Abstract
Autism spectrum disorder (ASD) is a heritable neurodevelopmental disorder affecting 1 in 59 children. While noncoding genetic variation has been shown to play a major role in many complex disorders, the contribution of these regions to ASD susceptibility remains unclear. Genetic analyses of ASD typically use unaffected family members as controls; however, we hypothesize that this method does not effectively elevate variant signal in the noncoding region due to family members having subclinical phenotypes arising from common genetic mechanisms. In this study, we use a separate, unrelated outgroup of individuals with progressive supranuclear palsy (PSP), a neurodegenerative condition with no known etiological overlap with ASD, as a control population. We use whole genome sequencing data from a large cohort of 2182 children with ASD and 379 controls with PSP, sequenced at the same facility with the same machines and variant calling pipeline, in order to investigate the role of noncoding variation in the ASD phenotype. We analyze seven major types of noncoding variants: microRNAs, human accelerated regions, hypersensitive sites, transcription factor binding sites, DNA repeat sequences, simple repeat sequences, and CpG islands. After identifying and removing batch effects between the two groups, we trained an ℓ1-regularized logistic regression classifier to predict ASD status from each set of variants. The classifier trained on simple repeat sequences performed well on a held-out test set (AUC-ROC = 0.960); this classifier was also able to differentiate ASD cases from controls when applied to a completely independent dataset (AUC-ROC = 0.960). This suggests that variation in simple repeat regions is predictive of the ASD phenotype and may contribute to ASD risk. Our results show the importance of the noncoding region and the utility of independent control groups in effectively linking genetic variation to disease phenotype for complex disorders.
Keywords: Autism Spectrum Disorder, noncoding region, tissue-specific microRNAs, human accelerated regions, hypersensitive sites, transcription factor binding sites, DNA repeat sequences, simple repeat sequences, CpG islands, batch effects
1. Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by social impairments, communication difficulties, and restricted and repetitive patterns of behavior. ASD usually manifests in infants and children and presents a wide range of symptoms that vary from person to person. Currently, 1 in 59 children in the United States are affected, and prevalence rates are expected to increase drastically over the next decade.1 ASD is known to be highly genetic with a concordance rate between monozygotic twins of 77–99%.2,3 The genetic architecture of the disorder is known to be complex, with an estimated 1000 genes involved in disease susceptibility, spanning common, rare, and de novo variants.4,5
Models exploring the genetic basis of ASD typically focus on protein-coding genes; however, coding sequences account for only 1.5% of human DNA. The remaining segments of DNA are comprised of noncoding regions, which have been shown to play an important role in many genetic disorders. For example, recessive mutations in the PTF1A gene enhancer can cause pancreatic agenesis,6 a common mutation in the RET enhancer increases risk for Hirschprung disease,7 and mutations in topologically associating chromatin domains can cause limb malformation.8 Furthermore, a meta-analysis of over a thousand genetic association studies showed that most of the disease-associated single nucleotide variants identified by genome wide association studies (GWAS) lie in the noncoding region.9
However, the contribution of noncoding variants to ASD still remains unclear. A recent analysis of whole genome sequences of 516 children with ASD and their unaffected family members concluded that individuals with ASD tend to have significantly more de novo mutations in noncoding regions. The study evaluated two noncoding regions: untranslated regions (UTRs) of genes and conserved transcription factor binding sites that map to sites of DNase I hypersensitivity.10 However, a separate evaluation of the same dataset concluded that although individuals with ASD possessed a small excess of de novo mutations in noncoding regions, there were no significant results across over 50,000 regulatory classes after multiple testing correction.11
As shown by these studies, population genetic analyses typically classify unaffected family members as controls. However, we hypothesize that this assumption does not effectively elevate variant signal from the genome for ASD cohorts. For example, close relatives of individuals with ASD often exhibit autistic behaviors, such as social deficits and delayed speech.12,13 Thus, it is possible that family members possess a subclinical phenotype of ASD that may arise from genomic features shared with their affected children. Also, the diagnostic criteria for ASD were modified in 2013 with the release of the fifth edition of the Diagnostic and Statistical Manual of Mental Disorders. Most parents would have been evaluated using an earlier version of diagnostic criteria, making it possible that some would qualify for an ASD diagnosis by modern clinical standards.
In order to address this issue and to exacerbate signal in the noncoding region, we introduce a separate outgroup of patients with progressive supranuclear palsy (PSP), a neurodegener ative condition that causes difficulty with movement and thought.14 We chose this group of control patients because there is no known etiological overlap or comorbidity between PSP and ASD, and PSP is generally not heritable. There are some familial cases caused by a mutation in at least one copy of the gene MAPT on chromosome 17, but this is the only gene currently known to be linked with PSP.15 No patients in the control group exhibit symptoms of ASD. In this work, we use whole genome sequencing data from 2182 children with ASD and 379 PSP controls to investigate the role of noncoding variants in ASD susceptibility.
This study focuses on seven major noncoding regions: tissue specific microRNAs, hu man accelerated regions, hypersensitive sites, transcription factor binding sites, DNA repeat sequences, simple repeat sequences, and CpG islands. Tissue-specific microRNAs play important roles in the regulation of mRNA expression and the development of neurons, and recent studies have implicated a total of 219 microRNAs in the development of ASD.16 Human accelerated regions, which consist of only 49 highly-conserved segments in DNA, have been shown to regulate neural activity, with de novo copy number variations in these regions enriched in individuals with ASD.17 Hypersensitive sites are regulatory regions that are sensitive to cleavage by nucleases, and de novo mutations in these regions are significantly enriched in ASD probands.18 Transcription-factor binding sites are located in the noncoding regions of genes and assist in the regulation of transcription; variants in binding sites in MEGF10 and TCF4 have been associated with ASD and other intellectual disabilities.19,20 DNA Repeat sequences and simple repeat sequences are sequences of repeating base pairs, distinguished by the length of the repeating pattern, that have been linked to neuronal differentiation and brain development.21 Finally, CpG islands, which consist of regions with high frequencies of the cytosine and guanine base pairs, can have higher rates of methylation in individuals with ASD.22
2. Methods
2. 1. Data and Preprocessing
We analyzed 30x-coverage whole genome sequencing data from the Hartwell Foundation’s Autism Research and Technology Initiative (iHART); iHART has amassed data from 1006 multiplex families, each with at least two ASD-affected children. We also analyzed 30x-coverage whole genome sequencing data from 379 patients diagnosed with PSP. In order to limit batch effects due to inconsistencies in sequencing methodologies, we sequenced both populations at the New York Genome Center with Illumina HiSeq X instruments and utilized the same GATK variant calling pipeline; in addition, there is no sample overlap between the cohorts.
Chromosome coordinate lists for the seven noncoding regions were downloaded from the UCSC Genome Browser and the Regulatory Elements Database.23,24 Quality control was performed on the variant call format (VCF) files by removing all variants with high excess heterozygosity scores, which typically indicate sequencing artifacts or consanguinity within the population. We then filtered the variant-call format files to extract all variants within these regions that were present in both the PSP and ASD populations. We also removed all variants with a large proportion (greater than 20%) of missing sites.
2. 2. Accounting for Batch Effects
Batch effects present a major challenge when combining whole genome sequencing data across cohorts, resulting in many false positive associations.25 Batch effects can result from almost any step in the whole genome sequencing procedure, including library preparation, sequencing machine or center, sequencing depth, and variant calling pipelines.26 Several methods have been developed to mitigate these effects, but these procedures focus on reducing batch effects for datasets collected and analyzed independently.27,28 In our case, care was taken to sequence our ASD case and PSP control samples at the same center with the same platform and to analyze them using identical variant calling pipelines. In order to detect the more subtle batch effects that may remain, we expand on the method used by the UK10K project, detecting batch effects using a genome-wide association test with batch (ASD and PSP) as the phenotype.29 To do this, we performed a chi-squared test for each variant, comparing the number of individuals with homozygous reference, heterozygous, homozygous alternate, and missing genotypes between the two datasets. Any variants with a batch association p-value below 0.05 after applying a Bonferroni multiple testing correction were discarded, resulting in the removal of approximately 5% of variants. Figure 1 shows the number of variants within each region that passed our preprocessing and batch effect filters.
Fig. 1.
Number of noncoding variants of each type after applying preprocessing filters and removing variants affected by batch effects.
2. 3. Feature Representation and Logistic Regression Classifier
We designed a machine learning approach to determine if variation within noncoding regions could be utilized to predict ASD. In order to capture variant information from both the ASD and PSP populations, we constructed binary feature matrices for each of the seven noncoding regions. Each matrix includes 2561 rows corresponding to the 379 PSP control patients and 2182 ASD case patients; the columns represent the variants (shown in Fig. 1) associated with the region. We set each cell of the matrix as 1 if the individual expressed an alteration at the variant site (either heterozygous or homozygous alternate) and as 0 if the variant matched the reference sequence. Since several of these feature matrices included over one billion elements, all matrices were encoded in a customized sparse representation to ensure that machine learning would remain computationally tractable.
We created a logistic regression classifier with ℓ1 regularization in order to encourage the use of the smallest possible number of relevant features. 80% of the individuals in the dataset were randomly selected for inclusion in the training set, and the remaining 20% were added to the held-out test set; train and test sets were divided by family, so there is no familial overlap between sets. In order to address class imbalance between the case and control populations, we adjusted classifier weights such that they are inversely proportional to class sizes. We ran 5-fold cross validation in order to tune the level of regularization (represented by λ). Then, we evaluated performance on the held-out test set by measuring F1 scores, precision, recall, and AUC-ROC.
We extracted the top-ranked variants from each of the seven noncoding regions for further analysis by selecting the five variants from each classifier with the highest positive regression coefficient values as well as the five variants with the lowest negative coefficient values. We also confirmed that these variants were highly-ranked across multiple folds in our cross-validation tests.
2. 4. Validation
We validated the performance of our classifier using a held-out test set composed of 20% of the individuals from both cohorts. To demonstrate that our classifier can generalize, we also measured performance of our trained models on a completely independent cohort consisting of 517 ASD patients from the Simons Simplex Collection30 and 2054 control individuals from the 1000 Genomes Project.31 These cohorts were sequenced at different depths on different machines; however, the same GATK variant calling pipeline was utilized. We use this cohort to show that our classifier can effectively generalize to new populations and that we have adequately addressed batch effects in our training data.
Next, we devised a bootstrap test in order to determine if the seven groups of features used in this analysis were relevant predictors of ASD status when compared to random variants. To do so, we randomly sampled from the set of variants called in both the PSP and ASD cohorts. Feature matrices were designed according to the same procedures outlined in sections 2.1 and 2.2, and classifiers were trained on the random variants using the procedure outlined in section 2.3. This process was repeated between 20 and 100 times to obtain 95% confidence intervals. We ran separate bootstrap tests using different numbers of variants in order to account for the wide range in sizes of our variant sets; bootstrap test sizes range from 102 to 106 variants.
We also ran several tests to ensure that our logistic regression classifier was not biased by population stratification. Ethnicity is responsible for much of the variation in human genomes, so to ensure that population substructure was not confounding our results, we examined performance separately for Europeans and non-Europeans in our test set. Autism is also sex-biased, with males about 4 times more likely to be affected than females; in order to verify that our results are robust to differences in the sex chromosomes, we also examined test performance on males and females separately.
Finally, we evaluated the biological functions of top-ranked variants in order to determine potential correlation with the ASD phenotype.
3. Results
3. 1. Classifier Performance
Results from the logistic regression classifier as well as top-ranked variants are summarized in Figure 3. The classifier was evaluated on a held-out test set and was able to differentiate between ASD and PSP with high accuracy, with AUC-ROC values ranging from 0.600 to 0.960. The logistic regression classifier trained on variants located in simple repeat sequences showed the best performance out of all seven variant sets.
Fig. 3. Machine learning results.
We performed ℓ1-regularized logistic regression for each noncoding region. AUC-ROC, precision, recall, and F1 score show performance evaluated on the held-out test set. λ values for each noncoding region, as well as the number of remaining variants with nonzero coefficients remaining after feature selection, are listed. The 10 top-ranked variants for each classifier are listed in GRCh37 coordinates; the presence of variants with positive coefficient scores and the absence of variants with negative coefficient scores are likely to suggest the ASD phenotype.
3. 2. Bootstrap Test
To determine whether the seven types of noncoding regions we tested are more predictive of ASD status than random sets of variants, we performed a bootstrap test. Figure 4 shows the 95% confidence interval for AUC-ROC performance of random variant sets of various sizes on the held-out test set. As the number of variants used for prediction increases, the AUC values achieved by the classifier also increase. This is expected because as we incorporate more variants into our classifier, we become increasingly likely to by chance include ASD-associated variants or variants in linkage-disequilibrium with autism-associated variants. Furthermore, as the number of variants included in the classifier increases, any subtle batch effects missed by our filtering procedure will begin to influence results.
Fig. 4. Evaluating prediction performance of noncoding regions.
The blue shaded region shows the 95% confidence interval for AUC-ROC performance of randomly selected sets of variants. As the number of variants provided to the model increases, performance increases as well. Six of the non-coding regions we studied performed at or below the bootstrapped models. However, the simple repeat sequences variants significantly outperformed the bootstrap, suggesting that these noncoding variants may be associated with ASD.
We see that after accounting for variant set size, the microRNA, human accelerated region, and CpG island variant sets perform within the bootstrapped 95% confidence interval. Hypersensitive sites, transcription factor binding sites, and DNA repeat sequences all perform worse than random variant sets. These noncoding regions may not be associated with ASD, or our batch effect correction procedure may have been too stringent and removed important autism-associated signal. The classifier trained on simple repeat sequences is the only variant set that significantly outperforms the random bootstrap with a Bonferonni corrected p-value (accounting for the 7 tests performed) of 0.0287. This suggests that genetic variation within simple repeats may be associated with ASD risk.
3. 3. Performance on an Independent Test Set
In order to measure generalization ability, all seven classifiers were evaluated on an independent test set consisting of ASD patients from the Simons Simplex Collection and control individuals from the 1000 Genomes Project. AUC-ROC values ranged from 0.361 to 0.960, with most of the models suffering from a degradation in performance. However, the model trained on simple repeat sequences maintained a large AUC-ROC, consistent with the hypothesis that this region contains relevant signal for differentiating ASD and neurotypical individuals. These results are in agreement with our bootstrap analysis.
3. 4. Accounting for Population Substructure and Sex Differences
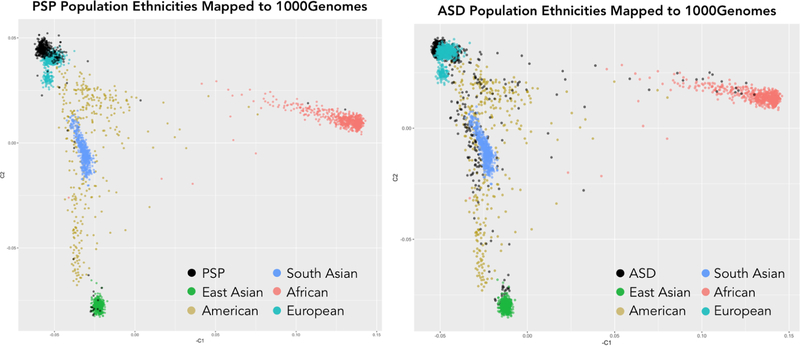
To show that our classifier trained on simple repeat sequences is robust to population substructure, we analyzed the population composition of our case and control groups. Figure 6 shows our case and control populations superimposed on ethnicity profiles from the 1000 Genomes Project. Our PSP population is predominantly of European descent, while the iHART population is more diverse.
Fig. 6. Population compositions of PSP and ASD cohorts.
These plots map the PSP and ASD populations to a principal components plot of the 1000 Genomes population in order to identify the ethnicity of individuals in our datasets.
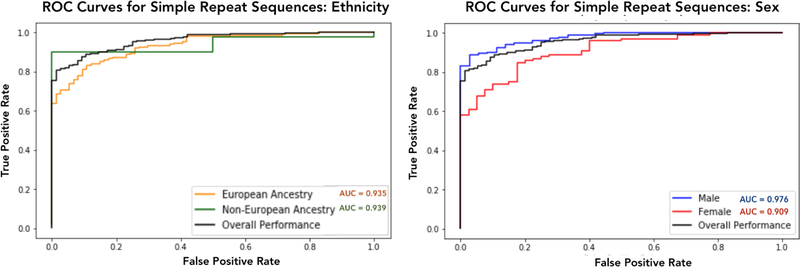
In order to ensure that this classifier is not biased by ethnicity, we evaluated its test performance on individuals of European and non-European descent separately. Figure 7 shows that it performs equally well on individuals of European or non-European ancestry, increasing our confidence that our results are not confounded by population substructure. We also evaluated differences in classification performance between males and females, also shown in Figure 7.
Fig. 7. ROC curves for the classifier trained on simple repeat sequences across four splits of the held-out test set.
The plots show that the classifier yields similar results on the European and non-European population. However, classifier performance is higher across males than females.
Our classifier is better able to predict ASD affected status in males than in females. This is interesting because ASD has a strong male bias with male children being four times more likely to develop autism than female children.32
3. 5. Biological Functions
We evaluated the biological functions of all 70 top-ranked variants in order to identify potential correlations with the ASD phenotype. Since each variant either occurs in the intronic region of a gene or in an intergenic region between two genes, we generated a comprehensive list of genes associated with top-ranked variants. This resulted in a set of 98 genes, which we utilized to evaluate biological evidence. In the tissue-specific microRNA regions, a variant at position 200,938,662 in chromosome 1 is located in the intronic region of KIF21B, a gene that regulates synapse function and morphology of neurons; this gene is also known to play a role in learning and memory.33 A variant at position 124,950,150 in chromosome 3 is located in ZNF148, which has been linked with developmental delays.34 A top-ranked variant in chromosome 12 is located in the intronic region of CD4, a gene expressed in regions of the brain that is known to be a mediator of neuronal damage.35 In noncoding regions containing DNA repeat sequences, gene GFOD1 contains a variant at location 13,509,234 on chromosome 6 and has been linked with Attention Deficit-Hyperactivity Disorder, a common comorbid condition of ASD.36 Similarly, a top-ranked variant in a simple repeat sequence in chromosome 7 is located within the intronic region of gene DGKI; this gene has been linked with dyslexia, which is also a comorbid condition of ASD.37 In addition, a variant at chromosome 17 in a simple repeat region is located within gene SHISA6, a regulator of synaptic transmission.38
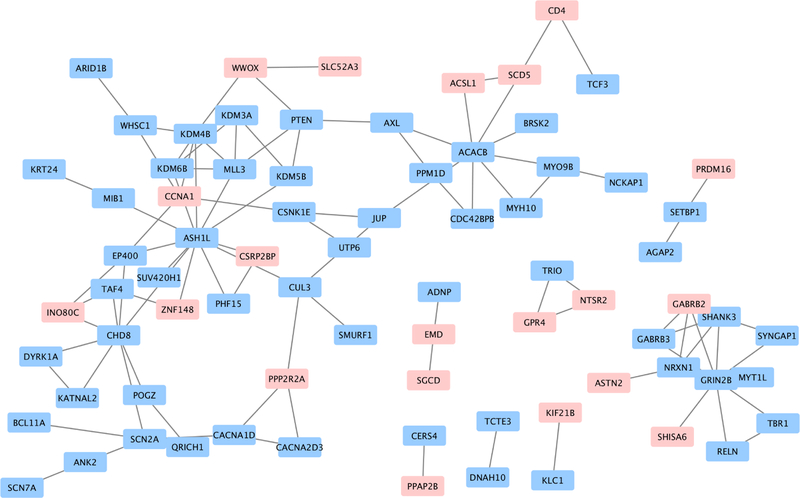
In order to analyze the relationship between the 98 identified genes and a set of 109 genes known to confer elevated ASD risk, we constructed a protein-protein interaction network in STRING, as shown in Figure 8.39 Edges are derived from text-mining, experiments, databases, co-expression, neighborhood, gene fusion, and co-occurrence. The network showed that twenty newly-identified genes are closely connected to known ASD-linked genes.
Fig. 8. Gene interaction network.
Interactions between genes previously linked with autism (in blue) and genes associated with the noncoding variants identified in this analysis (in pink) are shown in the figure. 20 identified genes interact closely with known ASD-risk genes. Notably, the gene CCNA1 is known to interact with 5 known ASD-linked genes.
4. Discussion
By utilizing outgroup machine learning to investigate the noncoding space, we were able to identify single nucleotide variants potentially associated with ASD. Biological validation of genes associated with top-ranked variants revealed a highly interconnected gene network, suggesting that identified genes interact closely with ASD-linked genes and may contribute to the ASD phenotype. Out of the seven regions analyzed in this work, the classifier trained on simple repeat regions demonstrated the strongest performance. Simple repeat sequences, also known as microsatellites, consist of repetitive sequences of one to ten base pairs; these regions are known to be extremely susceptible to mutations.40 More than twenty neurodevelopmental and neurodegenerative conditions, many of which are comorbid with ASD, have been linked to unstable expansion of repeat sequences and consequent loss of protein function.41 In addition, variation in promoter microsatellites of the gene AVPR1A has been implicated in increased susceptibility to ASD in an Irish population.42 In this work, the classifier trained on simple repeat sequences significantly outperformed the random bootstrap test, indicating a potential correlation between variants in this region and the ASD phenotype; this was further supported by a biological analysis of top-ranked variants in simple repeat regions that revealed two genes associated with neural function.
Thus, our outgroup machine learning approach to elevate hidden signal in ASD genomes can effectively evaluate feature representations of the noncoding space; however, this method has potential limitations, including batch effect correction and population stratification.
Current methods for addressing batch effects in whole genome sequencing data are meant to capture major differences in sequencing pipelines and are therefore quite stringent; the Type 2 Diabetes Consortium uses a series of quality control filters to identify batch effects resulting in a loss of 9.9% of called SNPs.43 Our method for batch effect correction, adapted from the algorithm used by the UK10K Project,29 is less conservative, discarding just under 5% of called SNPs. We believe this is appropriate since the batch effects in our dataset are much more subtle than those encountered by large consortia. Since our samples were sequenced at the same sequencing center with the same protocols and variant calling pipeline, we were able to control for many of the variables that could introduce batch effects. However, differences between populations in both cell type and the joint variant calling process could still create batch effect biases. The ASD samples were sequenced from lymphoblastoid cell lines while the PSP samples were sequenced from whole blood. Furthermore, while the same variant calling pipeline was used on both samples, GATK performs joint genotyping, a procedure that uses other samples in the cohort to resolve sequencing errors; since the two cohorts were run through the variant calling pipeline separately, subtle batch effects could have been introduced.
Regardless of batch effects, there remains the fundamental issue of population stratification in the merged dataset, especially since the initial cohorts were not drawn from the same ancestral or ethnic group. In order to establish a control for stratification, we created a null distribution by performing a bootstrap on successively larger variant sets, as reflected in Figure 4. High-performing null models likely do not reflect any neurological phenotype; rather, they represent the effect of divergent ancestry between the ASD and PSP cohorts. Interestingly, only the classifier trained on simple repeat sequences exceeded the null distribution for models of its size, suggesting a potential link with ASD.
Further analysis is needed to understand the biological consequences of these results. 40% of the top-ranked variants discovered in this analysis lie in intergenic regions; these may be enhancers to nearby genes, and we intend to explore associations between these variants and specific genes in a followup study. In addition, variants within simple repeat regions are challenging to call at low depth; in our current analysis, the top ten variants in simple repeat regions have an average read depth of 30.23 across the SSC dataset and an average read depth of 6.21 across the 1000 Genomes control dataset. In the future, we will validate our classifier using an independent test set sequenced at a higher depth of coverage.
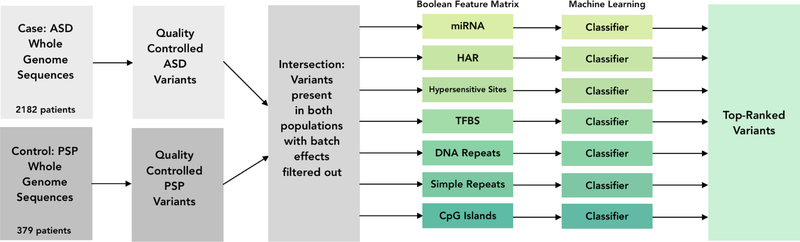
Fig. 2. Machine learning pipeline.
Variants were called separately for cases and controls. The variant calls were then merged and a batch-effect filter was applied. Feature matrices were created for each of the seven noncoding regions and served as input to A1-regularized logistic regression classifiers. Finally, the top-ranked features were extracted from each classifier.
Fig. 5. Performance on an independent test set.
This figure includes AUC-ROC values from validation on an independent cohort consisting of individuals from the Simon’s Simplex Collection and the 1000 Genomes Project. Only the classifier trained on simple repeat sequences is able to generalize.
5. Acknowledgments
This work was supported by the Hartwell Foundation award to D.P. Wall and the Hartwell Autism Research and Technology Initiative (iHART). This work was also supported by Bio-X and the Precision Health and Integrated Diagnostics (PHIND) Center at Stanford University.
References
- 1.Fombonne E, Journal of Child Psychology and Psychiatry 59, 717 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Hallmayer J, Cleveland S, Torres A et al. , Archives of General Psychiatry 68, 1095 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colvert E, Tick B, McEwen F, Stewart C et al. , JAMA Psychiatry 72, 415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geschwind DH et al. , The Lancet Neurology 14, 1109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramaswami G and Geschwind DH, Genetics of autism spectrum disorder, 1 edn. (Elsevier B.V, 2018). [DOI] [PubMed] [Google Scholar]
- 6.Weedon MN, Cebola I, Patch A-M, Flanagan SE, De Franco E, Caswell R, Rodríguez-Seguí SA, Shaw-Smith C, Cho CH, Allen HL et al. , Nature genetics 46, p. 61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emison ES, McCallion AS, Kashuk CS, Bush RT, Grice E, Lin S, Portnoy ME, Cutler DJ, Green ED and Chakravarti A, Nature 434, p. 857 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R et al. , Cell 161, 1012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leslie R, O’Donnell CJ and Johnson AD, Bioinformatics 30, 185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner T, Coe P, Dickel D, Hoekzema K, Nelson B, Zody M, Kronenberg Z, Hormozdiari F, Raja A, Pennacchio L, Darnell R and Eichler E, Cell 171, 710 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werling D, Brand H, An J et al. , Nature Genetics 50, p. 727736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piven J, American Journal of Medical Genetics 105, 34 (2001). [PubMed] [Google Scholar]
- 13.De Groot K and Van Strien JW, Advances in Neurodevelopmental Disorders 1, 129 (2017). [Google Scholar]
- 14.Morris HR, Neurodegeneration, p. 72 (2017).
- 15.Borroni B, Agosti C, Manani E, Luca MD and Padovani A, Current Medicinal Chemistry 18 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Hicks S and Middleton F, Front Psychiatry 7, p. 176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doan R, Bae B, Cubelos B, Nieto M and Walsh C, Cell 167 (2016). [Google Scholar]
- 18.Turner T, Hormozdiari F, Duzyend M, McClymont S et al. , American Journal of Human Genetics 98, 58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X, Qin J, You Y et al. , Scientific Reports 7 (2017). [Google Scholar]
- 20.Forrest M, Hill M, Kavanagh D et al. , Schizophrenia Bulletin (2017). [DOI] [PMC free article] [PubMed]
- 21.Fondon J, Hammock E, Hannan A and King D, Neuroscience Trends 8, 328 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Like Y, Hannan A and Craig J, Frontiers in Neurology 6, p. 107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent W, Sugnet C, Furey T et al. , Genome Research 12, 996 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheffield N, Thurman R, Song L, Safi A et al. , Genome Research 23, 777 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K and Irizarry RA, Nature Reviews Genetics 11, p. 733 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tom JA, Reeder J, Forrest WF et al. , BMC bioinformatics 18, p. 351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leek JT, Nucleic Acids Research 42, p. e161 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taub MA, Bravo HC and Irizarry RA, Genome medicine 2, p. 87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consortium U et al. , Nature 526, p. 82 (2015).26367797 [Google Scholar]
- 30.Chaste P, Klei L, Sanders SJ et al. , Biological Psychiatry 77, 775 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auton A, Abecasis GR, Altshuler DM et al. , Nature 526, 68 (2015).26432245 [Google Scholar]
- 32.Werling DM and Geschwind DH, Current opinion in neurology 26, p. 146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muhia M, Thies E, Labonte D et al. , Cell 15, 968 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens S, van Essen A, van Ravenswaiij C et al. , Genome Medicine 8, p. 131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byram SC, Carson MJ, DeBoy CA, Serpe CJ, Sanders VM and Jones KJ, Journal of Neuroscience 24, 4333 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lasky-Su J, Neale B, Franke B et al. , American Journal of Medicine 147B, 1345 (2008). [Google Scholar]
- 37.Matsson H, Tammimies K, Zucchelli M et al. , Behavioral Genetics 41, 134 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Klaassen R, Stroeder J, Coussen F et al. , Nature Communications 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szklarczyk D, Franceschini A, Wyder S et al. , Nucleic Acids Research 43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MLC Vieira ADCM, Santini L, Genetics and Molecular Biology 39, 312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gatchel J and Zoghbi H, Nature Reviews Genetics 6, 743 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Tansey K, Hill M, Cochrane L, Gill M, Anney R and Gallagher L, Molecular Autism 2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuchsberger C, Flannick J, Teslovich T et al. , Nature 536, p. 41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]