HRs; otherwise the random-effects model was performed. Funnel plots and the Egger test were utilized to determine the possible publication bias.[25] If the funnel plot was asymmetric and the Egger test reported a P value of less than .05, publication bias was deemed to probably exist. If publication bias was observed, we adjusted for the effect by the use of the trim-and-fill method. Sensitivity analysis was also conducted to find out if certain single article could influence the overall result. Statistical analyses were performed via the Stata 14.0 (StataCorp, College Station, TX). P values for all comparisons were 2-tailed.
2.6. Ethical approval
All analyses are based on previous published studies. Therefore, there is no need for ethical approval and patient consent.
3. Results
3.1. Study characteristics
Using the described searching strategy, 256 published studies were initially retrieved after duplicates were removed. Once 242 irrelevant abstracts were excluded, 14 full-text articles were reviewed for a more detailed evaluation. Of these, one article did not have sufficient data to allow for estimation of the HR and one was duplicate report. Finally, 12 studies were enrolled into the meta-analysis.[12–23] Details of the study selection process are shown in Fig. 1. The characteristics of eligible studies are listed in Table 1. All studies used immunohistochemistry techniques to assess the expression level of PBK/TOPK. A total of 1571 patients from China, Japan and Switzerland were diagnosed with a variety of cancers, including three studies evaluated lung adenocarcinoma,[12–14] 2 evaluated colorectal cancer,[15,16] and 1 each evaluated gastric carcinoma,[17] oral cancer,[23] prostate cancer,[18] esophageal squamous cell carcinoma,[19] nasopharygneal carcinoma,[20] ovarian cancer,[21] cholangiocarcinoma.[22] The endpoints OS and DFS were addressed in 11 and 5 studies, respectively. HRs were reported directly in 8 studies and estimated indirectly in the other 4 studies. The cut-off values were different in these studies.
Figure 1.
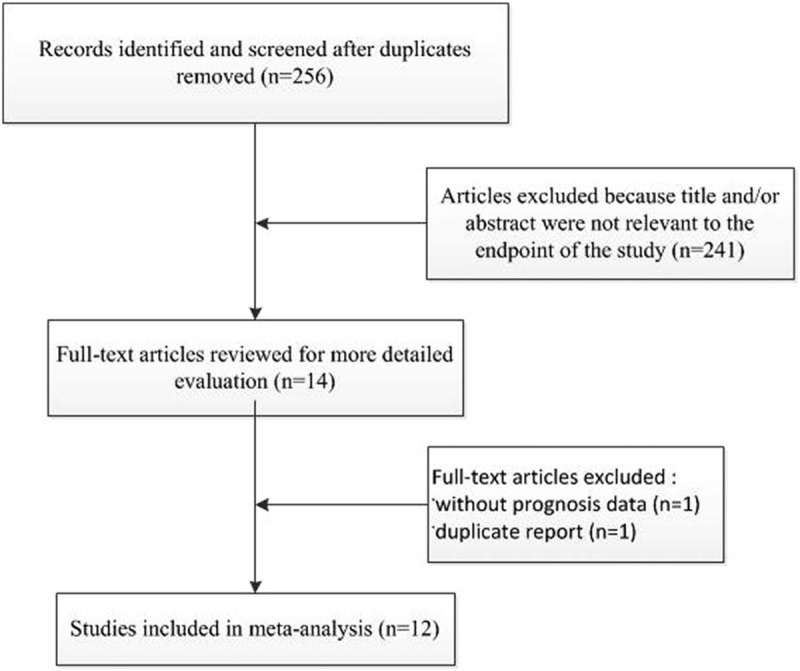
Flow chart depicting the selection of eligible studies.
3.2. Meta-analysis
Overall, 11 studies trials offered data on PBK/TOPK overexpression and OS in solid tumors. The synthesis indicated that over-expression of PBK/TOPK was significantly related to a poorer OS (pooled HR = 1.91, 95%CI = 1.22–3.00, P = .005) (Fig. 2). Because there was heterogeneity between studies (I2 = 82.6%, P = .000), we utilized a random-effects model to determine the pooled HR and 95% CI. In the stratified analysis based on tumor type, high levels of PBK/TOPK was significantly associated with worse OS in lung adenocarcinoma (pooled HR = 2.72, 95%CI = 1.93–3.83, P < .001) with low heterogeneity and colorectal cancer (pooled HR = 1.98, 95%CI = 1.17–3.37, P = .011) without any heterogeneity. There was only 1 study each evaluating the association between PBK/TOPK overexpression and OS in gastric carcinoma,[17] oral cancer,[23] prostate cancer,[18] esophageal squamous cell carcinoma,[19] nasopharygneal carcinoma,[20] ovarian cancer,[21] cholangiocarcinoma,[22] and therefore, the results related entirely to these studies; these tumors were defined as “other cancers”. Combined data from these studies showed that PBK/TOPK overexpression has no effect on OS (pooled HR = 1.58, 95%CI = 0.74–3.36, P = .239), along with a high heterogeneity (I2 = 87.3%, P = .000).
Figure 2.
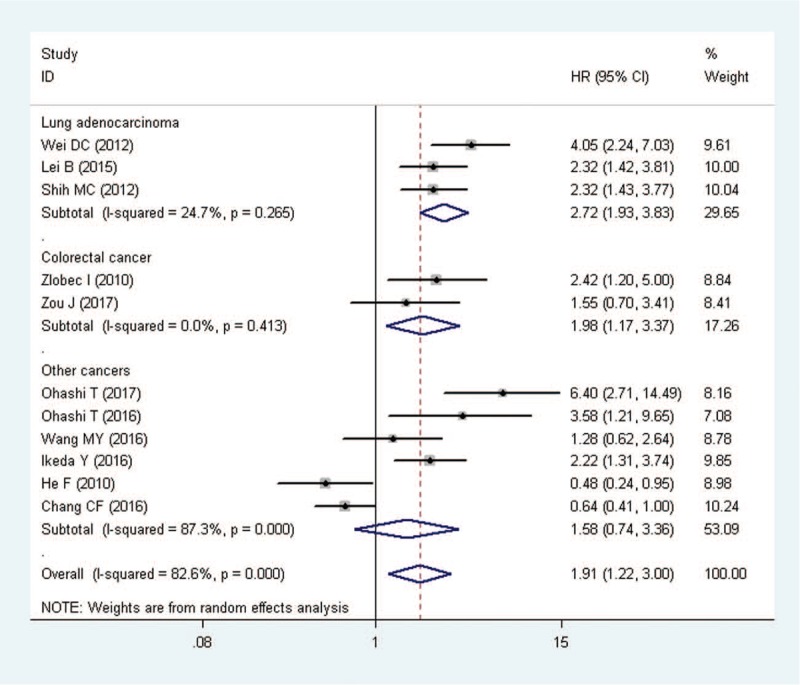
Meta-analysis of the association between PBK/TOPK overexpression and overall survival (OS) stratified by tumor types. Other cancers include gastric carcinoma, oral cancer, esophageal squamous cell carcinoma, nasopharygneal carcinoma, ovarian cancer and cholangio carcinoma. HR = hazard ratio, CI = confidence intervals.
Five studies were included in the meta-analysis of DFS. PBK/TOPK overexpression was significantly associated with poor DFS in all studies (pooled HR = 1.95, 95%CI = 1.46–2.58, P < .001) (Fig. 3). A fixed effects model was used because the heterogeneity test reported a P value of .517.
Figure 3.
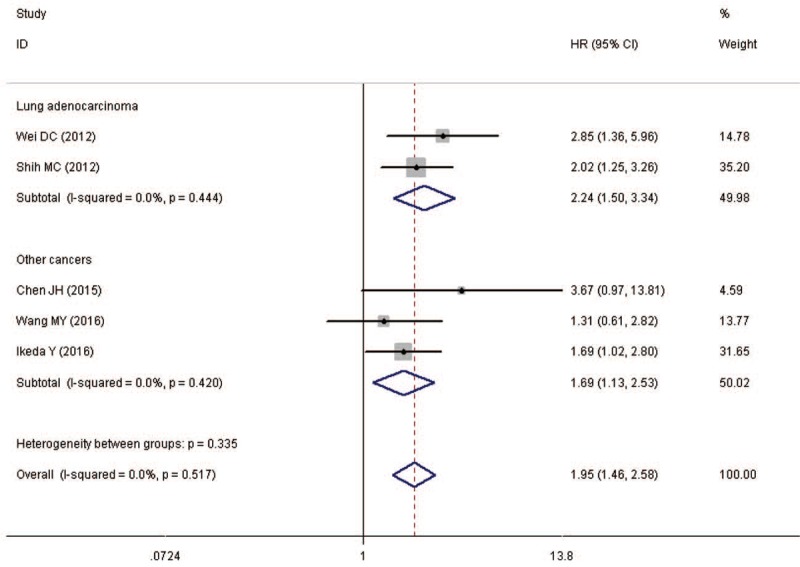
Meta-analysis of the association between PBK/TOPK overexpression and disease-free survival (DFS) stratified by tumor types. Other cancers include prostate cancer, nasopharygneal carcinoma and ovarian cancer. HR = hazard ratio, CI = confidence intervals.
3.3. Publication bias and sensitivity analysis
The Begg funnel plot shapes for the OS and DFS had no obvious asymmetry (Fig. 4) and Egger test showed there was no publication bias for DFS (P = .221). However, publication bias existed for OS (P = .000) in the analysis of high versus low PBK/TOPK expression. Therefore, we performed trim and fill method to make pooled HR more reliable, but the P value was not significant (random model: HR = 0.442, 95%CI = −0.012–0.896, P = .010), and with significant heterogeneity (P = .001). Sensitivity analyses were used to evaluate whether individual studies influenced the results of OS and DFS. The results showed that the overall conclusion was not significantly influenced after omitting any single study for the effect of PBK/TOPK expression on OS and DFS (Fig. 5).
Figure 4.
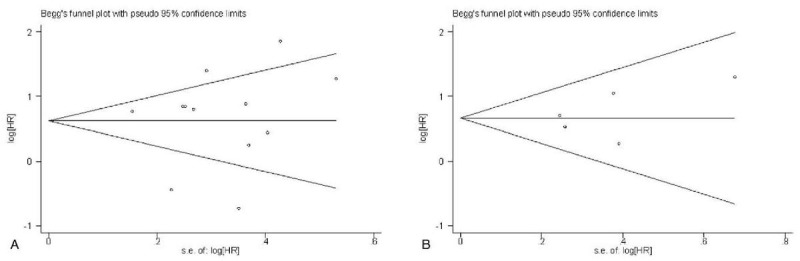
Begg funnel plots for the studies involved in the meta-analysis. (a) Overall survival. (b) disease free survival (DFS). loghr = logarithm of hazard ratios, s.e. = standard error.
Figure 5.
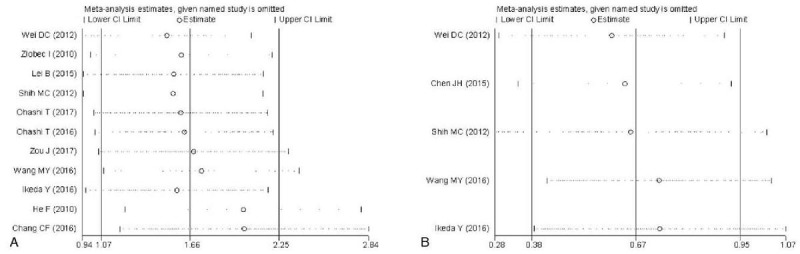
Sensitivity analysis of the meta-analysis. (A) Overall survival. (B) disease-free survival (DFS).
4. Discussions
The PBK/TOPK protein, a member of the MAPKK family, is a growth-factor-regulated kinase, which is constitutively high in tumor cells. PBK/TOPK which is phosphorylated by the cdc2/cyclin B complex and activated in a cell cycle-dependent manner during mitosis [1] may have a role in the regulation of cell proliferation and cell cycle. Growing evidence implicate PBK/TOPK expression in tumor development, cancer growth, and apoptosis.[6,26–28]
Many clinical studies investigated the prognostic value of PBK/TOPK. Most of these studies, however, include only limited number of patients, and the results remain not comprehensive. PBK/TOPK overexpression often predicts unfavorable outcome in many cancer, such as lung adenocarcinoma,[12–14] gastric carcinoma, [17] prostate cancer.[18] On the other hand, it is a favorable prognostic indicator in oral cancer [23] and cholangiocarcinoma.[22] To our knowledge, the present study is the first complete overview of all reported clinical studies exploring the possible prognostic role of PBK/TOPK up-regulation in solid tumors.
We systematically evaluated survival data of 1571 solid tumor patients included in 12 different studies. Overall, these results clearly show that high PBK/TOPK expression could be a poor prognostic factor of various solid tumors, with both results of poor OS (pooled HR = 1.91, 95%CI = 1.22–3.00, P = .005) and poor DFS (pooled HR = 1.95, 95%CI = 1.46–2.58, P < .001). Similarly, subgroup analysis based on tumor type, revealing that PBK/TOPK overexpression was significantly associated with worse OS and DFS in lung adenocarcinoma; thus, PBK/TOPK may serve as a novel prognostic marker and therapeutic target for lung adenocarcinoma. Further,
PBK/TOPK overexpression was also significantly related to poor OS in colorectal cancer patients. Apart from being a promising biomarker, PBK/TOPK may also provide a new target for anticancer therapy. HI-TOPK-032, a specific inhibitor for PBK/TOPK, reduces cell viability and colony formation via a dramatic increase in apoptotic cells in vitro and results in a significant decrease of tumor growth in vivo.[20,29,30]
This meta-analysis study involves several important implications. First, it reveals that PBK/TOPK overexpression is correlated to unfavorable outcome for a wide range of human cancers, which indicates that PBK/TOPK may be a promising therapeutic target. Second, it identifies a subgroup of tumors with adverse outcome in lung adenocarcinoma and colorectal cancer. Finally, it highlights the potential clinical application of PBK/TOPK as a valuable prognostic biomarker.
However, several limitations also exist in this study. First, most researches included in the meta-analysis were designed as retrospective studies, and such studies are more likely to be published if they have positive results than if they have negative results. Secondly, some of the studies were small sample size; even one study [22] included only 24 patients. Furthermore, the method for assessing PBK/TOPK expression and definition of PBK/TOPK positivity were inconsistent. Besides, some studies which neither provide complete data nor published in English were excluded in statistics. Therefore, we detected significant publication bias in the pooled analysis of OS. Lastly, substantial heterogeneity observed among included studies cannot be completely interpreted in spite of the use of appropriate meta-analytic techniques with random-effects models.
In conclusion, this meta-analysis indicates that PBK/TOPK expression is associated with unfavorable outcome in most solid tumors, suggesting that PBK/TOPK is a useful prognostic biomarker and a promising therapeutic target for solid tumors. Nevertheless, more clinical studies related to specific tumor types and perspectives are required to corroborate the clinical utility of PBK/TOPK expression in solid tumors.
Acknowledgments
The authors thank the study participants and research personnel for their helpful comments.
Author contributions
Data curation: Ming Xu, Song Xu.
Formal analysis: Ming Xu.
Project administration: Song Xu.
Software: Song Xu.
Writing – original draft: Ming Xu.
Writing – review and editing: Song Xu
Footnotes
Abbreviations: CI = confidence interval, DFS = disease-free survival, HR = hazard ratio, IHC = immunohistochemistry, OS = overall survival, PBK/TOPK = PDZ-binding kinase/T-LAK cell-originated protein kinase.
The authors declare no conflicts of interest.
References
- [1].Gaudet S, Branton D, Lue RA. Characterization of PDZ-binding kinase, a mitotic kinase. Proc Natl Acad Sci U S A 2000;97:5167–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhao S, Dai J, Zhao W, et al. PDZ-binding kinase participates in spermatogenesis. Int J Biochem Cell Biol 2001;33:631–6. [DOI] [PubMed] [Google Scholar]
- [3].Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Park JH, Jeong YJ, Won HK, et al. Activation of TOPK by lipopolysaccharide promotes induction of inducible nitric oxide synthase through NF-kappaB activity in leukemia cells. Cell Signal 2014;26:849–56. [DOI] [PubMed] [Google Scholar]
- [5].Li Y, Yang Z, Li W, et al. TOPK promotes lung cancer resistance to EGFR tyrosine kinase inhibitors by phosphorylating and activating c-Jun. Oncotarget 2016;7:6748–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liu Y, Liu H, Cao H, et al. PBK/TOPK mediates promyelocyte proliferation via Nrf2-regulated cell cycle progression and apoptosis. Oncol Rep 2015;34:3288–96. [DOI] [PubMed] [Google Scholar]
- [7].Park JH, Nishidate T, Nakamura Y, et al. Critical roles of T-LAK cell-originated protein kinase in cytokinesis. Cancer Sci 2010;101:403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Simons-Evelyn M, Bailey-Dell K, Toretsky JA, et al. PBK/TOPK is a novel mitotic kinase which is upregulated in Burkitt's lymphoma and other highly proliferative malignant cells. Blood Cells Mol Dis 2001;27:825–9. [DOI] [PubMed] [Google Scholar]
- [9].Matsumoto S, Abe Y, Fujibuchi T, et al. Characterization of a MAPKK-like protein kinase TOPK. Biochem Biophys Res Commun 2004;325:997–1004. [DOI] [PubMed] [Google Scholar]
- [10].Abe Y, Takeuchi T, Kagawa-Miki L, et al. A mitotic kinase TOPK enhances Cdk1/cyclin B1-dependent phosphorylation of PRC1 and promotes cytokinesis. J Mol Biol 2007;370:231–45. [DOI] [PubMed] [Google Scholar]
- [11].Chen TC, Lee SA, Hong TM, et al. From midbody protein-protein interaction network construction to novel regulators in cytokinesis. J Proteome Res 2009;8:4943–53. [DOI] [PubMed] [Google Scholar]
- [12].Wei DC, Yeh YC, Hung JJ, et al. Overexpression of T-LAK cell-originated protein kinase predicts poor prognosis in patients with stage I lung adenocarcinoma. Cancer Sci 2012;103:731–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lei B, Qi W, Zhao Y, et al. PBK/TOPK expression correlates with mutant p53 and affects patients’ prognosis and cell proliferation and viability in lung adenocarcinoma. Hum Pathol 2015;46:217–24. [DOI] [PubMed] [Google Scholar]
- [14].Shih MC, Chen JY, Wu YC, et al. TOPK/PBK promotes cell migration via modulation of the PI3K/PTEN/AKT pathway and is associated with poor prognosis in lung cancer. Oncogene 2012;31:2389–400. [DOI] [PubMed] [Google Scholar]
- [15].Zlobec I, Molinari F, Kovac M, et al. Prognostic and predictive value of TOPK stratified by KRAS and BRAF gene alterations in sporadic, hereditary and metastatic colorectal cancer patients. Br J Cancer 2010;102:151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zou J, Kuang W, Hu J, et al. miR-216b promotes cell growth and enhances chemosensitivity of colorectal cancer by suppressing PDZ-binding kinase. Biochem Biophys Res Commun. 2017. Mar 31. [DOI] [PubMed] [Google Scholar]
- [17].Ohashi T, Komatsu S, Ichikawa D, et al. Overexpression of PBK/TOPK relates to tumour malignant potential and poor outcome of gastric carcinoma. Br J Cancer 2017;116:218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen JH, Liang YX, He HC, et al. Overexpression of PDZ-binding kinase confers malignant phenotype in prostate cancer via the regulation of E2F1. Int J Biol Macromol 2015;81:615–23. [DOI] [PubMed] [Google Scholar]
- [19].Ohashi T, Komatsu S, Ichikawa D, et al. Overexpression of PBK/TOPK contributes to tumor development and poor outcome of esophageal squamous cell carcinoma. Anticancer Res 2016;36:6457–66. [DOI] [PubMed] [Google Scholar]
- [20].Wang MY, Lin ZR, Cao Y, et al. PDZ binding kinase (PBK) is a theranostic target for nasopharyngeal carcinoma: driving tumor growth via ROS signaling and correlating with patient survival. Oncotarget 2016;7:26604–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ikeda Y, Park JH, Miyamoto T, et al. T-LAK Cell-Originated Protein Kinase (TOPK) as a prognostic factor and a potential therapeutic target in ovarian cancer. Clin Cancer Res 2016;22:6110–7. [DOI] [PubMed] [Google Scholar]
- [22].He F, Yan Q, Fan L, et al. PBK/TOPK in the differential diagnosis of cholangiocarcinoma from hepatocellular carcinoma and its involvement in prognosis of human cholangiocarcinoma. Hum Pathol 2010;41:415–24. [DOI] [PubMed] [Google Scholar]
- [23].Chang CF, Chen SL, Sung WW, et al. PBK/TOPK expression predicts prognosis in oral cancer. Int J Mol Sci 2016;17: pii: E1007. doi: 10.3390/ijms17071007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ayllon V, O’Connor R. PBK/TOPK promotes tumour cell proliferation through p38 MAPK activity and regulation of the DNA damage response. Oncogene 2007;26:3451–61. [DOI] [PubMed] [Google Scholar]
- [27].Yang YF, Pan YH, Cao Y, et al. PDZ binding kinase, regulated by FoxM1, enhances malignant phenotype via activation of beta-Catenin signaling in hepatocellular carcinoma. Oncotarget 2017;8:47195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zou J, Kuang W, Hu J, et al. miR-216b promotes cell growth and enhances chemosensitivity of colorectal cancer by suppressing PDZ-binding kinase. Biochem Biophys Res Commun 2017;488:247–52. [DOI] [PubMed] [Google Scholar]
- [29].Joel M, Mughal AA, Grieg Z, et al. Targeting PBK/TOPK decreases growth and survival of glioma initiating cells in vitro and attenuates tumor growth in vivo. Mol Cancer 2015;14:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kim DJ, Li Y, Reddy K, et al. Novel TOPK inhibitor HI-TOPK-032 effectively suppresses colon cancer growth. Cancer Res 2012;72:3060–8. [DOI] [PubMed] [Google Scholar]


