Abstract
In Caenorhabditis elegans, RNA interference (RNAi) responses can transmit across generations via small RNAs. RNAi inheritance is associated with Histone-3-Lysine-9 tri-methylation (H3K9me3) of the targeted genes. In other organisms, maintenance of silencing requires a feed-forward loop between H3K9me3 and small RNAs. Here, we show that in C. elegans not only is H3K9me3 unnecessary for inheritance, the modification’s function depends on the identity of the RNAi-targeted gene. We found an asymmetry in the requirement for H3K9me3 and the main worm H3K9me3 methyltransferases, SET-25 and SET-32. Both methyltransferases promote heritable silencing of the foreign gene gfp, but are dispensable for silencing of the endogenous gene oma-1. Genome-wide examination of heritable endogenous small interfering RNAs (endo-siRNAs) revealed that endo-siRNAs that depend on SET-25 and SET-32 target newly acquired and highly H3K9me3 marked genes. Thus, ‘repressive’ chromatin marks could be important specifically for heritable silencing of genes which are flagged as ‘foreign’, such as gfp.
Editorial note: This article has been through an editorial process in which the authors decide how to respond to the issues raised during peer review. The Reviewing Editor's assessment is that all the issues have been addressed (see decision letter).
Research organism: C. elegans
Introduction
RNA interference (RNAi) responses are inherited in Caenorhabditis elegans nematodes across generations via heritable small RNAs (Alcazar et al., 2008; Buckley et al., 2012; Vastenhouw et al., 2006). In worms, exposure to a number of environmental challenges, such as viral infection (Gammon et al., 2017; Rechavi et al., 2011), starvation (Rechavi et al., 2014), heat (Klosin et al., 2017), and growth in liquid (Lev et al., 2018) induces heritable physiological responses that persist for multiple generations. Inheritance of such transmitted information was linked to inheritance of small RNAs and chromatin modifications, and hypothesized to protect and prepare the progeny for the environmental challenges that the ancestors met.
By base-pairing with complementary mRNA sequences, small RNAs in C. elegans control the expression of thousands of genes, and protect the genome from foreign elements (Luteijn and Ketting, 2013; Malone and Hannon, 2009). Via recruitment of RNA-binding proteins, small interfering RNAs (siRNAs) can induce gene silencing also by inhibiting transcription (Castel and Martienssen, 2013).
Small RNA-mediated transcription inhibition involves modification of histones, however the exact role that histone marks play in inheritance of RNAi and small RNA synthesis is still not entirely clear (Rechavi and Lev, 2017). In C. elegans small RNAs that enter the nucleus were shown to inhibit the elongation phase of Pol II (Guang et al., 2010); In addition, nuclear small RNAs are thought to recruit histone modifiers to the target’s chromatin, resulting in deposition of histone marks such as histone H3K9-tri methylation (H3K9me3) and H3K27me3 (Gu et al., 2012; Lev et al., 2017; Mao et al., 2015).
The interactions between small RNAs and repressive chromatin marks are reciprocal: in Arabidopsis thaliana (Holoch and Moazed, 2015; Molnar et al., 2010) and Schizosaccharomyces pombe (Moazed et al., 2006; Verdel et al., 2004; Hall et al., 2002) small RNAs and repressive histone marks form a self-reinforcing feed-forward loop, where nuclear small RNAs induce deposition of repressive histone marks, and in turn the repressive chromatin marks recruit the small RNA machinery to synthesize additional small RNAs. Whether a similar feedback operates in worms and other organisms, is still under investigation. In Neurospora crassa, transgene-induced small RNAs work independently of H3K9me3 (Chicas et al., 2005). In C. elegans, it was previously suggested that H3K9me is required for RNAi inheritance (Shirayama et al., 2012). However, studies from different groups have shown that the situation is more complex, and that H3K9me could be dispensable, and can even suppress heritable silencing of some targets (Kalinava et al., 2017; Lev et al., 2017; Minkina and Hunter, 2017).
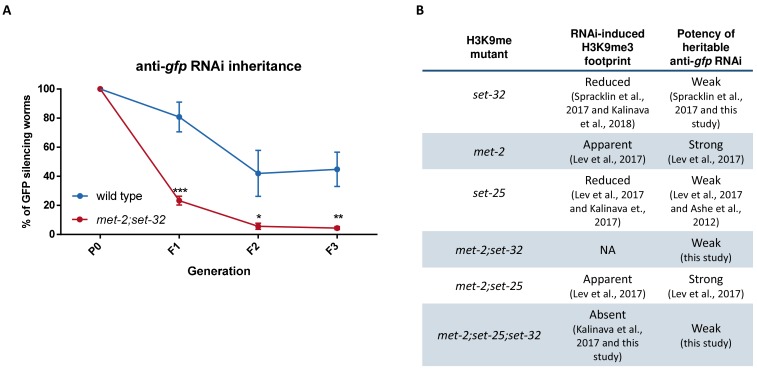
In C. elegans H3K9me is considered to depend mainly on the methyltransferases MET-2, SET-25, and SET-32 (Kalinava et al., 2017; Spracklin et al., 2017; Towbin et al., 2012). H3K9 methylation by MET-2 and SET-25 occurs in a step-wise fashion – after MET-2 deposits the first two methyl groups (H3K9me1/2), SET-25 can add the third methyl group (me3) (Towbin et al., 2012). In the germline, however, SET-25 is capable of tri-methylating H3K9 in a MET-2-independent manner (Bessler et al., 2010; Towbin et al., 2012). SET-32-dependent H3K9me3 is at least in part independent of the activity of SET-25 or MET-2 (Kalinava et al., 2017).
To study the roles of H3K9me3 in the maintenance of heritable small RNAs, we examined the inheritance of small RNAs in mutants defective in these histone methyltransferases. Although H3K9me3 was thought to be required for heritable RNAi (Ashe et al., 2012; Gu et al., 2012), we found the heritable RNAi-responses are greatly potentiated in met-2 mutant background (Lev et al., 2017). Our data indicated that the enhanced strength of the RNAi responses in met-2 mutants stems from a genome-wide massive loss of different endogenous small RNA (endo-siRNAs) species. In normal circumstances, these endo-siRNAs compete with exogenously derived siRNAs over shared biosynthesis components required for small RNA production or inheritance (Lev et al., 2017). In addition, we found that the accumulated sterility (or ‘Mortal Germline’, Mrt phenotype) of met-2 mutants results from dysfunctional small RNA inheritance (Lev et al., 2017).
However, our previous results regarding the role of H3K9me1/2 (deposited by MET-2) did not rule out the possibility that H3K9me3 is yet required for efficient heritable silencing of gfp transgenes: We found that RNAi responses in met-2 mutants nevertheless lead to marking of the target gene’s histones with a heritable H3K9me3 modification. Further, a comparison of the H3K9me3 signal on the gfp locus in different mutants has shown that anti-gfp RNAi responses were strongly inherited only in genetic backgrounds where some H3K9me3 trace could be detected (i.e, in wild type, met-2, and met-2;set-25 mutants). In set-25 single mutants, where no statistically significant H3K9me3 footprint could be detected, anti-gfp RNAi was only weakly inherited. Previously, set-25 mutants were reported to be deficient in heritable RNAi responses targeting different fluorescent transgenes (Ashe et al., 2012; Lev et al., 2017).
RNAi silencing of the endogenous oma-1 gene is also inherited transgenerationally. In contrast to anti-gfp heritable RNAi responses, for which H3K9me3 is important, we detected an enhancement in the inheritance potency of anti-oma-1 RNAi in set-25 mutants (Lev et al., 2017). However, in that study we did not examine whether an H3K9me3 footprint was deposited on the endogenous gene oma-1 in the set-25 background (Lev et al., 2017). The publication of a recent paper (Kalinava et al., 2017) which described strong anti-oma-1 RNAi inheritance in met-2;set-25;set-32 triple mutants, despite the absence of a detectable H3K9me3 footprint, prompted us to re-examine the inheritance of anti-gfp RNAi in this triple mutants. We hypothesized that gene-specific characteristics lead to contrasting requirements for H3K9me3 and specific methyltransferases. In this manuscript, we describe an asymmetry in the requirement for H3K9me3 and specific methyltransferases for heritable RNAi responses aimed against the endogenous gene oma-1 and the foreign gene gfp. These differences led us to perform a genome-wide analysis of H3K9 methyltransferase-dependent small RNAs, which revealed that the endo-siRNAs, which depend on H3K9me3 target newly acquired C. elegans genes that might be considered ‘foreign’, similarly to gfp.
Results
Recently Kalinava et al. examined the heritable RNAi responses against oma-1 also in a triple mutant, lacking the three main C. elegans H3K9 methyltransferases, SET-25, SET-32 and MET-2 (Kalinava et al., 2017). The authors reported that silencing of oma-1 was independent of H3K9me3, as in these mutants RNAi responses raised against the oma-1 gene were heritable despite the lack of an H3K9me3 trace (Kalinava et al., 2017).
We successfully replicated the results of Kalinava et al., and came to the same conclusion, that the met-2;set-25;set-32 triple mutant worms inherit RNAi responses against the oma-1 gene, also when we used a different assay for inheritance (Figure 1A and Figure 1B, upper panel). Unlike Kalinava et al., which used qPCR to score for downregulation of oma-1 expression, we targeted a redundant, temperature-sensitive and dominant oma-1 allele, that in the restrictive temperatures does not allow the development of embryos unless silenced (as previously described [Alcazar et al., 2008]). Upon shifting to 20 degrees, only worms that silence the oma-1 gene in a heritable manner survive.
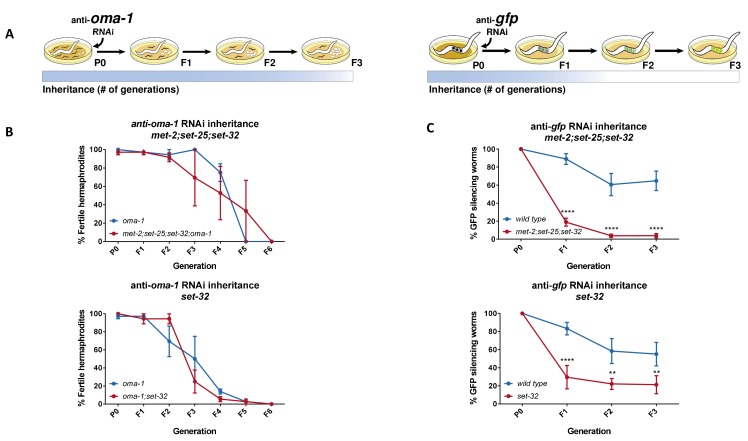
Figure 1. Heritable RNAi responses against the oma-1 and gfp genes have different requirements for H3K9me3 methyltransferases.
(A) Scheme depicting the different requirements for H3K9 methyltransferases in RNAi inheritance responses aimed at different genes. In the parental generation, worms are exposed to RNAi by growing on plates seeded with dsRNA-producing bacteria. Next, worms are transferred to plates seeded with control bacteria (that do not express dsRNA) to lay the eggs the next generation. (left) Only worms that inherit small RNAs that silence the temperature-sensitive dominant allele of oma-1 can hatch. Heritable RNAi responses aimed against the endogenous oma-1 gene do not require H3K9me3 methyltransferases. (right) Inheritance of anti-gfp small RNAs lead to heritable silencing of the gfp transgene (Pmex-5::gfp::h2b transgene). Heritable RNAi responses aimed against the foreign gfp gene strongly depends on H3K9me3 methyltransferases. (B) Inheritance of anti-oma-1 RNAi response in H3K9me3 methyltransferase mutants. The percentage of fertile worms per replicate and generation is presented (N = 12, three biological replicates). (upper panel) RNAi inheritance dynamics in met-2;set-25;set-32;oma-1 mutants compared to oma-1 mutants. (lower panel) RNAi inheritance dynamics in set-32;oma-1 mutants compared to oma-1 mutants. (C) Inheritance of anti-gfp RNAi response in H3K9me3 methyltransferase mutants. In each generation the percentage of worms silencing a germline expressed GFP transgene is presented (N > 60, five replicates). (upper panel) RNAi inheritance dynamics in met-2;set-25;set-32 triple mutants. (lower panel) RNAi inheritance dynamics in set-32 single mutants. Error bars represent standard error of mean. *p-value<0.05, **p-value<0.005, ***p-value<0.001, ****p-value<0.0001, Two-way ANOVA, Sidak's multiple comparisons test.
Figure 1—figure supplement 1. Weak anti-gfp heritable RNAi silencing of gfp in methyltransferases mutants.
Figure 1—figure supplement 2. SET-32 is required for the strong heritable RNAi-induced silencing of gfp in met-2 mutants.
Figure 1—figure supplement 3. RNAi-induced silencing of fog-2 or sup-35 is not inherited transgenerationally.
In parallel we discovered, surprisingly, that in contrast to anti-oma-1 inheritance, heritable silencing of a gfp transgene was defective in the same triple mutants (Figure 1C, upper panel, p=0.0014, 2-way ANOVA). In addition, we also confirmed (Spracklin et al., 2017) that while set-32 single mutants are deficient in inheriting RNAi responses raised against the gfp transgene (Figure 1C, lower panel, p=0.0026, 2-way ANOVA), they are capable (Kalinava et al., 2017) of inheriting responses raised against oma-1 (Figure 1B, lower panel, p=0.8487, 2-way ANOVA). Previously we have shown that while set-25 mutants are defective in inheritance of anti-gfp RNAi, weak inheritance responses can still be observed (Lev et al., 2017). Similarly, we were able to detect weak inheritance responses that last at least until the F3 generation also in met-2;set-25;set-32 and set-32 mutants (Figure 1—figure supplement 1, p-value < 0.0001 for met-2;set-25;set-32 and set-32 in the F3 generation, Two-way ANOVA). Together with our previous data, which showed that set-25 is required for inheriting anti-gfp RNAi, but not anti-oma-1 RNAi (Lev et al., 2017), these results suggested that heritable RNAi requires H3K9 methyltransferases in a gene-specific manner.
The levels of RNAi-induced H3K9me3 do not explain the gene-specific requirements of methyltransferases for heritable RNAi
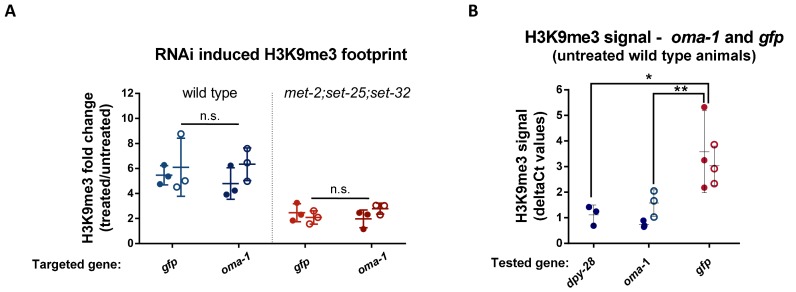
Histone methyltransferase mutants may affect RNAi-induced H3K9me3 levels in a gene-specific manner, thus leading to different inheritance dynamics for each gene. To test this possibility, we performed anti-H3K9me3 Chromatin Immunoprecipitation (ChIP) on F1 met-2;set-25;set-32 triple mutant progeny, that were derived from parents exposed to anti-oma-1 RNAi, anti-gfp RNAi, or untreated controls. Using qPCR we found, as was discovered before (Kalinava et al., 2017) that in met-2;set-25;set-32 triple mutants the RNAi-induced H3K9me3 signal was significantly reduced (p-value=0.0007 and 0.0009, Two-way ANOVA, for gfp and oma-1, respectively). Importantly, this was true for both the oma-1 and gfp loci (Figure 2A). Interestingly, in naive wild-type animals, that were not treated with RNAi, the levels of H3K9me3 on gfp were significantly higher than on oma-1 (Figure 2B, p-value = 0.0039, Two-Way ANOVA) and an additional germline-expressed gene dpy-28 (Figure 2B, p-value = 0.0176, student's t-test). We discuss the possible contribution of this RNAi-independent H3K9me3 signal below. Regardless, as no differences can be found in the RNAi-induced fold changes in H3K9me3 levels between gfp and oma-1 (Figure 2A), the levels of RNAi-induced H3K9me3 cannot explain the gene-specific requirements of methyltransferases for heritable RNAi.
Figure 2. The fold change in RNAi-induced H3K9me3 on oma-1 and gfp is comparable.
(A) The RNAi-induced H3K9me3 footprint on the RNAi-targeted genes. The fold change in H3K9me3 levels in F1 progeny of animals exposed to RNAi versus untreated control animals. The H3K9me3 footprint levels were assessed using a qPCR quantification of ChIP experiments conducted with both wild type (left) and met-2;set-25;set-32 mutants (right). Filled or empty circles represent qPCR data obtained using two different primer sets that span different parts of the examined locus. (B) H3K9me3 levels on the gfp and oma-1 genes in naive untreated wild type animals. The deltaCt numbers used to obtain the fold change values were calculated using the eft-3 gene as an endogenous control. The presented data were obtained from three biological replicates. The levels of gfp and dpy-28 H3K9me3 signal in wild type animals are adapted from raw data from our previous publication (Lev et al., 2017). Two-way ANOVA, Sidak's multiple comparisons test. **p-value<0.005. Error bars represent standard deviations.
SET-32 acts upstream to MET-2 and SET-25 to support RNAi inheritance
We previously found that in contrast to set-25 single mutants, which are deficient in RNAi-induced heritable H3K9me3 methylation (Lev et al., 2017; Mao et al., 2015), met-2;set-25 double mutants display a modest but robust H3K9me3 footprint following RNAi (Kalinava et al., 2017; Lev et al., 2017). We therefore hypothesized that in the met-2 background, an additional, perhaps otherwise inactive H3K9 methyltransferase, is expressed or activated, compensating for the absence of SET-25, to allow efficient heritable RNAi responses (see Figure 1—figure supplement 2 for summary). To test this hypothesis, we first examined whether met-2;set-32 double mutants can inherit RNAi responses raised against gfp. If SET-32 and SET-25 compensate for each other and are redundant, then met-2;set-32 double mutants are expected to strongly inherit RNAi responses, similar to met-2;set-25 double mutants (Lev et al., 2017). Our results show, that in contrast to met-2;set-25 double mutants, met-2;set-32 double mutants are defective in RNAi inheritance raised against gfp, since only a very weak response can be detected (Figure 1—figure supplement 2A). The potency of RNAi inheritance in met-2;set-32 double mutants is comparable to that of set-25 (Lev et al., 2017) and set-32 single mutants, or met-2;set-25;set-32 triple mutants (Figure 1C). These results suggest that SET-32 has a distinct role, and that it probably acts upstream to MET-2 and SET-25, in promoting RNAi inheritance. This conclusion is also consistent with the recent observation that SET-32, in contrast to MET-2 and SET-25, has an essential role in the establishment of RNAi-mediated nuclear silencing (Kalinava et al., 2018).
Unlike RNAi silencing of oma-1, silencing of sup-35 and fog-2 genes is not inherited transgenerationally
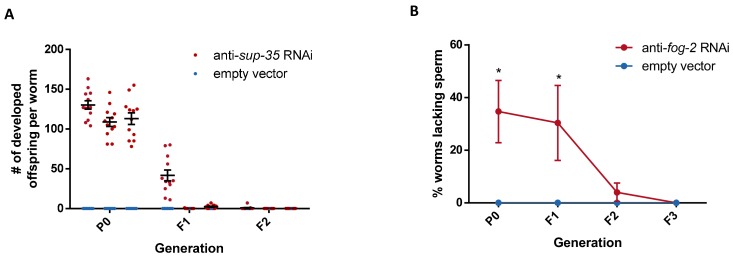
Currently, the only gene that serves to study heritable transgenerational (more than two generations) RNAi of endogenous genes is oma-1. Transgenerational RNAi inheritance requires the target gene to be expressed in the germline, and many germline genes are essential or do not have a phenotype that can be scored over many generations. The oma-1 gene can serve as a tool for studying RNAi inheritance owing to the availability of a temperature-sensitive, dominant-lethal and redundant allele that can be rescued by RNAi (Alcazar et al., 2008). In search of other endogenous target genes whose heritable silencing could be studied, we examined the inheritance of RNAi against the non-essential germline genes sup-35 and fog-2. SUP-35 is a maternally deposited toxin, expressed in the mother’s germline, suppressed by PHA-1, a zygotically expressed anti-toxin (Ben-David et al., 2017). Consequently, temperature-sensitive pha-1(e2123) mutants develop when grown at 15 degrees but arrest their development when grown in restrictive temperatures, unless exposed to anti-sup-35 RNAi. As previously described (Ben-David et al., 2017), RNAi silencing of sup-35 allowes pha-1 mutants to develop. However, we found this response was not inherited beyond the F1 generation (Figure 1—figure supplement 3A). Expression of the germline gene fog-2 is required for hermaphrodite worms to produce sperm, but is dispensable for sperm production in males (Schedl and Kimble, 1988). Silencing of fog-2 by RNAi lead to depletion of sperm (as evident by stacked oocytes), and the worms were unable to reproduce unless crossed with a male. While we found that this response was inherited to the F1 progeny, it was not inherited transgenerationally (Figure 1—figure supplement 3B). In conclusion, we could not find additional endogenous gene targets that can be transgenerationally silenced upon RNAi. Conveniently, many endo-siRNAs that target various endogenous genes are inherited transgenerationally, and such inheritance can be studied using RNA sequencing.
H3K9me3 methyltransferases are required for the biogenesis of a specific class of endo-siRNAs
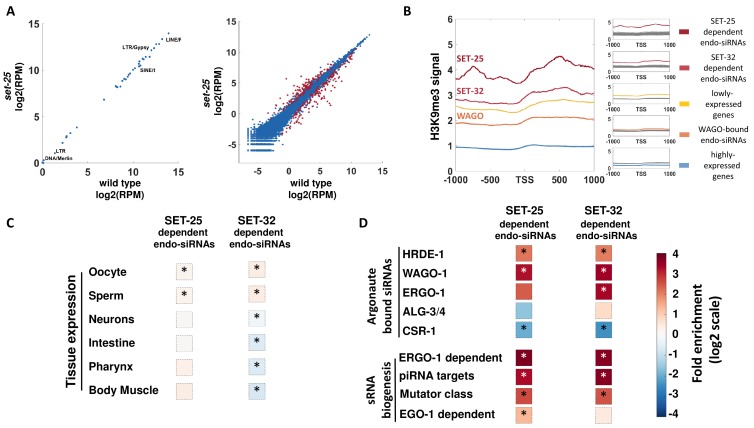
Certain germline small RNAs have evolved to confer immunity against foreign genetic elements, while sparing endogenous genes (Malone and Hannon, 2009). The different requirements for particular methyltransferases and H3K9me3 for heritable silencing of gfp and oma-1 may be connected to the fact that gfp is a ‘foreign’ gene, while oma-1 is an endogenous gene. We previously found that exogenous siRNAs that target gfp are lost in set-25 mutants, and hypothesized that endo-siRNAs that target other ‘foreign’ genes would be likewise affected. Therefore, we re-analyzed our previously published small RNA sequencing data, obtained from set-25 mutants (Lev et al., 2017). However, among the targets of these differentially expressed endo-siRNAs, we could not detect striking changes (fold change >1.2) in endo-siRNAs that target transposons and repetitive elements in set-25 mutants (Figure 3A, left panel). In contrast, a subset of endo-siRNAs that target 279 different protein-coding genes was found to exhibit significant changes in set-25 mutants (adj.p <0.1, DESeq2 Figure 3A, right panel). To understand why these small RNAs are uniquely affected by SET-25, we characterized this group and the endo-siRNAs that target them. To compare the endo-siRNA pools that depend on these two H3K9 tri-methyltrasferases, we re-analyzed the recently published small RNA-seq data obtained from set-32 mutants (Kalinava et al., 2018).
Figure 3. A genome-wide analyses of endo-siRNAs that depend on SET-25 or SET-32.
(A) An expression analysis of endo-siRNAs targeting transposons and repetitive elements classes (left panel) or protein-coding genes (right panel). Shown are the expression values as log2 of number of Reads per Million (RPM) in set-25 mutants (y-axis) compared to wild type animals (x-axis). Gene targets of endo-siRNAs, which display significant differential expression (analyzed with Deseq2, adjusted p-value<0.1) are marked in Red (B) An analysis of H3K9me3 signals (based on published data from McMurchy et al., 2017) on differet sets of gens: highly expressed genes (top 10%, Blue), lowly expressed genes (top 10%, Yellow) and gene targets of endo-siRNAs that depend on SET-25 (based on Lev et al., 2017, Red), SET-32 (based on Kalinava et al., 2018), Dark Red) or endo-siRNAs associated with WAGOs (HRDE-1,WAGO-1, and ERGO-1, Light Red). H3K9me3 signal is aligned according to gene's Transcription Start Sites (TSS), and the regions of 1000 base pairs upstream and downstream of the TSS are shown on the x axis. The y axis shows the averaged signal of the H3K9me3 modification as a function of distance from the TSS. For statistical analysis, control data sets (shown in Gray) were created by sampling the H3K9me3 levels of randomly selected gene sets of the same size as the examined gene list. (C and D) An enrichment analysis of genes with significantly lowered levels of endo-siRNAs targeting them in set-25 and set-32 mutants compared to wild type. Fold enrichment values (log2 scale) are color coded. (C) An enrichment analysis for expression in specific tissues. (D) An enrichment analysis for different small RNA pathways. The p-values were calculated using 10,000 random gene sets identical in their size to the examined endo-siRNA-target gene list. Asterisk denotes statistically significant enrichment values (p-value<0.05).
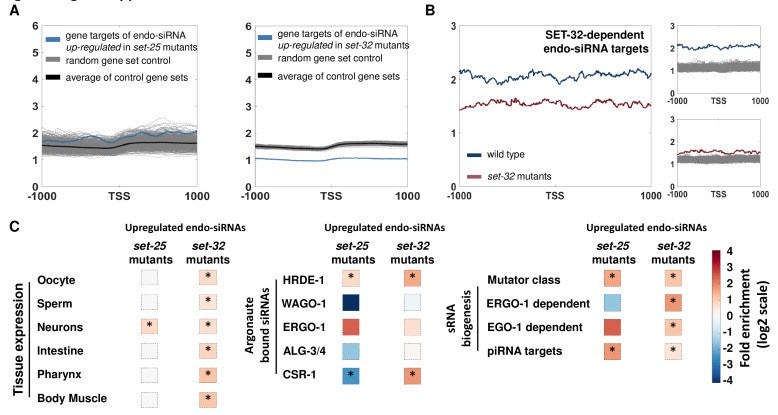
Figure 3—figure supplement 1. An analysis of genes targeted by endo-siRNAs upregulated in set-25 and set-32 mutants.
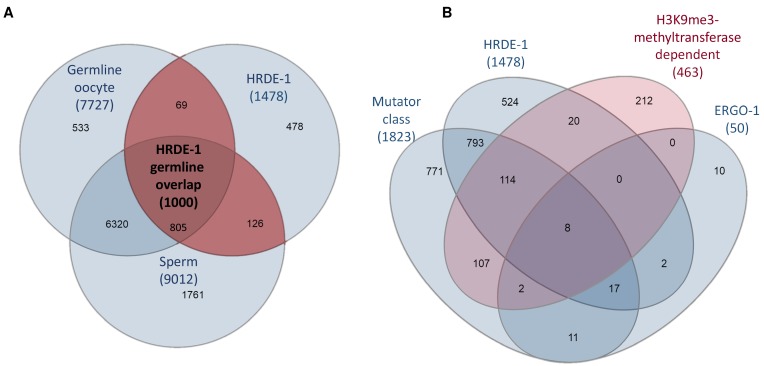
Figure 3—figure supplement 2. An analysis of overlap between different endo-siRNA target gene sets.
Since in set-25 the loss of exogenous siRNAs coincided with the loss of heritable RNAi-induced H3K9me3 (Lev et al., 2017), we first tested whether genes that were differentially targeted by endo-siRNAs in set-25 mutants were also marked by H3K9me3. By examining publicly available H3K9me3 data (McMurchy et al., 2017), we found that the 151 genes that lost the endo-siRNAs that target them in set-25 mutants were robustly marked by H3K9me3 in wild type animals (Figure 3B). We also found that in contrast, the 128 genes that had increased endo-siRNA levels that target them in set-25 and mutants were not significantly marked by H3K9me3 (Figure 3—figure supplement 1A). By analyzing an available mRNA-seq dataset (Klosin et al., 2017), we also found a significant enrichment for genes that were upregulated (at the mRNA level) in set-25 mutants amongst the list of SET-25-dependent endo-siRNA targets (1.93-fold enrichment, 18/151 genes, p-value=0.006). This suggests that endo-siRNAs that depend on SET-25 silence targeted gene. Recently, Kalinava et al. sequenced endo-siRNAs from set-32 mutants (Kalinava et al., 2018). Our analysis show that the 337 genes that had reduced levels of endo-siRNAs (fold change >2) in set-32 mutants, were also significantly marked by H3K9me3 (Figure 3B). As expected, these genes showed lower levels of H3K9me3 in set-32 mutants (Figure 3—figure supplement 1B), and genes having increased levels of endo-siRNAs were not significantly marked by H3K9me3 (Figure 3—figure supplement 1A). Together, these results support the hypothesis that H3K9me3 methyltransferases directly support the biogenesis of silencing endo-siRNAs by tri-methylating the H3K9 histones of the endo-siRNAs targeted genes.
Next we examined whether genes that display altered endo-siRNAs levels in set-25 and set-32 mutants are expressed in specific tissues. Genes that had significantly reduced levels of endo-siRNAs targeting them in set-25 or in set-32 mutants exhibited significant, but modest, enrichment for expression in the germline (Figure 3C and Figure 3—figure supplement 1C). No significant enrichment was found for other tissues (Figure 3C and Figure 3—figure supplement 1C).
To identify the small RNA pathways which are affected by set-25 and set-32, we tested whether the differentially expressed endo-siRNAs depend on particular argonautes, or associate with specific biosynthesis or functional pathways (Figure 3D). It was previously suggested that the CSR-1 argonaute carries heritable endo-siRNAs that mark endogenous genes (Claycomb et al., 2009), while the HRDE-1 argonaute carries heritable endo-siRNAs that silence foreign or aberrant elements, whose expression could be deleterious, such as transposons (Luteijn et al., 2012; Rechavi, 2014; Shirayama et al., 2012). A strong and significant enrichment (Figure 3D and Figure 3—figure supplement 1C) was found for endo-siRNAs which are carried in the germline by the argonautes WAGO-1 (Gu et al., 2009) and HRDE-1, which is required for inheritance of exogenous siRNAs (Buckley et al., 2012). Both argonautes were found to be involved in gene silencing (Buckley et al., 2012; Gu et al., 2009). Nevertheless, some of the targets of HRDE-1-bound endo-siRNAs are expressed in the germline (Figure 3—figure supplement 2A). This may explain the concurrent enrichment for both germline-expressed genes and targets of HRDE-1-bound endo-siRNAs amongst the gene targets of endo-siRNAs that depend on SET-25 or SET-32. A significant enrichment was also found for Mutator pathway small RNAs (Zhang et al., 2011), ERGO-1-dependent small RNAs, and putative piRNA targeted genes (Bagijn et al., 2012). On the contrary, a significant depletion was found for genes known to be targeted by CSR-1-carried small RNAs, a pathway that was suggested to support the expression of targeted genes (Claycomb et al., 2009; Shen et al., 2018). The helicase EMB-4 (Akay et al., 2017; Tyc et al., 2017) was shown to preferably bind introns of genes targeted by CSR-1; We could not detect a significant enrichment for genes whose introns are bound by EMB-4 (fold change = 1.07 and 0.79, p-value=0.26 and 0.002, for endo-siRNAs dependent on SET-25 or SET-32, respectively). All together, these results suggest that H3K9 methyltransferases are required for the maintenance of a specific sub-class of HRDE-1 and WAGO-1 small RNAs, that are associated with the Mutator and piRNA pathways, and that target protein-coding genes (Figure 3—figure supplement 2B).
Endo-siRNAs that depend on H3K9me3 methyltransferases target a distinctive subset of newly evolved genes
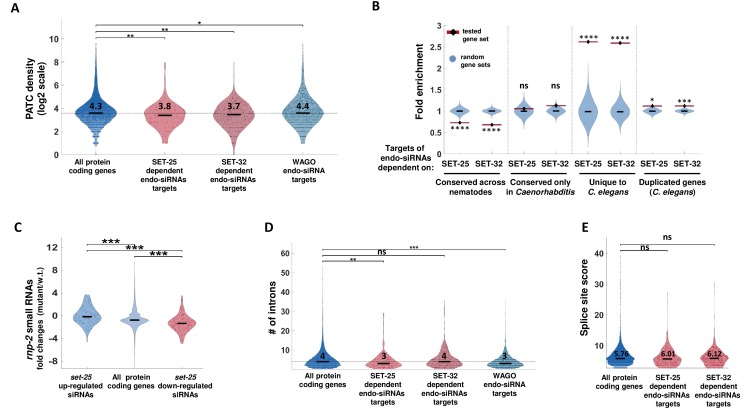
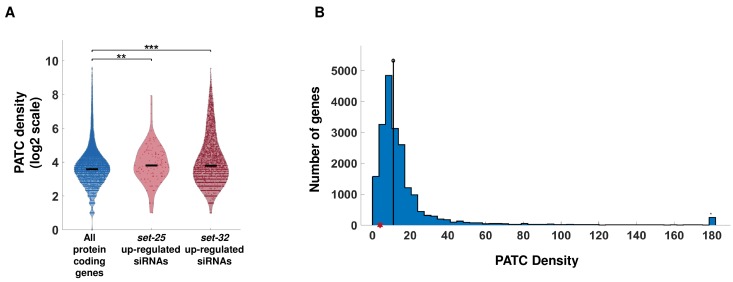
What distinguishes the target genes of endo-siRNAs that depend on SET-25 and SET-32 methyltransferases? It was recently found that periodic A/T (PATC) sequences can shield germline genes from piRNA-induced silencing and allow germline expression of genes in H3K9me3-rich genomic regions (Frøkjær-Jensen et al., 2016; Zhang et al., 2018). Fittingly, we found that genes targeted by SET-25-dependent and SET-32-dependent endo-siRNAs exhibit a moderate (~9–13% in median values) but significant reduction in PATC density compared to all protein coding genes (Figure 4A, p-value = 0.0026 and 0.0011 for SET-25 and SET-32, respectively). This feature is not general for genes targeted by WAGOs (Worm-specific Argonautes, HRDE-1, WAGO-1 and ERGO-1) associated endo-siRNAs, since these targeted genes have a higher PATC density (Figure 4A, 10% increase in average values, p-value=0.034). In addition, genes targeted by endo-siRNAs that are increased in set-25 or set-32 mutants exhibit significantly increased PATC density (Figure 4—figure supplement 1A). However, we posit that this feature is unlikely to be sufficient for distinguishing between oma-1 and gfp, since the oma-1 gene has a very low PATC density (Figure 4—figure supplement 1B).
Figure 4. SET-25-dependent endo-siRNAs target newly evolved genes.
(A) A PATC density analysis for SET-25 and SET-32-dependent endo-siRNAs gene targets. The PATC density values (obtained from Frøkjær-Jensen et al., 2016) are presented for all protein-coding genes, gene targets of endo-siRNAs that depend on SET-25 or SET-32 and gene targets of endo-siRNAs associated with WAGO small RNA pathways (HRDE-1,WAGO-1 or ERGO-1). **p-value<0.005, *p-value<0.05, Wilcoxon rank sum test. For clarity of display, values are shown in log2 scale (after addition of 1). The median (black line) and average levels (numbers) of PATC density levels of each plot are indicated (log2 scale). (B) An enrichment analysis of genes conserved at different levels and duplicated genes amongst gene targets of SET-25- and SET-32-dependent endo-siRNAs. The gene sets were generated based on the homology field in WormBase that details the orthologs and paralogs of each nematode gene. We defined a duplicated gene as a gene that has a paralog in C. elegans. We define genes unique to C. elegans as genes that lack an ortholog amongst the nematode species we examined (see Materials and methods). For statistical analysis, control enrichment values were obtained from 10,000 random gene sets with the same size as the examined endo-siRNA-target gene list. ****p-value<0.0001,***p-value<0.001, *p-value<0.05 (C) An analysis of endo-siRNAs fold changes in rnp-2 mutants for genes targets of endo-siRNAs downregulated or upregulated in set-25 mutants or all genes. All p-values<0.001, Wilcoxon rank sum test. (D) An analysis of intron numbers of gene targets of SET-25- and SET-32-dependent endo-siRNAs and WAGO-associated endo-siRNAs compared to all protein-coding genes. In cases of genes that have more than one transcript, the average intron value is used. The median intron number of each plot is indicated (log2 scale). ***p-value<0.001,**p-value<0.005, Wilcoxon rank sum test. (E) An analysis of splicing motif divergence score (based on Newman et al., 2018) of gene targets of SET-25 and SET-32-dependent endo-siRNAs, WAGO associated endo-siRNAs and all protein-coding genes. The median score levels of each plot are indicated. p-value>0.05, Wilcoxon rank sum test.
Figure 4—figure supplement 1. An analysis PATC density of the oma-1 gene and of targets of endo-siRNAs upregulated in set-25 and set-32 mutants.
Figure 4—figure supplement 2. Gene targets of endo-siRNAs that depend on SET-25 or SET-32 are preferably targeted by small RNAs on intron-exon junctions compared to all protein coding genes.
Figure 4—figure supplement 3. An analysis of endo-siRNAs targeting and H3K9me3 levels of newly evolved genes.
The lists of genes which are targeted by SET-25- and SET-32-dependent endo-siRNAs were enriched for genes targeted by ERGO-1-dependent endo-siRNAs (Figure 3D). Many of the genes that are targeted by ERGO-1-bound endo-siRNAs are duplicated genes (Fischer et al., 2011; Vasale et al., 2010). Accordingly, we found an enrichment for duplicated genes amongst the genes that had reduced endo-siRNA levels targeting them in set-25 and set-32 mutants (Figure 4B). An additional characteristic of the set of genes targeted by ERGO-1 endo-siRNAs is an enrichment for poorly conserved genes, that have fewer introns, and possess splicing site sequences that diverge from the consensus sequence (Fischer et al., 2011; Newman et al., 2018). It was recently suggested that these poorly conserved genes are targeted for silencing because their aberrant or”non-self-like’ splicing signals are detected by the splicing machinery (Newman et al., 2018).
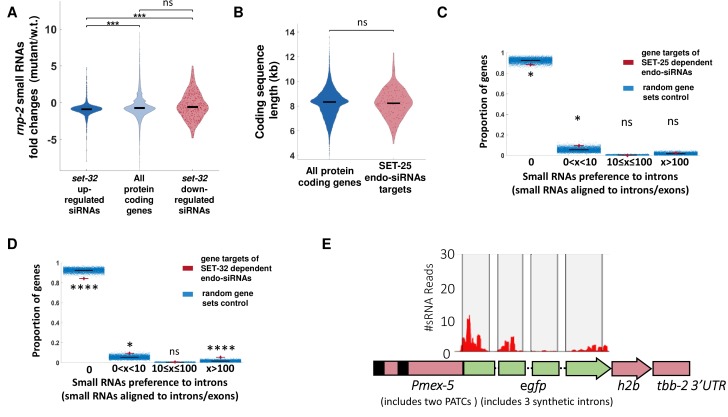
Therefore, we examined whether the targets of the endo-siRNAs that depend on SET-25 or SET-32 can be distinguished by their splicing signals. The changes in the endo-siRNA pool in mutants of small nuclear ribonucleoprotein-associated protein RNP-2/U1A (rnp-2) mirrored the endo-siRNA changes found in set-25 mutants (Figure 4C), but not that of set-32 mutants (Figure 4—figure supplement 2A). We also found that genes targeted by SET-25-dependent- but not SET-32-dependent endo-siRNAs bear fewer introns (Figure 4D, median of 3 and 4 compared to 4 of all protein coding genes, p-values=0.0047, and 0.42 for SET-25 and SET-32, respectively). No significant differences in the length of the coding sequences were found, hence, the difference in intron number does not simply derive from differences in gene lengths (Figure 4—figure supplement 2B, p-value = 0.8673). The lists of genes targeted by SET-25-dependent or SET-32-dependent endo-siRNAs were enriched with genes shown to be targeted by intron-targeting small RNAs (Figure 4—figure supplement 2C and D). We could not find, however, small RNAs aligning to the introns of the gfp transgene that we studied (Figure 4—figure supplement 2E, in most cases endo-siRNAs target only exons). We also did not find significant differences in the splicing motif divergence score (obtained from Newman et al., 2018). Since splicing also directly affects the RNAi machinery untangling its role in endogenous RNAi is challenging (Newman et al., 2018). In summary, splicing may be one of the factors that contribute to distinguishing genes targeted by SET-25-dependent endo-siRNAs, but not by SET-32-dependent endo-siRNAs.
In contrast, in the sets of genes targeted by either SET-25-dependent or SET-32-dependent small RNA we found a significant enrichment for newly evolved genes (Figure 4B, fold-change = 2.57 and p-value<0.0001 for both SET-25- and SET-32-dependent endo-siRNAs targets, respectively). We define newly evolved genes here as genes which had no orthologs outside C. elegans (35/151 and 78/337 of genes targeted by SET-25- or SET-32-dependent endo-siRNAs, respectively). Concordantly, in the same gene sets we also found a significant depletion for nematode-conserved genes (Figure 4B). Importantly, the sets of genes targeted by SET-25-dependent endo-siRNAs and SET-32-dependent endo-siRNAs show very small overlap (25 out of 465 genes). Thus, while SET-25 and SET-32 are required for the maintenance of endo-siRNAs that target different genes, the characteristics of these genes are very similar, that is they are distinctively newly evolved genes that have slightly lower levels of PATC sequences. Although the changes in PATC density and intron numbers that distinguish these target genes are moderate, it is possible that the cumulative effect of these small differences may result in the exposure of foreign genes that need to be silenced.
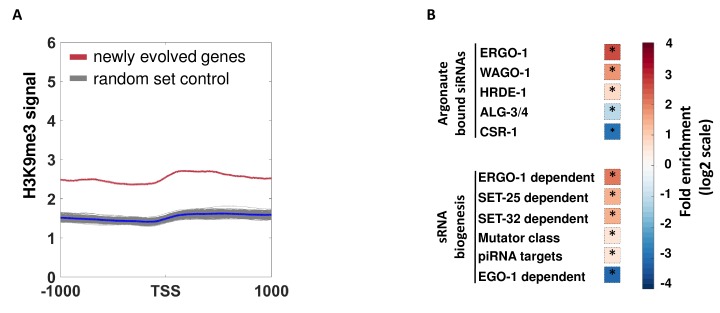
In general, we find that certain sub-classes of endo-siRNA, such as ERGO-1 and HRDE-1 bound small RNAs, target gene sets enriched for newly evolved genes (Figure 4—figure supplement 3). The significant enrichment for newly evolved genes among SET-25- and SET-32-dependent endo-siRNAs is maintained, however, even after excluding genes that are also targeted by HRDE-1, ERGO-1, WAGO-1 or Mutator endo-siRNAs (SET-25: 59/151 genes are not shared, fold enrichment = 2.97, p-value=0.0001, SET-32: 153/337 genes are not shared, fold-enrichment = 1.89, p-value=0.0012). Thus, the enrichment of newly evolved genes amongst the targets of SET-25- and SET-32-dependent endo-siRNAs is not simply due to a general preference for newly evolved genes by endo-siRNA pathways. Further, we find that newly evolved genes are marked by higher levels of H3K9me3 in comparison to the average level of H3K9me3 on protein coding genes (Figure 4—figure supplement 3). Likewise, in the absence of RNAi, in wild-type animals, gfp, the example for a foreign (non-nematode) gene that we investigated, has higher levels of H3K9me3, in comparison to the well-conserved oma-1 gene (Figure 2B). The fact that across the genome SET-25-dependent- and SET-32-dependent endo-siRNAs target newly evolved and H3K9me3 methylated genes (Figure 3B and Figure 4B), may explain why inheritance of RNAi responses raised against gfp, but not oma-1, depends on SET-25 and SET-32 (Figure 1).
In summary, our experiments reveal a specific role for histone modifications in small RNA inheritance. While in S. pombe and A. thaliana a feedback between H3K9me3 and small RNAs was suggested to be required for silencing, the worm’s RNAi inheritance machinery may use H3K9me3 as a mark that distinguishes genes identified as ‘new’. Since newly evolved genes can be disruptive, small RNAs survey these H3K9me3-flagged elements transgenerationally.
Discussion
Our study began from an investigation of a perplexing asymmetry in the requirement of specific H3K9 methyltransferases for heritable silencing of the endogenous gene oma-1 and the ‘foreign’ gene gfp. Single mutants of set-25 and set-32 and the met-2;set-25;set-32 triple mutant displayed different heritable dynamics when either the gfp or the oma-1 gene were targeted by RNAi. These results are not unique to the specific gfp transgene that was tested, since similar observations have been made with other transgenes (Klosin et al., 2017; Lev et al., 2017; Shirayama et al., 2012; Spracklin et al., 2017).
Unlike mutations in these histone methyltransferases, which negatively affect heritable silencing of gfp, but not oma-1, mutations in genes required for small RNA inheritance negatively affect heritable silencing of both oma-1 and gfp. For example, the argonaute HRDE-1 is required for inheritance of RNAi responses against both genes (Ashe et al., 2012; Buckley et al., 2012; Kalinava et al., 2017; Shirayama et al., 2012; Weiser et al., 2017). The fact that heritable RNAi responses aimed at different genes are affected by different proteins should be taken into account when studying transgenerational inheritance. Specifically, when screening for genes that affect such inheritance, one must acknowledge that heritable silencing of different targets requires different chromatin modifiers.
Future studies will hopefully reveal why some recently evolved genes, but not others, display high levels of H3K9me3 (in the absence of RNAi), and are targeted by endo-siRNAs. Recent studies examined why transgenes are sensitive to silencing by synthetic piRNAs, while endogenous germline expressed genes, including oma-1, are not. This protection was suggested to be conferred at least in part by PATC sequences, and to be independent of the genomic location of the gene (Zhang et al., 2018). PATC sequences were previously shown to allow expression of transgenes in the germline in heterochromatic areas (Frøkjær-Jensen et al., 2016). Similarly, our analysis revealed that the gene targets of SET-25-dependent and SET-32-dependent endo-siRNAs have lower levels of PATC density (Figure 4A). However, the oma-1 gene does not possess many PATC sequences (Figure 4—figure supplement 1B). An additional theory suggested that an intrinsic unknown coding-sequence feature confers resistance to silencing by piRNAs. Seth et al. have studied why a fusion between oma-1 and gfp can trans-activate silenced gfp transgenes (an effect known as ‘RNAa’,(Seth et al., 2013)). While unique ‘protective’ sequence features were not described in that work, the authors showed that an unknown coding-sequence feature, not related to the codon usage or the translation of the protein, grants the oma-1 gene with its ability to activate silenced transgenes (Seth et al., 2018). It is possible that the gene targets of SET-25- dependent and SET-32-dependent small RNAs that we describe here have unique intrinsic sequences that distinguish them as well. The different requirement of methyltransferases for heritable silencing of some genes but not others may be related to such intrinsic sequence features. Alternatively, it is possible, as was suggested in the past, that new genes are silenced because they are not licensed transgenerationally by heritable small RNAs for expression (Claycomb et al., 2009; Shen et al., 2018). If this is the case, future studies will hopefully reveal how such license is granted (See Figure 5 for Scheme).
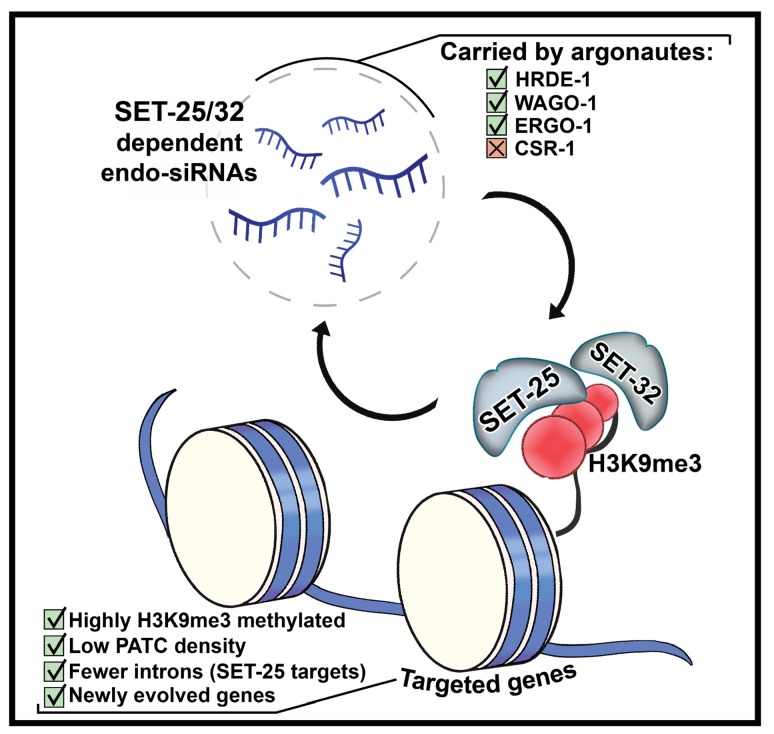
Figure 5. Scheme characterizing H3K9me3 methyltransferase-dependent endo-siRNAs and their targets.
SET-25-dependent and SET-32-dependent endo-siRNAs are enriched with small RNAs known to be carried by the argonautes HRDE-1, WAGO-1 and ERGO-1 but not CSR-1. SET-25-dependent and SET-32-dependent endo-siRNAs targets are enriched with newly evolved genes, have fewer PATC sequences, and are marked with higher levels of H3K9me3. Targets of SET-25-dependent but not SET-32-dependent endo-siRNAs bear fewer introns.
Materials and methods
Key resources table.
| Reagent type | Designation | Source of reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (E. coli) |
OP50 | (Brenner, 1974) | Op50 RRID:WB-STRAIN:OP50 |
|
| Strain (E. coli) |
anti-gfp RNAI bacteria | This study | ||
| Strain (E. coli) |
anti-oma-1 RNAi bacteria | Ahringer RNAi library (Kamath and Ahringer, 2003) |
4-4-3-C01 | |
| Strain (E. coli) |
anti-sup-35 RNAi bacteria | Vidal RNAi library (Rual et al., 2004) |
11006-E2 | |
| Strain (E. coli) |
anti-fog-2 RNAi bacteria | Vidal RNAi library (Rual et al., 2004) |
10011-C3 | |
| Strain (C. elegans) |
N2 | CGC | N2 RRID:WB-STRAIN:N2_(ancestral) |
|
| Strain (C. elegans) |
set-32(ok1457) ; oma-1(zu405) | CGC | BFF25 | |
| Strain (C. elegans) |
mjIs134[pmex-5::gfp::h2b::tbb-2] | Erik Miska's lab (Univeristy of Cambridge) | SX1263 | |
| Strain (C. elegans) |
oma-1(zu405) | CGC | TX20 | |
| Strain (C. elegans) |
pha-1(e2123) | CGC | GE24 | |
| Strain (C. elegans) |
set-32(ok1457);mjIs134[pmex-5::gfp::h2b::tbb-2] | This study | BFF24 | |
| Strain (C. elegans) |
met-2(n4256);set-25(n5021);set-32(ok1457);mjIs134[pmex-5::gfp::h2b::tbb-2] | This study | BFF26 | |
| Strain (C. elegans) |
met-2(n4256);set-25(n5021);set-32(ok1457);oma-1(zu405) | This study | BFF27 | |
| Strain (C. elegans) |
met-2(n4256);set-32(ok1457);mjIs134[pmex-5::gfp::h2b::tbb-2] | This study | BFF28 | |
| Antibody | H3K9me3 | Abcam | RRID:AB_306848 | |
| Software, algorithm |
GraphPad Prism | https://www.graphpad.com/scientific-software/prism/ | RRID:SCR_002798 | |
| Software, Algorithm |
Image J | Opensource: https://imagej.nih.gov/ij/ | RRID:SCR_003070 | |
| Software, Algorithm |
MATLAB MathWorks | https://www.mathworks.com/ | RRID:SCR_016651 |
Cultivation of the worms
Standard culture techniques were used to maintain the nematodes on nematode growth medium (NGM) plates seeded with OP50 bacteria. Extreme care was taken to avoid contamination or starvation, and contaminated plates were discarded from the analysis.
RNAi bacteria
HT115 Escherichia coli strains expressing dsRNAs were used: anti-oma-1 RNAi bacteria were obtained from the Ahringer RNAi library (Kamath and Ahringer, 2003). Anti-fog-2 and anti-sup-35 RNAi were obtained from the Vidal RNAi library (Rual et al., 2004). For the sequence of the anti-gfp RNAi see supplemental data.
RNAi experiments
RNAi HT115 E.coli bacteria were incubated in lysogeny broth (LB) containing Carbenicillin (25 μg/mL) at 37°C overnight with shaking. Bacterial cultures were seeded onto NGM plates containing isopropyl β-D-1-thiogalactopyranoside (IPTG; 1 mM) and Carbenicillin (25 μg/mL) and grown overnight in the dark at room temperature. Five L4 animals were placed on RNAi bacteria plates and control empty-vector bearing HT115 bacteria plates and maintained at 20°C for 2 days and then removed. The progeny hatching on these plates was termed the P0 generation. In the next generations the worms were grown on E.coli OP50 bacteria. For anti-gfp RNAi experiments, four L4 animals were placed on plates for two days to lay the next generation. In every generation approximately 60 one day adult worms were collected and photographed per condition (see below). For anti-oma-1 experiments, in each generation twelve individual L4 staged worms were placed in individual wells of a twelve well plate. Four days later the number of fertile worms was assessed (at least one progeny) and 12 individual L4 progeny worms were chosen from the most fertile well to continue to the next generation. For and anti-sup-35 RNAi experiments, in each generation 12 individual L4 staged worms were placed in individual wells of a 12-well plate. Two days later the adult worms were removed. Two days later the number of developing worms was counted and twelve individual L4 progeny worms were chosen from the well with the highest number of developing progeny to continue to the next generation. For and anti-fog-2 RNAi experiments, five L4 worms were crossed on RNAi bacteria for 24 hr. The crossed worms were transferred to fresh RNAi bacteria plates. In each generation, five resulting L4 progeny from the cross were crossed on control bacteria plates and ~40 L4 worms were picked to control bacteria plates and photographed a day later. The number of sterile worms with stacked oocytes was assayed.
Germline GFP expression analysis
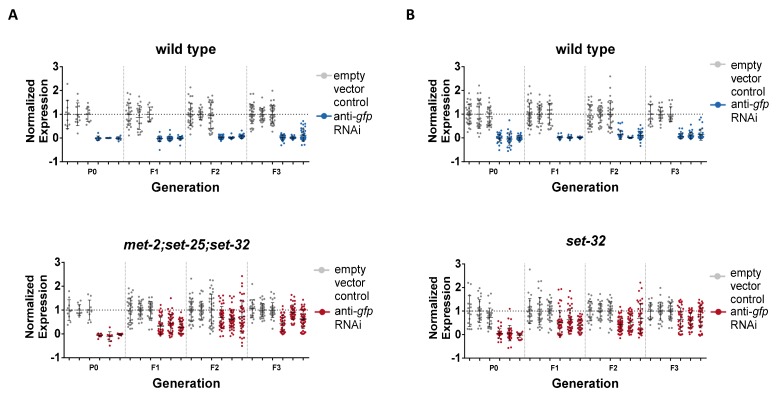
Percentage silencing analysis: for each condition, around 60 animals were mounted on 2% agarose slides and paralyzed in a drop of M9 with 0.01% levamisole/0.1% tricaine. The worms were photographed with 10x objective using a BX63 Olympus microscope (Exposure time of 200 ms, and gain of 2). The images were analyzed with ImageJ2 software, and the percentage of worms lacking any observable germline GFP signal was calculated.
GFP expression level analysis: for each condition, the GFP fluorescence level of the background and of oocyte nuclei of at least 30 worms was calculated using ImageJ2.
CTCF value was calculated as follows: CTCF = Integrated density of selected object X – (area of selected object X * mean fluorescence of background readings). The obtained CTCF value was normalized to the average CTCF value obtained from photographs of control animals of the same genotype, generation and age which were fed on control plates.
Chromatin immunoprecipitation
Chromatin immunoprecipitation experiments were conducted as described in Lev et al. (2017). For anti-H3K9me3 ChIP experiments the abcam, ab8898 antibodies were used.
qPCR reactions
All Real time PCR reactions were performed using the KAPA SYBR Fast qPCR and run in the Applied Biosystems 7300 Real Time PCR System.
The primer sequences used in qRT-PCR:
gfp set #1 FOR: ACACAACATTGAAGATGGAAGC
gfp set #1 REV: GACAGGTAATGGTTGTCTGG
gfp set #2 FOR: GTGAGAGTAGTGACAAGTGTTG
gfp set #2 REV: CTGGAAAACTACCTGTTCCATG
oma-1 set#1 FOR: AACTTTGCCCGTTTCACC
oma-1 set#1 REV: TCAAGTTAGCAGTTTGAGTAACC
oma-1 set#2 FOR: TTGTTAAGCATTCCCTGCAC
oma-1 set#2 REV: TCGATCTTCTCGTTGTTTTCA
(The above primer set was adapted from Spracklin et al., 2017)
dpy-28 FOR: CTGATGGATCCAGAGTTGG
dpy-28 REV: CTGCTATACGCATCCTGTTC
eft-3 FOR: CCAACATGATTAGTCAGATGACC
eft-3 REV: CTAGGAGTTAGATGTGCAGG.
Information on the sequencing libraries analyzed in this paper
All the studied publicly available sequencing libraries were prepared from synchronized young adult worms grown at 20 degrees (Kalinava et al., 2018; Klosin et al., 2017; Lev et al., 2017; McMurchy et al., 2017). For more information see the original publications and GEO information: A. set-25 small RNAs (Rechavi and Lev, 2017; GEO accession: GSE94798), B. set-25 mRNA (Klosin et al., 2017; GEO accession: GSE83528), C. set-32 small RNAs (Kalinava et al., 2018; GEO accession: GSE117662, the set-32 (red11) allele data were used)., D. set-32 and wild type H3K9me3 ChIP-seq (Kalinava et al., 2018; GEO accession: GSE117662). E. wild type H3K9me3 ChIP-seq (McMurchy et al., 2017; GEO accession: GSE87524).
Bioinformatic genome-wide endo-siRNAs analysis
Small RNA analysis was conducted as previously described (Lev et al., 2017). Briefly, adapters were cut from the reads using Cutadapt (Martin, 2011). Reads that were not cut or were less than 19 bp long, were removed. The quality of the libraries was assessed by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were mapped to the C. elegans genome (WS235) using Bowtie2 (Langmead and Salzberg, 2012). In total 31,053,062, 21,913,420 and 18,372,739 reads were mapped in the three wild type biological repeats and 21,258,241, 19,925,004 and 21,391,091 reads were mapped in the three set-25 biological repeats. The mapped reads were then counted using the python script HTseq_count (Langmead and Salzberg, 2012) using. gff feature file from wormbase.org (version WBcel235). Differential expression was analyzed using DESeq2 (Love et al., 2014). p-adjusted value <0.1 was regarded as statistically significant. The set-32 data from Kalinava et al (Kalinava et al., 2018) GEO was analyzed in a similar fashion. The reads the 5' barcode and 3' linker were trimmed using Cutadapt (Martin, 2011), in accordance the information supplied by Kalinava et al in GEO (accession number: GSE117662). Next, reads were filtered to lengths of 20–23 bp and aligned (not allowing mismatches) to the C. elegans genome (ce10) by Shortstack (Axtell, 2013). In total 1,342,884 and 1,006,568 reads were mapped in the wild type and set-32(red11) small RNA samples, respectively. The reads mapping to each genomic were counted by HTseq_count (Langmead and Salzberg, 2012). Since one biological sample was available, significantly altered small RNAs were defined as genes having fold change of larger than 2 (up-regulated) or smaller than 0.5 (downregulated).
Bioinformatic genome-wide analysis of H3K9me3 signal
For analysis of H3K9me3 signal on different genes in wild type worms, the processed H3K9me3 data (aligned and normalized) from the McMurchy et al. study was used (McMurchy et al., 2017; GEO accession GSE87524). The shown H3K9me3 signal represents the averaged H3K9me3 signal in two replicates of young adults. For analysis of the H3K9me3 levels in wild type and set-32 mutants the raw data from the Kalinava et al. study was used (Kalinava et al., 2018; GEO accession: GSE117662). The raw data were analyzed in a similar fashion to the analysis conducted by McMurchy et al. Briefly, adaptors were trimmed using Cutadapt (Martin, 2011) and aligned using Bowtie2 (Langmead and Salzberg, 2012). H3K9me3-enriched regions were identified using MACS2 (Lupien et al., 2008) and the H3K9me3 signal was corrected for biases using BEADS (Cheung et al., 2011).
Bioinformatic mRNA expression analysis
Processed files with raw counts of reads mapping to each gene were downloaded from GEO (Klosin et al., 2017; GEO accession: GSE83528). Differential expressed genes were detected using DESeq2 (adjusted p-value<0.1).
Bioinformatic gene enrichment analysis
The enrichment values denote the ratio between (A) the observed representations of a specific gene set within a defined differentially expressed genes group, to (B) the expected one, that is the representation of the examined gene set among all protein-coding genes in C. elegans. The analysis was done for 15 gene sets: (1) 7727 genes enriched in oocytes gonads (Ortiz et al., 2014) and 9012 genes enriched in spermatogenic gonads (Ortiz et al., 2014); we excluded genes with expression lower than 1 RPKM(2) 11427 genes expressed in isolated neurons (Kaletsky et al., 2016). (3) 7176 genes expressed in intestine (Gerstein et al., 2010) (4) 2957 genes expressed in pharynx (Gerstein et al., 2010) (5) 2526 genes expressed in body muscle (Gerstein et al., 2010) (6) 4146 targets of CSR-1 (Claycomb et al., 2009) (7) 1478 targets of HRDE-1 (Buckley et al., 2012) (8) 87 targets of WAGO-1 (Gu et al., 2009) (9) 399 targets of ALG-3/4 class small RNAs (Conine et al., 2010) (10) 1823 targets of mutator class small RNAs (11) 721 EGO-1 dependent small RNA gene targets (Maniar and Fire, 2011), (12) 23 gene targets of small RNAs up-regulated in ego-1 mutants (Maniar and Fire, 2011), (13) 49 genes targeted by 26G-RNAs enriched in ERGO-IP (Vasale et al., 2010) (14) 77 genes depleted of 22G-RNAs in ergo-1 mutants (Vasale et al., 2010), and (15) 348 putative piRNA gene targets (Bagijn et al., 2012). The putative piRNA gene targets were defined as genes for which, in at least one transcript, the ratio of the # 22G-RNA reads at piRNA target sites between wild type to prg-1 is at least 2 (linear scale). Note that the indicated number above achieved after intersection between the various published data sources and the records appears in the *.gff file used by us.
The enrichment value of a given gene set i in differentially expressed gene targeting small RNAs was calculated using the following formula:
Obtaining the observed-to-expected ratios, we then calculated the corresponding p-values using 10,000 random gene groups identical in size to that of the examined group of differentially expressed genes. Next, the enrichment values of the random sets are ranked and the p-value is determined by the ranking of the examined gene set amongst the ranking of all enrichment values of the random sets.
Gene sets by conservation
The classification of gene sets by conservation was done by mining the ‘Homology’ field of all the C. elegans protein-coding genes in WormBase (www.wormbase.com). We defined the following three gene sets (Figure 4B):
Unique to C. elegans – C. elegans genes which have no orthologues gene in any of the following species: B. malayi, C. brenneri, C. briggsae, C. japonica, C. remanei, O. volvulus, P. pacificus and S. ratti.
Caenorhabditis only - C. elegans genes which have at least one orthologues gene in one of the C. brenneri, C. briggsae, C. remanei and C. japonica species, and have no orthologues gene in any of the B. malayi, O. volvulus, P. pacificus and S. ratti species.
Conserved among nematodes - C. elegans genes which have at least one orthologues gene in one of the C. brenneri, C. briggsae, C. remanei and C. japonica species, and in addition have at least one orthologues gene in one of the B. malayi, O. volvulus, P. pacificus and S. ratti species.
Statistical analysis
For RNAi experiments, Two-way ANOVA tests were used to compare the percentages of the RNAi-affected worms (GFP silencing or fertility for the oma-1 assay) between the tested genotypes. In cases of multiple comparisons between genotypes and across generations, Sidak multiple comparison tests were applied. For GFP fluorescence experiments, Two-way ANOVA tests were used to compare the normalized GFP expression levels between the genotypes and across the biological repeats. For H3K9me3 qPCR-ChIP experiments Two-way ANOVA tests were used to compare the delta-delta-Ct (or delta-Ct) values between the gfp and the oma-1 loci obtained using two different primer sets. In cases of comparisons between genotypes and loci the Sidak multiple comparison tests were applied. Biological replicates were performed using separate populations of animals. Statistical tests were performed using GraphPad Prism software (Graphpad Prism) version 6. The statistical analysis used for each of the bioinformatics analyses is listed under the corresponding bioinformatics methods.
Acknowledgements
We thank all the Rechavi lab members for the helpful comments and fruitful discussions. We thank Yael Mor for the fruitful discussions and asistance with formulating the newly evolved genes hypothesis. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Yosef Shiloh, Yael Ziv, for their assistance and advice. Special thanks to Dror Cohen for the illustrations that he contributed. This work was supported by the ERC (grant #335624) and the Israel Science Foundation (grant #1339/17) and OR gratefully acknowledges the support of the Allen Discovery Center of the Paul G Allen Frontiers Group and the support of the Adelis foundation (no. 01430001000).
Funding Statement
The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Contributor Information
Itamar Lev, Email: itamai.et@gmail.com.
Oded Rechavi, Email: odedrechavi@gmail.com.
Patricia J Wittkopp, University of Michigan, United States.
Patricia J Wittkopp, University of Michigan, United States.
Funding Information
This paper was supported by the following grants:
Israel Science Foundation 1339/17 to Itamar Lev, Hila Gingold, Oded Rechavi.
European Research Council 335624 to Itamar Lev, Hila Gingold, Oded Rechavi.
Adelis Foundation 01430001000 to Oded Rechavi.
Paul G. Allen Family Foundation to Oded Rechavi.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Formal analysis, Investigation, Visualization, Methodology, Writing—original draft, Project administration, Writing—review and editing.
Conceptualization, Software, Formal analysis, Investigation, Methodology, Writing—original draft, Project administration, Writing—review and editing.
Conceptualization, Supervision, Funding acquisition, Writing—original draft, Project administration, Writing—review and editing.
Additional files
The table includes all the C. elegans genes we examined (20,447 genes). Each gene is marked for being: A. a target of SET-25 dependent endo-siRNAs, B. a target of SET-32 dependent endo-siRNAs. C. Conserved among nematodes, D. Conserved only in Caenorhabditis, E. Unique to C. elegans.
The table lists the fold change enrichment values and the matching p-values found in gene targets of SET-25 and SET-32 dependent endo-siRNAs for other genes sets discussed in this manuscript.
Data availability
All data generated or analyzed during this study are included in the manuscript and supporting files.
The following previously published datasets were used:
Lev I, Seroussi U, Gingold H, Bril R, Anava S, Rechavi O. 2017. MET-2-Dependent H3K9 Methylation Suppresses Transgenerational Small RNA Inheritance. NCBI Gene Expression Omnibus. GSE94798
McMurchy AN, Stempor P, Gaarenstroom T. 2017. A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. NCBI Gene Expression Omnibus. GSE87524
Kalinava N, Ni JZ, Gajic Z, Kim M, Ushakov H, Gu SG. 2018. C. elegans Heterochromatin Factor SET-32 Plays an Essential Role in Transgenerational Establishment of Nuclear RNAi-Mediated Epigenetic Silencing. NCBI Gene Expression Omnibus. GSE117662
Klosin A, Casas E, Hidalgo C, Vavouri T. 2017. Transgenerational transmission of environmental information in C. elegans. NCBI Gene Expression Omnibus. GSE83528
References
- Akay A, Di Domenico T, Suen KM, Nabih A, Parada GE, Larance M, Medhi R, Berkyurek AC, Zhang X, Wedeles CJ, Rudolph KLM, Engelhardt J, Hemberg M, Ma P, Lamond AI, Claycomb JM, Miska EA. The helicase aquarius/EMB-4 is required to overcome intronic barriers to allow nuclear RNAi pathways to heritably silence transcription. Developmental Cell. 2017;42:241–255. doi: 10.1016/j.devcel.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar RM, Lin R, Fire AZ. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis Elegans. Genetics. 2008;180:1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, Pintacuda G, Sakaguchi A, Sarkies P, Ahmed S, Miska EA. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA. 2013;19:740–751. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David E, Burga A, Kruglyak L. A maternal-effect selfish genetic element in Caenorhabditis Elegans. Science. 2017;356:1051–1055. doi: 10.1126/science.aan0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessler JB, Andersen EC, Villeneuve AM. Differential localization and independent acquisition of the H3K9me2 and H3K9me3 chromatin modifications in the caenorhabditis elegans adult germ line. PLOS Genetics. 2010;6:e1000830. doi: 10.1371/journal.pgen.1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis Elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nature Reviews Genetics. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung MS, Down TA, Latorre I, Ahringer J. Systematic bias in high-throughput sequencing data and its correction by BEADS. Nucleic Acids Research. 2011;39:e103. doi: 10.1093/nar/gkr425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Forrest EC, Sepich S, Cogoni C, Macino G. Small interfering RNAs that trigger posttranscriptional gene silencing are not required for the histone H3 Lys9 methylation necessary for transgenic tandem repeat stabilization in neurospora crassa. Molecular and Cellular Biology. 2005;25:3793–3801. doi: 10.1128/MCB.25.9.3793-3801.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, Conte D, Mello CC. The argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in caenorhabditis elegans. PNAS. 2010;107:3588–3593. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SE, Montgomery TA, Zhang C, Fahlgren N, Breen PC, Hwang A, Sullivan CM, Carrington JC, Ruvkun G. The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLOS Genetics. 2011;7:e1002369. doi: 10.1371/journal.pgen.1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Jain N, Hansen L, Davis MW, Li Y, Zhao D, Rebora K, Millet JRM, Liu X, Kim SK, Dupuy D, Jorgensen EM, Fire AZ. An abundant class of Non-coding DNA can prevent stochastic gene silencing in the C. elegans Germline. Cell. 2016;166:343–357. doi: 10.1016/j.cell.2016.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammon DB, Ishidate T, Li L, Gu W, Silverman N, Mello CC. The antiviral RNA interference response provides resistance to lethal arbovirus infection and vertical transmission in caenorhabditis elegans. Current Biology. 2017;27:795–806. doi: 10.1016/j.cub.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, Alves P, Chateigner A, Perry M, Morris M, Auerbach RK, Feng X, Leng J, Vielle A, Niu W, Rhrissorrakrai K, Agarwal A, Alexander RP, Barber G, Brdlik CM, Brennan J, Brouillet JJ, Carr A, Cheung MS, Clawson H, Contrino S, Dannenberg LO, Dernburg AF, Desai A, Dick L, Dosé AC, Du J, Egelhofer T, Ercan S, Euskirchen G, Ewing B, Feingold EA, Gassmann R, Good PJ, Green P, Gullier F, Gutwein M, Guyer MS, Habegger L, Han T, Henikoff JG, Henz SR, Hinrichs A, Holster H, Hyman T, Iniguez AL, Janette J, Jensen M, Kato M, Kent WJ, Kephart E, Khivansara V, Khurana E, Kim JK, Kolasinska-Zwierz P, Lai EC, Latorre I, Leahey A, Lewis S, Lloyd P, Lochovsky L, Lowdon RF, Lubling Y, Lyne R, MacCoss M, Mackowiak SD, Mangone M, McKay S, Mecenas D, Merrihew G, Miller DM, Muroyama A, Murray JI, Ooi SL, Pham H, Phippen T, Preston EA, Rajewsky N, Rätsch G, Rosenbaum H, Rozowsky J, Rutherford K, Ruzanov P, Sarov M, Sasidharan R, Sboner A, Scheid P, Segal E, Shin H, Shou C, Slack FJ, Slightam C, Smith R, Spencer WC, Stinson EO, Taing S, Takasaki T, Vafeados D, Voronina K, Wang G, Washington NL, Whittle CM, Wu B, Yan KK, Zeller G, Zha Z, Zhong M, Zhou X, Ahringer J, Strome S, Gunsalus KC, Micklem G, Liu XS, Reinke V, Kim SK, Hillier LW, Henikoff S, Piano F, Snyder M, Stein L, Lieb JD, Waterston RH, modENCODE Consortium Integrative analysis of the caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, Chen CC, Chaves DA, Duan S, Kasschau KD, Fahlgren N, Yates JR, Mitani S, Carrington JC, Mello CC. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Molecular Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis Elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nature Genetics. 2012;44:157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nature Reviews Genetics. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky R, Lakhina V, Arey R, Williams A, Landis J, Ashraf J, Murphy CT. The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature. 2016;529:92–96. doi: 10.1038/nature16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinava N, Ni JZ, Peterman K, Chen E, Gu SG. Decoupling the downstream effects of germline nuclear RNAi reveals that H3K9me3 is dispensable for heritable RNAi and the maintenance of endogenous siRNA-mediated transcriptional silencing in Caenorhabditis Elegans. Epigenetics & Chromatin. 2017;10:6. doi: 10.1186/s13072-017-0114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinava N, Ni JZ, Gajic Z, Kim M, Ushakov H, Gu SG. C. elegans Heterochromatin Factor SET-32 Plays an Essential Role in Transgenerational Establishment of Nuclear RNAi-Mediated Epigenetic Silencing. Cell Reports. 2018;25:2273–2284. doi: 10.1016/j.celrep.2018.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis Elegans. Methods. 2003;30:313–321. doi: 10.1016/S1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Klosin A, Casas E, Hidalgo-Carcedo C, Vavouri T, Lehner B. Transgenerational transmission of environmental information in C. elegans. Science. 2017;356:320–323. doi: 10.1126/science.aah6412. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev I, Seroussi U, Gingold H, Bril R, Anava S, Rechavi O. MET-2-Dependent H3K9 methylation suppresses transgenerational small RNA inheritance. Current Biology. 2017;27:1138–1147. doi: 10.1016/j.cub.2017.03.008. [DOI] [PubMed] [Google Scholar]
- Lev I, Bril R, Liu Y, Cere LI, Rechavi O. Inter-generational consequences for growing C. elegans in liquid. bioRxiv. 2018 doi: 10.1101/467837. [DOI] [PMC free article] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M, Li W, Liu XS, Lupien M, Eeckhoute J, Meyer C, Wang Q, Zhang Y, Li W, Carroll J, Liu X, Brown M, Kim T, Ren B, Iyer V, Horak C, Scafe C, Botstein D, Snyder M, Brown P, Ren B, Robert F, Wyrick J, Aparicio O, Jennings E, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert T, Wilson C, Bell S, Young R, Barski A, Cuddapah S, Cui K, Roh T, Schones D, Wang Z, Wei G, Chepelev I, Zhao K, Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith O, He A, Marra M, Snyder M, Jones S, Mikkelsen T, Ku M, Jaffe D, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T, Koche R, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander E, Bernstein B, Johnson D, Mortazavi A, Myers R, Wold B, Wei C, Wu Q, Vega V, Chiu K, Ng P, Zhang T, Shahab A, Yong H, Fu Y, Weng Z, Liu J, Zhao X, Chew J, Lee Y, Kuznetsov V, Sung W, Miller L, Lim B, Liu E, Yu Q, Ng H, Ruan Y, Redon R, Ishikawa S, Fitch K, Feuk L, Perry G, Andrews T, Fiegler H, Shapero M, Carson A, Chen W, Cho E, Dallaire S, Freeman J, Gonzalez J, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald J, Marshall C, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville M, Tchinda J, Valsesia A, Woodwark C, Yang F, Johnson W, Li W, Meyer C, Gottardo R, Carroll J, Brown M, Liu X, Song J, Johnson W, Zhu X, Zhang X, Li W, Manrai A, Liu J, Chen R, Liu X, Carroll J, Meyer C, Song J, Li W, Geistlinger T, Eeckhoute J, Brodsky A, Keeton E, Fertuck K, Hall G, Wang Q, Bekiranov S, Sementchenko V, Fox E, Silver P, Gingeras T, Liu X, Brown M. Model-based analysis of ChIP-Seq (MACS) Cell. 2008;132:958–970. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, van Bergeijk P, Kaaij LJ, Almeida MV, Roovers EF, Berezikov E, Ketting RF. Extremely stable Piwi-induced gene silencing in Caenorhabditis Elegans. The EMBO Journal. 2012;31:3422–3430. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nature Reviews Genetics. 2013;14:523–534. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniar JM, Fire AZ. EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Current Biology. 2011;21:449–459. doi: 10.1016/j.cub.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Zhu C, Zong D, Weng C, Yang X, Huang H, Liu D, Feng X, Guang S. The nrde pathway mediates Small-RNA-Directed histone H3 lysine 27 trimethylation in Caenorhabditis Elegans. Current Biology. 2015;25:2398–2403. doi: 10.1016/j.cub.2015.07.051. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- McMurchy AN, Stempor P, Gaarenstroom T, Wysolmerski B, Dong Y, Aussianikava D, Appert A, Huang N, Kolasinska-Zwierz P, Sapetschnig A, Miska EA, Ahringer J. A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. eLife. 2017;6:e21666. doi: 10.7554/eLife.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkina O, Hunter CP. Stable heritable germline silencing directs somatic silencing at an endogenous locus. Molecular Cell. 2017;65:659–670. doi: 10.1016/j.molcel.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Bühler M, Buker SM, Colmenares SU, Gerace EL, Gerber SA, Hong EJ, Motamedi MR, Verdel A, Villén J, Gygi SP. Studies on the mechanism of RNAi-dependent heterochromatin assembly. Cold Spring Harbor Symposia on Quantitative Biology. 2006;71:461–471. doi: 10.1101/sqb.2006.71.044. [DOI] [PubMed] [Google Scholar]
- Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- Newman MA, Ji F, Fischer SEJ, Anselmo A, Sadreyev RI, Ruvkun G. The surveillance of pre-mRNA splicing is an early step in C. elegans RNAi of endogenous genes. Genes & Development. 2018;32:670–681. doi: 10.1101/gad.311514.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz MA, Noble D, Sorokin EP, Kimble J. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode caenorhabditis elegans. G3: Genes|Genomes|Genetics. 2014;4:1765–1772. doi: 10.1534/g3.114.012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Montgomery BE, Breen PC, Roovers EF, Rim YS, Ohsumi TK, Newman MA, van Wolfswinkel JC, Ketting RF, Ruvkun G, Montgomery TA. MUT-14 and SMUT-1 DEAD box RNA helicases have overlapping roles in germline RNAi and endogenous siRNA formation. Current Biology. 2014;24:839–844. doi: 10.1016/j.cub.2014.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O, Minevich G, Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell. 2011;147:1248–1256. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O. Guest list or black list: heritable small RNAs as immunogenic memories. Trends in Cell Biology. 2014;24:212–220. doi: 10.1016/j.tcb.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O, Houri-Ze'evi L, Anava S, Goh WSS, Kerk SY, Hannon GJ, Hobert O. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O, Lev I. Principles of transgenerational small RNA inheritance in Caenorhabditis Elegans. Current Biology. 2017;27:R720–R730. doi: 10.1016/j.cub.2017.05.043. [DOI] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M. Toward improving Caenorhabditis Elegans phenome mapping with an ORFeome-based RNAi library. Genome Research. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T, Kimble J. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis Elegans. Genetics. 1988;119:43–61. doi: 10.1093/genetics/119.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M, Shirayama M, Gu W, Ishidate T, Conte D, Mello CC. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Developmental Cell. 2013;27:656–663. doi: 10.1016/j.devcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M, Shirayama M, Tang W, Shen EZ, Tu S, Lee HC, Weng Z, Mello CC. The coding regions of germline mRNAs confer sensitivity to argonaute regulation in C. elegans. Cell Reports. 2018;22:2254–2264. doi: 10.1016/j.celrep.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EZ, Chen H, Ozturk AR, Tu S, Shirayama M, Tang W, Ding YH, Dai SY, Weng Z, Mello CC. Identification of piRNA binding sites reveals the argonaute regulatory landscape of the C. elegans Germline. Cell. 2018;172:937–951. doi: 10.1016/j.cell.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklin G, Fields B, Wan G, Vijayendran D, Wallig A, Shukla A, Kennedy S. Identification and characterization of C. elegans RNAi inheritance machinery. Genetics. 2017:1–19. doi: 10.1534/genetics.116.198812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- Tyc KM, Nabih A, Wu MZ, Wedeles CJ, Sobotka JA, Claycomb JM. The conserved intron binding protein EMB-4 plays differential roles in germline small RNA pathways of C. elegans. Developmental Cell. 2017;42:256–270. doi: 10.1016/j.devcel.2017.07.003. [DOI] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. PNAS. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Brunschwig K, Okihara KL, Müller F, Tijsterman M, Plasterk RH. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser NE, Yang DX, Feng S, Kalinava N, Brown KC, Khanikar J, Freeberg MA, Snyder MJ, Csankovszki G, Chan RC, Gu SG, Montgomery TA, Jacobsen SE, Kim JK. MORC-1 integrates nuclear RNAi and transgenerational chromatin architecture to promote germline immortality. Developmental Cell. 2017;41:408–423. doi: 10.1016/j.devcel.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Montgomery TA, Gabel HW, Fischer SE, Phillips CM, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in caenorhabditis elegans. PNAS. 2011;108:1201–1208. doi: 10.1073/pnas.1018695108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tu S, Stubna M, Wu WS, Huang WC, Weng Z, Lee HC. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science. 2018;359:587–592. doi: 10.1126/science.aao2840. [DOI] [PMC free article] [PubMed] [Google Scholar]