Abstract
The increasing amount of scientific literature in biological and biomedical science research has created a challenge in continuous and reliable curation of the latest knowledge discovered, and automatic biomedical text-mining has been one of the answers to this challenge. In this paper, we aim to further improve the reliability of biomedical text-mining by training the system to directly simulate the human behaviors such as querying the PubMed, selecting articles from queried results, and reading selected articles for knowledge. We take advantage of the efficiency of biomedical text-mining, the flexibility of deep reinforcement learning, and the massive amount of knowledge collected in UMLS into an integrative artificial intelligent reader that can automatically identify the authentic articles and effectively acquire the knowledge conveyed in the articles. We construct a system, whose current primary task is to build the genetic association database between genes and complex traits of human. Our contributions in this paper are three-fold: 1) We propose to improve the reliability of text-mining by building a system that can directly simulate the behavior of a researcher, and we develop corresponding methods, such as Bi-directional LSTM for text mining and Deep Q-Network for organizing behaviors. 2) We demonstrate the effectiveness of our system with an example in constructing a genetic association database. 3) We release our implementation as a generic framework for researchers in the community to conveniently construct other databases.
Keywords: Biomedical text-mining, Deep Reinforcement Learning, Genetic Association
1. Introduction
Understanding the biological and biomedical science is one of the most fundamental goals of research and an essential step towards the realization of “precision medicine” in this era. Scientists all over the world are collaboratively contributing to this final goal, leading to an accompanying growth of the scientific literature. For example, PubMeda has seen exponential growth regarding the number of publications in recent years1 and has collected over 27 million abstracts.2 These massive amount of articles consequently bring in the challenge of integrating the information conveyed effectively and accurately.
Biomedical information extraction has been the answer to this challenge for a long time.3,4 However, due to the demand of high reliability in biomedical research, following a typical general-purpose information extraction protocol and examining every article in the corpus nondiscriminatorily may lead to falsely constructed knowledge because of the non-negligible number of scientific literature with the issues of reproducibility.5–7
To fulfill the need of reliability in text mining and knowledge-base construction, instead of requiring the system to scan the entire corpus uniformly, we propose to train the system to directly simulate the behavior of a scientist with a sequence of actions including 1) querying the web, 2) evaluating the article, 3) studying the article for knowledge if necessary, 4) rejecting the knowledge if necessary, and 5) storing the knowledge. The 2nd and 4th steps play the essential roles in maintaining the reliability in constructed databases in our proposed system. Boosted by the power of deep reinforcement learning in organizing these actions, the ability of deep Bidirectional long short-term memory (LSTM) in text mining, and massive amount of knowledge encoded in Unified Medical Language System (UMLS),8 we are able to present our humanlike system that can imitate the behaviors of a real scientist and construct the database of reliable and cutting-edge biomedical publications efficiently and endlessly. Therefore, we name our system the Everlasting Iatric Reader (Eir)b. We further apply our system to construct a genetic association database, where we can verify the performance of Eir with a manually crafted database of 167k gene-trait associations from high quality articles.9
The contributions of this paper are three-fold:
We propose to improve the reliability of text-mining by building a system that can directly simulate the behavior of a researcher, and we develop corresponding methods, such as Bi-directional LSTM for text mining and Deep Q-Network for organizing behaviors.
We demonstrate the effectiveness of our system with an example in constructing a genetic association database.
We release our implementation as a generic framework for researchers in the community to conveniently construct other databases.
The remainder of this paper is organized as follows. In Section 2, we will introduce the related works in biomedical text mining. In Section 3, we will systematically introduce our system, mainly with deep reinforcement learning module that organizes the actions, text mining module that extracts the information, and implementation specifications. In Section 4, we will compare the performance to validate the strategy of Eir. Finally, in Section 5, we will draw conclusions and discuss about the future work.
2. Related Work
Text mining from biomedical literature has been studied extensively for a long time with a variety of different applications, such as patient analysis from electronic health records,10–12 gene annotations from protein networks,13 and drug repositioning from literature.14 One can refer to comprehensive reviews4,15,16 and the references therein for more detailed discussions.
The text mining usually leads to automatic construction of knowledge bases. In recent years, Mallory et al.17 curated a database of gene-gene interactions. They applied the information extraction engine DeepDive18 to around 100k full text PLOS articles for extracting direct and indirect gene-gene interactions. Poon et al.19 introduced the Literome project, where they extracted directed genic interactions and genotype-phenotype associations from PubMed articles. Lossio-Ventura et al.20 introduced a pipeline to build an obesity and cancer knowledge base. Very recently, Lossio-Ventura et al. also noticed the reliability issue of knowledge base, so they further proposed to incorporate cross-sourcing process to improve the reliability of the their previously developed knowledge base.21
On the other hand, the boom of deep learning techniques has allowed many more advanced methods developed for biomedical applications.22–24 As a result, LSTM and its variants,25,26 and word embedding techniques27,28 have been studied extensively for a variety of applications.
In comparison, a difference between most of previous work and our work is that we aim to improve the reliability of the extracted knowledge by examining the source unstructured data (i.e. the PubMed literature in our case). To put in simpler words, while most previous work are extending human’s intelligence of comprehending the articles, our system aims to extend human’s intelligence of the entire research process that starts with querying the web and selecting the interesting article. To the best of our knowledge, this paper is the first one that simulates the entire research process in biomedical information extraction to improve the reliability of the constructed knowledge base. However, many similar concepts29–31 have been proposed previously. Most relevantly, Kanani et al32 utilized reinforcement learning to reduce computational bottlenecks, minimizing the number of queries, document downloads and extraction action, a similar strategy has been proposed independently for biomedical text mining with the concept “focused machine reading”,33 which is inspired by Narasimhan et al,34 who built an information extraction system that can query the web for extra information with reinforcement learning.
3. Method
In this section, we officially introduce the our system. We will start with the main framework, and continue to introduce the deep reinforcement learning module that organize different actions of the system, which is followed by the discussions of proprecessing module and biomedical text mining module. After a systematic introduction of the detailed algorithms, this section is concluded with implementation specifications.
3.1. Model Framework
Eir’s research process is a markov decision process (MDP), where the model learns to query the search engine for scientific articles to read for the knowledge. We represent the MDP as a tuple < S,A,T,R >, where S = s is the space of all possible states, A = a is the set of all actions, R(s, a) is the reward function, and T(s’|s, a) is the transition function.
We present the details of these components as following:
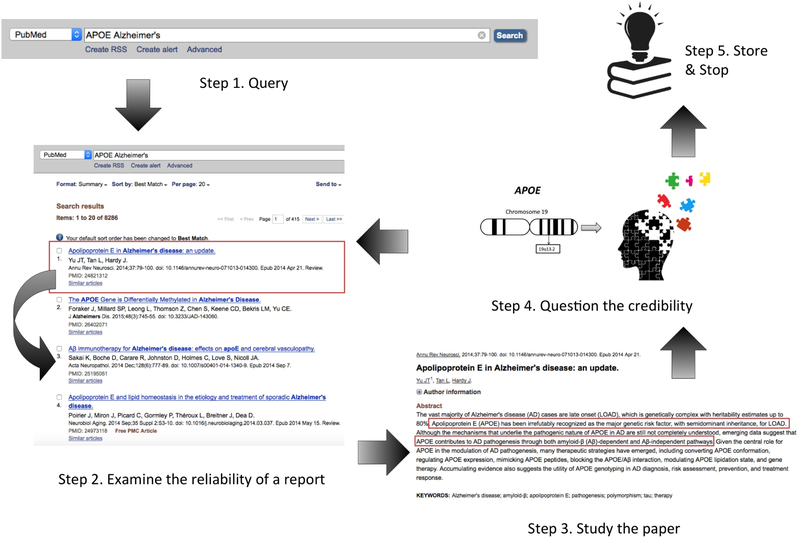
- Actions: Action (we use a to denote action throughout this paper) is a set of Eir’s behaviors to simulate a real researcher, including
- Query the search engine.
- Evaluate whether the article is reliable.
- Read the article for detailed information.
- Exam credibility of the information and querying again.
- Stop.
As shown in Figure 1, for every interesting query, Eir starts with the 1st action and then enters the loop from the 2nd action to the 4th action until Eir is satisfied with the finding of current research interest and ceases with the 5th action. Then Eir repeats the entire process with another query.
Fig. 1:
Overview of Eir’s possible behaviors
States: The state s in the MDP describes the research status of Eir, possible candidate states include the ones that are precedent or after each aforementioned action. There are only a countable number of actions, but we use continuous real-valued vector to represent each state so that we could have a better modeling power to distinguish Eir’s research status after each action. The state is constructed with a variety of information, including the embedding vector that the Bidirectional LSTM yields, the confidence of biomedical text mining module, the confidence of selecting an article to read, etc.
Rewards: The reward function is chosen to maximize the intermediate paper selection accuracy and final extraction accuracy together while minimizing the number of queries. The accuracy component is calculated using the difference between the accuracy of the current and the previous set of entity values.
Transitions: Transition T(s’|s, a) is modeled as a function of how the next state s’ is updated given the current state s and action a taken.
3.2. Deep Reinforcement Learning for Organizing Actions
As we have introduced previously, we utilized deep reinforcement learning to arrange the sequence of actions a to perform, given a state function denoted as Q(s, a). To update Q(s, a), we used the popular Q-learning,35: which iteratively updates Q(s; a) as following:
where r = R(s, a) is the reward and λis a discounting factor.
Because of the continuous nature of our state space S, we use a deep Q-network (DQN)36 as a function approximator Q(s, a) = Q(s, a;θ). The Q-function of DQN is approximated by a neural network, whose parameters (i.e. θ) are updated through stochastic gradient descent. We followed the detailed parameter learning strategies introduced previously.34
3.3. Preprocessing and Name Entity Recognition with UMLS
Before we feed in the texts into the text mining module, we notice that the literature is filled with alternative, idiosyncratic and arbitrary names and symbols. The text mining module will only exhibit its full power when the texts are processed into a uniform representation. Therefore, we utilize the rich information collected by the unified medical language system (UMLS).8 UMLS defines a unique concept for all the terms that are interchangeable. For example, “Alzheimer’s disease”, “Alzheimer’s”, and “alzheimer” will be mapped into the same concept. UMLS contains over one million biomedical concepts that are split into 133 broad categories (such as “Organisms”, “Anatomical structures”, “Biologic function”). With the help of MetaMap,37 we are able to translate the unstructured texts into a sequence of concepts, together with the category information, an associated confidence score, and two binary values to indicate whether the concept is in gene ontology, and in disease ontology respectively.
3.4. Bidirectional LSTM for Relation Classification
As Eir queries PubMed with a gene-trait pair, the text mining model only needs to classify whether the returned texts from PubMed can be seen as evidences to support that there is association between the queried gene-trait pair. Therefore, the text mining module can be conveniently regarded as a classification module. We use a Bidirectional LSTM38 for classifying whether the text describes as association relationship between the gene and the trait the system queried. We choose this Bidirectional LSTM architecture mainly because we notice that it is empirically the best performing method among other neural architecture for our specific task. We first treat the sequence of concepts as words in text and created a 512-dimension vector of continuous values to represent each concept. Further, we feed in this concept-embedding, together with an one-hot representation of the category information, and the two binary values into the Bidirectional LSTM, which is trained through Adam.
3.5. Algorithm
Algorithm 1 describes the overall algorithm of the MDP process of Eir, where g and t stands for gene and trait respectively, a stands for action, s stands for state, and r stands for reward. “Agent” refers to DQN, which organizes the sequence of actions given states and reward. Details including the methodology of updating (s, r) has been discussed in previous sections.
3.6. Implementation Specification
The Deep Reinforcement Learning component of Eir is implemented as an extension of Narasimhan et al,34 we also use a DQN consisting of two linear layers (20 hidden units each) followed by rectified linear units (ReLU), along with two separate output layers.
The web query component is built with a web crawling engine Scrapyc communicating with NCBI PubMed search engine. At this moment, we only query for the abstracts of the articles. We only work with abstracts for three reasons: 1) this allow us to conveniently access and scan a large amount literature, 2) we notice that a majority of articles disclose the most important findings in the abstract with a straightforward style of writing, 3) previous work notice that mining from full texts may lead to more false positives.39
The preprocessing module is built as a python script that runs MetaMap, which is a binary software that allows users to conveiently annotate words and phrases of texts with manually defined concepts in UMLS.
The sentences are truncated with max length of 300 concepts. We only consider the 30,000 most frequent concepts together with the specific defined ‘SOS’ (start of sentence), ‘EOS’ (end of sentence), ‘UNK’ (unknown) and ‘PAD’ (padding the sentences shorter than 300) concepts. We use a 2-layer Bidirectional LSTM with hidden dimension set to 1000, and feed 512 dimension concept embedding, one dimension gene ontology, one dimension disease ontology, and 136 dimension semantic type as the input of LSTM. The LSTM is trained jointly with the embedding matrix using Adam with step size set to 0.00004 and batch size set to 64.
Then We train the Eir models for 10000 steps every epoch using the Maxent classifier as the base extractor, and evaluate on the entire test set every epoch. The final accuracy reported are averaged over three independent runs; each runs score is averaged over 5 epochs after 45 epochs of training. The penalty per step is set to −0.001. We used a replay memory of size 500k, and a discount γ of 0.8. We set the learning rate to 2.5E5. The ϵ in ϵ-greedy exploration is annealed from 1 to 0.1 over 500k transitions. The target-Q network is updated every 5k steps. The whole framework was trained to optimize the reward function.
We release our implementationd for the community to use our system or build more advanced text mining module into our system for better performance.
| Algorithm 1 MDP framework of Eir |
|---|
| for epoch = 1,M do |
| for g, t in query list do |
| Query the search engine with g and t. |
| Update and send state (s, r) to agent |
| Get action a from agent |
| while a is not “stop” do |
| if a is “select” then |
| Update (s, r) with selection |
| else if a is “reject” then |
| Update (s, r) with rejection |
| else |
| Translate texts into sequence of concept embeddings. |
| Relation classification with Bidirectional LSTM |
| Update (s, r) with classified relation |
| end if |
| Send state (s, r) to agent |
| Get action a from agent |
| end while |
| end for |
| end for |
4. Experiments
In this section, we will verify the performance of Eir by showing that, with the same text mining module, the Eir system can help improve the performance of extracted associations. We will first discuss how we construct the experimental data sets then discuss the results.
4.1. Data
Within the scope of this paper, Eir focus on constructing the knowledge base for gene-trait association relationship of human. To enable Eir to learn the associations, we utilized the highquality data set of 167k association relationship that is manually crafted for over ten years.9
In addition to the gold-standard information of gene-trait association relationship, another contribution of this data set is the collection of high quality publications that report these associations. Every entry in the database is grounded by the authentic source of scientific paper that originally publishes the relationship. These detailed information grants us the possibility of directly training Eir to discriminate the reliable papers out of the less favourable papers that were not selected by GAD curators for some reasons.
Despite that Eir is designed for extracting latest information online, in order to test the effectiveness of Eir, we need to run the core functions on a local collections of articles with manually labelled true associations. Therefore, we query the PubMed with 54,041 queries of gene-trait pairs through our API and download 913,939 results with 305,651 distinct medical articles. After removing some invalid records (e.g. articles with invalid PMID), there are roughly 133,548 records (44,592 distinct articles) appear in the GAD database, which will serve as the reliable articles. As the construction of GAD ceased in 2014, we regard the articles that are published before 2014 but not in the GAD database as less favorable articles. To balance the data set for performance evaluation, we sampled 140,361 less favorable records before 2014 for comparison. Note that, these less favorable articles are not collected randomly, but are returned from PubMed search engine when we query with a pair of gene and trait. Besides, we delete the articles whose titles and abstracts do not contain the queried gene and trait explicitly to remove obviously irrelevant articles. Then, we random split the whole data set to sample 80% records as training data, and the rest as testing data. The training set consists of 55k records, the testing set consists of 219k records.
4.2. Evaluation
In order to show the effectiveness of the Eir system, we compare the system’s precision, recall, and F1 score with a conventional biomedical text mining strategy that scans all the documents nondiscriminatorily. As Eir uses the Bidirectional LSTM for text mining module, we use the same model as baseline method for fair comparison.
4.3. Results
4.3.1. Improved Reliability
We first train our baseline Bidirectional LSTM and the results are shown in the Table 1 (first row). The Bidirectional LSTM yields a precision of 91.25%, a recall of 96.55%, and an overall F1 of 93.80%. These numbers indicate that the Bidirectional LSTM is capable to capture the feature of authentic articles.
Table 1:
Results of Reliability Comparison
| precision | recall | F1 | |
|---|---|---|---|
| Bidirectional LSTM | 91.25% | 96.55% | 93.80% |
| Eir | 91.4% | 97.0% | 94.1% |
Further we add the Deep Reinforcement Learning component to train the overall Eir system. The results of Eir are shown as Table 1 (second row). We can see that the precision score is 91.4%, the recall score is 97.0%, the F1score is 94.1%. Compared to the baseline model, our Eir framework is better at extracting the features of valuable articles and utilizing the information and can retrieve the authentic articles more efficiently by employing the Deep Reinforcement Learning module.
4.3.2. Robustness in Real-world Situations
To better simulate the real-world situation that the researchers are in, we remove different percentage of authentic articles both in the training data set and in the testing data set, for the researchers get ample amount of less favorable articles. We randomly remove a certain percentage of authentic articles to do the ablation experiments. As the percentage of authentic articles decreases, the difficulty of our task increases. The results are shown in Table 2. We can see the Eir system is more robust than the baseline model under these situations. Eir reports higher precision, recall, and F1 score in all of these settings. More interestingly, we calculate the increments Eir achieves over baseline model. We notice that, as the difficulty increases, the increment also increases. Therefore, we believe Eir will be more helpful in the real-world situation when a large amount of articles are less favorable articles.
Table 2:
Results of Eir in real-world situations
| Full Data | 20% Authentic Articles | 10% Authentic Articles | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Prec | Recall | F1 | Prec | Recall | F1 | Prec | Recall | F1 | |
| Bi-LSTM | 91.25% | 96.55% | 93.80% | 87.7% | 95.7% | 91.5% | 86.9% | 92.2% | 89.4% |
| Eir | 91.4% | 97.0% | 94.1% | 87.9% | 96.9% | 92.2% | 87.8% | 96.9% | 92.1% |
| Increment | 0.16% | 0.47% | 0.32% | 0.23% | 1.25% | 0.77% | 1.04% | 5.10% | 3.02% |
4.3.3. Number of Articles Read
Finally, we examine Eir’s performance in the numbers of articles it needs to read to make a decision. Since Eir stops once it believes it has sufficient amount of information, we anticipate Eir will inspect less amount of articles than baseline models. To conduct this experiment, we exclude the gene-trait query pair with only one authentic articles. In the remaining data set, there is on average 2.54 articles for every query, and Eir reads only on average 2.46 articles. We further repeat this experiment with a data set that excludes all the articles with less than 4 articles per query, resulting in a data set with on average 6.23 articles for every query. Eir reads on average 6.10 articles.
5. Conclusions and Future Work
In this work, we introduced a system, namely Everlasting Iatric Reader (Eir), for biomedical text mining. A distinct difference between our system and previous biomedical text mining works is that our system is aimed to directly simulate the behaviors of scientists, including searching for scientific literature, examining the reliability of the manuscript, studying the paper for details, and continuing to search with suspicion of the learned knowledge.
In contrast to traditional biomedical text mining tools, the distinguishable advantage Eir has is the ability to discriminate reliable articles out of questionable articles and to shield the problems introduced by humans. This ability is particularly important in biomedical areas because in clinics, a falsely constructed knowledge may lead to fatal errors, while a missing piece of true knowledge will at most delay the cure of certain disease. Also, it is necessary to select trustworthy papers to read for information because it is known that there is a non-negligible number of publications with the troubles of reproducibility.
There are also limitations of the current Eir. For example, the action of Eir for evaluating the literature quality is trained supervisedly. The performance of our Eir can be greatly improved with a more cleaned data source, as now the false positives are introduced by some manuaaly crafted data that are labeled not correctly. Therefore, we will need a manually crafted data set first before we use Eir in some application. In this paper, we choose to construct the genetic association database because of the availability of GAD.9 However, there are still a large number of manually curated databases with information about which paper these information comes from, such as GWAS Catalog40 for SNP-phenotype association or UniProt41 for protein function annotation.
Looking into the future, a direct extension of our work is to broaden Eir vision to ask investigate into more biomedical topics in addition to gene-trait association relationships. Our immediate next-step plan is to train Eir for SNP-phenotype association with GWAS Catalog, then we can integrate these databases into GenAMap,42 a visual machine learning tool for GWASe, for validation purpose of GWAS results. On the method development side, we hope to upgrade the biomedical text mining module with state-of-the-art methods to improve the information extraction performance, so that Eir could serve the community better. As a longterm plan, we hope Eir could help the community to build the omini-biomedical knowledge base, therefore, we released the source code of Eir for others in the community to use.
6. Acknowledgement
This work is funded and supported by the Department of Defense under Contract No. FA8721-05-C-0003 with Carnegie Mellon University for the operation of the Software Engineering Institute, a federally funded research and development center. This work is also supported by the National Institutes of Health grants R01-GM093156 and P30-DA035778.
Footnotes
the database maintained by the National Center for Biotechnology Information (NCBI)
We name our system Everlasting Iatric Reader because it can endlessly construct the knowledge in the medical area, where the high reliability is an issue, and also because the acronym (Eir) shares the name of the goddess of medical knowledge in Norse mythology, which is related to the final goal of this and following-up projects.
References
- 1.Larsen PO and Von Ins M, The rate of growth in scientific publication and the decline in coverage provided by science citation index, Scientometrics 84, 575 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raja K, Patrick M, Gao Y, Madu D, Yang Y and Tsoi LC, A review of recent advancement in integrating omics data with literature mining towards biomedical discoveries, International journal of genomics 2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen AM and Hersh WR, A survey of current work in biomedical text mining, Briefings in bioinformatics 6, 57 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Cohen KB and Demner-Fushman D, Biomedical natural language processing (John Benjamins Publishing Company, 2014). [Google Scholar]
- 5.Poste G, Bring on the biomarkers, Nature 469, 156 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Shoenbill K, Fost N, Tachinardi U and Mendonca EA, Genetic data and electronic health records: a discussion of ethical, logistical and technological considerations, Journal of the American Medical Informatics Association 21, 171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kilicoglu H, Biomedical text mining for research rigor and integrity: tasks, challenges, directions, Briefings in bioinformatics (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodenreider O, The unified medical language system (umls): integrating biomedical terminology, Nucleic acids research 32, D267 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker KG, Barnes KC, Bright TJ and Wang SA, The genetic association database, Nature genetics 36, 431 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Hollister BM, Restrepo NA, Farber-Eger E, Crawford DC, Aldrich MC and Non A, Development and performance of text-mining algorithms to extract socioeconomic status from de-identified electronic health records, in PACIFIC SYMPOSIUM ON BIOCOMPUTING 2017, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beaulieu-Jones BK, Orzechowski P and Moore JH, Mapping patient trajectories using longitudinal extraction and deep learning in the mimic-iii critical care database, bioRxiv, p. 177428 (2017). [PubMed] [Google Scholar]
- 12.Glicksberg BS, Miotto R, Johnson KW, Shameer K, Li L, Chen R and Dudley JT, Automated disease cohort selection using word embeddings from electronic health records, in Pac Symp Biocomput, 2018. [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S, Ma J, Yu MK, Zheng F, Huang EW, Han J, Peng J and Ideker T, Annotating gene sets by mining large literature collections with protein networks, in Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing, 2018. [PMC free article] [PubMed] [Google Scholar]
- 14.Frijters R, Van Vugt M, Smeets R, Van Schaik R, De Vlieg J and Alkema W, Literature mining for the discovery of hidden connections between drugs, genes and diseases, PLoS computational biology 6, p. e1000943 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez GH, Tahsin T, Goodale BC, Greene AC and Greene CS, Recent advances and emerging applications in text and data mining for biomedical discovery, Briefings in bioinformatics 17, 33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F, Chen J, Jagannatha A and Yu H, Learning for biomedical information extraction: Methodological review of recent advances, arXiv preprint arXiv: 160607993 (2016). [Google Scholar]
- 17.Mallory EK, Zhang C, Re C and Altman RB, Large-scale extraction of gene interactions from full-text literature using deepdive, Bioinformatics 32, 106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niu F, Zhang C, Re C and Shavlik JW, Deepdive: Web-scale knowledge-base construction using statistical learning and inference., VLDS 12, 25 (2012). [Google Scholar]
- 19.Poon H, Quirk C, DeZiel C and Heckerman D, Literome: Pubmed-scale genomic knowledge base in the cloud, Bioinformatics 30, 2840 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Lossio-Ventura JA, Hogan W, Modave F, Guo Y, He Z, Hicks A and Bian J, Oc-2-kb: A software pipeline to build an evidence-based obesity and cancer knowledge base, in Bioinformatics and Biomedicine (BIBM), 2017 IEEE International Conference on, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lossio-Ventura JA, Hogan W, Modave F, Guo Y, He Z, Yang X, Zhang H and Bian J, Oc-2-kb: integrating crowdsourcing into an obesity and cancer knowledge base curation system, BMC Medical Informatics and Decision Making 18, p. 55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Min S, Lee B and Yoon S, Deep learning in bioinformatics, Briefings in bioinformatics 18, 851 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Wang H and Raj B, On the origin of deep learning, arXiv preprint arXiv: 170207800 (2017). [Google Scholar]
- 24.Yue T and Wang H, Deep learning for genomics: A concise overview, arXiv preprint arXiv: 180200810 (2018). [Google Scholar]
- 25.Li F, Zhang M, Fu G and Ji D, A neural joint model for entity and relation extraction from biomedical text, BMC bioinformatics 18, p. 198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng W, Lin H, Luo L, Zhao Z, Li Z, Zhang Y, Yang Z and Wang J, An attention-based effective neural model for drug-drug interactions extraction, BMC bioinformatics 18, p. 445 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Z, Li L, Huang D and Jin L, Training word embeddings for deep learning in biomedical text mining tasks, in Bioinformatics and Biomedicine (BIBM), 2015 IEEE International Conference on, 2015. [Google Scholar]
- 28.Habibi M, Weber L, Neves M, Wiegandt DL and Leser U, Deep learning with word embeddings improves biomedical named entity recognition, Bioinformatics 33, i37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Termehchy A, Vakilian A, Chodpathumwan Y and Winslett M, Which concepts are worth extracting?, in Proceedings of the 2014 ACM SIGMOD international conference on Management of data, 2014. [Google Scholar]
- 30.West R, Gabrilovich E, Murphy K, Sun S, Gupta R and Lin D, Knowledge base completion via search-based question answering, in Proceedings of the 23rd international conference on World wide web, 2014. [Google Scholar]
- 31.Samadi M, Talukdar PP, Veloso MM and Mitchell TM, Askworld: Budget-sensitive query evaluation for knowledge-on-demand., in IJCAI, 2015. [Google Scholar]
- 32.Kanani PH and McCallum AK, Selecting actions for resource-bounded information extraction using reinforcement learning, in Proceedings of the fifth ACM international conference on Web search and data mining, 2012. [Google Scholar]
- 33.Noriega-Atala E, Valenzuela-Escarcega MA, Morrison CT and Surdeanu M, Learning what to read: Focused machine reading, arXiv preprint arXiv: 170900149 (2017). [Google Scholar]
- 34.Narasimhan K, Yala A and Barzilay R, Improving information extraction by acquiring external evidence with reinforcement learning, arXiv preprint arXiv: 160307954 (2016). [Google Scholar]
- 35.Watkins CJ and Dayan P, Q-learning, Machine learning 8, 279 (1992). [Google Scholar]
- 36.Mnih V, Kavukcuoglu K, Silver D, Rusu AA, Veness J, Bellemare MG, Graves A, Ried-miller M, Fidjeland AK, Ostrovski G et al. , Human-level control through deep reinforcement learning, Nature 518, 529 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Aronson AR, Effective mapping of biomedical text to the umls metathesaurus: the metamap program., in Proceedings of the AMIA Symposium, 2001. [PMC free article] [PubMed] [Google Scholar]
- 38.Graves A and Schmidhuber J, Framewise phoneme classification with bidirectional lstm and other neural network architectures, Neural Networks 18, 602 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Fleuren WW and Alkema W, Application of text mining in the biomedical domain, Methods 74, 97 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L et al. , The nhgri gwas catalog, a curated resource of snp-trait associations, Nucleic acids research 42, D1001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Consortium U, Uniprot: the universal protein knowledgebase, Nucleic acids research 45, D158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, Lengerich BJ, Lee MK and Xing EP, Genamap on web: Visual machine learning for next-generation genome wide association studies, in preparation (2018). [Google Scholar]