Summary
The lipopeptide antibiotic daptomycin (DAP) is a key drug against serious enterococcal infections, but emergence of resistance in the clinical setting is a major concern. The LiaFSR system plays a prominent role in the development of DAP resistance (DAP-R) in enterococci, and blocking this stress response system has been proposed as a novel therapeutic strategy. In this work, we identify LiaR-independent pathways in E. faecalis that regulate cell membrane adaptation in response to antibiotics. We adapted E. faecalis OG1RF (a laboratory strain) and S613TM (a clinical strain) lacking liaR to increasing concentrations of DAP, leading to the development of DAP-R and elevated MICs to bacitracin and ceftriaxone. Whole genome sequencing identified changes in the YxdJK two-component regulatory system and a putative fatty acid kinase (dak) in both DAP-R strains. Deletion of the gene encoding the YxdJ response regulator in both the DAP-R mutant and wild type OG1RF decreased MICs to DAP, even when a functional LiaFSR system was present. Mutations in dak were associated with slower growth, decreased membrane fluidity and alterations of cell morphology. These findings suggest overlapping stress response pathways can provide protection against antimicrobial peptides in E. faecalis at a significant cost in bacterial fitness.
Keywords: Enterococcus faecalis, antibiotic resistance, daptomycin, stress response, two-component regulatory system, bacterial membrane
Graphical Abstract
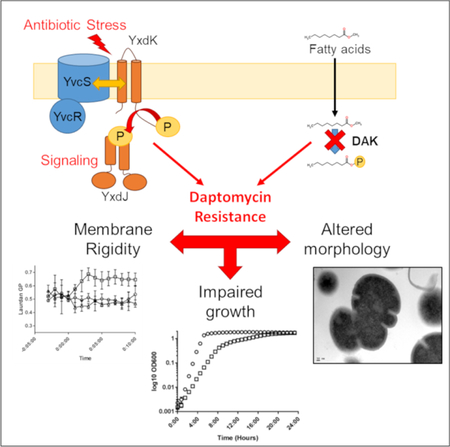
Abbreviated Summary
Resistance to daptomycin, a front line antibiotic for vancomycin-resistant enterococcal infections, is a growing clinical challenge. Through an adaptive evolution experiment, the two component signaling system YxdJK and a putative fatty acid kinase were linked to daptomycin resistance in Enterococcus faecalis lacking the primary stress response system LiaFSR. Resistance comes with a cost, as mutants displayed impaired growth with alterations in membrane fluidity, and in some cases distinct alterations of cell morphology.
Introduction
Enterococci are a leading cause of healthcare-associated infections (Weiner et al., 2016) and treatment is complicated by their resistance to a variety of antimicrobial agents. Indeed, the emergence of resistance to daptomycin (DAP), a lipopeptide antibiotic with dose-dependent bactericidal activity in vitro against enterococci, is an example of a pattern of successful adaptations against antimicrobials by these organisms (Arias and Murray, 2012). The mechanism of DAP action is not fully understood but the available evidence suggests that DAP binds to the anionic phospholipid phosphatidylglycerol (PG) in the outer leaflet of the cell membrane in a calcium dependent manner. After initial binding, as more molecules accumulate at the site of action, DAP forms oligomers and appears to transition across the outer to the inner leaflet of the membrane (Tran et al., 2015). The interactions of DAP with the cell membrane alter its structure and function, causing changes in membrane order and the displacement of membrane-associated proteins responsible for cell envelope biogenesis, ultimately leading to cell death (Müller et al., 2016).
Over 30 genes have been associated with DAP resistance (Arias et al., 2011; Palmer et al., 2011; Humphries et al., 2012; Diaz et al., 2014) in clinical and laboratory isolates of enterococci. A unifying theme associated with the resistance phenotype is the presence of mutations in genes encoding two- or three-component regulatory systems (TCS) and enzymes with predicted roles in phospholipid metabolism. Among the TCS, the LiaFSR system has been shown to play a major role in the maintenance of cell envelope stability under the DAP “attack” (Tran et al., 2013). For example, in an evolutionary model of development of DAP-R in E. faecalis, changes in the LiaFSR system occurred first, followed by major increases in the DAP MIC associated with substitutions in cardiolipin synthase (Miller et al., 2013). Similarly, changes in LiaFSR in clinical strains of E. faecium led to a loss of DAP bactericidal activity and preceded the emergence of clinical resistance (Munita et al., 2012; Munita et al., 2013). The major role of the LiaFSR system in DAP resistance has been confirmed by targeted deletions of liaR (encoding the response regulator of the system) in both E. faecalis and E. faecium, independent of the genetic background of the strain. These deletions have uniformly led to a DAP “hypersusceptible” phenotype with a decrease in DAP MICs over 100-fold (Panesso et al., 2015; Reyes et al., 2015).
Here, we aimed to discover and characterize new genes associated with the maintenance of cell envelope integrity in E. faecalis by adapting strains lacking LiaR to ascending concentrations of DAP. We found that resistance arising independent of LiaR is associated with changes in the TCS YxdJK and in a putative enzyme involved in extracellular lipid metabolism (a dihydroxyacetone kinase-like enzyme, designated DAK). The resistance phenotype was associated with major fitness costs for the cell, suggesting that the redundancy of genetic pathways is hierarchical and alternative pathways for resistance may not be evolutionary advantageous in some contexts.
Results
Emergence of DAP-resistance in the absence of LiaR.
We sought to elucidate whether additional networks can adapt to protect against DAP-triggered membrane damage when the primary mechanism for stress response is no longer available to E. faecalis. To test this hypothesis, we exposed both a modified clinical isolate that lacked liaR (S613TMΔliaR, a derivative of the clinical isolate S613 harboring the “resistance” alleles liaFΔIle177, gdpDΔIle170 and clsΔLys61) (Reyes et al., 2015), and a laboratory strain OG1RFΔliaR (Supplementary Table 1) to increasing sub-inhibitory concentrations of DAP in an in vitro adaptation experiment. The alleles liaFΔIle177, gdpDΔIle170 and clsΔLys61 were previously identified in a DAP-R clinical strain (Arias et al., 2011), and lead to activation of the LiaFSR system (in the case of liaFΔIle177) or are predicted to alter membrane phospholipid metabolism (gdpDΔIle170 and clsΔLys61). After 15 and 17 days, respectively, we selected derivatives of both parent strains in which the MIC had increased to 24 µg/ml (S613TMΔliaRyvcR219dak84asa1) and 6 µg/ml (OG1RFΔliaRyxdK202dak371ace) (Table 1). Unlike prior mutations associated with the LiaFSR pathway, resistance to DAP in the setting of the deletion of liaR resulted in a marked fitness cost, with an increase in the lag phase and longer time to reach stationary phase (Fig. 1, Table 2). This effect was seen in both brain heart infusion (BHI) and Mueller Hinton (MH) media. Since the resistant mutants displayed a small colony variant morphology, we performed colony counts with S613TMΔliaRyvcR219dak84asa1 and OG1RFΔliaRyxdK202dak371ace to confirm that OD600nm was proportional to the number of viable bacteria and correlated with spectrophotometer readings (Supplementary Fig. 1). The stability of the resistance phenotype was variable, with the emergence of a heterogeneous population with mixed MICs to DAP in OG1RFΔliaRyxdK202dak371ace after passage in antibiotic free media for 7 days. S613TMΔliaRyvcR219dak84asa1 remained resistant (Supplementary Fig. 2). Interestingly, OG1RFΔliaRyxdK202dak371ace also displayed major increases in the MIC to bacitracin (24 fold) and ceftriaxone (>16 fold). Neither derivative displayed major changes in susceptibility to ampicillin or vancomycin (Table 1).
Table 1.
Antimicrobial Minimum Inhibitory Concentrations of E. faecalis Strains.
| Strain | DAP | BAC | CRO | AMP | VAN |
|---|---|---|---|---|---|
| S613 | 0.5 | >256 | >256 | 0.38 | >256 |
| S613TM | 8 | >256 | >256 | 0.125 | >256 |
| S613TMΔliaR | 0.094 | >256 | >256 | 0.38 | >256 |
| S613TMΔliaRyvcR219dak84asa1 | 24 | >256 | >256 | 1 | >256 |
| OG1RF | 1.5 | 64 | 32 | 0.5 | 2 |
| OG1RFΔliaR | 0.047 | 4 | 16 | 0.5 | 2 |
| OG1RFΔliaRyxdK202dak371ace | 6 | 96 | >256 | 1 | 3 |
| OG1RFΔliaRyxdK202dak371aceΔyxdJ | 0.094 | 0.75 | >256 | 3 | 1.5 |
| OG1RFΔliaRyxdK202dak371aceΔyxdJ/pAT392::yxdJ | 3 | >256 | >256 | 0.38 | 1 |
| OG1RFΔliaRyxdK202dak371ace::dak | 2 | 128 | >256 | 3 | 3 |
| OG1RFΔyxdJ | 0.38 | 12 | 8 | 0.5 | 3 |
| OG1RFΔyxdJ/pAT392::yxdJ | 2 | 64 | 48 | 1 | 3 |
| OG1RFΔyxdJ/pAT392 | 0.5 | 12 | 8 | 1 | 3 |
| OG1RFΔliaRΔc-dak | 0.064 | 1.5 | >256 | 0.5 | 2 |
| OG1RFΔliaRΔc-dak::c-dak | 0.047 | 6 | 12 | 1.5 | 3 |
All values in µg/ml. DAP, daptomycin; BAC, bacitracin; CRO, ceftriaxone; AMP, ampicillin; VAN, vancomycin.
Figure 1. LiaR independent DAP-R is associated with impaired growth.
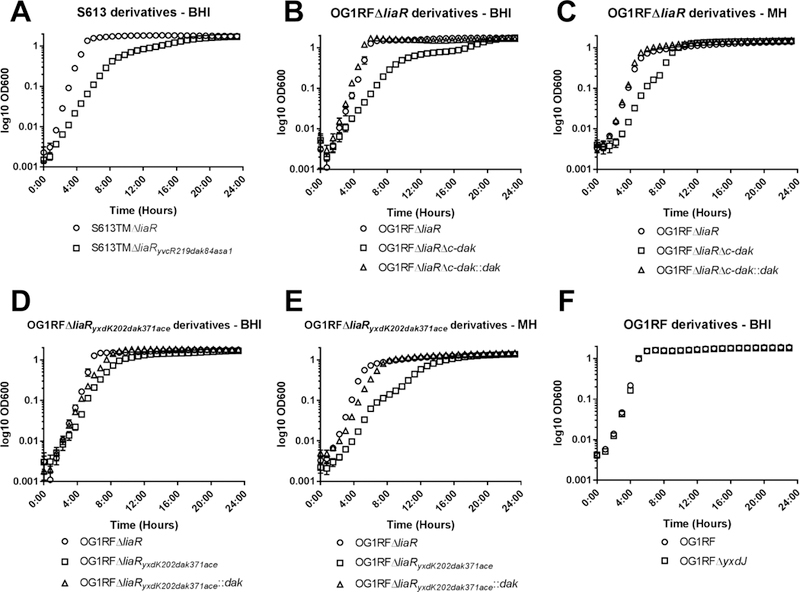
Growth curves in media without antibiotics for the (A) S613 background, (B and C) OG1RFΔliaR background, (D and E) OG1RFΔliaRyxdK202dak371ace background, and (F) wild type OG1RF background. Derivatives with mutations in dak showed alterations in growth (also see Table 2) independent of media, and reintroduction of the wild type dak allele complemented the growth phenotype. Error bars represent the standard error of the mean from three independent runs, n=10 measurements per strain. BHI, brain heart infusion; MH, Mueller Hinton.
Table 2.
Growth Parameters for E. faecalis Strains.
| BHI | MH | |||
|---|---|---|---|---|
| Lag Time† ± SD | Growth Rate§ ± SD | Lag Time† ± SD | Growth Rate§ ± SD | |
| OG1RFΔliaR | 76.2 ± 22 | 3.31 ± 0.58 | 83.7 ± 8 | 1.31 ± 0.03 |
| OG1RFΔliaRΔc-dak | 106.2 ± 42 | 0.77 ± 0.17 | 154.2 ± 9 | 0.88 ± 0.1 |
| OG1RFΔliaRΔc-dak::c-dak | 68.7 ± 25 | 4.70 ± 1.07 | 79.2 ± 9 | 1.49 ± 0.03 |
| OG1RFΔliaRyxdK202dak371ace | 80.7 ± 26 | 0.86 ± 0.07 | 158.7 ± 14 | 0.67 ± 0.04 |
| OG1RFΔliaRyxdK202dak371ace::dak | 73.2 ± 13 | 1.29 ± 0.05 | 86.7 ± 10 | 1.11 ± 0.04 |
| S613TMΔliaR | 47.7 ± 8 | 1.84 ± 0.07 | ND | ND |
| S613TMΔliaRyvcR219dak84asa1 | 73.2 ± 15 | 0.88 ± 0.02 | ND | ND |
| OG1RF | 58.2 ± 11 | 2.39 ± 0.06 | ND | ND |
| OG1RFΔyxdJ | 62.7 ± 8 | 2.81 ± 0.22 | ND | ND |
Minutes.
Maximal rate of change, OD per hour.
BHI, brain heart infusion broth; MH, Mueller Hinton broth; ND, not done; SD, standard deviation.
We performed whole genome sequencing of the DAP-R derivatives in both strain backgrounds in order to identify genetic changes associated with development of DAP-R (Table 3). Interestingly, both strains developed mutations in the sensing module of the two-component regulatory system YxdJK, a BceAB-like system previously linked to bacitracin (BAC) resistance in Bacillus subtilis and E. faecalis and chlorhexidine tolerance in E. faecium (Dintner et al., 2014; Gebhard et al., 2014; Prieto et al., 2017). The YxdJK system encodes a sensor histidine kinase (yxdK) and cognate response regulator (yxdJ), which positively regulates a putative ABC transporter (YvcRS, required for signal transduction along with YxdK) and a second transporter (YxdLM) responsible for the BAC resistance phenotype (Gebhard et al., 2014). Furthermore, both strains were also found to have changes in a gene (dak) which encodes a putative enzyme of the dihydroxyacetone kinase (DAK) family whose homolog in Staphylococcus aureus has been implicated in the metabolism of extracellular fatty acids (Parsons et al., 2014). Of note, neither gene has been previously associated with DAP-R. Each strain also had predicted substitutions in two major enterococcal virulence factors linked to extracellular matrix adhesion (aggregation substance and a collagen adhesin). We chose to further investigate the roles of the signal transduction system YxdJK and the putative lipid metabolism enzyme DAK in DAP resistance, since previous work has shown that activation of a stress response system coordinated with alteration of lipid metabolism is sufficient for DAP resistance (Miller et al., 2013; Diaz et al., 2014).
Table 3.
Nonsynonymous Single Nucleotide Polymorphisms Identified by Whole Genome Sequencing in S613TMΔliaRyvcR219dak84asa1 and OG1RFΔliaRyxdK202dak371ace.
| S613TMΔliaRyvcR219dak84asa1 | |||||
|---|---|---|---|---|---|
| Gene | Locus tag (V583 genome) | Locus tag (OG1RF genome) | NT Change | AA change | Predicted function |
| yvcR | EF2752 | OG1RF_12114 | 2443262, T>G | D219A | ABC Transporter, ATP binding subunit |
| dak | EF3114 | OG1RF_12374 | 2711659, A>G | S84P | DAK domain protein, fatty acid kinase |
| asa1 | EF0485 | - | 377891, G>A 378128, A>C 378268, C>A 378269, A>C 378335, A>C |
D1034N I1113L D1159E K1160Q N1182H |
Cell surface adhesin and virulence factor |
| OG1RFΔliaRyxdK202dak371ace | |||||
| Gene | Locus tag (V583 genome) | Locus tag (OG1RF genome) | NT Change | AA change | Predicted function |
| yxdK | EF0927 | OG1RF_10654 | 689129, C>A | A202E | Sensor histidine kinase |
| dak | EF3114 | OG1RF_12374 | 2512631 G>A |
Q371* | DAK domain protein, fatty acid kinase |
| ace | EF1099 | OG1RF_10878 | 918431, C>T 918448, A>G 918478, G>A |
A462V S468G A478T |
collagen adhesin, virulence factor |
NT, nucleotide; AA, amino acid; ABC, ATP-Binding Cassette; DAK, Dihydroxyacetone kinase
The two-component system YxdJK is associated with resistance to cell envelope antibiotics in the absence of a functional LiaFSR system.
Comparison of the antimicrobial susceptibility profiles of OG1RFΔliaR and its DAP-R derivative showed a 24-fold increase in BAC MIC, suggesting that the observed genetic changes resulted in activation of the YxdJK regulatory network, a system previously associated with BAC resistance in E. faecalis (Gebhard et al., 2014). No changes were seen in the BAC sensitivities of S613TMΔliaRyvcR219dak84asa1, likely due to the fact that this clinical strain also carries the bcrABDR operon, a transferable determinant which provides resistance to bacitracin through production of the BcrAB ATP-binding cassette (ABC) transporter and BcrD, an undecaprenol kinase (Manson et al., 2004).
To characterize the contributions of YxdJK to cell envelope active antimicrobials, we constructed a non-polar deletion of the gene encoding the response regulator YxdJ in the DAP resistant OG1RFΔliaRyxdK202dak371ace. We postulated that inactivation of the YxdJK system, via deletion of yxdJ, would eliminate the adaptive response and lead to significant decreases in the MICs of DAP, BAC and ceftriaxone (CRO). Indeed, loss of yxdJ resulted in a 64-fold drop in the MIC of DAP (6 to 0.094 µg/mL) and 128-fold drop in the MIC of BAC (96 to 0.75 µg/mL) confirming the importance of the YxdJK signaling system in resistance to these envelope targeting antimicrobials. Interestingly, we observed no change in the MIC of CRO (Table 1), a finding explained by our investigation of dak (see below). Complementation of OG1RFΔliaRyxdK202dak371aceΔyxdJ in trans using the plasmid pAT392 (Arthur et al., 1994) containing yxdJ increased the DAP MIC from 0.094 µg/mL in the deletion mutant to 3 µg/mL in the complemented strain. Complementation also led to an increase in the BAC MIC from 0.75 µg/mL to >256 µg/mL.
Next, we removed the gene encoding YxdJ from wild type OG1RF (OG1RFΔyxdJ) to examine the effect on membrane stress in E. faecalis with a functional LiaFSR system. Despite the presence of a functional LiaFSR operon, we observed a 4, 5 and 4-fold decrease in the MICs of DAP, BAC and CRO in the OG1RFΔyxdJ deletion mutant compared to OG1RF, respectively (Table 1). Importantly, this decrease in DAP MIC provides further evidence that the protective effect of YxdJK against the antimicrobial peptide attack is mediated by a LiaFSR-independent mechanism. Introduction of yxdJ on plasmid pAT392 (Arthur et al., 1994) (OG1RFΔyxdJ/pAT392::yxdJ) restored the MIC of the three above antibiotics to wild type levels, confirming the vital role of the YxdJK system in resistance against cell envelope antibiotics.
Mutations in a gene (dak) encoding a putative fatty-acid kinase, augment YxdJK mediated DAP susceptibility and markedly affect cell fitness.
The dak gene encodes a 558 amino acid protein with 53% amino acid identity to a fatty acid kinase previously described in Staphylococcus aureus (Parsons et al., 2014). This enzyme is capable of phosphorylating exogenous fatty acids for incorporation into membrane phospholipid synthesis. Our analysis identified two substitutions in DAK; the first resulted in a change of serine to proline at position 84 (in S613TMΔliaRyvcR219dak84asa1; Table 3), which is predicted to eliminate a hydrogen bond at the ATP binding site of DAK based on comparative modeling using the dihydroxyacetone kinase of Citrobacter freundii (Siebold et al., 2003; Kelley et al., 2015). The substitution of a proline into this position is also likely to distort the local main chain resulting in additional changes within the ligand-binding pocket. The second substitution introduces a premature stop codon at position 371 resulting in a truncation of the C-terminal 187 amino acids (in OG1RFΔliaRyxdK202dak371ace; Table 3). The predicted changes in the ATP binding site or the loss of the C-terminal end of the protein (containing the K domain involved in substrate phosphorylation) suggested that the enzymatic activity of DAK was likely to be impaired.
In order to confirm the role of DAK in DAP resistance, we complemented the wild type allele encoding the full length protein back into the DAP-R OG1RFΔliaRyxdK202dak371ace. The DAP MIC for the complemented strain was reduced from 6 to 2 µg/mL, which is within the susceptible range. This strain still carries the YxdKA202E amino acid substitution, thus the full resistance phenotype requires both yxdK and dak mutations to be present. There were no significant changes in the MICs of the other antimicrobials tested, and the DAP MIC remained above that of the OG1RFΔliaR parent strain, likely due to the presence of the mutant yxdK allele (Table 1).
We next attempted to produce a deletion mutant of the dak gene in OG1RFΔliaR. Our initial attempts to remove the entire reading frame in this strain were unsuccessful. Thus, we designed a strategy to remove the coding region from amino acid 371 to the stop codon at the C-terminal end (OG1RFΔliaRΔc-dak), recapitulating the effect of the frameshift seen in the evolved OG1RFΔliaRyxdK202dak371ace strain. We observed an increase in the lag phase and a slower growth rate in the OG1RFΔliaRΔc-dak mutant compared to the parental OG1RFΔliaR similar to that of OG1RFΔliaRyxdK202dak371ace (Fig. 1B, Table 2), suggesting that the change in DAK was likely responsible for the cell fitness effect. Table 1 shows that deletion of the C-terminal end of DAK alone did not have a significant impact on the DAP MIC. In contrast, it increased the ceftriaxone MIC >16 fold without changes in the ampicillin MIC, suggesting that the change influences cephalosporin susceptibility. We complemented the wild type dak allele encoding the full-length protein back into the deletion mutant in the native chromosomal location (OG1RFΔliaRΔc-dak::c-dak). Of note, restoration of the gene resulted in a change in the last C-terminal amino acid (E558G), but this did not affect the phenotype. The complemented strain OG1RFΔliaRΔc-dak::c-dak displayed a normal growth phenotype and reverted the ceftriaxone MIC from >256 µg/mL to 12 µg/mL (Fig. 1, Table 1, and Table 2).
Changes in DAK result in specific alterations of the cell envelope ultrastructure.
Prior studies have reported changes in the cell envelope in DAP-R E. faecalis (Arias et al., 2011). Using transmission electron microscopy (TEM), we evaluated the cell envelope structure of the DAP adapted mutants S613TMΔliaRyvcR219dak84asa1 and OG1RFΔliaRyxdK202dak371ace; and the mutants OG1RFΔyxdJ and OG1RFΔliaRΔc-dak. In both OG1RFΔliaRyxdK202dak371ace and OG1RFΔliaRΔc-dak, we observed cells with multiple septal events in which the orientation of the division plane was altered (Fig. 2, Supplemental Fig. 3). There was an increase in the thickness of the cellular envelope at the septal site, with cells displaying multiple protrusions of electron dense material from the cell surface. Complementation with the wild type dak allele restored the cell envelope structure in both OG1RFΔliaRyxdK202dak371ace and OG1RFΔliaRΔc-dak. To investigate if the changes in cell envelope structure varied with growth phase, we compared OG1RFΔliaR and OG1RFΔliaRΔc-dak at 8, 16 and 24 hours (Supplemental Fig. 4). The morphologic changes noted in OG1RFΔliaRΔc-dak were most prominent in late-exponential phase growth at 8 hours and less so at stationary phase (24 hours). Overall cell wall thickness of both OG1RFΔliaRyxdK202dak371ace and OG1RFΔliaRΔc-dak was increased from the parent strain OG1RFΔliaR at 16 hours, but this difference was statistically significant only for OG1RFΔliaRyxdK202dak371ace (p<0.01) (Supplemental Fig. 5A). The phenotype observed in the DAP-R derivative of S613TMΔliaR was less severe, without significant alterations of cell septum placement or cell wall thickness. This finding was supported by the lack of major ultrastructural changes observed in OG1RFΔyxdJ, suggesting that the C-terminal portion of DAK plays an important role in cell division homeostasis.
Figure 2. Loss of the C-terminal domain of DAK leads to altered cell morphology.
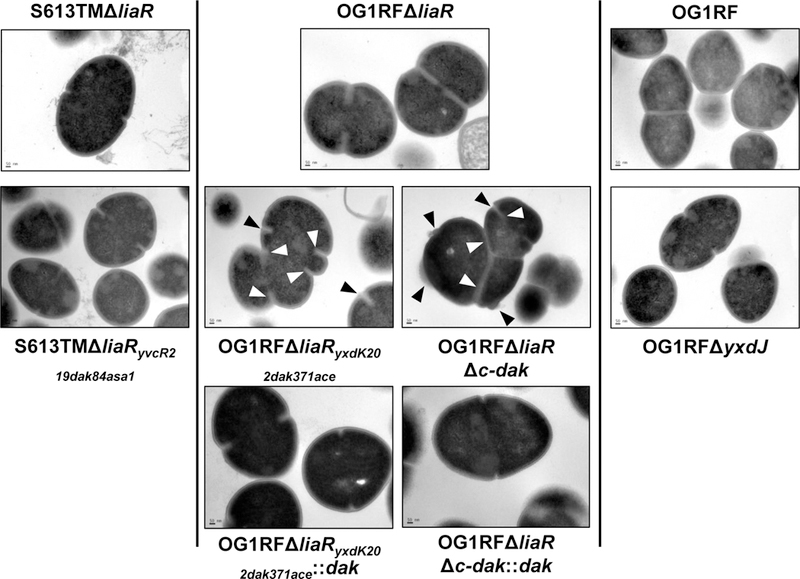
Transmission electron microscopy (TEM) of parent strains (top row), derivative strains (middle row), and complemented strains (bottom row) displaying changes in cell envelope structure. In OG1RFΔliaR septa are positioned at the middle of the cell, with one septal event per cell. In OG1RFΔliaRyxdK202dak371ace, multiple septal events per cell can be seen (white arrows), with loss of the division plane orientation, resulting in distorted cells. Cell envelope at the site of division is thickened, with small burs or protrusions from the cell surface (black arrows). A similar phenotype can be observed in OG1RFΔliaRΔc-dak, suggesting this domain plays an important role in cell division. These changes were not seen when the full length dak gene was complemented back to its chromosomal location. S613TMΔliaRyvcR219dak84asa1 and OG1RFΔyxdJ did not show structural changes. Scale bar for all images, 50 nm; Images are representative of over 25 different fields, wide field images are displayed in supplementary figure 3.
LiaR independent DAP-R is associated with alterations in membrane fluidity.
Resistance to DAP has been postulated to occur via multiple mechanisms, including alteration of cell surface charge (electrostatic repulsion) and variations in the content and architecture of the bacterial membrane, preventing DAP from reaching critical targets such as the division septum (diversion) (Tran et al., 2015). To rule out a repulsion-mediated mechanism of resistance, we performed a global assessment of cell surface charge using a poly-L-lysine assay and found no significant differences in cell surface charge between strain pairs (Supplemental Fig. 5B).
In E. faecalis, a distinct redistribution of membrane phospholipid microdomains has been observed as a major mediator of DAP resistance via the LiaFSR system (Tran et al., 2013). Thus, we sought to determine whether the observed mutations resulted in a similar alteration of the localization of membrane lipids. Using fluorescent microscopy with NAO, we were unable to detect any changes in the distribution of anionic phospholipid microdomains in either of the DAP adapted strains (Fig. 3A, Supplemental Figs. 6 and 7). Further, there was no decrease in the binding of BODIPY-DAP in the DAP resistant mutants as compared to their sensitive parental strains (Fig. 3B). In fact, OG1RFΔliaRyxdK202dak371ace bound more DAP than the sensitive OG1RFΔliaR, again implying that resistance is not mediated by repulsion of the antibiotic from the surface. Our data suggest that the mechanism of DAP-R mediated by changes in YxdJK and DAK appears to be distinct from that triggered by the LiaFSR system in E. faecalis.
Figure 3. DAP-R independent of LiaR does not result in phospholipid microdomain redistribution or a reduction in DAP binding.
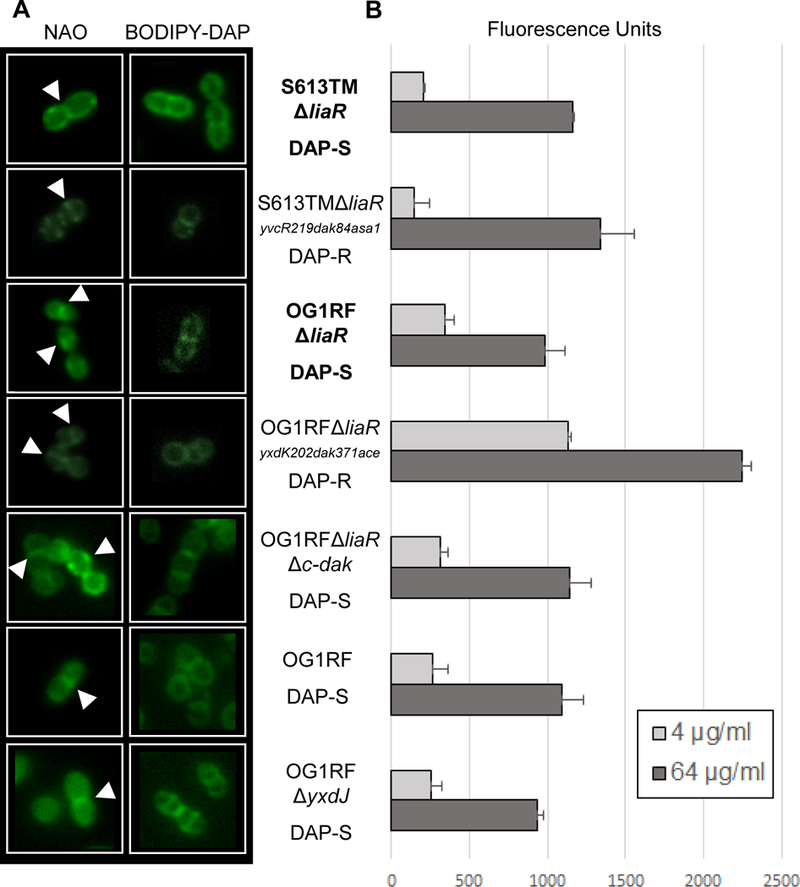
(A) N-nonyl acridine orange (NAO) and BODIPY-DAP staining of E. faecalis strains. NAO is a membrane dye that associates with anionic phospholipids, particularly cardiolipin. In all strains NAO and BODIPY-DAP can be seen localizing to the division septum (white arrows), without any areas of redistribution. (B) Quantitative BODIPY-DAP binding. Comparisons in binding were made between parental stains (bold) and derivatives. Increased DAP binding was seen in OG1RFΔliaRyxdK202dak371ace at both 4 and 64 µg/mL as compared to OG1RFΔliaR. No significant changes were seen between the other strain pairs. Representative results of two independent runs performed on different days.
DAP induced changes in membrane fluidity have recently been implicated in the antimicrobial action of the drug (Müller et al., 2016), with sequestration of fluid membrane lipids observed in B. subtilis treated with DAP, as compared to untreated cells. The membrane dye laurdan possesses a variable emission spectrum responsive to lipid order, allowing spectroscopic measurement of membrane fluidity. Since it does not appear that LiaR-independent mutations divert DAP from its site of action (as previously shown (Tran et al., 2013)), we postulated that they would interfere with membrane fluidity and potentially protect the cells against the antibiotic attack. In wild type OG1RF, we observed the characteristic shift towards a more rigid membrane after the addition of DAP at 10X the MIC and a more fluid membrane on incubation with ethanol (Fig. 4A). Interestingly, at lower DAP concentrations, we did not see a significant shift in rigidity in the DAP sensitive mutants OG1RFΔliaR (DAP MIC 0.047 µg/mL) and OG1RFΔliaRyxdK202dak371aceΔyxdJ (DAP MIC 0.094 µg/mL), despite the DAP concentration being ten times the MIC of the strain (Supplemental Fig. 8). Rigidity was induced in these strains at higher concentrations of DAP (200x MIC), thus the changes in rigidity did not appear to be due to the envelope stress response, and correlated with DAP concentration, but not strain MIC. Deletion of liaR did not alter baseline membrane fluidity (Fig. 4B). In OG1RFΔliaRyxdK202dak371ace baseline membrane rigidity in log phase growth was significantly increased as compared to either OG1RF or OG1RFΔliaR, and the membrane was more resistant to the induction of fluidity on addition of ethanol (Fig. 4C). This effect appears to be due to the changes in DAK, as complementation of the wild type dak allele, but not inactivation of the YxdJK system, restored membrane fluidity to wild type levels (Fig. 4D-F). Once the cells reached stationary phase there was no significant difference in baseline rigidity (Supplemental Fig. 9). These findings suggest an altered membrane environment during active cell growth facilitates DAP resistance, and that alterations of membrane fluidity are necessary, but not sufficient, for the DAP resistance phenotype in E. faecalis OG1RF lacking liaR.
Figure 4. Mutations in DAK and yxdJ alter membrane fluidity.
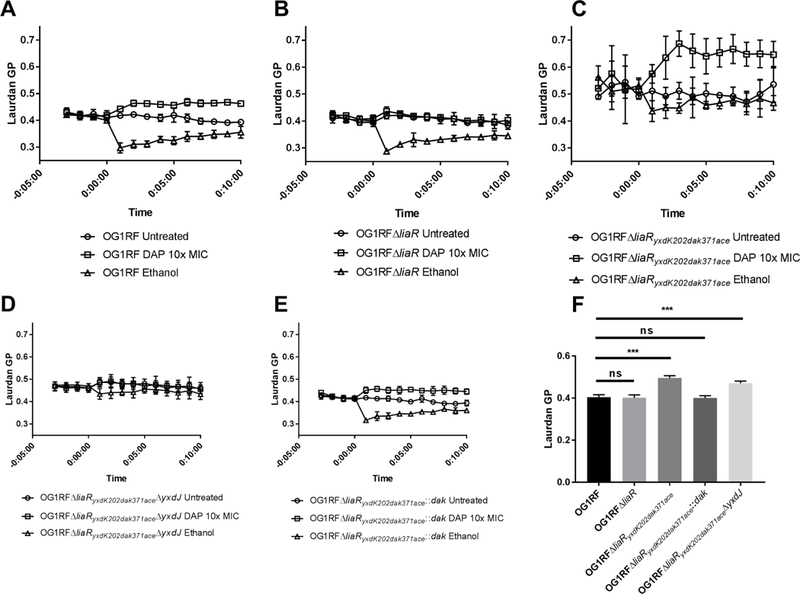
Membrane fluidity was assayed using the fluorescent dye laurdan, whose emission spectrum shifts in response to the order of the lipid bilayer (higher numbers indicate increasing rigidity). Representative results are displayed as the mean and standard deviation of two independent experiments run in technical quadruplicate, with DAP or ethanol added at time=0. (A) and (B) show that baseline fluidity and fluidity after the addition of ethanol are similar for both OG1RF and OG1RFΔliaR. (C) and (D) As compared to OG1RF wild type, mutants with changes in DAK had a significantly more rigid membrane at baseline, and were more resistant to the fluidity shift after addition of ethanol. (E) Complementation with the wild type dak allele restored membrane fluidity to wild type levels. (F) Comparison of mean baseline fluidity by two-way ANOVA with correction using Dunnett’s multiple comparisons test, using OG1RF as the control strain for comparison. ***p<0.001; ns, non-significant. Representative results of two independent assays for each strain performed on different days.
Discussion
We have previously shown that the LiaFSR system is crucial for development of antimicrobial peptide resistance in both E. faecalis and E. faecium (Panesso et al., 2015; Reyes et al., 2015). It has been proposed that this system could be targeted as a novel antimicrobial approach against these MDR organisms (anti-adaptation antibiotics). With that rationale, it becomes crucial to determine if resistance can develop in the absence of a functional LiaFSR system. Here, we demonstrate that DAP-R is able to arise independently of the presence of LiaR (the response regulator of the system), but with significant costs in cell fitness. More importantly, the LiaR-independent daptomycin resistance phenotype uncovered previously unknown genetic pathways leading to resistance to DAP and other cell envelope-targeting antimicrobials. In this work, we identified the TCS YxdJK (previously associated only with BAC resistance in E. faecalis) and a fatty acid metabolism enzyme harboring a DAK domain in association with DAP-R. The fact that mutations in genes encoding these proteins arose independently in two different strain backgrounds of E. faecalis lacking LiaR supports the concept that the cell membrane stress response network in enterococci is multi-layered as previously described in B. subtilis (Radeck et al., 2016).
The YxdJK stress response system is known to regulate two ABC transporters in Enterococcus faecalis, one involved in signaling (YvcRS) and the other as the effector of the BAC resistance phenotype (YxdLM) (Gebhard et al., 2014). These BceAB-type two component systems are conserved across the Firmicutes, with the YxdJK system present in E. faecium and B. subtilis, among others (Gebhard, 2012). We demonstrate that deletion of the response regulator YxdJ, responsible for activation of the ABC transporters, is sufficient to revert the resistant phenotype of OG1RFΔliaRyxdK202dak371ace. It is interesting to note that deletion of YxdJ in a background with a functional LiaFSR system leads to a 4-fold reduction in DAP MIC, suggesting an independent role of YxdJK in membrane homeostasis. Prior studies examining the cell wall response in E. faecalis OG1RF and JH2–2 showed that YxdJK was induced in response to BAC, mersacidin and cefalotin, but not ampicillin, nisin, gallidermin, vancomycin or teicoplanin (Abranches et al., 2013; Gebhard et al., 2014). We observed a similar phenomenon, with MIC shifts in BAC, CRO and DAP, but not ampicillin or vancomycin. This phenotype suggests that generalized responses to membrane induced stress, such as interference with lipid II or changes in cell wall homeostasis, are not the stimulus for YxdJK signaling. Instead, the system seems to sense antimicrobial compounds directly. The BceAB-type ABC transporters possess an extracellular domain postulated to interact with target molecules, a hypothesis supported by experiments in which extracellular domain swapping between transporters can alter substrate affinity (Gebhard, 2012). There is some evidence in vitro supporting a direct interaction between the permease subunit of the ABC transporter (YvcR homologue) and BAC in B. subtilis (Dintner et al., 2014). Elucidating the mechanism of antibiotic sensing and the specific downstream effects that bring about resistance is the basis of our future studies.
The DAK domain enzyme is conserved across the Firmicutes, with homology to the ATP-dependent dihyroxyacetone kinases of the glycolytic pathway (Siebold et al., 2003). They possess two functional domains, the L-domain involved in ATP binding and the K-domain implicated in substrate phosphorylation, and have been demonstrated to mediate incorporation of exogenous fatty acids into the bacterial phospholipid synthesis pathway in S. aureus (Erni et al., 2006; Parsons et al., 2014). The presence of mutations at both a conserved serine residue in the ATP binding pocket and a premature stop codon disrupting the predicted EDD fold in the K-domain point to a decrease of enzyme activity in the DAP adapted strains. Indeed, a similar disruption of the C-terminal domain of DAK in S. aureus was found to impair activity of the antimicrobial peptide dermcidin and decrease membrane cardiolipin levels (Li et al., 2009). Bacteria lacking DAK would be unable to utilize extracellular fatty acids, and would be dependent on the endogenous fatty acid synthesis pathway for membrane lipid biogenesis. This diversion of cellular metabolism into lipid biosynthesis may contribute to the DAK mutants’ impaired growth phenotype. Loss of the C-terminal domain of DAK in OG1RF was associated with a significant increase in membrane rigidity as compared to strains with a wild type DAK allele. This shift in rigidity alone did not increase the DAP MICs of DAK mutants, but was required for changes in the YxdJK system to confer a fully DAP-R phenotype. In addition to DAP, these changes to the membrane may also affect other integral membrane or membrane associated proteins, such as those involved in cell division, and may also explain why these cells display a growth defect. The specific effects of the loss of DAK activity on membrane lipid composition are not clear.
A unique aspect of our observations of the C-terminal deletion of DAK was the unexpected effect on cell morphology. The fact that the predicted ATP-binding mutation did not show altered septal genesis suggests that either the enzymatic activity is only partially impaired, or that the C-terminal end of DAK is important for septal synthesis. From published data, it does not seem that loss of FakA, the DAK homologue in S. aureus, leads to impaired growth or an alteration of cellular morphology (Li et al., 2009; Bose et al., 2014; Parsons et al., 2014). Conversely, screens for essential genes in Streptococcus pneumoniae (Zalacain et al., 2003; Opijnen and Camilli, 2012) and our unsuccessful attempts to make a full in frame deletion in E. faecalis OG1RFΔliaR, point to the N-terminal domain of the DAK protein as possessing an essential function in these bacteria. Further, the addition of exogenous fatty acids to growth media has been shown to confer protection against membrane stressors to E. faecalis, even those which lack LiaR (Saito et al., 2014; Harp et al., 2016). The mechanism of this protective effect is unknown, but may relate to changes in membrane composition. Given that membrane topology is important for correct localization of the division septum (Strahl and Errington, 2017) and metabolism and cell division are closely intertwined (Sperber and Herman, 2017), it is tempting to hypothesize that DAK mediated perturbation of membrane lipid composition impairs the bacteria’s ability to define the division point of the cell, with marked effects on bacterial morphogenesis.
In summary, we show that DAP-R in E. faecalis may arise independently of the LiaFSR system, and identify the YxdJK signaling network as a cell envelope defense against multiple classes of clinically relevant antimicrobials. This resistance phenotype comes with a fitness cost, and our experiments suggest that changes in the DAK enzyme are responsible for the alterations of growth, cell envelope morphology and membrane fluidity seen in the E. faecalis mutants. Our findings underscore the robust nature of the enterococcal stress response and the identification of new pathways may provide important insights into disarming a challenging clinical pathogen.
Experimental Procedures
Bacterial strains, plasmids and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table S1. We targeted two strains with previously generated non-polar deletions of liaR for mutagenesis, i) E. faecalis S613FΔliaF177gdpD170cls61 (S613TMΔliaR, DAP MIC 0.094 µg/mL) is a fusidic acid resistant derivative of clinical strain E. faecalis S613 containing three alleles associated with DAP-R (liaF, cls and gdpD) and a non-polar deletion of liaR, and ii) OG1RFΔliaR (DAP MIC 0.047 µg/mL) is a derivative of E. faecalis OG1RF (Reyes et al., 2015). Enterococcal strains were grown on Brain Heart Infusion Agar (BHI, Becton Dickinson) or in broth at 37˚ C with gentle agitation. The DAP adapted strain derivatives of S613TMΔliaR and OG1RFΔliaR were grown on BHI agar with DAP at 4 µg/mL and supplemented with 50 mg/L calcium chloride. E. coli strains EC1000 and TG-1 were grown on Luria-Bertani (LB) agar or in broth at 37˚ C with 25 µg/mL gentamicin added for propagation of pHOU1 or pAT392 containing constructs, respectively. Minimum inhibitory concentrations for each antibiotic were performed via Etest (bioMerieux) according to the manufacturer’s instructions. Briefly, 0.5 McFarland standards for respective strains were prepared and inoculated onto Muller-Hinton agar plates (Oxoid). Strips were placed on the agar surface after absorption of inoculum, plates were placed at 37˚C and MICs were read after 24 hours of incubation. Slower growing mutants with dak mutations were read at 36 to 48 hours to allow a discernable lawn to develop.
In vitro adaptation to daptomycin (DAP) of strains lacking liaR.
S613TMΔliaR and OG1RFΔliaR were grown overnight on BHI agar from glycerol stocks and single colonies were inoculated into 10 mL of Muller-Hinton II broth supplemented with 50 mg/L of calcium chloride (MHC). DAP was added at 0.5x and 1x the MIC for each strain. The population with growth (as assessed by turbidity on visual inspection) at the highest DAP concentration was diluted 1:100 in fresh MHC media daily. Population MIC to DAP was determined by E-test (bioMerieux) on MH agar every other day, and this value was used to adjust the DAP concentration. Serial passage was continued until DAP-R colonies were obtained. DAP-R was defined as an increase in the MIC above 4 μg/mL (former CLSI breakpoint). Colonies growing inside the ellipse delineating the 4 μg/mL cutoff were picked and streaked on MHC agar supplemented with 4 μg/mL DAP and 50 mg/L CaCl2 to isolate single colonies. The phenotype of purified colonies was verified by repeat DAP MIC determination via E-test.
Determination of stability and fitness.
Stability of the resistance phenotype for each strain was evaluated by DAP MIC determination after seven days of passage in antibiotic free media (BHI broth). Fitness of each strain was tested by comparing the growth curve of the DAP-R derivative strains to their DAP-S parents. Briefly, overnight cultures with BHI alone (parent strains) or BHI supplemented with 50 mg/L of CaCl2 and 4 μg/mL DAP (derivative strains), were washed with fresh BHI media to remove antibiotics and used to prepare a 0.5 McFarland standard. This inoculum was diluted 1:100 into 96 well plates with a final volume of 200 µL per well in fresh BHI or MH media (without antibiotics) to yield approximately 5 ×105 CFU/mL. Two blank wells on each row were used for background correction. The optical density (OD) at 600 nm was measured at inoculation and every 15 minutes with shaking for 24 hours, using a Synergy H1 spectrophotometer (BioTek). Every third reading was plotted for ease of visualization. Data sets for each growth curve were analyzed by logistic regression using the R package Growthcurver (Sprouffske and Wagner, 2016). Colony counts were performed on a single run with each strain to verify that OD reading correlated with viable bacterial colony forming units.
Whole genome sequencing (WGS) and mutational analysis.
WGS of both isolates was carried out on a MiSeq (Illumina). Briefly, genomic DNA was isolated using the DNeasy Blood & Tissue kit (Qiagen) after lysozyme treatment. DNA libraries were prepared using the Nextera XT DNA Sample Preparation kit and sequenced with 150 bp paired-end reads. De novo assembly for each genome was done using CLCGenomics Workbench version 8.0.1 (CLCBio). The reads were trimmed (phred score ≥ 30), removing adapter sequences, and discarding those reads with length lower than 50 nucleotides. Single nucleotide polymorphism (SNP) calling was made by aligning the contigs from the assembly of the DAP-R derivative strains using the corresponding parent strains S613 and OG1RF, which were previously sequenced (Bourgogne et al., 2008; Arias et al., 2011), with BWA6 (Li and Durbin, 2009). The variant calling was made with Samtools and Bfctools (Li, 2011). The annotation of the SNPs was made with the SNPeff program (Cingolani et al., 2012). The SNPs that were located on or upstream of coding regions were filtered for analysis and confirmed by Sanger sequencing.
Cloning and genetic manipulation of strains.
To investigate the contributions of genes identified via WGS, we constructed an isogeneic, non-polar deletion of yxdJ (the response regulator of the yxdJK system) by removing the sequence from the start codon to the stop codon of the open reading frame in both the OG1RF and OG1RFΔliaRyxdK202dak371ace backgrounds. A deletion of the C-terminal end of DAK (Δc-dak, removing the sequence encoding codon 371 to the 3’ end of the gene) was constructed in OG1RF. All mutants were made using the pheS* counterselection system, as previously described (Kristich et al., 2007; Panesso et al., 2011). Strains and primers used are listed in Table S1 and Table S2, respectively. Briefly, approximately 800 bp sequences flanking the regions of interest were amplified by PCR, cloned into pHOU1 (a shuttle vector that confers gentamicin resistance) using BamHI and EcoRI and propagated in E. coli EC1000 to generate the plasmids pHOU1::ΔyxdJ and pHOU1::Δc-dak. After sequencing each construct to verify integrity of the crossover region, plasmids were electroporated into E. faecalis CK111, then transferred by conjugation to the recipient strain OG1RF (for ΔyxdJ) or OG1RFΔliaR (for Δc-dak). Recipients were screened on BHI agar with 150 µg/mL of gentamicin, 25 µg/mL of fusidic acid and 125 µg/mL X-gal. The presence of the first crossover event was verified by PCR using primers in both flanks of the predicted insertion. Single colonies were suspended in 50 µL of normal saline and streaked on MM9YEG minimal media supplemented with 10mM p-chloro-phenylalanine to induce excision of the plasmid. White colonies, corresponding to bacteria in which pHOU1 have been excised, were screened for susceptibility to gentamicin and by PCR to assess for successful deletion of the target sequence. Mutants were confirmed with pulsed-field gel electrophoresis and sequencing of the crossover region to ensure that no additional changes were introduced.
Complementation in trans for the OG1RFΔyxdJ mutant was carried out using the pAT392 shuttle vector which has a constitutive P2 promoter and the aac(6’)-aph(2”) gene allowing selection with gentamicin (Arthur et al., 1994). Briefly, the region upstream of yxdJ containing the ribosomal binding site to the stop codon was amplified from OG1RF and cloned into pAT392 using the EcoRI and BamHI restriction sites. The resulting construct, pAT392::yxdJ, was transformed into E. coli TG-1, selected on LB agar plus 25 µg/mL gentamicin, screened by PCR and sequenced to verify the correct insert. OG1RFΔyxdJ was made competent via growth in BHI supplemented with glycine and sucrose (Wirth et al., 1986; Kristich et al., 2007) and transformants were generated via electroporation with pAT392::yxdJ and selection on BHI plus 200 µg/mL gentamicin and screened by PCR to confirm presence of the insert. For the dak complementation in cis, the missing 3’ end of the dak gene with flanking regions was amplified from OG1RF and cloned into pHOU1 using BamHI and EcoRI. Plasmid pHOU1 containing the intact c-dak gene for complementation was then electroporated into E. faecalis CK111 and mated to the OG1RFΔliaRyxdK202dak371ace or OG1RFΔliaRΔc-dak strain as described above. After induction of plasmid excision, colonies with normal growth were screened by PCR for the full-length dak gene. Candidates were confirmed by sequencing of the crossover region.
Cell envelope structure determination by transmission electron microscopy (TEM).
Single colonies of each strain were inoculated into BHI and grown for 16 hours overnight. Cells were pelleted, washed three times in Millonig’s phosphate buffer 0.1 M (13.85 g NaH2PO4·H2O, 3.56 g NaOH in 1 L H2O), and then fixed in 3% glutaraldehyde in Millonig’s phosphate buffer. Samples were processed, embedded in paraffin and sectioned at the University of Texas Health Science Center Electron Microscopy Core. Imaging was carried out on a JEOL JEM 1200 EX Electron Microscope. Cell wall (CW) thickness was determined by measuring from the outer cell membrane to the edge of the cell wall at the pole and midpoint and averaging the two values. A minimum of 25 cells imaged at 50,000x were used to calculate mean CW thickness for each strain.
Determination of cell surface charge by poly-L-lysine binding.
Surface charge was assessed using fluorescein isothiocyanate (FITC) conjugated poly-L-lysine (Mukhopadhyay et al., 2007). Cultures were grown overnight, washed three times with 5mL of 5 mM HEPES pH 7.2 plus 5 mM glucose, and then resuspended in this same buffer to an absorbance value at 578 nm of 0.1. For each strain, 1 mL of the suspension was incubated with 10 µg of FITC-poly-L-lysine for 10 minutes. HEPES buffer plus FITC-poly-L-lysine without cells and cells alone were run in parallel for each strain, for total and background fluorescence, respectively. The cells were pelleted and 100 µL of the supernatant was placed in a light protected 96-well plate and fluorescence intensity was measured on an Infinite M1000 fluorescent spectrophotometer (Tecan) in technical triplicate. Since poly-L-lysine is a positively charged molecule, the concentration remaining in the supernatant is proportional to the overall cell surface charge, and the fluorescent signal intensity was normalized to the total fluorescence of 10 µg/mL FITC-poly-L-lysine in HEPES alone. The assay was repeated three times for each strain on different days.
Membrane phospholipid distribution and BODIPY-DAP binding by fluorescence microscopy.
Visualization of membrane anionic phospholipids was performed with N-nonyl acridine orange (NAO), a membrane dye with preferential binding to anionic phospholipid (e.g., cardiolipin), as described previously (Tran et al., 2013). Briefly, cells grown to exponential phase were incubated with 500 nM NAO in the dark at 37˚ C for 3.5 hours. After staining, cells were washed with normal saline and immobilized on a poly-L-lysine coated coverslip (Sigma-Aldrich). Fluorescent images were taken on an Olympus IX71 or Keyence BX-700 microscope using a Nikon PlanApo N 100x objective, with a FITC filter set (excitation 490 nm, emission 528 nm).
Binding of BODIPY-FL labeled DAP (BODIPY-DAP, Cubist Pharmaceuticals) to cells in vitro and in vivo, was assessed as described previously (Tran et al., 2013). Bacteria were grown to exponential phase in LB media supplemented with 50 mg/L of CaCl2 at 37˚ C. BODIPY-DAP was then added at 4 µg/mL and 64 µg/mL and incubated for 10 minutes protected from light. Cells were washed three times in LB to remove unbound antibiotic, then immobilized on a poly-L-lysine coated coverslip. Cells were imaged using an Olympus IX71 or Keyence BX-700 microscope with a Nikon PlanApo N 100x objective and FITC filter set. Image processing was done using the Keyence software package and Adobe Photoshop CS5.
Membrane fluidity analysis by in vivo Laurdan generalized polarization spectroscopy.
Laurdan, 6-dodecanoyl-2-dimethylaminonaphthalene, is a membrane dye whose emission wavelength is responsive to changes in membrane phospholipid polarity, and, thus, alterations of membrane fluidity (Sanchez et al., 2007). Laurdan generalized polarization (GP) spectroscopy was carried out according to a previously published protocol (Scheinpflug et al., 2017), with the following modifications. Overnight cultures were inoculated into BHI supplemented with 0.1% glucose and grown at 37˚ C until an OD600 of 0.5 to 0.6, or 17 to 24 hours for stationary phase observations. A total of 1.9 mL of this culture was transferred to a 2 mL microcentrifuge tube and Laurdan (Molecular Probes, Thermo Fisher) was added to a final concentration of 10 µM, then incubated in a light protected box with shaking at 37˚ C for 10 minutes. The sample was then washed 4 times in 1 mL prewarmed (37˚ C) phosphate buffered saline (PBS) supplemented with 0.1% glucose and 50 mg/L CaCl2. After the final wash the sample was suspended in 1.9 mL of the PBS/glucose/calcium solution, and 4 technical replicates of 150 µL for each condition (untreated, DAP and ethanol) were placed into a light protected microtiter plate (Costar). An aliquot of 500 µL was taken from each sample, spun to pellet cells and two 150 µL aliquots of the supernatant were used to control for background fluorescence of Laurdan and Laurdan plus DAP. Baseline fluorescence intensity was measured with excitation at 350 nm and emission at 435 nm and 500 nm for five minutes at 37˚ C on a Synergy H1 spectrophotometer (BioTek) to allow equilibration of the cells. Cells were then treated with DAP at 10x the strain MIC, without antibiotics, or in the presence of 10% ethanol as a membrane fluidizer for 10 minutes, with measurements taken each minute as above. The Laurdan GP index was calculated with the formula (I435-I500)/(I435+I500), where I is the fluorescent intensity at the specified wavelength. Experiments were run in duplicate on separate days.
Statistical analysis.
Differences in cell surface charge and cell wall thickness between strain pairs was compared by two way t-test using Prism 7.0 (GraphPad). For membrane fluidity, differences in mean Laurdan GP were assessed by 2-way ANOVA with correction using Dunnett’s multiple comparisons test, using OG1RF as the control strain for each comparison. A difference in the mean value between samples was considered significant for p-values <0.05.
Accession Numbers.
Sequence data for the DAP-R strains was deposited with the National Center for Biotechnology Information (NCBI) GenBank, accession numbers NJIM00000000 (S613TMΔliaRyvcR219dak84asa1) and NJIN00000000 (OG1RFΔliaRyxdK202dak371ace).
Supplementary Material
Acknowledgements
Portions of this work were presented at ICAAC 2015, IDWeek 2015 and ASM 2017. Funding was provided by the National Institutes of Health, National Institute of Allergy and Infectious Diseases grants K08 AI135093 to WRM, K24-AI121296 and R01-AI134637 to CAA, R01AI080714 to YS and K08-AI113317 to TT. The funders had no role in study design, data collection or data interpretation. WRM has received grants, consulting fees and/or honoraria from Achaogen, Shionogi and Merck. TT has received grants from Merck. CAA has received grants from Merck and MeMed Diagnostics. All other authors have no reported conflicts.
References
- Abranches J, Tijerina P, Avilés-Reyes A, Gaca AO, Kajfasz JK, and Lemos JA (2013) The Cell Wall-Targeting Antibiotic Stimulon of Enterococcus faecalis. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias CA, and Murray BE (2012) The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10: 266–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, et al. (2011) Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med 365: 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M, Depardieu F, Snaith HA, Reynolds PE, and Courvalin P (1994) Contribution of vanY D,D-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob Agents Chemother 38: 1899–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Daly SM, Hall PR, and Bayles KW (2014) Identification of the Staphylococcus aureus vfrAB operon, a novel virulence factor regulatory locus. Infect Immun 82: 1813–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, Yerrapragada S, et al. (2008) Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol 9: R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz L, Tran TT, Munita JM, Miller WR, Rincon S, Carvajal LP, et al. (2014) Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob Agents Chemother 58: 4527–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dintner S, Heermann R, Fang C, Jung K, and Gebhard S (2014) A sensory complex consisting of an ATP-binding cassette transporter and a two-component regulatory system controls bacitracin resistance in Bacillus subtilis. J Biol Chem 289: 27899–27910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erni B, Siebold C, Christen S, Srinivas A, Oberholzer A, and Baumann U (2006) Small substrate, big surprise: Fold, function and phylogeny of dihydroxyacetone kinases. Cell Mol Life Sci 63: 890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard S (2012) ABC transporters of antimicrobial peptides in Firmicutes bacteria - phylogeny, function and regulation. Mol Microbiol 86: 1295–1317. [DOI] [PubMed] [Google Scholar]
- Gebhard S, Fang C, Shaaly A, Leslie DJ, Weimar MR, Kalamorz F, et al. (2014) Identification and characterization of a bacitracin resistance network in Enterococcus faecalis. Antimicrob Agents Chemother 58: 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harp JR, Saito HE, Bourdon AK, Reyes J, Arias CA, Campagna SR, and Fozo EM (2016) Exogenous fatty acids protect Enterococcus faecalis from daptomycin induced membrane stress independent of the response regulator LiaR. Appl Environ Microbiol 82: AEM.00933–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries RM, Kelesidis T, Tewhey R, Rose WE, Schork N, Nizet V, and Sakoulas G (2012) Genotypic and phenotypic evaluation of the evolution of high-level daptomycin nonsusceptibility in vancomycin-resistant Enterococcus faecium. Antimicrob Agents Chemother 56: 6051–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristich CJ, Chandler JR, and Dunny GM (2007) Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid 57: 131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Rigby K, Lai Y, Nair V, Peschel A, Schittek B, and Otto M (2009) Staphylococcus aureus mutant screen reveals interaction of the human antimicrobial peptide dermcidin with membrane phospholipids. Antimicrob Agents Chemother 53: 4200–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson JM, Keis S, Smith JMB, and Cook GM (2004) Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob Agents Chemother 48: 3743–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Kong J, Tran TT, Arias CA, Saxer G, and Shamoo Y (2013) Adaptation of Enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob Agents Chemother 57: 5373–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay K, Whitmire W, Xiong YQ, Molden J, Jones T, Peschel A, et al. (2007) In vitro susceptibility of Staphylococcus aureus to thrombin-induced platelet microbicidal protein-1 (tPMP-1) is influenced by cell membrane phospholipid composition and asymmetry. Microbiology 153: 1187–1197. [DOI] [PubMed] [Google Scholar]
- Müller A, Wenzel M, Strahl H, Grein F, Saaki TNV, Kohl B, et al. (2016) Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc Natl Acad Sci U S A 113: E7077–E7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munita JM, Panesso D, Diaz L, Tran TT, Reyes J, Wanger A, et al. (2012) Correlation between mutations in liaFSR of Enterococcus faecium and MIC of daptomycin: Revisiting daptomycin breakpoints. Antimicrob Agents Chemother 56: 4354–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munita JM, Tran TT, Diaz L, Panesso D, Reyes J, Murray BE, and Ariasa CA (2013) A liaf codon deletion abolishes daptomycin bactericidal activity against vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother 57: 2831–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opijnen T. Van, and Camilli A (2012) A fine scale phenotype − genotype virulence map of a bacterial pathogen. Genome Res 22: 2541–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Daniel A, Hardy C, Silverman J, and Gilmore MS (2011) Genetic basis for daptomycin resistance in enterococci. Antimicrob Agents Chemother 55: 3345–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panesso D, Montealegre MC, Rincón S, Mojica MF, Rice LB, Singh KV, et al. (2011) The hylEfm gene in pHylEfm of Enterococcus faecium is not required in pathogenesis of murine peritonitis. BMC Microbiol 11: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panesso D, Reyes J, Gaston E, Deal M, Londoño A, Nigo M, et al. (2015) Deletion of liaR Reverses Daptomycin Resistance in Enterococcus faecium Independent of the Genetic Background. Antimicrob Agents Chemother 59: AAC.01073–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JB, Broussard TC, Bose JL, Rosch JW, Jackson P, Subramanian C, and Rock CO (2014) Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc Natl Acad Sci U S A 111: 10532–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto AMG, Wijngaarden J, Braat JC, Rogers MRC, Majoor E, Brouwer EC, et al. (2017) The Two-Component System ChtRS Contributes to Chlorhexidine Tolerance in Enterococcus faecium. Antimicrob Agents Chemother 61: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeck J, Gebhard S, Orchard PS, Kirchner M, Bauer S, Mascher T, and Fritz G (2016) Anatomy of the bacitracin resistance network in Bacillus subtilis. Mol Microbiol 100: 607–620. [DOI] [PubMed] [Google Scholar]
- Reyes J, Panesso D, Tran TT, Mishra NN, Cruz MR, Munita JM, et al. (2015) A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis. J Infect Dis 211: 1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito HE, Harp JR, and Fozo EM (2014) Incorporation of exogenous fatty acids protects Enterococcus faecalis from membrane-damaging agents. Appl Environ Microbiol 80: 6527–6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez SA, Tricerri MA, Gunther G, and Gratton E (2007) Laurdan Generalized Polarization: from cuvette to microscope. Mod Res Educ Top Microsc 2: 1007–1014. [Google Scholar]
- Scheinpflug K, Krylova O, and Strahl H (2017) Measurement of Cell Membrane Fluidity by Laurdan GP: Fluorescence Spectroscopy and Microscopy. In Antibiotics: Methods and Protocols pp. 159–174. [DOI] [PubMed]
- Siebold C, Arnold I, Garcia-Alles LF, Baumann U, and Erni B (2003) Crystal Structure of the Citrobacter freundii Dihydroxyacetone Kinase Reveals an Eight-stranded alpha-Helical Barrel ATP-binding Domain. J Biol Chem 278: 48236–48244. [DOI] [PubMed] [Google Scholar]
- Sperber AM, and Herman JK (2017) Metabolism shapes the cell. J Bacteriol 199: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouffske K, and Wagner A (2016) Growthcurver: An R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17: 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl H, and Errington J (2017) Bacterial Membranes : Structure, Domains, and Function 519–538. [DOI] [PubMed]
- Tran TT, Munita JM, and Arias CA (2015) Mechanisms of drug resistance: Daptomycin resistance. Ann N Y Acad Sci 1354: 32–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TT, Panesso D, Mishra NN, Mileykovskaya E, Guan Z, Munita JM, et al. (2013) Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. MBio 4: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner LM, Webb AK, Limbago B, Dudeck M, Patel J, Kallen AJ, et al. (2016) Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol 37: 1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth R, An FY, and Clewell DB (1986) Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J Bacteriol 165: 831–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalacain M, Biswas S, Ingraham KA, Ambrad J, Bryant A, Chalker AF, et al. (2003) A global approach to identify novel broad-spectrum antibacterial targets among proteins of unknown function. J Mol Microbiol Biotechnol 6: 109–126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


