Abstract
Single-cell RNA sequencing (scRNA-seq) techniques have been very powerful in analyzing heterogeneous cell population and identifying cell types. Visualizing scRNA-seq data can help researchers effectively extract meaningful biological information and make new discoveries. While commonly used scRNA-seq visualization methods, such as t-SNE, are useful in detecting cell clusters, they often tear apart the intrinsic continuous structure in gene expression profiles. Topological Data Analysis (TDA) approaches like Mapper capture the shape of data by representing data as topological networks. TDA approaches are robust to noise and different platforms, while preserving the locality and data continuity. Moreover, instead of analyzing the whole dataset, Mapper allows researchers to explore biological meanings of specific pathways and genes by using different filter functions. In this paper, we applied Mapper to visualize scRNA-seq data. Our method can not only capture the clustering structure of cells, but also preserve the continuous gene expression topologies of cells. We demonstrated that by combining with gene co-expression network analysis, our method can reveal differential expression patterns of gene co-expression modules along the Mapper visualization.
Keywords: single-cell RNA sequencing, topological data analysis, Mapper
1. Introduction
Single-cell RNA sequencing (scRNA-seq) has provided an unprecedented view of heterogeneity in cell populations. While traditional bulk RNA-seq experiments quantify molecular states of cells by estimating mean expression profiles of millions of cells, scRNA-seq techniques can generate expression profiles of individual cells. Such improvement of resolution has made scRNA-seq a powerful tool to discover previously unknown cellular heterogeneity and functional diversity.1
However, the improvement of scRNA-seq techniques also provides new challenges in data analysis and interpretation. Firstly, the dimensionality of scRNA-seq data is very high. Typical scRNA-seq data usually contains RNA sequencing profile of over thousands of genes. Secondly, the number of cells is large. Recent high-throughput platforms are capable of generating data for thousands of cells. Thirdly, different scRNA-seq platforms and biological experiments may produce data with different biases or distributions, which introduces difficulty in comparing data across different platforms.
To address the aforementioned challenges, many computational tools have been developed to analyze and visualize high-dimensional scRNA-seq data, including Monocle,2 Wishbone,3 SMILE4 and FVFC.5 However, due to its advantage of detecting clusters in low dimensional space, t-distributed Stochastic Neighbor Embedding (t-SNE)6 has become the most commonly used technique in scRNA-seq data visualization to identify cell type clusters.7,8 However, cells in a population do not always form clustering structures. Oftentimes, they show continuous trajectories in space of gene expression profiles.3 Therefore there is a need for a scalable method to capture such continuous gene expression topologies of cells.
Mapper9 is a Topological Data Analysis (TDA) approach that extracts descriptions of high dimensional datasets in the form of simplicial complexes. As a method of representing data using topological networks, Mapper possesses several advantages when analyzing and visualizing scRNA-seq data. Firstly, similar to t-SNE, Mapper can preserve small-scale similarities among data points. However, while methods like t-SNE often tear apart the continuous structure in the original high dimensional space, Mapper can instead capture such continuous variation. Secondly, topological features are robust to small distortions of data, which makes Mapper robust to noise. Thirdly, Mapper captures the shape of the data by the distance functions chosen instead of depending on a specific coordinate system. Such coordinate-free approach gives Mapper the ability to compare data across different platforms.10 Fourthly, Mapper produces a compressed representation of the shape of the dataset using a graph, where each node represents a cluster of data points. While t-SNE relies on approximation approaches11 to scale to large datasets, Mapper is highly scalable to recent scRNA-seq datasets with large number of cells. Finally, Mapper can view data at multiple resolution.9 This means that Mapper is able to discover patterns at different scales and capture details in large datasets with complex structures. Mapper has been applied to many biomedical problems, including identifying patient subsets in breast cancer,12 analyzing murine embryonic stem cell (mESC) differentiation13 and studying dynamical organization of the brain.14
In this paper, we used Mapper to visualize scRNA-seq data in order to extract different cell types and understand the lineage relationship among them. Our approach is innovative in the following ways. Firstly, we visualize scRNA-seq data as combinatorial graphs through Mapper to capture topological features of the data. Mapper can visualize the continuous trajectory of cells over the space of gene expression profiles, which compliments the methods that recover the clusters of cells. Secondly, Mapper enables researchers to explore different biological meanings of scRNA-seq data by using different filter functions. In this paper, we took advantage of gene co-expression network analysis (GCNA) and focused on gene co-expression modules with biological functions. We further summarized gene modules into “eigengenes” and incorporated them into Mapper as filter functions or coloring of nodes. We applied our method on two large scRNA-seq datasets (melanoma and pancrease cell) and demontrated that our method can capture topological structures of scRNA-seq data. Combined with GCNA, Mapper also reveals that gene co-expression modules are differentially expressed between certain branches in the visualization and each is enriched with biological functions relevant to the corresponding cell types.
2. Methods
2.1. Data
In this paper, we applied our method on two large scRNA-seq datasets of melanoma tumor cells (GSE72056)7 and human pancreas cells (GSE85241).8 Details of datasets are summarized in Table 1 and both datasets can be accessed through NCBI Gene Expression Omnibus.
Table 1.
Summary of datasets used in this study.
| Dataset | Number of cells | Number of genes | Cell types(number of cells) |
|---|---|---|---|
| GSE72056 | 4645 | 23686 | unresolved(132), malignant(1257) non-malignant(3256: T(2040), B(512), Macro(119), Endo(62), CAF(56), NK(51), other(416)) |
| GSE85241 | 2126 | 19126 | acinar(219), alpha(812), beta(448), delta(193), ductal(245), endothelial(21), epsilon(3), mesenchymal(80), pp(101), unclear(4) |
The expression level of gene i in cell j was quantified as Gij = log2(TPMij/10 + 1), where TPMij is transcript-per-million (TPM) for gene i in cell j. In scRAN-seq, due to the low number of RNA transcriptomes, dropout events, where expression measurements of some random transcripts are missed as zeroes, often occur. To account for the dropout events, we filtered out genes with the lowest m_thr percent of mean expression level or the lowest v_thr percent of variance. We used m_thr = 95 and v_thr = 95 for the melanoma cell dataset and retained 775 genes after pre-processing. We used m_thr = 90 and v_thr = 90 for the pancreas cell dataset and retained 500 genes after pre-processing.
2.2. Mapper
Mapper, introduced by Singh et al.,9 is one of the most commonly used TDA approaches. Mapper contains four steps: filtering, binning, clustering and graph generation and we reiterate them as Algorithm 1.
| Algorithm 1 Mapper on scRNA-seq data |
|---|
| Input: a pre-processed gene expression matrix G |
| Output: a graph Grph capturing topological features of G |
| 1. filtering: apply a filter function f on G |
| 2. binning: fragment the range of f into overlapping intervals and separate G into overlapping bins {B1, B2, …, Bn} |
| 3. clustering: apply hierarchical clustering on each bin and get a series of overlapping clusters C |
| 4. graph generation: create a graph Grph to capture the shape of G based on C |
Filtering step uses a filter function f to project gene expression data G to a lower dimensional space, usually or . Different filter functions may generate networks with different shapes and researchers could view data from different perspectives by choosing different filter functions. One of the commonly used filter functions is eccentricity, which is a family of functions capturing the geometry of data. For cell ci ∈ G, given p with 1 ≤ p < +∞, we define the eccentricity of ci as
| (1) |
where ci,cj ∈ G. d(ci,cj) is the distance between ci and cj and N is the number of cells in G. When p = +∞, we define L∞ eccentricity as . L∞ eccentricity has being used as a filter function to identify patient subtypes in breast cancer.10 Dimension reduction methods such as Principle Component Analysis (PCA),10 Multi-Dimensional Scaling (MDS)13 and t-SNE14 can also be used as filter functions. Researchers can also choose their own pre-computed data as filter functions.
After applying f on G, range of f is fragmented into overlapping intervals S = {S1, S2, …, Sn}. The size of each interval is determined by several parameters: number of intervals n, fraction of overlap between adjacent intervals p and the interval generation method, which includes generating each interval with the same size or with the same number of cells. Cells in G are then put into a series of overlapping bins B = {B1,B2, … ,Bn} according to S.
Hierarchical clustering is used to cluster cells in each bin Bi and researchers could choose from different distance metrics and linkage functions. A histogram is plot with the threshold values for each transition in the hierarchical clustering dendrogram and the number of clusters ki is determined by the number of local maximas in the histogram.
After the clustering step, cells in G have been separated into a series of clusters . A graph Grph is constructed where each cluster Ci ∈ C is represented as a node and an edge is drawn between Ci and Cj if Ci ∩ Cj ≠ ∅. Grph is the output of Mapper and can capture the topological features of the original data G.
2.3. Gene co-expression network analysis
For GCNA, we applied local maximal Quasi-Clique Merger (lmQCM)15 to identify densely connected modules such as quasi-cliques in weighted gene co-expression networks. Different from methods like WGCNA,16 which partition genes into disjoint sets and do not allow overlap between clusters, lmQCM is a greedy approach that allows genes to be shared among multiple clusters. This is consistent with the fact that genes could participate in multiple biological processes. The lmQCM algorithm has four parameters: γ, α, t and β. γ determines if a new module can be initiated by setting the weight threshold for the first edge of the module, and has the largest influence on the result. We used γ = 0.2, α = 1, t = 1 and β = 0.4 in our experiments. After identifying gene co-expression modules, we further summarized them into “eigengenes” by taking the first principle component of gene expression profiles of the modules.
We used ToppGene Suite17 for gene set functional enrichment analysis to determine if gene modules detected by lmQCM are biologically meaningful. ToppGene finds biological annotations such as Gene Ontology (GO) items that are significant in a set of genes. To provide meaningful results, we only performed functional enrichment analysis on gene modules that contain at least 10 genes and at most 500 genes.
2.4. Visualizing networks
The output of Mapper on scRNA-seq data is a network where each node is a cluster of cells and each edge means that two clusters share some common cells. We used a force directed layout algorithm to calculate the position of each node, which means the positions of individual nodes do not have particular meanings and only the connections between nodes are informative.
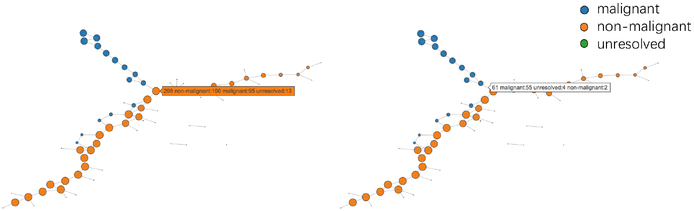
Each node contains several features of the cluster it represents. The size of a node is proportional to the number of cells in the node. The color of each node represents a specific property of cells, which could be determined by users. For quantitative features, such as the expression level of a gene or an eigengene, mean value is used to represent the cluster. For categorical features, such as types of cells, the majority category is used to represent the cluster. Pie charts is another option to visualize the category composition of the nodes, but it could clutter the visualization, making perception of composition difficult. However, to compensate the information loss by using the majority as representation, we utilized an interactive visualization technique that allows users to get the cell type composition of a node or an edge by hovering over it. An example of this is shown in Figure 1.
Figure 1.
Cursor hovering for detailed information: hovering over a node (left) and hovering over an edge(right).
3. Results
3.1. Visualizing melanoma cells using Mapper
We first compared Mapper with several commonly used dimensionality reduction algorithms (t-SNE,6 PCA, Isomap,18 LLE19 and Spectral Embedding20) by visualizing the melanoma cell dataset and the results are shown in Figure 2. We also compared Mapper with one of the state-of-the-art scRNA-seq visualization methods, Monocle 2.2 Each node in the Mapper visualization represents a cluster of cells while each point in other visualizations represents a cell.
Figure 2.
Visualization of melanoma cells.
We observe that all above algorithms are capable of separating malignant cells from non-malignant ones. Particularly, t-SNE separates malignant cells from different tumors into different clusters, which implies that t-SNE may be influenced by batch effects of different cell populations besides differentiating malignant and non-malignant cells. This also suggests that t-SNE often tends to break the continuous trajectory of cells in the space of gene expression profiles. On the other hand, by visualizing the shape of the data, Mapper not only separates malignant cells from non-malignant cells, but also preserves the continuous structure in scRNA-seq data by visualizing malignant cells as a branch separating from non-malignant cells. Monocle 2 also provides an interesting visualization, where non-malignant cells branch out into two clusters of malignant cells. However, further analysis did not find different patterns in expression levels of gene co-expression modules between the two malignant cell clusters.
Another advantage of Mapper is that it can view data under different resolutions and capture patterns of different scales. Figure 3 shows a series of visualizations of melanoma cell dataset with different number of bins (nbins) in the binning step. We observe that the graph representation of the data is coarse when the number of bins is low, which captures the global structure of the data. As the number of bins increases, more detailed structures are revealed and we can detect patterns at a higher resolution.
Figure 3.
Mapper visualization of melanoma cells with different number of bins.
Moreover, we could still take the advantage of t-SNE within the Mapper framework by using t-SNE as the filter function. Using t-SNE as the filter function can produce a compressed representation that captures the clustering structure of the t-SNE visualization.
3.2. Using eigengenes for node coloring in Mapper
GCNA can identify gene co-expression modules with potential biological meanings, which helps the interpretation of our visualizations. One way to utilize information from GCNA is to use expression profiles of eigengenes to color the nodes in graphs produced by Mapper, as shown in Figure 4.
Figure 4.
Mapper visualization of melanoma cells with coloring of eigengene expression profiles.
Two gene co-expression modules were identified in the melanoma dataset by applying lmQCM on the pre-processed scRNA-seq data. Gene set enrichment analysis results with false discovery rate corrected p values generated by ToppGene Suite are summarized in Table 2 Figure 4 shows obvious difference of the two eigengene expression profiles between the malignant branch and non-malignant cells. This is consistent with the fact that biological processes such as cell activation, immune response and regulation of cell migration are strongly associated with malignancy of cells.
Table 2.
Gene co-expression modules in the melanoma dataset.
| Module ID | Number of genes | Enriched GO items (p value) |
|---|---|---|
| 1 | 26 | G0:0001775 cell activation (1.983E-12) GO:0006955 immune response (1.983E-12) GO:0045321 leukocyte activation (1.983E-12) |
| 2 | 16 | GO:0042470 melanosome (7.681E-7) GO:0030334 regulation of cell migration (5.356E-4) GO:2000145 regulation of cell motility (5.356E-4) |
We further investigated genes in eigengene 2 and two non-overlapping sub-modules were discovered. One contains five genes (TYR, CTSB, MLANA, GPNMB, PMEL) which enriches with proteins associated with melanosome - a structure associated with melanocytes and potentially melanoma. The other contains seven genes (TIMP1, TMSB4X, SGK1, GSN, LGALS3, SERPINE2, APOD) and enriches with regulation of cell migration and extracellular matrix. Figure 5 shows that genes in both sub-modules have lower expression level in malignant cells than non-malignant cells, which indicates functions related to normal melanosome and cell migration activities may be disrupted in malignant melanoma cells.
Figure 5.
Using eigengene expression profiles of two sub-modules in eigengene 2 as coloring of nodes. The first sub-module is enriched in melanosome proteins and the second sub-module is enriched in cell migration and extracellular space.
3.3. Using eigengenes as filter functions in Mapper
By using different filter functions, researchers can rapidly explore different biological hypotheses in scRNA-seq data through Mapper. So, we can also incorporate GCNA into Mapper by using expression profiles of eigengenes as filter functions. Figure 6 shows L∞ eccentricity, a commonly used filter function, fails to separate different types of human pancreas cells. On the other hand, t-SNE completely separate different cell types into different clusters. Since similarities between points with long distances are not reliable in t-SNE visualization, we are not able to investigate the relationships between different cell types through t-SNE. By using the expression level of eigengene 2 as a filter function, Mapper can separate different types of pancreas cells with a branch-shape visualization, which preserves the continuity of cells at the same time. More specifically, the exocrine compartment of pancreas, including acinar cells and ductal cells, is visualized as a branch separating from the endocrine compartment. The shape of the visualization is consistent regardless of the linkage function in the clustering step (single or complete linkage). Enrichment analysis shows that eigengene 2 is associated with delta cells and PP cells of mouse adult pancreas in co-expression atlas, which indicates that eigengene 2 may contain genes conserved between species. This suggests that the eigengene 2 is worthy of further investigation for deeper understanding in pancreatic biology.
Figure 6.
Visualization of pancreas cells using t-SNE and Mapper.
Moreover, we can combine multiple eigengene profiles as filter functions. From Figure 7, we observe that using a single eigengene as the filter function can only differentiate some of the cell types in the melanoma cell dataset. However, combining two eigenegenes as the filter function can further differentiate different types such as macrophages and endothelial cells. Comparing to t-SNE, Mapper visualization using two eigenegenes not only preserves the similarities between B cells and T cells, but also reduces the batch effect by visualizing all malignant cells as a group of tightly connected clusters.
Figure 7.
Visualization of melanoma cells using t-SNE and Mapper with eigengene expression profiles as filter functions.
3.4. Mapper reveals potential functional relationships between exocrine cells in pancreas
To further investigate the biological significance of Mapper visualization, we used the expression levels of established marker genes for each of the six main pancreatic cell types in the human pancreas cell dataset to color the nodes in our visualization. From Figure 8, we observe that expression levels of marker genes in endocrine cells show significant difference in the corresponding cell types. However, KRT19 and PRSS1 could not well separate ductal cells and acinar cells in the exocrine branch, which indicates potential relationships within exocrine cells. We further applied GCNA on ductal cells and acinar cells separately, as well as combined together. Two gene co-expression modules were identified across all three cell populations. However, as shown in Figure 9, module 1 in the combined cell population shows very small overlap with all the gene modules identified from the ductal-only and acinar-only population. Enrichment analysis shows that module 1 in the combined cell population is associated with neuron part (GO:0097458, p = 1.141E-3) and extracellular space (GO:0005615, p = 5.735E-3), which could relate to enzymes production activities of acinar cells. Module 1 also enriches secretory granule (GO:0030141, p = 7.314E-3), which could relate to the production of bicarbonate-rich secretion in ductal cells.
Figure 8.
Mapper visualization of pancreas cells, with coloring of marker genes expression levels.
Figure 9.
Gene co-expression module detected only in combined cell population of acinar and ductal cells.
4. Conclusion
The scRNA-seq technology is becoming a common approach to study cellular heterogeneity and dynamic cellular process. Visualization techniques can help researchers effectively extract that information from scRNA-seq data. In this paper, we applied a TDA algorithm, Mapper, on two large scRNA-seq datasets. We showed that Mapper is able to preserve the continuous structure in gene expression profiles while effectively differentiate different cell types at the same time. This advantage allows us to investigate the relationships and connections between different cell types through visualization. Mapper also allows researchers to explore different biological hypotheses through different filter functions and generates results with rich biological information. We took this advantage by incorporating information from GCNA into our visualization. GCNA helps to differentiate different cell types more effectively and enrichment analysis of gene co-expression modules helps the interpretation of the visualization results. Moreover, our method provides various options for researchers to explore the data from different perspectives and is highly scalable to large number of cells.
While our method shows potential in effectively extracting biological insights from scRNA-seq data, some limitations still exists. Firstly, although different filter functions could produce networks with different structures, allowing researchers to explore data from different perspectives, not all filter functions could generate networks with meaningful shapes. Researchers need to work with the data experimentally in order to find informative visualizations. Secondly, enrichment analysis only provides preliminary results of potential biological significance and more rigorous experiments are needed to validate the findings. Finally, we plan to implement our method as a web tool so that more researchers can easily access our method.
Acknowledgements
This work is partially supported by IUSM startup fund, the NCI ITCR U01 (CA188547) and Data Science and Bioinformatics Program for Precision Health Initiative, Indiana University.
Contributor Information
Tongxin Wang, Department of Computer Science, Indiana University Bloomington, Bloomington, Indiana 47408, USA, tw11@iu.edu.
Travis Johnson, Department of Biomedical Informatics, The Ohio State University, Columbus, Ohio 43210, USA, Travis.Johnson@osumc.edu.
Jie Zhang, Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, Indiana 46202, USA, jizhan@iu.edu.
Kun Huang, Department of Medicine, Indiana University School of Medicine, Indianapolis, Indiana 46202, USA, Regenstrief Institute, Indianapolis, Indiana 46202, USA, kunhuang@iu.edu.
References
- 1.Shapiro E, Biezuner T and Linnarsson S, Nature Reviews Genetics 14, p. 618 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA and Trapnell C, Nature methods 14, p. 979 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Salame TM, Kathail P, Choi K, Ben-dall S, Friedman N and Pe’er D, Nature biotechnology 34, p. 637 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang B, Zhu J, Pierson E, Ramazzotti D and Batzoglou S, Nature methods 14, p. 414 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Han Z, Johnson T, Zhang J, Zhang X and Huang K, BioMed research international 2017 (2017). [Google Scholar]
- 6.Maaten L. v. d. and Hinton G, Journal of machine learning research 9, 2579 (2008). [Google Scholar]
- 7.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G et al. , Science 352, 189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muraro MJ, Dharmadhikari G, Grun D, Groen N, Dielen T, Jansen E, van Gurp L, Engelse MA, Carlotti F, de Koning EJ et al. , Cell systems 3, 385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh G, Memoli F and Carlsson GE, Topological methods for the analysis of high dimensional data sets and 3d object recognition., in SPBG, 2007. [Google Scholar]
- 10.Lum PY, Singh G, Lehman A, Ishkanov T, Vejdemo-Johansson M, Alagappan M, Carlsson J and Carlsson G, Scientific reports 3, p. srep01236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Der Maaten L, The Journal of Machine Learning Research 15, 3221 (2014). [Google Scholar]
- 12.Nicolau M, Levine AJ and Carlsson G, Proceedings of the National Academy of Sciences, p. 201102826 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rizvi AH, Camara PG, Kandror EK, Roberts TJ, Schieren I, Maniatis T and Rabadan R, Nature biotechnology 35, p. 551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saggar M, Sporns O, Gonzalez-Castillo J, Bandettini PA, Carlsson G, Glover G and Reiss AL, Nature communications 9, p. 1399 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J and Huang K, Cancer informatics 13, CIN (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langfelder P and Horvath S, BMC bioinformatics 9, p. 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Xu H, Aronow BJ and Jegga AG, BMC bioinformatics 8, p. 392 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balasubramanian M and Schwartz EL, Science 295, 7 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Roweis ST and Saul LK, science 290, 2323 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Von Luxburg U, Statistics and computing 17, 395 (2007). [Google Scholar]