Abstract
Fanconi anemia (FA) is a complex genetic disorder characterized by bone marrow failure (BMF), congenital defects, inability to repair DNA interstrand cross-links (ICLs), and cancer predisposition. FA presents two seemingly opposite characteristics: (a) massive cell death of the hematopoietic stem and progenitor cell (HSPC) compartment due to extensive genomic instability, leading to BMF, and (b) uncontrolled cell proliferation leading to FA-associated malignancies. The canonical function of the FA proteins is to collaborate with several other DNA repair proteins to eliminate clastogenic (chromosome-breaking) effects of DNA ICLs. Recent discoveries reveal that the FA pathway functions in a critical tumor-suppressor network to preserve genomic integrity by stabilizing replication forks, mitigating replication stress, and regulating cytokinesis. Homozygous germline mutations (biallelic) in 22 FANC genes cause FA, whereas heterozygous germline mutations in some of the FANC genes (monoallelic), such as BRCA1 and BRCA2, do not cause FA but significantly increase cancer susceptibility sporadically in the general population. In this review, we discuss our current understanding of the functions of the FA pathway in the maintenance of genomic stability, and we present an overview of the prevalence and clinical relevance of somatic mutations in FA genes.
Keywords: Fanconi anemia, DNA interstrand cross-links, DNA repair, genomic instability, somatic cancer
1. INTRODUCTION
Cells are constantly subjected to genomic insults from exogenous and endogenous sources. Cells are equipped with multiple specialized DNA repair mechanisms for detecting and repairing specific DNA damage lesions. Eradicating DNA damage is essential to the maintenance of genomic integrity. Unfaithful repair of DNA damage leads to genomic instability, which fuels cancer initiation and progression. Many chemotherapeutic drugs target the essential process of DNA replication of cancer cells by producing a wide range of DNA damage. To overcome these genotoxic effects and to enable their uncontrolled proliferation, cancer cells often rewire their DNA repair mechanisms, providing opportunities for targeted therapeutic approaches. Our understanding of complex DNA repair mechanisms, such as the Fanconi anemia (FA) pathway, has greatly increased in the past few years. Synthetic lethality approaches targeting one or more of these DNA repair pathways have been applied to resensitize cancer cells that are otherwise resistant to monotherapies.
2. MOLECULAR DETAILS OF THE FA/BRCA PATHWAY
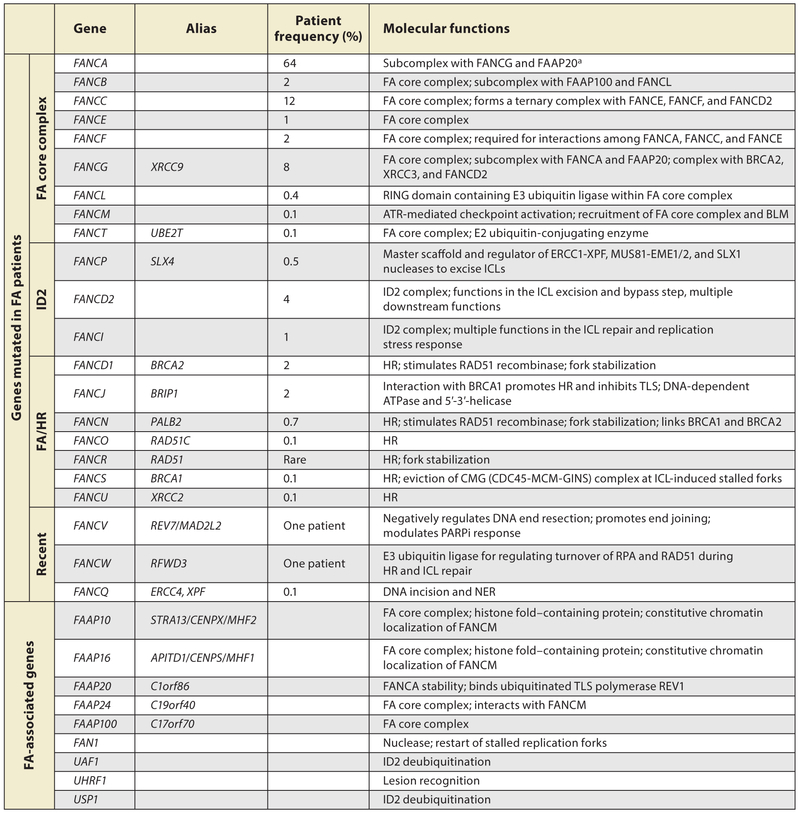
The inability to repair DNA interstrand cross-links (ICLs) is a key cellular feature of FA, a disorder first described by Swiss pediatrician Guido Fanconi in 1927 (Auerbach 2009). FA is a rare genetic syndrome (1 in 100,000) that is often diagnosed at the presentation of bone marrow failure (BMF) at a median age of 7 years (Rosenberg et al. 2011). The hypersensitivity to the clastogenic (chromosome-breaking) effects of ICL-inducing agents provides a reliable cellular marker for the diagnosis of FA (Auerbach 1993, Giampietro et al. 1993). Autosomal biallelic germline inactivation of any one of the 22 currently known FA genes (designated as complementation groups FANCA–FANCW) causes FA except for FANCB, which is X-chromosomal (Figure 1) (Auerbach 2009, Bluteau et al. 2016, Inano et al. 2017, McCauley et al. 2011, Park et al. 2016, Wang & Smogorzewska 2015) The protein products of these 22 FA genes, along with FA-associated proteins (FAAP), interact in a common cellular pathway to repair ICLs, known as the FA pathway or the FA/BRCA pathway (Figure 2). In eukaryotes, the FA pathway orchestrates the detection and removal of ICLs by the combined actions of nucleotide excision repair (NER) and homologous recombination (HR), with minor contributions from other DNA repair pathways.
Figure 1.
Classification of Fanconi anemia genes and their molecular functions. Data from Frohnmayer et al. (2014). Abbreviations: FA, Fanconi anemia; FAAP, FA-associated proteins; HR, homologous recombination; ICL, interstrand cross-link; NER, nucleotide excision repair; PARPi, poly (ADP-ribose) polymerase inhibitor; TLS, translesion synthesis.
a FAAPs are important for ICL repair, but to date no FA patient has been found harboring biallelic mutations of them.
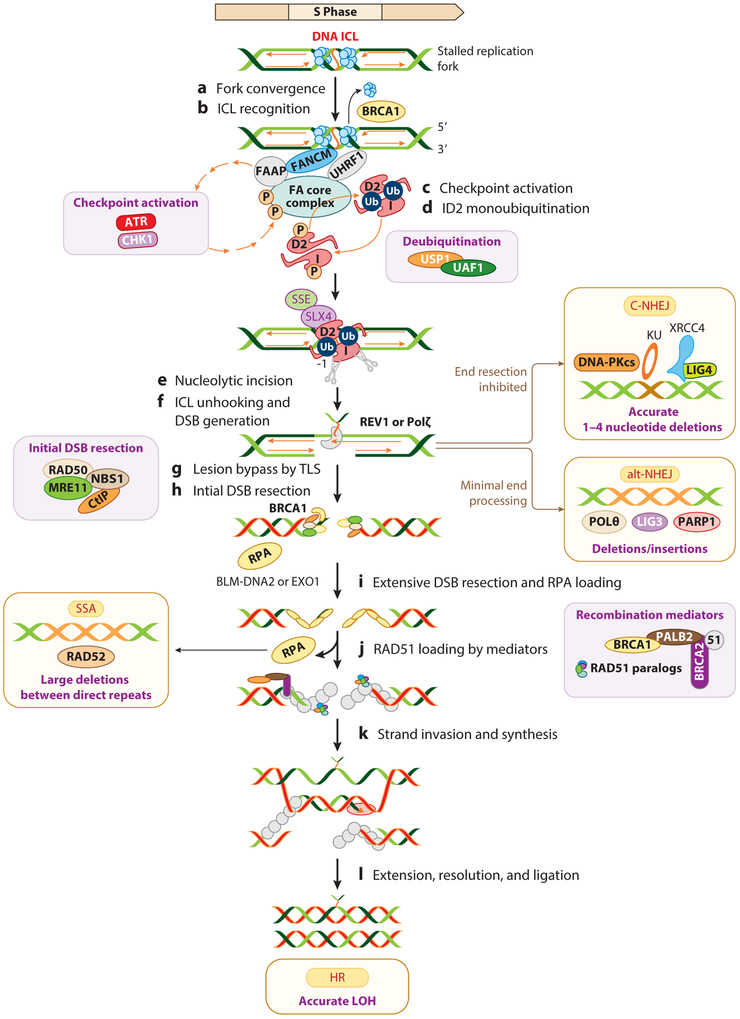
Figure 2.
Coordination of multiple DNA repair pathways in a common DNA ICL repair pathway. (a,b) Stalled replication forks at DNA ICLs are recognized by FANCM-FAAP24-MHF1-MFH2 (FAAPs) or UHRF1. Eviction of the replicative CMG helicase by BRCA1 allows one replication fork to approach the ICLs. (c) FANCM promotes the ATR kinase–dependent checkpoint response. (d) The FA core complex monoubiquitinates the FANCI-FANCD2 (ID2) complex. (e,f) FANCD2-Ub and SLX4/FANCP recruit SSEs to execute the unhooking step, generating DNA DSBs in the strand opposite to the strand on which the cross-linked nucleotide tethers. (g) DNA replication resumes by the bypass step, passing the tethered ICL by TLS polymerases, such as REV1 or Polζ. The USP1-UAF1 complex deubiquitinates the ID2 complex to efficiently execute the FA pathway. (h) The DSB ends are processed to generate single-strand DNA by the initial DSB resection machinery. The processed DSB ends can be repaired by alt-NHEJ. Alternatively, inhibition of end resection leads to direct ligation of the DNA ends by C-NHEJ. (i) Extensive DSB resection by EXO1 and the BLM-DNA2 complex generate longer stretches of RPA-coated ssDNA. (j) RPA is displaced by recombination mediators to load RAD51 to promote HR. (k,l) Alternatively, the repair is diverted to RAD52-mediated SSA. The different consequences of these DSB repair pathways are deletions, insertions, and LOH. The key players of each pathway are shown in the insets. Abbreviations: alt-NHEJ, alternative nonhomologous end joining; C-NHEJ, classical nonhomologous end joining; DNA-PKcs, DNA-dependent protein kinase catalytic subunit; DSB, double-strand break; FA, Fanconi anemia; FAAPs, Fanconi anemia–associated proteins; HR, homologous recombination; ICL, interstrand cross-link; LOH, loss of heterozygosity; SSA, single-strand annealing; ssDNA, single-stranded DNA; SSE, structure-specific endonuclease; TLS, translesion synthesis.
The relevance of FA to cancer in the general population came to light when biallelic mutations in the breast and ovarian cancer susceptibility genes BRCA1 (FANCS), PALB2 (FANCN), and BRCA2 (FANCD1) were identified in FA patients. Hence, the FA pathway is often as also called the FA/HR pathway (D’Andrea & Grompe 2003). Subsequently, large-scale genomic data revealed somatic monoallelic activation of FA genes in sporadic cancers. In line with these findings, FA patients are predisposed to various types of cancer (Garaycoechea & Patel 2014). For example, patients with FANCD1 (BRCA2) and FANCN (PALB2) mutations often present with acute myeloid leukemia (AML) and embryonic tumors (neuroblastoma, medulloblastoma, and Wilms tumors), while those with mutations in the other FA complementation groups develop AML and squamous cell carcinoma (Wang & Smogorzewska 2015). Intriguingly, FA shares many molecular features with other genetic syndromes such as Seckel and Nijmegen breakage syndromes, suggesting that FA proteins function in other converging DNA repair pathways (Andreassen et al. 2004, Gennery et al. 2004).
2.1. The Detection and Removal of DNA Interstrand Cross-Links by the FA Pathway
Cells deficient in the FA pathway are hypersensitive to ICL-inducing chemotherapeutic agents such as platinum compounds (e.g., cisplatin, carboplatin, etc.), nitrogen compounds (e.g., cyclophophamide), mitomycin C, and psoralen (Huang & Li 2013). Certain metabolic processes such as lipid peroxidation, histone demethylation, and alcohol metabolism produce intermediates such as formaldehyde and acetaldehyde that now are recognized as endogenous sources of ICLs (Ridpath et al. 2007, Stone et al. 2008). Double-knockout mice for Fancd2 and Aldh2 (enzyme-metabolizing acetaldehyde) genes show severe aplastic anemia along with increased DNA damage in hematopoietic stem cells and progenitor cells, thereby establishing acetaldehyde as a potent endogenous cross-linking agent (Garaycoechea et al. 2012, Hira et al. 2013, Langevin et al. 2011).
FA pathway–mediated ICL repair occurs primarily in S phase, when the DNA replication forks stall at the ICLs (Figure 2a). Contrarily, in nondividing cells, ICLs are repaired at actively transcribed regions by components of transcription-coupled NER (Enoiu et al. 2012, Hlavin et al. 2010). The NER or mismatch repair components can recognize ICLs throughout the cell cycle; however, repair is often futile with incomplete removal of ICLs. Polκ-mediated DNA replication and transcription–independent ICL repair was identified as essential for transcription in nondividing or slowly dividing cells (Williams et al. 2012). Nevertheless, complete ICL removal occurs upon elicitation of the FA pathway in S phase by the coordinated actions of the DNA replication and repair machineries (Figure 2).
Replication forks are stalled at ICLs due to the inability to separate covalently cross-linked DNA strands. ICL-induced stalled forks are the DNA intermediate structure recognized and stabilized by the FA pathway. The anchoring complex containing FANCM and some FAAPs recognize ICLs and play a pivotal role in the FA pathway activation (Figure 2b) (Huang et al. 2010, Walden & Deans 2014). Strikingly, most replication forks can traverse ICLs in a FANCM-, PCNA-, and RPA-dependent manner to resume DNA replication prior to postreplicative ICL repair (Rohleder et al. 2016). Alternatively, the NEIL3 DNA glycosylase can directly excise the psoralen-plus-UVA-induced ICLs, resulting in an abasic site that can presumably be repaired by base excision repair (Semlow et al. 2016). FANCM, a translocase, constitutively localizes to chromatin through its interaction with highly conserved histone fold–containing proteins MHF1 (or FAAP16/CENP-S) and MHF2 (or FAAP10/CENP-X) (Singh et al. 2010, Yan et al. 2010). The FANCM-FAAP24-MHF complex plays a major role in targeting the multisubunit FA core complex to ICLs (Figure 3) (Ciccia et al. 2007).
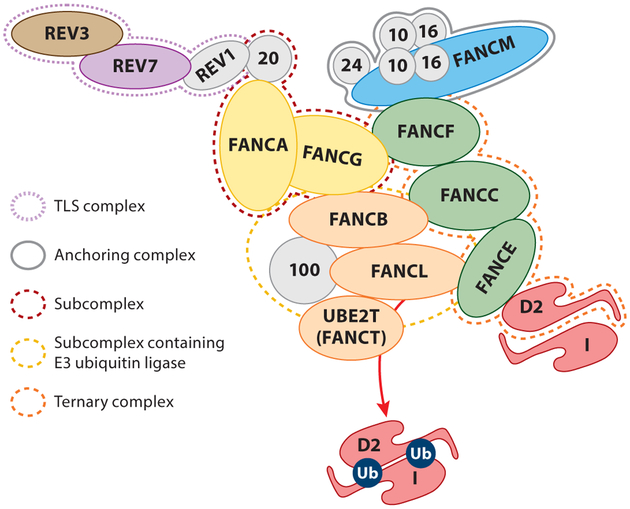
Figure 3.
Architecture of the Fanconi anemia (FA) core complex, with FAAPs (Fanconi anemia–associated proteins) indicated by numbers. The FAAP20-FANCG-FANCA subcomplex (red dotted line) is a link between the translesion synthesis (TLS) complex and the FA pathway through a direct interaction between FAAP20 and REV1, which interacts with REV3-REV7 (purple dotted line). FANCA gains its stability by binding to FAAP20, a small UBZ4-containing zinc finger protein that prevents its SUMOylation and RNF4-mediated degradation. The ternary complex FANCF-FANCC-FANCE (orange dotted line) bridges FANCD2, the substrate to the ICL-recognizing anchoring complex consisting of FANCM and FAAPs (gray solid line). Current understanding of mechanisms of FANCD2 monoubiquitination derived from biochemical and genetic approaches suggests that the FANCB-FANCL-FANC100-UBE2T complex (yellow dotted line) is a minimum module for FANCD2 and FANCI monoubiquitination.
The FA core complex harbors an enzymatic module containing FANCL, the E3 ubiquitin ligase and UBE2T (FANCT), the E2 ubiquitin-conjugating enzyme that catalyzes the monoubiquitination of FANCI and FANCD2 (ID2 complex) in response to ICLs and other genotoxic stresses (Meetei et al. 2003, Rickman et al. 2015). There are multiple autonomous modules within the FA core complex with incompletely dissected functions (Figure 3) (Medhurst et al. 2006). Many of the FA core complex proteins such as FANCE, FANCF, and FANCG possess coiled-coil or other repetitive domains (known as FANC or tetratricopeptide repeats) that might mediate extensive protein–protein interactions within and outside of the FA pathway (Alpi & Patel 2009, Walden & Deans 2014).
Intriguingly, cells depleted for FANCM, FAAP24, or MHF1 exhibit incomplete loss of ID2-ubiquitin (Wang et al. 2013, Yan et al. 2010). This has raised the possibility of alternative mechanisms by which the FA core complex is recruited to the sites of DNA damage. Accordingly, UHRF1 was shown to be involved in ICL sensing and required for the recruitment of FANCD2 to ICLs (Figure 2b) (Liang et al. 2015). Recently, FANCI but not FANCD2 was shown to be involved in recruiting the FA core complex to the damage, suggesting that FANCI could have possible roles upstream of the FA core complex (Castella et al. 2015). Once monoubiquitinated within the FA core complex, the ID2 complex accumulates at ICLs and colocalizes with additional downstream FA/HR proteins (Moldovan & D’Andrea 2009). A key function of the downstream FA/HR protein BRCA1 (FANCS) within the FA pathway is to evict the CMG helicase from stalled replication forks resulting from ICLs (Figure 2b) (Long et al. 2014). Despite a great deal of research, we know little about the other functions of the FA core complex proteins or the identity of its other monoubiquitination substrates.
2.2. Monoubiquitination of the FANCI and FANCD2 Complex
FANCD2 and FANCI are paralogs and form a saxophone-shaped heterodimeric complex, with their target monoubiquitination lysine buried in a solvent-inaccessible tunnel; their monoubiquitination requires a conformational change induced by DNA (Niraj et al. 2017, Sobeck et al. 2007). The monoubiquitination and localization of FANCD2 and FANCI to the DNA damage sites are interdependent (Sims et al. 2007, Smogorzewska et al. 2007). Ubiquitin carboxy-terminal hydrolase 1 (USP1) with USP1-associated factor (UAF1) are critical to ID2 deubiquitination for the completion of FA pathway (Figure 2d) (Cohn et al. 2007). However, the dynamics of monoubiquitination and its reversibility are incompletely understood.
The concurrent activation of a checkpoint response is important for the eradication of ICLs (Figure 2c). Long stretches of single-strand DNA (ssDNA) generated from uncoupling of the helicase and polymerase are rapidly coated and stabilized by RPA, thereby activating the ATR/CHK1 pathway (Zou & Elledge 2003). However, how long stretches of ssDNA are generated at the ICLs is unclear, as the helicase is also blocked at the ICLs. The ATR/CHK1 and ATM/CHK2 signaling cascades result in phosphorylation of chromatin-bound factors that promote fork stability, maintain the intra-S-phase checkpoint and promote repair. Direct FANCI phosphorylation by the ATR kinase and its dephosphorylation are components of a critical molecular switch in the FA pathway. This event promotes ID2 monoubiquitination by inducing dissociation of the ID2 complex (Ishiai et al. 2008, Sareen et al. 2012). Cumulatively, the ATR/CHK1 kinases play pivotal roles in the FA pathway at different levels by executing checkpoint responses and promoting ID2 monoubiquitination.
2.3. The Functional Consequences of ID2 Monoubiquitination: The Interstrand Cross-Links Unhooking and Bypass Steps
The ID2-Ub acts as a molecular platform to which various other DNA repair proteins, such as structure-specific nucleases (SSEs) and translesion synthesis (TLS) polymerases, are recruited and docked (Knipscheer et al. 2009). Chromatin binding of the monoubiquitinated ID2 complex controls nucleolytic cleavage at stalled forks to incise the ICL from one of the parental strands by a process known as unhooking (Figure 2e,f).
SSEs are recruited for the unhooking step by the interaction of SLX4/FANCP and FANCD2-Ub (Kim et al. 2011, Stoepker et al. 2011). SLX4 interacts and activates several SSEs, such as the XPF (FANCQ/ERCC4)-ERCC1 heterodimer, MUS81-EME1, and SLX1(Fekairi et al. 2009). FANCP/SLX4 and XPF/FANCQ form a complex in which the endonuclease XPF makes an incision to unhook the ICL. These results are consistent with the recent identification of XPF as the FANCQ complementation group (Bogliolo et al. 2013). FAN1 (Fanconi anemia–associated nuclease 1) was implicated in the unhooking step; however, its role in the ICL repair is enigmatic, as FAN1−/− mice develop chronic kidney disease rather than FA (Zhou et al. 2012).
After unhooking, reminiscents of ICLs still remain on one of the parental DNA strands because they are incompletely removed (Raschle et al. 2008). The nucleotide containing the damaged base needs to be bypassed for DNA replication to resume. The bypass step accounts for the point mutations at the ICL site (Figure 2g). The nascent DNA strand is then extended by an error-free process of extension. The bypass step is executed by REV1 (deoxy-cytidyl transferase inserts deoxycytidine across a guanine or an abasic site), and the extension step is executed by REV3 and REV7 (subunits of Polζ) (Roy & Scharer 2016). The damage bypass by REV1 requires its interaction with FAAP20 and an intact FA core complex but not with FANCD2-Ub. This indicates that the TLS step is autonomously regulated by the FA core complex and does not require FANCD2-Ub (Kim et al. 2012). The unhooking step also generates DNA double-strand breaks (DSBs) that are preferably repaired by HR and the downstream FANC proteins (Figure 2i–l), as many of the downstream FA/BRCA proteins were primarily identified as HR proteins.
3. THE FATE OF DOUBLE-STRAND BREAKS DURING INTERSTRAND CROSS-LINK REPAIR: THE ORCHESTRA OF MULTIPLE DNA REPAIR PATHWAYS
Nucleolytic processing of ICLs by the unhooking step generates DSBs that can be repaired by four major pathways (Figure 2). End resection at DSBs, which is restricted to the S phase, generates ssDNA overhangs that dictate DSB repair pathway choice and repair outcome (Ceccaldi et al. 2016a). In the initial phase of end resection, end clipping of the DSB ends by the MRE11 and CtIP nucleases generates 3′ ssDNA (Figure 2h). The minimally processed ends can be repaired by an error-prone POLθ-dependent alternative nonhomologous end joining (alt-NHEJ) (Figure 2) (Ceccaldi et al. 2015). In a subsequent step, extensive end resection by helicases and exonucleases (BLM, EXO1, and DNA2) generates longer ssDNA lengths required for single-strand annealing (SSA) or HR (Figure 2i) (Daley et al. 2017, Nimonkar et al. 2011). DSB resection and formation of 3′ ssDNA prompts the accumulation of RPA. SSA involves annealing of nucleotide repeats flanking the DSB in a RAD52-dependent manner, as well as the loss of sequences between the intervening repeats (Figure 2). (Bhargava et al. 2016). HR is an accurate templated pathway that is dominant in S phase, where classical nonhomologous end joining (C-NHEJ) is inhibited.
HR is inhibited in G1 phase and is reactivated as the cells enter S phase. HR involves the strand invasion and a homology search step and requires the formation of a RAD51 nucleofilaments, a function provided by the recombination mediators BRCA2 and PALB2 (Buisson et al. 2010). PALB2 binds directly to both BRCA1 and BRCA2, thereby physically linking these two major HR proteins (Figure 2) (Zhang et al. 2009). C-NHEJ can operate throughout the cell cycle, but it is more efficiently executed when end resection is blocked, predominantly in the G0/G1 and G2 phases of the cell cycle. In C-NHEJ, DNA ends are held together by the KU70-KU80 heterodimer, followed by a direct end ligation step catalyzed by the XRCC4/LIG4 ligase complex (Figure 2) (Mahaney et al. 2009). Despite its higher rate of mutagenicity compared to HR, C-NHEJ remains a safeguard against genome instability by suppressing chromosomal translocations at major DSB sites. The interplay between these pathways is not well understood, and SSA and alt-NHEJ can lead to oncogenic transformation due to their inaccuracy.
The hypersensitivity of human, nematode, and chicken DT40 cells mutated for the FA pathway to ICL-inducing agents can be partially rescued by knockdown, inhibition, or deletion of components of C-NHEJ (reviewed in Kottemann & Smogorzewska 2013). In contrast, the ICL sensitivity of FANCD2-depleted mouse embryonic fibroblast cells is aggravated by the deletion of either KU or 53BP1 (Bunting et al. 2012). The contribution of C-NHEJ to the molecular defects of the FA cells is debatable, and extending these findings with other FA proteins might shed light on this topic. Mutations of downstream FA/HR proteins may not interfere with the ICL incision and DSB generation steps. These DSBs can be subject to mutagenic repair by SSA or alternative end joining (alt-EJ), once end resection is promoted, and may significantly contribute to the pathogenicity of the FA cells and associated tumors. The contribution of alt-EJ and SSA to FA-associated genomic instability is poorly understood. Hence, it will be very interesting to determine whether knockdown of the key components of alt-EJ and SSA can rescue these FA cells’ ICL sensitivity.
Biallelic germline mutations in many HR genes result in an FA-like syndrome in which the cells are proficient for ID2 monoubiquitination but are sensitive to cross-linking agents. These mutations are rare in accordance with their role in viability, and patients with these mutations do not develop BMF for unidentified reasons. The homozygous germline mutations in HR genes BRCA1, BRCA2, and PALB2 are often hypomorphic, with residual activity capable of establishing an equilibrium between survival and diminished cellular function. Patients with biallelic BRCA1 exhibit congenital abnormalities, early-onset breast and ovarian cancer, and significant chemotherapy-associated toxicity (Domchek et al. 2013, Sawyer et al. 2015). Patients with biallelic BRCA2 mutations have classical FA pathologies, including cross-linker hypersensitivity, congenital abnormalities, and abnormal skin pigmentation. (Howlett et al. 2002). Homozygous BRCA2 mutations are also associated with a high risk of leukemia during early childhood and in women who received chemotherapy for breast or ovarian cancer (Iqbal et al. 2016, Wagner et al. 2004).
RAD51 is required for HR associated with ICL repair (Long et al. 2011). Cells derived from an FA patient with a pathogenic codominant-negative mutant of RAD51 have exhibited ICL sensitivity, indicating an abrogated ICL repair, but were HR proficient (Wang et al. 2015). The mutant RAD51 protein triggered extensive DNA2-/WRN-dependent end resection at the DNA ICLs, indicating additional roles of RAD51 beyond HR in protecting ICL-induced stalled replication forks. Moreover, the RAD51 nucleofilaments are stabilized by BOD1L, a newly identified player within ICL repair pathway that protects stalled replication forks from DNA2-mediated degradation (Ceccaldi et al. 2016b).
The roles of newer downstream FA genes in the coordination of the FA pathway are less well known. Biallelic mutations in the RAD51 paralogs RAD51C/FANCO and XRCC2/FANCU, in addition to PALB2 and BRCA2, cause FA (Park et al. 2016, Vaz et al. 2010). A patient-derived XRCC2 mutant cell line exhibited reduced levels of the XRCC2-RAD51B-C-D complex (RAD51 paralog complex) and FANCD2 monoubiquitination; however, cells expressing this mutant protein were proficient in the assembly of RAD51 foci (Park et al. 2016). Thus, XRCC2 might operate after the formation of RAD51-ssDNA nucleofilament. The FANCJ helicase, also known as BRIP1 or BACH1, is mutated in hereditary breast cancer and is required for HR. FANCJ functions in ICL repair by interacting with mismatch proteins MLH1 and PMS2 to promote the TLS step and inhibit HR. The interaction of FANCJ with BRCA1 appears to be required to promote HR but not ICL repair; readers are referred to Ceccaldi et al. (2016b) and the references therein. Finally, RFWD3/FANCW an E3 ligase, has been identified as a new FA gene (Knies et al. 2017). RFWD3 polyubiquitnates RPA and RAD51 in an ATM- and ATR-dependent manner. RFWD3 was shown to mediate timely turnover of RPA, and RAD51 is required to progress to late-phase HR, promote repair of stalled replication forks, and suppress the FA phenotype (Elia et al. 2015, Feeney et al. 2017, Inano et al. 2017).
3.1. DNA Resection and the FA Pathway
CtIP and DNA2 are required for end resection at ICL-induced DSBs, as their depletion exacerbates the genomic instability in response to ICL-inducing agents (Karanja et al. 2012, Murina et al. 2014, Unno et al. 2014). Contrarily, loss of expression of DNA2 provides a survival advantage to FANCD2-deficient cells by preventing deleterious resection at stalled replication forks (Karanja et al. 2014). Moreover, FA proteins are required to prevent unwanted digestion of stalled replication forks by DNA2 or MRE11. Excessive end resection at stalled replication forks can be deleterious; however, end resection is required to precipitate HR at the ICL-induced DSBs. Thus, FA seems to be a biphasic pathway in which an initial phase where replication forks are stalled at ICLs requires low activity of the end resection pathway to prevent unwanted degradation of stalled forks, and a later phase then requires it to promote HR of DSBs produced by the ICL excision step.
FANCY/REV7, a newly identified FA gene, promotes end joining contrary to other FA genes by inhibiting DNA end resection at DSBs and unprotected telomeres (Bluteau et al. 2016, Boersma et al. 2015, Xu et al. 2015). The role of REV7 in the DSB repair pathway choice is independent of its interaction with REV1 and REV3, which together form a TLS complex. Depletion of proteins that negatively regulate DNA end resection, such as 53BP1, REV7, and HELB, promotes the survival of BRCA1-mutated cells. REV7, being an FA protein, promotes NHEJ but not HR (Gupta et al. 2018); however, whether the resection-inhibiting property of REV7 (downstream of D2-Ub) is implicated in the FA pathway is unclear. These findings suggest that the regulation of end resection in the FA pathway is complex and still poorly understood.
4. THE ROLE OF FA PROTEINS IN REPLICATION STRESS
Intriguingly, monoubiquitinated FANCI and FANCD2 are involved in the maintenance of the genetically unstable common fragile sites (CFSs) FRA3B and FRA16D (Howlett et al. 2005). These sites are late-replicating hotspots for chromosomal translocations and sister chromatid exchange, and they are frequently associated with malignancies (Figure 4). In mitosis, under-replicated CFSs on different chromatids are linked by ultrafine bridges (UFBs). Failure to appropriately resolve the UFBs leads to chromosomal breakage and micronuclei formation, resulting in chromosomal instability. FANCI and FANCD2 were shown to colocalize at UFBs and are required for targeting the BLM complex to enable their processing and thus to prevent micronucleation (Naim & Rosselli 2009). Many secondary structures in DNA, such as G quadraplexes, RNA-DNA hybrids (R-loops), and stable complexes formed by protein to DNA, are physical obstructions to faithful replication. R-loop-mediated replication stress can activate the FA pathway (Garcia-Rubio et al. 2015, Schwab et al. 2015). Moreover, in FA/HR-deficient cells, abolishing R-loops can rescue replication fork arrest and DNA damage accumulation. In FANCJ-mutated patients, cells exhibited large deletions near the sequences with a high propensity to form G4 motifs (telomeric DNA) (London et al. 2008). However, FANCJ has not yet shown to be directly involved in G4 metabolism.
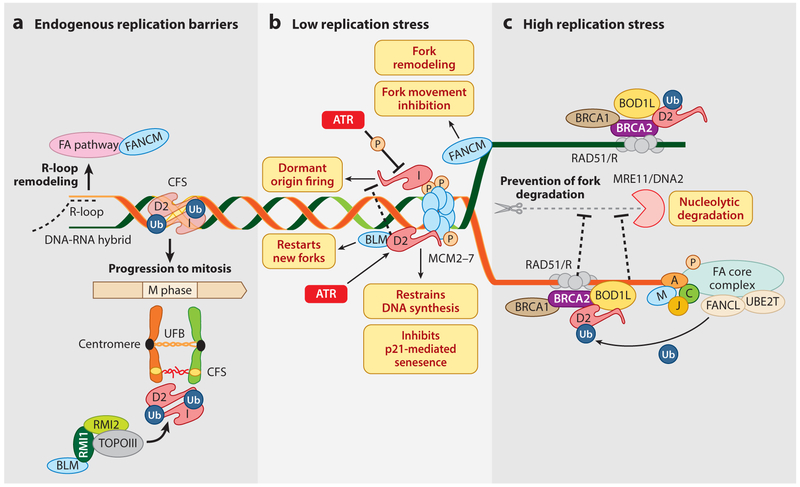
Figure 4.
Roles of FA proteins in replication stress. (a) The FANCI-FANCD2-Ub complex stabilizes the extracentromeric CFSs and mediates loading of the Bloom complex (BLM, RMI1, RMI2, and TOPOIII) on these under-replicated CFSs to ensure their protection, repair, and unperturbed mitosis. The endogenously produced R-loops (RNA-DNA hybrids) at some susceptible genetic loci are remodeled by the components of the FA pathway. (b) At low doses of replication stress, nonubiquitinated FANCD2 binds and inhibits the MCM2–7 helicase complex to restrain DNA synthesis. ATR stimulates binding of FANCD2 to MCM 2–7 to prevent p21-mediated cellular senescence by precluding the accumulation of ssDNA. FANCI also binds to MCM2–7 to fire dormant origins. The dormant origin firing by FANCI is inhibited by its phosphorylation by ATR kinase and the FANCD2-BLM complex. FANCM opposes fork movement, possibly by remodeling the stalled replication forks. (c) High doses of replication stress elicit the classical FA pathway. FANCA, FANCC, FANCJ, and FANCM, together with BOD1L, bind to nascent DNA strands to protect them from MRE11- or DNA2-mediated unwanted nucleolytic degradation. RAD51-ssDNA filaments on stalled replication forks are protected by BRCA1, BRCA2, and FANCD2-Ub by nucleases. Abbreviations: CFS, chromosomal fragile site; FA, Fanconi anemia; ssDNA, single-strand DNA; UFB, ultrafine bridge.
Seminal studies have demonstrated that the FA pathway is activated in response to hydroxyurea (HU), which generates replication stress by depleting the deoxyribonucleotide pool. The functions of the FA proteins in the presence of low and high levels of replication stress are quite different (Figure 4) (Chen et al. 2015, Lossaint et al. 2013, Michl et al. 2016b). Under low levels of replication stress, nonubiquitinated FANCD2, independent of FANCI, interacts and recruits the BLM helicase complex to restart stalled replication forks and suppress the firing of new and dormant origins (Chaudhury et al. 2013). Independent of the FA pathway, FANCD2 and FANCI also associate with the replicative helicase MCM2–7 complex upon ATR-mediated replication stress with different outcomes, as summarized in Figure 4 (Lossaint et al. 2013). FANCD2 and FANCI, which are believed to form a complex for ICL repair, clearly have distinct and independent roles in response to low levels of replication stress. At high levels of replication stress, FANCD2, FANCI, and the FA core complex proteins function cumulatively to confer fork stability and promote replication restart. FANCA-, BRCA1-, BRCA2-, PALB2-, and FANCD2-deficient human cells exhibit genomic instability at stalled replication forks (Figure 4). FANCD2-depleted cells fail to protect stalled replication forks from undesired digestion by Mre11, and this could be rescued by fork protection by BRCA2-stabilized RAD51 (Schlacher et al. 2011, 2012). Strikingly, FANCD2 was shown to play a role in stabilizing the replication forks in BRCA1/2-deficient cells, thus limiting the replication stress in these cells (Kais et al. 2016, Michl et al. 2016a).
Cumulatively, FA proteins play a central role in mitigating replication stress by suppressing dormant origin firing, promoting replication fork stability, and stabilizing CFSs. Interestingly, FA-derived patient cells are mildly sensitive to HU despite their role in coping with replication stress (Lossaint et al. 2013). Thus, deletions or loss-off-function mutations in the FA genes could lead to the accumulation of a chromosomal instability that does not lead directly to cellular demise. Instead, these changes may contribute to an increased risk of malignant transformation in the long term, as evident in FA patients. Gaining a better understanding of the mechanistic details of the FA pathways will also have wide impacts on the prevention, diagnosis, and treatment of somatic cancers in the general patient population.
5. THE RELEVANCE OF THE FA PATHWAY TO CANCER IN THE NON-FA GENERAL POPULATION
5.1. Germline Monoallelic FA Gene Alterations Cause Cancer Predisposition
Germline monoallelic mutations or promoter hypermethylations of FA genes in non-FA patients confer increased risk for multiple cancers. The greatest risk for the development of breast and ovarian cancer is inheritance of mutations in one of the breast cancer susceptibility genes, BRCA1 and BRCA2, leading to a clinical autosomal dominant hereditary breast and ovarian cancer (HBOC) syndrome (Kuchenbaecker et al. 2017) and fallopian tube/ovarian cancers (Burke et al. 1997, Levine et al. 2003). HBOC also increases the risk of pancreatic (Ferrone et al. 2009), stomach, and prostate cancers (Cavanagh & Rogers 2015). In high-grade ovarian carcinoma, BRCA1 and BRCA2 function as classic tumor suppressors, and the cancer development usually associates with loss of heterozygosity (LOH) of the other allele (Merajver et al. 1995).
Other than BRCA1 and BRCA2, germline monoallelic mutations in other FA pathway genes have increasingly been implicated in increased risk of multiple cancer types. Germline mutations of BRIP1/FANCJ have increased risk for ovarian cancer (Rafnar et al. 2011) but not for breast cancer (Easton et al. 2016). Germline mutations in PALB2 have also been implicated in a cumulative 2–4-fold risk increase for breast cancer (Hofstatter et al. 2011, Southey et al. 2010) and an increased prevalence of familial pancreatic cancer (Tischkowitz et al. 2009). Inactivating variants of FANCM increases the risk of triple-negative breast cancer 3.5-fold or more (Kiiski et al. 2014). FANCA deletions are associated with familial breast cancer (Solyom et al. 2011), and mutations in FANCO (RAD51C) are associated with increased prevalence of familial breast and ovarian cancers (Vaz et al. 2010). Similarly, LOH in FANCC or FANCG is associated with early-onset pancreatic cancer (van der Heijden et al. 2003). A monoallelic FANCT (UBE2T) truncation was found in 1 of 450 patients with high-risk breast cancer (Virts et al. 2015). Although some of these findings have not reached population-level statistical significance, the detection and functional validation of new genes and mutations leading to genetic predisposition to cancer are critical for early detection and counseling of patients and their families and for the design of effective preventive measures (Finch et al. 2014). Accumulating evidence suggests that LOH contributes to tumorigenesis among the patients with these germline monoallelic alterations in the FA tumor suppressor pathway (Kanchi et al. 2014, Pelttari et al. 2011).
5.2. FA Genes are Commonly Altered in Somatic Cancers
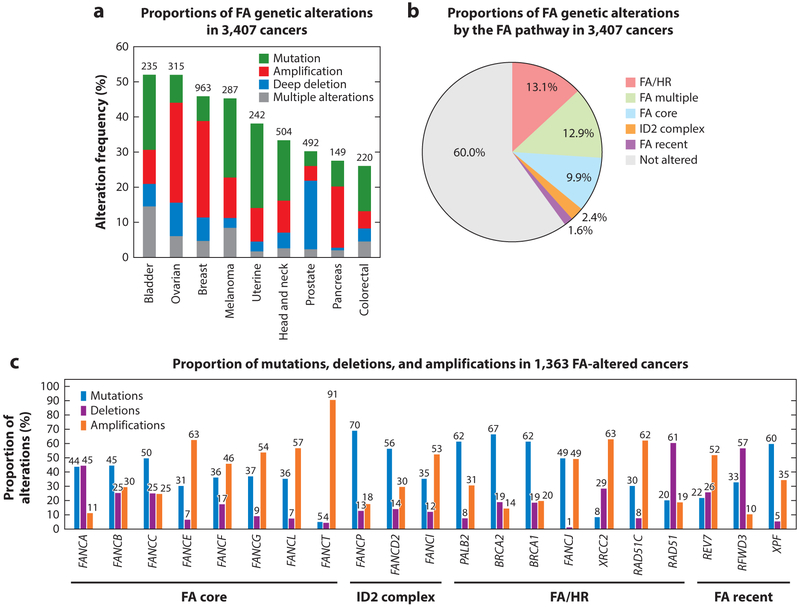
In addition to germline alterations, FA genes are commonly somatically mutated in multiple cancers (Figure 5). In a genomic analysis of nine common cancer types from The Cancer Genome Atlas (TCGA), FA genes were altered in 40% of the tumors, with the majority belonging to the FA/HR pathway (Figure 5a,b) (Duan et al. 2013). Of the single alterations in FA genes, the proportions of functionally different alterations (mutations, deletions, and amplifications) differ across the different complementation groups. For instance, the majority (75%) of the FA/HR pathway gene alterations are characterized by mutations or deep deletions, whereas FA core complex alterations are predominantly amplifications. The spectrum of alterations likely has functional and therapeutic implications. Deletions and loss-of-function mutations induce genomic instability responsible for malignant transformation and cancer progression, but at the same time they confer sensitivity to DNA-damaging treatments. Conversely, amplification and gain-of-function mutations in FA genes may offer an advantage to cancer cells by alleviating replication stress and mitigating DNA damage induced by chemotherapeutics.
Figure 5.
Genetic alterations of the Fanconi anemia (FA) genes (Figure 1) in somatic cancers. (a) Proportions of FA gene mutations and copy number variations in 3,407 cancers of nine common cancer types. (b) Proportions of FA genetic alterations by the FA pathway in 3,407 cancers. FA genes were divided into groups based on their functions listed in Figure 1. At least one FA gene alteration was detected in 40% of the cancers, FA/HR (homologous recombination) being the most commonly altered pathway. (c) Proportions of mutations, deletions, and amplifications in 1,363 FA-altered cancers. Data were generated by The Cancer Genome Atlas and were downloaded from cBioPortal on (a) March 28,2018 and (b,c) May3, 2018.
5.3. FA/HR-Deficient Cancers are Vulnerable to DSB Repair- and DNA Damage Response–Targeted Therapies
FA/HR-deficient cancers commonly respond to non-ICL, DSB-inducing agents, such as topoisomerase I (topotecan) and topoisomerase II (doxorubicin, etoposide) inhibitors (Gordon et al. 2001). These drugs induce DNA adducts that are converted to DSBs toxic in FA/HR-deficient cells. Poly (ADP-ribose) polymerase (PARP) inhibitors are a classical example of a synthetic lethality relationship of diverging DNA repair mechanisms involving the HR pathway. PARP1 inhibition kills HR-deficient cells by several mechanisms (e.g., destabilizing the replication fork and trapping PARP1-PARylation adducts onto DNA at sites of endogenous damage, causing toxicity in HR-deficient cells) (reviewed in Ceccaldi et al. 2015). Clinically, the PARP inhibitor olaparib was the first to show a durable antitumor response in breast and ovarian cancers (Kaufman et al. 2015, Ledermann et al. 2014, Tutt et al. 2010). These cancers commonly have underlying mutations in BRCA1/2 or other FA genes and are generally more sensitive to PARP inhibitors. Currently, three PARP inhibitors, olaparib, rucaparib, and niraparib, are FDA approved for the treatment of relapsed breast and ovarian cancers.
DNA damage response coordinates the appropriate cellular responses to DNA damage, including transcriptional changes, cell cycle checkpoint activation, and DNA damage repair pathway engagement. Importantly, FA deficient cancers are vulnerable to drugs targeting these processes. Of the DSB DNA damage response proteins, inhibitors of DNA-dependent protein kinase, ATM, and ATR are in early-phase clinical trials (Dohmen et al. 2017, Dong et al. 2017, Kondrashova et al. 2017). Similarly, inhibitors of the cell cycle checkpoints CHK1, CHK2, and WEE1 have shown promising antitumor activity in phase I and II trials (Lee et al. 2018, Leijen et al. 2016). Further, new promising preclinical therapeutic targets are also emerging, for example, inhibitors of DNA polymerases, such as POLQ (Higgins & Boulton 2018), or agents targeting deubiquitinating enzymes, such as USP1 (Guervilly et al. 2011).
5.4. Biomarkers for FA Pathway Alterations
The clinical relevance of these specific vulnerabilities to DNA-damaging agents is dependent on reliable biomarkers to detect functional FA defects. Several genomic approaches have been utilized, including identifying (a) single genetic mutations leading to predicted DNA repair/FA deficiency by targeted sequencing of DNA repair mutations (Wagle et al. 2012), (b) gene expression profiles of DNA repair deficiency (Kang et al. 2012, Konstantinopoulos et al. 2010), or (c) specific structural chromosomal aberrations or mutation scars (Abkevich et al. 2012, Birkbak et al. 2012, Polak et al. 2017, Popova et al. 2012, Wang et al. 2017). These genomic features have been implemented either alone or in combinations in clinical testing for DNA repair deficiency, which has profound therapeutic implications (Swisher et al. 2017). This so-called BRCAness phenotype (Turner et al. 2004), detected either by gene expression (Konstantinopoulos et al. 2010) or genomic signatures (Davies et al. 2017), identifies a larger patient population compared to single FA pathway alterations that is likely to benefit from platinum agents and PARP inhibitors. The limitations of these approaches are due to the lack of knowledge about the functionality of DNA repair. First, a deleterious mutation in an individual FA/HR gene can be compensated by rewiring the DNA damage response, leading to at least partial FA/HR DNA repair proficiency (Jaspers et al. 2013). Second, genomic scars are only reflective of the cumulative defects that have occurred in the cancer genome and do not reflect the current functional DNA repair status. Thus, dynamic and functional biomarkers are critically needed for the reliable identification of targetable vulnerabilities in DNA repair pathways.
The most promising functional approaches include assays where the DNA repair deficiency/proficiency can be mechanistically verified within tumor tissue or patient-derived cancer cells, or by assessing, for instance, the formation of RAD51 (Graeser et al. 2010, Mukhopadhyay et al. 2010, Naipal et al. 2014) or FANCD2 foci or FANCD2 monoubiquitination (Duan et al. 2013, Van Der Heijden et al. 2004). Further, patient-derived tumor cells in two-dimensional (2D), 3D, or organoid/tumoroid cultures can be assayed for their sensitivities to different DNA-damaging agents (Finnberg et al. 2017, Mukhopadhyay et al. 2010, van de Wetering et al. 2015, Vlachogiannis et al. 2018). Importantly, functional evaluation of key DNA repair dynamics, such as replication fork protection, in patient-derived models can reveal new targetable vulnerabilities (Yazinski et al. 2017). The challenge in these approaches lies in obtaining clinically relevant tumor tissue and in developing rapid, reproducible assays that functionally match the original tumor and patient treatment responses. The development and validation of functional biomarkers for FA-altered somatic cancers are areas of active research and will be even more important for stratification of patients in clinical trials with novel agents that target DNA damage repair/checkpoint proteins.
5.5. Mechanisms of Resistance to DNA-Damaging Therapies
Resistance to DNA-damaging therapies is common and constitutes a significant barrier to improving patient outcomes. Moreover, the mechanisms of resistance arising from the high cellular adaptability due to DNA repair deficiency and genomic instability are greatly variable. Mechanisms of resistance to ICL-inducing agents (e.g., platinum) range from reducing the bioavailability of the compound to transcriptional and genetic DNA repair alterations and modulation of the tumor microenvironment, as reviewed in Galluzzi et al. (2012), Pogge von Strandmann et al. (2017), and Shen et al. (2012). Clinically, the best characterized mechanism of genetic resistance to platinum and PARP inhibitors in FA/HR-deficient cancers is the somatic reversion of the original mutation, which can be detected from both tumor tissue and circulating cell-free DNA (Goodall et al. 2017, Kondrashova et al. 2017, Norquist et al. 2011, Weigelt et al. 2017). The restoration of DNA repair function can also be achieved by removing hypermethylation or by clonal selection (Schwarz et al. 2015). Rewiring DNA damage repair is a recently discovered mechanism of PARP inhibitor resistance, leading to modifications in the DNA repair pathway choice (Gupta et al. 2018, Jaspers et al. 2013) or replication fork protection (Ray Chaudhuri et al. 2016, Rondinelli et al. 2017), although the clinical relevance of these mechanisms needs to be further established. Uncovering the mechanisms and biomarkers of resistance is especially important when considering future combination therapies for these patients.
6. CONCLUSIONS
The FA pathway preserves genomic instability and is extensively connected with other DNA repair pathways. Despite being a rare disease, FA is important to study for two reasons. First, a better understanding of the molecular pathogenesis of FA can improve the treatment of BMF and associated malignancies. Second, somatic mutations in the FA genes can have profound effects on cancer progression and its treatment and can affect patient survival. The onset and progression of BMF and AML in FA patients is clinically variable, and the underlying molecular mechanisms are poorly understood. Moreover, FA proteins can elicit exclusive tumor-suppressing functions, and the severity of the phenotype is highly dependent on the mutation spectrum and the genetic background. Recent large-scale sequencing efforts on cancers in the general (non-FA) population have revealed somatic mutations in FA genes. The presence of these mutations and the corresponding functional defects in the FA pathway suggest specific therapeutic vulnerabilities of these tumors. For instance, biomarkers of the FA pathway are useful in predicting the PARP inhibitor sensitivity of these tumors. Functional validation of the alterations requires robust molecular research, which can lead to the development of rational biomarkers and novel therapies to improve treatment outcomes and the survival of not only FA patients but also patients with FA-altered somatic cancers in the general population.
ACKNOWLEDGMENTS
We thank Alfredo Rodriquéz, Connor Clairmont, Jia Zhou, and Prabha Sarangi for critical reading of the manuscript.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Abkevich V, Timms KM, Hennessy BT, Potter J, Carey MS, et al. 2012. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer 107:1776–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpi AF, Patel KJ. 2009. Monoubiquitylation in the Fanconi anemia DNA damage response pathway. DNA Repair 8:430–35 [DOI] [PubMed] [Google Scholar]
- Andreassen PR, D’Andrea AD, Taniguchi T. 2004. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 18:1958–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD. 1993. Fanconi anemia diagnosis and the diepoxybutane (DEB) test. Exp. Hematol. 21:731–33 [PubMed] [Google Scholar]
- Auerbach AD. 2009. Fanconi anemia and its diagnosis. Mutat. Res 668:4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava R, Onyango DO, Stark JM. 2016. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 32:566–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkbak NJ, Wang ZC, Kim JY, Eklund AC, Li Q, et al. 2012. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2:366–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluteau D, Masliah-Planchon J, Clairmont C, Rousseau A, Ceccaldi R, et al. 2016. Biallelic inactivation of REV7 is associated with Fanconi anemia. J. Clin. Investig. 126:3580–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma V, Moatti N, Segura-Bayona S, Peuscher MH, van der Torre J, et al. 2015. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature 521:537–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, et al. 2013. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am. J. Hum. Genet 92:800–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson R, Dion-Cote AM, Coulombe Y, Launay H, Cai H, et al. 2010. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 17:1247–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, et al. 2012. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell 46:125–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke W, Daly M, Garber J, Botkin J, Kahn MJ, et al. 1997. Recommendations for follow-up care of individuals with an inherited predisposition to cancer: II. BRCA1 and BRCA2. JAMA 277:997–1003 [PubMed] [Google Scholar]
- Castella M, Jacquemont C, Thompson EL, Yeo JE, Cheung RS, et al. 2015. FANCI regulates recruitment of the FA core complex at sites of DNA damage independently of FANCD2. PLOS Genet. 11:e1005563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh H, Rogers KM. 2015. The role of BRCA1 and BRCA2 mutations in prostate, pancreatic and stomach cancers. Hered Cancer Clin. Pract 13:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, et al. 2015. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518:258–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Rondinelli B, D’Andrea AD. 2016a. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26:52–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Sarangi P, D’Andrea AD. 2016b. The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol 17:337–49 [DOI] [PubMed] [Google Scholar]
- Chaudhury I, Sareen A, Raghunandan M, Sobeck A. 2013. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res. 41:6444–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Jones MJ, Yin Y, Crist SB, Colnaghi L, et al. 2015. ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol. Cell 58:323–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, et al. 2007. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell 25:331–43 [DOI] [PubMed] [Google Scholar]
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, et al. 2007. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell 28:786–97 [DOI] [PubMed] [Google Scholar]
- D’Andrea AD, Grompe M. 2003. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 3:23–34 [DOI] [PubMed] [Google Scholar]
- Daley JM, Jimenez-Sainz J, Wang W, Miller AS, Xue X, et al. 2017. Enhancement of BLM-DNA2-mediated long-range DNA end resection by CtIP. Cell Rep. 21:324–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, et al. 2017. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med 23:517–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen AJC, Qiao X, Duursma A, Wijdeven RH, Lieftink C, et al. 2017. Identification of a novel ATM inhibitor with cancer cell specific radiosensitization activity. Oncotarget 8:73925–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domchek SM, Tang J, Stopfer J, Lilli DR, Hamel N, et al. 2013. Biallelic deleterious BRCA1 mutations in a woman with early-onset ovarian cancer. Cancer Discov. 3:399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Zhang T, Ren Y, Wang Z, Ling CC, et al. 2017. Inhibiting DNA-PKcs in a non-homologous end-joining pathway in response to DNA double-strand breaks. Oncotarget 8:22662–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W, Gao L, Zhao W, Leon M, Sadee W, et al. 2013. Assessment of FANCD2 nuclear foci formation in paraffin-embedded tumors: a potential patient-enrichment strategy for treatment with DNA interstrand crosslinking agents. Transl. Res 161:156–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Lesueur F, Decker B, Michailidou K, Li J, et al. 2016. No evidence that protein truncating variants in BRIP1 are associated with breast cancer risk: implications for gene panel testing. J. Med. Genet 53:298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia AE, Wang DC, Willis NA, Boardman AP, Hajdu I, et al. 2015. RFWD3-dependent ubiquitination of RPA regulates repair at stalled replication forks. Mol. Cell 60:280–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoiu M, Jiricny J, Scharer OD. 2012. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 40:8953–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney L, Munoz IM, Lachaud C, Toth R, Appleton PL, et al. 2017. RPA-mediated recruitment of the E3 ligase RFWD3 is vital for interstrand crosslink repair and human health. Mol. Cell 66:610–21.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, et al. 2009. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138:78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrone CR, Levine DA, Tang LH, Allen PJ, Jarnagin W, et al. 2009. BRCA germline mutations in Jewish patients with pancreatic adenocarcinoma. J. Clin. Oncol 27:433–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch AP, Lubinski J, Moller P, Singer CF, Karlan B, et al. 2014. Impact of oophorectomy on cancer incidence and mortalityin women with a BRCA1 or BRCA2 mutation. J. Clin. Oncol 32:1547–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnberg NK, Gokare P, Lev A, Grivennikov SI, MacFarlane AWT, et al. 2017. Application of 3D tumoroid systems to define immune and cytotoxic therapeutic responses based on tumoroid and tissue slice culture molecular signatures. Oncotarget 8:66747–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohnmayer D, Frohnmayer L, Guinan E, Kennedy T, Larsen K, eds. 2014. Fanconi Anemia: Guidelines for Diagnosis and Management. Eugene, OR: Fanconi Anemia Res. Fund; 4th ed. [Google Scholar]
- Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, et al. 2012. Molecular mechanisms of cisplatin resistance. Oncogene 31:1869–83 [DOI] [PubMed] [Google Scholar]
- Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. 2012. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature 489:571–75 [DOI] [PubMed] [Google Scholar]
- Garaycoechea JI, Patel KJ. 2014. Why does the bone marrow fail in Fanconi anemia? Blood 123:26–34 [DOI] [PubMed] [Google Scholar]
- Garcia-Rubio ML, Perez-Calero C, Barroso SI, Tumini E, Herrera-Moyano E, et al. 2015. The Fanconi Anemia pathway protects genome integrity from R-loops. PLOS Genet. 11:e1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennery AR, Slatter MA, Bhattacharya A, Barge D, Haigh S, et al. 2004. The clinical and biological overlap between Nijmegen Breakage Syndrome and Fanconi anemia. Clin. Immunol 113:214–19 [DOI] [PubMed] [Google Scholar]
- Giampietro PF, Adler-Brecher B, Verlander PC, Pavlakis SG, Davis JG, Auerbach AD. 1993. The need for more accurate and timely diagnosis in Fanconi anemia: a report from the International Fanconi Anemia Registry. Pediatrics 91:1116–20 [PubMed] [Google Scholar]
- Goodall J, Mateo J, Yuan W, Mossop H, Porta N, et al. 2017. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 7:1006–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AN, Fleagle JT, Guthrie D, Parkin DE, Gore ME, Lacave AJ. 2001. Recurrent epithelial ovarian carcinoma: a randomized phase III study of pegylated liposomal doxorubicin versus topotecan. J. Clin. Oncol 19:3312–22 [DOI] [PubMed] [Google Scholar]
- Graeser M, McCarthy A, Lord CJ, Savage K, Hills M, et al. 2010. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin. Cancer Res 16:6159–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guervilly JH, Renaud E, Takata M, Rosselli F. 2011. USP1 deubiquitinase maintains phosphorylated CHK1 by limiting its DDB1-dependent degradation. Hum. Mol. Genet 20:2171–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Somyajit K, Narita T, Maskey E, Stanlie A, et al. 2018. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell 173:972–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins GS, Boulton SJ. 2018. Beyond PARP–POLθ as an anticancer target. Science 359:1217–18 [DOI] [PubMed] [Google Scholar]
- Hira A, Yabe H, Yoshida K, Okuno Y, Shiraishi Y, et al. 2013. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood 122:3206–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavin EM, Smeaton MB, Noronha AM, Wilds CJ, Miller PS. 2010. Cross-link structure affects replication-independent DNA interstrand cross-link repair in mammalian cells. Biochemistry 49:3977–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstatter EW, Domchek SM, Miron A, Garber J, Wang M, et al. 2011. PALB2 mutations in familial breast and pancreatic cancer. Fam. Cancer 10:225–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD, Glover TW. 2005. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum. Mol. Genet 14:693–701 [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, et al. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297:606–9 [DOI] [PubMed] [Google Scholar]
- Huang M, Kim JM, Shiotani B, Yang K, Zou L, D’Andrea AD. 2010. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol. Cell 39:259–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li L. 2013. DNA crosslinking damage and cancer–a tale of friend and foe. Transl. Cancer Res 2:144–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inano S, Sato K, Katsuki Y, Kobayashi W, Tanaka H, et al. 2017. RFWD3-mediated ubiquitination promotes timely removal of both RPA and RAD51 from DNA damage sites to facilitate homologous recombination. Mol. Cell 66:622–34.e8 [DOI] [PubMed] [Google Scholar]
- Iqbal J, Nussenzweig A, Lubinski J, Byrski T, Eisen A, et al. 2016. The incidence of leukaemia in women with BRCA1 and BRCA2 mutations: an International Prospective Cohort Study. Br. J. Cancer 114:1160–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, et al. 2008. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat. Struct. Mol. Biol 15:1138–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers JE, Kersbergen A, Boon U, Sol W, van Deemter L, et al. 2013. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 3:68–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kais Z, Rondinelli B, Holmes A, O’Leary C, Kozono D, et al. 2016. FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep. 15:2488–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchi KL, Johnson KJ, Lu C, McLellan MD, Leiserson MDM, et al. 2014. Integrated analysis of germline and somatic variants in ovarian cancer. Nat. Commun 5:3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, D’Andrea AD, Kozono D. 2012. A DNA repair pathway–focused score for prediction of outcomes in ovarian cancer treated with platinum-based chemotherapy. J. Natl. Cancer Inst 104:670–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanja KK, Cox SW, Duxin JP, Stewart SA, Campbell JL. 2012. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle 11:3983–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanja KK, Lee EH, Hendrickson EA, Campbell JL. 2014. Preventing over-resection by DNA2 helicase/nuclease suppresses repair defects in Fanconi anemia cells. Cell Cycle 13:1540–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman B, Shapira-Frommer R, Schmutzler RK, Audeh MW, Friedlander M, et al. 2015. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol 33:244–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiiski JI, Pelttari LM, Khan S, Freysteinsdottir ES, Reynisdottir I, et al. 2014. Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer. PNAS 111:15172–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Yang K, Dejsuphong D, D’Andrea AD. 2012. Regulation of Rev1 by the Fanconi anemia core complex. Nat. Struct. Mol. Biol 19:164–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. 2011. Mutations of the SLX4 gene in Fanconi anemia. Nat. Genet. 43:142–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knies K, Inano S, Ramirez MJ, Ishiai M, Surralles J, et al. 2017. Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. J. Clin. Investig 127:3013–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, et al. 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326:1698–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashova O, Nguyen M, Shield-Artin K, Tinker AV, Teng NNH, et al. 2017. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 7:984–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinopoulos PA, Spentzos D, Karlan BY, Taniguchi T, Fountzilas E, et al. 2010. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J. Clin. Oncol 28:3555–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottemann MC, Smogorzewska A. 2013. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493:356–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, et al. 2017. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317:2402–16 [DOI] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. 2011. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475:53–58 [DOI] [PubMed] [Google Scholar]
- Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, et al. 2014. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 15:852–61 [DOI] [PubMed] [Google Scholar]
- Lee JM, Nair J, Zimmer A, Lipkowitz S, Annunziata CM, et al. 2018. Prexasertib, a cell cycle checkpoint kinase 1 and 2 inhibitor, in BRCA wild-type recurrent high-grade serous ovarian cancer: a first-in-class proof-of-concept phase 2 study. Lancet Oncol. 19:207–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leijen S, van Geel RM, Sonke GS, de Jong D, Rosenberg EH, et al. 2016. Phase II study of WEE1 inhibitor AZD1775 plus carboplatin in patients with TP53-mutated ovarian cancer refractory or resistant to first-line therapy within 3 months. J. Clin. Oncol 34:4354–61 [DOI] [PubMed] [Google Scholar]
- Levine DA, Argenta PA, Yee CJ, Marshall DS, Olvera N, et al. 2003. Fallopian tube and primary peritoneal carcinomas associated with BRCA mutations. J. Clin. Oncol 21:4222–27 [DOI] [PubMed] [Google Scholar]
- Liang CC, Zhan B, Yoshikawa Y, Haas W, Gygi SP, Cohn MA. 2015. UHRF1 is a sensor for DNA interstrand crosslinks and recruits FANCD2 to initiate the Fanconi anemia pathway. Cell Rep. 10:1947–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London TB, Barber LJ, Mosedale G, Kelly GP, Balasubramanian S, et al. 2008. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem 283:36132–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DT, Joukov V, Budzowska M, Walter JC. 2014. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol. Cell 56:174–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DT, Raschle M, Joukov V, Walter JC. 2011. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333:84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossaint G, Larroque M, Ribeyre C, Bec N, Larroque C, et al. 2013. FANCD2 binds MCM proteins and controls replisome function upon activation of S phase checkpoint signaling. Mol. Cell 51:678–90 [DOI] [PubMed] [Google Scholar]
- Mahaney BL, Meek K, Lees-Miller SP. 2009. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J 417:639–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley J, Masand N, McGowan R, Rajagopalan S, Hunter A, et al. 2011. X-linked VACTERL with hydrocephalus syndrome: further delineation of the phenotype caused by FANCB mutations. Am. J. Med. Genet. A 155:2370–80 [DOI] [PubMed] [Google Scholar]
- Medhurst AL, Laghmani EH, Steltenpool J, Ferrer M, Fontaine C, et al. 2006. Evidence for subcomplexes in the Fanconi anemia pathway. Blood 108:2072–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, et al. 2003. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet 35:165–70 [DOI] [PubMed] [Google Scholar]
- Merajver SD, Frank TS, Xu J, Pham TM, Calzone KA, et al. 1995. Germline BRCA1 mutations and loss of the wild-type allele in tumors from families with early onset breast and ovarian cancer. Clin. Cancer Res 1(5):539–44 [PubMed] [Google Scholar]
- Michl J, Zimmer J, Buffa FM, McDermott U, Tarsounas M. 2016a. FANCD2 limits replication stress and genome instability in cells lacking BRCA2. Nat. Strut. Mol. Biol 23:755–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl J, Zimmer J, Tarsounas M. 2016b. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J. 35:909–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, D’Andrea AD. 2009. How the Fanconi anemia pathway guards the genome. Annu. Rev. Genet 43:223–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Elattar A, Cerbinskaite A Wilkinson SJ, Drew Y, et al. 2010. Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. Clin. Cancer Res 16:2344–51 [DOI] [PubMed] [Google Scholar]
- Murina O, von Aesch C, Karakus U, Ferretti LP, Bolck HA, et al. 2014. FANCD2 and CtIP cooperate to repair DNA interstrand crosslinks. Cell Rep. 7:1030–38 [DOI] [PubMed] [Google Scholar]
- Naim V, Rosselli F. 2009. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat. Cell Biol 11:761–68 [DOI] [PubMed] [Google Scholar]
- Naipal KA, Verkaik NS, Ameziane N, van Deurzen CH, Ter Brugge P, et al. 2014. Functional ex vivo assay to select homologous recombination-deficient breast tumors for PARP inhibitor treatment. Clin. Cancer Res 20:4816–26 [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, et al. 2011. BLM–DNA2–RPA–MRN and EXO1–BLM–RPA–MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25:350–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niraj J, Caron MC, Drapeau K, Berube S, Guitton-Sert L, et al. 2017. The identification of FANCD2 DNA binding domains reveals nuclear localization sequences. Nucleic Acids Res. 45:8341–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norquist B, Wurz KA, Pennil CC, Garcia R, Gross J, et al. 2011. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J. Clin. Oncol 29:3008–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Virts EL, Jankowska A, Wiek C, Othman M, et al. 2016. Complementation of hypersensitivity to DNA interstrand crosslinking agents demonstrates that XRCC2 is a Fanconi anaemia gene. J. Med. Genet. 53:672–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelttari LM, Heikkinen T, Thompson D, Kallioniemi A, Schleutker J, et al. 2011. RAD51C is a susceptibility gene for ovarian cancer. Hum. Mol. Genet 20(16):3278–88 [DOI] [PubMed] [Google Scholar]
- Pogge von Strandmann E, Reinartz S, Wager U, Muller R. 2017. Tumor–host cell interactions in ovarian cancer: pathways to therapy failure. Trends Cancer 3:137–48 [DOI] [PubMed] [Google Scholar]
- Polak P Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, et al. 2017. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat. Genet 49:1476–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova T, Manie E, Rieunier G, Caux-Moncoutier V, Tirapo C, et al. 2012. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 72:5454–62 [DOI] [PubMed] [Google Scholar]
- Rafnar T, Gudbjartsson DF, Sulem P, Jonasdottir A, Sigurdsson A, et al. 2011. Mutations in BRIP1 confer high risk of ovarian cancer. Nat. Genet. 43:1104–7 [DOI] [PubMed] [Google Scholar]
- Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, et al. 2008. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134:969–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, et al. 2016. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535:382–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman KA, Lach FP, Abhyankar A, Donovan FX, Sanborn EM, et al. 2015. Deficiency of UBE2T, the E2 ubiquitin ligase necessary for FANCD2 and FANCI ubiquitination, causes FA-T subtype of Fanconi anemia. Cell Rep. 12:35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, et al. 2007. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 67:11117–22 [DOI] [PubMed] [Google Scholar]
- Rohleder F, Huang J, Xue Y, Kuper J, Round A, et al. 2016. FANCM interacts with PCNA to promote replication traverse of DNA interstrand crosslinks. Nucleic Acids Res. 44:3219–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondinelli B, Gogola E, Yucel H, Duarte AA, van de Ven M, et al. 2017. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat. Cell Biol 19:1371–78 [DOI] [PubMed] [Google Scholar]
- Rosenberg PS, Tamary H, Alter BP. 2011. How high are carrier frequencies of rare recessive syndromes? Contemporary estimates for Fanconi Anemia in the United States and Israel. Am. J. Med. Genet. A 155:1877–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy U, Scharer OD. 2016. Involvement of translesion synthesis DNA polymerases in DNA interstrand crosslink repair. DNA Repair 44:33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen A, Chaudhury I, Adams N, Sobeck A. 2012. Fanconi anemia proteins FANCD2 and FANCI exhibit different DNA damage responses during S-phase. Nucleic Acids Res 40:8425–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Tian L, Kahkonen M, Schwartzentruber J, Kircher M, et al. 2015. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 5:135–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145:529–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22:106–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, et al. 2015. The Fanconi Anemia pathway maintains genome stability by coordinating replication and transcription. Mol. Cell 60:351–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz RF, Ng CK, Cooke SL, Newman S, Temple J, et al. 2015. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLOS Med. 12:e1001789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semlow DR, Zhang J, Budzowska M, Drohat AC, Walter JC. 2016. Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell 167:498–511.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen DW, Pouliot LM, Hall MD, Gottesman MM. 2012. Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol. Rev 64:706–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims AE, Spiteri E, Sims RJ 3rd, Arita AG, Lach FP, et al. 2007. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol 14:564–67 [DOI] [PubMed] [Google Scholar]
- Singh TR, Saro D, Ali AM, Zheng XF, Du CH, et al. 2010. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol. Cell 37:879–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER 3rd, Hurov KE, et al. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129:289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobeck A, Stone S, Hoatlin ME. 2007. DNA structure-induced recruitment and activation of the Fanconi anemia pathway protein FANCD2. Mol. Cell Biol 27:4283–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solyom S, Winqvist R, Nikkila J, Rapakko K, Hirvikoski P, et al. 2011. Screening for large genomic rearrangements in the FANCA gene reveals extensive deletion in a Finnish breast cancer family. Cancer Lett. 302:113–18 [DOI] [PubMed] [Google Scholar]
- Southey MC, Teo ZL, Dowty JG, Odefrey FA, Park DJ, et al. 2010. A PALB2 mutation associated with high risk of breast cancer. Breast Cancer Res 12:R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, et al. 2011. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat. Genet 43:138–41 [DOI] [PubMed] [Google Scholar]
- Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, et al. 2008. Interstrand DNA cross-links induced by α,β-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc. Chem. Res 41:793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, et al. 2017. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 18:75–87 [DOI] [PubMed] [Google Scholar]
- Tischkowitz MD, Sabbaghian N, Hamel N, Borgida A, Rosner C, et al. 2009. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology 137:1183–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Tutt A, Ashworth A. 2004. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat. Rev. Cancer 4:814–19 [DOI] [PubMed] [Google Scholar]
- Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, et al. 2010. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 376:235–44 [DOI] [PubMed] [Google Scholar]
- Unno J, Itaya A, Taoka M, Sato K, Tomida J, et al. 2014. FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep. 7:1039–47 [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, et al. 2015. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161:933–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden MS, Brody JR, Kern SE. 2004. Functional screen of the Fanconi anemia pathway in cancer cells by Fancd2 immunoblot. Cancer Biol. Ther. 3:534–37 [DOI] [PubMed] [Google Scholar]
- van der Heijden MS, Yeo CJ, Hruban RH, Kern SE. 2003. Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 63:2585–88 [PubMed] [Google Scholar]
- Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, et al. 2010. Mutation of the RAD51C gene in a Fanconi anemia–like disorder. Nat. Genet 42:406–9 [DOI] [PubMed] [Google Scholar]
- Virts EL, Jankowska A, Mackay C, Glaas MF, Wiek C, et al. 2015. AluY-mediated germline deletion, duplication and somatic stem cell reversion in UBE2T defines a new subtype of Fanconi anemia. Hum. Mol. Genet 24:5093–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, et al. 2018. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359:920–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, et al. 2012. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2:82–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JE, Tolar J, Levran O, Scholl T, Deffenbaugh A, et al. 2004. Germline mutations in BRCA2: shared genetic susceptibility to breast cancer, early onset leukemia, and Fanconi anemia. Blood 103:3226–29 [DOI] [PubMed] [Google Scholar]
- Walden H, Deans AJ. 2014. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu. Rev. Biophys 43:257–78 [DOI] [PubMed] [Google Scholar]
- Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, et al. 2015. A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol. Cell 59:478–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AT, Smogorzewska A. 2015. SnapShot: Fanconi anemia and associated proteins. Cell 160:354.e1 [DOI] [PubMed] [Google Scholar]
- Wang Y, Leung JW, Jiang Y, Lowery MG, Do H, et al. 2013. FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions. Mol. Cell 49:997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YK, Bashashati A, Anglesio MS, Cochrane DR, Grewal DS, et al. 2017. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat. Genet 49:856–65 [DOI] [PubMed] [Google Scholar]
- Weigelt B, Comino-Mendez I, de Bruijn I, Tian L, Meisel JL,et al. 2017. Diverse BRCA1 and BRCA2 reversion mutations in circulating cell-free DNA of therapy-resistant breast or ovarian cancer. Clin. Cancer Res. 23:6708–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HL, Gottesman ME, Gautier J. 2012. Replication-independent repair of DNA interstrand crosslinks. Mol. Cell 47:140–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Chapman JR, Brandsma I, Yuan J, Mistrik M, et al. 2015. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521:541–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Delannoy M, Ling C, Daee D, Osman F, et al. 2010. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell 37:865–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazinski SA, Comaills V, Buisson R, Genois MM, Nguyen HD, et al. 2017. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 31:318–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Fan Q, Ren K, Andreassen PR. 2009. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol. Cancer Res 7:1110–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZhouW Otto EA, Cluckey A, Airik R, Hurd TW, et al. 2012. FAN1 mutations cause karyomegalicinterstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat. Genet 44:910–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–48 [DOI] [PubMed] [Google Scholar]