Abstract
Transcriptome-wide association studies (TWAS) have recently gained great attention due to their ability to prioritize complex trait-associated genes and promote potential therapeutics development for complex human diseases. TWAS integrates genotypic data with expression quantitative trait loci (eQTLs) to predict genetically regulated gene expression components and associates predictions with a trait of interest. As such, TWAS can prioritize genes whose differential expressions contribute to the trait of interest and provide mechanistic explanation of complex trait(s). Tissue-specific eQTL information grants TWAS the ability to perform association analysis on tissues whose gene expression profiles are otherwise hard to obtain, such as liver and heart. However, as eQTLs are tissue context-dependent, whether and how the tissue-specificity of eQTLs influences TWAS gene prioritization has not been fully investigated. In this study, we addressed this question by adopting two distinct TWAS methods, PrediXcan and UTMOST, which assume single tissue and integrative tissue effects of eQTLs, respectively. Thirty-eight baseline laboratory traits in 4,360 antiretroviral treatment-naïve individuals from the AIDS Clinical Trials Group (ACTG) studies comprised the input dataset for TWAS. We performed TWAS in a tissue-specific manner and obtained a total of 430 significant gene-trait associations (q-value < 0.05) across multiple tissues. Single tissue-based analysis by PrediXcan contributed 116 of the 430 associations including 64 unique gene-trait pairs in 28 tissues. Integrative tissue-based analysis by UTMOST found the other 314 significant associations that include 50 unique gene-trait pairs across all 44 tissues. Both analyses were able to replicate some associations identified in past variant-based genome-wide association studies (GWAS), such as high-density lipoprotein (HDL) and CETP (PrediXcan, q-value = 3.2e-16). Both analyses also identified novel associations. Moreover, single tissue-based and integrative tissue-based analysis shared 11 of 103 unique gene-trait pairs, for example, PSRC1-low-density lipoprotein (PrediXcan’s lowest q-value = 8.5e-06; UTMOST’s lowest q-value = 1.8e-05). This study suggests that single tissue-based analysis may have performed better at discovering gene-trait associations when combining results from all tissues. Integrative tissue-based analysis was better at prioritizing genes in multiple tissues and in trait-related tissue. Additional exploration is needed to confirm this conclusion. Finally, although single tissue-based and integrative tissue-based analysis shared significant novel discoveries, tissue context-dependency of eQTLs impacted TWAS gene prioritization. This study provides preliminary data to support continued work on tissue context-dependency of eQTL studies and TWAS.
Keywords: TWAS, integrative, context, PrediXcan, UTMOST
1. Introduction
Improving antiretroviral therapy (ART) efficacy and safety is an ongoing goal for addressing the HIV pandemic. According to the Joint United Nations Programme on HIV and AIDS (UNAIDS) (http://aidsinfo.unaids.org/), approximately 36.7 million people worldwide were living with human immunodeficiency virus (HIV) in 2016. Over the past three decades there has been immense progress on HIV care and treatment, and in 2017 there were about 20.9 million HIV-positive people who had access to ART. The connection of genomics with pharmacology has led to the discovery of numerous single nucleotide polymorphisms (SNPs) in drug absorption, distribution, metabolism, and elimination (ADME) genes and off-target genes. Many SNPs have been related to effects and/or pharmacokinetics of antiretroviral drugs1–6. However, most trait-related SNPs lack connections to actual functional genes, which suggests the need for alternative analysis approaches.
The emerging field of transcriptome-wide association studies (TWAS) offer a new way to directly identify gene-trait associations via integration of genotypic data and expression quantitative trait loci (eQTLs). eQTLs are an important class of genetic functional elements, which affect transcriptional regulation on target genes. Integration of eQTL information with genotypic data allows TWAS to estimate the extent to which a gene’s expression level is regulated by genetic variants and how this correlates with traits of interest8. The Genotype Tissue Expression Project (GTEx7) provides the data and the opportunity to identify eQTLs and estimate effect sizes for multiple human tissues (44 tissues in GTEx v6p). With GTEx, TWAS can explore gene-trait associations on tissues whose gene expression profiles are otherwise hard to obtain, such as liver and heart. However, current TWAS focuses primarily on eQTLs identified in a tissue-by-tissue manner, while many studies have either acknowledged or supported the power of an integrative tissue context in identifying single-tissue and multi-tissue eQTLs9,10.
In this study, we aimed to address whether and how single tissue and integrative tissue context of eQTLs influence TWAS gene prioritization by comparing two distinct TWAS methods, PrediXcan11 and Unified Test for MOlecular SignaTures (UTMOST12). PrediXcan uses elastic-net regression model and identifies eQTLs in a tissue-by-tissue manner. UTMOST adopts group-lasso and search through all tissues at once to spot eQTLs of a certain gene. This strategy allows UTMOST to identify single-tissue specific eQTLs similar to PrediXcan but increase the chance of detecting multi-tissue eQTLs. Here, 38 baseline (i.e. pre-ART) laboratory values and genotypic data of 4,360 ACTG clinical trials participants from multiple previous studies13–19 comprised the input for TWAS. Genotyping had been previously generated in multiple phases with Illumina assays: 650Y (phase I), 1M Duo (phase II and III), or Human Core Exome (phase IV). We performed the two TWAS methods separately in a tissue-specific manner (i.e. 44 tissues) (Figure 1). If tissue context-dependency of eQTLs did not affect TWAS gene prioritization, we expected to observe shared gene-trait associations between single tissue-based analysis (PrediXcan) and integrative tissue-based analysis (UTMOST). The results partially supported this hypothesis, but also suggested varied gene prioritization abilities of single tissue-based and integrative tissue-based approaches respectively. The former found more unique gene-trait pairs, while the latter tended to prioritize genes expressed in multiple tissues. This study provides supportive evidence for tissue context-dependency of eQTLs and its impact on TWAS gene prioritization.
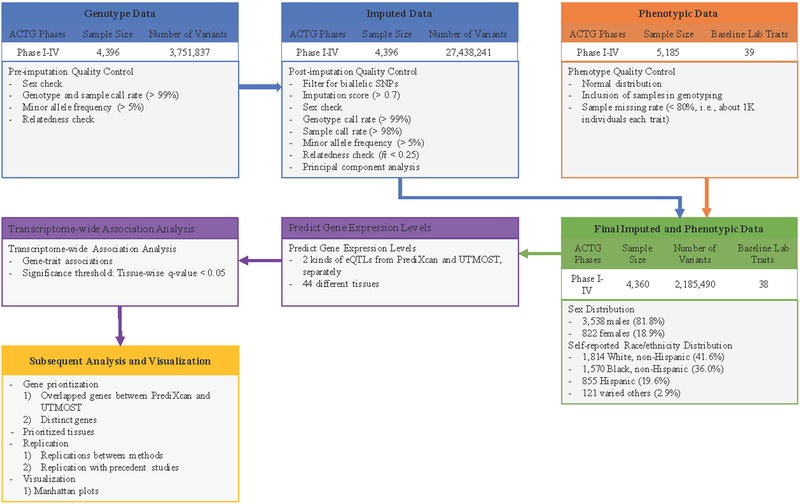
Figure 1.
This study investigates the influence of tissue context-dependency of eQTLs on TWAS gene prioritization by comparing two distinct TWAS methods, PrediXcan and UTMOST. PrediXcan assumes single tissue context of eQTLs, while UTMOST assumes eQTLs to possibly have effects in multiple tissues.
2. Methods
2.1. Data and Study Participants
In this study, we used four different genotyping phases of ACTG studies in a combined dataset that included samples and data from participants in prospective, randomized ART-naïve treatment trials13–19. Clinical trial designs and results, and results of a genome-wide pleiotropic study results for baseline laboratory values have been described elsewhere13–21.
2.2. Quality Control
2.2.1. Genotypic data
A total of 4,393 individuals were genotyped in four phases. Phase I was genotyped using Illumina 650Y array; Phase II and III were genotyped using Illumina 1M duo array; Phase IV was genotyped using Illumina HumanCoreExome BeadChip.
The computational preparation of genotypic data included pre-imputation quality control (QC), imputation, and post-imputation quality control. Pre- and post-imputation quality control followed the same guidelines22 and used PLINK1.9023 and R programming language. Imputation was performed on ACTG phase I-IV combined genotype data. Genotyped variants surviving the preimputation quality control comprised the input datasets for imputation, which used IMPUTE224 with 1000 Genomes25 Phase 1 v3 as the reference panel. ACTG phase I-IV combined imputed data had 4,941 individuals and 27,438,241 variants. The following procedures/parameters were used in the post-imputation quality control by PLINK1.90: sample inclusion in phase I-IV phenotype collection, biallelic SNP check, imputation score (> 0.7), sex check, genotype call rate (> 99%), sample call rate (> 98%), and minor allele frequency (MAF > 5%), and relatedness check . Subsequent principal component analysis (EIGENSOFT26) projected remaining individuals onto the 1000 Genomes Project sample space to examine for population stratification. The first three principal components were used as covariates to adjust for population structure in the subsequent analysis. The final QC’ed ACTG phase I-IV combined imputed data contained 2,185,490 genotyped and imputed biallelic SNPs for 4,360 individuals (Figure 1).
2.2.2. Phenotypic data
The ACTG clinical trials included in this analysis collected baseline (i.e., pre-ART) laboratory traits from 5,185 ART-naïve individuals. We only retained individuals who have been genotyped and traits that were normally distributed and met a criterion of phenotype missing rate < 80%. The final combined phenotype dataset of ACTG genotyping phase I-IV retained 38 traits and the same number of individuals as the QC’ed imputed dataset (Figure 1).
2.3. Predict Unmeasured Gene Expression Levels
We adopted two TWAS methods, PrediXcan and UTMOST, to predict unmeasured gene expression levels in a tissue-specific manner. PrediXcan and UTMOST have estimated SNP effect sizes on gene expression levels in 44 tissues, which are available at http://predictdb.org/ and https://github.com/Joker-Jerome/UTMOST, respectively. The PrediXcan and UTMOST scripts were pulled from their GitHub project repositories on April 23rd and Jun 6th, 2018, respectively.
PrediXcan and UTMOST followed the same multivariate models. Let N denote the sample size and M denote the number of eQTLs in a certain gene. A gene’s expression level can be predicted using the multivariate model as follows:
| (1) |
where E is the N × 1 vector of predicted gene expression levels of the gene, X is the N × M matrix of genotypes, and β is the M × 1 vector of eQTLs’ estimated regulatory effects on the gene.
Predicted gene expression levels were likely to differ between the two methods as each has a different hypothesis of eQTL regulatory mechanisms in terms of tissue context-dependency. To discover trait-related tissues without assumptions, we predicted gene expression levels in 44 tissues.
2.4. Transcriptome-wide Association Analysis
We tested for gene-trait associations by performing transcriptome-wide association tests on predicted gene expression levels and ACTG baseline lab traits using PLATO27,28. All baseline labtraits included in this study were continuous and thus were modeled using linear regression. Age, sex, and the first three principal components calculated by EIGENSOFT were included as covariates in linear models to adjust for sampling biases and underlying population structure. PrediXcan and UTMOST have different degrees of diversity in the number of eGenes and gene-trait associations among tissues. To avoid biases due to an uneven number of associations among tissues, p-values were adjusted using FDR with using Benjamini–Hochberg procedure29 in a tissue-specific manner. For this study, we consider gene-trait associations significant if they had single tissue-wise q-value < 0.05.
3. Results
We compared the influence of tissue context-dependency of eQTLs on TWAS gene prioritization by comparing single tissue-based analysis (PrediXcan) and integrative tissue-based analysis (UTMOST). We performed TWAS on ACTG phase I-IV combined datasets. The data aggregation of ACTG phase I-IV provided a larger sample size to ensure the power of identifying gene-trait association. QC procedures left the ACTG phase I-IV combined imputed data with 4,360 individuals and 2,185,490 SNPs. There were 38 baseline lab traits in the final phenotypic datasets.
Single tissue-based and integrative tissue-based analysis identified a total of 430 significant gene-trait associations (103 unique gene-trait pairs regardless of tissue, q-value < 0.05) and share 11 unique gene-trait pairs. Single tissue-based analysis identified 116 of the 430 significant associations (64 unique gene-trait pairs), encompassing 41 genes, 17 traits, and 28 tissues. Integrative tissue-based analysis identified the remaining 314 significant associations (50 unique gene trait pairs), encompassing 38 genes, 20 traits, and all 44 tissues.
3.1. Tissue Context-dependency Influenced TWAS Gene Prioritization
Gene prioritization results from single tissue-based analysis (PrediXcan) and integrative tissue-based analysis (UTMOST) were compared to evaluate the influence of tissue context-dependency of eQTLs on TWAS. Single and integrative tissue-based analyses shared 11 of 103 unique gene-trait pairs regardless of tissue (Table 1). Several of these replicated the findings of previous studies (Table 2). The lowest p-value by integrative tissue-based analysis was for MROH2A-total bilirubin levels20 (UTMOST, q-value = 6.0e-27), which had a moderate p-value from single tissue-based analysis (q-value = 0.005). Another replication was between PSRC1 and two lipid-related traits, cholesterol and LDL, which have been reported in other studies30–33. Although it was SORT1, which neighbors PSRC1, that has been functionally related to LDL via mice knockdown experiments34. ALDH5A1 and GPLD1 have been associated with the liver function test, alkaline phosphatase (ALP)35. In the cases of PSRC1, ALDH5A1, and GPLD1, integrative tissue-based analysis (UTMOST) prioritized the genes in their biological function-related organ, liver, which was not always the case for single tissue-based analysis (PrediXcan). Possible novel associations were observed between absolute neutrophil count and C1orf20436, ATF6, and VANGL237.
Table 1.
Significant gene-trait associations (q-value < 0.05) shared by single and integrative tissue-based analysis. The two different analyses shared 11 out of 103 unique significant gene-trait pairs.
| Traits | Genes | Methods | #Tissues | Major Tissue Types* |
|---|---|---|---|---|
| Absolute neutrophil count | ATF6 | PrediXcan | 1 | Brain |
| ATF6 | UTMOST | 2 | Brain, Transformed Fibroblasts | |
| Clorf204 | PrediXcan | 1 | Brain | |
| Clorf204 | UTMOST | 5 | Brain, Ovary, Pituitary | |
| VANGL2 | PrediXcan | 1 | Brain | |
| VANGL2 | UTMOST | 1 | Brain | |
| Alkaline phosphatase | ALDH5A1 | PrediXcan | 9 | Artery, Colon, Liver, Lung, Nerve, Pancreas, Skin, Thyroid, Transformed Lymphocytes |
| ALDH5A1 | UTMOST | 39 | Adipose, Adrenal Gland, Artery, Brain, Breast, Colon, Esophagus, Heart, Liver, Lung, Nerve, Ovary, Pancreas, Pituitary, Prostate, Skeletal Muscle, Skin, Small Intestine, Spleen, Stomach, Test’s, Thyroid, Transformed Lymphocytes, Uterus, Vagina | |
| GPLD1 | PrediXcan | 2 | Artery, Thyroid | |
| GPLD1 | UTMOST | 24 | Adipose, Artery, Brain, Esophagus, Heart, Liver, Lung, Nerve, Pituitary, Prostate, Skeletal Muscle, Skin, Small Intestine, Stomach, Test’s, Thyroid, Transformed Lymphocytes, Vagina, Whole Blood | |
| Cholesterol | PSRC1 | PrediXcan | 9 | Brain, Esophagus, Lung, Pancreas, Pituitary, Skeletal Muscle, Skin, Whole Blood |
| PSRC1 | UTMOST | 25 | Adipose, Brain, Breast, Colon, Esophagus, Heart, Liver, Lung, Nerve, Ovary, Pancreas, Pituitary, Prostate, Skeletal Muscle, Skin, Tests, Uterus, Whole Blood | |
| Fasting cholesterol | PSRC1 | PrediXcan | 9 | Brain, Esophagus, Lung, Pancreas, Pituitary, Skeletal Muscle, Skin, Whole Blood |
| PSRC1 | UTMOST | 22 | Adipose, Brain, Breast, Colon, Esophagus, Heart, Liver, Lung, Nerve, Ovary, Pituitary, Prostate, Skeletal Muscle, Skin, Tests, Uterus, Whole Blood | |
| Fasting LDL | PSRC1 | PrediXcan | 11 | Brain, Esophagus, Lung, Pancreas, Pituitary, Skeletal Muscle, Skin, Tests, Thyroid, Whole Blood |
| PSRC1 | UTMOST | 27 | Adipose, Brain, Breast, Colon, Esophagus, Heart, Liver, Lung, Nerve, Ovary, Pancreas, Pituitary, Prostate, Skeletal Muscle, Skin, Tests, Thyroid, Uterus, Whole Blood | |
| Hemoglobin | CAMSAP1 | PrediXcan | 1 | Nerve |
| CAMSAP1 | UTMOST | 31 | Adipose, Artery, Brain, Breast, Colon, Esophagus, Heart, Liver, Lung, Nerve, Ovary, Prostate, Skeletal Muscle, Skin, Small Intestne, Spleen, Thyroid, Transformed Fibroblasts, Transformed Lymphocytes, Whole Blood | |
| LDL | PSRC1 | PrediXcan | 11 | Brain, Esophagus, Lung, Pancreas, Pituitary, Skeletal Muscle, Skin, Tests, Thyroid, Whole Blood |
| PSRC1 | UTMOST | 27 | Adipose, Brain, Breast, Colon, Esophagus, Heart, Liver, Lung, Nerve, Ovary, Pancreas, Pituitary, Prostate, Skeletal Muscle, Skin, Tests, Thyroid, Uterus, Whole Blood | |
| Total bilirubin | MROH2A | PrediXcan | 1 | Adipose |
| MROH2A | UTMOST | 1 | Stomach |
For simplicity, only major tissue types were shown. Skin, heart, esophagus, colon, brain, artery, and adipose have subtypes.
Table 2.
Validation of some of the TWAS prioritized genes.
| GENES | METHODS | TISSUES | Q-VALUE+ | ACTG TRAITS | GWAS CATALOG REPORTED TRAITS | PMID |
|---|---|---|---|---|---|---|
| ATF6 | PrediXcan | Brain | 1.30E-02 | Absolute neutrophil count | White blood cell count | 28158719 |
| UTMOST | Transformed Fibroblasts*, Brain | 1.63E-02 | ||||
| VANGL2 | PrediXcan | Brain | 1.30E-02 | Absolute neutrophil count | Multiple sclerosis | 24076602 |
| UTMOST | Brain | 4.70E-02 | ||||
| ADAMTS4 | UTMOST | Artery | 1.50E-04 | Absolute neutrophil count*, White blood cell count | Monocyte percentage of white cells | 27863252 |
| ALDH5A1 | PrediXcan | Colon*, Artery, Liver, Lung, Nerve, Pancreas, Skin, Thyroid, Transformed Lymphocytes | 1.57E-05 | Alkaline phosphatase | Liver enzyme levels (alkaline phosphatase) | 22001757 |
| UTMOST | Artery*, Adipose, Adrenal Gland, Brain, Breast, Colon, Esophagus, Heart, Liver, Lung, Nerve, Ovary, Pancreas, Pituitary, Prostate, Skeletal Muscle, Skin, Small Intestine, Spleen, Stomach, Testis, Thyroid, Transformed Lymphocytes, Uterus, Vagina | 6.58E-03 | Alkaline phosphatase | |||
| ITLN1 | PrediXcan | Stomach | 1.04E-05 | Alkaline phosphatase, Absolute basophil count, Triglyceride, Viral load | Crohn’s disease | 18587394 |
| CELSR2 | PrediXcan | Brain*, Skeletal Muscle | 6.67E-06 | Cholesterol, Fasting cholesterol, Fasting LDL, LDL | Total cholesterol, LDL | 20686565, 17903299 |
| PSRC1 | PrediXcan | Lung*, Brain, Esophagus, Pancreas, Pituitary, Skeletal Muscle, Skin, Whole Blood | 8.47E-06 | LDL*, Cholesterol, Fasting cholesterol, Fasting LDL | Total cholesterol, LDL | 20686565, 17903299, 19936222, 17903299, 25101658 |
| UTMOST | Heart*, Adipose, Brain, Breast, Colon, Esophagus, Liver, Lung, Nerve, Ovary, Pancreas, Pituitary, Prostate, Skeletal Muscle, Skin, Testis, Thyroid, Uterus, Whole Blood | 1.75E-05 | ||||
| CETP | PrediXcan | Colon | 3.24E-17 | HDL*, Fasting HDL | HDL cholesterol | 25884002, 20686565 |
| MROH2A | PrediXcan | Adipose | 5.23E-03 | Total bilirubin | Bilirubin levels | 25884002,21646302 |
| UTMOST | Stomach | 5.97E-27 | ||||
| UGT1A1 | PrediXcan | Skin | 7.13E-07 | Total bilirubin | Bilirubin levels | 25884002,21646302 |
| UGT1A7 | UTMOST | Skin*, Adrenal Gland, Colon, Esophagus, Liver, Stomach | 5.15E-40 | Total bilirubin | Bilirubin levels | 25884002,21646302 |
| APOA1 | PrediXcan | Brain | 2.93E-02 | Triglyceride | Total cholesterol, Triglyceride, LDL, HDL | 20686565,17903299 |
| APOC3 | PrediXcan | Heart | 1.61E-02 | Triglyceride | Total cholesterol, Triglyceride, LDL, HDL | 20686565,17903299 |
Bolded tissues are known trait-related tissues.
denotes the most significant tissue and/or trait that were associated with genes.
q-value in the most significant tissue denoted by asterisk.
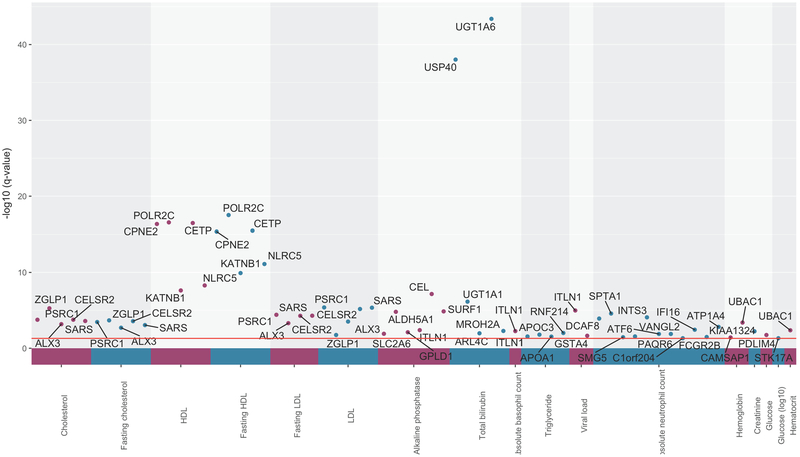
3.2. Single Tissue-based Analysis Found a Greater Number of Unique Gene-trait Associations
Single tissue-based analysis using PrediXcan identified 64 unique gene-trait association across different tissues (Figure 2). Some associations have been reported previously (Table 2). PrediXcan associated total bilirubin levels with UGT1A120 (skin, q-value = 7.1e-07) and MROH2A20 (adipose, q-value = 0.005), and LDL and cholesterol to CELSR230,38,39 (most significant with LDL in brain, q-value = 6.7e-06). HDL was associated with CETP20,32 (most significant in colon with q-value = 3.2e-17) and NLRC538 (adrenal gland, q-value = 7.8e-12). Triglyceride was associated with APOA130,39 (brain, q-value = 0.029) and APOC330,39 (heart, q-value = 0.016).
Figure 2.
Manhattan plot of gene-trait associations identified by PrediXcan. X-axis showed only significant traits. Y-axis was the q-value transformed by -log10. For simplicity, the plot only shows the lowest p-value of a gene-trait pair, which may appear in multiple tissues.
Single tissue-based analysis identified novel gene-trait associations, which warrants further investigation. One interesting example was the association of ITLN1 with multiple traits, including HIV-1 viral load, triglyceride, and total neutrophil count. As ITLN1 was reported in a previous Crohn’s disease study40, our result suggested an potential relationship between Crohn’s disease and HIV infection41.
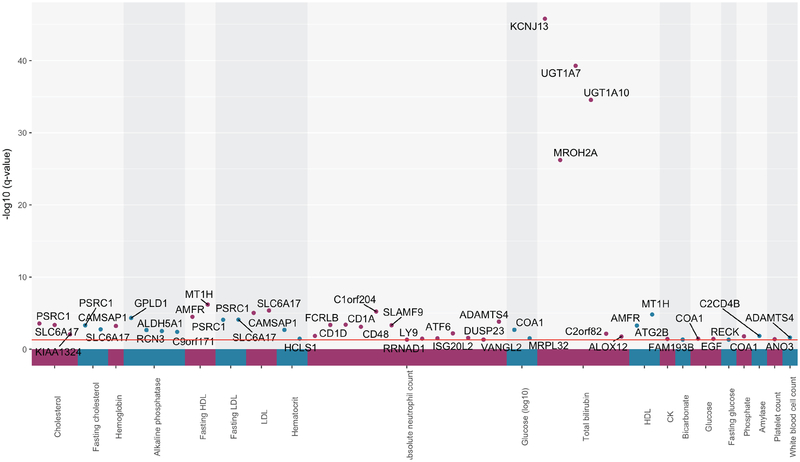
3.3. Integrative Tissue-based Analysis Found Multi-tissue Gene-trait Associations
Regardless of tissue, integrative tissue-based analysis using UTMOST identified 50 unique gene-trait pairs (Figure 3). Although it prioritized fewer genes, the integrative tissue-based analysis was more likely to prioritize multiple tissues where genes are expressed. For instance, PSRC1 is highly expressed in almost all tissues7. PSRC1-LDL and cholesterol associations were prioritized in at least ten more tissues by integrative tissue-based analysis Most importantly, they were found consistently in the liver which is critically involved in lipid regulation. There was some evidence for distinct associations identified via integrative tissue-based approach (Table 2), such as ADAMTS442 with white blood cell count (artery, q-value = 0.023), and AMFR43 with fasting HDL (most significant in heart, q-value = 3.2e-05).
Figure 3.
Manhattan plot of gene-trait associations identified by UTMOST. X-axis showed only significant traits. Y-axis was the q-value transformed by -log10. For simplicity, the plot only showed the most significant p-value of a gene-trait pair, which may appear in multiple tissues.
Other prioritized genes suggested novel associations and potential pleiotropy. Most prioritized genes have been associated with other traits by GWAS according to GWAS Catalog44. Similar to the single tissue-based approach, integrative tissue-based analysis prioritized total bilirubin-associated genes from the UGT1A45 gene locus (UGT1A7 and UGT1A10) across multiple tissues.
4. Discussions
This study investigated whether and how TWAS gene prioritization was influenced by tissue context-dependency of eQTLs by comparing two approaches, single tissue-based TWAS (implemented in PrediXcan) and integrative tissue-based TWAS (implemented in UTMOST). PrediXcan evaluated eQTLs’ effects in the context of a single tissue, which did not consider potential multi-tissue effects of eQTLs UTMOST estimated eQTLs’ effect in an integrative tissue setting and increase the chance of identifying multi-tissue eQTLs. We found that both types of analyses could replicate associations discovered by previous studies and identify novel ones. While there were a fair number of overlaps, the two types of analyses prioritized different sets of genes. Single tissue-based analysis identified more unique gene-trait associations. Integrative tissue-based analysis tended to prioritize the same associations in multiple tissues and most importantly association were found in tissues critically related to traits of interest. Results suggest that tissue context-dependency of eQTLs influenced TWAS gene prioritization results.
The comparison raised questions of power and type I error rate of tested TWAS approaches. Integrative tissue context has shown an improved power in identifying eQTLs. As such, integrative tissue-based analysis might have universally greater power in identifying trait-associated genes than single tissue-based analysis. However, in this study, single tissue-based analysis found more validated associations (Table 2). It is hard to tell if integrative tissue-based analysis has universally greater power as expected, whereas single tissue-based analysis happened to identify more false positives. It is also possible that one type of analysis outperformed the other at certain scenarios. A simulation study is necessary to discern these possibilities.
Similar to GWAS, prioritized genes might merely be tag genes for causal ones. Both kinds of analyses prioritized genes at the chromosome 1p13.3 locus where a lipid-related gene, SORT1, is located. Single tissue-based analysis associated multiple lipid-related traits with genes that neighbor SORT1, such as SARS, CELSR2, PSRC1, and ALX3, which all are in the 1p13.3 locus and the same topologically associating domain (TAD46,47). Besides PSRC1, integrative tissue-based analysis repetitively identified SLC6A17. Even though it is not adjacent to SORT1, this gene is in the 1p13.3 locus and might serve as a tag gene for causal one(s). Hence, for TWAS, prioritized genes might be merely tag genes and fine-mapping of causal genes may need a larger search boundary than GWAS, such as TADs.
Future investigation or validation experiments may be needed to explain the prioritized genes and/or tissues. For example, UGT1A1 glucuronidates bilirubin in the liver48, but single tissue-based analysis only identified a UGT1A1-total bilirubin association in skin. Further analysis found that there was no single UGT1A1 eQTL identified in liver by either PrediXcan or UTMOST trained on GTEx v6p or v7 data. It is likely that identification of UGT1A1 eQTLs is limited by tissue sample size (Nliver = 175) or genetic variants may regulate UGT1A1 via mechanisms other than transcriptional regulation. Another observation of this study was that genes adjacent to UGT1A1 sporadically showed up as significant in either single tissue-based or integrative tissue-based analysis, including USP40, UGT1A6, UGT1A7, UGT1A10, KCNJ13, and also MROH2A20. These genes span 1Mbp in chromosome 2 and locate within the same TAD46,47. The repetitive pattern may suggest a specific regulatory activity that targets the whole genetic region of KCNJ13-USP40-UGT1A-MROH2A.
TWAS can prioritize trait-related genes, which may be important for HIV-positive patients regarding genetically informed therapeutic development and drug safety. This study showed that TWAS were able to not only replicate known associations, but also identify novel gene-trait associations. It also suggested the importance of biological context in eQTL studies, and the ensemble of TWAS methods with different transcriptional regulation assumptions gave a more comprehensive picture of gene-trait relationships. In the future, we would like to perform cross-tissue TWAS analysis12,49, which aggregate gene-trait association information across all tissues and even across different consortia to further prioritize the trait-related genes and better describe the genetic architecture of complex diseases.
5. Acknowledgments
The authors are grateful to the many persons living with HIV who volunteered for A5095, A5142, ACTG 384, A5202, and A5257. In addition, they acknowledge the contributions of study teams and site staff for these protocols. We thank Paul J. McLaren, PhD (Public Health Agency of Canada, Winnipeg, Canada) for prior involvement and collaborations that used these genome-wide genotype data. Study drugs were provided by DuPont Pharmaceutical Company, Bristol-Myers Squibb, Inc., Agouron Pharmaceuticals, Inc., GlaxoWellcome, Inc., Merck and Co., Inc. Boehringer-Ingelheim Pharmaceuticals, Inc., Gilead Sciences, Inc., GlaxoSmithKline, Inc., Abbott Laboratories, Inc., Tibotec Therapeutics. The clinical trials were ACTG 384 (ClinicalTrials.gov: NCT00000919), A5095 (NCT00013520), A5142 (NCT00050895), A5202 (NCT00118898), and A5257 (NCT00811954). We also thank Yiming Hu from Yale University and Alvaro Barbeira and Dr. Hae Kyung Im from the University of Chicago for their support.
The project described was supported by Award Number U01AI068636 from the National Institute of Allergy and Infectious Diseases (NIAID) and supported by National Institute of Mental Health (NIMH), National Institute of Dental and Craniofacial Research (NIDCR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. This work was supported by the AIDS Clinical Trials Group funded by the National Institute of Allergy and Infectious Diseases (AI068636, AI038858, AI068634, AI038855). Additional grant support included AI077505, AI069439, TR000445, AI054999, AI116794 and AI110527.
Clinical Research Sites that participated in ACTG protocol A5202, and collected DNA under protocol A5128, were supported by the following grants from NIAID: AI069477, AI027675, AI073961, AI069474, AI069432, AI069513, AI069423, AI050410, AI069452, AI069450, AI054907, AI069428, AI069439, AI069467, AI045008, AI069495, AI069415, AI069556, AI069484, AI069424, AI069532, AI069419, AI069471, AI025859, AI069418, AI050409, AI069423, AI069501, AI069502, AI069511, AI069481, AI069465, AI069494, AI069472, AI069470, AI046376, AI072626, AI027661, AI034853, AI069447, AI032782, AI027658, AI-27666, AI058740, and AI046370, and by the following grants from the National Center for Research Resources (NCRR): RR00051, RR00046, RR025747, RR025777, RR024160, RR024996, RR024156, RR024160, and RR024160. Study drugs were provided by Bristol-Myers Squibb Co., Gilead Sciences, and GlaxoSmithKline, Inc.
Contributor Information
Binglan Li, Genomics and Computational Biology Program, University of Pennsylvania Philadelphia, PA 19104, USA.
Yogasudha Veturi, Department of Genetics, University of Pennsylvania Philadelphia, PA 19104, USA.
Yuki Bradford, Department of Genetics, University of Pennsylvania Philadelphia, PA 19104, USA.
Shefali S. Verma, Department of Genetics, University of Pennsylvania Philadelphia, PA 19104, USA
Anurag Verma, Department of Genetics, University of Pennsylvania Philadelphia, PA 19104, USA.
Anastasia M. Lucas, Department of Genetics, University of Pennsylvania Philadelphia, PA 19104, USA
David W. Haas, Departments of Medicine, Pharmacology, Pathology, Microbiology & Immunology, Vanderbilt University School of Medicine, Nashville, TN, and Department of Internal Medicine, Meharry Medical College, Nashville, TN, USA
Marylyn D. Ritchie, Department of Genetics, Institute for Biomedical Informatics, University of Pennsylvania Philadelphia, PA 19104, USA.
References
- 1.Mallal S et al. HLA-B*5701 screening for hypersensitivity to abacavir. N. Engl. J. Med 358, 568–579 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Rotger M et al. Gilbert syndrome and the development of antiretroviral therapy-associated hyperbilirubinemia. J. Infect. Dis 192, 1381–1386 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Holzinger ER et al. Genome-wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols implicates several CYP2B6 variants. Pharmacogenetics and Genomics 22, 858–867 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haas DW et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS 18, 2391–2400 (2004). [PubMed] [Google Scholar]
- 5.Lubomirov R et al. ADME pharmacogenetics: investigation of the pharmacokinetics of the antiretroviral agent lopinavir coformulated with ritonavir. Pharmacogenetics and Genomics 20, 217 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Yuan J et al. Toxicogenomics of nevirapine-associated cutaneous and hepatic adverse events among populations of African, Asian, and European descent. AIDS 25, 1271–1280 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consortium GTEx et al. Genetic effects on gene expression across human tissues. Nature Publishing Group 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B et al. Evaluation of PrediXcan for prioritizing GWAS associations and predicting gene expression. in 448–459 (WORLD SCIENTIFIC, 2017). doi: 10.1142/9789813235533_0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X et al. Functional Architectures of Local and Distal Regulation of Gene Expression in Multiple Human Tissues. American journal of human genetics 100, 605–616 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sul JH, Han B, Ye C, Choi T & Eskin E Effectively identifying eQTLs from multiple tissues by combining mixed model and meta-analytic approaches. PLoS Genet 9, e1003491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamazon ER et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 47, 1091–1098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Y et al. A statistical framework for cross-tissue transcriptome-wide association analysis. bioRxiv 286013 (2018). doi: 10.1101/286013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbins GK et al. Comparison of sequential three-drug regimens as initial therapy for HIV-1 infection. N. Engl. J. Med 349, 2293–2303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gulick RM et al. Triple-nucleoside regimens versus efavirenz-containing regimens for the initial treatment of HIV-1 infection. N. Engl. J. Med 350, 1850–1861 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Gulick RM et al. Three- vs four-drug antiretroviral regimens for the initial treatment of HIV-1 infection: a randomized controlled trial. JAMA 296, 769–781 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Riddler SA et al. Class-sparing regimens for initial treatment of HIV-1 infection. N. Engl. J. Med 358, 2095–2106 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sax PE et al. Abacavir-lamivudine versus tenofovir-emtricitabine for initial HIV-1 therapy. N. Engl. J. Med 361, 2230–2240 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daar ES et al. Atazanavir Plus Ritonavir or Efavirenz as Part of a 3-Drug Regimen for Initial Treatment of HIV-1: A Randomized Trial. Ann Intern Med 154, 445–456 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lennox JL et al. A Phase III Comparative Study of the Efficacy and Tolerability of Three Non-Nucleoside Reverse Transcriptase Inhibitor-Sparing Antiretroviral Regimens for Treatment-Naïve HIV-1-Infected Volunteers: A Randomized, Controlled Trial. Ann Intern Med 161, 461–471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore CB et al. Phenome-wide Association Study Relating Pretreatment Laboratory Parameters With Human Genetic Variants in AIDS Clinical Trials Group Protocols. Open Forum Infectious Diseases 2, ofu113–ofu113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verma A et al. Multiphenotype association study of patients randomized to initiate antiretroviral regimens in AIDS Clinical Trials Group protocol A5202. Pharmacogenetics and Genomics 27, 101–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner S et al. Quality control procedures for genome-wide association studies. Curr Protoc Hum Genet Chapter 1, Unit1.19–1.19.18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howie BN, Donnelly P & Marchini J A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genet 5, e1000529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.1000 Genomes Project Consortium et al. A map of human genome variation from population-scale sequencing. Nature Publishing Group 467, 1061–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Hall MA et al. PLATO software provides analytic framework for investigating complexity beyond genome-wide association studies. Nature Communications 8, 1167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grady BJ et al. Finding unique filter sets in PLATO: a precursor to efficient interaction analysis in GWAS data. Pac Symp Biocomput 315–326 (2010). doi: 10.1142/7628;page:string:Article/Chapter [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y & Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300 (1995). [Google Scholar]
- 30.Teslovich TM et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chasman DI et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet 5, e1000730 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willer CJ et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 45, 1274–1283 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kathiresan S et al. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med. Genet. 8 Suppl 1, S17 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strong A, Patel K & Rader DJ Sortilin and lipoprotein metabolism: making sense out of complexity. Curr. Opin. Lipidol 25, 350–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chambers JC et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet 43, 1131–1138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mero I-L et al. Oligoclonal band status in Scandinavian multiple sclerosis patients is associated with specific genetic risk alleles. PLoS ONE 8, e58352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.International Multiple Sclerosis Genetics Consortium (IMSGC) et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 45, 1353–1360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weissglas-Volkov D et al. Genomic study in Mexicans identifies a new locus for triglycerides and refines European lipid loci. J. Med. Genet 50, 298–308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer CJ et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 45, 1274–1283 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett JC et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet 40, 955–962 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lautenbach E & Lichtenstein GR Human immunodeficiency virus infection and Crohn’s disease: the role of the CD4 cell in inflammatory bowel disease. J. Clin. Gastroenterol 25, 456–459 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Astle WJ et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell 167, 1415–1429. e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benton MC et al. Mapping eQTLs in the Norfolk Island genetic isolate identifies candidate genes for CVD risk traits. American journal of human genetics 93, 1087–1099 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacArthur J et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Research 45, D896–D901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bielinski SJ et al. Mayo Genome Consortia: a genotype-phenotype resource for genome-wide association studies with an application to the analysis of circulating bilirubin levels. Mayo Clin. Proc 86, 606–614 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dixon JR et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature Publishing Group 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. bioRxiv 112268 (2017). doi: 10.1101/112268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tukey RH & Strassburg CP Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu. Rev. Pharmacol. Toxicol 40, 581–616 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Barbeira AN et al. Integrating Predicted Transcriptome From Multiple Tissues Improves Association Detection. bioRxiv 292649 (2018). doi: 10.1101/292649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jain D et al. Genome-wide association of white blood cell counts in Hispanic/Latino Americans: the Hispanic Community Health Study/Study of Latinos. Hum. Mol. Genet 26, 1193–1204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]